Abstract
Sunlight provides energy for photosynthesis and is essential for nearly all life on earth. However, too much or too little light or rapidly fluctuating light conditions cause stress to plants. Rapid changes in the amount of light are perceived as a change in the reduced/oxidized (redox) state of photosynthetic electron transport components in chloroplasts. However, how this generates a signal that is relayed to changes in nuclear gene expression is not well understood. We modified redox state in the reference plant, Arabidopsis thaliana, using either excess light or low light plus the herbicide DBMIB (2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone), a well-known inhibitor of photosynthetic electron transport. Modification of redox state caused a change in expression of a common set of about 750 genes, many of which are known stress-responsive genes. Among the most highly enriched promoter elements in the induced gene set were heat-shock elements (HSEs), known motifs that change gene expression in response to high temperature in many systems. We show that HSEs from the promoter of the ASCORBATE PEROXIDASE 2 (APX2) gene were necessary and sufficient for APX2 expression in conditions of excess light, or under low light plus the herbicide. We tested APX2 expression phenotypes in overexpression and loss-of-function mutants of 15 Arabidopsis A-type heat-shock transcription factors (HSFs), and identified HSFA1D, HSFA2, and HSFA3 as key factors regulating APX2 expression in diverse stress conditions. Excess light regulates both the subcellular location of HSFA1D and its biochemical properties, making it a key early component of the excess light stress network of plants.
Photosynthesis is the process by which light is converted to chemical energy; yet, despite its critical and central role for the sustenance of life, photosynthetic performance of plants is not optimal. Under light intensities of higher than optimal growth (termed excess light or EL), light energy absorption increases linearly as light intensity increases. However, carbon assimilation becomes saturated at a certain light intensity, termed the light saturation point. Thus, in EL, most of the absorbed light energy is not used for carbon metabolism. Instead, the EL energy causes damage to proteins involved in photosynthesis, a process called photoinhibition (1–3).
Plants have evolved multiple ways to sense and respond to EL (4). Photoreceptors, such as cryptochrome, sense light directly and lead to changes in nuclear gene expression and chloroplast avoidance responses. Excess light energy can also be dissipated as heat via nonphotochemical quenching, thereby avoiding damage to the photosynthetic apparatus. In addition to inactivating photosynthesis (3), it has been reported that EL brings about accumulation of a number of metabolic intermediates in plastids that act as signals of EL stress (5–7). Finally, EL causes the accumulation of multiple reactive oxygen species, including, singlet oxygen (1O2), hydrogen peroxide (H2O2), and superoxide anion (O2-) (8) in plastids, which are neutralized by reactive oxygen species detoxifying enzymes and by the synthesis of antioxidant molecules, such as carotenoids, ascorbic acid, and tocopherol (1).
Several studies have provided evidence for a role of photosynthetic electron transport (PET) components as sensors of EL and regulators of nuclear gene expression (9, 10). A stress-response gene encoding ASCORBATE PEROXIDASE 2 (APX2) is controlled by the reduced/oxidized (redox) state of the plastoquionone (PQ) pool in Arabidopsis (11, 12). The expression of APX2 is significantly induced by the reduced PQ pool that is generated by either EL or low light (LL) plus DBMIB (2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone), a herbicide that blocks electron transport between the PQ pool and the cytochrome b6f complex (11, 13, 14). EL-driven APX2 expression requires H2O2 and functional PET (11, 13).
The molecular mechanism by which the PQ redox state triggers the induction of APX2 is unknown, although clues to some possible mechanisms have been reported. Despite APX2’s fast and very strong induction upon exposure to EL (11), genetic screens have identified only constitutive APX2 expression mutants, but no mutants affected specifically in EL-driven APX2 induction have been found (15, 16). There is evidence linking APX2 induction by EL to the zinc-finger transcription factor ZAT10, to the plant hormone abscisic acid, and to 3′-phosphoadenosine 5′-phosphate, a phosphonucleotide (PAP) (5, 17, 18); however, a signal transduction pathway linking gene expression in the nucleus to redox of the PQ pool remains elusive.
Here, we took advantage of reporters of EL-induced gene expression and DBMIB to identify nuclear proteins specific to a chloroplast-to-nucleus signaling process generated by signals from the PQ pool. We determine that a reduced state of the PQ pool changes the expression of about 750 nuclear genes, many of which contain heat-shock elements (HSEs). A hierarchy of interactions among a subset of heat-shock transcription factors (HSFs)—HSFA1D, HSFA2, and HSFA3—is responsible for the early gene-expression response of Arabidopsis to EL.
Results
Defining Stress Conditions.
Recent studies have linked the accumulation of a number of different metabolic intermediates with EL stress, leading to the proposal that the accumulation of these intermediates is the initiating event for multiple stress signaling pathways (5–7). Other articles have reported that a signal originates from PET, specifically the redox state of the PQ pool (11, 19, 20). To better define the nature of the signals related to the PQ redox state, we treated plants with a number of different light stresses, as well as with LL plus the herbicide DBMIB.
First, we assessed the effectiveness of our DBMIB treatment in generating the PQ pool in a reduced state. Following incubation of leaf discs in a DBMIB (24 µM) solution for 2 h under LL conditions, chlorophyll fluorescence and APX2 transcript levels were determined. As expected, the DBMIB treatment increased the excitation pressure on PSII (1-qP) (21) (Fig. S1A) and APX2 induction levels (Fig. S1B); in contrast, the 3-3,4-dichlorophenyl-1,1-dimethylurea (DCMU) treatment under LL did not induce APX2 expression (Fig. S1B), despite its effect on increased 1-qP (Fig. S1A). These results confirmed that generation our conditions for reducing the PQ pool with DBMIB worked as expected (11). However, both DBMIB and DCMU treatments failed to generate H2O2 in leaf tissue, whereras “hot” EL (hEL) treatment resulted in H2O2 generation (Fig. S1C).
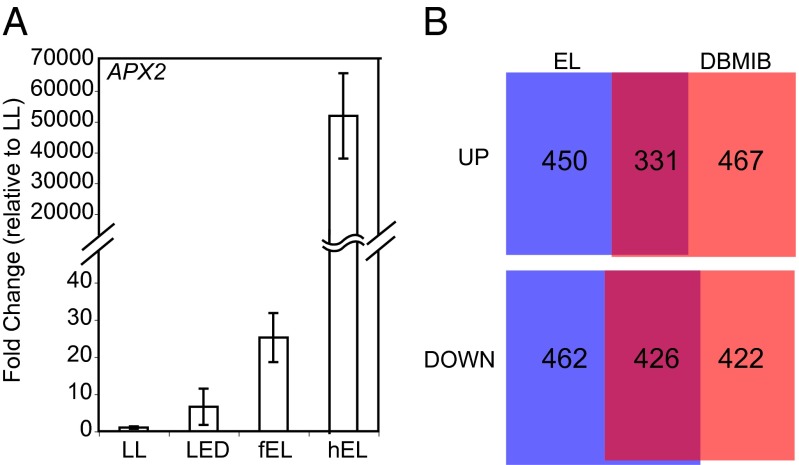
We further explored APX2 expression by light, high temperature, or both by exposing plant leaves to a range of EL conditions: hEL, “filtered” EL (fEL), which was achieved by using a mixture of water and ice layer (13, 22), and “cold” light (LED) (Fig. 1A). Although all EL conditions significantly increased leaf temperature, the effects of hEL on leaf temperature increases were more drastic than those of fEL and LED (Fig. S1D). All EL conditions up-regulated APX2 expression, although APX2 induction levels were lower in fEL and LED (Fig. 1A). In addition, it seems like APX2 induction is positively correlated with the amount of H2O2 in leaves (Fig. S1E). Based on these observations, it is very likely that APX2 expression is regulated by both light and high temperature in a synergistic way. In addition, it seems like there is a heat-independent APX2 induction pathway. For the rest of this report, all EL experiments described were conducted using hEL and presented as EL.
Fig. 1.
EL and DBMIB change expression levels of nuclear genes including APX2. (A) APX2 transcript abundance under LL, LED, fEL, and hEL. Plants were exposed to the EL treatments for 1 h. Light qualities have a total of ∼1,000 µmol photons m−2⋅s−1. hEL was filtered using a 2-cm height column of water and ice mixture in a clear perspex tray to generate fEL. For LED treatments, only the EL exposed tissue was harvested. Average values of fold-change relative to LL were from three biological replicates. Error bars represent SD. (B) The number of genes up- and down-regulated by EL and DBMIB treatment. The areas are proportional to the number of genes in each category.
EL Triggers Redox Stress and Changes the Expression of Hundreds of Genes.
To understand the effect of the reduced PQ pool on global nuclear gene expression, we harvested mRNA from Arabidopsis leaf tissue treated with EL (for 30 min or 2 h), or LL plus DBMIB (for 30 min or 2 h), and generated expression profiles using ATH1 microarrays. We found ∼1,250 genes up-regulated and ∼1,300 genes down-regulated by either treatment at either time point compared with before treatment, with 331 genes commonly up-regulated and 426 genes down-regulated by both treatments (Fig. 1B). Among the top genes up-regulated by both DBMIB and EL were genes encoding several heat-shock factors and small heat-shock proteins (some induced up to several thousand-fold), as well as APX2, expected from previous studies (Dataset S1) (13). Of the ∼20 heat-shock factor protein family members, 10 are significantly (P < 0.01) up-regulated in at least one condition, and at least 30 heat-shock proteins are also significantly induced (Dataset S1). Gene Ontology term-enrichment analysis revealed nearly 20 processes overrepresented (P < 0.01 using the hypergeometric test) among the core up-regulated gene set, including response to heat, response to high light intensity, and response to oxidative stress, consistent with the treatments applied (Dataset S2). Inspection of promoter sequences (defined as the 500-bp upstream from the predicted ATG) from the 331 commonly up-regulated genes revealed that a high proportion of genes contained at least one of four variants of the known HSEs (e.g., GAAnnTTC) (Fig. S1F). Genes down-regulated by EL and DBMIB include many biosynthetic enzymes, cytochrome P450 enzymes, and transcription factors (Dataset S1), although the magnitude of change of down-regulated genes is smaller than for up-regulated transcripts. Gene Ontology enrichment for this gene set indicates a high overrepresentation for metabolic enzymes, especially those involved in glucosinolate biosynthesis (Dataset S2). In summary, ∼750 genes are significantly altered by PQ redox state, which most notably induces expression of APX2 and heat-shock factors, indicating a strong redox stress response by EL and DBMIB.
In addition to HSEs, we mined the predicted promoters of up-regulated genes for other previously known and novel overrepresented motifs using the ELEMENT algorithm (23) (Fig. S1F). Among genes up-regulated by 30 min of EL, we found a number of known light-regulatory elements, including the G-box (CACGTG) (24), and a GCCAC motif previously identified as overrepresented in light-induced promoters (SORLIPs) (25). We also found an ACGT motif (26), a CCGAC motif (27), and an ATCCAAT motif (28) (Fig. S1F).
APX2 Induction by EL or DBMIB Does Not Require New Protein Synthesis.
About 50% of the genes induced by either EL or DBMIB were common to both conditions, but induction by EL appeared to be faster than in DBMIB. In agreement with previous reports (11, 13), APX2 induction was seen within 5 min of exposure to EL, achieved peak levels after 1 h of exposure to EL, and then decreased (Fig. S2A). Although APX2 mRNA levels also increased in the presence of DBMIB, the induction profile was different as the fold-change was smaller than that with EL (Fig. S2 A and B). Furthermore, APX2 mRNA levels never peaked during the 2 h of DBMIB treatment (Fig. S2B), indicating that the effects of DBMIB were slower than those of EL (11). The rapid induction kinetics of APX2 by EL or DBMIB led us to test whether new protein synthesis is required for APX2 induction. Leaf tissue was pretreated with 200 µM cycloheximide (CHX) for 30 min, then exposed to EL for 2 h, and APX2 mRNA accumulation was determined (Fig. S2A). APX2 induction was observed in the presence of CHX for the first hour of EL treatment. After 2 h of EL, APX2 transcript levels were higher in the presence of CHX than in the absence of CHX (Fig. S2A). One explanation for this observation is that an unidentified factor, synthesized under EL, might negatively regulate EL-driven APX2 induction. By a pretreatment with CHX followed by 2 h of DBMIB treatment, we did not see an effect of CHX on APX2 induction (Fig. S2B). As a positive control for the CHX treatment, we generated a transgenic line, A2P, in which LUCIFERASE (LUC) gene expression is under the control of the APX2 gene promoter [A2PAPX2(-1507)] (Fig. 2A). After 2 h of EL treatment, we observed about a 15-fold change in relative luciferase units (RLU) (Fig. S2C) in the absence of CHX, whereas in the presence of CHX, there was no change in RLU, indicating that the CHX treatment effectively inhibited synthesis of the LUC protein (Fig. S2C). Therefore, we conclude that EL or DBMIB induced APX2 expression in the absence of new protein synthesis.
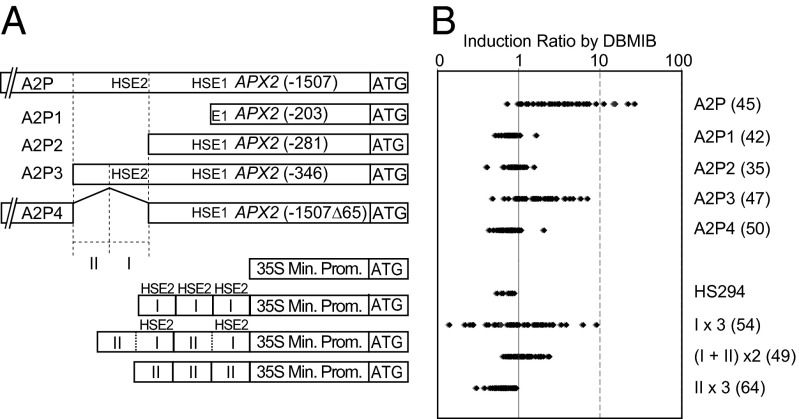
Fig. 2.
HSE2 is responsible for APX2 Induction by DBMIB. (A) Constructs generated to identify cis-acting elements responsible for DBMIB-driven APX2 induction. “ATG” is the translational start site for the LUC gene, and bars located in front of ATG represent the APX2 promoter regions. The numbers in parentheses following construct names are the size of the APX2 promoter regions from the translation start site of APX2. HSE1 and HSE2 are inverted repeat of HSE dimmers, and the numbers “1” and “2” are added for this study. The Roman numerals “I” and “II” were arbitrarily assigned to represent the portion with (I) and without (II) HSE2 in the segment from −282 to −346. “35S Min Prom” stands for the 35S minimal promoter in the vector of pHS294. (B) LUC induction ratio by DBMIB in transgenic plants transformed by constructs in A. Each dot represents an induction ratio (after/before DBMIB treatment) after 2 h of DBMIB treatment in a single transgenic plant in its T1 generation. The numbers of T1 lines tested per construct are in parentheses.
Heat-Shock Transcription Elements Are Necessary and Sufficient for APX2 Gene Induction by DBMIB.
Previously reported genetic screens failed to identify positive regulators of APX2 gene expression (15, 16), suggesting that these regulators may be redundantly encoded in the Arabidopsis genome. In addition, the rapid kinetics of the response, its independence of new protein synthesis, and the preponderance of HSEs predicted in the promoters of up-regulated genes was reminiscent of the response to heat shock in many organisms. To remove any effect of heat on APX2 expression (29) and to investigate a heat-independent APX2 induction mechanism, we decided to use DBMIB under LL in a temperature-controlled growth chamber. Under this condition, we were able to observe strong APX2 induction (Fig. S2B).
To identify cis-acting elements responsible for APX2 induction by DBMIB, we generated a series of transcriptional fusion constructs made with various lengths of the APX2 promoter region fused to the LUC gene (Fig. 2A). Almost all transgenic lines containing the A2P construct, which encompasses the 1,507 bp upstream from the APX2 start codon (Fig. 2A), showed an increase in LUC activity after DBMIB treatment. Several lines had greater than 10-fold LUC induction by DBMIB (Fig. 2B). The A2P3 construct, containing 346 bp upstream of the APX2 start site (Fig. 2A), also showed strong induction by DBMIB, although the levels of LUC activity were not as high as those from the A2P construct (Fig. 2B). LUC in the A2P1 and A2P2 constructs, which contained 203 bp and 281 bp upstream of the APX2 ATG, respectively (Fig. 2A), did not respond to DBMIB treatment (Fig. 2B), indicating that the 65-bp region from −282 to −346 was necessary for DBMIB-induced APX2 induction (Fig. 2A). Deletion of this region (−282 to −346) in the A2P4 construct resulted in transgenic plants (49/50 T1 lines) with little to no LUC activity (Fig. 2A), indicating that the 65-bp region from −282 to −346 is necessary for DBMIB-induced APX2 induction (Fig. 2B).
We scanned the interval between −282 and −346 of the APX2 promoter for known cis-acting elements, and found two HSEs in tail-to-tail arrangement (tTTCtgGAAg: HSE2 in Fig. 2A). These same elements have been reported to be responsible for heat-induced APX2 expression in a tobacco leaf protoplast transient expression assay (29), and the elements are known to be the minimally required sequence for active HSF binding (30). There are also HSEs in head-to-head arrangement, another functional type (30), adjacent to the start codon (aGAAgcTTCa: HSE1 in Fig. 2A); however, HSE1 does not appear to be necessary for APX2 induction by DBMIB (Fig. 2B).
To investigate whether the 65-bp interval containing HSE2 is sufficient for DBMIB-driven APX2 expression, we generated transgenic plants (HS294), in which LUC expression is under the control of the 35S cauliflower mosaic virus (35S) minimal promoter (Fig. 2A). None of the HS294-containing plants showed an increase in LUC activity after exposure to DBMIB (Fig. 2B). However, when two copies of the 65-bp interval were placed in front of the 35S minimal promoter and LUC fusion [(I + II) × 2] (Fig. 2A), 63% (31 of 49) of T1 transgenic plants showed LUC induction by DBMIB (Fig. 2B). We further subdivided the 65-bp region into two fragments: I (29 bp) and II (36 bp) (Fig. 2A). When three copies of fragment I encompassing HSE2 (I × 3) were attached to the 35S minimal promoter and translationally fused to LUC (Fig. 2A), about half (26 of 54) of the transgenic plants (T1) showed DBMIB-induced LUC induction, and the highest induction ratios in I × 3 were greater than those of two copies of the 65-bp interval [(I+II) × 2] (Fig. 2B). Three copies of fragment II (II × 3), lacking HSE2 (Fig. 2A), failed to show any DBMIB-induced LUC expression in the 64 T1 plants tested (Fig. 2B). Taking these data together, we conclude that a 29-bp element containing HSE2 is both necessary and sufficient to induce gene expression after short DBMIB treatments (Fig. 2B).
Dynamics of APX2 Induction by HSFs Under Stress Conditions.
The inverted repeats of HSEs, such as HSE2, are bound by HSFs (30), thus making HSFs the primary candidates mediating APX2 up-regulation in response to EL. In Arabidopsis, there are 21 HSFs (31): 15 type A, 5 type B, and 1 type C, characterized by their domain architectures. Because only type A HSFs contain transcriptional activation domains (31), and because it was reported that overexpression of HSFA2 and HSFA3 could induce APX2 expression in the absence of stress (32–34), we focused on type A HSFs as possible regulators of APX2 expression.
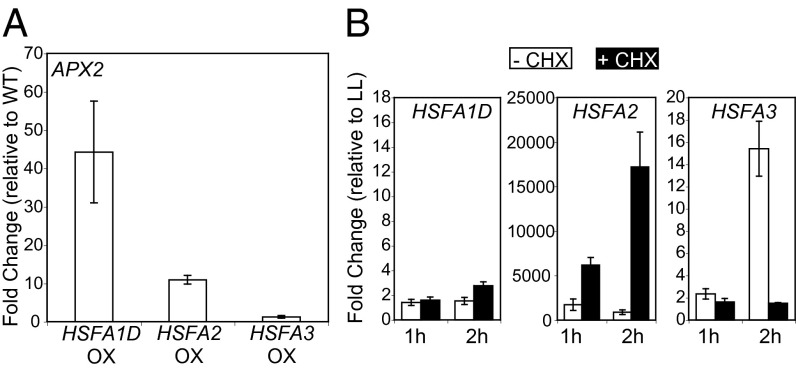
Constructs expressing each individual type A HSF under the control of the 2 × 35S promoter were introduced into wild-type plants. We collected leaf tissue from T1 plants, and monitored APX2 transcript levels using real-time RT-PCR (Fig. S3A). Besides HSFA2 and HSFA3 overexpression, HSFA1B, HSFA1D, HSFA4A, and HSFA6B overexpression induced APX2 (fold-change >10) compared with transgenic control plants harboring only the binary vector or wild-type Col-0 (Fig. S3A). Because knockout mutant lines of HSFA1D, HSFA2, and HSFA3 were available in the same genetic background from the SALK T-DNA collection, we focused on these three HSFs. The overexpression of HSFA1D, HSFA2, and HSFA3 were confirmed in the next generation (Fig. S3B). There was a hierarchy of APX2 expressions levels with HSFA1D > HSFA2 > HSFA3 (Fig. 3A).
Fig. 3.
Overexpression of HSFA1D, HSFA2, and HSFA3 could induce APX2 expression under LL. (A) Fold-changes of APX2 under LL in HSFA1D-, HSFA2-, and HSFA3-overexpression lines, relative to wild-type Col-0 plants, were determined. The average and SEs were calculated from real-time RT-PCR of two or three independent T2 lines and three technical replicates. (B) Fold-changes of HSFA1D, HSFA2, and HSFA3 by the indicated time of EL were determined using real-time RT-PCR. Fold-changes, relative to untreated LL samples, in the presence of 200 µM CHX and in mock-treated (ethanol) samples were displayed using filled and blank bars, respectively. Two biological and four technical replicates were tested to calculate the average and SEs.
We checked whether HSFA1D, HSFA2, and HSFA3 genes were up-regulated in EL. HSFA1D mRNA was detectable in LL, but its levels remained low after exposure to EL for 1 h (Fig. 3B and Fig. S3C); moreover, between 1- and 2-h treatment, no further increase was observed (Fig. 3B). mRNA levels of both HSFA2 and HSFA3 were low in LL, but dramatically increased in response to EL (Fig. 3B and Fig. S3C). A time-course of HSFA2 mRNA levels in EL revealed an induction pattern that is very similar to that of APX2 (Fig. 3B, and Figs. S2A and S3C). In contrast, HSFA3 mRNA levels increased continuously during 2 h of EL treatment (Fig. 3B and Fig. S3C). The EL-driven induction patterns of HSFA1D, HSFA2, and HSFA3 were similar to those elicited by a combined treatment of moderate light stress and heat stress (32).
The similarity of induction kinetics in EL between APX2 and HSFA2 prompted us to test whether EL-driven HSFA2 induction required new protein synthesis. We determined the induction levels of HSFA1D, HSFA2, and HSFA3 by EL in the presence of CHX and compared induction levels to LL samples (Fig. 3B). Both HSFA1D and HSFA2 were induced in the presence of CHX; they accumulated to even higher levels in CHX. In contrast, the induction of HSFA3 was completely abolished by the CHX treatment (Fig. 3B). Thus, the induction of HSFA1D and HSFA2 by EL does not require new protein synthesis, whereas new protein synthesis is required for HSFA3 induction by EL.
We obtained homozygous T-DNA insertion lines from the SALK collection for HSFA1D (SALK_022404), HSFA2 (SALK_008978), and HSFA3 (SALK_011107). After backcrossing at least twice, we confirmed the absence of HSFA1D, HSFA2, and HSFA3 transcripts in the T-DNA insertion mutants by semiquantitative RT-PCR (Fig. S3D). The mutants were used to test whether HSFA1D is required for EL-driven HSFA2 and HSFA3 induction, and whether HSFA2 or both HSFA1D and HSFA2 are essential for HSFA3 induction by EL (Fig. S3 E–G). In an hsfa1d-null mutant, HSFA2 and HSFA3 mRNA accumulated in a similar manner to wild-type. Induction of HSFA3 by EL was even faster in hsfa1d than wild-type (Fig. S3E). Similarly, we observed no difference from wild-type in EL-driven HSFA3 induction levels in either the hsfa2 or hsfa1d hsfa2 double mutant (Fig. S3 F and G).
Homozygous null-mutants were exposed to EL and the levels of APX2 expression were quantified. Although both hsfa1d and hsfa2 mutant plants exhibited a reduction of APX2 transcript accumulation in response to EL, (Fig. 4A, Left), there was still a significant increase in APX2 RNA after EL treatment. Moreover, after 1 h of EL, APX2 induction in an hsfa3-null mutant was indistinguishable from wild-type.
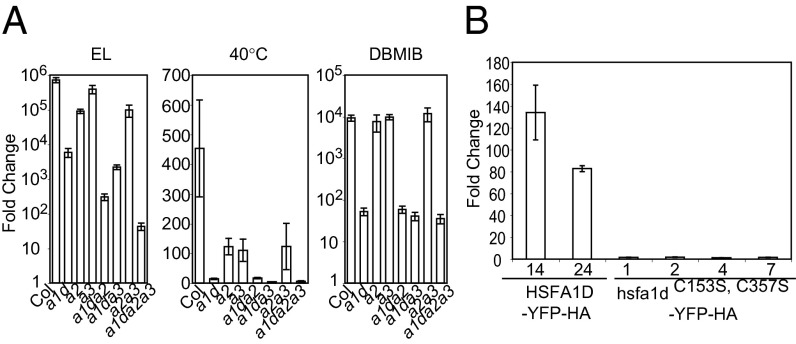
Fig. 4.
APX2 induction by EL is mediated by HSFs and their posttranslational modification. (A) Fold-changes of the APX2 gene by EL, heat shock (40 °C), or DBMIB in the single- (a1d, a2, a3), double- (a1da2, a1da3, and a2a3), and triple- (a1da2a3) knockout mutants of HSFA1D, HSFA2, and HSFA3. Real-time RT-PCR was used to determine the fold changes relative to before the treatment. Leaf tissue was treated with EL for 1 h and with heat shock or DBMIB for 2 h. Error bars indicate SE of two biological and two technical replicates. The y axes of EL and DBMIB are on log10 scales. (B) Real-time RT-PCR determined fold changes of APX2 by 1 h of EL exposure in transgenic plants expressing HSFA1D-YFP-HA and hsfa1dC153S, C357S-YFP-HA under HSFA1D native promoter. Both constructs were introduced to hsfa1d hsfa2 hsfa3. Two biological and two technical replicates were used to calculate the average and SEs.
To better understand the individual contributions of HSFA1D, HSFA2, and HSFA3 to APX2 expression, we exposed single- and higher-order mutants of hsfa1d, hsfa2, and hsfa3 to three environmental stresses: EL, heat (40 °C), or DBMIB (Fig. 4A). In EL, the hsfa1d hsfa2 hsfa3 triple-mutant had drastically lower APX2 induction levels compared with all other genotypes (Fig. 4A). The role of HSFA1D, HSFA2, and HSFA3 in EL-driven APX2 expression was also confirmed by the near elimination of LUC activity in the triple-mutant compared with wild-type after 2 h of EL treatment (Fig. S3H).
In EL, the contribution of HSFA1D for APX2 expression is greater than HSFA2, which is greater than HSFA3 (Fig. 4A, Left). Although overall induction levels caused by heat were lower than those caused by EL, HSFA1D was also the major contributor to heat-induced APX2 expression (Fig. 4A, Center). Under DBMIB treatment, HSFA1D again played the major role in APX2 expression and the contribution of HSFA2 and HSFA3 for DBMIB-induced APX2 expression was marginal (Fig. 4A, Right). In summary, HSFA1D is the major regulator of APX2 induction by multiple stresses, whereas the relative contribution of HSFA2 and HSFA3 to APX2 gene expression varied with the stress.
The contributions of the three HSFs to growth and development were assessed by comparing the triple-mutant to wild-type Col-0 in LL vs. EL (Fig. S4). Under a 12-h LL/12-h dark cycle, 3-wk-old hsfa1d hsfa2 hsfa3 plants were smaller than wild-type (Fig. S4A, LL) and flowered early (Fig. S4B). After 2 d of EL 12-h/Dark 12-h, we observed that emergent leaves of hsfa1d hsfa2 hsfa3 were yellow; however, over time, older leaves appeared to recover (Fig. S4A, EL).
Posttranslational Regulation of HSFA1D by EL.
HSFA1D transcription did not appear to be regulated by EL. Thus, we tested whether this protein is regulated posttranscriptionally in response to EL. The subcellular localization of HSFA1D under LL and EL was determined using transgenic lines expressing a HSFA1D-YFP-HA fusion protein in the A2PAPX2(-1507)/hsfa1d hafa2 hsfa3 background. We isolated 12 transgenic lines harboring the HSFA1D-YFP-HA construct (Fig. S5A). Each line had various amounts of the fusion protein, and the basal LUC activities under LL were relatively proportional to the amount of the fusion protein (Fig. S5 A and B). After exposure to 1 h of EL, all transgenic lines showed increased APX2 expression as measured by LUC activities (EL1h/LL) (Fig. S5C) but untransformed hsfa1d hsfa2 hsfa3 plants did not. These data indicate that the HSFA1D-YFP-HA fusion protein is biologically functional for EL-driven APX2 induction, and that HSFA1D could functionally replace HSFA2 and HSFA3 in APX2 induction under EL.
In transgenic line 24, in which basal LUC activity was similar to that in wild-type, HSFA1D-YFP-HA localized mainly in the cytoplasm (Fig. 5 A–C). In contrast, in transgenic line 14, which overexpressed the fusion protein and had the highest basal LUC activity in LL (Fig. S5 A and B), HSFA1D-YFP-HA was localized both in the cytoplasm and the nucleus (Fig. S5D). After 30 min of EL treatment, signals were observed exclusively in the nucleus in both lines (Fig. 5 D–F and Fig. S5E). Taken together, these data show that nuclear accumulation of HSFA1D is correlated with EL-driven APX2 expression. In addition to nuclear localization, activation of HSFA1D might be required to fully induce APX2 gene expression under EL, because LUC activity increased disproportionately in line 14 (Fig. S5C).
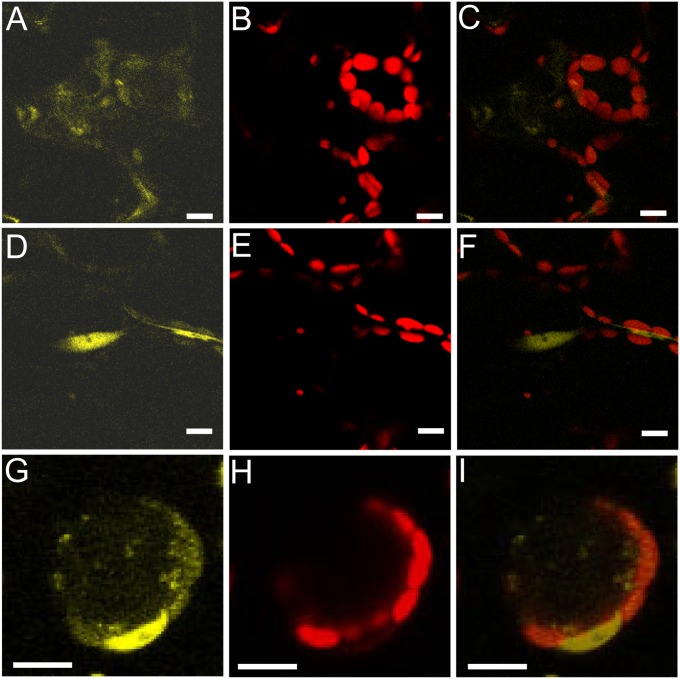
Fig. 5.
Translocation of HSFA1D from cytoplasm to the nucleus under EL. (A–C) Cytoplasmic localization of yellow fluorescence signals from the HSFA1D-YFP-HA fusion proteins in a LL condition. (D–I) After 30 min of EL treatment, the fluorescence signals from HSFA1D-YFP-HA were detected in the nucleus (D–F), and the fluorescence signals from hsfa1dC153S, C357S-YFP-HA (G–I) localized in cytoplasm and in the nucleus, with much higher signals in the nucleus (G–I). The merged images (C, F, and I) were generated by overlaying yellow fluorescence signal images (A, D, and G) and chlorophyll fluorescence images (B, E, and H) together. (Scale bars, 10 µm.)
In mammals, disulfide bond formation between three HSF monomers is important for its activity as a transcriptional regulator, presumably because of stabilization of the trimer (35, 36). To understand whether this function is conserved in Arabidopsis, we tested the effects of disulfide bonding in EL-driven APX2 expression. HSFA1D contains two cysteine residues at amino acid positions 153 and 357. These two cysteine residues were mutated to serines and then a construct expressing the mutant protein, hsfa1d C153S, C357S-YFP-HA (under the control of the HSFA1D promoter) was introduced to hsfa1d hsfa2 hsfa3. Four transgenic lines were isolated and their expression levels were determined. Protein levels of hsfa1dC153S, C357S-YFP-HA were comparable to those of HSFA1D-YFP-HA lines 14 and 24 (Fig. S5F); however, APX2 induction levels by EL in hsfa1dC153S, C357S-YFP-HA lines were significantly lower than HSFA1D-YFP-HA (Fig. 4B and Fig. S5G), indicating that disulfide bonding is critical for transcriptional activity of HSFA1D for EL-driven APX2 expression. Of note, Arabidopsis hsfa1dC153S, C357S-YFP-HA was still able to localize to the nucleus (Fig. 5 G–I), in contrast to the cysteine-to-serine mutant form of mammalian HSFs, which remain in the cytoplasm (35). Taken together, these data show that nuclear translocation of HSFA1D is coupled to its transcriptional activation in EL. These two processes then control the degree of APX2 induction.
Discussion
Our studies demonstrate a role for a subset of HSFs in rapid changes in nuclear gene expression in response to a redox-generated plastid stress signal. We have shown a role for three Arabidopsis HSFs—HSFA1D, HSFA2 and HSFA3—regulating APX2 expression in response to a change in redox generated by either EL or DBMIB, although HSFA6B may also have a minor role. HSFA1D plays a dominant role in the rapid response of plants to EL stress. In EL, preexisting HSFA1D accumulates in the nucleus, where it likely becomes activated by changes in disulfide bonding. Although no new protein synthesis is required for deployment of this rapid response, the response is likely to be sustained through the transcriptional activation of genes for HSFs and other transcription factors.
Multiple intracellular signaling pathways have been shown to play a role in the genomic response of plants to stress. Remarkably, these stress signals are almost always generated in chloroplasts, a highly active metabolic compartment where oxygen is generated during photosynthesis. Recent reports have identified several metabolic intermediates or degradation products whose accumulation signals changes in nuclear gene expression during EL stress, including: β-cyclocitral, a breakdown product of the carotenoid β-carotene (6); MEcPP, a precursor of isoprenoids (7); and PAP (5). Although all of these stresses were created after EL treatments of plants, it is not clear at this time how to compare conditions from laboratory to laboratory. For example, nuclear gene expression in response to β-cyclocitral accumulation shares about 80% common genes with genes whose expression is altered by the 1O2 signaling pathway (conditions in which APX2 may play no role). Similarly, signaling by MEcPP may be condition-specific. Contrary to the results of Xiao et al. (7), our global expression studies indicated that HPL1 gene expression is down-regulated (Dataset S1), rather than up-regulated. No obvious candidates exist for identification of the activator of APX2 gene expression in response to nuclear accumulation of PAP except for ZAT10, which is also up-regulated in PAP overaccumulators, such as altered expression of apx2 8 (5). In the case of PAP, nuclear localization of the phosphonucleotide can regulate the activity of exoribonuclease, thereby directly altering RNA pools; however, this is likely to indirectly affect APX2 and as yet it is unclear whether this pathway intersects with the HSF response.
Although there appears to be a number of signals that initiate EL stress signaling pathways, the transcription factors that rapidly regulate expression of hundreds of nuclear genes are mostly unknown. Previous studies reported that during heat stress, HSFA2 binds to the HSE2 element involved in APX2 induction by EL (29). In addition to heat, we showed that APX2 was induced by the reduced PQ pool in the absence of heat stress. APX2 induction by LED (Fig. 1A), fEL (Fig. 1A), and LL plus DBMIB (Figs. S1B and S2B) reflects that there are at least two pathways for APX2 expression: heat-dependent and heat-independent pathways.
It is also possible that these conditions generate a shared secondary signal that is transduced to the nucleus to regulate APX2 gene expression. A good candidate for this molecule is H2O2, which is generated during both heat and EL stress conditions (8, 37) (Fig. S1 C and E). However, the relationship between reduced PQ pool and H2O2 generation needs additional investigation because we could not—nor would we expect to—observe significant differences in the amount of H2O2 by our DBMIB treatment (Fig. S1C), although the DBMIB treatment could induce APX2 expression (Figs. S1B and S2B).
A model of the signaling network would predict that in conditions of fluctuating light intensity or prolonged high-intensity light, the PQ pool becomes reduced, and generates unidentified signaling molecules in chloroplasts (Fig. S6). The extent to which the different chloroplast signals (PAP, β-cycocitral, MEcPP) function in this HSF-mediated APX2 expression in parallel or independent pathways remains to be investigated. Indeed, given that there are substantial differences in the relative induction of APX2 under different experimental conditions (Fig. 1A), it is possible that multiple pathways function in its regulation. The unidentified molecule in this PQ-mediated pathway possibly facilitates HSFA1D movement to the nucleus. Disulfide bond formation among HSFA1Ds activates the HSF transcription complex to regulate genes such as APX2 and HSFA2. HSFA2 together with HSFA1D further increases APX2 transcription. If the EL condition is prolonged, newly synthesized unknown protein factors may induce expression of HSFA3 and other transcription factors, which in turn play a role in induction or repression of hundreds of nuclear genes involved in growth and metabolism. This transcription program may then protect the plant from numerous environmental stresses.
In conclusion, the identification of the gene networks involved in plants’ responses to stress is an area of considerable interest to plant biotechnologists. Our nascent studies identifying elements within the common set of EL and DBMIB up-regulated promoters indicate a convergence of plant responses to light and other abiotic stresses, including cold, dehydration, and heat at the level of transcriptional regulation, and may help to decipher this complex genetic program.
Materials and Methods
See Dataset S3 for a list of primers used in this study.
Plant Growth Conditions and Stress Treatments.
Arabidopsis plants were grown on a Linsmaier and Skoog medum (Caisson Laboratories) in growth chambers, or SALK Institute greenhouse number 3 (22 °C) for 3 to ∼4 wk before collection for the LUC assay or RNA extraction. Plant leaf tissue was incubated for at least 6 h in a LL growth chamber (light intensity was 65 µmol photons m−2⋅s−1, 22 °C) and then collected as an LL sample. The leaf tissue was exposed for the indicated time to a light intensity of 1,300 µmol photons m−2⋅s−1 (22 °C) (EL) or to 24 µM DBMIB (Aldrich) solution following 3 min of vacuum infiltration. hEL and fEL were generated by the metal halogen bulb lamp. For fEL, a perspex box containing a 2-cm height column of water and ice mixture was placed below the light source. The LED array system consists of nine individual white Luxeon-III star LEDs (Lumileds Lighting) controlled by current limiters and focusing lenses.
Leaf Disk Luciferase Assay.
Leaf disks were collected from rosette leaves using a 5-mm diameter cork borer. The leaf disks were floated on 2 mM luciferin dissolved in 0.002% (vol/vol) Triton X-100 solution in 96-well plates. LUC activities were measured using the GLOMAX luminometer (Promega) following a 2-min incubation in the dark condition to dissipate any possible luminescence from activated chlorophylls. To get baseline LUC activities, the leaf disks on 2 mM luciferin were incubated in a LL growth chamber for 6 h. Before the EL treatment, LL RLU was measured with a 4-s exposure per well, and immediately after the EL treatment, EL RLU was measured in the same way.
Supplementary Material
Acknowledgments
We thank Diep Ganguly (Australian National University) for technical assistance. These studies were supported by Grant FG02-04-ER15540 from the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, Office of Science, US Department of Energy, and the Howard Hughes Medical Institute (to J.C.); Australian Research Council Centre of Excellence in Plant Energy Biology Grant CE0561495; and Australian Grains Research and Development Corporation Scholarship GRS184 (to P.A.C.). Later stages of H.-S.J.’s work were supported by start-up funds from Dartmouth College.
Footnotes
The authors declare no conflict of interest.
Data deposition: The Affymetrix ATH1 CEL files reported in this paper has been deposited in the ArrayExpress, www.ebi.ac.uk/arrayexpress (accession no. E-MTAB-403).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1311632110/-/DCSupplemental.
References
- 1.Niyogi KK. Photoprotection revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50(1):333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 2.Barber J, Andersson B. Too much of a good thing: Light can be bad for photosynthesis. Trends Biochem Sci. 1992;17(2):61–66. doi: 10.1016/0968-0004(92)90503-2. [DOI] [PubMed] [Google Scholar]
- 3.Takahashi S, Murata N. How do environmental stresses accelerate photoinhibition? Trends Plant Sci. 2008;13(4):178–182. doi: 10.1016/j.tplants.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 5.Estavillo GM, et al. Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell. 2011;23(11):3992–4012. doi: 10.1105/tpc.111.091033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramel F, et al. Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc Natl Acad Sci USA. 2012;109(14):5535–5540. doi: 10.1073/pnas.1115982109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao Y, et al. Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell. 2012;149(7):1525–1535. doi: 10.1016/j.cell.2012.04.038. [DOI] [PubMed] [Google Scholar]
- 8.Apel K, Hirt H. Reactive oxygen species: Metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 9.Dietz K-J. Redox signal integration: From stimulus to networks and genes. Physiol Plant. 2008;133(3):459–468. doi: 10.1111/j.1399-3054.2008.01120.x. [DOI] [PubMed] [Google Scholar]
- 10.Pfalz J, et al. Environmental control of plant nuclear gene expression by chloroplast redox signals. Front. Plant Sci. 2012;3:257. doi: 10.3389/fpls.2012.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karpinski S, et al. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284(5414):654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- 12.Pogson BJ, Woo NS, Förster B, Small ID. Plastid signalling to the nucleus and beyond. Trends Plant Sci. 2008;13(11):602–609. doi: 10.1016/j.tplants.2008.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Rossel JB, Wilson IW, Pogson BJ. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 2002;130(3):1109–1120. doi: 10.1104/pp.005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trebst A. Inhibitors in electron flow: Tools for the functional and structural localization of carriers and energy conservation sites. Methods Enzymol. 1980;69:675–715. [Google Scholar]
- 15.Ball L, et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. Plant Cell. 2004;16(9):2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossel JB, et al. A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ. 2006;29(2):269–281. doi: 10.1111/j.1365-3040.2005.01419.x. [DOI] [PubMed] [Google Scholar]
- 17.Rossel JB, et al. Systemic and intracellular responses to photooxidative stress in Arabidopsis. Plant Cell. 2007;19(12):4091–4110. doi: 10.1105/tpc.106.045898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galvez-Valdivieso G, et al. The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell. 2009;21(7):2143–2162. doi: 10.1105/tpc.108.061507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfannschmidt T, Schütze K, Brost M, Oelmüller R. A novel mechanism of nuclear photosynthesis gene regulation by redox signals from the chloroplast during photosystem stoichiometry adjustment. J Biol Chem. 2001;276(39):36125–36130. doi: 10.1074/jbc.M105701200. [DOI] [PubMed] [Google Scholar]
- 20.Fey V, et al. Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J Biol Chem. 2005;280(7):5318–5328. doi: 10.1074/jbc.M406358200. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell K, Johnson GN. Chlorophyll fluorescence—A practical guide. J Exp Bot. 2000;51(345):659–668. doi: 10.1093/jxb/51.345.659. [DOI] [PubMed] [Google Scholar]
- 22.Gordon MJ, Carmody ME, Albrecht V, Pogson B. Systemic and local responses to repeated HL stress-induced retrograde signaling in Arabidopsis. Front. Plant Sci. 2013;3:303. doi: 10.3389/fpls.2012.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 24.Menkens A, Schindler U, Cashmore A. The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci. 1995;20(12):506–510. doi: 10.1016/s0968-0004(00)89118-5. [DOI] [PubMed] [Google Scholar]
- 25.Hudson ME, Quail PH. Identification of promoter motifs involved in the network of phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 2003;133(4):1605–1616. doi: 10.1104/pp.103.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simpson SD, et al. Two different novel cis-acting elements of erd1, a clpA homologous Arabidopsis gene function in induction by dehydration stress and dark-induced senescence. Plant J. 2003;33(2):259–270. doi: 10.1046/j.1365-313x.2003.01624.x. [DOI] [PubMed] [Google Scholar]
- 27.Jiang C, Iu B, Singh J. Requirement of a CCGAC cis-acting element for cold induction of the BN115 gene from winter Brassica napus. Plant Mol Biol. 1996;30(3):679–684. doi: 10.1007/BF00049344. [DOI] [PubMed] [Google Scholar]
- 28.Rieping M, Schoffl F. Synergistic effect of upstream sequences, CCAAT box elements, and HSE sequences for enhanced expression of chimaeric heat shock genes in transgenic tobacco. Mol Gen Genet. 1992;231(2):226–232. doi: 10.1007/BF00279795. [DOI] [PubMed] [Google Scholar]
- 29.Schramm F, et al. The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol. 2006;60(5):759–772. doi: 10.1007/s11103-005-5750-x. [DOI] [PubMed] [Google Scholar]
- 30.Sorger PK. Heat shock factor and the heat shock response. Cell. 1991;65(3):363–366. doi: 10.1016/0092-8674(91)90452-5. [DOI] [PubMed] [Google Scholar]
- 31.Nover L, et al. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need? Cell Stress Chaperones. 2001;6(3):177–189. doi: 10.1379/1466-1268(2001)006<0177:aathst>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishizawa A, et al. Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J. 2006;48(4):535–547. doi: 10.1111/j.1365-313X.2006.02889.x. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida T, et al. Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun. 2008;368(3):515–521. doi: 10.1016/j.bbrc.2008.01.134. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa D, Yamaguchi K, Nishiuchi T. High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased themotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot. 2007;58(12):3373–3383. doi: 10.1093/jxb/erm184. [DOI] [PubMed] [Google Scholar]
- 35.Ahn S-G, Thiele DJ. Redox regulation of mammalian heat shock factor 1 is essential for Hsp gene activation and protection from stress. Genes Dev. 2003;17(4):516–528. doi: 10.1101/gad.1044503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neef DW, Jaeger AM, Thiele DJ. Heat shock transcription factor 1 as a therapeutic target in neurodegenerative diseases. Nat Rev Drug Discov. 2011;10(12):930–944. doi: 10.1038/nrd3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Volkov RA, Panchuk II, Mullineaux PM, Schöffl F. Heat stress-induced H2O2 is required for effective expression of heat shock genes in Arabidopsis. Plant Mol Biol. 2006;61(4-5):733–746. doi: 10.1007/s11103-006-0045-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.