Abstract
BACKGROUND
In 2008, the US Food and Drug Administration (FDA) issued a Guidance for Industry statement formally recognizing (during drug development) the conjoined nature of type 2 diabetes (T2D) and cardiovascular disease (CVD), which has precipitated an urgent need for panels of markers (and means of analysis) that are able to differentiate subtypes of CVD in the context of T2D. Here, we explore the possibility of creating such panels using the working hypothesis that proteins, in addition to carrying time-cumulative marks of hyperglycemia (e.g., protein glycation in the form of Hb A1c), may carry analogous information with regard to systemic oxidative stress and aberrant enzymatic signaling related to underlying pathobiologies involved in T2D and/or CVD.
METHODS
We used mass spectrometric immunoassay to quantify, in targeted fashion, relative differences in the glycation, oxidation, and truncation of 11 specific proteins.
RESULTS
Protein oxidation and truncation (owing to modified enzymatic activity) are able to distinguish between subsets of diabetic patients with or without a history of myocardial infarction and/or congestive heart failure where markers of glycation alone cannot.
CONCLUSION
Markers based on protein modifications aligned with the known pathobiologies of T2D represent a reservoir of potential cardiovascular markers that are needed to develop the next generation of antidiabetes medications.
The clinical definition of type 2 diabetes (T2D)5 hinges on blood glucose control. Traditionally, T2D has been indicated by measuring the absolute concentration of blood glucose (1), and more recently by measuring the relative abundance of glycohemoglobin (Hb A1c) (2). Although measurement of increased blood glucose by either of these markers is the definition of T2D, most T2D-related deaths are attributed to coronary artery disease (3). Consequently, a large portion of T2D research over the past 2 decades has centered on the connection between poor glucose control and negative cardiovascular outcomes, with particular attention focused on the relationship between patient levels of Hb A1c and cardiovascular disease (CVD). Although epidemiological connections remain under debate (1, 4–6), recent large randomized trials [Action to Control Cardiovascular Disease in Diabetes (ACCORD), Action in Diabetes and Vascular Disease: Preterax and Diamicron MR Controlled Evaluation (ADVANCE), and VA Diabetes Trial (VADT)] have failed to translate tight control of Hb A1c into cardiovascular benefit (7–9). Strikingly, the risk of myocardial infarction has been reported to increase in association with certain classes of antidiabetic therapies (10).
In response, the US Food and Drug Administration (FDA) issued a Guidance for Industry suggesting that developers of new antidiabetes drugs demonstrate that therapies will not result in an unacceptable increase in cardiovascular risk (11). Thus, the therapeutic definition of diabetes is beginning to extend beyond that of increased blood glucose and further into downstream comorbidities. This expanded definition creates serious challenges in the drug development industry by requiring the concurrent monitoring of markers for both CVD (risk and/or outcome) and T2D (Hb A1c as an efficacy marker of lowered blood glucose) during drug trials. In effect, these events have broadened the working definition of T2D, with immediate impact in the therapeutic industry and, in all likelihood, throughout the clinical community over the longer term. Consequently, there is an urgent need for panels of markers used in the synergistic monitoring of T2D and related cardiovascular complications.
Here, we report on biomarker development studies undertaken to characterize protein microheterogeneity and evaluate its use in creating multidimensional biomarker views related to the pathobiologies of T2D and CVD comorbidities. We used standardized mass spectrometric immunoassays (MSIAs) to characterize and quantify microheterogeneity in 7 nonoverlapping patient subgroups totaling 212 individuals, including healthy plasma (HP; n = 37) and healthy serum (HS; n = 29), plus plasma from patients with diagnosed type 2 diabetes (T2D; n = 50), diabetes with history of congestive heart failure (CHF) and previous myocardial infarction (MI) (T2D/MI/CHF; n = 17), well-controlled diabetes with a history of congestive heart failure (T2D/CHF; n = 25), nondiabetes with a history of congestive heart failure and previous myocardial infarction (MI/CHF; n = 25), and nondiabetes with congestive heart failure and no previous myocardial infarction (CHF; n = 29). We selected proteins for investigation based on (a) well-established clinical roles in relation to diabetes and/or cardiovascular disease and/or (b) prior population proteomics studies where microheterogeneity was evident in disease populations (12–20). They included albumin, apolipoprotein A–I (apoAI), apoCI, apoCII, vitamin D binding protein (VDBP), transthyretin (TTR), β2-microglobulin (B2M), cystatin C (CysC), serum amyloid P (SAP), C-reactive protein (CRP), and the chemokine RANTES. Microheterogeneity (i.e., posttranslational modifications or point mutations) evident in each protein was recorded in terms of frequency and relative abundance, as described (16, 20). In total, we performed >2300 assays, during which 41 different molecular species (12, 13, 16–23) were recorded for each of the 212 individuals, producing a total of >8600 data points that were subjected to both unsupervised and biologically supervised modeling to produce multidimensional biomarker views of T2D and CVD.
Materials and Methods
MATERIALS
We obtained polyclonal rabbit antihuman antibodies against VDBP, albumin, TTR, CysC, B2M, and SAP from Dako. According to the manufacturer’s specifications, many of these antibodies are for in vitro diagnostic use and are intended for determining the respective protein in gel immunoprecipitation and other techniques. We obtained goat anti-CRP antibody from Immunology Consultants Laboratory and purchased antihuman apolipoprotein antibodies from Academy Biomedical. Premixed 2-(N-morpholino) ethanesulfonic acid (MES)-buffered saline powder packets were from Pierce, and monoclonal antibody against human RANTES from R&D Systems. We isolated specific proteins from plasma/serum using carboxyl-functionalized MSIA pipette tips from Intrinsic Bioprobes derivatized with antibodies via 1,1′-carbonyldiimidazole (CDI) chemistry as described in the Data Supplement, which accompanies the online version of this article at http://www.clinchem.org/content/vol57/issue5. We obtained protein Captrap cartridges for LC-MS from Michrom Bioresources and purchased premade 10 mmol/L HEPES-buffered saline (HBS) from Biacore. We acquired MALDI-TOF mass spectral calibrants from Bruker Daltonics and all other chemicals from Sigma-Aldrich.
SAMPLES
We obtained blood samples (>2 mL in EDTA plasma or serum collection tubes) from volunteers under institutional review board protocols after informed consent. Samples were processed immediately by use of standard plasma or serum preparation protocols, then promptly placed in a freezer at −80 °C, where they were kept until aliquoting and analysis. Samples were analyzed within 6 months of acquisition. We investigated plasma samples from controls and 5 disease subgroups, including healthy individuals [healthy plasma (HP); n = 37] and patients with type 2 diabetes (T2D; n = 50), well-controlled diabetes with a history of congestive heart failure (T2D/CHF; n = 25), diabetes with a history of congestive heart failure and previous myocardial infarction (T2D/MI/CHF; n = 17), nondiabetes with a history of congestive heart failure and previous myocardial infarction (MI/CHF; n = 25), and nondiabetes with congestive heart failure and no previous myocardial infarction (CHF; n = 29). We also included a subgroup of serum samples from healthy individuals to investigate potential differences between sample presentations [healthy serum (HS); n = 29]. Cohorts were of roughly equal distributions of African American, white, and Hispanic donors at proportions typically observed in the US. Disease cohorts were sex-and age-matched with healthy controls. Additional patient information is available in the online Data Supplement.
SAMPLE PREPARATION FOR THE ANALYSIS OF INTACT PROTEINS BY MSIA
In preparation for MSIA (for all proteins other than RANTES), we mixed 25–100 µL sample 1:1 to 1:4 with HBS (with or without 0.05% Tween 20) and/or MES-buffered saline. Detailed methods for the analysis of individual proteins have been described in detail (12, 13, 16–24). In preparation for RANTES, we mixed 230 µL sample with 115 µL of a detergent solution containing 4.5% Tween 20, 150 mmol/L octyl-β-glucopyranoside, 1.5 mol/L ammonium acetate, and concentrated PBS (0.67 mol/L sodium phosphate, 1 mol/L sodium chloride), for a total analytical volume of 345 µL, as described in detail (20). Antibody-linked tips were stored in HBS at 4 °C until the day of use, at which time they were loaded onto a 96-well pipetting robot and prerinsed (400 µL/well; 150 µL aspirate and dispense cycles; 10 cycles) with HBS and then used to extract specific proteins from individual samples at room temperature (85 µL aspirate and dispense cycles; 250 cycles). After extraction, tips were washed (by drawing from a fresh reservoir of liquid and dispensing to waste) as follows. Five cycles of 200 µL HBS, 5 cycles of 200 µL distilled water, 5 cycles of 200 µL of 2 mol/L ammonium acetate/acetonitrile (3:1 vol/vol), 10 cycles of 200 µL distilled water, and 5 cycles of air (to remove any residual water). Retained proteins were eluted for MALDI-TOF-MS by drawing 4 µL of matrix solution (33% acetonitrile in water containing 0.4% trifluoroacetic acid saturated with sinapinic acid), allowing the solution to dwell in the tip for 20 s, and then dispensing onto a MALDI target (13). For electrospray ionization time-of-flight mass spectrometry (ESI-TOF-MS), we replaced the matrix solution with 10 µL of 0.4% trifluoroacetic acid and deposited the eluent into a 96-well (conical) polypropylene autosampler tray.
MALDI-TOF MS
We performed MALDI-TOF mass spectrometry for TTR, CysC, B2M, SAP, apoCI, apoCII, and RANTES using a Bruker Autoflex III or Ultraflex III operating in positive-ion, delayed-extraction linear mode. Instrument settings for each analyte are provided as a table in the online Data Supplement. We used a laser (Nd: YAG) repetition rate of 100–200 Hz to sum 20 000 laser shots (into an individual spectrum) for each sample, resulting in typical mass resolutions [full width at half maximum (FWHM)] ≥1000 and signal-to-noise ratios (S/N) >3 for low-level signals [i.e., relative ion signals (integral) of 1% or less of total ion signal]. Spectra were mass calibrated externally (from calibration spots placed on each sample target) with a mixture of ubiquitin (calculated MH+avg 8565.76), cytochrome C (calculated MH+avg 12360.97), and trypsinogen (calculated MH+avg 23982) (MH+avg: isotopically averaged molecular mass of a protonated molecule). For semiquantitative data, we processed spectra by baseline subtraction followed by signal integration (Zebra, Beavis Informatics) of each signal corresponding to the protein of interest. For each sample, we measured and recorded the relative abundances of the variants by normalizing the integral of each variant to the summed integrals of all observed forms of the protein.
ESI-TOF MS
We performed ESI-TOF MS for albumin, VDBP, CRP, and apoAI using a column-free trap-and-elute approach (rather than traditional liquid chromatography gradient methods). ESI rather than MALDI was used for these larger proteins because of its ability to achieve greater resolving power than MALDI at masses >25 kDa. Eluent (5 µL) was injected by a Spark Holland Endurance autosampler in microliter pickup mode and loaded by an Eksigent nanoLC⋆1D at 10 µL/min [90/10 water/acetonitrile containing 0.1% formic acid (solvent A)] onto a protein Captrap (polymeric/reversed phase sorbent, Michrom Biore-sources) configured for unidirectional flow on a 6-port divert valve. After 2 min, the divert valve position was automatically toggled and flow over the cartridge changed to 1 µL/min solvent A (running directly to the ESI inlet), which was ramped by use of acetonitrile (solvent B) over 8 min to 1/99 water/acetonitrile. By 10.2 min, the run was completed and the flow back to 100% solvent A. All proteins targets eluted between 5 and 7.5 min into a Bruker MicrOTOF-Q (quadrupole time-of-flight) mass spectrometer operating in positive-ion, TOF-only mode, acquiring spectra in the m/z range of 50–3000. ESI settings for the Agilent G1385A capillary nebulizer ion source were as follows: end plate offset −500 V, capillary −4500 V, nebulizer nitrogen 2 bar, dry gas nitrogen 3.0 L/min at 225 °C. Data were acquired in profile mode at a digitizer sampling rate of 2 GHz, with spectra rate control by summation at 1 Hz. Spectra were mass calibrated by use of a multipoint calibration curve generated from Agilent’s proprietary ESI-MS tuning mix. For semiquantitative data, 1.5 min of recorded spectra were averaged across the chromatographic peak apex of protein elution. The ESI charge-state envelope was deconvoluted with Bruker DataAnalysis v3.4 software to a mass range of 1000 Da on either side of any identified peak. Deconvoluted ESI mass spectra were baseline subtracted, and all peaks were integrated. For each sample, we measured and recorded the relative abundances of the variants by normalizing the integral of each variant to the summed integrals of all observed forms of the protein.
DATA PREPARATION
Each assay analyzed >1 molecular species, resulting in a total of 41 values for each of the 212 individuals. Data for each molecular species were tabulated as relative abundance per individual. For all species, the lower limit of quantification was approximately 1% while maintaining standard errors of <10% [i.e., 1% (0.1%)], as illustrated elsewhere (16, 20). The relative abundance data for each analyte were then mean-centered and normalized by the SD of the entire population, so that each molecular species was treated (weighted) equally during subsequent analyses.
DATA ANALYSIS: UNSUPERVISED PRINCIPAL COMPONENT ANALYSIS
We applied principal component analysis (PCA) (25) to the entire data set of 41 variables observed in 212 individuals. The first 3 PC scores were extracted and used to create a 3-dimensional plot, after which color coding was superimposed on each point corresponding to the subgroups to which the point belonged: HP (dark green), HS (light green), T2D (red), T2D/MI/ CHF (dark blue), CHF/T2D (orange), MI/CHF (light blue), and CHF (yellow). To obtain ROC area under the curve (AUC) values between each group, a best-separating plane was constructed in the 3D space by use of a linear kernel support vector machine classifier which places, orthogonal to this plane,a single line representing a new univariate scale that cuts across the 3-dimensional space (see online Supplemental Fig. 1 for a graphical representation of this process). Using this new 1-dimensional scale, ROC curves were constructed for every possible pair of cohorts and the area underneath them obtained in the traditional manner (Table 1). For readers interested in further discussion of the significance of ROC curves, Zweig and Campbell (26) provide an excellent review on the use of ROC plots in the clinical laboratory. Matlab software was used to implement the support vector machine classifier.
Table 1.
| Healthy serum |
T2D | CHF | CHF/MI/T2D | CHF/T2D | CHF/MI | ||
|---|---|---|---|---|---|---|---|
| All healthy | 1.00 | 0.98 | 1.00 | 0.98 | 1.00 | Conventional PCA | |
| 1.00 | 0.95 | 1.00 | 0.99 | 1.00 | Biologically supervised PCAb | ||
| Healthy plasma | 0.99 | 1.00 | 0.99 | 1.00 | 0.98 | 1.00 | Conventional PCA |
| 0.80 | 1.00 | 0.97 | 1.00 | 1.00 | 1.00 | Biologically supervised PCA | |
| Healthy serum | 1.00 | 0.99 | 1.00 | 0.98 | 1.00 | Conventional PCA | |
| 1.00 | 0.95 | 1.00 | 0.99 | 1.00 | Biologically supervised PCA | ||
| T2D | 1.00 | 0.89 | 1.00 | 0.98 | Conventional PCA | ||
| 1.00 | 0.76 | 1.00 | 0.95 | Biologically supervised PCA | |||
| CHF | 0.98 | 0.77 | 0.98 | Conventional PCA | |||
| 1.00 | 0.84 | 1.00 | Biologically supervised PCA | ||||
| CHF/MI/T2D | 0.97 | 0.81 | Conventional PCA | ||||
| 0.97 | 0.94 | Biologically supervised PCA | |||||
| CHF/T2D | 0.97 | Conventional PCA | |||||
| 0.99 | Biologically supervised PCA |
For each paired cohort comparison in this pivot table, linear kernel support vector machine was used to draw a best-separating plane across 3 dimensions to generate distributions along an orthogonal axis, which became a new univariate scale by which to produce ROC curves.
x Axis is PC1 of albumin, DBP, CRP, B2M, and CysC glycation; yaxis is PC1 of apoAI and apoCI oxidation; z axis is PC1 of native apoC-I, apoC-I [3–57], native RANTES, and RANTES [3–68].
DATA ANALYSIS: BIOLOGICALLY SUPERVISED PCA
Protein variants were grouped by identity to align with common themes of (a) protein glycation (glycemic control), (b) protein oxidation (oxidative stress), or (c) protein truncation (alterations in enzymatic and signaling pathways). Using PCA, each marker was evaluated in combination with the other markers within a common pathobiology to find the most efficient and significant combination of markers—i.e., the minimum number of markers that provided the greatest degree of separation between disease subgroups. In this manner, glycation markers under consideration were reduced to (glycated) albumin, B2M, CysC, VDBP, and CRP. Oxidation markers under consideration were sulfoxidized apoAI and apoC1. Truncation markers were found as N-terminal di-peptide truncations on RANTES and apoC1. We used PCA to condense multimarker data in each category into a single metric (PC1), resulting in 3 numerical descriptors (1 for each of the pathobiologies) for each sample, which were plotted in 3D with color coding as described above. We ascertained ROC AUCs between all possible pairwise combinations of subgroups as described above (Table 1).
Results
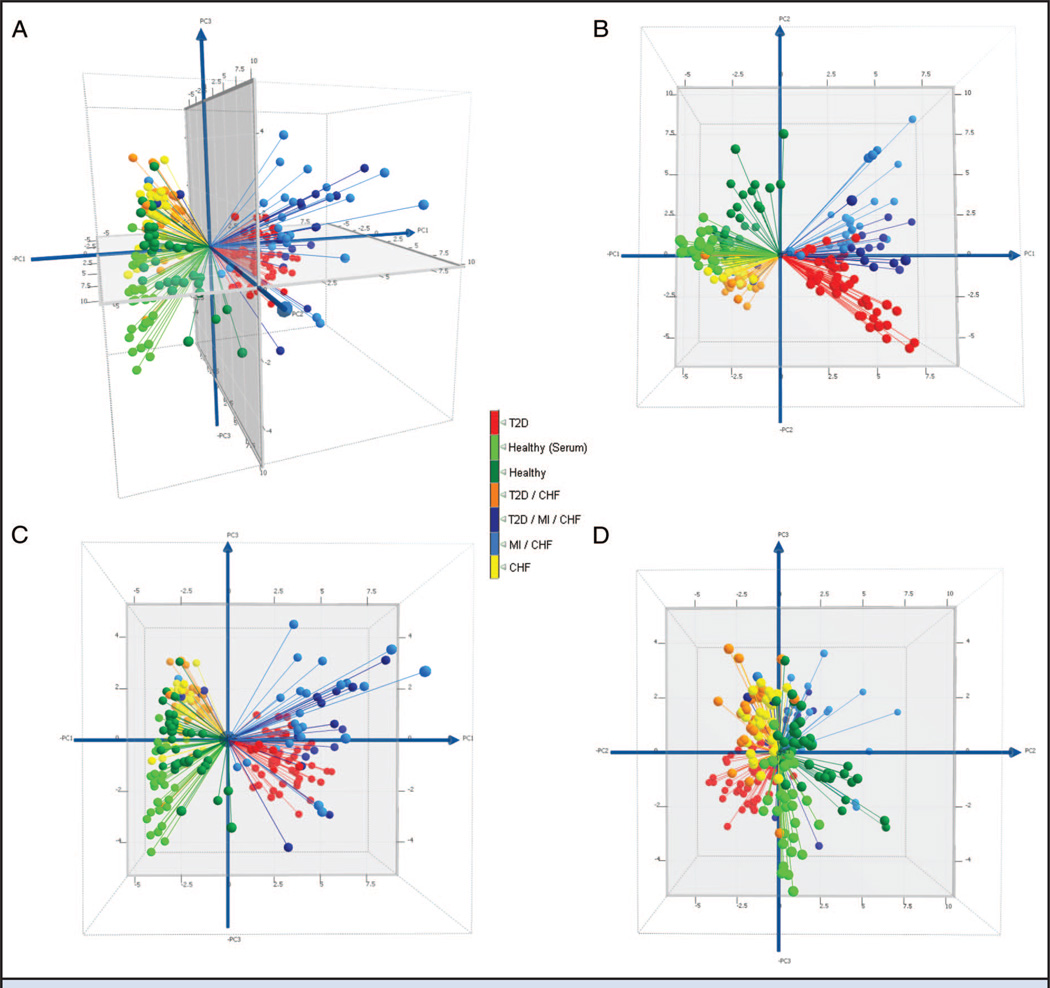
We evaluated summary data using unsupervised and supervised multidimensional approaches for usefulness in differentiating the different subclasses within the population. We applied variable normalized PCA to the entire data set (212 individuals × 41 molecular species; see online Data Supplement) to reveal whether the collective data were able to differentiate the subgroups in an unsupervised (unbiased) mode, and if so, for use as a point of comparison for subsequent supervised approaches. Fig. 1A shows a 3D plot of the first 3 PC scores (with bisecting planes shown in Fig. 1, B–D). Two clusters were observed for healthy samples, indicating that the approach is sensitive enough to differentiate between the sample presentation of serum (light green) and plasma (dark green). Individuals with T2D were also apparent as a separate cluster (red). The CHF (yellow) and CHF with T2D (orange) cohorts were clustered together with moderate visual separation between the 2 subgroups. Similarly, individuals with CHF and MI (light blue), and those with all 3 conditions (dark blue), were clustered in 1 group with moderate separation. On evaluation of the loading values contributing most significantly to the principal components, we found PC1 to be most influenced by oxidation of apolipoproteins (sulfoxide formation), and PC2 and PC3 were influenced most significantly by protein truncation. Interestingly, protein glycation was not in the top 5 loading values for any of PC1–3, despite the fact that one eighth of the variables in the data set stem from protein glycation.
Fig. 1. Conventional PCA score plots for′ the entire data set (PC1 vs PC2 vs PC3) (A); PC2 vs PC1 (B); PC3 vs PC1 (C); and PC3 vs PC2 (D).
PC1 was found to be most influenced by oxidation of apolipoproteins (sulfoxide formation), and PC2 and PC3 were influenced most significantly by protein truncation. ROC AUCs between all possible pairwise cohort combinations are shown in Table 1.
ROC AUC values between the subgroups are reported in Table 1. A significant ROC value was observed between healthy plasma and serum (0.99). In contrast, the ROC AUC values of the disease subgroups did not change significantly compared with either of the healthy sample presentations. These findings indicate that the molecular differences present in the disease subgroups were more pronounced than those found in the healthy controls (regardless of sample presentation). As a note of protocol, biomarker development should be undertaken with standardized (singular) sample presentation. Accordingly, all subgroups under investigation were presented in plasma, which comparatively yielded ROC AUC values of 0.98–1.0 between healthy groups and any of the disease subgroups. ROC AUC values between the disease subgroups ranged from 0.77 to 1.0, indicating moderate to excellent separation of the nonhealthy subcohorts.
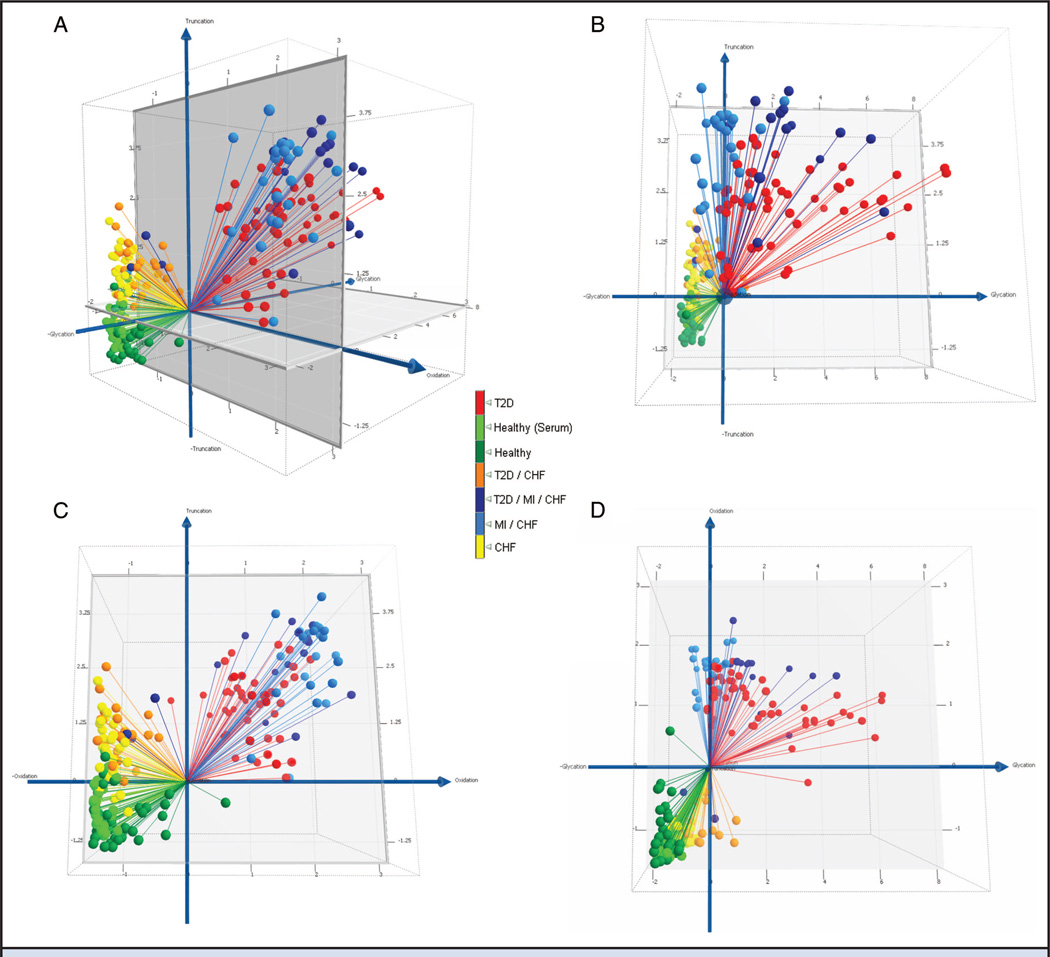
We subjected the data to a second modeling treatment based on the identity of the molecular variants. Differential glycation patterns, although not heavy contributors to cohort separation in the unsupervised PCA model, were observed consistently across the subgroups in albumin, VDBP, CRP, B2M, and CysC. Consistent with the unsupervised PCA model, differential oxidation was observed most prominently as methionine sulfoxidation of apoAI and apoCI. Also consistent was the observation of N-terminal truncations for RANTES and apoCI (Fig. 2). We grouped these markers into categories of protein glycation, oxidation, and truncation and performed PCA separately on each category. We then created a 3D plot of glycation vs oxidation vs truncation by plotting the PC1 score for each category of protein modification along a separate axis (Fig. 3A). All disease subgroups were observed to differentiate from healthy serum and/or plasma (light and dark green, respectively) along the truncation (z) axis. Separation of related disease subgroups then occurred primarily in planes. CHF without T2D (yellow) were separated from CHF with T2D (orange) primarily on the truncation-vs-glycation plane (Fig. 3B). Individuals with CHF (yellow) were separated from their counterparts with MI (light blue) in the truncation-vs-oxidation plane (Fig. 3C). Differentiation of T2D- and MI-containing subgroups occurred primarily in the glycation-vs-oxidation plane (Fig. 3D). The MI-containing subgroups were observed at the extreme of oxidation defined by the stark-T2D subgroups (red), and were separated into 2 clusters on the glycation axis dependent on the presence or absence of T2D (dark and light blue, respectively). Treated in these 3 dimensions, ROC values of 0.97–1.0 were observed between the healthy (plasma) group and any disease subgroup, and 0.76–1.0 between the disease subgroups (Table 1).
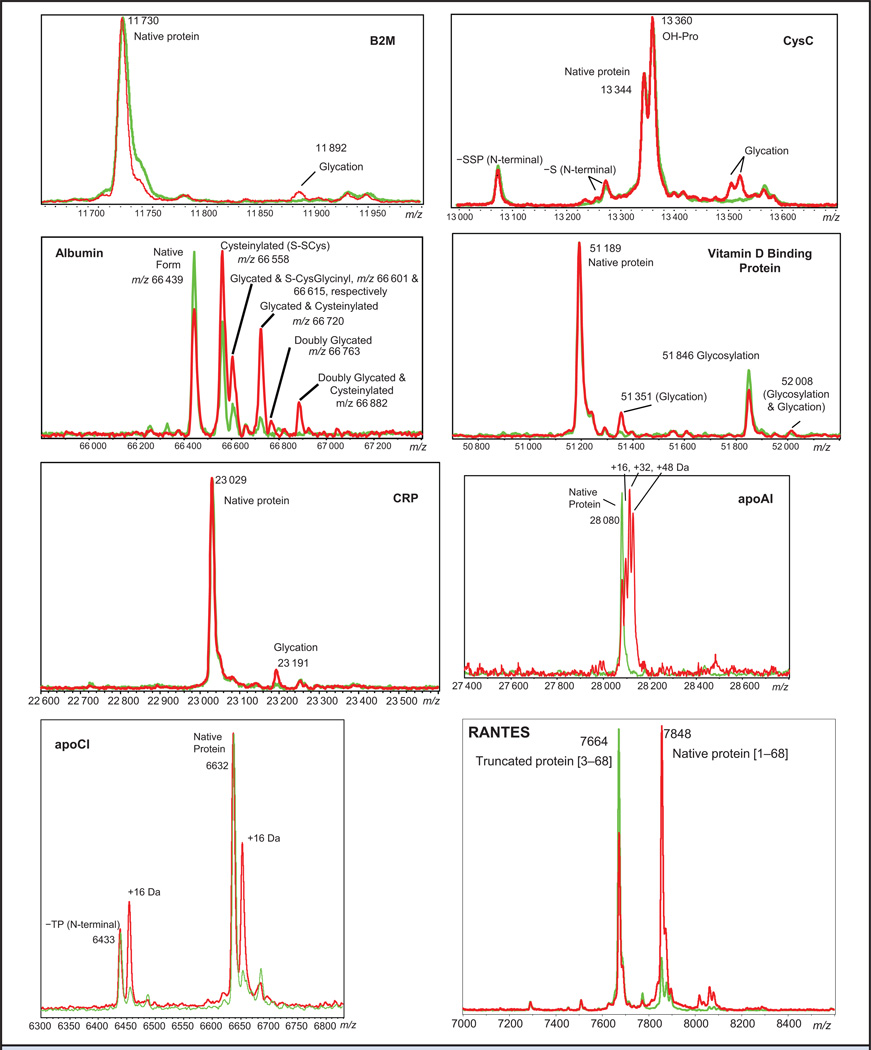
Fig. 2. Representative mass spectra of glycated, oxidized, and/or truncated proteins.
Spectra from healthy patients are shown in green, and spectra from diabetic patients are shown in red. Differences in protein glycation, oxidation, and truncation between the patient cohorts are apparent in the mass spectra. Proteins with a nominal molecular weight >20 kDa were analyzed by ESI-MS.
Fig. 3. Biologically supervised multidimensional analysis.
(A), 3D representation of T2D/CVD pathobiology manifested as protein modifications. The x axis is PC1 of a PCA in which only albumin, VDBP, CRP, B2M, and CysC glycation were included as variables. Likewise, the y axis is PC1 for apoAI and apoCI oxidation, and the zaxis, PC1 for RANTES and apoCI truncation. (B), Truncation vs. glycation. (C), Truncation vs. oxidation. (D), Oxidation vs. glycation. ROC AUCs between all possible pairwise cohort combinations are shown in Table 1.
Discussion
Based on the complex pathobiologies involved, it is reasonable to anticipate that neither a single marker, nor even multiple markers focused on a single biological pathway, will be able to adequately indicate developing CVD in the context of T2D. From a biomarker development point of view, this paradigm requires unifying multiple markers aligned with common and specific pathobiologies of metabolic syndrome (27), diabetes (28), and CVD (29)—i.e., the upstream causes and downstream effects of T2D and CVD. Surveying multiple markers that align commonly with a particular theme amplifies the detection of a specific pathobiology, and with the progressive addition of multiple pathobiologies, builds additional axes relevant to diabetes/comorbidity detection and differentiation. Here, we evaluated multiple markers in this manner, foremost by using unsupervised data evaluation, which confirmed, in an unbiased mode, that determinant data was present within the microheterogeneity evident in the target proteins. Subsequent understanding of the chemical nature of this microheterogeneity led to rationally combining multiple markers to reflect and amplify pathophysiologies common to cardiometabolic disorders.
Specifically, increased proportions of glycation were observed in 5 proteins. Notably, this is the same post-translational modification that is currently used to evaluate diabetes [i.e., glycohemoglobin B-chain (Hb A1c)]. Thus, in the context of increased blood glucose, the additional glycated-protein markers make sense; albeit now with 5 blood-borne protein precursors instead of 1. Other than a monitor of glucose concentrations, glycation is a precursor to the formation of intracellular and plasmaborne advanced glycation endproducts (AGEs) that are found to accumulate in arterial and microvascular plaques (28). Thus, the monitor of glycated-protein to AGE transitions focuses on molecular transformations involved in both diabetes and CVD.
Differential oxidation was observed most significantly in select apolipoproteins (e.g., apoAI and apoCI). Hyperglycemia-induced production of superoxide begins to explain this phenomenon (28), as it forms oxidized lipids that are known to oxidize apolipoproteins by direct oxidation at methionine groups (30). Consequences of apolipoprotein oxidation range from reduction of cholesterol efflux, to foam cell formation (as a precursor to arterial plaques), to attenuation of antiinflammatory and antioxidant defensive properties of HDL (31, 32). In these manners, analysis of oxidized HDL apolipoproteins serves as a monitor of both antagonistic and defense mechanisms contributing to CVD.
The third axis reflects truncated variants of proteins that can serve as substrates for dipeptidyl peptidase IV (DPP-IV; EC 3.4.14.5), which, incidentally, is a target for new drug candidates (inhibitors) for T2D treatment (33). Both RANTES and apoCI contain DPP-IV recognition motifs and exhibit truncations similar to those previously observed in other signaling peptides involved in T2D and CVD (34–37). In this manner, the ratio of truncated-to-intact variants of these 2 proteins serves simply as a surrogate monitor of abnormal DPP-IV activity in the disease subpopulations. In future studies, however, other correlations— such as attenuated activity of truncated RANTES as a chemokine and influences on ApoCI activation of lecithin cholesterol acyltransferase (38–40)—may be considered.
These studies have produced multidimensional “views” based on microheterogeneity aligned with disease pathobiologies, which are able to accurately differentiate between small scouting cohorts of different subtypes of T2D/CVD. We consider these findings a promising starting point for expanded studies—i.e., addition of more markers reflective of other pathobiologies and application to large longitudinal cohorts— aimed at verifying multimarker panels for use in monitoring across the T2D-CVD continuum.
Supplementary Material
Acknowledgments
Research Funding: R.W. Nelson, NIH grants R24DK083948 and R01DK082542.
Role of Sponsor: The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, or preparation or approval of manuscript.
Footnotes
Nonstandard abbreviations: T2D, type 2 diabetes; Hb A1c, glycohemoglobin; CVD, cardiovascular disease; ACCORD, Action to Control Cardiovascular Disease in Diabetes; ADVANCE, Action in Diabetes and Vascular Disease: Preterax and Diamicron MR Controlled Evaluation; VADT, VA Diabetes Trial; FDA, US Food and Drug Administration; MSIA, mass spectrometric immunoassay; CHF, congestive heart failure; MI, myocardial infarction; apo, apolipoprotein; VDBP, vitamin D binding protein; TTR, transthyretin; B2M, β2-microglobulin; CysC, cystatin C; SAP, serum amyloid P; CRP, C-reactive protein; MES, 2-(N-morpholino)ethanesulfonic acid; CDI, 1,1′-carbonyldiimidazole; HBS, HEPES-buffered saline; HP, healthy plasma; HS, healthy serum; ESI-TOF-MS, electrospray ionization time-of-flight mass spectrometry; FWHM, full width at half maximum; PCA, principal component analysis; AUC, area under the curve; AGE, advanced glycation endproduct; DPP-IV, dipeptidyl peptidase IV.
Author Contributions:All authors confirmed they have contributed to the intellectual content of this paper and have met the following 3 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; and (c) final approval of the published article.
Authors’ Disclosures or Potential Conflicts of Interest: Upon manuscript submission, all authors completed the Disclosures of Potential Conflict of Interest form. Potential conflicts of interest:
Employment or Leadership: R.W. Nelson, President, Intrinsic Bio-probes, Inc.
Consultant or Advisory Role: R.W. Nelson, Intrinsic Bioprobes, Inc.
Stock Ownership: R.W. Nelson, Intrinsic Bioprobes, Inc.
Honoraria: None declared.
Expert Testimony: None declared.
References
- 1.Brunner EJ, Shipley MJ, Witte DR, Fuller JH, Marmot MG. Relation between blood glucose and coronary mortality over 33 years in the Whitehall Study. Diabetes Care. 2006;29:26–31. doi: 10.2337/diacare.29.01.06.dc05-1405. [DOI] [PubMed] [Google Scholar]
- 2.Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Suppl 1):S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bierman EL. George Lyman Duff Memorial Lecture: Atherogenesis in diabetes. Arterioscler Thromb. 1992;12:647–656. doi: 10.1161/01.atv.12.6.647. [DOI] [PubMed] [Google Scholar]
- 4.Libby P, Plutzky J. Diabetic macrovascular disease: the glucose paradox? Circulation. 2002;106:2760–2763. doi: 10.1161/01.cir.0000037282.92395.ae. [DOI] [PubMed] [Google Scholar]
- 5.Selvin E, Marinopoulos S, Berkenblit G, Rami T, Brancati FL, Powe NR, Golden SH. Meta-analysis: glycosylated hemoglobin and cardiovascular disease in diabetes mellitus. Ann Intern Med. 2004;141:421–431. doi: 10.7326/0003-4819-141-6-200409210-00007. [DOI] [PubMed] [Google Scholar]
- 6.Selvin E, Steffes MW, Zhu H, Matsushita K, Wagenknecht L, Pankow J, et al. Glycated hemoglobin, diabetes, and cardiovascular risk in non-diabetic adults. N Engl J Med. 2010;362:800–811. doi: 10.1056/NEJMoa0908359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerstein HC, Miller ME, Byington RP, Goff DC, Jr, Bigger JT, Buse JB, et al. Effects of intensive glucose lowering in type 2 diabetes. N Engl J Med. 2008;358:2545–2559. doi: 10.1056/NEJMoa0802743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duckworth W, Abraira C, Moritz T, Reda D, Emanuele N, Reaven PD, et al. Glucose control and vascular complications in veterans with type 2 diabetes. N Engl J Med. 2009;360:129–139. doi: 10.1056/NEJMoa0808431. [DOI] [PubMed] [Google Scholar]
- 9.Patel A, MacMahon S, Chalmers J, Neal B, Billot L, Woodward M, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358:2560–2572. doi: 10.1056/NEJMoa0802987. [DOI] [PubMed] [Google Scholar]
- 10.Nissen SE, Wolski K. Rosiglitazone revisited: an updated meta-analysis of risk for myocardial infarction and cardiovascular mortality. Arch Intern Med. 2010;170:1191–1201. doi: 10.1001/archinternmed.2010.207. [DOI] [PubMed] [Google Scholar]
- 11.FOA. FDA guidance for industry: diabetes mellitus: evaluating cardiovascular risk in new antidiabetic therapies to treat type 2 diabetes. U.S. Department of Health and Human Services, Food and Drug Administration, Center for Drug Evaluation and Research (CDER) 2008 [Google Scholar]
- 12.Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. Investigating diversity in human plasma proteins. Proc Natl Acad Sci U S A. 2005;102:10852–10857. doi: 10.1073/pnas.0500426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niederkofler EE, Tubbs KA, Gruber K, Nedelkov D, Kiernan UA, Williams P, Nelson RW. Determination of beta-2 microglobulin levels in plasma using a high-throughput mass spectrometric immunoassay system. Anal Chem. 2001;73:3294–3299. doi: 10.1021/ac010143j. [DOI] [PubMed] [Google Scholar]
- 14.Niederkofler EE, Tubbs KA, Kiernan UA, Nedelkov D, Nelson RW. Novel mass spectrometric immunoassays for the rapid structural characterization of plasma apolipoproteins. J Lipid Res. 2003;44:630–639. doi: 10.1194/jlr.D200034-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Borges CR, Jarvis JW, Oran PE, Rogers SP, Nelson RW. Population studies of intact vitamin D binding protein by affinity capture ESI-TOF-MS. J Biomol Tech. 2008;19:167–176. [PMC free article] [PubMed] [Google Scholar]
- 16.Borges CR, Rehder DS, Jarvis JW, Schaab MR, Oran PE, Nelson RW. Full-length characterization of proteins in human populations. Clin Chem. 2010;56:202–211. doi: 10.1373/clinchem.2009.134858. [DOI] [PubMed] [Google Scholar]
- 17.Kiernan UA, Addobbati R, Nedelkov D, Nelson RW. Quantitative multiplexed C-reactive protein mass spectrometric immunoassay. J Proteome Res. 2006;5:1682–1687. doi: 10.1021/pr0601133. [DOI] [PubMed] [Google Scholar]
- 18.Kiernan UA, Nedelkov D, Nelson RW. Multiplexed mass spectrometric immunoassay in biomarker research: a novel approach to the determination of a myocardial infarct. J Proteome Res. 2006;5:2928–2934. doi: 10.1021/pr060062+. [DOI] [PubMed] [Google Scholar]
- 19.Kiernan UA, Nedelkov D, Tubbs KA, Niederkofler EE, Nelson RW. Proteomic characterization of novel serum amyloid P component variants from human plasma and urine. Proteomics. 2004;4:1825–1829. doi: 10.1002/pmic.200300690. [DOI] [PubMed] [Google Scholar]
- 20.Oran PE, Sherma ND, Borges CR, Jarvis JW, Nelson RW. Intrapersonal and populational heterogeneity of the chemokine RANTES. Clin Chem. 2010;56:1432–1441. doi: 10.1373/clinchem.2010.147884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nedelkov D, Tubbs KA, Niederkofler EE, Kiernan UA, Nelson RW. High-throughput comprehensive analysis of human plasma proteins: a step toward population proteomics. Anal Chem. 2004;76:1733–1737. doi: 10.1021/ac035105+. [DOI] [PubMed] [Google Scholar]
- 22.Jaleel A, Halvatsiotis P, Williamson B, Juhasz P, Martin S, Nair KS. Identification of Amadori-modified plasma proteins in type 2 diabetes and the effect of short-term intensive insulin treatment. Diabetes Care. 2005;28:645–652. doi: 10.2337/diacare.28.3.645. [DOI] [PubMed] [Google Scholar]
- 23.Bar-Or D, Heyborne KD, Bar-Or R, Rael LT, Winkler JV, Navot D. Cysteinylation of maternal plasma albumin and its association with intrauterine growth restriction. Prenat Diagn. 2005;25:245–249. doi: 10.1002/pd.1122. [DOI] [PubMed] [Google Scholar]
- 24.Kiernan UA, Nedelkov D, Niederkofler EE, Tubbs KA, Nelson RW. High-throughput affinity mass spectrometry. Methods Mol Biol. 2006;328:141–150. doi: 10.1385/1-59745-026-X:141. [DOI] [PubMed] [Google Scholar]
- 25.Jolliffe IT. Principal component analysis. Springer-Verlag; 1986. pp. 78–108. [Google Scholar]
- 26.Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]
- 27.Reaven GM. Banting Lecture 1988: Role of insulin resistance in human disease. Diabetes. 1988;37:1595–1607. doi: 10.2337/diab.37.12.1595. [DOI] [PubMed] [Google Scholar]
- 28.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 29.Stump CS, Clark SE, Sowers JR. Oxidative stress in insulin-resistant conditions: cardiovascular implications. Treat Endocrinol. 2005;4:343–351. doi: 10.2165/00024677-200504060-00003. [DOI] [PubMed] [Google Scholar]
- 30.Mashima R, Yamamoto Y, Yoshimura S. Reduction of phosphatidylcholine hydroperoxide by apolipoprotein A–I: purification of the hydroperoxide-reducing proteins from human blood plasma. J Lipid Res. 1998;39:1133–1140. [PubMed] [Google Scholar]
- 31.Shao B, Cavigiolio G, Brot N, Oda MN, Heinecke JW. Methionine oxidation impairs reverse cholesterol transport by apolipoprotein A–I. Proc Natl Acad Sci U S A. 2008;105:12224–12229. doi: 10.1073/pnas.0802025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barter PJ, Nicholls S, Rye KA, Anantharamaiah GM, Navab M, Fogelman AM. Antiinflammatory properties of HDL. Circ Res. 2004;95:764–772. doi: 10.1161/01.RES.0000146094.59640.13. [DOI] [PubMed] [Google Scholar]
- 33.McIntosh CH, Demuth HU, Kim SJ, Pospisilik JA, Pederson RA. Applications of dipeptidyl peptidase IV inhibitors in diabetes mellitus. Int J Biochem Cell Biol. 2006;38:860–872. doi: 10.1016/j.biocel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 34.Oran PE, Jarvis JW, Borges CR, Nelson RW. C-peptide microheterogeneity in type 2 diabetes populations. Proteomics Clin Appl. 2010;4:1–6. doi: 10.1002/prca.200800249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niederkofler EE, Kiernan UA, O’Rear J, Menon S, Saghir S, Protter AA, et al. Detection of endogenous B-type natriuretic peptide at very low concentrations in patients with heart failure. Circ Heart Fail. 2008;1:258–264. doi: 10.1161/CIRCHEARTFAILURE.108.790774. [DOI] [PubMed] [Google Scholar]
- 36.Rasmussen HB, Branner S, Wiberg FC, Wagtmann N. Crystal structure of human dipeptidyl peptidase IV/CD26 in complex with a substrate analog. Nat Struct Biol. 2003;10:19–25. doi: 10.1038/nsb882. [DOI] [PubMed] [Google Scholar]
- 37.Brandt I, Lambeir AM, Maes MB, Scharpe S, De Meester I. Peptide substrates of dipeptidyl peptidases. In: Lendeckel U, Reinhold D, Bank U, editors. Dipeptidyl aminopeptidases: basic science and clinical applications. Springer Science+Business Media; 2006. pp. 3–18. [Google Scholar]
- 38.Jong MC, Hofker MH, Havekes LM. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol. 1999;19:472–484. doi: 10.1161/01.atv.19.3.472. [DOI] [PubMed] [Google Scholar]
- 39.Tall AR. An overview of reverse cholesterol transport. Eur Heart J. 1998;19(Suppl A):A31–A35. [PubMed] [Google Scholar]
- 40.Spady DK. Reverse cholesterol transport and atherosclerosis regression. Circulation. 1999;100:576–578. doi: 10.1161/01.cir.100.6.576. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.