Abstract
Sterile alpha motif and HD-domain containing protein 1 (SAMHD1) is a triphosphohydrolase converting deoxynucleoside triphosphates (dNTPs) to deoxynucleosides. The enzyme was recently identified as a component of the human innate immune system that restricts HIV-1 infection by removing dNTPs required for viral DNA synthesis. SAMHD1 has deep evolutionary roots and is ubiquitous in human organs. Here we identify a general function of SAMHD1 in the regulation of dNTP pools in cultured human cells. The protein was nuclear and variably expressed during the cell cycle, maximally during quiescence and minimally during S-phase. Treatment of lung or skin fibroblasts with specific siRNAs resulted in the disappearence of SAMHD1 accompanied by loss of the cell-cycle regulation of dNTP pool sizes and dNTP imbalance. Cells accumulated in G1 phase with oversized pools and stopped growing. Following removal of the siRNA, the pools were normalized and cell growth restarted, but only after SAMHD1 had reappeared. In quiescent cultures SAMHD1 down-regulation leads to a marked expansion of dNTP pools. In all cases the largest effect was on dGTP, the preferred substrate of SAMHD1. Ribonucleotide reductase, responsible for the de novo synthesis of dNTPs, is a cytosolic enzyme maximally induced in S-phase cells. Thus, in mammalian cells the cell cycle regulation of the two main enzymes controlling dNTP pool sizes is adjusted to the requirements of DNA replication. Synthesis by the reductase peaks during S-phase, and catabolism by SAMHD1 is maximal during G1 phase when large dNTP pools would prevent cells from preparing for a new round of DNA replication.
Keywords: dNTP regulation, cell cycle arrest, dGTP pool
SAMHD1 was first described in the year 2000 as a component of the human innate immune system under the name dendritic cell–derived IFN-γ induced protein (DCIP) (1). DCIP mRNA was expressed constitutively in most human organs with particularly large signals in skeletal muscle and heart. Sequences of distantly related genes were found in a wide spectrum of Eubacteria and Archea as well as in Eukaryotes, from Nematodes to humans. The protein was later renamed SAMHD1 (2) when it was realized that its structure contains two previously known protein modules: an N-terminal short sterile-alpha (SAM) domain (3) and a central longer HD domain (4) with a conserved doublet of histidine (H) and aspartate (D). The SAM domain is a putative protein–protein and protein–nucleic acid interaction module, and the HD domain is found in diverse families of phosphohydrolases (4).
Mutations in SAMHD1 are responsible for Aicardi–Goutières syndrome, a genetic neurodegenerative disorder with a defective innate immune response (5). Furthermore, SAMHD1 serves as a restriction factor in HIV-1 infection (2). Other lentiviruses escape restriction by coinfection with a protein (Vpx) targeting SAMHD1 for ubiquitin-dependent degradation (6, 7).
Restriction required a functioning HD domain. Both the pure protein and a shorter fragment containing the HD domain were found to be an unusual triphosphohydrolase, degrading dNTPs to deoxynucleosides + triphosphate and thereby depleting the infected cells of the deoxynucleotides required for viral DNA synthesis (8–11).
A distantly related bacterial triphosphohydrolase was discovered in Escherichia coli more than 50 y ago (12). This enzyme is a dGTPase that hydrolyzes dGTP to deoxyguanosine + triphosphate. Other more recently discovered homologous microbial enzymes show a wider substrate specificity for dNTPs. Thus, the structure of an Enterococcus faecalis oligomeric enzyme able to hydrolyze all four canonical dNTPs (13) contained in addition to the substrate sites allosteric sites specific for dGTP. An enzyme from Thermus thermophilus also hydrolyzes all four dNTPs but only with dTTP + dATP as allosteric effectors (14).
The substrate specificity of mammalian SAMHD1 is similar to that of the E. faecalis dNTPase. Both enzymes have the capacity to hydrolyze all four dNTPs, but only in the presence of dGTP. The structural basis for the specificity has not yet been analyzed in detail. The hydrolysis of dNTPs by SAMHD1 and its regulation are conceptually related to the synthesis of deoxynucleotides and its regulation by ribonucleotide reductase (RNR) (15). In both cases oligomeric enzymes with the potential to operate with four separate substrates use dNTPs as allosteric effectors to direct their substrate specificity.
Imbalanced dNTP pools decrease the fidelity of DNA polymerases and increase mutation rates (16–19). Surprisingly, a surplus of dNTPs may create problems for DNA replication, both in E. coli (20) and in Eukaryotes (21). In Saccharomyces cerevisiae a large constitutive expansion of the dNTP pools led to a block in the G1 phase of the cell cycle (22). Also correct proportions are important. Specific substitutions of amino acids in the allosteric site of yeast RNR affecting its substrate specificity changed the relative proportions of the dNTP pools and the mutation pattern of the cells (19).
Our laboratory has as long-standing interest in the enzymes that supply and regulate dNTP pools. Mammalian cells contain two distinct pathways for dNTP synthesis: (i) in the cytosol, RNR catalyzes deoxynucleotide de novo synthesis, and (ii) in the cytosol and mitochondria, deoxynucleoside kinases phosphorylate deoxynucleosides to their 5′-phosphates that are further phosphorylated to dNTPs. RNR and several other synthetic enzymes are cell cycle regulated, with highest activity in S-phase. Catabolic 5′-nucleotidases in the cytosol and in mitochondria oppose the synthetic deoxynucleoside kinases fine-tuning the intracellular concentrations of deoxyribonucleoside 5′-phosphates. Additional catabolic enzymes (deaminases and phosphorylases) contribute to the establishment of final pool levels by removing deoxynucleosides from the cycle. Thus, a combination of interlocked synthetic and catabolic enzymes sets the intracellular concentrations of each dNTP (23).
The occurrence of a gene coding for SAMHD1-related triphosphohydrolases in a large variety of bacteria and Archea suggests a deep evolutionary origin with a wider function for the enzyme than to be a guardian against viral infections. Here we describe the presence of SAMHD1 in the nuclei of human lung and skin fibroblasts, with lowest abundance during S-phase and a large increase of the protein in quiescent cells. Down-regulation of SAMHD1 by siRNAs increased intracellular dNTP pools, in particular dGTP, leading to changes in the proportions of the four pools. In proliferating cells the treatment interfered with the normal cell-cycle–related regulation of dNTP pool sizes and caused a delay of the cell cycle in the G1 phase. Pool expansion depended on RNR activity, as quiescent mutant skin fibroblasts with an inactive p53R2 subunit of RNR were deficient in accumulating dNTPs. We propose that SAMHD1 is a unique participant in the enzymatic network regulating cellular dNTP concentrations and prevents overproduction of dNTPs, exerting its activity mostly outside S-phase.
Results
Expression of SAMHD1 in Cultured Cells.
We found that SAMHD1 protein and mRNA were present in extracts from a variety of human cell lines (Fig. 1A). Monocytic THP1 cells had the highest SAMHD1 content, and Jurkat T cells were negative. Nontransformed lung and skin fibroblasts showed an intermediate expression of SAMHD1, and we chose these cells for our experiments. In proliferating cell extracts, SAMHD1 appeared as a doublet, possibly due to posttranslational modification (Fig. 1B). The concentration of the protein changed considerably with cell proliferation, with confluent and quiescent fibroblasts containing much more SAMHD1 than cycling cells. By fluorescence microscopy we detected in agreement with earlier work (5) SAMHD1 exclusively in the cell nucleus (Fig. 1C), in contrast to the known cytoplasmic location of proteins R1 and R2, the two subunits of RNR catalyzing the synthesis of dNTPs de novo (24, 25). The fluorescent signal of SAMHD1 was present in all cells, but its intensity varied markedly, suggesting different concentrations of the protein in dependence of the cell position along the cell cycle (Fig. 1C). Comparing the abundance of R2 and SAMHD1 in a same culture (Fig. 1C), we found that cells with a strong SAMHD1 signal were negative for R2, whereas cells with a strong R2 signal had only weak SAMHD1 fluorescence. The two proteins not only were present in separate cellular compartments but also expressed differently during the cell cycle. RNR is specifically induced during S-phase to supply dNTPs for nuclear DNA replication (26), but its R2 subunit is degraded after completion of DNA replication and absent from quiescent cells (27, 28). The fluorescence data, as well as the results shown in Fig. lB, suggest that the largest expression of SAMHD1 instead takes place outside S-phase and the enzyme degrades dNTPs mostly when DNA replication does not occur. Indeed, in cycling cultures with varying amounts of S-phase cells, we found a clear inverse relation between the concentration of SAMHD1 and the frequency of S-phase cells (Fig. 1D). Moreover, in cultures of quiescent lung fibroblasts, all nuclei had a strong SAMHD1 signal (Fig. 1C), and immunoblotting analysis showed that SAMHD1 progressively accumulates in the cultures during serum starvation (Fig. 1E).
Fig. 1.
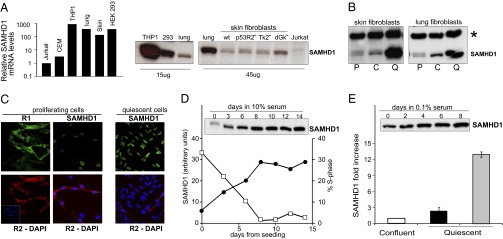
Expression of SAMHD1 in cultured human cells. (A) Relative levels of SAMHD1 mRNA and protein in different cell lines. The relative amount of mRNA was measured by RT real-time PCR in transformed cell lines (THP1, HEK293, and Jurkat) and in nontransformed lung and skin fibroblasts. SAMHD1 protein was detected by immunoblotting in extracts from transformed cells, lung and wild-type skin fibroblasts, and skin fibroblasts mutated for p53R2 or the mitochondrial thymidine (TK2) or deoxyguanosine (dGK) kinases. (B) Immunoblots of SAMHD1 from proliferating (P), confluent (C), and quiescent (Q) WT skin and lung fibroblasts. Asterisk marks an unspecific band (loading control). (C) Immunofluorescence shows nuclear localization of SAMHD1 (green) and cytosolic localization of the R1 (green) and R2 (red) subunits of RNR in lung fibroblasts. (D) Inverse relation between frequency of S-phase cells (filled circle) and abundance of SAMHD1 protein (open square) in proliferating cultures of lung fibroblasts. (E) Content of SAMHD1 in quiescent lung (black) and skin (gray) fibroblasts relative to the amount at confluency. The immunoblot shows the increasing SAMHD1 signal in lung fibroblasts.
siRNA Silencing of SAMHD1 in Proliferating Cells Inhibits Cell Cycle Progression and the Cell-Cycle–Dependent Regulation of dNTP Pools.
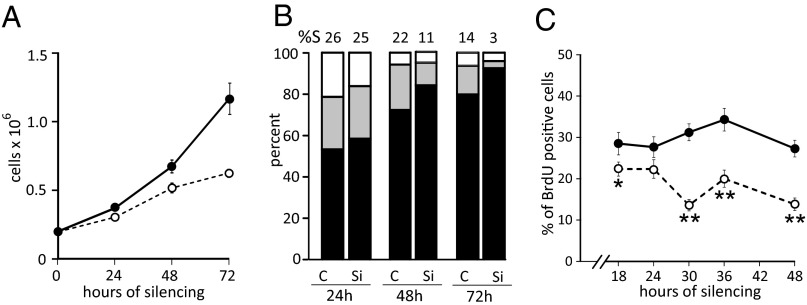
Transfection of cycling lung (Fig. 2) or skin fibroblasts (Fig. S1) with siRNAs directed against SAMHD1 effectively depleted the cells of the mRNA in 24 h, whereas the protein declined more gradually. When after 48 h we transferred the silenced cells to new plates in siRNA-free medium, the mRNA reappeared after a lag of several days and the protein still later (Fig. 2 and Fig. S1). During siRNA transfection the SAMHD1-silenced cultures grew more slowly than the nonsilenced controls, became growth-arrested at a lower density, and contained fewer S-phase cells with a concomitant increase of G1 cells (Fig. 3 A and B and Fig. S2 A and B). To follow more closely the alterations of the cell cycle produced by the decline of SAMHD1, we pulsed cycling lung fibroblasts for 30 min with BrdU at different time points between 18 and 48 h of transfection with siRNA and determined the percentages of BrdU-positive cells by immunofluorescence (Fig. 3C). The frequency of positive cells in the control cultures remained at around 30% at all time points. Instead, a small but significant decrease of BrdU-positive S-phase cells was already apparent after 18 h of silencing and became stronger after 30 h, when the silenced cultures contained only half as many S-phase cells as the controls. Even an incomplete down-regulation of SAMHD1 (Fig. 2) affected the progression of the cell cycle in nontransformed fibroblasts.
Fig. 2.
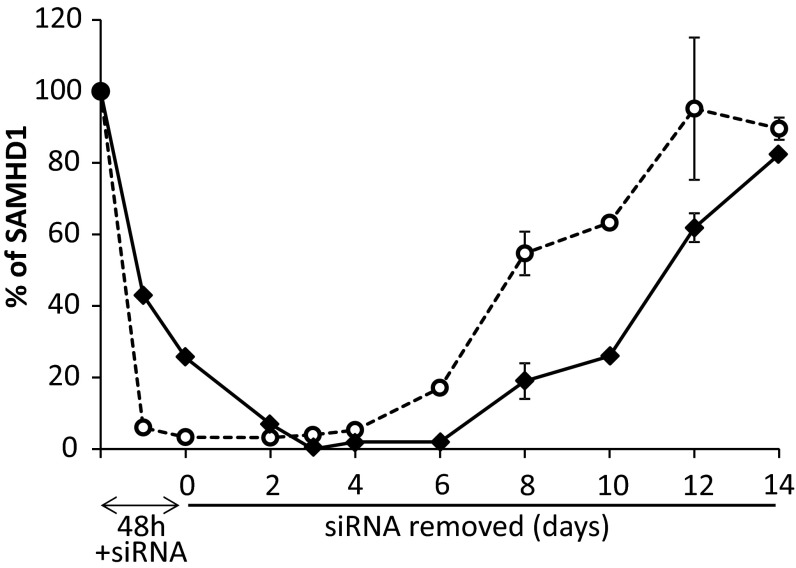
Decline and recovery of SAMHD1 mRNA and protein during siRNA transfection and subsequent removal of siRNA. Proliferating cultures of lung fibroblasts were transfected for 48 h with anti-SAMHD1 siRNA 24 h after seeding. Cells were then replated in siRNA-free medium and grown for 14 d with medium changes every 3–4 d. SAMHD1 mRNA (open circle) and protein (filled diamond) were measured at the indicated times. Bars indicate range of values in two similar experiments.
Fig. 3.
Effect of SAMHD1 silencing on the growth of lung fibroblasts. (A) Cell growth in cultures transfected for 24–72 h with anti-SAMHD1 (open circle) or control (filled circle) siRNAs. Data from three experiments. Bars are SEs. (B) Percent of G1 (black), S (gray), and G2/M (white) cells in transfected cultures. The frequency of S-phase cells is indicated above each control (C) and silenced (Si) sample. (C) Frequency of BrdU-positive cells in cultures transfected with control (filled circle) or anti-SAMHD1 (open circle) siRNAs for 18–48 h and incubated with BrdU during the last 30 min. Data are from two identical experiments. At least 500 cells were scored for each time point. The frequencies were compared by 2 × 2 χ2 analysis. *P < 0.05; **P < 0.001. Bars are SEs.
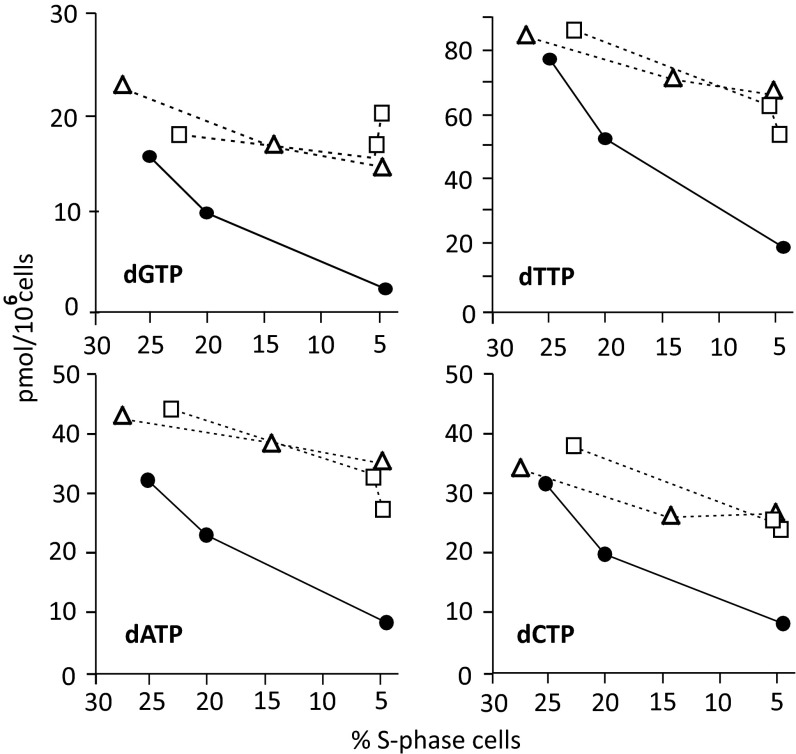
In proliferating cell populations, the concentrations of the four dNTPs depend on the frequency of S-phase cells in which ribonucleotide reduction is strongly induced (26). During the silencing experiments, the growing control cultures became progressively more crowded and their percentage of S-phase cells declined (Fig. 3 A and B). Accordingly, also the size of the dNTP pools declined, with dGTP always representing the smallest pool (Fig. 4). In SAMHD1-silenced cultures S-phase cells decreased faster than in the controls (Fig. 3B), but the dNTP pools did not show a corresponding decrease. In Fig. 4 we transfected lung fibroblasts with two different anti-SAMHD1 siRNAs or control siRNA and during silencing related pool sizes to the frequency of S-phase cells, which reached its lowest value after 72 h of transfection. In the silenced cultures the progressive loss of SAMHD1 stabilized the pools, interfering with their normal cell-cycle–dependent down-regulation outside S-phase (Fig. 4). Similar results were obtained by silencing SAMHD1 in proliferating WT skin fibroblasts (Fig. S2C). The ratios between the dNTP pools of silenced and control cells increased (Fig. 5A and Fig. S2D) because of the slower decay of the dNTPs in the absence of SAMHD1 rather than of a real accumulation of dNTPs. A different situation was observed when SAMHD1 was silenced in quiescent cells.
Fig. 4.
Effect of SAMHD1 silencing on dNTP pools of cycling lung fibroblasts. Relation between dNTP pool sizes and loss of S-phase cells in cycling cultures during transfection with two separate anti-SAMHD1 siRNAs (open square, open triangle) or with control siRNA (closed circle).
Fig. 5.
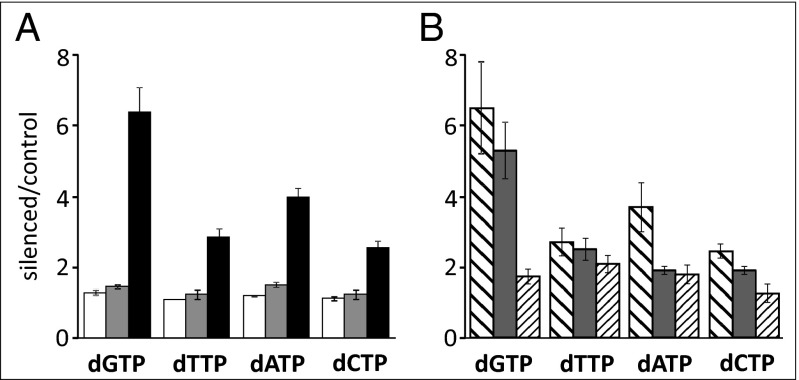
Ratios between pool sizes of SAMHD1-silenced and control lung fibroblasts. (A) Proliferating cultures transfected for 24 (white), 48 (gray), or 72 (black) h with control or anti-SAMHD1 siRNAs. Data are from three independent experiments. (B) Quiescent cultures of lung (broad stripes), WT (gray), or p53R2-mutated (thin stripes) skin fibroblasts transfected for 10 d with siRNAs during serum starvation. Data are from four to six experiments per cell line. All bars are SEMs.
Silencing of SAMHD1 in Quiescent Fibroblasts Increases dNTP Pool Sizes.
The above experiments show results of SAMHD1 depletion in cycling cultures containing cells at various stages of the cell cycle. We now turn to results from more homogeneous cultures of SAMHD1-silenced and control lung fibroblasts, wild-type skin fibroblasts, and mutant skin fibroblasts devoid of p53R2 activity (29), made quiescent by serum starvation. These cultures contained less than 3% S-phase cells and after transfection with SAMHD1 siRNAs retained less than 10% mRNA and only minimal SAMHD1 protein. In four to six independent similar experiments, per fibroblast line, we measured the four dNTP pools (Table S1) and calculated the increases in pool sizes caused by silencing (Fig. 5B). In lung and wild-type skin fibroblasts, the loss of SAMHD1 increased all four pools, in particular the dGTP pool. In the p53R2 mutant fibroblasts, the increases relative to the WT skin fibroblasts were smaller, with a highly significant difference for dGTP (P < 0.01), less significant for dCTP (P < 0.05), and not significant for dATP and dTTP. As protein R2 is degraded during the attainment of quiescence, dNTP synthesis depended on p53R2 activity, defective in the mutant cells. The inability of the mutant fibroblasts to accumulate dNTPs in the absence of SAMHD1 reflects the key role of p53R2-dependent ribonucleotide reduction for the synthesis of dNTPs during quiescence (30, 31). The expansion of the pools in the silenced fibroblasts with an active p53R2 enzyme demonstrates the involvement of SAMHD1 in a continuous turnover of dNTP pools also in the absence of DNA replication.
Recovery of SAMHD1 After siRNA Silencing.
How fast do SAMHD1-silenced fibroblasts regain balanced dNTP pools and normal growth after removal of the siRNA? First we transfected cultures of wild-type lung or skin fibroblasts with SAMHD1 or control siRNAs for 48 h (inhibition period). We then replated the cells at low density in fresh medium lacking siRNAs and continued the incubation for 12–14 d, during which time the medium was changed every 3–4 d (release period). During the inhibition period, the effects of SAMHD1 down-regulation on cell growth, cell cycle progression, and the dNTP pools were those described above (Figs. 3 and 4 and Fig. S2).
During the release period the control lung fibroblasts restarted their growth, with the highest percentage of S-phase cells immediately after seeding. In Fig. 6 A and B we have combined the results of two experiments run with the same basic protocol, in which the cultures were analyzed at partly different time points. In these experiments the growth of the silenced cells remained strongly inhibited for 4–6 d (Fig. 6A), with a peak of S-phase cells appearing around 6 d from the start of the recovery period (Fig. 6B). The silencing caused a similar growth delay in skin fibroblasts, but they grew at a slower rate and their final growth was lower (Fig. S3). They also presented a small peak of S-phase cells after 6 d (Fig. S3B). Both cell lines had the capacity to resume growth after removal of siRNA, but their recovery only occurred after an extended lag phase.
Fig. 6.
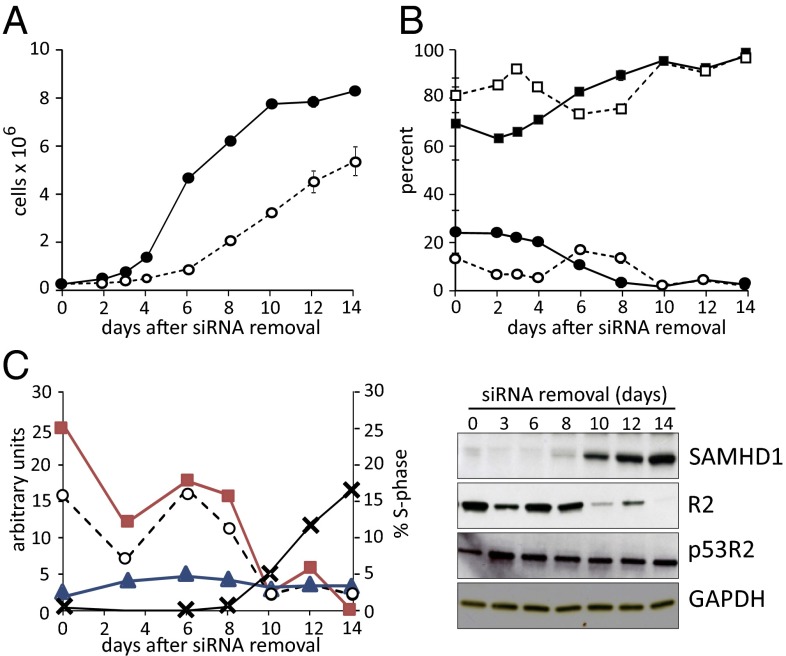
Slow recovery of growth and cell cycle progression in lung fibroblasts after SAMHD1-silencing. (A) Cell growth of control (filled circle) and SAMHD1-silenced (open circle) lung fibroblasts replated in siRNA-free medium after 48 h transfections. (B) Frequencies of G1- (squares) and S-phase (circles) cells in control (filled square, filled circle) and silenced (open square, open circle) cultures. Bars in A and B show range of values at identical time points in two independent experiments. (C) Abundance of SAMHD1 (x), R2 (filled square), and p53R2 (filled triangle) at the indicated days of culture without siRNA. Left reports in aribitrary units the quantification of the immunoblots on the right. Notice the parallelism between R2 expression and frequency of S-phase cells (open circle) in the cultures.
The long lag period paralleled the slow recovery of SAMHD1 mRNA and protein after the shift to siRNA-free conditions (Fig. 2 and Fig. S1) and was probably due to the stability of the siRNA internalized by the cells during the 48 h transfection. We followed the reappearance of SAMHD1 in lung fibroblasts by Western blotting and at the same time measured the two small subunits of RNR, i.e., proteins R2 and p53R2 (Fig. 6C). The pattern of R2 variations coincided with that of S-phase cells, whereas p53R2 was more stable during the whole release period.
We determined the effect of the preceding siRNA treatment on the size of the dNTP pools during the release period in both lung and skin fibroblasts (Fig. S4 A and B). In both cases there was a nearly identical dramatic pool increase in all four pools, with a peak at the third to sixth day of recovery, when the cells contained some SAMHD1 mRNA but SAMHD1 protein had not yet returned. The dNTP peaks coincided in time with a small peak of S-phase cells.
The two cell lines showed highly similar behavior both for the timing of pool changes and for their increases. We therefore combined the results from Fig. S4 A and B and divided the average pool sizes of inhibited and control cells to calculate the increase in size for each pool (Fig. 7). Most impressive is the peak value for dGTP after 6 d. At that point dGTP accounted for more than 20% of the total cellular pools, more than twice its percentage in a cycling cell population. When SAMHD1 at later times became available, the pool values normalized and cell growth resumed. These results clearly demonstrate that SAMHD1 is not only required for the maintenance of normal-sized dNTP pools but also for their normal proportions.
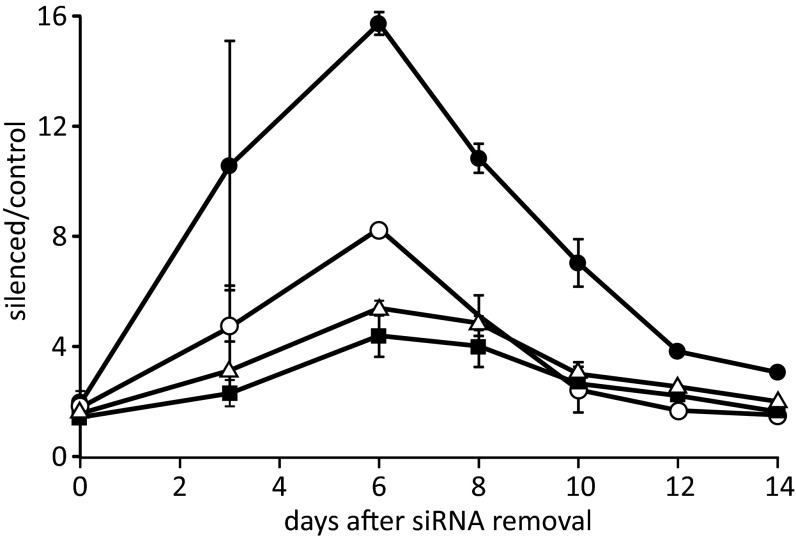
Fig. 7.
Persistence of large dNTP pools in lung and skin fibroblasts after siRNA silencing of SAMHD1. Mean ratios between sizes of dGTP (filled circle), dCTP (open circle), dTTP (filled square), and dATP (open triangle) pools in silenced and control cells during 14 d without siRNA. Data from experiments with lung fibroblasts (Fig. 6 and Fig. S4A) and skin fibroblasts (Fig. S3 and Fig. S4B). Bars show range of values.
Discussion
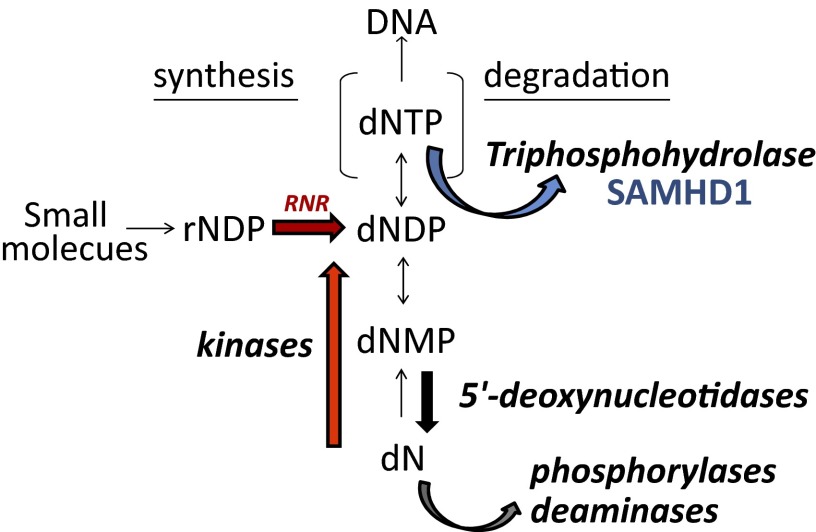
The main interest for SAMHD1 has so far been concentrated on the mechanism of its antilentiviral action. To us its presence in most mammalian organs and in many cell lines suggested a more general role in the regulation of DNA precursors in mammalian cells. We have previously shown how in these cells the balance of dNTP pools results from the interplay of the synthetic and catabolic activities summarized in Fig. 8 and demonstrated the regulatory function of substrate cycles between deoxynucleoside kinases and 5′-deoxynucleotidases (32, 33). We now postulate that SAMHD1 provides an additional unique level of catabolic intervention. The enzyme removes in the nucleus a potentially dangerous surplus of DNA precursors during the G1 phase of the cell cycle by their total dephosphorylation. The cell-cycle–related regulation of the enzyme minimizes its activity during S-phase when large quantities of dNTPs are required for DNA replication. These are provided from the cytosol mainly by RNR. The cell cycle regulation of RNR is opposite to that of SAMHD1 and provides maximal activity during S phase.
Fig. 8.
The pools of the dNTPs required for DNA replication are regulated by synthetic and catabolic enzymes. Synthesis occurs by (i) de novo synthesis by RNR in the cytosol and (ii) salvage of deoxynucleosides by deoxynucleoside kinases in the cytosol and mitochondria. A stepwise catabolism of dNTPs to deoxynucleosides occurs with the final intervention of 5′-deoxynucleotidases. Phosphorylases and deaminases further degrade deoxynucleosides in the cytosol. The triphosphohydrolase SAMHD1 is located in the nucleus and degrades dNTPs directly to deoxynucleosides.
We obtained evidence for this concept by investigating how SAMHD1 participates in the regulation of cellular dNTP pools in nontransformed human fibroblasts that retain a normal control of cell cycle progression. When we down-regulated SAMHD1 expression by RNA interference in proliferating fibroblasts, we detected an early disturbance of the G1/S transition, with progressive accumulation of cells in G1 and reduction of S-phase cells, resulting in slower cell growth (Fig. 3). The decline of S-phase cells was not accompanied by a parallel decline of dNTP pool sizes, in marked contrast to the control cells with normal SAMHD1 expression (Fig. 4). A G1 delay produced by constitutively high dNTP pools was reported earlier in budding yeasts and was attributed to a disturbance in the loading of Cdc45, a component of preinitiation complexes at the DNA replication origins (22). In proliferating fibroblasts, SAMHD1 silencing produced a slightly lower but comparable expansion of dNTP pools relative to the controls only after 72 h (Fig. 5A), yet a decrease in the frequency of S-phase cells started to appear when the total cellular pools were hardly changed (Fig. 3 B and C). In contrast to the yeast experiments, our experiments were done with unsynchronized cell populations and we could not measure the pool alterations occurring specifically in G1 cells. Nevertheless, it appears likely that the down-regulation of SAMHD1 affected the small G1 pools at an early stage and raised them enough to trigger the G1 arrest, by a mechanism at present still undefined.
When cells enter the S phase, ribonucleotide reduction is induced to synthesize the dNTPs required for nuclear DNA replication (26). Here the cellular content of the S-phase–specific R2 subunit of RNR changed in parallel with the frequency of S-phase cells (Fig. 6C). Remarkably, SAMHD1 variations showed the opposite behavior, increasing as S-phase cells declined and accumulating in quiescent cultures (Fig. 1). This difference suggests that the enzyme has the specific role of keeping the pools low when the cells are not replicating their nuclear DNA. Indeed, when SAMHD1 was silenced in cells made quiescent by serum starvation, the pools underwent large actual increases (Fig. 5B and Table S1), revealing that the enzyme normally curbs the pools of nonproliferating cells. The loss of SAMHD1 not only enlarged the sizes of the four dNTP pools but also modified their relative proportions, more than doubling the percentage of dGTP compared with the original pool composition. A similar albeit smaller effect on the relative ratios of pool sizes was produced when SAMHD1 was silenced in proliferating cells (Fig. 5A).
It is interesting that both in growing and resting cells the main change caused by SAMHD1 knockdown concerns the dGTP pool, which is normally the smallest pool. These results reflect the preference of SAMHD1 for dGTP as a substrate and suggest that the small dGTP pool size may not be due to a low dGTP synthesis by ribonucleotide reduction but to a more efficient catabolism by SAMHD1. Considering that dGTP acts as an allosteric effector of both RNR and SAMHD1, the restraint operated by the latter enzyme on dGTP concentration may have interesting and still unidentified regulatory implications for both enzymes.
The data available so far suggest that the extra-S phase activity of SAMHD1 fulfils two main functions: (i) during cell proliferation it maintains the G1 pools at the correct level for a regular transition into S, possibly through the normal assembly of the preinitiation complexes, and (ii) by lowering the dNTP pools, it deprives invading viruses of the necessary DNA precursors. Already in the early 1990s, the concept that the cell-cycle–related variations of dNTP pools may be a defensive adaptation against viral infections had been formulated by McIntosh (34). The recent discoveries on the antilentiviral activity of SAMHD1 directly confirm that earlier suggestion. A further role of SAMHD1 might be the control of dNTP pool sizes in the cells of multicellular animals that undergo cyclic fluctuations in response to sudden peaks of deoxynucleoside concentrations in the blood in connection with food intake.
A regulated dNTP pool outside S phase is important for the fidelity of DNA repair and mitochondrial DNA replication. The destabilizing effects of imbalanced dNTP pools on the nuclear and mitochondrial genomes are well known and exemplified by severe human diseases linked to genetic deficiencies of catabolic enzymes (35–37). The mechanism underlying the phenotype of the Acardi–Goutières syndrome caused by SAMHD1 mutations is still unknown, but the occasional occurrence of mitochondrial DNA deletions might be linked to dNTP pool abnormalities. The recently reported specific influence of the dGTP pool on telomere length homeostasis (38) further emphasizes the importance of keeping dGTP under control.
Recent studies have shown that methylation of the SAMHD1 promoter regulates gene expression in human T cells (39) and that the protein is phosphorylated by the cyclin A2/CDK1 complex in cycling cells (40, 41). The doublet SAMHD1 signal observed in immunoblots (Fig. 1B) may depend on the latter phenomenon. The inverse relation between SAMHD1 abundance and frequency of S-phase cells, together with the variable levels of SAMHD1 in different cells and tissues (1) (Fig. 1), is likely the result of these and other still unknown regulatory mechanisms. In contrast to SAMHD1, the 5′-nucleotidases contributing to the balance of dNTP pools are constitutively expressed proteins whose experimental down-regulation does not induce significant alterations of dNTP pool sizes (33, 42). SAMHD1 is clearly a major regulator of dNTP pool turnover, whose loss has large effects on the composition of the cellular dNTP pool both during quiescence and proliferation.
Materials and Methods
Cell Lines and Cell Growth.
Three human control skin fibroblast lines and one line derived from a patient with an inactivating mutation in RRM2B, the gene for p53R2, used by us previously (29, 43) were immortalized with human telomerase coded by plasmid CMV–hTERT/PGK–Puro as described (44). An established line of lung fibroblasts (CCD34Lu) was from the American Type Culture Collection. We cultured skin fibroblasts in minimal Eagle’s medium (MEM), 10% (vol/vol) FCS + nonessential amino acids, and lung fibroblasts in Dulbecco’s MEM (DMEM) with 4.5 g glucose per liter + 10% (vol/vol) FCS + nonessential amino acids + 20 mM Hepes buffer pH 7.4. We determined the cell cycle distribution by flow cytometry after propidium iodide staining with a BD FACSCanto II Flow cytometer (BD Biosciences).
Methods.
Details of other methodologies used in this study, including sources of materials, transfection protocols, determination of dNTPs, quantification of mRNA by RT real-time PCR, immunoblotting, and immunofluorescence, are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Cristiano Salata for help with the immortalization of some fibroblast lines. This work was supported by Italian Telethon Grant GGP09019 (to V.B.), Italian Association for Cancer Research Grant 1091 (to V.B.), and the University of Padova, Strategic Project 2008 “Models of Mitochondrial Diseases.”
Footnotes
The authors declare no conflict of interest.
See Commentary on page 14120.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1312033110/-/DCSupplemental.
References
- 1.Li N, Zhang W, Cao X. Identification of human homologue of mouse IFN-gamma induced protein from human dendritic cells. Immunol Lett. 2000;74(3):221–224. doi: 10.1016/s0165-2478(00)00276-5. [DOI] [PubMed] [Google Scholar]
- 2.Laguette N, et al. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature. 2011;474(7353):654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qiao F, Bowie JU. The many faces of SAM. Sci STKE. 2005;2005(286):re7. doi: 10.1126/stke.2862005re7. [DOI] [PubMed] [Google Scholar]
- 4.Aravind L, Koonin EV. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem Sci. 1998;23(12):469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 5.Rice GI, et al. Mutations involved in Aicardi-Goutières syndrome implicate SAMHD1 as regulator of the innate immune response. Nat Genet. 2009;41(7):829–832. doi: 10.1038/ng.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hrecka K, et al. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature. 2011;474(7353):658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schaller T, Goujon C, Malim MH. AIDS/HIV. HIV interplay with SAMHD1. Science. 2012;335(6074):1313–1314. doi: 10.1126/science.1221057. [DOI] [PubMed] [Google Scholar]
- 8.Goldstone DC, et al. HIV-1 restriction factor SAMHD1 is a deoxynucleoside triphosphate triphosphohydrolase. Nature. 2011;480(7377):379–382. doi: 10.1038/nature10623. [DOI] [PubMed] [Google Scholar]
- 9.Powell RD, Holland PJ, Hollis T, Perrino FW. Aicardi-Goutieres syndrome gene and HIV-1 restriction factor SAMHD1 is a dGTP-regulated deoxynucleotide triphosphohydrolase. J Biol Chem. 2011;286(51):43596–43600. doi: 10.1074/jbc.C111.317628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahouassa H, et al. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol. 2012;13(3):223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim B, Nguyen LA, Daddacha W, Hollenbaugh JA. Tight interplay among SAMHD1 protein level, cellular dNTP levels, and HIV-1 proviral DNA synthesis kinetics in human primary monocyte-derived macrophages. J Biol Chem. 2012;287(26):21570–21574. doi: 10.1074/jbc.C112.374843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kornberg SR, Lehman IR, Bessman MJ, Simms ES, Kornberg A. Enzymatic cleavage of deoxyguanosine triphosphate to deoxyguanosine and tripolyphosphate. J Biol Chem. 1958;233(1):159–162. [PubMed] [Google Scholar]
- 13.Vorontsov II, et al. Characterization of the deoxynucleotide triphosphate triphosphohydrolase (dNTPase) activity of the EF1143 protein from Enterococcus faecalis and crystal structure of the activator-substrate complex. J Biol Chem. 2011;286(38):33158–33166. doi: 10.1074/jbc.M111.250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo N, Kuramitsu S, Masui R. Biochemical characterization of TT1383 from Thermus thermophilus identifies a novel dNTP triphosphohydrolase activity stimulated by dATP and dTTP. J Biochem. 2004;136(2):221–231. doi: 10.1093/jb/mvh115. [DOI] [PubMed] [Google Scholar]
- 15.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 16.Meuth M, L’Heureux-Huard N, Trudel M. Characterization of a mutator gene in Chinese hamster ovary cells. Proc Natl Acad Sci USA. 1979;76(12):6505–6509. doi: 10.1073/pnas.76.12.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg G, Ullman B, Martin DW., Jr Mutator phenotypes in mammalian cell mutants with distinct biochemical defects and abnormal deoxyribonucleoside triphosphate pools. Proc Natl Acad Sci USA. 1981;78(4):2447–2451. doi: 10.1073/pnas.78.4.2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kunkel TA. DNA replication fidelity. J Biol Chem. 1992;267(26):18251–18254. [PubMed] [Google Scholar]
- 19.Kumar D, et al. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39(4):1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. Proc Natl Acad Sci USA. 2011;108(48):19311–19316. doi: 10.1073/pnas.1113664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davidson MB, et al. Endogenous DNA replication stress results in expansion of dNTP pools and a mutator phenotype. EMBO J. 2012;31(4):895–907. doi: 10.1038/emboj.2011.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chabes A, Stillman B. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2007;104(4):1183–1188. doi: 10.1073/pnas.0610585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rampazzo C, et al. Regulation by degradation, a cellular defense against deoxyribonucleotide pool imbalances. Mutat Res. 2010;703(1):2–10. doi: 10.1016/j.mrgentox.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Engström Y, Rozell B, Hansson HA, Stemme S, Thelander L. Localization of ribonucleotide reductase in mammalian cells. EMBO J. 1984;3(4):863–867. doi: 10.1002/j.1460-2075.1984.tb01897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engström Y, Rozell B. Immunocytochemical evidence for the cytoplasmic localization and differential expression during the cell cycle of the M1 and M2 subunits of mammalian ribonucleotide reductase. EMBO J. 1988;7(6):1615–1620. doi: 10.1002/j.1460-2075.1988.tb02987.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 27.Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA. 2003;100(7):3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Angiolella V, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149(5):1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pontarin G, et al. Deoxyribonucleotide metabolism in cycling and resting human fibroblasts with a missense mutation in p53R2, a subunit of ribonucleotide reductase. J Biol Chem. 2011;286(13):11132–11140. doi: 10.1074/jbc.M110.202283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Håkansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem. 2006;281(12):7834–7841. doi: 10.1074/jbc.M512894200. [DOI] [PubMed] [Google Scholar]
- 31.Pontarin G, et al. p53R2-dependent ribonucleotide reduction provides deoxyribonucleotides in quiescent human fibroblasts in the absence of induced DNA damage. J Biol Chem. 2007;282(23):16820–16828. doi: 10.1074/jbc.M701310200. [DOI] [PubMed] [Google Scholar]
- 32.Gazziola C, Ferraro P, Moras M, Reichard P, Bianchi V. Cytosolic high K(m) 5′-nucleotidase and 5′(3′)-deoxyribonucleotidase in substrate cycles involved in nucleotide metabolism. J Biol Chem. 2001;276(9):6185–6190. doi: 10.1074/jbc.M007623200. [DOI] [PubMed] [Google Scholar]
- 33.Rampazzo C, et al. Mitochondrial deoxyribonucleotides, pool sizes, synthesis, and regulation. J Biol Chem. 2004;279(17):17019–17026. doi: 10.1074/jbc.M313957200. [DOI] [PubMed] [Google Scholar]
- 34.McIntosh EM. MCB elements and the regulation of DNA replication genes in yeast. Curr Genet. 1993;24(3):185–192. doi: 10.1007/BF00351790. [DOI] [PubMed] [Google Scholar]
- 35.Nishino I, Spinazzola A, Hirano M. Thymidine phosphorylase gene mutations in MNGIE, a human mitochondrial disorder. Science. 1999;283(5402):689–692. doi: 10.1126/science.283.5402.689. [DOI] [PubMed] [Google Scholar]
- 36.Daddona PE, et al. Human adenosine deaminase. cDNA and complete primary amino acid sequence. J Biol Chem. 1984;259(19):12101–12106. [PubMed] [Google Scholar]
- 37.Markert ML. Purine nucleoside phosphorylase deficiency. Immunodefic Rev. 1991;3(1):45–81. [PubMed] [Google Scholar]
- 38.Gupta A, et al. Telomere length homeostasis responds to changes in intracellular dNTP pools. Genetics. 2013;193(4):1095–1105. doi: 10.1534/genetics.112.149120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Silva S, et al. Promoter methylation regulates SAMHD1 gene expression in human CD4+ T cells. J Biol Chem. 2013;288(13):9284–9292. doi: 10.1074/jbc.M112.447201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cribier A, Descours B, Valadão AL, Laguette N, Benkirane M. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep. 2013;3(4):1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 41.White TE, et al. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe. 2013;13(4):441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Careddu MG, et al. Knockdown of cytosolic 5′-nucleotidase II (cN-II) reveals that its activity is essential for survival in astrocytoma cells. Biochim Biophys Acta. 2008;1783(8):1529–1535. doi: 10.1016/j.bbamcr.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 43.Frangini M, et al. Unchanged thymidine triphosphate pools and thymidine metabolism in two lines of thymidine kinase 2-mutated fibroblasts. FEBS J. 2009;276(4):1104–1113. doi: 10.1111/j.1742-4658.2008.06853.x. [DOI] [PubMed] [Google Scholar]
- 44.Pontarin G, Ferraro P, Bee L, Reichard P, Bianchi V. Mammalian ribonucleotide reductase subunit p53R2 is required for mitochondrial DNA replication and DNA repair in quiescent cells. Proc Natl Acad Sci USA. 2012;109(33):13302–13307. doi: 10.1073/pnas.1211289109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.