Significance
Sec1/Munc18 (SM) proteins are essential for every vesicle fusion pathway, but their molecular mechanisms remain poorly understood. Our comparative studies of two functionally distinct SM proteins, Munc18c and Munc18-1, suggest that one conserved function of SM proteins is to recognize their cognate trans-SNARE complexes and accelerate fusion kinetics. The “closed” syntaxin binding mode of Munc18-1, however, is not conserved in Munc18c. Unexpectedly, we discovered that the architecture of the SNARE/SM complex differs across fusion pathways. Together, these findings reveal conserved as well as divergent functions of SM proteins in vesicle fusion.
Keywords: membrane fusion, vesicle transport, exocytosis
Abstract
Sec1/Munc18 (SM) family proteins are essential for every vesicle fusion pathway. The best-characterized SM protein is the synaptic factor Munc18-1, but it remains unclear whether its functions represent conserved mechanisms of SM proteins or specialized activities in neurotransmitter release. To address this question, we dissected Munc18c, a functionally distinct SM protein involved in nonsynaptic exocytic pathways. We discovered that Munc18c binds to the trans-SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) complex and strongly accelerates the fusion rate. Further analysis suggests that Munc18c recognizes both vesicle-rooted SNARE and target membrane-associated SNAREs, and promotes trans-SNARE zippering at the postdocking stage of the fusion reaction. The stimulation of fusion by Munc18c is specific to its cognate SNARE isoforms. Because Munc18-1 regulates fusion in a similar manner, we conclude that one conserved function of SM proteins is to bind their cognate trans-SNARE complexes and accelerate fusion kinetics. Munc18c also binds syntaxin-4 monomer but does not block target membrane-associated SNARE assembly, in agreement with our observation that six- to eightfold increases in Munc18c expression do not inhibit insulin-stimulated glucose uptake in adipocytes. Thus, the inhibitory “closed” syntaxin binding mode demonstrated for Munc18-1 is not conserved in Munc18c. Unexpectedly, we found that Munc18c recognizes the N-terminal region of the vesicle-rooted SNARE, whereas Munc18-1 requires the C-terminal sequences, suggesting that the architecture of the SNARE/SM complex likely differs across fusion pathways. Together, these comparative studies of two distinct SM proteins reveal conserved as well as divergent mechanisms of SM family proteins in intracellular vesicle fusion.
The fusion of intracellular vesicles with target membranes requires two classes of conserved proteins: SNAREs and SM (Sec1/Munc18) proteins (1, 2). SNAREs are membrane-associated proteins that contain characteristic stretches of 60–70 amino acids known as core domains or SNARE motifs. Fusion is initiated when the core domains of the vesicle-rooted SNARE (v-SNARE) and the target membrane-associated SNAREs (t-SNAREs) zipper into a four-helix trans-SNARE complex between two apposed bilayers (2–5). N- to C-terminal zippering of the trans-SNARE complex brings the two membranes into close apposition to fuse (6–8).
First isolated in genetic screens in yeast and nematodes (9, 10), SM proteins are hydrophilic factors of 60–70 kDa that regulate membrane fusion through binding to their cognate SNAREs (11–13). SM proteins exhibit a similar loss-of-function phenotype as that of SNAREs (i.e., abrogation of fusion) and are essential for every pathway of intracellular vesicle fusion (14–16). Mutations of SM proteins give rise to a number of human diseases, including epilepsy and inflammatory disorders, as well as arthrogryposis, renal dysfunction, and cholestasis (ARC) syndrome (17–21). Although the mechanism of SNAREs is well established, we are only beginning to understand how SM proteins regulate vesicle fusion.
The best-characterized SM protein is the synaptic factor Munc18-1 (also known as nSec1 or STXBP1), which is required for the fusion of neurotransmitter-filled synaptic vesicles with the plasma membrane (1, 22). Synaptic neurotransmitter release serves as the nervous system’s major form of cell-to-cell communication and requires three SNARE proteins: syntaxin-1, SNAP-25, and VAMP2/synaptobrevin (3, 23, 24). Munc18-1 has been shown to play dual roles in synaptic vesicle fusion. First, Munc18-1 positively regulates the SNARE-dependent fusion reaction by interacting with the trans-SNARE complex and accelerating the fusion kinetics (12, 25–33). Second, Munc18-1 binds to syntaxin-1 monomer and locks the latter into a “closed” configuration that prevents SNARE complex formation (34–36). This closed syntaxin binding mode can promote syntaxin trafficking and guide the SNAREs down a specific assembly route with the assistance of Munc13 (27, 37–39). In view of the highly specialized nature of neurotransmitter release, however, it remains to be determined whether these functions constitute conserved mechanisms of the SM family proteins or represent specialized activities of Munc18-1 at the synapse. To address this question, it is imperative to dissect another member of the SM protein family and compare its functions with those of Munc18-1.
In this study, we chose to characterize Munc18c (also known as Munc18-3), a ubiquitously expressed SM protein involved in nonsynaptic exocytic pathways (40, 41). Munc18c is not functionally interchangeable with the synaptic SM protein Munc18-1, indicating that they regulate distinct vesicle fusion pathways (16). Munc18c has been shown to regulate the exocytosis of the glucose transporter GLUT4 in body glucose homeostasis. Under basal conditions, GLUT4 is sequestered in intracellular vesicles in adipocytes and skeletal muscles. On insulin stimulation, GLUT4-containing vesicles fuse with the plasma membrane, delivering GLUT4 to the cell surface to facilitate glucose uptake. GLUT4 vesicle fusion requires syntaxin-4 and SNAP-23 as the t-SNAREs, VAMP2 as the primary v-SNARE, and Munc18c as the cognate SM protein (40, 42). Mutations in Munc18c interfere with GLUT4 vesicle fusion and disrupt insulin-stimulated glucose transport into the cell (41, 43, 44). Importantly, Munc13 and synaptotagmins appear to be absent in adipocytes and are not known to be involved in the GLUT4 trafficking pathway (45, 46), highlighting major functional differences between GLUT4 exocytosis and synaptic release. In addition to GLUT4 exocytosis, Munc18c regulates a range of other exocytic pathways, including neutrophil secretion, amylase release, platelet exocytosis, and the sustained phase of insulin secretion (47–51).
Although the physiological role of Munc18c in vesicle exocytosis is clear, its molecular mechanism remains to be established. Here, we sought to define the mechanisms underlying Munc18c function by reconstituting it into a defined fusion reaction containing GLUT4 exocytic SNAREs. We observed that Munc18c bound to the ternary trans-SNARE complex and strongly accelerated the fusion rate. Munc18c recognizes both the v- and t-SNAREs, and it potently promotes trans-SNARE zippering at the postdocking stage of the fusion reaction. The stimulatory activity of Munc18c was specific to the fusion reactions reconstituted with its cognate SNAREs. These data, in combination with previous findings of Munc18-1, suggest a conserved mechanism of SM proteins in intracellular vesicle fusion: to interact with their cognate trans-SNARE complex and accelerate the fusion kinetics. Like Munc18-1, Munc18c also binds to the syntaxin monomer. However, the binding of Munc18c to syntaxin did not block SNARE assembly or the fusion reaction, in agreement with our observation that six- to eightfold increases in Munc18c expression do not inhibit insulin-stimulated glucose uptake in adipocytes. These data indicate that Munc18c does not adopt the inhibitory closed syntaxin binding mode as shown for Munc18-1. Therefore, the closed syntaxin binding mode may not be a general feature of SM proteins. Unexpectedly, we found that the stimulation of fusion by Munc18c requires the N-terminal regions of the v-SNARE, whereas Munc18-1 recognizes the C-terminal motifs. These results suggest that although the conserved function of SM proteins involves binding to trans-SNAREs, the architecture of the SNARE/SM complexes likely differs across fusion pathways. Together, these findings establish the conserved as well as divergent functions of SM family proteins in intracellular vesicle fusion.
Results
Munc18c Binds Stoichiometrically to GLUT4 Exocytic SNARE Complex.
Although studies in intact cells have emphasized the physiological importance of Munc18c, the complexity of the cellular environment precludes further mechanistic insights. We addressed this problem by functionally reconstituting SNARE-dependent GLUT4 vesicle fusion in a defined system, in which SNAREs and regulatory factors can be added or altered individually in the absence of other potentially confounding factors.
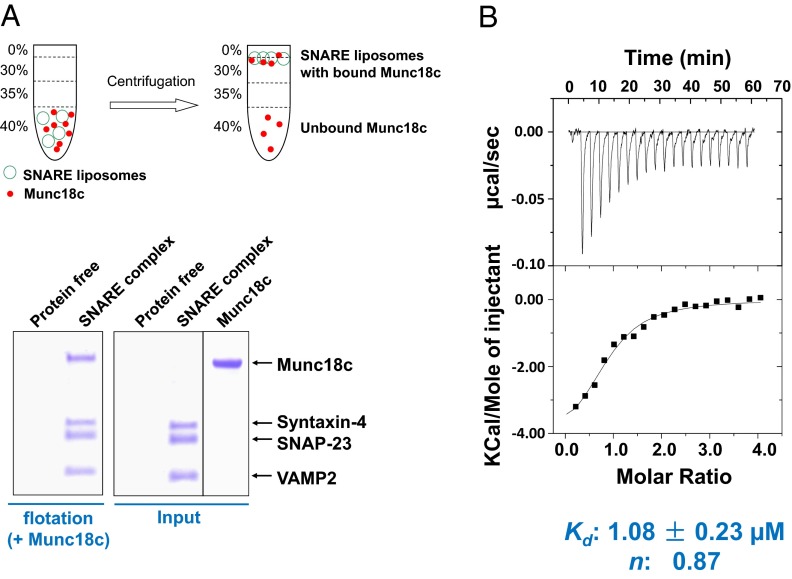
We expressed and purified recombinant Munc18c from Sf9 insect cells using baculovirus. In a liposome coflotation assay, Munc18c bound to proteoliposomes reconstituted with GLUT4 exocytic SNAREs: syntaxin-4, SNAP-23, and VAMP2 (Fig. 1A). Munc18c did not bind to protein-free liposomes (Fig. 1A), indicating that the Munc18c/SNARE interaction was specific. Next, we quantitatively analyzed the Munc18c/SNARE interaction using isothermal titration calorimetry (ITC). We found that Munc18c bound to the soluble form of the ternary SNARE complex with a Kd of ∼1 μM (Fig. 1B), similar to the association of Munc18-1 with soluble synaptic SNARE complexes (34, 52). ITC measurements showed that Munc18c bound to the SNARE complex at a ratio close to 1:1 (Fig. 1B), in agreement with the liposome coflotation results (Fig. 1A).
Fig. 1.
Munc18c binds stoichiometrically to the GLUT4 exocytic SNARE complex. (A) Coomassie blue-stained SDS/PAGE gel showing the binding of Munc18c to protein-free or SNARE liposomes. Liposomes containing the ternary GLUT4 SNARE complex were prepared by incubating VAMP2 CD with WT t-SNARE liposomes (containing syntaxin-4 and SNAP-23) overnight at 4 °C. (B) To assemble the ternary SNARE complex, the preassembled t-SNAREs, composed of syntaxin-4 (aa 1–273) and SNAP-23, were incubated overnight with VAMP2 CD (aa 1–95). The SNARE complex was injected into the sample cell of a VP-ITC instrument (Microcal) containing recombinant Munc18c. The Kd and stoichiometry of the interaction were calculated by fitting the data with a nonlinear least squares routine using Microcal Origin software.
Munc18c Accelerates the Kinetics of SNARE-Dependent Membrane Fusion in a Reconstituted Fusion system.
Next, we sought to determine how Munc18c regulates the SNARE-dependent fusion reaction. Membrane fusion is a highly dynamic process in which the zippering of the trans-SNARE complex pulls two membrane bilayers into close proximity to fuse (1). This dynamic fusion reaction can only be recapitulated using reconstituted fusion assays in which the v- and t-SNAREs are anchored in separate membrane bilayers.
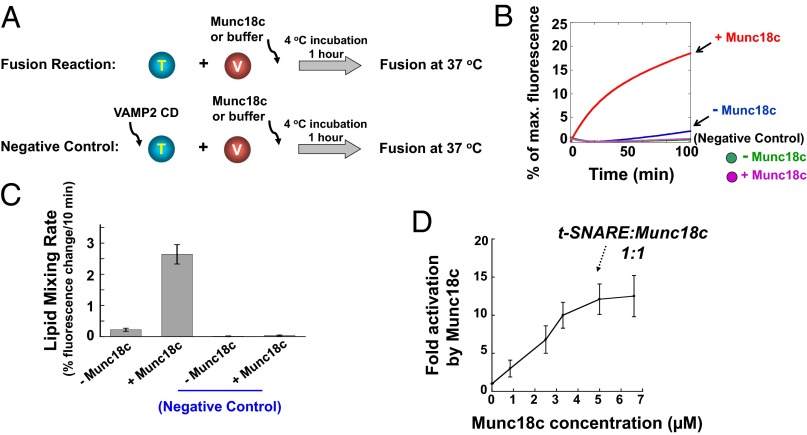
The fusion of v- and t-SNARE liposomes was first monitored by a FRET-based lipid-mixing assay (26). GLUT4 exocytic SNAREs alone drove a basal level of lipid mixing (Fig. 2 A and B). Munc18c strongly accelerated the lipid-mixing kinetics with an increase in the initial rate of ∼12-fold (Fig. 2 B and C). Munc18c robustly accelerated lipid mixing without preincubation, but its stimulatory activity was enhanced by preincubation at 4 °C (Fig. S1). We postulate that the preincubation step may facilitate the binding of Munc18c to a metastable intermediate of SNARE assembly. Lipid mixing was completely blocked by inclusion of the VAMP2 cytoplasmic domain (CD) (Fig. 2 B and C), a dominant negative inhibitor of SNARE complex assembly (4). Maximum stimulation of lipid mixing was reached using Munc18c at a concentration of 5 μM, similar to the concentration of t-SNAREs present on liposomes (Fig. 2D).
Fig. 2.
Munc18c strongly accelerates the kinetics of SNARE-dependent membrane fusion. (A) Illustrations of the liposome fusion procedures. The t-SNARE liposomes were reconstituted with syntaxin-4 and SNAP-23, whereas the v-SNARE liposomes contained VAMP2. The v- and t-SNARE liposomes were preincubated at 4 °C for 1 h before the temperature was elevated to 37 °C to start fusion. (B) Fusion of the reconstituted proteoliposomes in the absence or presence of 5 μM Munc18c. Each fusion reaction contained 5 μM t-SNAREs and 1.5 μM v-SNARE. The fusion reactions were measured by a FRET-based lipid-mixing assay. Twenty micromolar VAMP2 CD (aa 1–95) was added at the beginning of the preincubation as a negative control. max., maximum. (C) Initial lipid-mixing rates of the fusion reactions shown in B. Data are presented as the percentage of fluorescence change per 10 min. Error bars indicate SD. (D) Dose dependence of Munc18c activity in the SNARE-dependent fusion reaction. Munc18c was added to the reconstituted fusion reaction at the indicated concentrations. Fold increases in the initial lipid-mixing rates of the fusion reactions are shown. Error bars indicate SD.
It was observed that under certain experimental conditions, squid Munc18-1 (but not mammalian Munc18-1) could undergo denaturation and increase the emission intensity of lipid-conjugated fluorescent dyes independent of membrane fusion (53). The recombinant mammalian Munc18-1 and Munc18c proteins used in this study were highly soluble and exhibited no noticeable denaturation. Indeed, in the absence of functional SNAREs, Munc18c was unable to elicit fluorescence emission in the reconstituted lipid-mixing assay (Fig. 2B). Hence, Munc18c acts by facilitating the SNARE-mediated fusion pathway, rather than by causing fusion via an alternative mechanism. We further addressed the issue by examining the content mixing of the liposomes. The soluble dye sulforhodamine B was encapsulated in the VAMP2 liposomes in which its fluorescence was inhibited by self-quenching. Fusion of the v-SNARE liposomes with unlabeled t-SNARE liposomes led to the dilution and dequenching of sulforhodamine B fluorescence (Fig. S2A). Using this assay, we observed that GLUT4 exocytic SNAREs drove a basal level of content mixing that was markedly accelerated by Munc18c (Fig. S2 B and C). The content mixing was blocked by inclusion of the VAMP2 CD (Fig. S2 B and C). In dequenching controls, the sulforhodamine B dye was included in both v- and t-SNARE liposomes. If content leakage did not occur, no sulforhodamine B dequenching would be observed (Fig. S2A). We observed that the sulforhodamine B emission was not increased in the dequenching control reactions, indicating that content leakage did not occur appreciably in the liposome fusion reactions (Fig. S2B). Together, these results demonstrate that Munc18c promotes both lipid and content mixing of the SNARE-mediated fusion reaction.
Munc18c Promotes trans-SNARE Zippering at the Postdocking Stage of the Fusion Reaction.
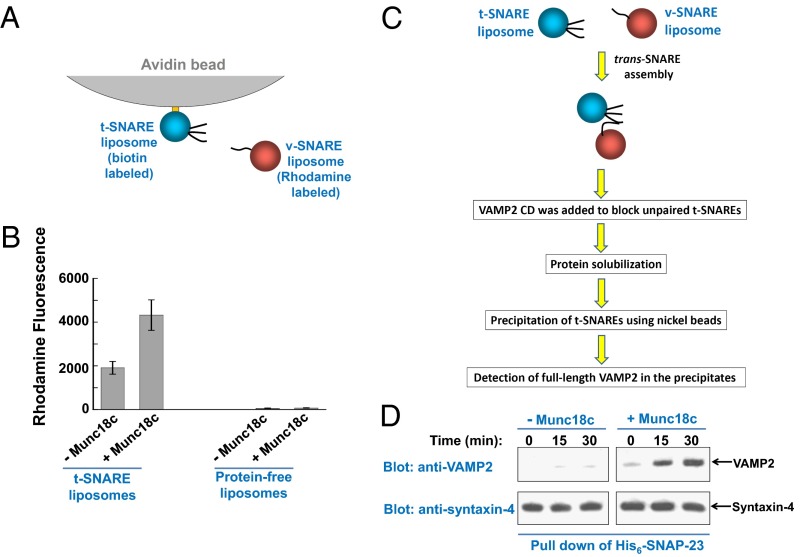
Next, we sought to dissect how Munc18c regulates SNARE-dependent membrane fusion. We began by testing how Munc18c influences the docking of liposome membranes. The t-SNARE liposomes were anchored to avidin beads through biotinylated phospholipids and were used to pull down v-SNARE liposomes (Fig. 3A). Pairing of v- and t-SNAREs allowed the v-SNARE liposomes to dock onto the bead-anchored t-SNARE liposomes (Fig. 3B). This SNARE-mediated liposome docking was mildly enhanced by Munc18c (Fig. 3B). We also examined liposome docking using cryo-EM. We found that the SNARE-dependent liposome docking was only slightly increased in the presence of Munc18c (Fig. S3), consistent with the results using the liposome pull-down assay (Fig. 3B).
Fig. 3.
Munc18c promotes trans-SNARE zippering at the postdocking stage of the fusion reaction. (A) Diagram of the liposome pull-down assay. (B) Measurements of the docking of t- and v-SNARE liposomes using the liposome pull-down assay. Biotin-labeled t-SNARE liposomes were anchored to avidin agarose beads and were used to pull down rhodamine-labeled v-SNARE liposomes. The binding reactions were performed at 4 °C for 1 h in the absence or presence of 5 μM Munc18c. Data are presented as rhodamine fluorescence intensity. In the negative control, t-SNARE liposomes were substituted with protein-free liposomes. Error bars indicate SD. (C) Diagram of the trans-SNARE formation assay. (D) Reconstituted t- and v-SNARE liposomes were incubated at 4 °C for indicated periods in the presence or absence of 5 μM Munc18c before a 10-fold excess amount of inhibitory VAMP2 CD was added to block unpaired t-SNAREs. The liposomes were subsequently solubilized, and the t-SNAREs were precipitated using nickel Sepharose beads. The presence of full-length VAMP2 in the precipitates was probed by Western blotting, which was used as an indicator for trans-SNARE assembly between liposomes.
We then determined how Munc18c regulates the zippering of trans-SNARE complex, a postdocking step of SNARE-dependent membrane fusion. Incubation of v- and t-SNARE liposomes at 4 °C led to progressive formation of trans-SNARE complexes between apposed lipid bilayers. VAMP2 CD was subsequently added to block unpaired t-SNAREs, whereas assembled trans-SNARE complexes were resistant to VAMP2 CD. After solubilization of the membrane-bound SNARE proteins, the t-SNAREs were precipitated. We then selectively probed full-length VAMP2 in the precipitates, which was indicative of trans-SNARE complex assembly (Fig. 3C). Using this trans-SNARE formation assay, we observed that Munc18c strongly promoted the assembly of the trans-SNARE complex (Fig. 3D). We estimated that in the presence of Munc18c, the amount of VAMP2 CD-resistant trans-SNAREs increased approximately eightfold (Fig. 3D). No lipid mixing occurred during these trans-SNARE assembly reactions (Fig. S4). Altogether, these data suggest that Munc18c promotes fusion primarily by facilitating trans-SNARE zippering at the postdocking stage of the membrane fusion reaction.
Stimulatory Function of Munc18c in Membrane Fusion Is Specific to Its Cognate SNARE Isoforms.
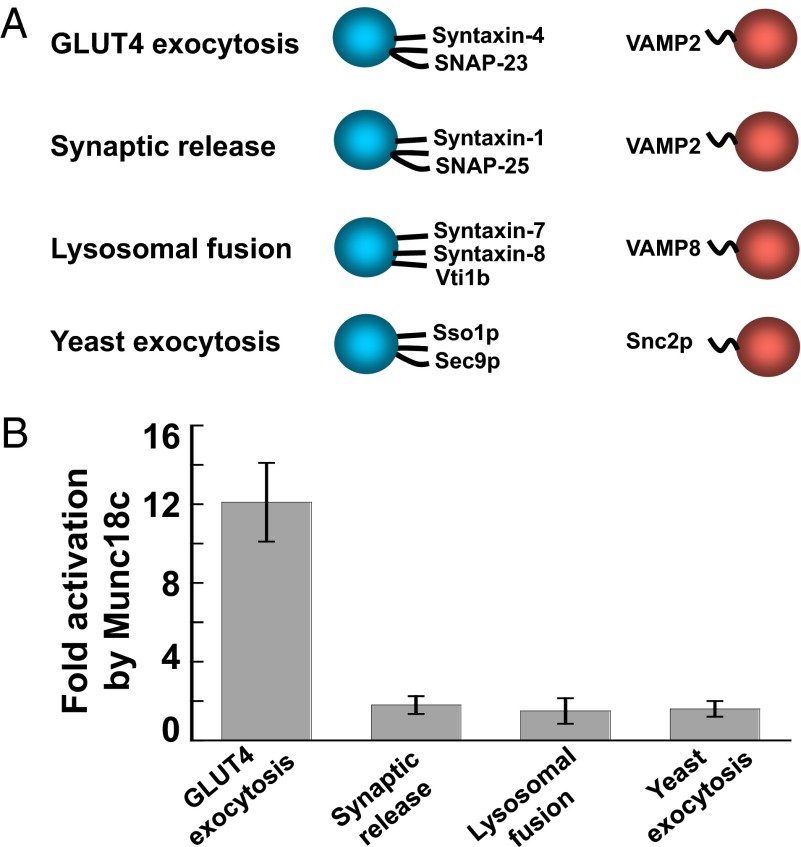
In the cell, SM proteins only regulate the fusion reactions mediated by their cognate SNARE isoforms (14, 16), which likely contributes to the overall precision of intracellular vesicle trafficking. Next, we explored the intrinsic specificity of Munc18c function in the reconstituted fusion reaction. Proteoliposomes were reconstituted using noncognate SNAREs involved in synaptic release (syntaxin-1, SNAP-25, and VAMP2), lysosomal/late-endosomal fusion (syntaxin-7, syntaxin-8, Vti1b, and VAMP8), or yeast exocytosis (Sso1p, Sec9p, and Snc2p) (Fig. 4A). We found that the lipid-mixing reactions mediated by these noncognate SNAREs were not activated by Munc18c (Fig. 4B). This compartmental specificity of Munc18c function is in agreement with the pathway-specific activities of SM proteins in vesicle fusion, and it supports that our reconstituted system has recapitulated the physiological function of Munc18c.
Fig. 4.
Munc18c selectively activates its cognate SNAREs. (A) Illustrations of the liposome fusion pairs. (B) Activation of the indicated SNARE-dependent fusion reactions by Munc18c. Each fusion reaction contained 5 μM t-SNAREs and 1.5 μM v-SNARE. The fusion reactions were measured by a FRET-based lipid-mixing assay. Fold increases in the initial lipid-mixing rates of the fusion reactions are shown. Error bars indicate SD.
Munc18c Can Activate Multiple SNARE Pairs.
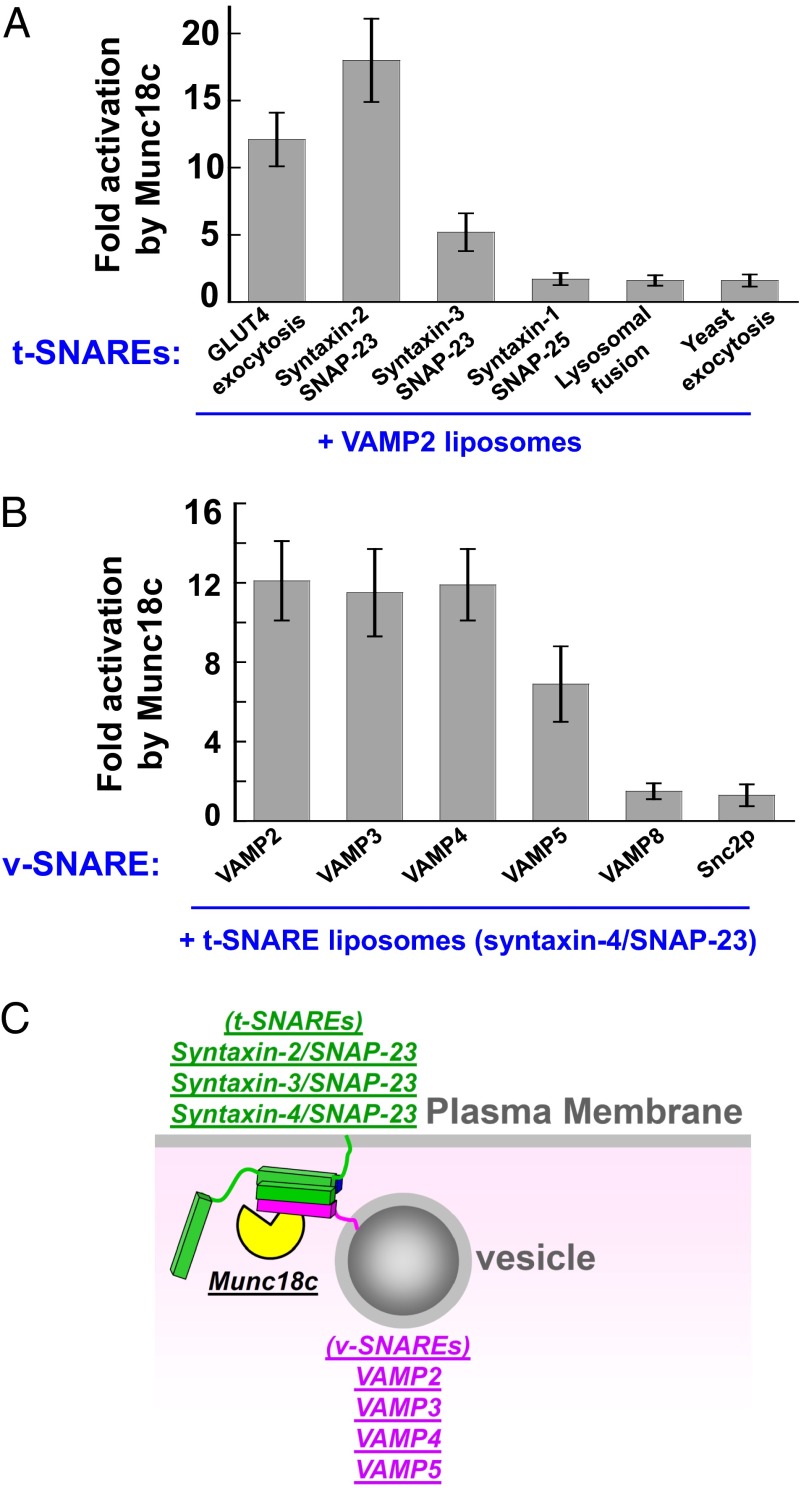
The reconstitution of Munc18c into the defined system enabled us to dissect its regulatory mechanism further in SNARE-dependent membrane fusion. In contrast to the highly specialized SM protein Munc18-1, Munc18c is ubiquitously expressed and is thought to participate in a range of exocytic fusion pathways (54). Because mammalian exocytosis involves multiple SNARE pairs (5), we reasoned that Munc18c might be capable of regulating SNARE isoforms beyond those of GLUT4 exocytosis. To test this, we first reconstituted proteoliposomes bearing each of the four exocytic t-SNARE complexes found on the plasma membrane: syntaxin-1/SNAP-25, syntaxin-2/SNAP-23, syntaxin-3/SNAP-23, and syntaxin-4/SNAP-23 (5). Each of these t-SNARE liposomes was directed to fuse with VAMP2 liposomes (Fig. 5A). We observed that in addition to its known cognate t-SNAREs (syntaxin-4/SNAP-23), Munc18c strongly stimulated the lipid-mixing reaction driven by syntaxin-2/SNAP-23 (Fig. 5A). Munc18c also activated the fusion reaction reconstituted with the t-SNARE syntaxin-3/SNAP-23, albeit the stimulation was weaker than that of syntaxin-2/SNAP-23 and syntaxin-4/SNAP-23 (Fig. 5A). Interestingly, the synaptic t-SNAREs (syntaxin-1/SNAP-25) could not support the stimulatory activity of Munc18c (Fig. 5A). Thus, three exocytic t-SNARE complexes, syntaxin-2/SNAP-23, syntaxin-3/SNAP-23, and syntaxin-4/SNAP-23, can potentially serve as the cognate t-SNAREs of Munc18c.
Fig. 5.
Munc18c regulates multiple v- and t-SNAREs. (A) Activation of the indicated SNARE-dependent fusion reactions by Munc18c. The v-SNARE liposomes of the fusion reactions contained VAMP2, whereas the t-SNARE liposomes were reconstituted with the indicated t-SNAREs. Each fusion reaction contained 5 μM t-SNAREs and 1.5 μM v-SNARE. The fusion reactions were measured by a FRET-based lipid-mixing assay. Fold increases in the initial lipid-mixing rates of the fusion reactions are shown. Error bars indicate SD. (B) Activation of the indicated SNARE-dependent fusion reactions by Munc18c. The t-SNARE liposomes of the fusion reactions contained syntaxin-4 and SNAP-23, whereas the v-SNARE liposomes were reconstituted with the indicated v-SNAREs. Fold increases in the initial lipid-mixing rates of the fusion reactions are shown. Error bars indicate SD. (C) Model showing the potential cognate v- and t-SNAREs of Munc18c in vesicle fusion. Munc18c (SM protein) is colored yellow, t-SNAREs are colored green, and v-SNAREs are colored pink.
We then examined the selection of v-SNAREs by Munc18c. The t-SNARE liposomes containing syntaxin-4/SNAP-23 were directed to fuse with liposomes bearing v-SNAREs found at the late secretory pathway, including VAMP2, VAMP3/cellubrevin, VAMP4, VAMP5, VAMP8/endobrevin, and the yeast exocytic v-SNARE Snc2p. Interestingly, we found that multiple v-SNARE isoforms, including VAMP2, VAMP3, VAMP4, and VAMP5, were able to support the stimulatory activity of Munc18c (Fig. 5B). By contrast, Munc18c was unable to stimulate lipid mixing in reactions reconstituted with VAMP8 or Snc2p (Fig. 5B). Together, these data reveal the intrinsic cognate v- and t-SNAREs of Munc18c (Fig. 5C), most of which were not previously known to be regulated by Munc18c. Combinations of these v- and t-SNAREs form 12 possible cognate trans-SNARE pairs that can potentially support Munc18c function in membrane fusion.
Munc18c Does Not Possess the Inhibitory Closed Syntaxin Binding Mode.
In addition to its stimulatory activity, the synaptic SM protein Munc18-1 negatively regulates fusion by binding to the syntaxin-1 monomer (31, 55, 56). In vitro evidence suggests that Munc18-1 binding locks syntaxin-1 into a closed configuration incompatible with SNARE complex assembly (34, 35, 56). Previous solution binding assays reached contradictory conclusions regarding whether the closed syntaxin binding mode is conserved in other SM proteins, such as Munc18c (41, 57). It has been proposed that the Munc18c/syntaxin-4 dimer precludes the assembly of the t-SNARE complex (41). However, solution structural data suggest that Munc18c-bound syntaxin-4 does not adopt a closed conformation (58). Here, we sought to determine whether Munc18c negatively regulates SNARE-mediated fusion using full-length proteins in a membrane environment.
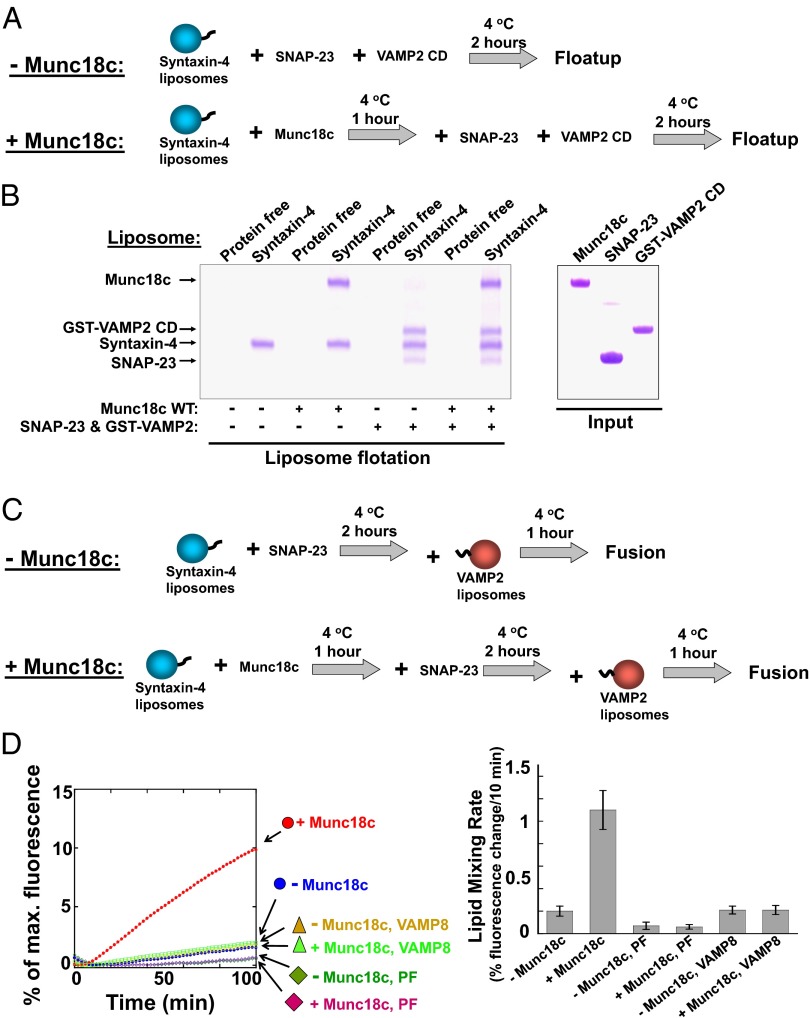
In our standard reconstituted fusion reactions, the t-SNAREs were preassembled to reveal the downstream trans-SNARE binding function of Munc18c (Fig. 2A). To assess the role of Munc18c in the upstream step of t-SNARE assembly, we reconstituted syntaxin-4 monomer into proteoliposomes, whereas other SNARE subunits were subsequently added as soluble proteins (Fig. 6A). In the liposome coflotation assay, we observed that membrane-anchored syntaxin-4 efficiently assembled with SNAP-23 and VAMP2 CD into the ternary SNARE complex (Fig. 6B), suggesting that syntaxin-4 does not normally adopt an autoinhibitory closed configuration. Next, Munc18c was added to syntaxin-4 liposomes to form the syntaxin-4/Munc18c dimer (Fig. 6B). SNAP-23 and VAMP2 CD were then introduced to examine the assembly of the ternary SNARE complex (Fig. 6A). We found that Munc18c binding did not prevent the pairing of syntaxin-4 with SNAP-23 to form the binary t-SNARE complex (Fig. S5) or with SNAP-23 and VAMP2 CD to form the ternary SNARE complex (Fig. 6B and Fig. S6). Therefore, the Munc18c/syntaxin-4 dimer does not inhibit SNARE complex assembly.
Fig. 6.
Munc18c does not harbor an inhibitory closed syntaxin binding mode. (A) Diagram illustrating the experimental procedures for liposome coflotation assays. (B) Munc18c did not inhibit SNARE assembly in the liposome coflotation assay. Syntaxin-4 liposomes were incubated with or without Munc18c at 4 °C for 1 h. Subsequently, SNAP-23 and VAMP2 CD were added. After 2 h of incubation at 4 °C, the samples were floated up on a Nycodenz gradient. To visualize VAMP2 CD better by Coomassie blue staining, a GST tag was included at its N terminus. (C) Diagram illustrating the experimental procedures for the reconstituted fusion reactions. (D, Left) Munc18c binding to syntaxin-4 did not inhibit the SNARE-dependent fusion reaction. Syntaxin-4 liposomes were incubated with or without 5 μM Munc18c at 4 °C for 1 h before 5 μM SNAP-23 was added. After 2 h at 4 °C, VAMP2 liposomes were introduced. After another hour of incubation at 4 °C, the temperature was raised to 37 °C to initiate fusion. In the negative control experiments, t-SNARE liposomes were substituted with protein-free (PF) liposomes. Each fusion reaction contained 5 μM t-SNAREs and 1.5 μM v-SNARE. The fusion reactions were measured by a FRET-based lipid-mixing assay. (Right) Fold increases in the initial lipid-mixing rates of the reconstituted fusion reactions (Left) are shown. Error bars indicate SD.
Next, we examined the functional role of the Munc18c/syntaxin-4 interaction in the dynamic membrane fusion reaction. In agreement with the liposome binding results, the interaction of Munc18c with syntaxin-4 did not inhibit the reconstituted SNARE-mediated lipid-mixing reaction (Fig. 6 C and D). Instead, Munc18c strongly accelerated the lipid-mixing rate (Fig. 6D and Fig. S7). The stimulation of lipid mixing by Munc18c was abolished when VAMP2 was substituted with VAMP8, a noncognate v-SNARE (Fig. 6D). We also tested a different fusion condition in which v-SNARE liposomes were not preincubated with Munc18c or t-SNARE liposomes before fusion. Again, Munc18c did not inhibit the lipid-mixing reaction (Fig. S8). Hence, the binding of Munc18c to the syntaxin-4 monomer does not inhibit SNARE assembly or fusion kinetics, suggesting that Munc18c-bound syntaxin-4 adopts an “open” conformation fully competent for SNARE complex assembly.
It was previously reported that overexpression of Munc18c in adipocytes inhibited insulin-stimulated glucose transport (59, 60), which implies that Munc18c has a negative regulatory role in vesicle transport. However, those Munc18c overexpression studies did not directly quantify Munc18c levels in control and Munc18c-overexpressing cells. We therefore sought to determine quantitatively the effects of Munc18c overexpression on glucose uptake in 3T3-L1 adipocytes. Importantly, a 100% transduction efficiency was achieved in adipocytes using a lentiviral expression system, allowing us to assess the functional consequences of Munc18c overexpression directly. Immunoblotting using anti-Munc18c antibodies indicates that Munc18c expression was increased by six- to eightfold over endogenous levels in Munc18c-overexpressing cells (Fig. S9A). Munc18c was found to colocalize with syntaxin-4 at the plasma membrane in these cells (Fig. S9B). Insulin strongly promoted the uptake of glucose by control cells, and this insulin-stimulated glucose transport was unaffected in cells overexpressing Munc18c (Fig. S9C). Based on these findings, we found no evidence for an inhibitory role of Munc18c in adipocytes, in agreement with the results of our reconstitution studies (Fig. 6). Collectively, these data demonstrate that the inhibitory closed syntaxin binding mode is not conserved in Munc18c.
Munc18c and Munc18-1 Recognize Different Motifs of the v-SNARE.
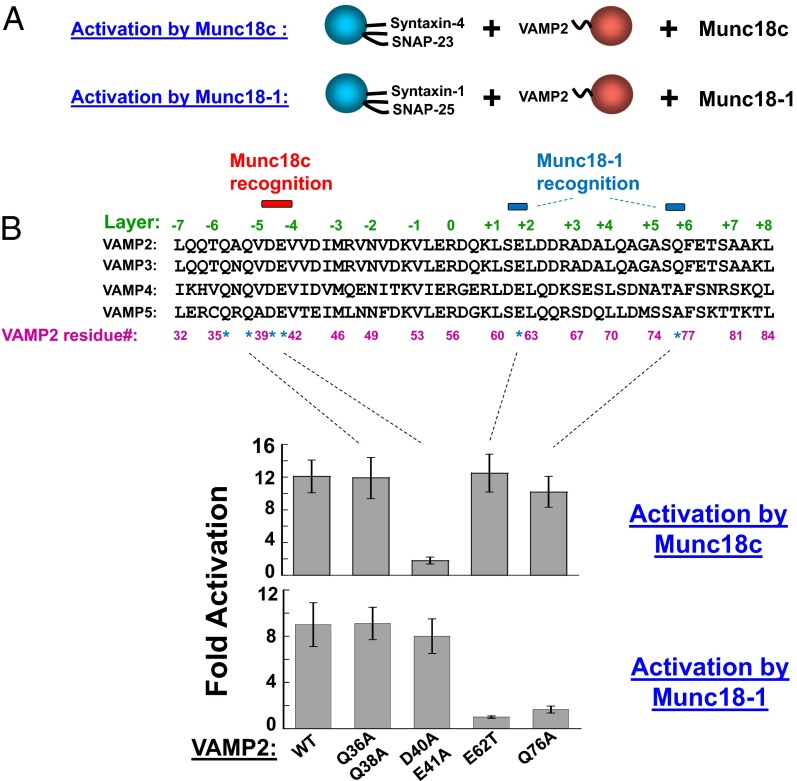
Although Munc18c and Munc18-1 recognize different t-SNARE isoforms, both use VAMP2 as the cognate v-SNARE. We previously showed that stimulation of fusion by Munc18-1 requires two C-terminal regions in the core domain of VAMP2 (12). Further analysis showed that mutation of either E62 or Q76 of VAMP2 abolished the stimulatory activity of Munc18-1 in fusion (Fig. 7 A and B). Unexpectedly, mutations of these same residues had little effect on the stimulatory function of Munc18c (Fig. 7B), indicating that Munc18c does not rely on these C-terminal motifs to promote SNARE assembly.
Fig. 7.
Munc18-1 and Munc18c recognize different motifs on the v-SNARE. (A) Illustrations of the reconstituted fusion reactions. (B, Upper) Sequence alignment of the core domains of mouse VAMP2, rat VAMP3, mouse VAMP4, and mouse VAMP5. Residues tested here are indicated with asterisks. Residues required for Munc18c or Munc18-1 activation are marked by colored bars. (Lower) Activation of the indicated SNARE-dependent fusion reactions by Munc18c or Munc18-1. Each fusion reaction contained 5 μM t-SNAREs and 1.5 μM v-SNARE. The fusion reactions were measured by a FRET-based lipid-mixing assay. Fold increases in the initial lipid-mixing rates of the fusion reactions are shown. Error bars indicate SD.
To determine how Munc18c recognizes the v-SNARE, we tested two N-terminal motifs of VAMP2, D40/E41 and Q36/Q38, based on multiple criteria. These VAMP2 motifs should be (i) distributed between the helical bundle-forming layer residues such that they are exposed on the surface of SNAREs to interact with regulatory proteins (Fig. 7B) and (ii) conserved in VAMP2, VAMP3, VAMP4, and VAMP5, which are v-SNAREs that can be activated by Munc18c (Fig. 5). We found that mutations of D40/E41 abolished the stimulatory activity of Munc18c (Fig. 7B). By contrast, the promotion of fusion by Munc18c was not reduced by mutating Q36/Q38 in VAMP2 (Fig. 7B). Intriguingly, mutations within these N-terminal motifs (Q36/Q38 or D40/E41) did not affect the stimulatory function of the synaptic SM protein Munc18-1 (Fig. 7B). None of the VAMP2 mutations affected the basal SNARE-mediated lipid-mixing reactions (Fig. S10). Although it remains possible that these VAMP2 mutations interfere with certain aspects of SNARE function, our data suggest that they do not significantly reduce the capacity of VAMP2 to assemble with the t-SNAREs.
Together, these data suggest that Munc18c and Munc18-1 recognize different motifs on the same v-SNARE protein when activating their respective fusion reactions. Munc18-1 preferentially recognizes the C-terminal regions of the VAMP2 core domain, whereas Munc18c requires the N-terminal motifs. Thus, the architecture of SNARE/SM complexes likely differs across vesicle fusion pathways.
Discussion
Molecular Mechanism of Munc18c in Vesicle Fusion.
It has long been known that the exocytic SM protein Munc18c positively regulates vesicle fusion, but its exact molecular mechanism has been unclear. In this study, we characterized Munc18c in a defined fusion system reconstituted with GLUT4 exocytic SNAREs. We discovered that Munc18c binds to the trans-SNARE complex and strongly accelerates the fusion kinetics. We identified the cognate SNARE targets of Munc18c in membrane fusion. In addition to the known GLUT4 exocytic SNAREs, Munc18c activates multiple v- and t-SNAREs not previously known to be its molecular targets. Although SNARE pairing is also influenced by other cellular factors, such as Rabs (2), our findings suggest that the trans-SNARE complexes formed by these v- and t-SNAREs can potentially serve as the cognate targets of Munc18c in vesicle fusion. Together, these findings provide a molecular explanation for the positive role of Munc18c observed in vesicle fusion pathways, such as insulin-controlled GLUT4 exocytosis (41, 43, 61).
By definition, all reconstituted systems are artificial such that the physiological relevance of reconstitution studies needs to be verified. A strong connection of this study to physiology, however, is established by the stringent compartmental specificity Munc18c exhibited in the reconstituted fusion assays. Munc18c selectively activated the fusion reactions reconstituted with its cognate SNARE isoforms, in agreement with the pathway-specific activities of SM proteins observed in vivo (16).
Insights into the Conserved Functions of SM Proteins in Intracellular Vesicle Fusion.
SM family proteins are known to regulate vesicle fusion by interacting with SNAREs (14, 16). The SM/SNARE binding modes, however, exhibit significant degrees of heterogeneity across pathways or species (62–67), which may argue against a universal mechanism for SM protein function in vesicle fusion. On the other hand, each vesicle transport step in eukaryotes requires the activity of an SM protein, and different SM proteins display similar structures and loss-of-function phenotypes (1). Thus, despite the seeming heterogeneity of SM/SNARE binding modes, the core mechanism of SM proteins is likely conserved. Based on our comparative studies of two functionally distinct SM proteins, Munc18c and Munc18-1, we propose that one conserved target of SM proteins in vesicle fusion is the trans-SNARE complex. By interacting with their cognate trans-SNARE complexes, SM proteins accelerate the kinetics of the membrane fusion reaction. This model is supported by multiple lines of evidence.
First, previous studies demonstrated that the synaptic SM protein Munc18-1 stimulates SNARE-dependent vesicle fusion by binding to the trans-SNARE complex (12, 26, 27, 30, 31). However, it was unclear whether this stimulatory function represents a conserved mechanism of SM proteins. Now, with the characterization of a second and nonredundant SM protein (Munc18c), this appears to be the case.
Second, the specificity of SM proteins is defined by their combinatorial partnering with both the v- and t-SNAREs. Substitutions of either the v- or t-SNAREs with noncognate isoforms result in loss of the stimulatory activities of Munc18c and Munc18-1, further supporting the notion that they act on the SNARE complex.
Third, the stimulatory functions of SM proteins are abolished by mutations on either the v- or t-SNAREs. The v-SNARE mutations are most informative in this regard because they do not affect the interaction of SM proteins with syntaxin monomer, a widely studied binding partner of SM proteins (22).
Finally, binding to the SNARE complex has also been demonstrated for other SM proteins, including the yeast exocytic SM protein Sec1p, the endocytic SM protein Vps45, the lysosomal/vacuolar SM protein Vps33, and the endoplasmic reticulum/Golgi apparatus SM protein Sly1 (62–66, 68, 69). Although the detailed mechanisms of these SM proteins remain to be defined, they all appear to associate with the SNARE complex formed by v- and t-SNAREs. Thus, the SNARE complex is likely the conserved target of SM family proteins.
How do SM proteins promote fusion kinetics? Because SM proteins recognize both the v- and t-SNAREs, it is conceivable that they can facilitate the zippering of the trans-SNARE complex. Indeed, our data demonstrate that Munc18c promotes trans-SNARE zippering at the postdocking stage of the fusion reaction. SM proteins may also orchestrate multiple trans-SNARE complexes into a ring-like structure at the fusion site to drive fusion cooperatively (22). It should be noted that the trans-SNARE complex is a highly dynamic structure markedly distinct from the static, postfusion cis-SNARE configuration (70). As previously proposed (71), the fully zippered cis-SNARE complex is unlikely the biological target of SM proteins in driving membrane fusion. Commonly used solution binding assays (e.g., ITC measurements) only examine the binding of SM proteins to the cis-SNARE complex, and thus cannot reflect the dynamic SM/SNARE interactions en route to fusion. The binding of SM proteins to the trans-SNARE involves multiple motifs on each of the distinct subunits in the context of the SNARE complex (12, 25, 26, 30). Therefore, measurements of SM binding to a single SNARE subunit in isolation may not be representative of the interaction in the content of the trans-SNARE complex, which likely underlies the observed heterogeneity in the SM/SNARE binding modes.
In addition to its trans-SNARE binding stimulatory function described in this study, SM proteins may possess other conserved functions in membrane fusion. SM proteins may interact with other SNARE assemblies to regulate fusion kinetics in a positive manner. It is also possible that they act in concert with additional regulators (e.g., tethering factors) to mediate and regulate vesicle fusion.
Divergent Functions of SM Family Proteins.
The structures of SNARE complexes are conserved across vesicle fusion pathways (72–74). Similarly, all SM proteins adopt a compact, arch-like configuration (34, 35, 58, 75, 76). However, the architecture of the SNARE/SM complex appears to differ significantly across vesicle fusion pathways. We observed that Munc18c and Munc18-1 recognize distinct motifs of VAMP2 in the context of the trans-SNARE complex. Munc18c preferentially binds to the N-terminal region of the VAMP2 core domain, whereas Munc18-1 requires the C-terminal sequences. We suggest that this divergent binding allows the SM/SNARE complex to adapt to the specific requirements of a fusion pathway. For instance, the synaptic fusion regulator complexin binds to the N terminus of VAMP2 (77, 78), a region not associated with Munc18-1. This raises the intriguing possibility that Munc18-1 and complexin may form a supracomplex in regulating synaptic release.
In addition to the conserved stimulatory function, the synaptic SM protein Munc18-1 (as well as its counterparts in lower organisms) interacts with the syntaxin monomer to lock the latter into a closed state incompatible with SNARE complex formation (34, 35, 56, 79). This inhibitory binding mode plays critical roles in regulating synaptic neurotransmitter release (27, 37–39, 56). Our data demonstrate that unlike Munc18-1, Munc18c binding to syntaxin does not block SNARE complex assembly or the fusion kinetics. When bound to Munc18c, syntaxin-4 adopts an open conformation fully competent for assembly with other SNARE subunits, in agreement with Munc18c structural and binding studies performed in solution (57, 58). Therefore, the closed syntaxin binding mode is not conserved even among exocytic SM proteins. This conclusion is further supported by previous studies of the yeast exocytic SM protein Sec1p, which does not bind appreciably to its cognate syntaxin Sso1p (14).
It has been shown that Munc18c inhibited insulin-regulated glucose uptake when overexpressed in adipocytes (59, 60). However, our quantitative analysis indicates that Munc18c does not play an inhibitory role in adipocytes when its expression level is increased six- to eightfold. Although the reason for this discrepancy is unclear, our Munc18c overexpression findings are in agreement with the lack of inhibitory activity of Munc18c in reconstituted fusion assays. Nevertheless, it remains possible that Munc18c acts in concert with other cellular factors to arrest the fusion reaction reversibly, allowing vesicle exocytosis to be coupled to intracellular signaling (41).
Our findings regarding Munc18c/syntaxin-4 association are in line with a mechanistic model proposed for synaptic neurotransmitter release (37, 38). The closed syntaxin binding mode of Munc18-1 in synaptic release can direct the SNAREs along a specific assembly pathway, beginning with the binary syntaxin/Munc18 complex. The Munc13 protein then relieves the inhibitory Munc18/syntaxin binding, allowing SNARE complex assembly to proceed efficiently (37, 38). In GLUT4 vesicle fusion, the Munc18c/syntaxin heterodimer may still serve as the initiation point for SNARE complex assembly. However, because Munc18c binding does not inhibit the pairing of syntaxin-4 with other SNARE subunits, Munc13 would not be needed to modulate the Munc18c/syntaxin-4 interaction in SNARE complex assembly. Indeed, Munc13 proteins appear to be absent in adipocytes (45), and they are not known to be involved in GLUT4 exocytosis (78–80). Hence, a requirement for Munc13 likely correlates with the ability of the SM protein to adopt the inhibitory closed syntaxin binding mode in a specific fusion pathway. Similarly, synaptotagmins are not implicated in GLUT4 trafficking (45), in contrast to their well-established roles in synaptic neurotransmitter release (80). These observations demonstrate major functional differences across exocytic vesicle fusion pathways.
SNAREs and SM proteins represent two conserved families of molecules required for every vesicle fusion pathway in the cell, from yeast to humans. In regulated exocytosis, SNAREs and SM proteins are superimposed by pathway-specific regulators (e.g., synaptotagmins in synaptic release and insulin secretion) or coupled to intracellular signaling cascades (e.g., insulin signaling in GLUT4 exocytosis). The defined systems reconstituted with SNAREs and SM proteins described in this study will serve as a foundation upon which regulatory or signaling molecules can be added, individually and in combination, to establish how they act in concert to achieve an integrated response.
Materials and Methods
Reconstitution of Proteoliposomes.
All lipids were obtained from Avanti Polar Lipids, Inc. For t-SNARE reconstitution, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine (POPS), and cholesterol were mixed in a molar ratio of 60:20:10:10. For v-SNARE reconstitution, POPC, POPE, POPS, cholesterol, N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE), and N-(Lissamine rhodamine B sulfonyl)-DPPE (rhodamine-DPPE) were mixed at a molar ratio of 60:17:10:10:1.5:1.5. SNARE proteoliposomes were prepared by detergent dilution and isolated on a Nycodenz (Axis-Shield) density gradient (81, 82). Detergent was removed by overnight dialysis using Novagen dialysis tubes against the reconstitution buffer [25 mM Hepes (pH 7.4), 100 mM KCl, 10% (vol/vol) glycerol, and 1 mM DTT]. To prepare sulforhodamine-loaded liposomes, SNARE liposomes were reconstituted in the presence of 50 mM sulforhodamine B (Sigma). Free sulforhodamine B was removed by overnight dialysis, followed by liposome flotation on a Nycodenz gradient. The protein/lipid ratio was at 1:200 for v-SNAREs and at 1:500 for t-SNARE liposomes. To ensure the consistency in SNARE liposome preparations, we routinely monitored the sizes and morphologies of reconstituted liposomes using dynamic light scattering and EM.
FRET-Based Lipid- and Content-Mixing Assays.
A standard lipid-mixing reaction contained 45 μL of unlabeled t-SNARE liposomes and 5 μL of v-SNARE liposomes labeled with NBD and rhodamine, and it was conducted in a 96-well Nunc plate at 37 °C. The fusion reactions were carried out in the reaction buffer [25 mM Hepes (pH 7.4), 50 mM KCl, and 1 mM DTT]. Before fusion, an NBD emission from the v-SNARE liposomes was quenched by neighboring rhodamine molecules through FRET. After fusion, the NBD dyes were diluted such that their emission was increased. The increase in NBD fluorescence at 538 nm (excitation = 460 nm) was measured every 2 min in a BioTek Synergy HT microplate reader. At the end of the reaction, 10 μL of 2.5% (wt/vol) dodecyl-maltoside was added to the liposomes. Fusion data were presented as the percentage of maximum fluorescence change. The maximum fusion rate within the first 10 min of the reaction was used to represent the initial rate of a fusion reaction. In fusion reactions with decreases in initial fluorescence (due to temperature change), the phase of fluorescence decrease was omitted from the calculation.
In content-mixing assays, unlabeled t-SNARE liposomes were directed to fuse with sulforhodamine B-loaded v-SNARE liposomes in which the sulforhodamine B fluorescence was inhibited by self-quenching. The fusion of the liposomes led to the mixing of their contents and the dequenching of sulforhodamine B fluorescence. The increase of sulforhodamine B fluorescence at 585 nm (excitation = 565 nm) was measured every 2 min. To assess Munc18c function, recombinant Munc18c protein was incubated with v- and t-SNARE liposomes at 4 °C for 1 h. The samples were subsequently heated to 37 °C to initiate the fusion reactions. Full accounting of statistical significance was included for each figure based on at least three independent experiments. Additional experimental procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Gustav Lienhard (Dartmouth University) for helpful discussions on GLUT4 transport and Yan Ouyang (University of Colorado Boulder) for technical assistance. We are grateful to Dr. Andy Hoenger and Cindi Schwartz (Boulder Laboratory for 3D EM) for assistance on cryo-EM and Dr. Brooke Hirsch and Shaun Bevers (University of Colorado School of Medicine) for assistance on ITC measurements. We thank Drs. Wanjin Hong (Institute of Molecular and Cell Biology, Singapore), Paul Roche [National Institutes of Health (NIH)], Thomas Weimbs (University of California, Santa Barbara), and Jeffrey Pessin (Albert Einstein College of Medicine) for sending us plasmids. This work was supported by an NIH Pathway to Independence Award (DK080080) and NIH Grant DK095367 (both to J.S.). D.E.J. and J.L.M. are supported by a National Health and Medical Research Council (NHMRC) program grant. D.E.J. is an NHMRC Senior Principal Research Fellow, and J.L.M. is an Australian Research Council Australian Laureate Fellow. J.S. is a Pew Scholar in the Biomedical Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 14116.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311232110/-/DCSupplemental.
References
- 1.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323(5913):474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15(7):658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Söllner T, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362(6418):318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 4.Weber T, et al. SNAREpins: Minimal machinery for membrane fusion. Cell. 1998;92(6):759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 5.Jahn R, Scheller RH. SNAREs—Engines for membrane fusion. Nat Rev Mol Cell Biol. 2006;7(9):631–643. doi: 10.1038/nrm2002. [DOI] [PubMed] [Google Scholar]
- 6.Melia TJ, et al. Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J Cell Biol. 2002;158(5):929–940. doi: 10.1083/jcb.200112081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pobbati AV, Stein A, Fasshauer D. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 2006;313(5787):673–676. doi: 10.1126/science.1129486. [DOI] [PubMed] [Google Scholar]
- 8.Gao Y, et al. Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science. 2012;337(6100):1340–1343. doi: 10.1126/science.1224492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Novick P, Schekman R. Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1979;76(4):1858–1862. doi: 10.1073/pnas.76.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hata Y, Slaughter CA, Südhof TC. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature. 1993;366(6453):347–351. doi: 10.1038/366347a0. [DOI] [PubMed] [Google Scholar]
- 12.Shen J, Tareste DC, Paumet F, Rothman JE, Melia TJ. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell. 2007;128(1):183–195. doi: 10.1016/j.cell.2006.12.016. [DOI] [PubMed] [Google Scholar]
- 13.Dulubova I, et al. Munc18-1 binds directly to the neuronal SNARE complex. Proc Natl Acad Sci USA. 2007;104(8):2697–2702. doi: 10.1073/pnas.0611318104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carr CM, Rizo J. At the junction of SNARE and SM protein function. Curr Opin Cell Biol. 2010;22(4):488–495. doi: 10.1016/j.ceb.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burgoyne RD, et al. The functions of Munc18-1 in regulated exocytosis. Ann N Y Acad Sci. 2009;1152:76–86. doi: 10.1111/j.1749-6632.2008.03987.x. [DOI] [PubMed] [Google Scholar]
- 16.Toonen RF, Verhage M. Munc18-1 in secretion: Lonely Munc joins SNARE team and takes control. Trends Neurosci. 2007;30(11):564–572. doi: 10.1016/j.tins.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 17.Gissen P, et al. Mutations in VPS33B, encoding a regulator of SNARE-dependent membrane fusion, cause arthrogryposis-renal dysfunction-cholestasis (ARC) syndrome. Nat Genet. 2004;36(4):400–404. doi: 10.1038/ng1325. [DOI] [PubMed] [Google Scholar]
- 18.Saitsu H, et al. De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet. 2008;40(6):782–788. doi: 10.1038/ng.150. [DOI] [PubMed] [Google Scholar]
- 19.Cetica V, et al. STXBP2 mutations in children with familial haemophagocytic lymphohistiocytosis type 5. J Med Genet. 2010;47(9):595–600. doi: 10.1136/jmg.2009.075341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Côte M, et al. Munc18-2 deficiency causes familial hemophagocytic lymphohistiocytosis type 5 and impairs cytotoxic granule exocytosis in patient NK cells. J Clin Invest. 2009;119(12):3765–3773. doi: 10.1172/JCI40732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meeths M, et al. Spectrum of clinical presentations in familial hemophagocytic lymphohistiocytosis type 5 patients with mutations in STXBP2. Blood. 2010;116(15):2635–2643. doi: 10.1182/blood-2010-05-282541. [DOI] [PubMed] [Google Scholar]
- 22.Rizo J, Südhof TC. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices—Guilty as charged? Annu Rev Cell Dev Biol. 2012;28:279–308. doi: 10.1146/annurev-cellbio-101011-155818. [DOI] [PubMed] [Google Scholar]
- 23.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 24.Brandie FM, et al. Negative regulation of syntaxin4/SNAP-23/VAMP2-mediated membrane fusion by Munc18c in vitro. PLoS ONE. 2008;3(12):e4074. doi: 10.1371/journal.pone.0004074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190(1):55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rathore SS, et al. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci USA. 2010;107(52):22399–22406. doi: 10.1073/pnas.1012997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L, Kümmel D, Coleman J, Melia TJ, Giraudo CG. Dual roles of Munc18-1 rely on distinct binding modes of the central cavity with Stx1A and SNARE complex. Mol Biol Cell. 2011;22(21):4150–4160. doi: 10.1091/mbc.E11-02-0150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tareste D, Shen J, Melia TJ, Rothman JE. SNAREpin/Munc18 promotes adhesion and fusion of large vesicles to giant membranes. Proc Natl Acad Sci USA. 2008;105(7):2380–2385. doi: 10.1073/pnas.0712125105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rathore SS, Ghosh N, Ouyang Y, Shen J. Topological arrangement of the intracellular membrane fusion machinery. Mol Biol Cell. 2011;22(14):2612–2619. doi: 10.1091/mbc.E11-03-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodkey TL, Liu S, Barry M, McNew JA. Munc18a scaffolds SNARE assembly to promote membrane fusion. Mol Biol Cell. 2008;19(12):5422–5434. doi: 10.1091/mbc.E08-05-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schollmeier Y, Krause JM, Kreye S, Malsam J, Söllner TH. Resolving the function of distinct Munc18-1/SNARE protein interaction modes in a reconstituted membrane fusion assay. J Biol Chem. 2011;286(35):30582–30590. doi: 10.1074/jbc.M111.269886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Diao J, et al. Single-Vesicle Fusion Assay Reveals Munc18-1 Binding to the SNARE Core Is Sufficient for Stimulating Membrane Fusion. ACS Chem Neurosci. 2010;1(3):168–174. doi: 10.1021/cn900034p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou P, et al. Syntaxin-1 N-peptide and Habc-domain perform distinct essential functions in synaptic vesicle fusion. EMBO J. 2012;32(1):159–171. doi: 10.1038/emboj.2012.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burkhardt P, Hattendorf DA, Weis WI, Fasshauer D. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide. EMBO J. 2008;27(7):923–933. doi: 10.1038/emboj.2008.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Misura KM, Scheller RH, Weis WI. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex. Nature. 2000;404(6776):355–362. doi: 10.1038/35006120. [DOI] [PubMed] [Google Scholar]
- 36.Latham CF, Osborne SL, Cryle MJ, Meunier FA. Arachidonic acid potentiates exocytosis and allows neuronal SNARE complex to interact with Munc18a. J Neurochem. 2007;100(6):1543–1554. doi: 10.1111/j.1471-4159.2006.04286.x. [DOI] [PubMed] [Google Scholar]
- 37.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339(6118):421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma C, Li W, Xu Y, Rizo J. Munc13 mediates the transition from the closed syntaxin-Munc18 complex to the SNARE complex. Nat Struct Mol Biol. 2011;18(5):542–549. doi: 10.1038/nsmb.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arunachalam L, et al. Munc18-1 is critical for plasma membrane localization of syntaxin1 but not of SNAP-25 in PC12 cells. Mol Biol Cell. 2008;19(2):722–734. doi: 10.1091/mbc.E07-07-0662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tellam JT, McIntosh S, James DE. Molecular identification of two novel Munc-18 isoforms expressed in non-neuronal tissues. J Biol Chem. 1995;270(11):5857–5863. doi: 10.1074/jbc.270.11.5857. [DOI] [PubMed] [Google Scholar]
- 41.Jewell JL, et al. Munc18c phosphorylation by the insulin receptor links cell signaling directly to SNARE exocytosis. J Cell Biol. 2011;193(1):185–199. doi: 10.1083/jcb.201007176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aran V, et al. Characterization of two distinct binding modes between syntaxin 4 and Munc18c. Biochem J. 2009;419(3):655–660. doi: 10.1042/BJ20082293. [DOI] [PubMed] [Google Scholar]
- 43.Oh E, Spurlin BA, Pessin JE, Thurmond DC. Munc18c heterozygous knockout mice display increased susceptibility for severe glucose intolerance. Diabetes. 2005;54(3):638–647. doi: 10.2337/diabetes.54.3.638. [DOI] [PubMed] [Google Scholar]
- 44.Thurmond DC, Pessin JE. Discrimination of GLUT4 vesicle trafficking from fusion using a temperature-sensitive Munc18c mutant. EMBO J. 2000;19(14):3565–3575. doi: 10.1093/emboj/19.14.3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Humphrey SJ, et al. Dynamic Adipocyte Phosphoproteome Reveals that Akt Directly Regulates mTORC2. Cell Metab. 2013;17(6):1009–1020. doi: 10.1016/j.cmet.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rowland AF, Fazakerley DJ, James DE. Mapping insulin/GLUT4 circuitry. Traffic. 2011;12(6):672–681. doi: 10.1111/j.1600-0854.2011.01178.x. [DOI] [PubMed] [Google Scholar]
- 47.Oh E, Thurmond DC. Munc18c depletion selectively impairs the sustained phase of insulin release. Diabetes. 2009;58(5):1165–1174. doi: 10.2337/db08-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brochetta C, et al. Involvement of Munc18 isoforms in the regulation of granule exocytosis in neutrophils. Biochim Biophys Acta. 2008;1783(10):1781–1791. doi: 10.1016/j.bbamcr.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 49.Imai A, Nashida T, Shimomura H. Roles of Munc18-3 in amylase release from rat parotid acinar cells. Arch Biochem Biophys. 2004;422(2):175–182. doi: 10.1016/j.abb.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 50.Schraw TD, Lemons PP, Dean WL, Whiteheart SW. A role for Sec1/Munc18 proteins in platelet exocytosis. Biochem J. 2003;374(Pt 1):207–217. doi: 10.1042/BJ20030610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Houng A, Polgar J, Reed GL. Munc18-syntaxin complexes and exocytosis in human platelets. J Biol Chem. 2003;278(22):19627–19633. doi: 10.1074/jbc.M212465200. [DOI] [PubMed] [Google Scholar]
- 52.Deák F, et al. Munc18-1 binding to the neuronal SNARE complex controls synaptic vesicle priming. J Cell Biol. 2009;184(5):751–764. doi: 10.1083/jcb.200812026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y, Seven AB, Su L, Jiang QX, Rizo J. Membrane bridging and hemifusion by denaturated Munc18. PLoS ONE. 2011;6(7):e22012. doi: 10.1371/journal.pone.0022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bryant NJ, Govers R, James DE. Regulated transport of the glucose transporter GLUT4. Nat Rev Mol Cell Biol. 2002;3(4):267–277. doi: 10.1038/nrm782. [DOI] [PubMed] [Google Scholar]
- 55.Dulubova I, et al. A conformational switch in syntaxin during exocytosis: Role of munc18. EMBO J. 1999;18(16):4372–4382. doi: 10.1093/emboj/18.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gerber SH, et al. Conformational switch of syntaxin-1 controls synaptic vesicle fusion. Science. 2008;321(5895):1507–1510. doi: 10.1126/science.1163174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Latham CF, et al. Molecular dissection of the Munc18c/syntaxin4 interaction: Implications for regulation of membrane trafficking. Traffic. 2006;7(10):1408–1419. doi: 10.1111/j.1600-0854.2006.00474.x. [DOI] [PubMed] [Google Scholar]
- 58.Christie MP, et al. Low-resolution solution structures of Munc18:Syntaxin protein complexes indicate an open binding mode driven by the Syntaxin N-peptide. Proc Natl Acad Sci USA. 2012;109(25):9816–9821. doi: 10.1073/pnas.1116975109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tamori Y, et al. Inhibition of insulin-induced GLUT4 translocation by Munc18c through interaction with syntaxin4 in 3T3-L1 adipocytes. J Biol Chem. 1998;273(31):19740–19746. doi: 10.1074/jbc.273.31.19740. [DOI] [PubMed] [Google Scholar]
- 60.Thurmond DC, et al. Regulation of insulin-stimulated GLUT4 translocation by Munc18c in 3T3L1 adipocytes. J Biol Chem. 1998;273(50):33876–33883. doi: 10.1074/jbc.273.50.33876. [DOI] [PubMed] [Google Scholar]
- 61.Tellam JT, et al. Characterization of Munc-18c and syntaxin-4 in 3T3-L1 adipocytes. Putative role in insulin-dependent movement of GLUT-4. J Biol Chem. 1997;272(10):6179–6186. doi: 10.1074/jbc.272.10.6179. [DOI] [PubMed] [Google Scholar]
- 62.Carr CM, Grote E, Munson M, Hughson FM, Novick PJ. Sec1p binds to SNARE complexes and concentrates at sites of secretion. J Cell Biol. 1999;146(2):333–344. doi: 10.1083/jcb.146.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Scott BL, et al. Sec1p directly stimulates SNARE-mediated membrane fusion in vitro. J Cell Biol. 2004;167(1):75–85. doi: 10.1083/jcb.200405018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carpp LN, Ciufo LF, Shanks SG, Boyd A, Bryant NJ. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes. J Cell Biol. 2006;173(6):927–936. doi: 10.1083/jcb.200512024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Krämer L, Ungermann C. HOPS drives vacuole fusion by binding the vacuolar SNARE complex and the Vam7 PX domain via two distinct sites. Mol Biol Cell. 2011;22(14):2601–2611. doi: 10.1091/mbc.E11-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pieren M, Schmidt A, Mayer A. The SM protein Vps33 and the t-SNARE H(abc) domain promote fusion pore opening. Nat Struct Mol Biol. 2010;17(6):710–717. doi: 10.1038/nsmb.1809. [DOI] [PubMed] [Google Scholar]
- 67.Furgason ML, et al. The N-terminal peptide of the syntaxin Tlg2p modulates binding of its closed conformation to Vps45p. Proc Natl Acad Sci USA. 2009;106(34):14303–14308. doi: 10.1073/pnas.0902976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Munson M, Chen X, Cocina AE, Schultz SM, Hughson FM. Interactions within the yeast t-SNARE Sso1p that control SNARE complex assembly. Nat Struct Biol. 2000;7(10):894–902. doi: 10.1038/79659. [DOI] [PubMed] [Google Scholar]
- 69.Lobingier BT, Merz AJ. Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex. Mol Biol Cell. 2012;23(23):4611–4622. doi: 10.1091/mbc.E12-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jahn R. Principles of exocytosis and membrane fusion. Ann N Y Acad Sci. 2004;1014:170–178. doi: 10.1196/annals.1294.018. [DOI] [PubMed] [Google Scholar]
- 71.Meijer M, et al. Munc18-1 mutations that strongly impair SNARE-complex binding support normal synaptic transmission. EMBO J. 2012;31(9):2156–2168. doi: 10.1038/emboj.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Antonin W, et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19(23):6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395(6700):347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 74.Fasshauer D, Sutton RB, Brunger AT, Jahn R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci USA. 1998;95(26):15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hu SH, Latham CF, Gee CL, James DE, Martin JL. Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 2007;104(21):8773–8778. doi: 10.1073/pnas.0701124104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bracher A, Kadlec J, Betz H, Weissenhorn W. X-ray structure of a neuronal complexin-SNARE complex from squid. J Biol Chem. 2002;277(29):26517–26523. doi: 10.1074/jbc.M203460200. [DOI] [PubMed] [Google Scholar]
- 77.Krishnakumar SS, et al. A conformational switch in complexin is required for synaptotagmin to trigger synaptic fusion. Nat Struct Mol Biol. 2011;18(8):934–940. doi: 10.1038/nsmb.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kümmel D, et al. Complexin cross-links prefusion SNAREs into a zigzag array. Nat Struct Mol Biol. 2011;18(8):927–933. doi: 10.1038/nsmb.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burkhardt P, et al. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci USA. 2011;108(37):15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sudhof TC. The synaptic vesicle cycle. Annu Rev Neurosci. 2004;27:509–547. doi: 10.1146/annurev.neuro.26.041002.131412. [DOI] [PubMed] [Google Scholar]
- 81.Yu H, Rathore SS, Shen J. Synip Arrests Soluble N-Ethylmaleimide-sensitive Factor Attachment Protein Receptor (SNARE)-dependent Membrane Fusion as a Selective Target Membrane SNARE-binding Inhibitor. J Biol Chem. 2013;288(26):18885–18895. doi: 10.1074/jbc.M113.465450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yu H, Rathore SS, Davis EM, Ouyang Y, Shen J. Doc2b promotes GLUT4 exocytosis by activating the SNARE-mediated fusion reaction in a calcium- and membrane bending-dependent manner. Mol Biol Cell. 2013;24(8):1176–1184. doi: 10.1091/mbc.E12-11-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.