Abstract
Secreted by the placental trophoblast, human chorionic gonadotropin (hCG) is an important hormone during pregnancy and is required for the maintenance of pregnancy. Previous studies have shown that dys-regulation of hCG expression is associated with preeclampsia. However, the exact relationship between altered hCG levels and development of preeclampsia is unknown. Metastasis associated protein 3 (MTA3), a chromatin remodeling protein, is abundantly expressed in the placental trophoblasts, but its function is unknown. In breast cancer, MTA3 has been shown to repress the expression of Snail and cell migration. However, whether MTA3 acts similarly in the trophoblast has not been investigated. In the present study, we examined the role of MTA3 in regulating the hCG β-subunit gene (gene name: CGB5) and Snail expression in the trophoblast cell line, BeWo, as well as its relevance to the high hCG expression levels seen in preeclampsia. First, we investigated MTA3 expression in preeclamptic placenta as compared to normal control placenta via gene expression microarray and qRT-PCR and found that MTA3 was significantly down-regulated, whereas both CGB5 and Snail were up-regulated in preeclamptic placenta. Secondly, we knocked down MTA3 gene in trophoblast cell line BeWo and found Snail and hCG were both up-regulated, suggesting that MTA3 represses Snail and hCG gene expression in trophoblasts. Next, we cloned the CGB5 and Snail promoters into the pGL3-basic vector individually and found that silencing of MTA3 by siRNA resulted in an increase of both CGB5 and Snail promoter activities. To confirm that this MTA3 inhibition is a direct effect, we performed a chromatin immune-precipitation (ChIP) assay and found that MTA3 occupied the proximal promoter regions of both Snail and hCG within BeWo cells. Furthermore, we examined MTA3 expression in placental trophoblast by immunohistochemistry and found that MTA3 expression was higher in villous cytotrophoblasts versus syncytiotrophoblasts, which supports an inverse association of MTA3 with hCG expression. Lastly, using the well-characterized trophoblast fusion model, we examined MTA3 and hCG levels in forskolin-treated BeWo cells and found that MTA3 down-regulation was accompanied by an up-regulation of hCG. These data further suggest that MTA3 is repressing placental hCG expression. In summary, MTA3 plays a critical role in repressing hCG and Snail in placenta trophoblast and its deregulation is associated with preeclampsia.
Keywords: Chromatin remodeling, Trophoblast, Preeclampsia
1. Introduction
The placenta is an endocrine organ and secretes several hormones required for successful pregnancy, such as human placental lactogen (hPL) and human chorionic gonadotropin (hCG). Previous research has shown that hCG is required for embryo implantation and placental development. hCG is composed of a shared α-subunit and a specific β-subunit coded by a lists of CGB genes with 99% sequence similarity [1,2]. The four genes which encode the β-subunit (CGB8, CGB5, CGB, CGB7) together with the ancestral LHβ (LHB) and the hCGβ noncoding genes CGB1 and CGB2 all map to a gene cluster located at chromosome 19q13.33 . Given that CGB5 and CGB8 together account for 62–82% of the total pool of hCGβ transcripts, CGB5 or CGB8 genes are used to study CGB gene regulation [3,4]. Proper serum hCG levels during pregnancy are critical for a healthy pregnancy, and abnormally high levels in the late of 2nd trimester are considered an indicator of preelampsia, a pregnancy complication associated with shallow placentation [5,6].
DNA methylation, histone modification and chromatin remodeling are important epigenetic mechanisms of gene regulation. It has been reported that DNA methylation of the CGB5 promoter is associated with repressed CGB5 gene expression and first trimester pregnancy loss [7] but the mechanism is unknown. Metastasis associated protein 3 (MTA3) is one subunit of the Nucleosome Remodeling and Histone Deacetylase (NuRD) complex, which couples histone deacetylase with nuclesome stimulated ATPase-activity and plays a major role in gene repression. MTA3 has been well studied in breast cancer cells where it has been shown to repress Snail (an inducer of cell movement) gene transcription in a histone deacetylase (HDAC)-dependent manner [8]. Furthermore, via its regulation of Snail, MTA3 subsequently alters the target genes downstream of Snail such as E-Cadherin and, therefore, has been implicated in cancer cell migration and metastasis as well as the mediate epithelial to mesenchymal transition [8].
In the placenta, MTA3 is abundantly expressed in trophoblast cells [9]; however, its function and its potential role in the pathology/etiology of pregnancy complications such as preeclampsia has not been investigated . In the present study, we investigated the relevance of MTA3 to preeclampsia and its role in the regulation of CGB5 and Snail genes in the placental trophoblast using the human trophoblast cell line BeWo.
2. Material and method
2.1. Preeclamptic placental samples recruitment
This study protocol was approved by the Ethical Review Board for Human Genome Studies at Fujita Health University. All placental samples both from preeclamptic and normal pregnancies were obtained at the department of Obstetrics and Gynecology, Fujita Health University, Japan, with patients' Informed consent. Preeclampsia was defined as a blood pressure of higher than 140/90 mmHg, with proteinuria of more than 0.3 g in a 24-h collection.
2.2. Analysis of mRNA expression by microarray and quantitative RT-PCR
The transcriptome database of preelamptic and normal placenta, which was deposited at GEO database (http://www.ncbi.nlm.nih.gov/gds/) (GSE24129) and included 8 independent samples in each group [10], was used in this study. Gene expression based on probe signal intensity was extracted and analyzed using the Student' T-Test.
A total of 83 placenta samples were collected (preeclamptic placenta samples n = 44 and normal placenta samples n = 39) in the qRT-PCR assay. The clinic information of these samples is listed in Table 1. In each placenta, chorionic villous tissue was collected randomly from four areas, and then total RNA were extracted and mixed together. These RNA samples were reverse-transcribed into cDNA with Superscript III First-Strand Synthesis System for quantitative RT-PCR (Invitrogen, CA). The power SYBR PCR regent (ABI, CA) was used for qRT-PCR. Delta Ct method was conducted to analyze results. Primers are listed in Table 2.
Table 1.
Clinic parameters of the preelamptic (PE) and normal (NL) pregnancy.
| Maternal age | Gestational Age at testing | BMI (pre-pregnancy) | Systolic BP | Diastlic BP | Gestational age at delivery (wks) | Birth weight | Placental weight | |
|---|---|---|---|---|---|---|---|---|
| NL (n = 39) | 31.7 ± 5.4 | 35 ± 4.8 | 22 ± 4.2 | 114 ± 9.9 | 67.2 ± 9.8 | 35.1 ± 4.9 | 2408.1 ± 852.3 | 529.9 ± 150.9 |
| PE (n = 44) | 30.9 ± 4.5 | 33.4 ± 3.2 | 22.2 ± 4.4 | 164.5 ± 13.7 | 102.7 ± 12.9 | 33.7 ± 3 | 1531.4 ± 559.1 | 323.6 ± 92.7 |
| P value (PE vs NL) | 0.502 | 0.061 | 0.869 | <0.001 | <0.001 | 0.118 | <0.001 | <0.001 |
Date are shown as the mean ± standard deviation (SD).
Table 2.
Primers for qRT-PCR and Luciferase reporter assays and ChIP assay.
| Gene symbol | NCBI GI/ID | Primer sequence (5′ to 3′) | Position to TSS or in cDNA | |
|---|---|---|---|---|
| MTA3 | GI:50878291 | F | AAGCCTGGTGCTGTGAATGG | 1281 |
| R | CTGGTGAGACTGTGTAGCATAG | 1382 | ||
| Actin | GI:168480144 | F | CAGCAGATGTGGATCAGCAAG | 1141 |
| R | TTGTCAAGAAAGGGTGTAACGC | 1252 | ||
| Snail Promoter | 6615 | F | AAGCGCTCAGACCACCGGGC | −350 |
| R | AGTGGTCGAGGCACTGGGGT | +84 | ||
| CGB5 promoter | 93,659 | F | ATAATGCCTGACCTGTAGTTGT | −2000 |
| R | CCTTGGTGCGTCCCCTGCCT | +364 | ||
| Snail ChIP | 6615 | F | GAACGGGTGCTCTTGGCTAG | −600 |
| R | ACGAGGGAAACGCACATCACTGG | −500 | ||
| CGB5 ChIP | 93,659 | F | CCTGGCTTGAGGGTAGAGTG | −219 |
| R | GAAGCCACTTGACCCAGATGC | −126 |
2.3. Cell culture, lentivirus mediated gene knock-down and forskolin treatment
The trophoblast cell line BeWo (ATCC) was cultured in F12K supplemented with 10% FBS, 2 mmol/L L-glutamine and 1% Pen/Strep. Medium was refreshed every other day. The lentivirus particles expressing MTA3 shRNA or control shRNA (titer: 108−109) were purchased from Openbiosystem (Thermo Scientific). 2 days after infection with these viruses, BeWo cells were cultured in media containing 5 μg/ml puromycin (Invitrogen) for 3 weeks prior to harvesting cells for protein analysis. In forskolin treatment, 50 μM forskolin (R&D) was added into culture media.
In the tissue culture of full term placenta, we obtained fresh normal placental tissue from Michigan State University's Center for Women's Health Research , Human Female Reproductive Tract Bio-repository with the donors' written informed consent and the approval of the MSU Institutional Review Board. After being minced with scissors, placenta tissues were cultured with the same media as the BeWo cells. After one week in culture, the trophoblast outgrowth appeared beside tissue clump.
2.4. Luciferase reporter assay
Luciferase reporter assay was performed as previously published [11]. Briefly, the CGB5 gene promoter region, −2000 to +364 bps relative to its transcription start site (TSS) and Snail gene promoter region, −350 to +84 bps relative to TSS, were cloned into pGL3-Basic vector (Promega), respectively, upstream of firefly luciferase coding sequence via PCR and subsequent ligation. Primers are listed in Table 2. Next, BeWo cells plated at 5 × 104 cells per well in a 96-well plate were transiently transfected with 0.1 μg of plasmid and 0.005 μg of the internal control plasmid (pRL-TK vector, expressing Renilla luciferase), along with MTA3 siRNA or control siRNA using the Lipofectimine 2000 transfection reagent (Invitrogen, CA). 5 h post-transfection, cells were treated with 100 nM trichostatin-A (TSA, Sigma) or 50 μM forskolin for 18 h. DMSO treatment was used as vehicle control. The activities of both luciferases were determined using the Dual-Luciferase Reporter System (Promega) according to the manufacturer's instructions. Assays were performed 3 times each in duplicate and promoter activity determined by the ratio of the two luciferase activities.
2.5. Chromatin-immunoprecipitation (ChIP) assay
BeWo cells were harvested, chemically crosslinked with 1% formaldehyde (Sigma–Aldrich) for 10 min at room temperature, pelleted, washed with PBS and flash frozen in liquid nitrogen. Pellets were resuspended in ChIP lysis buffer and sonicated using a Branson Sonifier 450D (Branson, Danbury, CT) at 20% amplitude, with 3 × 30-s pulses in ice water, given an average DNA size of about 200–800 bp. Post-sonication, samples were centrifuged, flash frozen in liquid nitrogen until future use. Sonicated extracts (equivalent to 1 × 106 cells) were used in each ChIP reaction according to manufacturer's protocol (LowCell ChIP kit, Diagenode). 2 μg of MTA3 antibody (Rabbit) and non-specific IgG (Rabbit) were added per reaction (antibody sources are listed in Table 3) and quantitative PCR was used to confirm enrichment of MTA3 in the Snail and CGB5 promoters (Primers listed in Table 2). ChIP DNA and IgG control DNA were normalized to input DNA.
Table 3.
Antibody list used for IHC and Western blot and ChIP.
| Antibody | Company | Catalog | Used in |
|---|---|---|---|
| E-Cadherin | R&D | MAB1838 | IHC and Western blot |
| hCG | Abcam | Ab9376 | Western blot |
| Snail | ABgent | AP2054a | Western blot |
| Actin | Sigma | A5441 | Western blot |
| MTA3 | Abcam | Ab87275 | IHC and Western blot and ChIP |
| IgG | ABgent | P1200-101 | ChIP |
2.6. Immunocytochemistry (ICC), Immunohistochemistry (IHC) and Western blot
Cells were fixed in 4% paraformaldehyde (PFA) for 20 min. Then, the cells were permeabilized with 0.3% Triton X-100 in PBS for 20 min, blocked with PBS with 4% BSA for half hour, and incubated with antibodies (antibody source listed in Table 3) for two hours. Next, cells were incubated one hour with either a FITC- or Rhodamine-conjugated secondary antibody, mounted with an anti-fade mounting solution containing DAPI (Invitrogen) and examined using fluorescence optics (Nikon TS100).
Placental tissue was fixed in 4% PFA for 20 min, paraffin embedded and 10 μM sections were subjected to immunohistochemistry using the same ICC protocol listed above.
For Western analysis, total protein was isolated from frozen placental tissue using RIPA buffer supplemented with protease inhibitor cocktail (Invitrogen) and quantified using BCA kit (Thermo Scientific) according to manufacturer's protocol. 10 μg total protein was loaded per well and subjected to electrophoresis and immunoblotting using the antibodies listed in Table 3.
2.7. Statistical analysis
The Student' T-Test was used for the statistical analysis of clinic information, microarray gene expression data, real-time PCR, luciferase reporter assay, and ChIP assays. A p-value of less than 0.05 was considered statistically significant.
3. Results
3.1. Repressed MTA3 expression correlates with high CGB5 and Snail expression in preeclamptic placenta
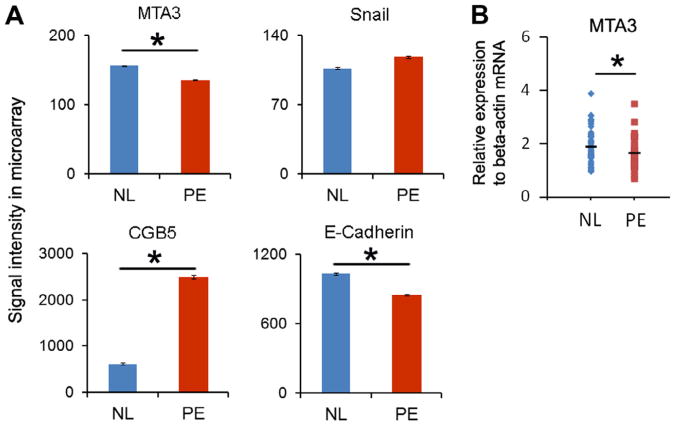
To verify the expression of MTA3 in placental trophoblast , we examined the MTA3 expression in the trophoblast outgrowth of in vitro cultured placental villi tissue and trophoblast cell line, BeWo, and found that MTA3 was expressed in the trophoblast nuclei (Fig. S1). To investigate the relationship between MTA3 with preeclampsia and the interaction of MTA3 and CGB5 genes in the placenta, we first compared the expression levels of MTA3, Snail, E-Cadherin and CGB5 in our previous transcriptome data of preeclamptic and normal placenta (n = 8 each group) (GSE24129) [10]. We found that in preeclamptic placenta MTA3 and E-Cadherin were significantly down-regulated, Snail was up-regulated (a trend which did not reach statistical significance, p = 0.13) and CGB5 was significantly up-regulated as compared with normal placenta (Fig. 1A). To confirm this microarray data, we performed qRT-PCR to measure MTA3 expression in preeclamptic and normal placenta (n = 44 in preeclamptic group and 39 in normal group). We found that, consistent with the microarray data, MTA3 was significantly down-regulated in preeclampsia (Fig. 1B).
Fig. 1.
Down-regulation of MTA3 and up-regulation of CGB5 and Snail are associated with preeclampsia. (A) Gene expression of MTA3, Snail, CGB5 and E-Cadherin in preeclamptic and normal placenta in microarray data (GSE24129). (B) Gene expression of MTA3 using qRT-PCR normalized to beta-Actin mRNA. Data is presented as mean ± SEM.
3.2. Knock down of MTA3 increases Snail and HCG expression in trophoblast cell line, BeWo
The trophoblast cell line, BeWo, is a well-characterized in vitro model for the placental cytotrophoblasts, which is known to undergo cell fusion as well as to highly express and secrete hCG upon forskolin treatment [12]. To further investigate the role of MTA3 in trophoblast function, we knocked down MTA3 gene expression in BeWo cells using a lentiviral-mediated MTA3 shRNA and found that, upon MTA3 silencing, hCG and Snail protein levels were up-regulated and E-Cadherin was down-regulated (based upon normalization to beta-tubulin levels). These results indicate that MTA3 represses Snail and hCG gene expression in the trophoblast (Fig. 2A). To confirm the regulation of hCG and Snail by MTA3, we cloned the CGB5 (the major contributing member of the CGB gene family) promoter and Snail promoter into pGL3-basic up-stream of the luciferase gene, respectively (pGL3-CGB5 and pGL3-Snail). After co-transfection with MTA3 siRNA (Promega) or control siRNA in BeWo cells, transcriptional activity of the CGB5 promoter was up-regulated in MTA3 siRNA group compared with control siRNA group with forskolin treatment (significant) or vehicle treatment (a trend which did not reach statistical significance). Similar with the result in CGB5 promoter, transcriptional activity of the Snail promoter was also significantly up-regulated, indicating that MTA3 repressed CGB5 and Snail transcription (Fig. 2B and C). The effectiveness of MTA3 siRNA and control siRNA in knocking-down MTA3 protein were examined by Western blot after transfection of these siRNA into BeWo cells. Results showed that MTA3 siRNA efficiently down-regulated MTA3 protein level compared with control siRNA (Fig. S2). MTA3 is one component of NuRD complex, and the cellular function of MTA3 is mediated via the histone deacetylace 1/2, (HDAC1/2), another component of NuRD. HDAC1/2 can deacetylate histones to wrap the DNA more tightly and silence gene expression. Trichostatin-A (TSA) is a known inhibitor of HDAC1/2 activity which has previously been shown to increase Snail promoter activity in breast cancer cells [8]. To examine whether HDAC1/2 has a potential role in MTA3 regulation of CGB5 and Snail promoter activities in this model, we used TSA, forskolin and DMSO (vehicle control) to treat pGL3-CGB5 or pGL3-Snail transfected BeWo cells, and found that TSA significantly increased CGB5 and Snail transcription activities, whereas forskolin only significantly increased CGB5 transcriptional activity but not increase Snail transcriptional activity (Fig. 2D). This result suggests that the regulation of CGB5 and Snail promoter activities by MTA3 is mediated via the HDAC1/2 component of NuRD complex.
Fig. 2.
MTA3 represses of CGB5 gene in BeWo cells. (A) Knock-down of MTA3 increases Snail and HCG expression in the trophoblast cell line, BeWo, as detected by Western blot. (B) Schematic diagrams of pGL3-CGB5 and pGL3-Snail used in luciferase reporter assay. (C) Knock-down of MTA3 via MTA3 siRNA caused significantly up-regulation of CGB5 and Snail promoter activity in BeWo cells. (D) Treatment with Trichostatin-A (TSA), an inhibitor of HDAC1/2, significantly increased CGB5 and Snail promoter activities in BeWo cells. (E) Schematic diagram of CGB5 and Snail promoters and primer positions used in ChIP assay. (F) MTA3 occupation of CGB5 and Snail promoters using ChIP assay. Occupation level of MTA3 and Control IgG is demonstrated relative to input DNA. Data is presented as mean ± SEM.
3.3. MTA3 complex occupies the CGB5 and Snail promoter
It is well established that MTA3 regulates Snail gene expression by occupying its promoter in breast cancer cells [8]. Accordingly, we hypothesized that MTA3 could occupy CGB5 and Snail promoter in BeWo cells. To test this hypothesis, we performed chromatin immune-precipitation (ChIP) assay and found that the MTA3 antibody bound to the proximal promoter regions of both Snail (−600 to −500 bp to transcription start site, TSS) and hCG (−219 to −126 bp to TSS) in BeWo cells as compared with the binding level of IgG control, suggesting that MTA3 could directly regulates these two genes.
3.4. MTA3 is inversely associated with hCG expression in trophoblasts
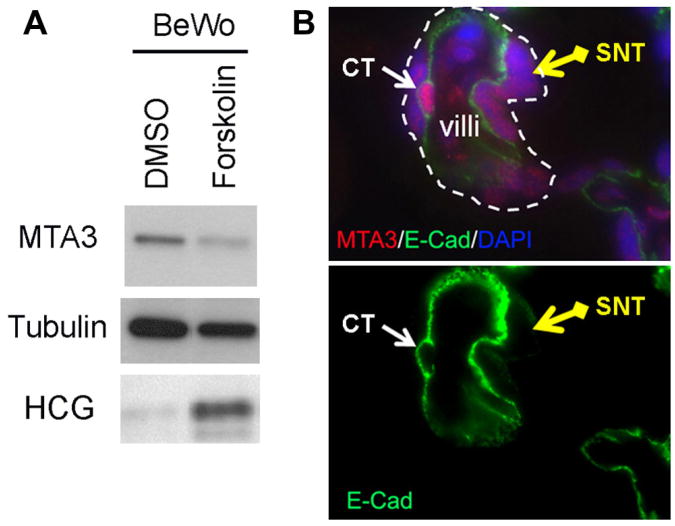
To examine the role of MTA3 in trophoblast , we investigated MTA3's dynamic expression during forskolin-induced expression of hCG in BeWo cells. After 2 days of forskolin treatment, we examined MTA3 and hCG protein levels by Western blot, and found that forskolin treatment decreased MTA3 and increased hCG expression (Fig. 3A) as compared to vehicle-treated cells. These data suggest an inverse relationship between MTA3 and hCG expression in trophoblasts. It has been shown that hCG is more highly expressed in syncytiotrophoblasts as compared to cytotrophoblasts within placental villi [13]. E-Cadherin is a known cytotrophoblast marker gene which is not expressed by synctiotrophoblasts. To compare the expression of MTA3 in both cytotrophoblasts and syncytiotrophoblasts, we co-stained MTA3 and E-Cadherin in fixed full term placental sections via IHC Our results show that MTA3 signal intensity is greater in the cytotrophoblasts as compare d to syncytiotrophoblasts (Fig. 3B), suggesting an inverse correlation between MTA3 and hCG expression in the placenta.
Fig. 3.
MTA3 expression is inversely associated with hCG expression in the trophoblast. (A) Forskolin treatment increases hCG protein levels and decreased MTA3 protein levels in BeWo cells as detected by Western blot. (B) MTA3 expression is higher in cytotrophoblasts (CT) than syncytiotrophoblasts (SNT) as detected by IHC using MTA3 and E-Cadherin antibodies. The white dash line shows representative placental villi, the white arrow shows a representative cytotrophoblast and the yellow arrow shows a representative syncytiotrophoblast.
4. Discussion
Preeclampsia is a pregnancy complication characterized by high blood pressure (more than 140/90 mmHg) and significant amounts of protein in the urine in a pregnant woman, affecting 6–8% of all pregnancies [14]. Compromised trophoblast function, such as decreased trophoblast invasion and under-grown villi is directly linked to preeclampsia, resulting in decreased blood perfusion and hypoxia within the placental environment. Previous studies have also shown that altered levels of Snail [15,16] and abnormally high levels of hCG [5] are linked with this disease. As a chromatin modifier, MTA3 is an important component of the nucleosome remodeling and deacetylation complex (NuRD). NuRD can repress gene expression by deacetylating chromatin via its component, histone deacetylases 1/2 (HDAC1/2). MTA3 has previously been shown to mediate breast cancer cell invasion via regulation of Snail gene expression [8]. In this research, we have shown that, consistent with its function in cancer cells, MTA3 also represses Snail expression in the trophoblast. Furthermore, we have shown that MTA3 regulates hCG expression in the trophoblast in a HDAC1/2-dependent manner. Lastly, we have shown that MTA3 expression is reduced in preeclamptic placenta, which may account for previous reports of dys-regulation of hCG and Snail expression in this disease.
Trophoblast fusion (cytotrophoblasts fuses into syncytiotrophoblasts) is important for placental villi maintenance and growth. Syncytiotrophoblast is the epithelia which covered the placenta villi with multiple nuclei, and maintenance of syncytiotrophoblasts physiological function need cytotrophoblasts continuously to fuse with them. Previous studies have shown that forskolin treatment can induce BeWo cell fusion in vitro, making this cell line an ideal model for examining the cellular events during trophoblast fusion [12]. Following forskolin treatment, BeWo cells undergo significant changes in cell size and morphology (from uni- to multi-nuclear) and hCG expression is highly induced; hence expression of hCG is universally considered a marker of trophoblast fusion. In this study, we observed that MTA3 repressed hCG gene expression in BeWo cells (Fig. 2), and higher expression in un-fused than in fused trophoblast in trophoblast fusion models (Fig. 3). These results suggest that MTA3 could play an important role in the trophoblast fusion process.
A deeper understanding of the regulatory mechanisms of MTA3 in the placenta will enhance our understanding of pregnancy complications such as preeclampsia. For example, in cancer cells, MTA3 is directly regulated by estrogen and estrogen receptor alpha (Erα) binding to its promoter [17]. In the trophoblast cell line, JAR, recent research has shown that over-expression of estrogen receptor beta (ERβ) increased MTA3 expression and decreased expression of its target gene, Snail [18]. Furthermore, dys-regulation of estrogen and ERα expression has been reported in preeclamptic placenta [19,20]. These studies suggest that estrogen, ER dependent regulation of the MTA3 gene and dys-regulation of estrogen/ER/M TA3 signaling axis may participate in the impaired MTA3 expression seen in preelamptic placenta. Therefore, in our future studies, we will investigate the molecular mechanism of estrogen/ER regulation on MTA3 in trophoblasts. In summary, the present study has indicated that MTA3 regulates hCG and Snail expression in trophoblasts, and its dys-regulation is associated with preeclampsia.
Supplementary Material
Acknowledgments
We acknowledge Dr. Trixie Smith, Dr. Sok Kean Khoo, Ms. Susan Ferguson and Mr. Mark Nelson for critically reading the manuscript and providing constructive criticisms.
Funding: This work was supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development/National Institute of Health cooperative agreement (U54 HD 40093 to R.E.L) as part of the Specialized Cooperative Centers Program in Reproduction and Infertility Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest: There is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2013.02.102.
Contributor Information
Richard Leach, Email: Richard.Leach@hc.msu.edu.
Kai Wang, Email: Kai.Wang@hc.msu.edu.
References
- 1.Hallast P, Nagirnaja L, Margus T, Laan M. Segmental duplications and gene conversion: human luteinizing hormone/chorionic gonadotropin beta gene cluster. Genome Res. 2005;15:1535–1546. doi: 10.1101/gr.4270505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pierce JG, Parsons TF. Glycoprotein hormones: structure and function. Annu Rev Biochem. 1981;50:465–495. doi: 10.1146/annurev.bi.50.070181.002341. [DOI] [PubMed] [Google Scholar]
- 3.Rull K, Laan M. Expression of beta-subunit of HCG genes during normal and failed pregnancy. Hum Reprod. 2005;20:3360–3368. doi: 10.1093/humrep/dei261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller-Lindholm AK, LaBenz CJ, Ramey J, Bedows E, Ruddon RW. Human chorionic gonadotropin-beta gene expression in first trimester placenta. Endocrinology. 1997;138:5459–5465. doi: 10.1210/endo.138.12.5618. [DOI] [PubMed] [Google Scholar]
- 5.Brouillet S, Hoffmann P, Chauvet S, Salomon A, Chamboredon S, Sergent F, Benharouga M, Feige JJ, Alfaidy N. Revisiting the role of hCG: new regulation of the angiogenic factor EG-VEGF and its receptors. Cell Mol Life Sci. 2012;69:1537–1550. doi: 10.1007/s00018-011-0889-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basirat Z, Barat S, Hajiahmadi M. Serum beta human chorionic gonadotropin levels and preeclampsia. Saudi Med J. 2006;27:1001–1004. [PubMed] [Google Scholar]
- 7.Uuskula L, Rull K, Nagirnaja L, Laan M. Methylation allelic polymorphism (MAP) in chorionic gonadotropin beta5 (CGB5) and its association with pregnancy success. J Clin Endocrinol Metab. 2011;96:E199–E207. doi: 10.1210/jc.2010-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- 9.Bruning A, Makovitzky J, Gingelmaier A, Friese K, Mylonas I. The metastasis-associated genes MTA1 and MTA3 are abundantly expressed in human placenta and chorionic carcinoma cells. Histochem Cell Biol. 2009;132:33–38. doi: 10.1007/s00418-009-0595-z. [DOI] [PubMed] [Google Scholar]
- 10.Nishizawa H, Ota S, Suzuki M, Kato T, Sekiya T, Kurahashi H, Udagawa Y. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod Biol Endocrinol. 2011;9:107. doi: 10.1186/1477-7827-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hattori N, Imao Y, Nishino K, Hattori N, Ohgane J, Yagi S, Tanaka S, Shiota K. Epigenetic regulation of Nanog gene in embryonic stem and trophoblast stem cells. Genes Cells. 2007;12:387–396. doi: 10.1111/j.1365-2443.2007.01058.x. [DOI] [PubMed] [Google Scholar]
- 12.Taylor RN, Newman ED, Chen SA. Forskolin and methotrexate induce an intermediate trophoblast phenotype in cultured human choriocarcinoma cells. Am J Obstet Gynecol. 1991;164:204–210. doi: 10.1016/0002-9378(91)90654-a. [DOI] [PubMed] [Google Scholar]
- 13.Handwerger S, Aronow B. Dynamic changes in gene expression during human trophoblast differentiation. Recent Prog Horm Res. 2003;58:263–281. doi: 10.1210/rp.58.1.263. [DOI] [PubMed] [Google Scholar]
- 14.Matthys LA, Coppage KH, Lambers DS, Barton JR, Sibai BM. Delayed postpartum preeclampsia: an experience of 151 cases. Am J Obstet Gynecol. 2004;190:1464–1466. doi: 10.1016/j.ajog.2004.02.037. [DOI] [PubMed] [Google Scholar]
- 15.Brown LM, Lacey HA, Baker PN, Crocker IP. E-cadherin in the assessment of aberrant placental cytotrophoblast turnover in pregnancies complicated by pre-eclampsia. Histochem Cell Biol. 2005;124:499–506. doi: 10.1007/s00418-005-0051-7. [DOI] [PubMed] [Google Scholar]
- 16.Blechschmidt K, Mylonas I, Mayr D, Schiessl B, Schulze S, Becker KF, Jeschke U. Expression of E-cadherin and its repressor Snail in placental tissue of normal, preeclamptic and HELLP pregnancies. Virchows Arch. 2007;450:195–202. doi: 10.1007/s00428-006-0343-x. [DOI] [PubMed] [Google Scholar]
- 17.Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Li X, Yin L, Ding J, Jin H, Feng Y. Genistein inhibits placental choriocarcinoma cell line JAR invasion through ERbeta/MTA3/Snail/E-cadherin pathway. Oncol Lett. 2011;2:891–897. doi: 10.3892/ol.2011.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Molvarec A, Ver A, Fekete A, Rosta K, Derzbach L, Derzsy Z, Karadi I, Rigo J., Jr Association between estrogen receptor alpha (ESR1) gene polymorphisms and severe preeclampsia. Hypertens Res. 2007;30:205–211. doi: 10.1291/hypres.30.205. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, Bai H, Liu X, Fan P, Liu R, Huang Y, Wang X, He G, Liu Y, Liu B. Genotype distribution of estrogen receptor alpha polymorphisms in pregnant women from healthy and preeclampsia populations and its relation to blood pres sure levels. Clin Chem Lab Med. 2009;47:391–397. doi: 10.1515/CCLM.2009.096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.