Abstract
Mass spectrometry is used routinely for large-scale protein identification from complex biological mixtures. Recently, relative quantitation approach on the basis of spectra count has been applied in several cancer proteomic studies. In this review, we examine the mechanism of this technique and highlight several important parameters associated with its application.
Keywords: Mass spectrometry, label-free quantitation, spectra count, proteomics, review
Introduction
Today, liquid chromatography-coupled tandem mass spectrometry (LC-MS/MS) is used routinely for large-scale protein identification and global profiling of post-translational modifications (PTMs) from complex biological mixtures (1–4). To quantitatively characterize and compare two or more proteomes by MS, a variety of methods involving the incorporation of stable isotope labels have been developed (5). Because the stable isotope-labeled peptides possess similar physical and chemical properties as their unlabeled equivalents but with a different mass that can be recognized by a mass spectrometer, quantification is achieved by comparing their respective signal intensities. The labels can be introduced into samples by methods such as isotope coded affinity tag (ICAT) (6), isotope tags for relative and absolute quantitation (iTRAQ) (7), and stable isotope labeling by amino acids in cell culture (SILAC) (8). However, in many cases, clinical samples and most animal based samples may not be suitable for metabolic labeling. Moreover, chemical isotopic labeling of many samples can be extremely expensive and time prohibitive (9). As a result, alternative label-free quantitation approaches, either by measuring and comparing the MS signal intensity of peptide precursor ions or by counting and comparing the number of matched MS2 spectra of a given protein, have gained increasing popularity over the past several years. Investigators have validated this approach as a viable method for MS-based differential display and shown excellent correlation between spectral counts and relative quantitation (10–12). The pros and cons of these methods have been reported in previous publications (9, 13, 14). Here, we attempt to discuss the features of the commonly used spectra count label-free quantitation technique based on the authors’ experience using ion trap mass spectrometers, focusing on both theoretical and practical aspects of the technique. alternative label-free quantitation approaches, either by measuring and comparing the MS signal intensity of peptide precursor ions or by counting and comparing the number of matched MS2 spectra of a given protein, have gained increasing popularity over the past several years. Investigators have validated this approach as a viable method for MS-based differential display and shown excellent correlation between spectral counts and relative quantitation (10–12). The pros and cons of these methods have been reported in previous publications (9, 13, 14). Here, we attempt to discuss the features of the commonly used spectra count label-free quantitation technique based on the authors’ experience using ion trap mass spectrometers, focusing on both theoretical and practical aspects of the technique.
CID versus ETD
The shotgun MS strategy consists of enzymatic digestion of proteins by trypsin to yield peptides, which are analyzed by mass spectrometer either directly or after enrichment using a data-dependent MS/MS method (15, 16). Collision-induced dissociation (CID) and Electron-transfer dissociation (ETD) are the most widely used techniques to fragment peptide ions in a mass spectrometer and have proven to be extremely useful for amino acid sequence assignment and for PTMs studies. Notably, low-energy CID of peptides with labile PTMs such as phosphorylation and glycosylation typically results in the predominant neutral loss with inadequate fragmentation of the peptide backbone, whereby only limited sequence information is obtained, and large peptides (>2500 Da) are difficult to fragment by CID. On the other hand, ETD has the advantage of enabling efficient fragmentation of peptides with labile PTMs and larger peptides with charge states of 3+ or higher (17, 18). However, ETD can yield poorer fragmentation efficiency for smaller peptides with lower (1+, 2+) charge states (19), and it takes longer time to fragment peptides and acquire an MS2 spectrum by ETD. If the same complex biological mixture is analyzed by either CID or ETD, more CID spectra than ETD spectra can be collected within the same amount of time for peptide identification. As a result, CID is still the preferred method for general proteomic study with the goal to identify unique peptides, as many as possible and as quickly as possible, whereas ETD is mostly used as a complementary technique for characterization of multiple charged peptides or peptides with labile PTMs that could be missed by CID. The recent development of new mass spectrometers, such as LTQ-Orbitrap Velos which provides even faster CID scan (20, 21), further consolidates CID as a major player in general proteomic research.
Trypsin is commonly used for LC-MS/MS analysis, because tryptic peptides can be positively charged at both N and C-terminus, and most abundant fragmented ions are b and y ions that are well-suited to database search for peptide matching (22). In contrast, the C-terminus of non-tryptic peptides generated by enzyme, such as Glu-C, are not positively charged and the obtained CID spectra are not favored for database search algorithm such as Sequest, due to lack of abundant y series ions. Furthermore, some peptides generated by enzyme, such as Lys-C, are likely longer than tryptic peptides, and the long peptide does not fragment well by CID. Nonetheless, proteolytic digests by trypsin together with other proteases will help to increase the proteome coverage (23–25).
MS2 Spectra Count
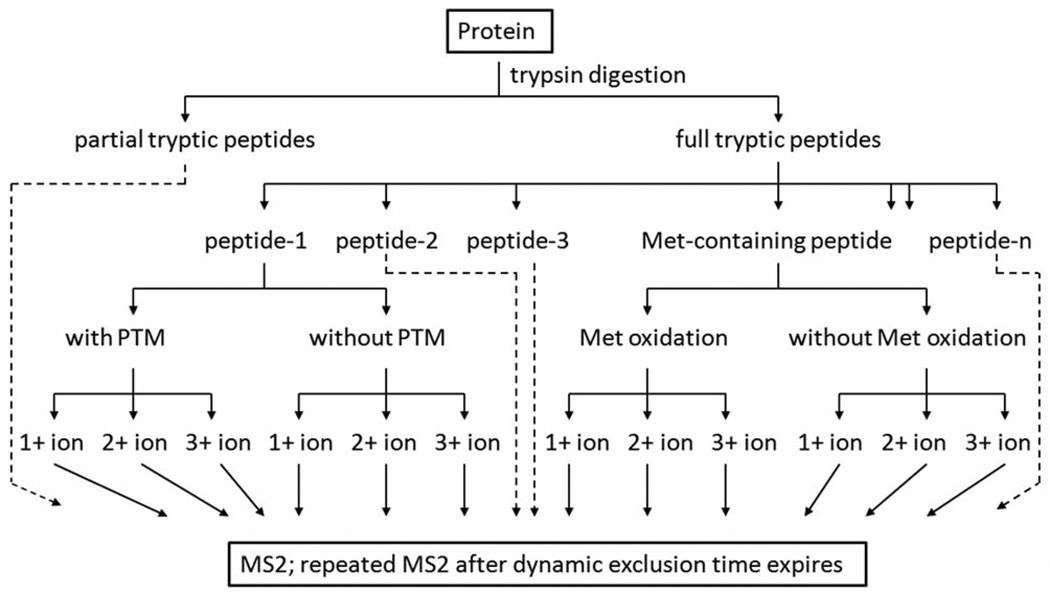
The mass spectrometer, such as ion trap from ThermoFisher, is set to perform a full-scan (survey scan) to determine the m/z values and intensities of ionized peptides, and subsequently perform several MS/MS scans on the most intense ions in the full-scan spectrum with “Dynamic Exclusion” enabled. The Dynamic Exclusion function contains several important parameters, such as Repeat Count, Exclusion Mass Width, Exclusion List Size, and Exclusion Duration. Dynamic Exclusion temporarily puts a mass into an exclusion list after its MS2 spectrum is acquired, providing the opportunity to collect MS2 information on less intense ions in the next cycle. The obtained MS2 spectra are then processed by computer algorithms to deduce peptide sequence. It is noteworthy that (a) although thousands of MS2 spectra can be collected during a typical LC-MS/MS experiment, this number is small in relation to the number of peptides generated by tryptic digestion of a large proteome from human. The signal intensities of the tryptic peptides from an abundant protein are more likely to be above the threshold to trigger CID events; (b) by electrospray ionization (ESI), one tryptic peptide presents multiple charged-forms in the sample solution, carrying one or more protons. If their signal intensities are above the threshold and their m/z values are within full scan mass range, they all can trigger CID events. Consequently, for an abundant peptide, multiple MS2 spectra corresponding to different chargedforms can be obtained; (c) many proteins have PTMs, and the tryptic peptides are subject to variable modifications. In addition, if the peptide contains methionine (Met), variable methionine oxidation during sample preparation and MS analysis is not unusual; (d) abundant peptides have bigger peak width on liquid chromatogram. If its peak width is bigger than dynamic exclusion duration time, the peptide can be selected again for CID after its exclusion time expires, and more than one MS 2 spectra from the same peptide can be acquired. Hence, the total MS2 spectra that matched to a protein is a combination of spectra from different partial tryptic peptides and full tryptic peptides, spectra from the same peptide with different charges, spectra of the same peptide with variable modifications, and repeated spectra from the same peptide due to expired dynamic exclusion (Figure 1).
Figure 1.
The origin of spectra count. Fully digested tryptic peptides can be positively charged at both N and C-terminus upon electrospray ionization. Some tryptic peptides, containing histidine or extra arginine/lysine due to missed cleavage, can be 3+ charged. The MS2 spectra that matched to a protein is a combination of spectra from different partial tryptic peptides and full tryptic peptides, spectra from the same peptide with different charges, spectra of the same peptide with variable modifications, and repeated spectra from the same peptide due to expired dynamic exclusion. At some situations, spectra that are potentially matched to partial tryptic peptides or peptides with PTMs are not included during data analysis by spectral interpretation software.
Spectra Count Label-free Relative Quantitation
Based on the empirical observation that proteins existing in higher concentration are associated with a larger number of MS2 spectra for peptides, relative quantitation of the identified proteins can be achieved (5, 9). In general, the abundant proteins have more than 10 spectra counts, the medium-abundance proteins from 2 to 10, and lowabundance proteins less than 2. Although there is an approximate correlation between the number of spectra per protein and the amount of protein present in the mixture, such a correlation should be viewed in the context of the following sources of error: (a) if a protein was not identified (zero count), it still could be in the sample with low abundance; (b) a great deal of caution should be exercised in assessing the absolute quantity of two different proteins, because for the same number of molecules, larger protein can give rise to more tryptic peptides for CID; (c) the MS signal for any given peptide is determined by many factors, such as ionizability in electrospray and fragmentation efficiency, that have effect on the number of spectra matched with a protein. Frequently, the number of MS2 spectra contributing to the identification of a protein can be used as an indication of that protein’s abundance in the samples from one dimensional or two dimensional electrophoresis gel pieces in which proteins have similar molecular weight (26). In order to quantify the proteins that have different molecular weight from complex biological mixture such as cell lysate, some researchers proposed methods using normalized spectra count that takes into account the length of the protein (27).
For LC-MS/MS analysis, the mass spectrometer is configured to sequentially select and fragment ions from high-intensity to low-intensity with dynamic exclusion; however, in very complex peptide mixtures the number of ions co-eluting can significantly exceed the number of ions for which tandem mass spectra can be acquired. As a result, the high-abundance and medium-abundance proteins will be repeatedly identified from multiple iterations, whereas some low-abundance proteins cannot. The newly identified proteins from succeeding iterations are mainly from relative low-abundance proteins, and the total number of identified proteins will be increased somewhat with repeated iterations (28). Since the obtained MS2 spectra count of each protein could be different through multiple iterations, measurement variability can be reduced by using the average spectra count. Based on the observation that the degree of spectra count variation from one iteration to another is relatively higher for low-abundance proteins, a great deal of caution should be exercised in assessing the relative quantity of a protein that is identified as low-abundance in both groups (28). Nevertheless, reliable relative quantitation can be obtained if the protein is identified as low-abundance in one group but high-abundance in another group, or if the protein is identified as high-abundance in both groups but the difference of spectra count is dramatic. For the proteins that are identified as low-abundance in both groups and are possibly differentially expressed, it is highly recommended to further verify the difference by targeted quantitative MS analysis after enrichment or by more sensitive technique such as Western blot.
The Factors Affecting MS2 Spectra Count
Many LC-MS/MS instrument parameters, such as “ion injection time”, “automatic gain control”, “micro scan” and “dynamic exclusion”, have a significant impact on the number of identified proteins and their spectra counts, and some of them have been described in previous publications (29–34).
Microscans
The parameter displays the number of microscans per scan. Each microscan is one mass analysis, containing the steps of ion injection, ion storage and ion detection. Microscans are summed, to produce one scan, to improve the signal-to-noise ratio of the mass spectral data. The number of microscans per scan is an important factor in determining the overall scan time. Apparently, increasing the number of microscans results longer scan time, and the instrument will acquire less MS2 spectra for peptide identification. For proteomic analysis, the number of “Microscans” is generally set to be “1” in order to increase proteome coverage (Table I), which has been confirmed by study from Kalli et al. (32).
Table I.
Recommended values for some parameters in LC-MS/MS analysis.
| Parameter | Proteomic analysis favoring abundant proteins |
Proteomic analysis covering both abundant and low-abundance proteins |
|---|---|---|
| Microscans | =1, or >1 | =1 |
| Repeat count | =1, or >1 | =1 |
| Exclusion mass width | 1 amu for LTQ, 0.1 amu for LTQ-Orbitrap | 1 amu for LTQ, 0.1 amu for LTQ-Orbitrap |
| Exclusion list size | <250 | >250 |
| Exclusion duration | <60 s | >60 s |
| HPLC gradient | 1–2 h | 2–4 h |
Repeat count and repeat duration
When Dynamic Exclusion function is enabled, the mass of the ion that has been chosen for data dependent fragmentation is put on a “pre-exclusion list”. The Repeat Count displays the number of times (counts) that a mass may be selected as a data dependent mass before it goes to the dynamic exclusion list; the Repeat Duration displays the amount of time an ion stays on the “pre-exclusion list”. If an ion triggers a data dependent scan the number of times specified by Repeat Count within the repeat duration time, then it is removed from the pre-exclusion list and is added to the dynamic exclusion list. Obviously, if the Repeat Count is bigger than “1” and if the peptide’s chromatographic peak width is bigger than MS analysis cycle time (the cycle time is ∼3 s for commonly used instrument method such as “Top 8” by LTQ-Orbitrap), it is very likely that the instrument will perform a repeated MS2 scan of the same precursor ion before putting the mass on the dynamic exclusion list, favoring analysis of the abundant ions in the sample; however, the less abundant ions may lose the chance to be picked for fragmentation. On the contrary, if the Repeat Count is set to be “1”, the mass of an ion that has triggered a data dependent scan will be directly added to the dynamic exclusion list, allowing instrument to analyze less abundant ions and increasing the proteome coverage.
Exclusion mass width
This parameter displays the window for determining whether the mass of an ion matches a mass on the dynamic exclusion list. An ion will not trigger a dependent scan if its mass is within the window. For ion trap mass spectrometer LTQ-XL that has low mass accuracy, the width of the window is ∼1 amu, excluding both the analyzed peptide and the likely isotopic forms from the peptide, whereas for hybrid mass spectrometer LTQ-Orbitrap that has high mass accuracy, the width of the window can be set to ∼0.1 amu since isotopic forms will be rejected for CID by activation of “Monoisotopic precursor selection” mode (Figure 2). The merit of Dynamic Exclusion is that it temporarily puts the recently analyzed peptides into an exclusion list and allows the mass spectrometer to obtain CID spectra from other less intense peptides; however, the co-eluted other peptides that have similar m/z can fall into the “black window” and will be rejected for CID, decreasing and the spectra count of the corresponding proteins. If the Exclusion Mass Width is too big, the odd for other peptides to fall into these “black windows” will be higher and those peptides will be skipped for CID. Therefore, depending on the type of mass spectrometers, appropriate Exclusion Mass Width should be applied.
Figure 2.
Exclusion Mass Width and the “black window” effect. The mass of tryptic peptide is relatively small and the intensity of its monoisotopic peak is generally higher than that of other isotopic peaks. At normal scan rate and in centroid data type, low mass accuracy mass spectrometer such as LTQ-XL cannot resolve these isotopic peaks and deduce the charge state of the peptide; Dynamic Exclusion temporarily puts the mass, close to the average mass of the analyzed peptide, into an exclusion list. High mass accuracy mass spectrometer such as LTQ-Orbitrap can distinguish the isotopic peaks and deduce the charge state of the peptide; Dynamic Exclusion temporarily puts the mass, close to the monoisotopic mass of the analyzed peptide, into an exclusion list. Exclusion Mass Width is applied in order to avoid repeated analysis of the same peptide and its isotopic peaks, thus creating a “black window ”.
Exclusion list size
This parameter defines the maximum number of masses that can be on the dynamic exclusion list. It is usually in the order of 100–500. The mass of the newly analyzed peptide will queue in and the mass of previously analyzed peptide will queue out when the list is full. Since each mass on the list will create a “black window”, this parameter will also determine how many “black windows” may exist during MS analysis. A relatively large Exclusion List Size can ensure that the analyzed peptide will stay on the dynamic exclusion list before its exclusion duration time expires. If the Exclusion List Size is too big, the odd for other peptides to fall into these “black windows” will be higher and those peptides will be skipped for CID. In contrast, a relatively small Exclusion List Size can potentially remove the analyzed peptide from dynamic exclusion list when the list is full, even though its exclusion duration time does not expire. Thus, a relatively larger number of this parameter will be considered in favor of qualitative proteomic analysis, while a relatively small number can be selected in favor of quantitative analysis by allowing repeated analysis of the abundant peptides.
Exclusion duration
This parameter determines how long the analyzed peptide will stay on the dynamic exclusion list after its MS2 spectrum is acquired, and it determines the lifetime of each “black window” as well. After the exclusion time expires, the peptide can be selected again for CID if its intensity is above the threshold to trigger CID. Consequently, the longer exclusion duration time will prevent the mass spectrometer from repeatedly collecting MS2 spectrum from the same peptide, providing opportunity to obtain CID spectra from other unique peptides. As a result, the relatively longer exclusion duration time will be in favor of qualitative proteomic analysis by identifying as many unique peptides as possible, whereas it will be disadvantageous for quantification due to its unbiasedness on abundant and lowabundance peptide ions. In contrast, shorter exclusion duration time will favor quantitative analysis by collecting multiple MS2 spectra from the same peptide, whereas it will be disadvantageous for general proteomic analysis since the mass spectrometer will spend less time to collect MS2 spectra from other low-abundance peptides (Figure 3). Thus, a medium number of this parameter should be considered in order to achieve satisfactory result of both qualitative and quantitative MS analysis from complex mixture. Given that chromatographic peak widths of most peptides are in the order of 10–300 s by nano-flow liquid chromatography separations, the Exclusion Duration is generally set to 20–200 s. Notably, two research groups observed that the optimal Exclusion Duration time was around 90 s in their MS method development (29, 30).
Figure 3.
The effect of dynamic exclusion duration time on spectra count. The exclusion duration time is generally bigger than the liquid chromatography retention time of the peptide to avoid the repeated analysis of the same peptide. Since it is unnecessary to keep the “black window” alive after the peptide is completely eluted from the analysis column, it is not recommended to set the parameter to a number that is much bigger than the retention time of most abundant peptides. A large exclusion duration time will be unbiased to abundant and low-abundance peptides since both of them will be analyzed once; small exclusion duration time will allow multiple analysis of the abundant peptide.
HPLC gradient
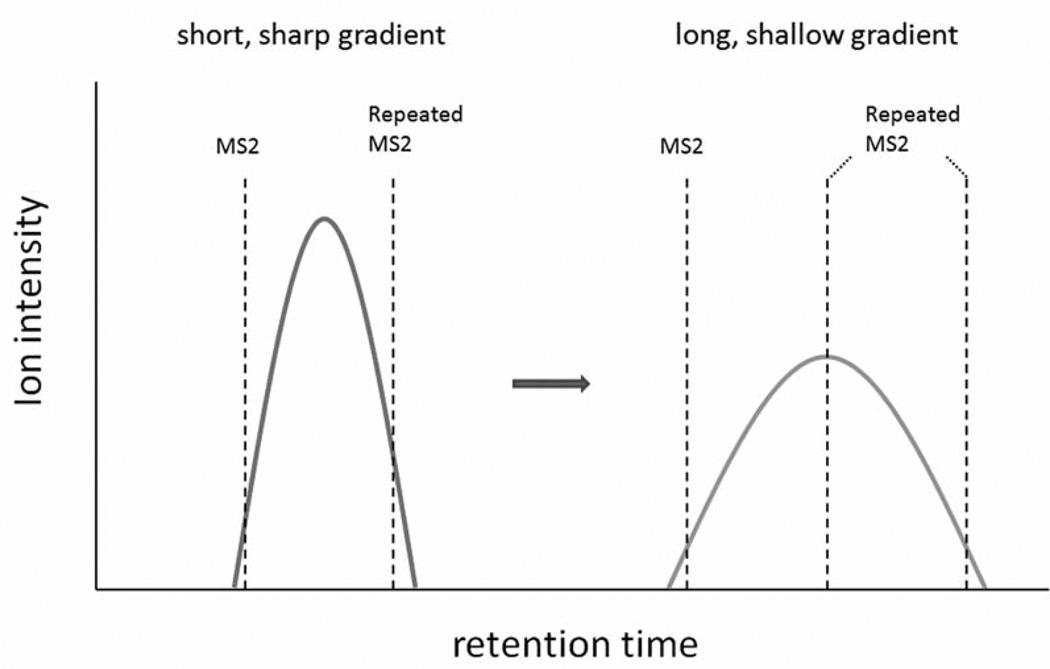
High performance liquid chromatography (HPLC) is coupled to tandem mass spectrometry to separate the peptide mixture from trypsin digestion, and reproducible chromatography is essential for relative quantitation. For analyzing complex biological samples by LC-MS/MS, a relatively longer/shallower HPLC gradient is generally applied in order to improve resolution and acquire more MS2 spectra for protein identification; however, if the gradient is too long, it is challengeable to obtain reproducible elution profiles from multiple samples, and the MS sensitivity will drop due to bigger peak width and lower peak intensity of the eluted peptide. Given the same dynamic exclusion duration time, it is expected that more repeated MS2 spectra count from a peptide will be collected if a longer/shallower HPLC gradient is applied (Figure 4).
Figure 4.
The effect of HPLC gradient on spectra count. Peptides eluted by short/sharp gradient have high peak intensity and narrow peak width, whereas peptides eluted by long/hallow gradient have low peak intensity and wide peak width. If the same dynamic exclusion duration time is maintained, while HPLC gradient is changed during MS analysis, it is expected that more repeated MS2 spectra count from a peptide will be collected with longer/shallower HPLC gradient.
Other factors
The spectra count associated with the identified proteins varies depending on instrumentation, protein database, spectral interpretation software and algorithm to estimate false discovery rate (FDR) (35–38). Due to differential fragmentation efficiency of various peptides, the high quality MS2 spectrum, containing abundant b and y ions, will be matched to the same peptide by different spectral interpretation software such as Sequest and Mascot, whereas the relatively low quality MS2 spectrum, containing more fragment ions from neutral loss or fragment ions other than b and y ions, may be assigned to different peptides by these software, thus affecting the spectra count of identified proteins. Separately, a filter criterion is generally applied after database search to remove candidate peptides with low scores that could be false positive. More often than not, a loose filter criterion will generate more identified proteins with higher spectra count and higher FDR, whereas the stringent filter criterion will generate less identified proteins with lower spectra count and lower FDR. Furthermore, many human proteins have splicing variants and PTMs, and the spectra count of these identified proteins may be incomplete if the information is not included during database search for peptide matching.
Conclusion
Quantitative and qualitative methods are complementary, not mutually exclusive. Indeed, all assays have limits in regard to analytical sensitivity and precision, the dynamic range for quantitation and the lowest level of detection. An ongoing challenge for MS practitioners is to identify all the expressed proteins and their PTMs in a complex proteome such as human cell or body fluid. Frequently some proteins and PTMs are detected in one sample, but not in the other, due to low abundance. Thus, absolute quantitation can be hampered since this calculation requires that the analyte can be detected with measurable quantities in both samples, whereas relative quantitation by spectra count and qualitative comparison are not limited by that. For instance, on the basis of the spectra count label-free quantitation approach, we recently characterized the proteomes of pancreatic ductal adenocarcinoma (PDAC) cells and normal pancreatic duct cells and identified a large number of differentially expressed metabolic enzymes. Identification of this differential level of abundance can facilitate our understanding of cancer cell’s survival in hypoxic and metabolic stresses (28). We also qualitatively characterized the phosphoproteomes of PDAC cells and normal duct cells. The analysis revealed differential phosphorylation of cell adhesion, cell junction, and structural proteins, providing clues to the complex dynamics of tumor invasion and metastasis in pancreatic cancer (39). Notably, more and more studies based on MS2 spectra count labelfree relative quantitation have been carried out in the field of molecular cell biology and cancer research (40–43). At situations that there is no immediate need for absolute quantitation, relative quantitation technique by spectra count shows its merits to fulfill the objective of comparative analysis since it is fast, direct and simple.
Acknowledgements
The authors wish to thank the support from the College of Science in George Mason University.
References
- 1.Aebersold R, Mann M. Mass spectroMetry-based proteoMics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 3.Cravatt BF, Simon GM, Yates JR3rd. The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 4.Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- 5.Domon B, Aebersold R. Options and considerations when selecting a quantitative proteomics strategy. Nat Biotechnol. 2010;28:710–721. doi: 10.1038/nbt.1661. [DOI] [PubMed] [Google Scholar]
- 6.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 7.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Quantitative mass spectrometry in proteomics: a critical review. Anal Bioanal Chem. 2007;389:1017–1031. doi: 10.1007/s00216-007-1486-6. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Sadygov R, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 11.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Albrethsen J, Knol JC, Piersma SR, Pham TV, de Wit M, Mongera S, Carvalho B, Verheul HM, Fijneman RJ, Meijer GA, Jimenez CR. Subnuclear proteomics in colorectal cancer: identification of proteins enriched in the nuclear matrix fraction and regulation in adenoma to carcinoma progression. Mol Cell Proteomics. 2010;9:988–1005. doi: 10.1074/mcp.M900546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neilson KA, Ali NA, Muralidharan S, Mirzaei M, Mariani M, Assadourian G, Lee A, van Sluyter SC, Haynes PA. Less label, more free: approaches in label-free quantitative mass spectrometry. Proteomics. 2011;11:535–553. doi: 10.1002/pmic.201000553. [DOI] [PubMed] [Google Scholar]
- 14.Li Z, Adams RM, Chourey K, Hurst GB, Hettich RL, Pan C. Systematic Comparison of Label-Free, Metabolic Labeling, and Isobaric Chemical Labeling for Quantitative Proteomics on LTQ Orbitrap Velos. J Proteome Res. 2012;11:1582–1590. doi: 10.1021/pr200748h. [DOI] [PubMed] [Google Scholar]
- 15.Yates JR3rd, Morgan SF, Gatlin CL, Griffin PR, Eng JK. Method to compare collision-induced dissociation spectra of peptides: potential for library searching and subtractive analysis. Anal Chem. 1998;70:3557–3565. doi: 10.1021/ac980122y. [DOI] [PubMed] [Google Scholar]
- 16.Link AJ, Eng J, Schieltz DM, Carmack E, Mize GJ, Morris DR, Garvik BM, Yates JR., 3rd Direct analysis of protein complexes using mass spectrometry. Nat Biotechnol. 1999;17:676–682. doi: 10.1038/10890. [DOI] [PubMed] [Google Scholar]
- 17.Chi A, Huttenhower C, Geer LY, Coon JJ, Syka JE, Bai DL, Shabanowitz J, Burke DJ, Troyanskaya OG, Hunt DF. Analysis of phosphorylation site on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc Natl Acad Sci USA. 2007;104:2193–2198. doi: 10.1073/pnas.0607084104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiesner J, Premsler T, Sickmann A. Application of electron transfer dissociation (ETD) for the analysis of posttranslational modifications. Proteomics. 2008;8:4466–4483. doi: 10.1002/pmic.200800329. [DOI] [PubMed] [Google Scholar]
- 19.Molina H, Matthiesen R, Kandasamy K, Pandey A. Comprehensive comparison of collision induced dissociation and electron transfer dissociation. Anal Chem. 2008;80:4825–4835. doi: 10.1021/ac8007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Second TP, Blethrow JD, Schwartz JC, Merrihew GE, MacCoss MJ, Swaney DL, Russell JD, Coon JJ, Zabrouskov V. Dual-pressure linear ion trap mass spectrometer improving the analysis of complex protein mixtures. Anal Chem. 2009;81:7757–7765. doi: 10.1021/ac901278y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen JV, Schwartz JC, Griep-Raming J, Nielsen ML, Damoc E, Denisov E, Lange O, Remes P, Taylor D, Splendore M, Wouters ER, Senko M, Makarov A, Mann M, Horning S. A dual pressure linear ion trap Orbitrap instrument with very high sequencing speed. Mol Cell Proteomics. 2009;8:2759–2769. doi: 10.1074/mcp.M900375-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabb DL, Huang Y, Wysocki VH, Yates JR3rd. Influence of basic residue content on fragment ion peak intensities in low-energy collision-induced dissociation spectra of peptides. Anal Chem. 2004;76:1243–1248. doi: 10.1021/ac0351163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Running WE, Ravipaty S, Karty JA, Reilly JP. A top-down/bottom-up study of the ribosomal proteins of Caulobacter crescentus. J Proteome Res. 2007;6:337–347. doi: 10.1021/pr060306q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammed S, Lorenzen K, Kerkhoven R, van Breukelen B, Vannini A, Cramer P, Heck AJ. Multiplexed proteomics mapping of yeast RNA polymerase II and III allows near-complete sequence coverage and reveals several novel phosphorylation sites. Anal Chem. 2008;80:3584–3592. doi: 10.1021/ac7024283. [DOI] [PubMed] [Google Scholar]
- 25.Priego-Capote F, Scherl A, Müller M, Waridel P, Lisacek F, Sanchez JC. Glycation isotopic labeling with 13C-reducing sugars for quantitative analysis of glycated proteins in human plasma. Mol Cell Proteomics. 2010;9:579–592. doi: 10.1074/mcp.M900439-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou W, Capello M, Fredolini C, Piemonti L, Liotta LA, Novelli F, Petricoin EF. Mass spectrometry analysis of the posttranslational modifications of a-enolase from pancreatic ductal adenocarcinoma cells. J Proteome Res. 2010;9:2929–2936. doi: 10.1021/pr901109w. [DOI] [PubMed] [Google Scholar]
- 27.Florens L, Carozza MJ, Swanson SK, Fournier M, Coleman MK, Workman JL, Washburn MP. Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods. 2006;40:303–311. doi: 10.1016/j.ymeth.2006.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou W, Capello M, Fredolini C, Racanicchi L, Piemonti L, Liotta LA, Novelli F, Petricoin EF. Proteomic analysis reveals Warburg effect and anomalous metabolism of glutamine in pancreatic cancer cells. J Proteome Res. 2012;11:554–563. doi: 10.1021/pr2009274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Wen Z, Washburn MP, Florens L. Effect of dynamic exclusion duration on spectral count based quantitative proteomics. Anal Chem. 2009;81:6317–6326. doi: 10.1021/ac9004887. [DOI] [PubMed] [Google Scholar]
- 30.Hoehenwarter W, Wienkoop S. Spectral counting robust on high mass accuracy mass spectrometers. Rapid Commun Mass Spectrom. 2010;24:3609–3614. doi: 10.1002/rcm.4818. [DOI] [PubMed] [Google Scholar]
- 31.Andrews GL, Dean RA, Hawkridge AM, Muddiman DC. Improving proteome coverage on a LTQ-Orbitrap using design of experiments. J Am Soc Mass Spectrom. 2011;22:773–783. doi: 10.1007/s13361-011-0075-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalli A, Hess S. Effect of mass spectrometric parameters on peptide and protein identification rates for shotgun proteomic experiments on an LTQ-Orbitrap mass analyzer. Proteomics. 2012;12:21–31. doi: 10.1002/pmic.201100464. [DOI] [PubMed] [Google Scholar]
- 33.Xu P, Duong DM, Peng J. Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res. 2009;8:3944–3950. doi: 10.1021/pr900251d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou W, Petricoin EF, Longo C. Mass spectrometry-based biomarker discovery. Methods Mol Biol. 2012;823:251–264. doi: 10.1007/978-1-60327-216-2_16. [DOI] [PubMed] [Google Scholar]
- 35.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 36.Kapp EA, Schütz F, Connolly LM, Chakel JA, Meza JE, Miller CA, Fenyo D, Eng JK, Adkins JN, Omenn GS, Simpson RJ. An evaluation, comparison, and accurate benchmarking of several publicly available MS/MS search algorithms: sensitivity and specificity analysis. Proteomics. 2005;5:3475–3490. doi: 10.1002/pmic.200500126. [DOI] [PubMed] [Google Scholar]
- 37.Carr S, Aebersold R, Baldwin M, Burlingame A, Clauser K, Nesvizhskii A. The need for guidelines in publication of peptide and protein identification data. Mol Cell Proteomics. 2004;(3):531–533. doi: 10.1074/mcp.T400006-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Reiter L, Claassen M, Schrimpf SP, Jovanovic M, Schmidt A, Buhmann JM, Hengartner MO, Aebersold R. Protein identification false discovery rates for very large proteomics data sets generated by tandem mass spectrometry. Mol Cell Proteomics. 2009;8:2405–2417. doi: 10.1074/mcp.M900317-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W, Capello M, Fredolini C, Racanicchi L, Piemonti L, Liotta LA, Novelli F, Petricoin EF. Phosphoproteomic analysis of pancreatic ductal adenocarcinoma cells reveals differential phosphorylation of cell adhesion, cell junction and structural proteins. J Proteomics Bioinform. 2011;4:170–178. [Google Scholar]
- 40.Paulo JA, Kadiyala V, Lee LS, Banks PA, Conwell DL, Steen H. Proteomic analysis (GeLC-MS/MS) of ePFT-collected pancreatic fluid in chronic pancreatitis. J Proteome Res. 2012;11:1897–1912. doi: 10.1021/pr2011022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warmoes M, Jaspers JE, Pham TV, Piersma SR, Oudgenoeg G, Massink MP, Waisfisz Q, Rottenberg S, Boven E, Jonkers J, Jimenez CR. Proteomics of mouse BRCAl-deficient mammary tumors identifies DNA repair proteins with diagnostic and prognostic value in human breast cancer. Mol Cell Proteomics [Epub ahead of print] doi: 10.1074/mcp.M111.013334. Published online Feb 24, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowman JP, Hages E, Nilsson RE, Kocharunchitt C, Ross T. Investigation of the Listeria monocytogenes Scott A acid tolerance response and associated physiological and phenotypic features via whole proteome analysis. J Proteome Res [Epub ahead of print] doi: 10.1021/pr201137c. Publsihed online Feb 29, 2012. [DOI] [PubMed] [Google Scholar]
- 43.Sigdel TK, Kaushal A, Gritsenko M, Norbeck AD, Qian WJ, Xiao W, Camp DG, 2nd, Smith RD, Sarwal MM. Shotgun proteomics identifies proteins specific for acute renal transplant rejection. Proteomics Clin App1. 2010;4:32–47. doi: 10.1002/prca.200900124. [DOI] [PMC free article] [PubMed] [Google Scholar]