Significance
Hybrid rice developed in China has been contributing greatly to the world’s food production. The pioneer super hybrid rice developed by crossing 93–11 and Peiai 64s, Liang–You–Pei–Jiu has been widely grown in China and other Asia-Pacific regions for its high yield. Here, the quality genome sequences for both parental lines were presented and updated, and a high-resolution map of genome-wide graphic genotypes was constructed by deep resequencing a core population of 132 Liang–You–Pei–Jiu recombinant inbred lines. A series of yield-associated loci were fine-mapped, and two of them were delimited to regions each covering one candidate gene with the large recombinant inbred line population. The study provided an ideal platform for molecular breeding by quantitative trait loci cloning in rice.
Keywords: Oryza sativa, QTL dissection, genome sequence update
Abstract
The growing world population and shrinkage of arable land demand yield improvement of rice, one of the most important staple crops. To elucidate the genetic basis of yield and uncover its associated loci in rice, we resequenced the core recombinant inbred lines of Liang–You–Pei–Jiu, the widely cultivated super hybrid rice, and constructed a high-resolution linkage map. We detected 43 yield-associated quantitative trait loci, of which 20 are unique. Based on the high-density physical map, the genome sequences of paternal variety 93–11 and maternal cultivar PA64s of Liang–You–Pei–Jiu were significantly improved. The large recombinant inbred line population combined with plentiful high-quality single nucleotide polymorphisms and insertions/deletions between parental genomes allowed us to fine-map two quantitative trait loci, qSN8 and qSPB1, and to identify days to heading8 and lax panicle1 as candidate genes, respectively. The quantitative trait locus qSN8 was further confirmed to be days to heading8 by a complementation test. Our study provided an ideal platform for molecular breeding by targeting and dissecting yield-associated loci in rice.
Rice is one of the most important staple crops in the world and serves as a model for monocots (1). Currently, rice breeding faces the challenge of overcoming the yield plateau. All important agronomic traits would ultimately need to consider their impacts on the yield, which is linked to various growth and developmental components, such as tiller number, seed number and set, and grain weight, to name a few. A number of quantitative trait loci (QTLs) have been reported to control these components, including those revealed by map-based cloning studies, such as IPA1/WFP for tiller and spikelet numbers (2, 3); days to heading8 (DTH8)/Ghd8 and Ghd7 for heading date, plant height, and spikelet number (4, 5); Gn1 for spikelet number (6); GIF1 for seed set (7); and grain size3 (GS3) and GW5 for grain size and weight (8, 9). Although a series of QTLs for yield components have been cloned, elucidation of the genetic mechanisms underlying the inheritance of superior yield in super hybrid rice still has a long way to go.
Hybrid rice has a notable contribution to yield improvement. Various commercialized hybrids are derived by crossing different varieties within or between two subspecies, Oryza sativa ssp. indica and ssp. japonica (10, 11). As a pioneer super hybrid rice, Liang–You–Pei–Jiu (LYP9) realized the target of 10.5 tons/ha in 2000 (12). LYP9 was developed by a cross of the paternal 93–11, an indica variety widely grown in China (13), and the maternal PA64s cultivar with a mixed genetic background of indica and javanica. To date, it has been widely cultivated for commercial production in China. Such a feature was thought to make LYP9 recombinant inbred lines (RILs) ideal materials for exploring molecular mechanisms underlying rice yield.
Here, we constructed a high-density linkage map by resequencing the parents of LYP9 and 132 core RILs. As a result, we finished the chromosome-scale genome sequence of PA64s and updated the 93–11 genome sequence by anchoring to chromosomes, filling up gaps, and correcting single-base errors. Twenty-five unique QTLs related to rice production were identified. One QTL, qSN8 for spikelet number, was fine-mapped with a large RIL population and confirmed as DTH8 by a complementation test.
Results
Population Sequencing and Linkage Map Construction.
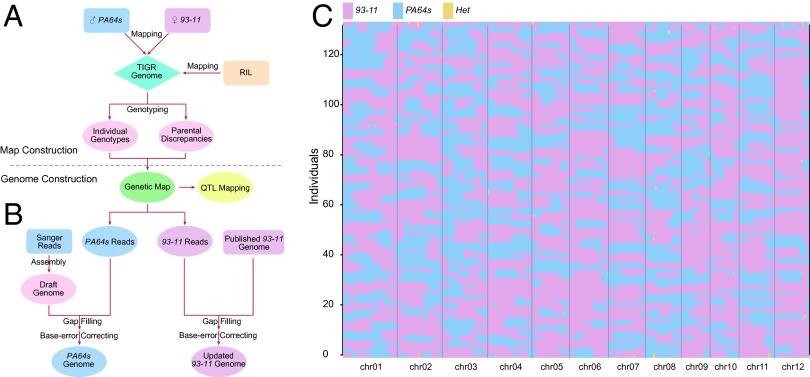
A strategy of sequencing-based map construction was developed with LYP9 core RILs and applied in parental genome assembly and QTL mapping (Fig. 1 A and B). We sequenced a segregating population of 132 LYP9 RILs together with PA64s and 93–11 on Illumina HiSeq2000 platform. A total of 244 GB of raw data were generated for all of the RILs, with approximately fourfold depth for each RIL, 48-fold depth for PA64s, and 36-fold depth for 93–11 (SI Appendix, Table S1). These data were aligned to the Nipponbare genome (Os-Nipponbare-Reference-IRGSP-1.0, MSU release 7) using SOAP2 (14). We identified 501,499 single nucleotide polymorphisms (SNPs) with homozygous genotypes between both parents using SOAPsnp (15). These polymorphisms were used as potential SNP markers, at which each RIL was genotyped. Some markers with low genotyping scores and those located in highly repeated regions were removed to avoid ambiguity in the following map construction. The missing genotypes of each RIL caused by low-coverage sequencing were imputed by using an effective imputation model, k-nearest neighbor algorithm (16). Finally, we used 171,847 high-quality polymorphic SNP markers, of which 39.21% were located in genic regions, to construct a recombinant bin map (SI Appendix, Table S2 and Fig. S1). In addition, distortions in segregation were found at 4,799 loci based on χ2 analysis (P < 0.01), among which two large regions on chromosome 01 (Chr01) (39–41 Mb) and Chr12 (23–25 Mb) might be associated with selective fertilization of 93–11 gametal genotypes (SI Appendix, Fig. S2).
Fig. 1.
Schematic overview of the parents–RIL system construction and the map of genome-wide graphic genotypes. (A) SNP markers were developed between PA64s and 93–11, at which point each RIL was genotyped. A map of genome-wide graphic genotypes with high resolution was constructed to dissect QTL. (B) Improved genome sequence assembly of both parents, including gap filling and base-error correction. (C) Graphic genotypes of 132 RILs were identified by a sliding window approach along each chromosome. Different colors represent different genotypes: purple, 93–11; sapphire, PA64s; maroon, heterozygous blocks.
A modified sliding window approach was adopted to determine recombinant breakpoints along chromosomes of each individual (17). Based on the reported recombination rate (18), the 92.4 kb minimum recombinant distance was calculated to estimate the minimum window size. The error-prone genotypes of RILs at polymorphic loci were corrected as the window sliding SNP-by-SNP along chromosomes. The frequently transient genotypes of adjacent windows were merged into a heterozygous block. By integrating adjacent windows with the same genotype, a map of genome-wide graphic genotypes was obtained in high resolution (Fig. 1C). The map contained 3,524 recombinant blocks with the average length of 105.6 kb, which was in parallel to the expected minimum observation. On average, Chr01 and Chr09 had the highest and lowest frequency of the crossover event, 6.40 and 2.76 times for each RIL, respectively (SI Appendix, Table S3). The graphic genotypes were converted into a genetic map with total genetic distance of 1381.9 cM and an average of ∼0.392 cM between adjacent recombinant blocks. The average region of heterozygous graphic genotypes in the RILs occupied 1.79% of the genome, which might be caused by gene conversions (19) or structure variations between indica and japonica. We identified 386 recombinant hotspots in the map using sequenceLDhot (20), of which 74.7% were located in the heterozygous regions, and the gene density near hotspots was relatively high compared with that at the whole genome level (SI Appendix, Table S4 and Fig. S3). We also observed that the polymorphic rate near the hotspots was significantly higher than that on the whole genome level (t test, P < 0.001) (SI Appendix, Table S5). Multiple EM for Motif Elicitation (MEME) analysis revealed that some conserved motifs preferentially appeared surrounding the hotspots, such as the CpG islands (SI Appendix, Fig. S4). The recombination was suppressed in the region surrounding the centromere on every chromosome, resulting in an S-shape curve, which reflected the overall quality of the map (SI Appendix, Fig. S5).
Assembly of Parental Genomes Using RIL Population.
To obtain the genome sequence of PA64s, we first generated 6.0× high-quality reads by the Sanger method using the whole genome shotgun strategy. The assembly, performed with RePS software (13, 21), reached 347 Mb with the contig N50 length of 17.0 Kb. Integrated with developed SNP markers, the scaffolds were anchored to chromosomes to create a 322 Mb chromosome-scale genome sequence. However, there remained a mass of gaps because of genomic repeats, sequencing depth and coverage, and representativeness and randomness of genomic libraries. Therefore, two approaches were taken to fill up the remaining gaps in the PA64s genome sequence (Fig. 1B). Using SOAPdenovo (22) we assembled the maternally derived reads (MDRs) into scaffolds, which were then located to gaps in the PA64s genome with SNP markers. And gaps were filled with 24.4 Mb of new sequences. We then realigned the MDRs to the genome sequence of PA64s. Depending on the relationship of mate reads, we accumulated the reads located in gaps when one of the paired reads was mapped well at the edge of a gap and the other one was unmapped. The overlap information of the reads in the same gap was used to fill up the gap. By this approach, we introduced 4.9 Mb of new sequences to the gaps so that the final genome sequence of PA64s reached 382 Mb, with 351.5 Mb of nongaped sequences on chromosomes.
To evaluate the coverage and quality of the genome assembly, we aligned the nonredundant collection of 26,278 full-length cDNAs and mRNAs in japonica to the PA64s genome (23, 24). Among them, 82.90% and 91.29% were mapped with 95% coverage in the genome, respectively (SI Appendix, Table S6). Forty-six filled gaps were randomly selected for evaluation of the introduced sequences by PCR. A pair of primers were designed with one located on the previous scaffold and the other on the new sequence, and 93.5% of filled gaps were confirmed to be correct as revealed by 43 expected PCR products (SI Appendix, Table S7). The PA64s genome sequence was also polished by correcting single nucleotide errors, possibly introduced by sequencing mistakes. Theoretically, all polymorphic SNPs can be mapped by linkage with the RIL population. A single-base error was claimed when a SNP marker was polymorphic between the parents but could not be mapped, with the exception of extremely distorted markers. Based on the two criteria, 36,033 loci with homozygous genotypes were identified as single-base errors with a frequency of 1 bp/10 kb. We sequenced 29 PCR products with single corrected base in the middle. Except for one sequence aligned nonspecifically, all of the remaining 28 sequences of the PCR products were uniquely aligned to the PA64s genome with the expected genotypes, testifying to the effectiveness of single nucleotide correction (SI Appendix, Table S8).
The available genome sequence of 93–11 (13) was also improved using the same strategy (Fig. 1B). Approximately 3.8 Mb of new sequences in 1,493 gaps were introduced across the genome and the updated 93–11 genome sequence reached 423.0 Mb, including 369.8 Mb of quality sequence anchored to chromosomes using the linkage map. Besides, 62,650 single-base errors were corrected by aligning paternally derived reads (PDRs) to the 93–11 genome. We also removed 23.6 Mb of falsely assembled sequences that lacked support by linkage or synteny analysis (SI Appendix, Fig. S6). Therefore, the reference-guided and improved assemblies of PA64s and 93–11 provide an important reference for indica rice.
Comparative Analysis of Three Rice Genomes.
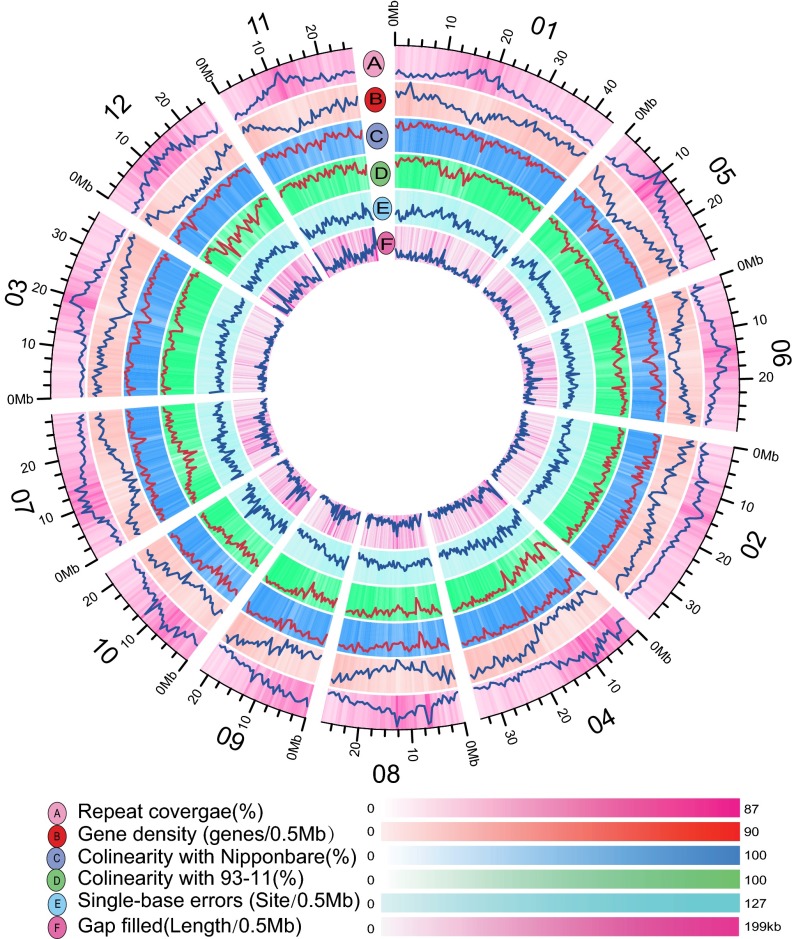
The genome sequences of PA64s and 93–11 provided the basis for detailed genomic analysis. Genome-wide synteny analyses among the three genomes of PA64s, 93–11, and Nipponbare revealed extensive distribution of tandem and segmental duplications (SI Appendix, Fig. S7). A total of 280,055 insertions/deletions (InDels) (<20 bp) and 323 large inversions were identified between parental genomes (SI Appendix, Table S9 and Fig. S8). To facilitate the genome annotation, we generated an average of 7.1 Gb RNA-Seq reads (49 bp) from panicles of each parent in the booting stage. A total of 36,909 high-confident genes were predicted in the PA64s genome with both evidence-based and de novo prediction (Fig. 2, SI Appendix, Table S10), each gene supported by more than one protein from rice, maize, and sorghum (SI Appendix, Table S11). Among them, 97% were functionally annotated (SI Appendix, Table S12), and 70% were covered by RNA-Seq reads. As observed, gene density is relatively low surrounding the centromeres, where the repetitive contents are inversely high (Fig. 2). The updated 93–11 genome was also annotated to contain 40,464 high-confident genes using the same genome annotation pipeline (SI Appendix, Table S13).
Fig. 2.
Concentric circles showing different features of the PA64s genome using the Circos program (36). (A) Sequence coverage of transposable elements (TEs) with all identified intact elements in PA64s. (B) Density of annotated genes counted per 0.5-MB sliding window. (C and D) Graphical view of the collinear blocks of genome sequence in PA64s compared with Nipponbare (C) and with 93–11 (D). (E) Density of single-base errors per 0.5-Mb sliding window. (F) The distribution of filled gaps.
To reveal variation in the gene content, we studied homology between the putative coding sequences of PA64s and 93–11. Similarity searches by BLAST revealed that 90.30% of all predicted genes in 93–11 showed substantial similarity with those of the PA64s. In total, 62% of protein-coding genes were expressed in at least one parent. Comparison of those genes among three genomes showed that 20,699 gene families were shared by all three varieties. In total, 725 (0.81%) and 803 (0.89%) unique genes were identified for PA64s and 93–11, respectively (SI Appendix, Fig. S9). Interestingly, the genes related to environmental adaptation were significantly abundant among specific genes in 93–11 through Kyoto Encyclopedia of Genes and Genomes pathway analysis (SI Appendix, Fig. S10).
QTL Detection and Analysis Using the LYP9 RIL Population.
To elucidate the genetic mechanism underlying yield, we primarily focused on 12 traits known to be important for rice yield (SI Appendix, Table S14). Both the 93–11 and PA64s showed significant differences in all traits but effective tiller number and primary panicle branch number. Phenotypic values of the yield-associated traits in the LYP9 RILs were all found to be continuous and exhibited normal or skewed distribution patterns (SI Appendix, Fig. S11). Therefore, the characters in the population inherited quantitatively and met the requirements for QTL mapping. SI Appendix, Tables S15 and S16 present the correlations between the traits of 132 RILs under two environments, Hangzhou and Hainan, respectively. A positive correlation was observed in Hangzhou between yield per plant and plant height, spikelet number per panicle, seed set, and 1,000-grain weight.
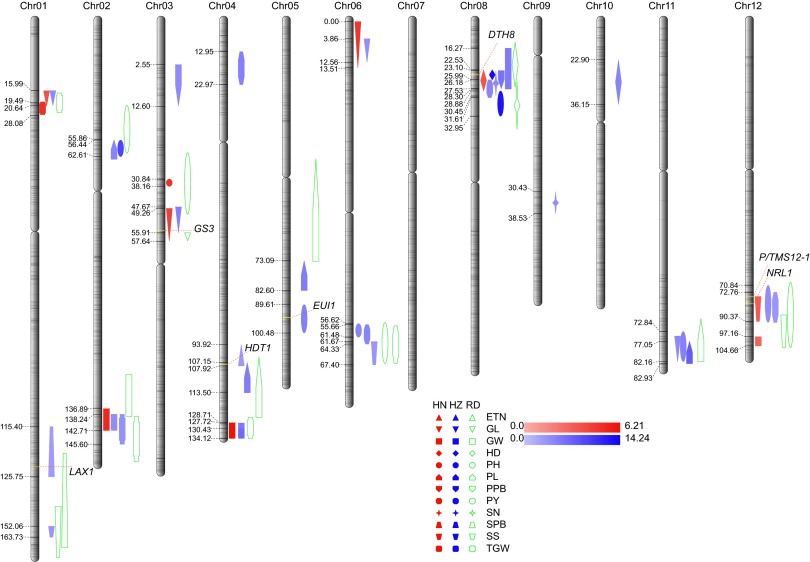
Except for unique peak for qETN4 on Chr04 (logarithm of odds (LOD) = 2.82) and qSPB1 on Chr01 (LOD = 2.70), respectively, detected in Hangzhou, QTLs with LOD > 3.0 were declared. Based on the linkage map, 20 unique QTLs and 23 reported QTLs were detected, with 1∼7 QTLs for each trait (Fig. 3, SI Appendix, Table S17). The regions of previously identified QTLs were effectively narrowed down from 4,052 kb to 1,844 kb on average. Among the 33 QTLs detected in Hangzhou and 10 QTLs in Hainan, six QTLs were detected at both locations for the same phenotype, whereas the remaining 31 QTLs were environmentally dependent. Eight QTL clusters were identified, suggesting that the pleiotropic effect or linkage of genes was likely responsible for the trait correlation. Positive alleles of 37 QTLs were from 93–11 and those of six QTLs were from PA64s. Genotypic analysis on five QTLs demonstrated that haplotype status in RIL lines was responsible for corresponding traits, whereas combination of favorable QTLs contributed to relatively high yield per plant (SI Appendix, Fig. S12). One QTL for seed set detected in Hangzhou, qSS1, was located in the significantly distorted regions (SI Appendix, Fig. S2 and Table S17). Therefore, these identified QTLs and their haplotypes may be used in plant breeding with marker-assisted selection (MAS).
Fig. 3.
The positions of QTLs located on each chromosome. The number on the left of each chromosome is the marker’s genetic distance (cM). The red patterns represent phenotypes collected in Hainan, the blue ones those in Hangzhou, and the green ones represent reported QTL. The texts of different shapes are abbreviations of different phenotypes: ETN, effective tiller number; GL, grain length; GW, grain width; HD, heading date; PH, plant height; PL, panicle length; PPB, primary panicle branch number; PY, yield per plant; SN, spikelet number per panicle; SPB, secondary panicle branch number; SS, seed set; TGW, 1,000-grain weight. The gradual change of colors represents different LOD values.
Fine-Mapping of QTLs for Yield-Associated Traits.
With a relatively high mapping resolution, 10 QTLs were mapped to small genomic intervals. The largest effect of QTL on heading date, qHD8, was mapped at the same locus as qSN8 detected in Hangzhou at 3.95∼4.75 Mb on Chr08 (SI Appendix, Fig. S13 A and B), where DTH8 previously cloned was the candidate gene (4). The major QTL for grain length identified in both conditions, qGL3, was positioned to 2.0∼2.6 Mb on Chr03 (SI Appendix, Fig. S13 C and D), where GS3 was previously cloned as a grain length and weight controller gene (8). The qPH5 gene for plant height had its LOD peak in the region of 2.3 Mb on Chr05 (SI Appendix, Fig. S13E), covering EUI1, a major QTL associated with first internode length and plant height in rice (25). Another QTL on Chr12 for plant height, qPH12, was located at the same locus as NRL1 (26), a gene regulating leaf morphology and plant architecture in rice (SI Appendix, Fig. S13F). The region of qETN4 detected in Hangzhou for effective tiller number on Chr04 (SI Appendix, Fig. S13G) included HTD1, a major locus for tillering and dwarfism (27). The qSPB1 locus for secondary panicle branch number was mapped within the region on Chr01 (SI Appendix, Fig. S13H), where lax panicle1 (LAX1), a gene responsible for rachis-branch and spikelet development in rice, was located (28). Finally, the QTL for seed set qSS12 detected in Hainan had its LOD peak in the region of ∼2 Mb on Chr12 (SI Appendix, Fig. S13I), where a noncoding small RNA gene, P/TMS12-1, served as an important regulator of the development of male reproductive organs in rice (29).
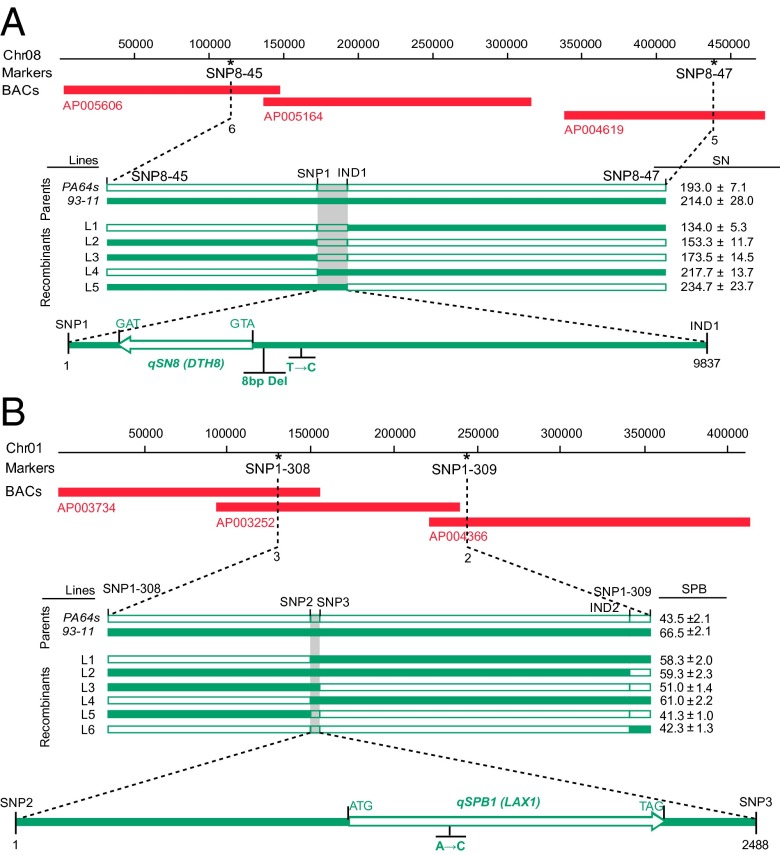
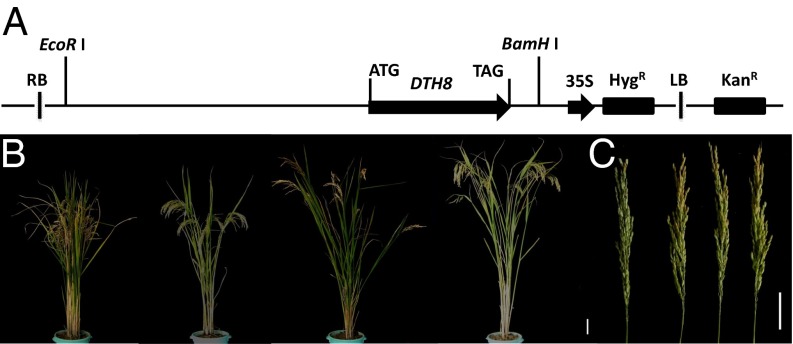
To further fine-map qSN8 and qSPB1, a larger LYP9 RIL population with 1,709 lines was adopted. Through linkage analysis using developed SNP and InDel markers (SI Appendix, Table S18), the QTL regions were narrowed down to 9.83 kb for qSN8 and 2.50 kb for qSPB1, respectively, where single candidate genes, DTH8 and LAX1, were located. Sequence comparison of the two candidate genes between 93–11 and PA64s revealed one SNP (T–873→C–873) and one InDel (–478TATCATTG–471→null) in the promoter region of DTH8 and one SNP causing amino acid substitution in the coding region (A+222→C+222) of LAX1 (Fig. 4 A and B). These results in part explained the differences in spikelet number per panicle and secondary panicle branch number between the two parents, respectively. Real-time PCR analysis of candidate genes showed the expressions of GS3 and EUI1 significantly enhanced, whereas those of DTH8, HTD1, and LAX1 were reduced dramatically in PA64s compared with 93–11 (SI Appendix, Fig. S14 and Table S19). Up-regulation of DTH8 in 93–11 compared with PA64s may attribute to the InDel and/or SNP in the promoter region of the gene. Genetic complementation tests showed that all T1 plants with DTH8 alleles of 93–11 were taller to different degrees than PA64s and exhibited phenotypes of late heading with increased spikelet number (Fig. 5, SI Appendix, Table S20).
Fig. 4.
(A) Fine-mapping of qSN8. The qSN8 locus was mapped between the SNP markers SNP8-45 and SNP8-47 on Chr08, and was covered by three discontinuous BACs. The qSN8 locus was narrowed down to a 9.83-kb genomic region between SNP marker SNP1 and InDel marker IND1. T→C, 93–11 (C) to PA64s (T) at 873 from A+1TG in DTH8. 8 bp Del, 8 bp deletion (TATCATTG) at 471–478-bp from A+1TG in PA64s. (B) Fine-mapping of qSPB1. The qSPB1 locus was mapped between the SNP markers SNP1-308 and SNP1-309 on Chr01, and was covered by three BAC contigs. The qSPB1 locus was narrowed down to a 2.50-kb genomic DNA region between the SNP markers SNP2 and SNP3. A→C, 93–11 (A) to PA64s (C) at +222 from A+1TG in LAX1. The numerals indicate those of recombinants identified from 1,708 RIL lines. All lines displayed are the recombinants from the RIL population. Closed boxes indicate the coding sequence. The white and black bars denote the molecular marker genotype of PA64s and 93–11, respectively.
Fig. 5.
Genetic complementation test. (A) Schematic structure of the complementation construct pCAMBIA1300-DTH8, containing the entire DTH8 gene, the 2,627-bp upstream sequence, and the 234-bp downstream sequence. (B) Comparison of plant height of the DTH8 transformants and PA64s at the mature stage. (Scale bar 10 cm.) (C) Comparison of panicle of the DTH8 transformants and PA64s. (Scale bar, 5 cm.) From left to right are PA64s, T1-1, T1-2, and T1-3.
Discussion
Improvement of Genome Sequence by Resequencing the Core RILs.
A set of high-quality SNP markers was developed using the sequencing-based genotyping approach. In contrast to the requirement of genome sequences of the mapping parents (17) or maximum parsimonious inference as parent-independent genotyping method (30), we genotyped each parent using high-depth resequencing data to avoid potential errors caused by reference. More accurate fine structures were revealed in the map of genome-wide graphic genotypes, such as heterozygosis and distortion. Our modeling method of individual and graphic genotyping has potential applications in other species and mapping populations.
Two updated genomes of both parents are presented here. By making full use of the sequencing data of RILs, we were able to improve the genome sequences by filling up gaps and correcting single-base errors. Here, we also proved an alternative approach to improving the genome sequence of agriculturally important crops. Besides, the improvement will be accumulated as more RILs, developed from the same parents, are sequenced. Thus, we recommend that, for each crop, at least one segregating population and its parents should be sequenced. Core and larger mapping populations probably should be sequenced step by step upon the reduction of cost.
QTL Fine-Mapping with the Large RILs and SNP Markers.
Complementary to population mapping, family mapping can detect low-frequency alleles and small-effect QTLs with less false positives (31). It is encouraging that sequencing-based SNP markers could be made useful for constructing a genetic map. The improved quality and resolution of the linkage map greatly facilitated QTL dissection. Based on the map, a total of 43 yield-associated QTLs were detected by family mapping, including 20 unique QTLs. Multiple QTLs were mapped to the same region, such as qPH2 and qPL2 on Chr02 and qPH11, qPL11, and qGL11 on Chr11 detected in Hangzhou (Fig. 3), suggesting that the relationship among these quantitative traits was intricate. The resolution of QTL mapping, determined by the size of the confidence interval of the QTL, depends on both the population size and marker density. To fine-map candidate genes, a large RIL population was adopted here, with which qSPB1 and qSN8 were fine-mapped. With the advantage of simultaneous mapping for multiple traits, the large RIL population has been and is now being served as genetically stable material for fine-mapping a series of unique yield-associated QTLs. As a persuasive example for the feasibility of the strategy, qSN8 was confirmed to be DTH8, which controls heading date and spikelet number.
In our previous study, 120 polymorphic markers were screened out between 93–11 and PA64s among 336 markers (including 287 SSR and 49 STS markers), only reaching a ∼35.7% polymorphic rate (32). Owing to plenty of accurate SNPs (4 SNPs/10 kb) and InDels (8 InDels/10 kb) identified between the two parents, unique reliable SNP and InDel markers were developed for dissecting yield-associated QTLs. By comparing the physical regions of all QTLs detected here and recombinant hotspots, we found that 15 regions were shared (SI Appendix, Table S21), involving 23 QTLs and 42 recombinant hotspots. The results will facilitate fine-mapping of these QTLs. SNPs and InDels within or adjacent to genes especially can also be used in functional MAS in hybrid rice. Combination of advantageous alleles has created the RIL lines exhibiting agronomical traits, especially plant yield superior to 93–11, which has better traits than PA64s (SI Appendix, Fig. S12). Therefore, it is an effective platform for dissection and isolation of yield-associated QTL, which will certainly advance our knowledge on the genetic mechanisms of yield and facilitate molecular breeding in rice.
Materials and Methods
Sample Preparation and Sequencing.
The genomic DNA of LYP9 RILs together with their parents was collected with DNeasy Plant Mini Kit (QIAGEN). Total RNA from panicles of each parent in the booting stage was extracted with TRIzol reagent (Invitrogen). Sequencing libraries with a 500 bp insert size were constructed and sequenced on HiSeq2000 according to the manufacturer’s instructions (Illumina). Raw pair-end reads were generated by applying the base-calling pipeline, SolexaPipeline-0.3 (Illumina). The mapped reads were classified into three categories based on the alignment results: “uniquely aligned,” “repeatedly aligned,” and “unaligned” reads. The trimming strategy for dealing with mismatches was as described previously (33). Duplicated reads caused by the PCR process were removed by a PERL script. For each RIL, about 60% of the reads were properly and uniquely aligned to the reference genome.
Construction of Graphic Genotypes and Linkage Map.
We modified the sliding window approach developed by Han and colleagues (17) to construct the recombinant bin map based on the SNP markers developed. For our mapping population of LYP9 RILs, the minimum observable recombination fraction is 1/132. Therefore, we could estimate the minimum length of the distinguishable recombinant interval of each chromosome according to the reported recombination rate (18). From the equation r = 1/2 × σ × 1/132, we determined the window size and the marker numbers in a window along the chromosomes of each RIL. Based on the high-resolution map of genome-wide graphic genotypes, we constructed a linkage map using the Kosambi map function: θ = 0.25 ln[(1 + 2r)/(1 – 2r)] (34), with r representing the value of recombination rate. Four primary steps adopted in the graphic genotyping procedure are available in SI Appendix, Experimental Procedures.
Genome Sequence Gap Filling.
Based on the constructed map of graphic genotypes and the alignment results by SOAP2 (14), the reads for each RIL were classified into three categories using a PERL script: “MDRs,” “PDRs,” and “undistinguished reads.” Two approaches were used to fill the remaining sequence gaps on the chromosomes of two parents. Briefly, we describe the gap filling procedure in the PA64s genome as an example. In the first approach, we assembled the MDRs into scaffolds based on a de Bruijn graph using short-read sequence assembler SOAPdenovo (22). The same genomic markers developed for chromosome anchoring again were used to locate the new assembled scaffolds on the chromosomes of PA64s. We kept the uniquely and accurately mapped markers for further analysis. Some newly assembled scaffolds were properly located in the gaps, whereas some had overlaps with the assembled sequences of the chromosomes. By this way, any amount of new sequences filled the gaps of PA64s. In the second approach, we accumulated the MDR reads located in the gaps when only one of the paired reads was uniquely mapped at the edge of a gap and the other one was unmapped, depending on the relationship of the pair-end reads. By taking full advantage of the information of the overlapped reads in the same gaps, new sequences were introduced into the gaps. Some gaps, which were shorter than the insert size of the resequencing library, were filled up by this approach.
Correction of Single-Base Errors.
Based on the classified reads, we corrected single base errors in the genomes of two parents. Taking the PA64s genome as an example, we first realigned MDR to the gap-filled genome of PA64s. Then SOAPsnp (15) was used to calculate the posterior probability of each genotype and construct a consensus sequence of PA64s by locating the allele type with the highest probability at each position. Homozygous sites were treated as potential single-base errors in the genome when satisfying the following criteria: sequencing depth between 10 and 400, base quality greater than 20, and copy number smaller than 2. These sites were corrected in the final PA64s genome sequence. The same approach was used to correct single-base errors in the genome sequence of 93–11.
Mapping Population.
The core mapping population of 132 LYP9 RILs was randomly chosen from 1,840 RILs derived by single-seed descendants from a cross between an elite paternal inbred Oryza sativa ssp. indica cv. 93–11 and the maternal inbred Oryza sativa ssp. indica cv. PA64s, a photo-thermosensitive male sterile line. The population was developed in the experimental fields at China National Rice Research Institute in Hangzhou, Zhejiang Province, and in Sanya, Hainan Province, China. After 12 generations of self-fertilization following the initial cross, genomic DNA samples of the F13 RILs were isolated for genotyping.
QTL Analysis.
QTL analysis was performed with the MultiQTL package (www.multiqtl.com) using the maximum likelihood interval mapping approach for the RIL-selfing population. For major-effect QTLs, the LOD threshold was obtained based on a permutation test (1,000 permutations, P = 0.05) for each dataset. QTLs were named according to McCouch et al. (35).
Additional Experimental Procedures.
A detailed description of SNP markers screening individual genotyping, PA64s genome annotation, phenotyping methods, correlation analysis for 12 traits, InDel/SNP marker development for fine-mapping (primer sequences are shown in SI Appendix, Table S18), RNA extraction, real-time RCR analysis (primer sequences are shown in SI Appendix, Table S19), vector construction, and plant transformation can be found in SI Appendix, Experimental Procedures.
Supplementary Material
Acknowledgments
We sincerely thank Dr. Yunbi Xu and Dr. Bo Liu for their critical reading of and suggestions for our manuscript. This work was supported by grants from the National Natural Science Foundation of China (Grants 31221004 and 30921140408), the Hi-Tech Research and Development (863) Program of China (Grant 2012AA10A300), Zhejiang Provincial Natural Science Foundation of China (Grant LR12C06001), and the Key Laboratory Projects supported by Shenzhen City (Grant CXB200903110066A) and Shenzhen Key Laboratory of Transomics Biotechnologies (Grant CXB201108250096A).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper has been deposited in the GenBank database\European Molecular Biology Laboratory/DNA Data Base in Japan [accession nos. PRJNA208877 (genome sequence and annotation of PA64s BioProject), SRA091322 (Sanger and Hiseq sequencing data of PA64s), SRA091762 (RNA-seq data of PA64s), SRA093225 (RNA-seq data of 93-11), SRA093226 (raw resequencing data of 93-11), and SRA091270 (short reads of RILs)]. Genomic data including the updated 93-11 sequences and annotations are also available at our Rice Information System database, http://rice.genomics.org.cn/rice.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306579110/-/DCSupplemental.
References
- 1.Guo LB, Cheng SH, Qian Q. Highlights in sequencing and analysis of rice genome. Chinese J Rice Sci. 2004;18(6):557–562. [Google Scholar]
- 2.Jiao Y, et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat Genet. 2010;42(6):541–544. doi: 10.1038/ng.591. [DOI] [PubMed] [Google Scholar]
- 3.Miura K, et al. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat Genet. 2010;42(6):545–549. doi: 10.1038/ng.592. [DOI] [PubMed] [Google Scholar]
- 4.Wei X, et al. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153(4):1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xue W, et al. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 6.Ashikari M, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–745. doi: 10.1126/science.1113373. [DOI] [PubMed] [Google Scholar]
- 7.Wang ET, et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008;40(11):1370–1374. doi: 10.1038/ng.220. [DOI] [PubMed] [Google Scholar]
- 8.Fan C, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112(6):1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 9.Weng J, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18(12):1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZJ. Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci. 2010;15(2):57–71. doi: 10.1016/j.tplants.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng SH, Zhuang JY, Fan YY, Du JH, Cao LY. Progress in research and development on hybrid rice: A super-domesticate in China. Ann Bot (Lond) 2007;100(5):959–966. doi: 10.1093/aob/mcm121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan LP, editor. Super Hybrid Rice Research. China: Shanghai Scientific & Technical Publishers, Shanghai; 2006. pp. 2–3. [Google Scholar]
- 13.Yu J, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. indica) Science. 2002;296(5565):79–92. doi: 10.1126/science.1068037. [DOI] [PubMed] [Google Scholar]
- 14.Li R, et al. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics. 2009;25(15):1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 15.Li R, et al. SNP detection for massively parallel whole-genome resequencing. Genome Res. 2009;19(6):1124–1132. doi: 10.1101/gr.088013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet. 2010;42(11):961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 17.Huang X, et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19(6):1068–1076. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M, et al. An integrated physical and genetic map of the rice genome. Plant Cell. 2002;14(3):537–545. doi: 10.1105/tpc.010485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S, et al. Great majority of recombination events in Arabidopsis are gene conversion events. Proc Natl Acad Sci USA. 2012;109(51):20992–20997. doi: 10.1073/pnas.1211827110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fearnhead P. SequenceLDhot: Detecting recombination hotspots. Bioinformatics. 2006;22(24):3061–3066. doi: 10.1093/bioinformatics/btl540. [DOI] [PubMed] [Google Scholar]
- 21.Wang J, et al. RePS: A sequence assembler that masks exact repeats identified from the shotgun data. Genome Res. 2002;12(5):824–831. doi: 10.1101/gr.165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li R, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20(2):265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikuchi S, et al. Rice Full-Length cDNA Consortium National Institute of Agrobiological Sciences Rice Full-Length cDNA Project Team Foundation of Advancement of International Science Genome Sequencing & Analysis Group RIKEN Collection, mapping, and annotation of over 28,000 cDNA clones from japonica rice. Science. 2003;301(5631):376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- 24.International Rice Genome Sequencing Project The map-based sequence of the rice genome. Nature. 2005;436(7052):793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 25.Luo A, et al. EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol. 2006;47(2):181–191. doi: 10.1093/pcp/pci233. [DOI] [PubMed] [Google Scholar]
- 26.Hu J, et al. Identification and characterization of NARROW AND ROLLED LEAF 1, a novel gene regulating leaf morphology and plant architecture in rice. Plant Mol Biol. 2010;73(3):283–292. doi: 10.1007/s11103-010-9614-7. [DOI] [PubMed] [Google Scholar]
- 27.Zou J, et al. Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.) Planta. 2005;222(4):604–612. doi: 10.1007/s00425-005-0007-0. [DOI] [PubMed] [Google Scholar]
- 28.Komatsu M, Maekawa M, Shimamoto K, Kyozuka J. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Dev Biol. 2001;231(2):364–373. doi: 10.1006/dbio.2000.9988. [DOI] [PubMed] [Google Scholar]
- 29.Zhou H, et al. Photoperiod- and thermo-sensitive genic male sterility in rice are caused by a point mutation in a novel noncoding RNA that produces a small RNA. Cell Res. 2012;22(4):649–660. doi: 10.1038/cr.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie W, et al. Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc Natl Acad Sci USA. 2010;107(23):10578–10583. doi: 10.1073/pnas.1005931107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myles S, et al. Association mapping: Critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21(8):2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian FK, et al. Genetic analysis and QTL mapping of mature seed culturability in indica rice. Rice Sci. 2013;20(5):313–319. [Google Scholar]
- 33.Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456(7218):60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kosambi DD. The estimation of map distances from recombination values. Ann Hum Genet. 1943;12(1):172–175. [Google Scholar]
- 35.McCouch SR, et al. Microsatellite marker development, mapping and applications in rice genetics and breeding. Plant Mol Biol. 1997;35(1-2):89–99. [PubMed] [Google Scholar]
- 36.Krzywinski M, et al. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19(9):1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.