Summary
DNA damage is linked to multiple human diseases, such as cancer, neurodegeneration and senescence. Little is known about the role of chromatin accessibility in DNA repair. Here, we find that the histone deacetylase SIRT6 is one of the earliest factors recruited to sites of Double-Strand Breaks (DSBs). SIRT6 recruits the ISWI-chromatin remodeler SNF2H to DSBs, and deacetylates focally histone H3K56. Lack of SIRT6 and SNF2H impairs chromatin remodeling, increasing sensitivity to genotoxic damage and recruitment of downstream factors, such as 53BP1, BRCA1 and RPA. Remarkably, SIRT6 deficient mice exhibit lower levels of chromatin-associated SNF2H in specific tissues, a phenotype accompanied by increased DNA damage. We demonstrate that SIRT6 is critical for recruitment of a chromatin remodeler as an early step in the DNA damage response, indicating that proper unfolding of chromatin plays a rate-limiting role. We present a novel crosstalk between a histone modifier and a chromatin remodeler, regulating a coordinated response to prevent DNA damage.
Introduction
Preservation of DNA integrity is critical to ensure accurate inheritance of the genetic material, as well as proper cellular and organismal function. Intrinsic and extrinsic processes such as DNA transcription and replication, cellular metabolism and environmental challenges, represent persistent genotoxic threats. Indeed, unrepaired DNA damage can frequently lead to cell senescence, apoptosis or tumorigenesis, and thus jeopardize organismal well-being (Papamichos-Chronakis and Peterson, 2012). Multiple mechanisms have thus evolved to protect and repair damaged DNA. Probably the most critical DNA lesions are double-strand breaks (DSBs), which can result in loss of genetic material, mutations and deleterious translocations. Consequently, cells have evolved two primary DSB repair mechanisms: Non-Homologous End Joining (NHEJ), a mutation-prone pathway that repairs DSBs by joining two ends together, and the error-free Homologous Recombination (HR) pathway, which operates only when sister chromatids are paired together (Chapman et al., 2012). A number of factors are involved in the recognition, amplification and repair cascade that is triggered by DSBs, a process known as the DNA Damage Response (DDR) (Ciccia and Elledge, 2010). In this orchestrated response that is set in motion, breaks are “sensed” by members of the PARP family, which activates PI3K-related kinases including ATM, ATR and DNA-PK. These proteins in turn recruit sensors that amplify the signal, including the MRN complex and multiple histone modifiers (such as Tip60, RNF8 and RNF168), which orchestrate a broad spectrum of histone post-translational modifications, including methylation, acetylation, ubiquitylation, and phosphorylation. In turn, these modifications work in concert to recruit DNA repair factors, such as 53BP1, Rad51 and DNA ligases to faithfully repair the broken DNA (Lukas et al., 2011).
Eukaryotic DNA is packaged within nucleosomes, which represents an additional physical barrier for DDR factors to access damaged DNA. Only in recent years the role of chromatin accessibility in DNA repair has begun to emerge. Various chromatin remodelers, including INO80 (Gospodinov et al., 2011; Kashiwaba et al., 2010; Neumann et al., 2012; Papamichos-Chronakis et al., 2011), SMARCAD1 (Fun 30 in yeast) (Chen et al., 2012; Costelloe et al., 2012); (Lee et al., 2010), p400 (Xu et al., 2012; Xu et al., 2010), CHD4 (Larsen et al., 2010; Polo et al., 2010), and the NuRD complex (Smeenk et al., 2010) were shown to be recruited to sites of damage, suggesting the need of chromatin relaxation and remodeling in order to allow repair (Papamichos-Chronakis and Peterson, 2012). Of note, most of the above factors have been mainly characterized in yeast, and whether mammalian cells exhibit alterations in chromatin structure during DSB repair as well as the precise mechanisms regulating chromatin dynamics in the context of DNA repair remain poorly understood. Interestingly, the ISWI family member SNF2H, an ATP-dependent chromatin remodeler with roles in transcription and replication (Erdel and Rippe, 2011), has been proposed to be recruited to sites of DNA damage downstream of the ubiquitin ligase RNF20 (Erdel et al., 2010; Lan et al., 2010; Nakamura et al., 2011; Smeenk et al., 2010). However, the specific signals recognized by SNF2H for its targeting to DNA damage sites remain unknown.
In this study, we have uncovered a new role for the histone deacetylase SIRT6 as a scaffold protein in DDR. SIRT6 is a chromatin-bound protein that belongs to the highly conserved sirtuin family of NAD(+)-dependent deacetylases with various roles in DNA damage, metabolism and cancer (Finkel et al., 2009; Mostoslavsky et al., 2006; Toiber et al., 2011; Zhong et al., 2010). Following DNA damage, SIRT6 is recruited to sites of DSBs within seconds, specifically recruiting the chromatin remodeler SNF2H to open up condensed chromatin, and deacetylating H3K56, both critical steps required for proper recruitment of downstream DDR factors and efficient DNA repair.
Results
SIRT6 interacts with the chromatin remodeler SNF2H
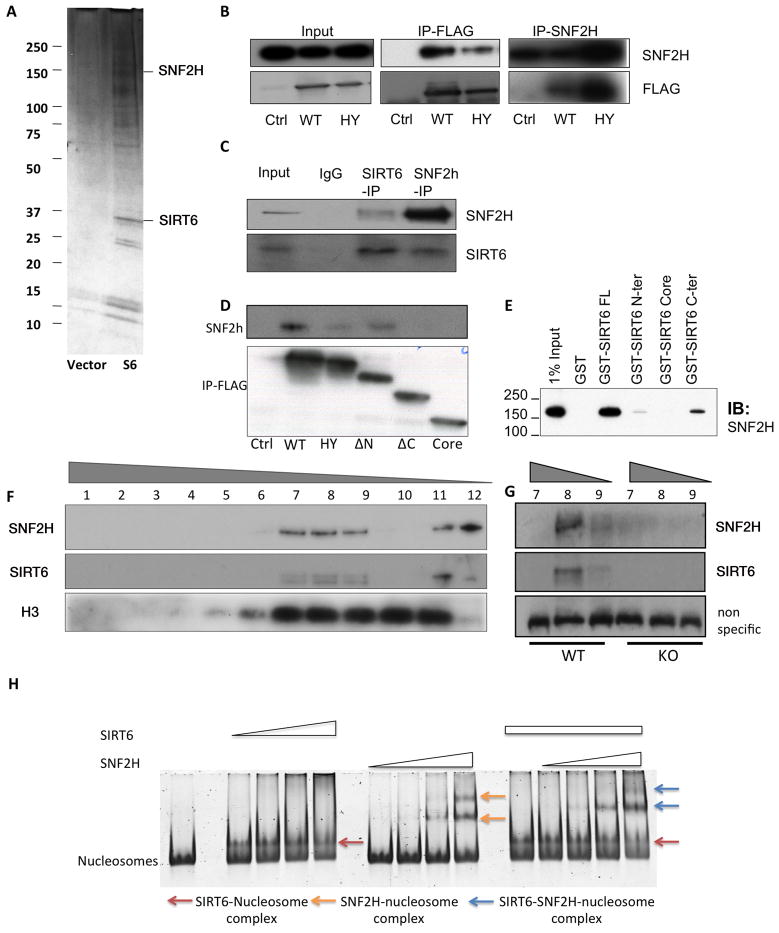
In previous studies, we have shown that SIRT6 deficient cells accumulate genomic instability (Mostoslavsky et al., 2006), yet the precise molecular mechanism behind this defect remained unclear. In order to gain further insights into the molecular functions of SIRT6, we employed affinity purification to identify associated proteins. Flag-tagged SIRT6 was expressed in human embryonal kidney 293T cells and was used as a bait to identify interacting proteins by mass spectrometry. One of the top 5 most abundant SIRT6-interacting proteins was the ISWI-family member SNF2H (Figure 1A; see also Methods). We first confirmed this interaction by reciprocal immunoprecipitation (IP) following Flag-SIRT6 expression in 293T cells (Figure 1B), which demonstrates clear interaction between these proteins. To determine whether such interaction is physiologically relevant, we performed reciprocal co-IP assays with endogenous proteins. Figure 1C shows that endogenous SIRT6 associated with SNF2H in mouse embryonic stem (ES) cells, indicating that these two proteins interact under physiological conditions. To interrogate the specific domains in SIRT6 that were required for this interaction, we expressed SIRT6 lacking the amino-terminus (N), carboxyl-terminus (C) or both domains (Tennen et al., 2010). As shown in Figure 1D, lack of the C-terminus completely abolished the interaction, indicating that this domain was necessary for SNF2H binding. To further confirm whether SIRT6 interacts directly with SNF2H, we generated bacterial recombinant, purified GST-SIRT6 proteins encompassing the full-length protein (334 amino acids), the C-terminus, the catalytic core or the N-terminus of SIRT6, and measured interaction with purified-recombinant SNF2H. In vitro binding assays indicated that GST-SIRT6 interacts directly with SNF2H, and confirmed that SNF2H binds preferentially to the C-terminus of SIRT6 (Figure 1E and S1). Notably, SIRT6 also interacted with multiple proteins described earlier as SNF2H partners (Figure S2A), including ACF1 and WSTF, two-additional subunits of SNF2H-containing chromatin remodeling complexes (Figure S2A–B), indicating that SIRT6 may associate with one of these complexes. Indeed, glycerol-gradient fractionation clearly showed that SNF2H and SIRT6 co-purified in the same molecular weight (MW) fractions (Fractions 7–9, 11 and 12, Figure 1F). Notably, SNF2H was absent in the higher MW fractions of SIRT6-deficient samples (fractions 7–9, Figure 1G), indicating that SIRT6 is required for the association of SNF2H into these higher-molecular weight complexes.
Figure 1. SIRT6 interacts with SNF2H and recruits it to chromatin.
A) Flag-IP of protein extracts from Flag-SIRT6- or empty vector-expressing cells was used for mass spectrometry analysis where SNF2H was identified as SIRT6 interactor. Silver-staining of extracts from Flag-control (Vector) or SIRT6-Flag transfected cells is shown. B–C) Exogenous (B) and endogenous (C) co-IPs were performed, and western blots were developed with the indicated antibodies. Flag-vector (Ctrl), Flag-WT-SIRT6 (WT) and Flag-HY-SIRT6 (HY) transfected cells. D) IP for SIRT6 Flag (WT), SIRT6-HY (HY), SIRT6 fragments lacking N (ΔN), C (ΔC) or both termini (Core). E) GST-Full Length SIRT6 and SIRT6 C-terminus, N-terminus or Core domain were tested in vitro for interaction with baculovirus purified SNF2H. F) Glycerol gradient fractionation for WT cells showing SIRT6, SNF2H and H3 bands. G) Fractions 7–9 from WT and 7–9 from SIRT6-KO cells. H) Nucleosome shift assay was performed with 75nM nucleosomes, SIRT6 (μM range) and SNF2H (nM range). Marked with the arrows are the different complexes formed with the nucleosomes under SIRT6/SNF2H incubation, or both proteins. See also Fig. S1.
SIRT6 enhances SNF2H nucleosome binding
Given the interaction of SIRT6 and SNF2H, we evaluated whether SIRT6 may modulate SNF2H activity and nucleosome binding in vitro. We first measured SNF2H remodeling activity through the Restriction Enzyme Accessibility assay. No differences in SNF2H-dependent chromatin remodeling were observed in the presence or absence of SIRT6 (data not shown), indicating that SIRT6 does not modulate SNF2H remodeling activity. We then tested whether SIRT6 may modulate SNF2H nucleosome binding in vitro. First, we found that SIRT6 binds to nucleosomes even at nM concentrations, and when added together, SIRT6 and SNF2H are able to bind to the same nucleosome, resulting in a different nucleosome shift compared to the shift observed with the individual enzymes. Importantly, in the presence of SIRT6, SNF2H bound nucleosomes at lower concentrations than when incubated alone (Figure 1H). When we quantified levels of free nucleosomes and nucleosomes bound under the different conditions (Figure S1B) SIRT6 reduced the levels of free nucleosomes in the presence of SNF2H by at least 2-fold, indicating that, even in vitro, the presence of SIRT6 increased the ability of SNF2H to bind nucleosomes.
SIRT6 recruits SNF2H to chromatin
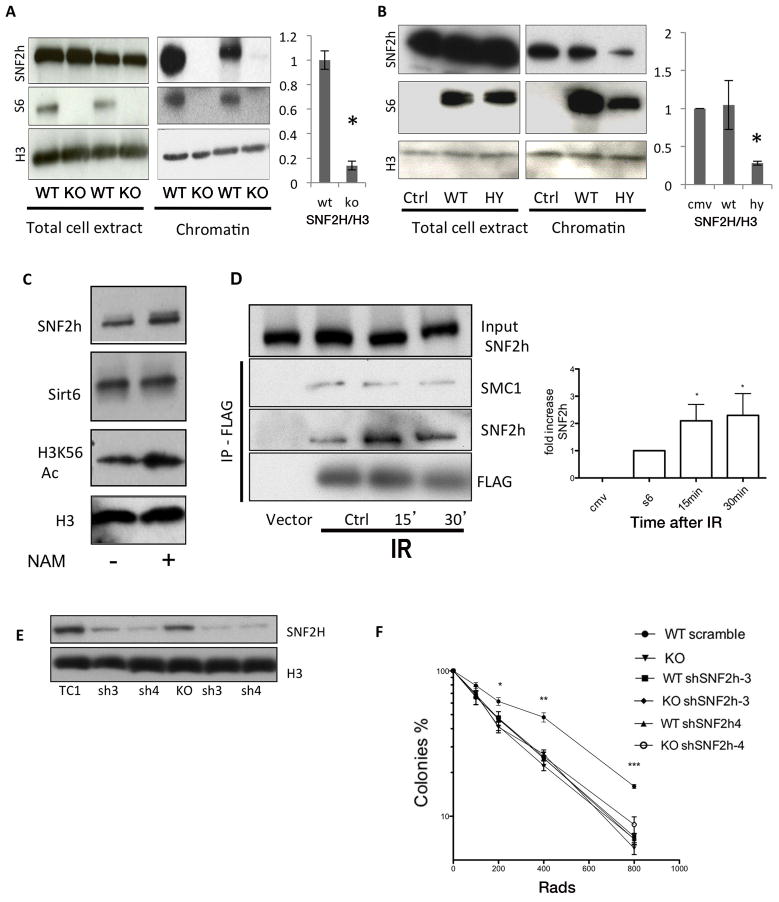
Given the disappearance of SNF2H from the glycerol gradient heavier fractions in SIRT6 deficient cells, we first evaluated whether SIRT6 could modulate SNF2H stability or expression. We did not observe any difference in total SNF2H protein levels in whole-cell extracts (WCE) from SIRT6-deficient mouse ES cells (Figure 2A). Thus, SIRT6 does not affect total SNF2H protein levels. Given their tighter interaction with nucleosomes (Figure 1H), a second possibility would be that SIRT6 and SNF2H interact specifically on chromatin, and lack of SIRT6 might affect chromatin binding of SNF2H. Indeed, biochemical fractionation assays revealed a striking reduction of SNF2H in SIRT6-deficient chromatin fractions (Figure 2A). In order to validate these results, we silenced SIRT6 in two other cell types, U2OS and 293T cells. Again, SNF2H was significantly reduced in chromatin following SIRT6 RNAi-mediated knockdown in these cells (Figure S2D, E). In addition, we took advantage of a catalytic domain mutant form of SIRT6, SIRT6-HY, which remains mainly in the cytoplasm, but still interacts with SNF2H. When SIRT6-HY was overexpressed, SNF2H was significantly decreased in the chromatin fraction, further supporting the conclusion that SIRT6 is required to recruit SNF2H to chromatin (Figure 2B). Finally, we tested whether inhibition of the catalytic activity of chromatin-bound SIRT6 could impair SNF2H recruitment. For this purpose we treated wildtype (WT) cells with nicotinamide (NAM), a sirtuin inhibitor. Inhibition of SIRT6 was confirmed, as H3K56 acetylation (a SIRT6 substrate) was clearly elevated. However, SNF2H recruitment to chromatin was unaffected (Figure 2C), indicating that SIRT6 catalytic activity and H3K56 acetylation are not required for SNF2H recruitment when SIRT6 is already present on chromatin. Together, the above studies clearly demonstrate that SIRT6 is required both in vitro and in mammalian cells to recruit and maintain SNF2H on chromatin.
Figure 2. SIRT6 and SNF2H act together in DNA repair.
A) Whole cell extracts (WCE) and chromatin fractions from WT and SIRT6 KO cells, showing impaired SNF2H recruitment to chromatin. Quantification of SNF2H levels within the chromatin fraction in three independent experiments is shown, B) SIRT6-WT and SIRT6-HY mutant were expressed in 293T cells and the presence of SNF2H in WCE and chromatin was analyzed by western blot. Quantification of SNF2H levels within the chromatin fraction in three independent experiments is shown. C) Western blot with the indicated antibodies was performed using chromatin fractions of NAM-treated or untreated cells. D) Cells were irradiated and Flag-SIRT6 immunoprecipitated 15 or 30 minutes after IR exposure. Cohesin protein SMC1 is shown as a SIRT6 interactor that was not affected by damage as a control. E) SNF2H expression was silenced in Sirt6+/+ and Sirt6−/− ES cells using two different shRNA sequences. F) Survival assays upon IR of ES cells of the indicated genotypes. Data is represented as mean +/− SEM. (right panel, * p< 0.01). See also Fig. S2.
SIRT6 and SNF2H act as epistatic genes
Previous studies have indicated that SNF2H may play a role in promoting genome integrity (Lan et al., 2010). Interestingly, we found that the interaction between SIRT6 and SNF2H increased rapidly upon ionizing radiation (IR) treatment (Figure 2D), suggesting that SIRT6 may recruit SNF2H to sites of DNA damage. To test whether these proteins may act in concert to maintain proper DNA repair, we silenced SNF2H in both WT and SIRT6-deficient ES cells (Figure 2E), and measured survival rates in clonogenic assays. As we showed previously (Mostoslavsky et al., 2006), SIRT6-deficient cells exhibited hypersensitivity to IR. The same phenotype was observed in WT cells in which SNF2H was silenced. However, when SNF2H was silenced in SIRT6 deficient cells, there was no additive effect on survival (Figure 2F). In this context, SIRT6-deficient cells exhibited increased basal levels of phosphorylated p53, a marker of DNA-damage checkpoint activation. SNF2H silencing in these cells did not cause any further increase in p53 phosphorylation, whereas SNF2H silencing in WT cells causes elevated p53 phosphorylation (Figure S2F). Together with the repair assays discussed below, these results indicate that SIRT6 and SNF2H may work through a common pathway.
SIRT6 recruits SNF2H to the sites of DNA damage
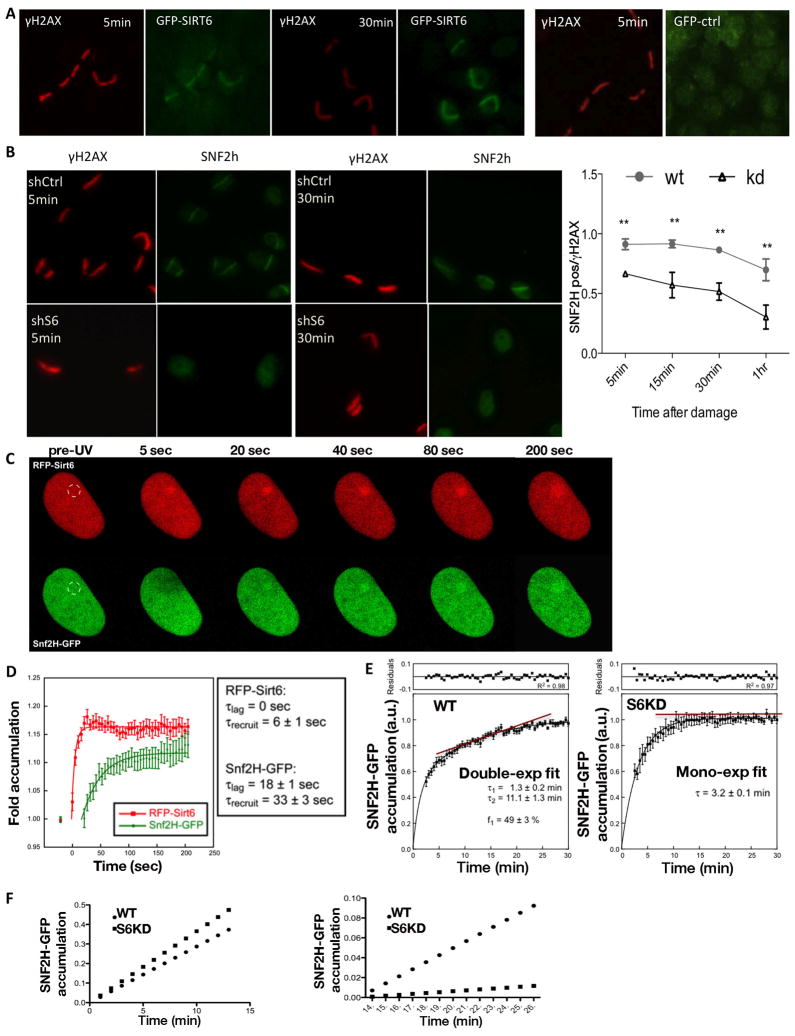
Several lines of evidence suggest that SNF2H might be recruited specifically to DSBs through its interaction with SIRT6: (i) Both SIRT6 and SNF2H are chromatin-bound proteins, and under basal conditions SIRT6 enhanced SNF2H binding to chromatin (Figure 1H, 2A–D). (ii) SIRT6 is mobilized to sites of DNA damage (Kaidi et al., 2010; McCord et al., 2009). (iii) Interaction between SIRT6 and SNF2H was enhanced upon IR (Figure 2D). To further explore this possibility, we measured the recruitment kinetics of SNF2H to sites of DNA breaks, taking advantage of a laser-induced DNA breaks assay. Specifically, we scored for cells with staining for both the DSB marker γH2AX and SNF2H, and measure which proportion of γH2AX-stained cells exhibit SNF2H staining. Notably, SNF2H readily accumulated at sites of DNA damage within 5 minutes in control cells, while such recruitment in SIRT6 down-regulated (shSIRT6) cells was less efficient, and clearly diminished over time (Figure 3B), pointing to a defect in recruitment and/or stabilization of SNF2H at DNA damage sites.
Figure 3. Kinetics of SIRT6 and SNF2H recruitment to DSBs.
A) Laser-induced DNA damage was performed in cells transfected with GFP or GFP-SIRT6, fixed at indicated time-points, and immunostained with anti-γH2AX and anti-GFP antibodies. B) Laser induced DNA damage in U2OS shCtrl or shSIRT6 cells immunostained with γH2AX and SNF2H at different time points. Graph shows time-course analysis. SNF2H-positive cells among γH2AX positive cells were quantified. C) RFP-SIRT6 and SNF2H-GFP recruitment was measured on sites of damage, and followed over time with live cell imaging. A representative series is shown. D) Quantitative analysis of the recruitment experiments described in (C). E) Analysis of SNF2H recruitment in shCtrl versus shSIRT6 cells. Fluorescence intensity is normalized as 1=maximum intensity reached in each case after 30 minutes. F) Graphic representation of the early- and late-recruitment phases of the graphs shown in (E). Data is represented as mean +/− SEM.. *p< 0.05, **p<0.001. See also Fig. S3.
To further characterize SIRT6 and SNF2H kinetics upon DNA damage in real-time, we took advantage of an RFP-SIRT6/SNF2H-GFP system analyzed with live-cell microscopy. Consistent with previous publications (Kaidi et al., 2010; McCord et al., 2009), we observed recruitment of SIRT6 to DNA breaks, however with much faster kinetics than previously reported, arriving at breaks five seconds after DNA damage, and reaching a plateau after 30 seconds. This finding places SIRT6 within the fastest enzymes recruited to sites of DNA damage (Figure 3C, D). SNF2H mobilization to DNA damage sites, although rapid, occurred after SIRT6 recruitment (Figure 3C, D). The time-course plot shows that the time constant for reaching 50% of the plateau value is ~ 40 seconds lower for SIRT6 with respect to SNF2H, indicating a recruitment rate ~5-times faster for RFP-SIRT6 than for SNF2H-GFP. As noted, RFP-SIRT6 recruitment starts earlier after damage induction than that of SNF2H-GFP, consistent with SIRT6 bringing SNF2H to sites of breaks.
SIRT6 accelerates SNF2H binding to DSBs
Our results suggest that SIRT6 brings SNF2H to sites of damage. In order to further dissect this process, we generated shSIRT6 or scramble shRNA-control (shCtrl) cell lines stably expressing a SNF2H-GFP protein (Figure 3E S4A) (Erdel et al., 2010). In this system, we measured SNF2H-GFP recruitment to sites of damage induced by BrdU presensitization and UV laser irradiation (a treatment that primarily generates DSBs). While shCtrl cells exhibited a clear biphasic SNF2H recruitment (i.e. rapid recruitment within seconds, followed by a slower phase of accumulation starting around 7 min), shSIRT6 cells were monophasic, presenting only the early SNF2H recruitment phase (Figure 3C, D). These results indicate that the second phase of SNF2H recruitment is dependent on SIRT6 (Figure 3C, D). To distinguish between active recruitment versus stabilization of SNF2H by SIRT6, we used Fluorescence Loss in Photobleaching (FLIP) experiments -where the decay of SNF2H-GFP signal was measured during both early and late phases- and Fluorescence Recovery After Bleaching (FRAP) experiment -where damage sites were bleached and the SNF2H-GFP recovery was measured-. In these assays, the decay and the recovery of SNF2H-GFP were identical in shCtrl and shSIRT6 U2OS cells at both time points (early and late) (Figure S4B), indicating that stability of SNF2H binding is independent of SIRT6. Interestingly, our FLIP and FRAP results revealed that SNF2H turnover during the early accumulation phase was fast, indicating labile SNF2H binding at the site of breaks in this phase. On the contrary, during the late accumulation phase, SNF2H-GFP remained bound at DNA damage sites for a longer period of time. Since no differences in the SNF2H dissociation kinetics were observed between shCtrl and shSIRT6 cells, we concluded that SIRT6 does not stabilize chromatin-bound SNF2H (i.e. it does not reduces its kinetics dissociation rate), but rather accelerates SNF2H association to sites of damage.
To evaluate whether the SIRT6-independent recruitment (rapid and unstable) of SNF2H occurs at double-strand breaks or else at other types of DNA lesions, we used non-presensitized cells (UV-irradiation only), a treatment that causes mainly pyrimidine dimers, abasic sites and single-strand breaks (resembling oxidative damage) (Kielbassa et al., 1997; Pierce et al., 2001). In this experiment, both shCtrl and shSIRT6 cells exhibited the early SNF2H recruitment phase (Figure S4C), while the late SNF2H recruitment phase was absent, and this unstable SNF2H signal decayed early in both cell types (Figure S4D). Overall, these results strongly suggest that DNA single-strand breaks and other non-DSB lesions cause a brief and unstable SNF2H recruitment that is independent of SIRT6. On the other hand, stable SNF2H recruitment, specific to DSBs, occurs in a SIRT6-dependent fashion.
SIRT6 and SNF2H are required for efficient DNA repair
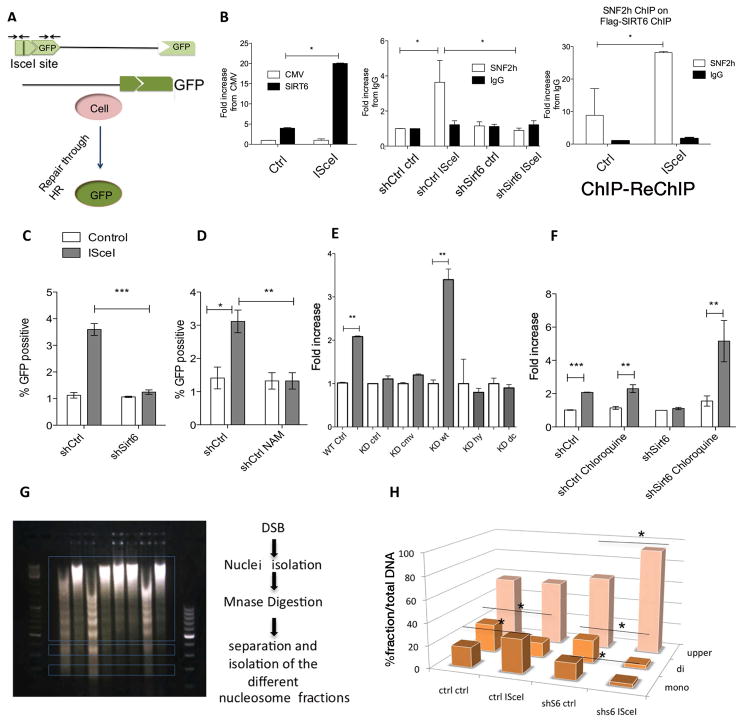
To confirm that SIRT6-dependent SNF2H recruitment is essential for DSB repair, we took advantage of U2OS cells that were engineered to induce a unique DNA cut upon expression of the I-SceI endonuclease (Pierce et al., 2001)(Figure 4A). First, we measured SIRT6 recruitment to sites of damage, confirming its enrichment after DSB induction (Figure 4B). Next, we stably knocked down SIRT6 in these cells and performed chromatin immunoprecipitation (ChIP) of SNF2H to analyze its enrichment at the DNA break site using specific primers. In shCtrl cells, SNF2H was clearly recruited to the I-SceI break (Figure 4B). Strikingly, SNF2H recruitment was completely abolished in SIRT6 knockdown cells (Figure 4B). In order to confirm that SIRT6 is required to bring SNF2H to sites of breaks, we performed a sequential ChIP-ReChIP assay following I-SceI induction. In this assay, the SIRT6-chipped chromatin exhibited more than ~25 fold-enrichment for SNF2H binding following induction of the break (Figure 4B). These studies are consistent with our immunofluorescence and live-imaging results, and further demonstrate that SIRT6 is necessary for SNF2H recruitment to specific DSBs.
Figure 4. SIRT6 modulates SNF2H-dependent DSBs chromatin opening and repair.
A) Schematic representation of the DR-GFP/ISce-I system. B) Left panel: ChIP of cells transfected with Flag-cmv empty vector, or Flag-S6, with or without I-SceI transfection. Middle panel: ChIP of SNF2H in shCtrl or shSIRT6 cells with or without Isce-I transfection. Right panel: Sequential ChIP from Flag-SIRT6 eluted chromatin (ctrl or I-sceI treated), where SNF2H or IgG were used for the second ChIP. C-F) HR efficiency measured by GFP positive cells in (C) shCtrl vs shSIRT6, (D) NAM treated cells (10mg/ml, 12hrs). E) SIRT6-KD cells transfected with either WT-SIRT6, the catalytic mutant SIRT6-HY (catalytically inactive) or the C-terminus deleted (non-SNF2H interacting) SIRT6 fragments, and (F) shCtrl and shSIRT6 cells pre-treated for two hours with chloroquine. G) Chromatin accessibility at DNA breaks. Scheme of the experiment: DSBs were induced with the I-SceI endonuclease, nuclei were isolated and digested with MNase. Different nucleosomal fractions (mono, di and upper) were separated on an agarose gel, and the abundance of the I-SceI site in the isolated DNA of each fraction was quantified using specific primers adjacent to the breaks. (H) qPCR of the isolated DNA from the nucleosomes with primers adjacent to the site of damage in shCtrl cells vs shSIRT6 cells. Data is represented as mean +/− SEM. *p< 0.05, **p<0.01, ***p<0.001. See also Fig. S4.
Additionally, the I-SceI system allowed us to analyze homologous recombination (HR) efficiency (Figure 4A). In these cells, a truncated GFP protein is restored to its functional form when DSBs are repaired. A fraction of ~4–7% of control cells was GFP positive after induction of DNA breaks, while in the absence of SIRT6 such repair activity was completely lost (Figure 4C). Similar results were observed when SIRT6 activity was inhibited by nicotinamide (NAM) (Fig 4D). Re-expression of WT SIRT6 in shSIRT6 cells rescued the repair phenotype, whereas neither the SIRT6-HY nor the SIRT6- C (catalytically active but unable to bind SNF2H) rescued the defects in DNA repair (Figure 4E). Overall, these results indicate that both SIRT6-dependent SNF2H recruitment and SIRT6 catalytic activity are necessary for DNA repair.
To further investigate the roles of SIRT6 and SNF2H in DNA repair, we used a transient DR-GFP/I-SceI system where both HR and NHEJ could be tested, following silencing of SIRT6, SNF2H or both. As previously shown, shSIRT6 completely abolished HR, and similar results were observed in the shSNF2H or double-knock-down cells (Figure S4A–B). Notably, we also observed a significant defect in NHEJ repair (Figure S4C), indicating that chromatin remodeling is required for both HR and NHEJ pathways.
Chromatin relaxation is necessary for proper DSB repair
Previous studies have shown that defects in DNA damage repair in SNF2H knockdown cells could be rescued with chloroquine treatment, a drug that causes chromatin relaxation (Murr et al., 2006; Nakamura et al., 2011). Strikingly, DNA damage repair was fully rescued when we treated shSIRT6 cells with chloroquine (Figure 4F), suggesting that SIRT6 and SNF2H open chromatin at sites of DNA damage, and opening of chromatin by chemical means was sufficient to bypass the requirement for either SIRT6 or SNF2H.
We predicted that SIRT6-dependent SNF2H recruitment was required in order to catalyze nucleosome remodeling, and therefore chromatin would remain compacted at DSBs in the absence of SIRT6. To test this hypothesis, we developed an assay using a similar system to the I-SceI system described above. Following I-SceI induction, DNA was digested with Micrococcal Nuclease (MNase), and the digested DNA was resolved on an agarose gel to separate nucleosomal fragments (Figure 4G). Enrichment for DSBs was measured by qPCR near the site of damage using specific primers. shCtrl cells exhibited a clear increase in chromatin relaxation following DNA damage, as reflected by the enrichment of the I-SceI locus in the mononucleosome fraction and its concomitant depletion from dinucleosomes (Figure 4H). In contrast, the amount of DSBs in mononucleosomes did not increase in shSIRT6 cells upon DNA damage, indicating that the chromatin flanking the breaks remained less accessible to MNase digestion following DNA damage in these cells (Figure 4H). No difference was observed when we measured accessibility at a downstream region located 2kb from the I-SceI site (Figure S4D), indicating that opening of chromatin occurs only locally at the DSB site. Consistent with these results, radial expansion of SNF2H at the sites of damage was moderate and did not change in SIRT6 deficient cells (Figure S4F). As in our previous experiment, chloroquine treatment rescued the chromatin relaxation defect in SIRT6 knockdown cells, (Figure S4E), further confirming our conclusion that increased local chromatin accessibility is required for proper DNA repair.
SIRT6 deacetylates histone H3K56 at DNA damage sites
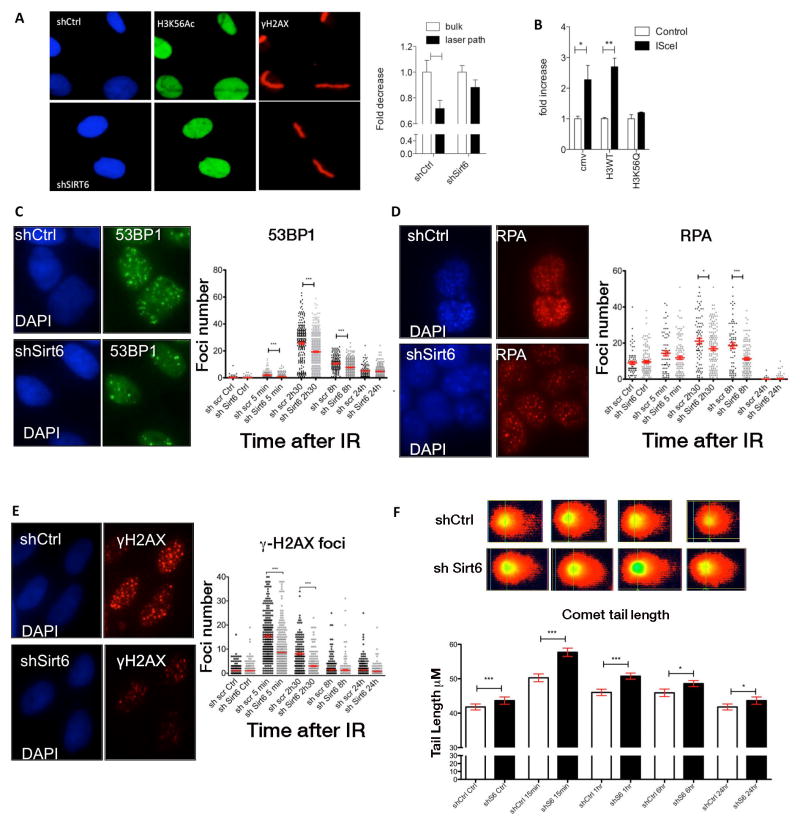
SIRT6 is known to function as a histone H3K56 deacetylase (Michishita et al., 2009; Yang et al., 2009). In addition, previous studies have indicated that H3K56 is actively deacetylated at sites of DNA breaks (Miller et al., 2010; Tjeertes et al., 2009; Yuan et al., 2009). Therefore we tested whether SIRT6 functions as a H3K56 deacetylase at sites of damage. ChIP with an anti-H3K56Ac antibody showed increased overall levels of H3K56 acetylation in shSIRT6 cells at sites of damage (data not shown). Thus, we took advantage of our laser-induced DNA damage approach to determine whether SIRT6 was required to deacetylate H3K56 at sites of breaks. We observed that H3K56Ac was clearly reduced in control cells within 7 minutes following laser-induced damage (Figure 5A), confirming previously published results (Miller et al., 2010; Tjeertes et al., 2009; Yuan et al., 2009). In contrast, such H3K56 deacetylation was barely observed in shSIRT6 cells (Figure 5A). To evaluate the impact of this modification on DSB repair, we transfected U2OS DR-GFP cells with a vector expressing a mutant form of histone H3 (H3K56Q) where the K56 lysine residue was mutated to glutamine, an acetyl mimetic, and measured HR efficiency. Expression of the H3K56Q mutant impaired repair compared to WT H3K56-transfected cells (Figure 5B). These results suggest that SNF2H is unable to open chromatin when H3K56 cannot be deacetylated, highlighting a critical role for SIRT6 activity at sites of damage, both as a recruiter of SNF2H and as a histone H3K56 deacetylase.
Figure 5. SIRT6 modulates H3K56 deacetylation and recruitment of repair factors at DSBs.
A) Immunofluorescence showing H3K56ac at sites of damage after laser induced damage in shCtrl and shSIRT6 U2OS cells. H3K56Ac levels were measured at and besides damage sites, data is represented as mean +/− SEM.. B) Quantification of GFP positive U2OS-DR-GFP cells transfected with H3-WT or H3K56Q mutant. C–E) High-throughput analysis of foci number showing (C) 53BP1, (D) RPA and (E) γH2AX foci number per cell at different time points after IR in shCtrl vs shSIRT6 U2OS cells. F) Comet tail length for shCtrl and shSIRT6 is quantified at the indicated time-points. Representative pictures are shown (15 min. time-point). Data is represented as mean +/− SEM. p-values are abbreviated as in Figure 4. See also Fig. S5.
Lack of SIRT6 impairs downstream DDR signaling
SIRT6 recruitment of SNF2H and H3K56 deacetylation appear to represent very early events in the DSBs repair process. To evaluate downstream effects of SIRT6 deficiency, we analyzed recruitment of known DNA repair factors, taking advantage of a high-throughput microscope kinetic analysis of foci number (Figure S5). Recruitment kinetics were measured in shCtrl and shSIRT6 cells by immunostaining DNA repair signaling factors, including 53BP1, γH2AX and RPA, at different time points after damage induction by IR. In U2OS shSIRT6 cells, formation of 53BP1 and γH2AX foci was impaired starting at 5 min, while a reduced number of RPA-foci appeared at a later time-point (30 min) (Figure 5C, D). In addition, we performed a comet assay in non-alkaline conditions (to measure double-strand break repair), and followed repair kinetics. In this assay, we observed a statistically significant increase in the tail length following damage in SIRT6 deficient cells at every time-point we measured (Figure 5F), supporting our previous results demonstrating decreased DNA repair in shSIRT6 cells. Further, these results indicate that the reduced foci formation in these cells is not due to less damage but rather reflects inefficient signaling at DSB.
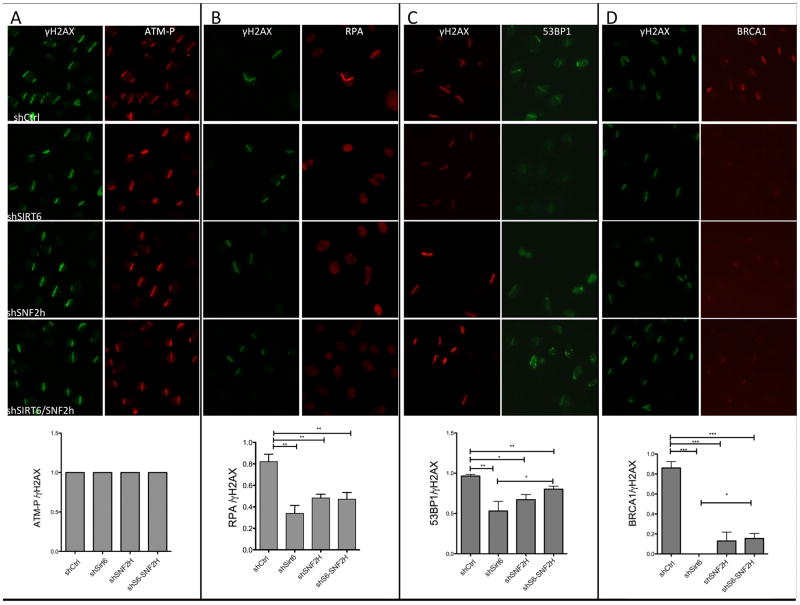
Using the laser-induced DNA damage assay, we showed that shSIRT6 cells exhibit a clear reduction in recruitment of RPA, 53BP1 and BRCA1 to laser-induced breaks, consistent with the above results (Figure 6A–D), and the few positive cells denoted significantly less recruitment. As a control, no changes were seen for ATM phosphorylation at sites of breaks (Figure 6A). Notably, shSNF2H cells showed a similar decrease in DDR factor recruitment, a phenotype that was not further reduced when both proteins were silenced (Figure 6A–D). Overall, these results indicate that lack of SIRT6 profoundly impacts downstream recruitment of DNA repair factors, suggesting that both H3K56 deacetylation and SNF2H chromatin remodeling play critical roles in the DSB DNA repair pathway.
Figure 6. Decreased recruitment of repair factors to laser-induced breaks in the absence of SIRT6.
Following laser-induced damage, recruitment of (A) ATM-P, (B) RPA, (C) 53BP1 and (D) BRCA1 was quantified in the indicated genotypes, data is represented as mean +/− SEM. See also Fig. S6.
Lack of SIRT6 increases DNA damage in vivo in a SNF2H dependent manner
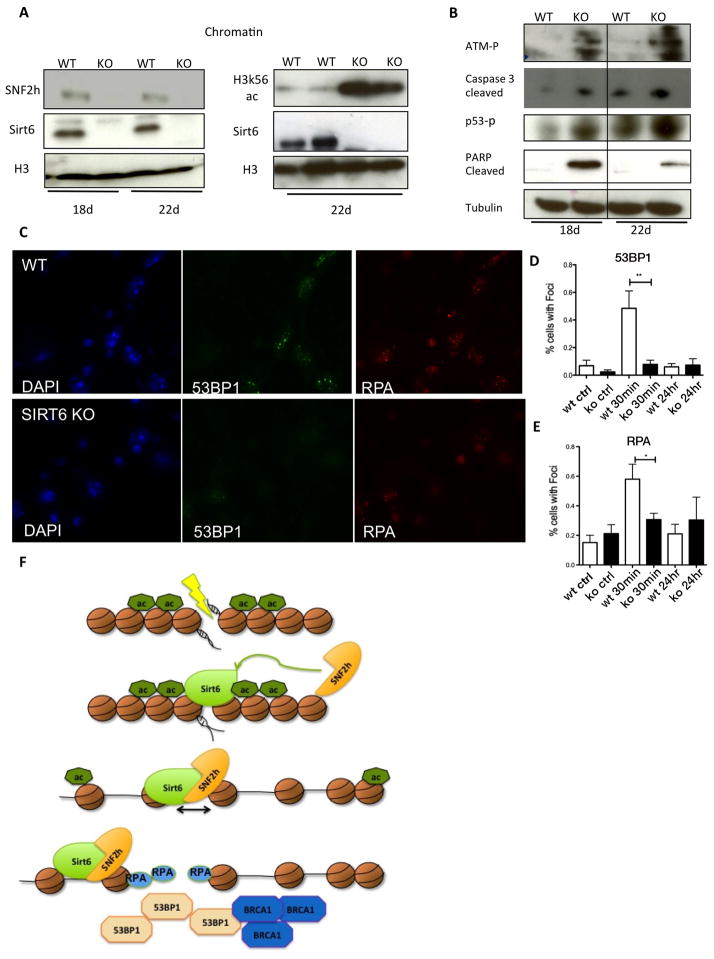
In order to test whether the effect of SIRT6 on DNA repair was physiologically relevant, we analyzed SNF2H and DNA damage responses in SIRT6-deficient mice. For this purpose, we isolated chromatin fractions from different tissues, including liver, heart, pancreas and brain. Remarkably, we observed significantly decreased SNF2H chromatin localization in SIRT6-deficient brain and pancreas (Figure 6A and Figure S7A), but not liver or heart (Figure S7B–C). Thus, SIRT6-dependent recruitment of SNF2H to chromatin appears to be tissue-specific. To determine the importance of SIRT6 in DNA damage repair in vivo, we focused on brain, where DNA damage may accumulate in post-mitotic neurons. In addition, previous studies reported that brain-specific SIRT6KO mice exhibit growth defects (Schwer et al., 2010), indicating that SIRT6 appears to play an important role in brain function. Notably, SIRT6-deficient brains (and pancreas) exhibited increased caspase-3 cleavage, p53 phosphorylation, PARP cleavage, ATM phosphorylation and, consistently, increased H3K56Ac levels (Figure 7A, B and S7A-C). We then generated primary brain cultures from WT and SIRT6KO mice, and measured the presence of RPA and 53BP1 foci following irradiation. Primary brain cultures from SIRT6-KO mice had reduced 53BP1 and RPA foci at 30 minutes (Figure 7C–E), similar to the effect observed in shSIRT6 U2OS cells (Figure 5C,D and 6B,C). In summary, these results indicate that SIRT6-deficient animals experience increased DNA damage in brain, a phenotype strongly correlated with defective H3K56 deacetylation and recruitment of SNF2H to chromatin.
Figure 7. SIRT6 modulates SNF2H recruitment and DNA repair in vivo.
A–B) Chromatin fractions or whole cell extracts from brains of 18 or 22 days old mice from WT or SIRT6 KO animals with the noted antibodies C–E) Primary brain cells cultured for 14 days before damage and collected at different time points (shown is 30min post IR) for RPA and 53BP1. F) Proposed model: SIRT6 is mobilized very early to the sites of damage, recruiting SNF2H and deacetylating H3K56, allowing opening of chromatin and recruitment of downstream repair factors, such as 53BP1, RPA and BRCA1. Data is represented as mean +/− SEM. *p< 0.05, **p<0.001. See also Fig. S7.
DISCUSSION
SIRT6 is a chromatin-bound enzyme that was first described as a suppressor of genomic instability by regulating base excision DNA repair (BER) (Mostoslavsky et al., 2006). Recent studies have demonstrated that SIRT6 is involved in DNA double-strand break resection through deacetylation of C-terminal binding protein (CtBP) interacting protein (CtIP) (Kaidi et al., 2010), increase in repair capacity under oxidative stress through poly[adenosine diphosphate (ADP)-ribose] polymerase 1 (PARP1) (Mao et al., 2011), and stabilization of DNA-dependent Protein Kinase (DNA-PK) at sites of damage (McCord et al., 2009). Here, we demonstrate that SIRT6 is one of the most rapidly recruited factors to sites of DNA damage. Such fast recruitment may allow SIRT6 to coordinate proper DNA repair through a complex and step-wise response (model Figure 7F). Our results extend previously proposed roles for SIRT6 in DDR, and reveal that SIRT6 plays a critical role in regulating chromatin accessibility as a very early event during the DNA damage response. This activity involves direct recruitment of the ATP-dependent chromatin remodeler SNF2H to sites of DNA breaks and in parallel rapid histone deacetylation at histone H3K56, to allow recruitment of downstream DNA repair factors. To the best of our knowledge, our results represent the first example of a sirtuin deacetylase functioning as a specific scaffold for recruiting a chromatin remodeler to sites of DNA damage, to open chromatin and repair DNA breaks in a coordinated manner.
SIRT6 and SNF2H are recruited early to DNA damage sites to modulate chromatin accessibility
Our results show that the interaction between SNF2H and SIRT6 is important to bind SNF2H to chromatin, even at basal levels (Fig 2A). It has been previously proposed that SNF2H “probes” the chromatin through continuous sampling, until it “recognizes” an anchoring signal which in turn increases SNF2H binding affinity (Erdel and Rippe, 2011). We believe that SIRT6 is one of those signals, as it increases SNF2H binding to nucleosomes (Figure 1H), and this chromatin-bound complex might have additional functions beyond DNA repair. However, the interaction of both proteins and thus the abundance of this complex are clearly increased upon DNA damage. Our results indicate that SNF2H and SIRT6 work in an epistatic manner to prevent genomic instability (Figure 2F–G and Figure 6A–D). Remarkably, recruitment of SIRT6 appears extremely early (~5 sec with ~30 sec plateau), positioning SIRT6 as one of the earliest factors to accumulate at DNA damage sites, followed by SNF2H, which is only recruited to DSBs in the presence of SIRT6 (Figure 3A–E, 4B). Our experiments in living cells demonstrate that SIRT6 and SNF2H are both recruited very early, where SIRT6 appears to modulate the second, more stable phase of SNF2H binding specifically at DSBs. Early, transient binding of SNF2H occurs independently of SIRT6, and may represent SNF2H recruitment to other types of DNA lesions.
SIRT6 activity is required for proper DNA repair
Although SIRT6 activity is not required to bring SNF2H to chromatin (Figure 2C), nor to control SNF2H remodeling activity in vitro, both SIRT6 deacetylase activity and recruitment of SNF2H were required for efficient DNA repair (Figure 4D,E). The role of H3K56 acetylation in DNA damage has been controversial, with some studies reporting hypoacetylation of H3K56 at sites of breaks (Miller et al., 2010; Tjeertes et al., 2009) whereas others showed hyperacetylation following DNA damage (Das et al., 2009). In addition, both class I HDACs and sirtuins are capable of deacetylating H3K56 (Michishita et al., 2009; Miller et al., 2010; Yang et al., 2009). Our results indicate that SIRT6 is critical for proper deacetylation of H3K56 at sites of damage in our experimental conditions (Figure 5A). Some of the discrepancies may be due to the time scale of the measurement, type of DNA damage, and specific cells tested. In our system, we observed clear hypoacetylation at early time-points (~7 min.), but detected bulk H3K56 hyperacetylation at very late time points (6–8 hr., data not shown), a potential requirement for proper refolding of chromatin following repair, as seen in yeast (Chen et al., 2008). Of note, several HDACs have been shown to play roles in DNA repair. Notably, we observed increase H3K56Ac following TSA treatment, however this increase did not influence recruitment of SNF2H to chromatin (data not shown). These results indicate that SNF2H recruitment specifically depends on SIRT6, and is not regulated by other HDACs or the H3K56Ac modification per se. Furthermore, the fact that we observe a clear repair defect in the absence of SIRT6 strongly suggests non-overlapping or redundant functions with other HDACs.
SIRT6 affects downstream DDR signaling
Our results show that lack of SIRT6 influences recruitment of downstream repair factors such as 53BP1, γH2AX, RPA and BRCA1. In our shCtrl and shSIRT6 cells this was independent of the cell cycle stage, in agreement with previous findings by Kaidi et al (Kaidi et al., 2010). Taken together, our results indicate that chromatin remodeling and early histone modifications have a profound impact on recruitment of downstream effectors. Previous work has shown that lack of 53BP1 increases end-resection and RPA foci (Bunting et al., 2010), therefore one may hypothesize that decreased 53BP1 foci in SIRT6-deficient cells should increase RPA foci. However, given the previous published effect of SIRT6 on CtIP (Kaidi et al., 2010), it is likely that end-resection is inhibited in the absence of SIRT6, and thus both RPA and 53BP1 cannot be recruited. Consistently, cells deficient in the remodelers INO80 and SMARCAD1 exhibited defective DNA end resection and concomitant lack of both RPA and 53BP1 foci (Costelloe et al., 2012). Previous studies have shown that SNF2H recruitment occurs downstream of histone H2B K120 ubiquitylation (H2B K120Ub) by RNF20 (Nakamura et al., 2011). Interestingly, in SIRT6 KO cells and tissues, RNF20 recruitment to chromatin was clearly impaired. Further, H2B K120Ub was severely diminished in SIRT6-deficient cells (Figure S6 A, B), suggesting that these events occur downstream of SIRT6. Of note, Smeenk et al showed that SNF2H spreading on chromatin was dependent on PARP1 activity (Smeenk et al., 2012), and SIRT6 was recently shown to activate PARP1 under oxidative damage conditions (Mao et al., 2011). These results suggest that SNF2H spreading could be affected also by lack of SIRT6 through a PARP1-dependent mechanism. However, in our system, we did not observe defects in SNF2H spreading (Figure S4F), nor on PAR levels at sites of DNA damage (Figure S6D, E) in the absence of SIRT6, indicating that the previously published role for SIRT6 in modulating PARP1 activity might be specific for oxidative DNA damage.
Lack of SIRT6 increases vulnerability to DNA damage in a tissue-specific manner
The roles of SIRT6 in DNA damage repair are physiologically relevant, since SIRT6-deficient brains exhibited a clear decrease in chromatin-localized SNF2H and increased DNA damage (Fig 7A–E), a phenotype also observed in other tissues such as pancreas (Figure S7A). Proper DNA repair is critical to prevent neurodegeneration, cancer and premature aging. Indeed, recent studies have shown that SIRT6 overexpression extends lifespan (Kanfi et al., 2012), and deleting SIRT6 specifically in the brain causes metabolic abnormalities (Schwer et al., 2010). Further, recent studies indicate that SIRT6 can function as a tumor suppressor, at least in part through modulation of metabolism (Sebastian et al., 2012). Intriguingly, SIRT6 protein levels in these different tissues do not correlate with the DNA damage phenotype (Liszt et al., 2005; Mostoslavsky et al., 2006), suggesting that tissue specificity is not based on SIRT6 levels, but rather on either as yet unknown co-factors or redundancy with other proteins able to recruit SNF2H. Based on the results presented here, we conclude that the SIRT6 chromatin scaffolding function is essential for preventing genomic instability, and may explain several of the phenotypes previously associated to SIRT6.
Experimental Procedure
Immunoprecipitation (IP)
Cells were lysed and sonicated for 15 minutes (with intervals of 1 second ON- 3 seconds OFF) in Lysis buffer (10mMTris pH7.9, 150 mM KCL, and protease, deacetylase and phosphatase inhibitors). Protein was precleared for 2 hr. with beads at 4°C, and left overnight with blocked beads (5% BSA) and the desired antibody. Beads were washed 2 times with 150mM KCL, 2 times with 300mM KCL and 1 time with 150 mM KCL, and proteins were eluted either by flag peptide/or boiling.
MNase Assay-qPCR
2×106 shControl or shSIRT6 U2OS cells were transfected with a plasmid carrying the I-SceI site (I-SceI-pBSK, a kind gift from F. Alt (Wang et al., 2009). 48 hr. later, cells with or without the I-SceI enzyme were homogenized with a Dounce homogenizer in RSB Buffer (10mM Tris-HCl pH 7.4, 10mM NaCl, 3mM MgCl2, 0.5% NP-40, 1mM PMSF, 1mM DTT, Protease inhibitors). Cells were incubated on ice for 15 min and nuclei were collected by centrifugation (1000g), 10 min, 4°C, washed twice with RSB Buffer and digested with MNase in Digestion Buffer at room temperature (RT) (140 units, time course from 5–20 minutes). Reaction was stopped adding 1 volume of Stop Solution (50mM Tris-HCl pH 7.5,150mM NaCl, 50mM EDTA, 0.3% SDS). Samples were centrifuged at 10,000g for 10min at 4°C. DNA was purified by phenol-chloroform extraction and resolved in a 2% agarose gel. Bands were isolated and purified using a Gel Extraction Kit (Qiagen). Extracted DNA was used in q-PCR reactions as indicated.
Nuclear IP
Nuclei were isolated from cells and treated with 2 Units of Micrococcal nuclease (MNase) for 1 hour at room temperature. MNase reaction was stopped and protein was measured with Bradford to proceed with the IP as described before.
Primary cortical cell culture
Cortex and hippocampus were separated from 18-days old SIRT6 WT and KO embryos, minced, and cells plated on poly-L-ornithine coated coverslips in Neurobasal medium, B27 supplement, Glutamax and Penicillin/streptomycin (Invitrogen). Tail DNA was used to genotype the embryos.
Supplementary Material
Highlights.
SIRT6 arrives to sites of DNA breaks within 5 seconds.
Lack of SIRT6 and SNF2H increases sensitivity to genotoxic damage.
SIRT6 directly recruit SNF2H, which in turn open chromatin at sites of breaks.
SIRT6 and SNF2h are necessary to recruit 53BP1, RPA, BRCA1 to the sites of damage.
Acknowledgments
This work was supported in part by NIH grant GM093072-01 (R.M.). R.M is a Howard Goodman Scholar Awardee and an MGH Research Scholar. D.T. is the recipient of the Brain Power for Israel Foundation grant. C.C. is supported by a Fellowship from the Fondazione Umberto Veronesi. C.S. is the recipient of a Beatriu de Pinos Postdoctoral Fellowship (Generalitat de Catalunya). B.M-P is the recipient of a postdoctoral fellowship from the Spanish Ministry of Education. We would like to thank Steve Jackson for the SIRT6-GFP and SIRT6-RFP plasmids, and Laura Prickett-Rice and Kate Folz-Donahue for technical assistance with the FACS analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141:243–254. doi: 10.1016/j.cell.2010.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Molecular cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Acetylated lysine 56 on histone H3 drives chromatin assembly after repair and signals for the completion of repair. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Cui D, Papusha A, Zhang X, Chu CD, Tang J, Chen K, Pan X, Ira G. The Fun30 nucleosome remodeller promotes resection of DNA double-strand break ends. Nature. 2012;489:576–580. doi: 10.1038/nature11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costelloe T, Louge R, Tomimatsu N, Mukherjee B, Martini E, Khadaroo B, Dubois K, Wiegant WW, Thierry A, Burma S, et al. The yeast Fun30 and human SMARCAD1 chromatin remodellers promote DNA end resection. Nature. 2012;489:581–584. doi: 10.1038/nature11353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das C, Lucia MS, Hansen KC, Tyler JK. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature. 2009;459:113–117. doi: 10.1038/nature07861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdel F, Rippe K. Chromatin remodelling in mammalian cells by ISWI-type complexes--where, when and why? The FEBS journal. 2011;278:3608–3618. doi: 10.1111/j.1742-4658.2011.08282.x. [DOI] [PubMed] [Google Scholar]
- Erdel F, Schubert T, Marth C, Langst G, Rippe K. Human ISWI chromatin-remodeling complexes sample nucleosomes via transient binding reactions and become immobilized at active sites. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:19873–19878. doi: 10.1073/pnas.1003438107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gospodinov A, Vaissiere T, Krastev DB, Legube G, Anachkova B, Herceg Z. Mammalian Ino80 mediates double-strand break repair through its role in DNA end strand resection. Molecular and cellular biology. 2011;31:4735–4745. doi: 10.1128/MCB.06182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kanfi Y, Naiman S, Amir G, Peshti V, Zinman G, Nahum L, Bar-Joseph Z, Cohen HY. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Kashiwaba S, Kitahashi K, Watanabe T, Onoda F, Ohtsu M, Murakami Y. The mammalian INO80 complex is recruited to DNA damage sites in an ARP8 dependent manner. Biochemical and biophysical research communications. 2010;402:619–625. doi: 10.1016/j.bbrc.2010.10.066. [DOI] [PubMed] [Google Scholar]
- Kielbassa C, Roza L, Epe B. Wavelength dependence of oxidative DNA damage induced by UV and visible light. Carcinogenesis. 1997;18:811–816. doi: 10.1093/carcin/18.4.811. [DOI] [PubMed] [Google Scholar]
- Lan L, Ui A, Nakajima S, Hatakeyama K, Hoshi M, Watanabe R, Janicki SM, Ogiwara H, Kohno T, Kanno S, et al. The ACF1 complex is required for DNA double-strand break repair in human cells. Molecular cell. 2010;40:976–987. doi: 10.1016/j.molcel.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Larsen DH, Poinsignon C, Gudjonsson T, Dinant C, Payne MR, Hari FJ, Rendtlew Danielsen JM, Menard P, Sand JC, Stucki M, et al. The chromatin-remodeling factor CHD4 coordinates signaling and repair after DNA damage. The Journal of cell biology. 2010;190:731–740. doi: 10.1083/jcb.200912135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Park JH, Kim SJ, Kwon SJ, Kwon J. A cooperative activation loop among SWI/SNF, gamma-H2AX and H3 acetylation for DNA double-strand break repair. The EMBO journal. 2010;29:1434–1445. doi: 10.1038/emboj.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L. Mouse Sir2 homolog Sirt6 is a nuclear ADP-ribosyltransferase. J Biol Chem. 2005;280:21313–21320. doi: 10.1074/jbc.M413296200. [DOI] [PubMed] [Google Scholar]
- Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13:1161–1169. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCord RA, Michishita E, Hong T, Berber E, Boxer LD, Kusumoto R, Guan S, Shi X, Gozani O, Burlingame AL, et al. SIRT6 stabilizes DNA-dependent protein kinase at chromatin for DNA double-strand break repair. Aging (Albany NY) 2009;1:109–121. doi: 10.18632/aging.100011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, Barber MF, Hong T, Gozani O, Chua KF. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nature cell biology. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira DV, Shimada M, Tauchi H, Suzuki H, Tashiro S, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Molecular cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- Neumann FR, Dion V, Gehlen LR, Tsai-Pflugfelder M, Schmid R, Taddei A, Gasser SM. Targeted INO80 enhances subnuclear chromatin movement and ectopic homologous recombination. Genes & development. 2012;26:369–383. doi: 10.1101/gad.176156.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nature reviews Genetics. 2012;14:62–75. doi: 10.1038/nrg3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global regulation of H2A.Z localization by the INO80 chromatin-remodeling enzyme is essential for genome integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce AJ, Hu P, Han M, Ellis N, Jasin M. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes & development. 2001;15:3237–3242. doi: 10.1101/gad.946401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Kaidi A, Baskcomb L, Galanty Y, Jackson SP. Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4. The EMBO journal. 2010;29:3130–3139. doi: 10.1038/emboj.2010.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Schumacher B, Lombard DB, Xiao C, Kurtev MV, Gao J, Schneider JI, Chai H, Bronson RT, Tsai LH, et al. Neural sirtuin 6 (Sirt6) ablation attenuates somatic growth and causes obesity. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21790–21794. doi: 10.1073/pnas.1016306107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM, Gymrek M, Goren A, Zhong L, Ram O, Truelove J, Guimaraes AR, Toiber D, et al. The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell. 2012;151:1185–1199. doi: 10.1016/j.cell.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Marteijn JA, Luijsterburg MS, Sroczynski N, Costelloe T, Romeijn RJ, Pastink A, Mailand N, Vermeulen W, et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. Journal of cell science. 2012 doi: 10.1242/jcs.109413. [DOI] [PubMed] [Google Scholar]
- Smeenk G, Wiegant WW, Vrolijk H, Solari AP, Pastink A, van Attikum H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. The Journal of cell biology. 2010;190:741–749. doi: 10.1083/jcb.201001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennen RI, Berber E, Chua KF. Functional dissection of SIRT6: identification of domains that regulate histone deacetylase activity and chromatin localization. Mech Ageing Dev. 2010;131:185–192. doi: 10.1016/j.mad.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjeertes JV, Miller KM, Jackson SP. Screen for DNA-damage-responsive histone modifications identifies H3K9Ac and H3K56Ac in human cells. The EMBO journal. 2009;28:1878–1889. doi: 10.1038/emboj.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toiber D, Sebastian C, Mostoslavsky R. Characterization of nuclear sirtuins: molecular mechanisms and physiological relevance. Handb Exp Pharmacol. 2011;206:189–224. doi: 10.1007/978-3-642-21631-2_9. [DOI] [PubMed] [Google Scholar]
- Wang JH, Gostissa M, Yan CT, Goff P, Hickernell T, Hansen E, Difilippantonio S, Wesemann DR, Zarrin AA, Rajewsky K, et al. Mechanisms promoting translocations in editing and switching peripheral B cells. Nature. 2009;460:231–236. doi: 10.1038/nature08159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Ayrapetov MK, Xu C, Gursoy-Yuzugullu O, Hu Y, Price BD. Histone H2A.Z Controls a Critical Chromatin Remodeling Step Required for DNA Double-Strand Break Repair. Molecular cell. 2012;48:723–733. doi: 10.1016/j.molcel.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Sun Y, Jiang X, Ayrapetov MK, Moskwa P, Yang S, Weinstock DM, Price BD. The p400 ATPase regulates nucleosome stability and chromatin ubiquitination during DNA repair. The Journal of cell biology. 2010;191:31–43. doi: 10.1083/jcb.201001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell cycle. 2009;8:1747–1753. doi: 10.4161/cc.8.11.8620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong L, D’Urso A, Toiber D, Sebastian C, Henry RE, Vadysirisack DD, Guimaraes A, Marinelli B, Wikstrom JD, Nir T, et al. The histone deacetylase Sirt6 regulates glucose homeostasis via Hif1alpha. Cell. 2010;140:280–293. doi: 10.1016/j.cell.2009.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.