Abstract
Human epidermal growth factor receptor 2 (HER2; ERBB2) amplification and phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA) mutations often co-occur in breast cancer. Aberrant activation of the phosphatidylinositol 3-kinase (PI3K) pathway has been shown to correlate with a diminished response to HER2-directed therapies. We generated a mouse model of HER2-overexpressing (HER2+), PIK3CAH1047R-mutant breast cancer. Mice expressing both human HER2 and mutant PIK3CA in the mammary epithelium developed tumors with shorter latencies compared with mice expressing either oncogene alone. HER2 and mutant PIK3CA also cooperated to promote lung metastases. By microarray analysis, HER2-driven tumors clustered with luminal breast cancers, whereas mutant PIK3CA tumors were associated with claudin-low breast cancers. PIK3CA and HER2+/PIK3CA tumors expressed elevated transcripts encoding markers of epithelial-to-mesenchymal transition and stem cells. Cells from HER2+/PIK3CA tumors more efficiently formed mammospheres and lung metastases. Finally, HER2+/PIK3CA tumors were resistant to trastuzumab alone and in combination with lapatinib or pertuzumab. Both drug resistance and enhanced mammosphere formation were reversed by treatment with a PI3K inhibitor. In sum, PIK3CAH1047R accelerates HER2-mediated breast epithelial transformation and metastatic progression, alters the intrinsic phenotype of HER2-overexpressing cancers, and generates resistance to approved combinations of anti-HER2 therapies.
The human epidermal growth factor receptor 2 (HER2) oncogene, which encodes the HER2 receptor tyrosine kinase, is amplified in 20% of breast cancers (1). HER2 activation triggers signal transduction through oncogenic signaling pathways, such as the PI3K–Akt survival pathway, which is arguably the most commonly mutated pathway in human cancer. In breast cancer, hyperactivation of the PI3K pathway can occur through a number of mechanisms, including activating mutations in phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha (PIK3CA, the gene encoding the p110α catalytic subunit) (2). More than 80% of PIK3CA mutations occur in two “hot spots”: E452K and E545K (exon 9) in the helical domain and H1047R (exon 20) in the kinase domain. The PI3K–Akt survival pathway has attracted considerable interest as a target for therapeutic intervention. Indeed, PI3K inhibitors are currently in clinical trials for the treatment of breast and other cancers (2).
Approximately 40% of HER2+ breast cancers harbor activating mutations in PIK3CA (3), consistent with the notion that these two oncogenes have nonoverlapping functions and may cooperate to promote tumor growth. In addition, mutant PIK3CA enhances HER2-mediated transformation of MCF10A breast epithelial cells in vitro (4). Trastuzumab, an antibody directed against the ectodomain of HER2, pertuzumab, an antibody that blocks dimerization of HER2 with other ERBB receptors, and lapatinib, an ATP-competitive tyrosine kinase inhibitor of EGFR and HER2, are approved for the treatment of patients with HER2-overexpressing breast cancer. However, both de novo and acquired resistance to anti-HER2 therapies are not uncommon (5). Several studies suggest that PIK3CA mutations confer resistance to these therapies (6–8), but confirmation of a causal association between aberrant activation of the PI3K pathway and resistance to HER2-directed therapies in the clinic is still missing.
To further study the role of PIK3CA mutations in HER2+ breast cancer, we generated a genetically engineered mouse model expressing human HER2 and PIK3CAH1047R, the most common PIK3CA mutation found in human breast cancers. We used this model to determine whether HER2 and PIK3CAH1047R cooperate to promote mammary tumor formation and progression and whether mutant PI3K promotes innate resistance to anti-HER2 therapies in a defined genetic background.
Results
HER2 and Mutant PIK3CA Induce Distinct Premalignant Changes in the Mammary Gland.
To generate a mouse model of human HER2+/PIK3CA-mutant breast cancer, we crossed mice bearing three transgenes (Fig. S1A). MMTV-HER2 drives high levels of human HER2 in the mammary epithelium, resulting in tumor formation (9). MMTV-rtTA, together with TetOp-HA-PIK3CAH1047R-IRES-luciferase, allows for the doxycycline (DOX)-inducible expression of mutant PI3K in the mammary gland (10). For simplicity, we refer to mice bearing all three oncogenes as HER2+PIK3CA; mice expressing either oncogene alone are referred to as HER2 or PIK3CA. Starting at 4 wk of age, female mice were treated with DOX in their drinking water. DOX-induced expression of luciferase in the mammary glands was observed within 1 wk (Fig. S1B). Luciferase expression was not detected in the absence of DOX, or in MMTV-HER2.rtTA mice treated with DOX (Fig. S1B).
We asked first if the HER2 and PIK3CAH1047R transgene products synergistically transformed mammary epithelial cells. We performed whole mount staining of mammary glands from 12-wk-old virgin transgenic mice and found that expression of HER2 increased ductal side branching, whereas mutant PI3K increased growth of lobuloalveolar structures (Fig. S2A). The combined expression of both HER2 and PI3KH1047R resulted in a combination of both phenotypes, such that side branching and lobulo-alveolar growth were markedly increased. Immunohistochemistry (IHC) showed elevated levels of the PI3K target p-AKT and the proliferation marker Ki67 in oncogene-expressing glands (Fig. S2 A–C). Terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) staining indicated higher rates of apoptosis in PIK3CA-expressing mammary glands relative to wild type (Fig. S2D). However, the differences in Ki67 and TUNEL staining between the three oncogene-expressing models were not statistically significant.
HER2 and Mutant PIK3CA Accelerate Mammary Tumor Formation and Lung Metastases.
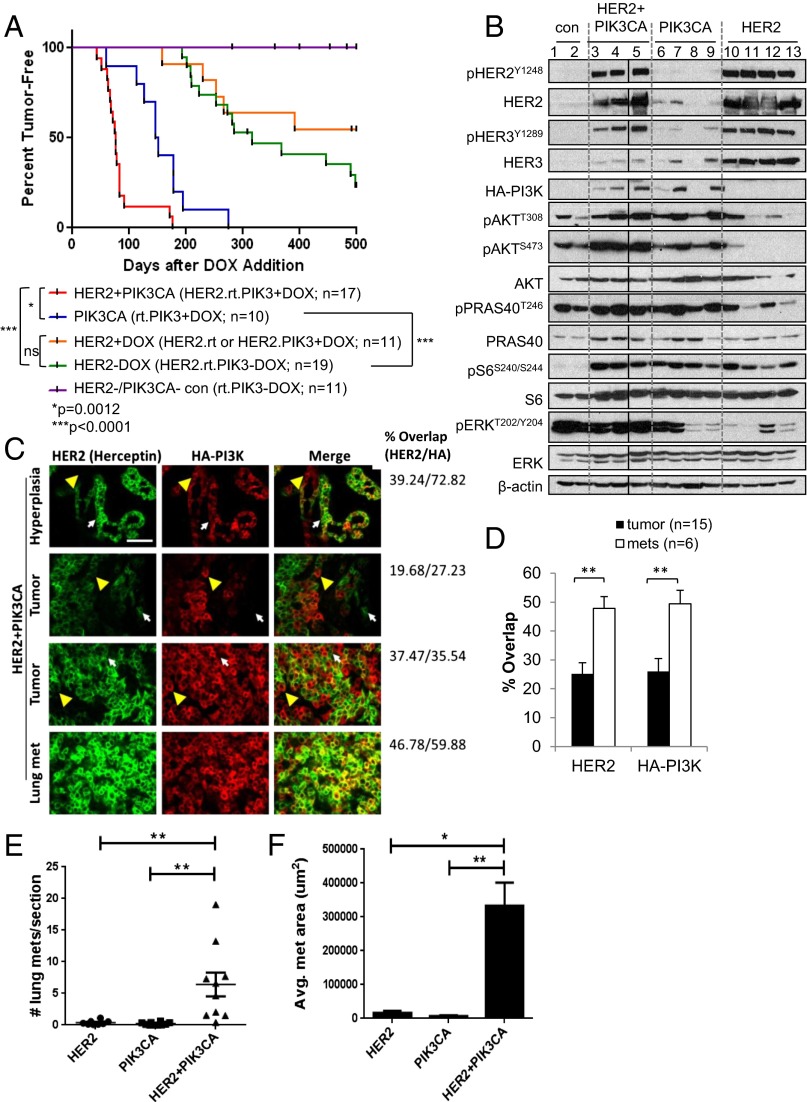
We next treated mice expressing HER2 alone, PIK3CA alone, or both oncogenes with DOX beginning at 4 wk of age, and serially palpated mammary glands to detect tumor formation. In the absence of DOX, rtTA.PIK3CA mice failed to develop tumors up to 500 d of follow-up (Fig. 1A). HER2-expressing mice (HER2.rtTA or HER2.PIK3CA +DOX, or uninduced HER2.rtTA.PIK3CA –DOX) formed tumors with a median latency of 368 d (∼12 mo). PIK3CA mice formed tumors with a median latency of 149 d (∼5 mo), whereas HER2+PIK3CA mice did so at 76 d (∼2.5 mo) postinduction. This latency period was significantly shorter than that seen in mice expressing either oncogene alone (P = 0.0012 vs. PIK3CA; P < 0.0001 vs. HER2; log-rank test; Fig. 1A). When corrected for age at time of killing, the HER2+PIK3CA mice developed significantly more tumors per mouse, and carried a larger tumor burden, measured as total tumor volume and tumors per mouse (Fig. S3 A and B). In addition, HER2+PIK3CA tumors grew faster than tumors expressing only PIK3CA (Fig. S3C). Suggestive of faster growth outpacing tumor angiogenesis, HER2+PIK3CA tumors and lung metastases were frequently hemorrhagic (Fig. S3 D–G).
Fig. 1.
HER2 and mutant PIK3CA cooperate to promote mammary tumor formation and metastasis. (A) Female mice of the indicated genotypes were continuously administered DOX or vehicle control beginning at 4 wk of age. Mice were palpated twice weekly for the presence of mammary tumors. HER2.rtTA.PIK3CA + DOX tumors that lacked human HER2 or HA-PI3K expression by immunofluorescence were excluded from analysis. *P = 0.0012; **P < 0.0001, log-rank test. (B) Mammary glands from MMTV-rtTA mice (control) or mammary tumors from HER2+PIK3CA, PIK3CA, or HER2 mice were snap frozen in liquid nitrogen, homogenized, and lysed in Nonidet P-40–containing lysis buffer. Tissue lysates were subjected to immunoblot analysis with the indicated antibodies. β-Actin was used as a loading control. Scans are all from the same gel/film; the vertical black line indicates an irrelevant lane that was removed from the figure for clarity. (C) FFPE tumor and lung tissues were subjected to IF with the indicated antibodies. (Scale bar, 50 µm.) White arrows indicate HER2-expressing cells that fail to express HA, whereas yellow arrowheads indicate HA-expressing cells that fail to express human HER2. The percent overlap refers to the percentage of green pixels (HER2) that contain red (HA) and vice versa. (D) Percent overlap of HER2 and HA-PI3K staining was quantified from immunofluorescent images of HER2.rtTA.PIK3CA + DOX primary tumors using MetaMorph software. Data shown are the average ± SEM **P < 0.01, Student t test. (E and F) Nude mice (n = 8–10 per group) were injected in the tail vein with dissociated tumor cells of the indicated genotypes. Lungs were harvested 32 d later. (E) Metastases were scored from H&E sections taken at 50-µm intervals throughout the lungs (13–26 sections per mouse; **P < 0.01, Tukey’s multiple comparisons test). (F) Area of lung metastases was quantifed using LP2 software. Data shown are the average ± SEM (*P < 0.01; **P < 0.001, Student t test).
HER2+PIK3CA tumors expressed both HER2 and HA-tagged p110α by Western blot analysis (Fig. 1B). PIK3CA tumors also expressed elevated levels of p-AKT and its substrate p-PRAS40, suggesting that the PI3K pathway is highly activated in these tumors. All tumors exhibited elevated p-S6 compared with control mammary glands. Although HER2-expressing tumors exhibited elevated p-HER3, the HER2+PIK3CA tumors showed reduced expression of total HER3 in comparison, consistent with PI3K-mediated feedback repression of HER3 transcription (11–13).
We next determined if human HER2 and HA-p110αH1047R were coexpressed within the same cells in HER2+PIK3CA tumors. We performed immunofluorescence (IF) of mouse mammary tumors using trastuzumab as the primary antibody, followed by a fluorescently conjugated anti-human secondary antibody. Trastuzumab only recognizes human HER2 and not mouse erbb2. We costained with an HA antibody to detect HA-p110αH1047R. We observed heterogeneous expression of both transgenes in primary tumors; surprisingly, not all tumor cells coexpressed both oncogene products as measure by IF (Fig. 1C). In contrast, detectable coexpression was significantly higher in lung metastases (Fig. 1 C and D), suggesting that high coexpression of both oncogenes selects for tumor cells with an increased capacity to metastasize.
We further studied the metastatic phenotype of the mammary tumors. Lung metastases were rare in our original cohort, probably because these mice were killed when total tumor burden reached ∼1,500 mm3. Therefore, we injected cells isolated from HER2, PIK3CA, or HER2+PIK3CA tumors in the tail vein of nude mice. HER2+PIK3CA tumor cells formed significantly more lung metastases than cells from tumors expressing either oncogene alone (Fig. 1E). The HER2+PIK3CA lung metastases were also significantly larger (Fig. 1F and Fig. S3 H–J). These data suggest that HER2 and PI3KH1047R strongly cooperate to promote metastatic colonization and growth.
Mutant PIK3CA Confers Features of Epithelial-to-Mesenchymal Transition and Stem-Like Cells.
We next examined the histopathological features of tumors expressing HER2, mutant PI3K, or both. HER2-expressing tumors were histologically homogeneous, consisting mostly of solid adenocarcinomas. By IHC, they expressed luminal cytokeratin 18 but were negative for the basal markers p63 and cytokeratin 14 (Fig. S4A). In accordance with previous studies (10), PIK3CA tumors exhibited heterogeneous cancer histologies, including a large proportion of metaplastic carcinomas. HER2+PIK3CA tumors were also heterogeneous, although the histological spectrum was distinct from PIK3CA-only tumors (Fig. S4 A and B). Like the PIK3CA tumors, the HER2+PIK3CA tumors expressed basal and luminal markers (Fig. S4A), suggesting that these tumors may arise from pluripotent progenitor cells and/or that mutant PI3K enhances tumor cell plasticity.
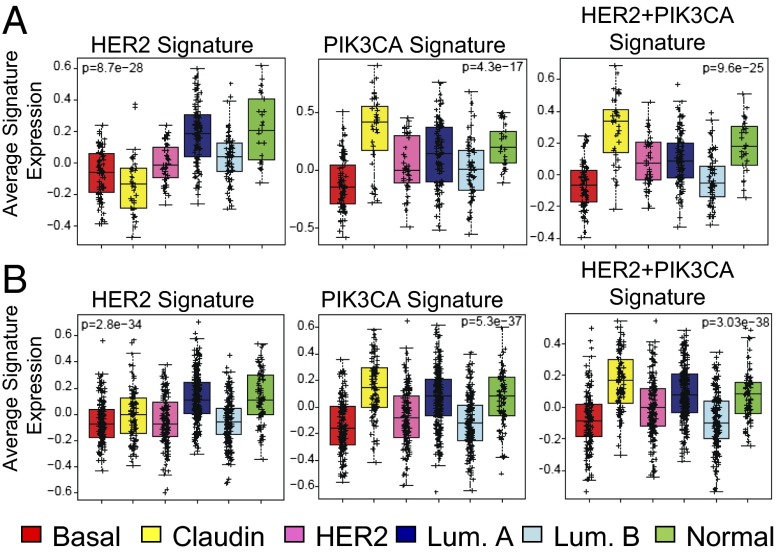
Next, we analyzed gene expression using microarrays. Mammary tumors from each genotype (HER2, PIK3CA, or HER2+PIK3CA) contained distinct clusters of genes, further suggesting that HER2+PIK3CA tumors are phenotypically distinct (Fig. S5A). Tumors from HER2 mice treated with or without DOX exhibited similar gene expression patterns. Microarray data were used to determine whether these mouse tumors are associated with a particular subtype of human breast cancer. We derived a gene signature for each of our models by identifying differentially expressed genes in comparison with a large series of mouse models of breast cancer (14, 15). The average of each signature was scored in the UNC337 dataset (Fig. 2A) (16) and the combined 855 dataset (Fig. 2B) (17) of human breast tumors. Like the MMTV-Neu model, the MMTV-HER2 signature was enriched in the luminal breast cancer subtype (Fig. 2) (14). In both datasets, the PIK3CA and HER2+PIK3CA signatures were most strongly associated with the claudin-low breast cancer subtype (Fig. 2).
Fig. 2.
Gene expression profiles of PIK3CA and HER2+PIK3CA tumors resemble the claudin-low subtype of human breast cancer. RNA isolated from mouse mammary tumors of the indicated genotypes was hybridized to Agilent 4 × 180 k mouse microarrays. Gene signatures were obtained by determining all genes that were up-regulated or down-regulated in tumors of the shown genotypes relative to other genetically engineered mouse models of breast cancer (false discovery rate, 0%). The average of each gene signature was calculated for each array in the UNC337 dataset (A) or combined 855 dataset (B).
Claudin-low breast cancer is a subtype of triple-negative breast cancer characterized by poor differentiation, elevated expression of markers of an epithelial-to-mesenchymal transition (EMT) and enrichment for cancer stem cells (CSCs). This subtype also shares similarities with metaplastic breast cancer (16, 18), consistent with our histological findings. In agreement, gene set analysis (GSA) showed that gene sets associated with EMT, metastasis, spherical vs. adherent growth, and metaplastic breast carcinoma, were elevated in the PIK3CA and HER2+PIK3CA expression profiles (Table S1 and Fig. S5B). We then used a recently described metric to calculate a “differentiation score” (D score) (16), with high scores representing a likeness to mature luminal cells, intermediate scores to luminal progenitor cells, and low scores to adult mammary stem cells. Whereas the HER2 tumors were more similar to mature luminal cells, PIK3CA tumors had intermediate D scores on average (Fig. S6C), suggesting that cells from PIK3CA tumors are most similar to luminal progenitors (16).
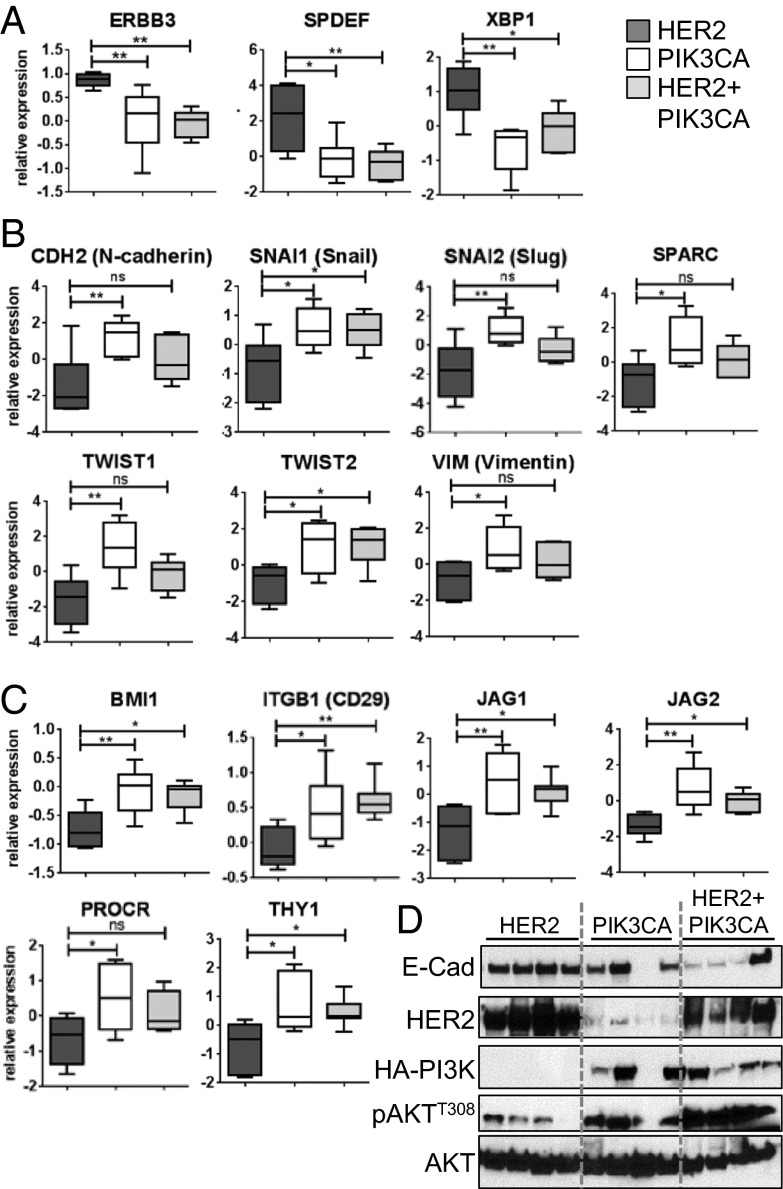
To further test the association of PIK3CA tumors with the claudin-low subtype, we examined our microarray data for expression of luminal and EMT markers and putative markers of mammary stem-like cells. Fig. 3A shows that expression of the luminal markers Xbp1, Erbb3, and Spdef were reduced in the PIK3CA and HER2+PIK3CA tumors relative to the HER2 tumors. In addition, the EMT markers Snai1 (Snail), Snai2 (Slug), Twist1, Twist2, Vim (Vimentin), Cdh2 (N-cadherin), and Sparc were elevated in PIK3CA and HER2+PIK3CA tumors (Fig. 3B). Finally, expression of CSC markers, including Bmi1, Itgb1 (CD29), Procr, Thy1, and the Notch ligands Jag1 and Jag2 (19–21) were significantly up-regulated in PIK3CA and HER2+PIK3CA tumors (Fig. 3C). Consistent with these results, protein levels of the epithelial cell marker E-cadherin were reduced in HER2+PIK3CA tumors relative to HER2 tumors (Fig. 3D). Further, IHC staining of Vimentin and Snail/Slug was elevated in HER2+PIK3CA tumors and lung metastases relative to corresponding tissues from HER2 transgenics (Fig. S7 A–C). These results suggest that mutant PIK3CA induces mammary tumors with EMT and stem-like features.
Fig. 3.
Expression of EMT and CSC markers is increased in PIK3CA and HER2+PIK3CA tumors, whereas expression of luminal markers is reduced. (A–C) Relative gene expression of luminal breast cancer markers (A), EMT markers (B), or CSC markers (C) was quantified from the microarray expression data described above using GraphPad PRISM software (ANOVA P < 0.05). *P < 0.05, **P < 0.01, ***P < 0.001, Tukey’s multiple comparison test. (D) Mouse mammary tumor lysates were subjected to immunoblot analysis with the indicated antibodies.
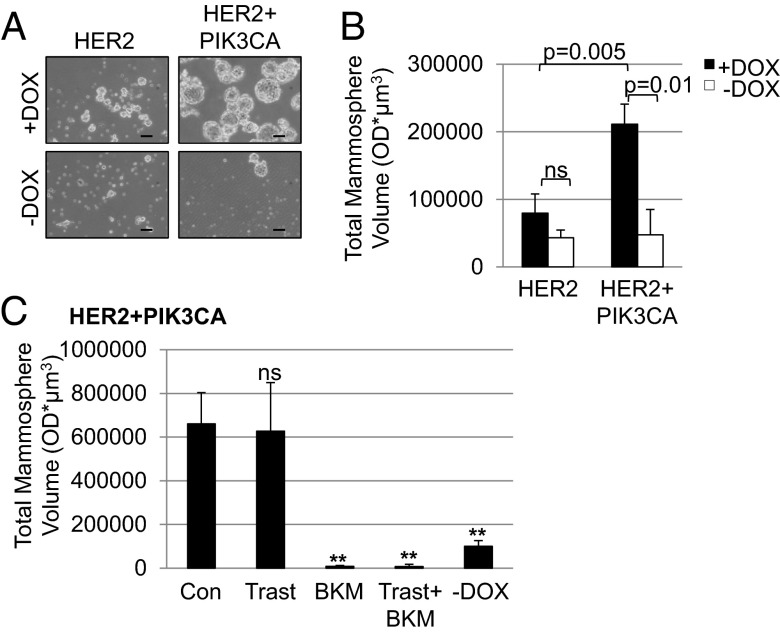
To further address if mutant PIK3CA tumors are enriched with CSCs, we harvested primary tumor cells and performed mammosphere-forming assays. This method is used to measure the tumor-initiating capacity of cancer cells. HER2+PIK3CA mammosphere cultures retained luciferase expression only in the presence of DOX (Fig. S8). DOX-treated cells from HER2+PIK3CA tumors formed significantly more and larger mammospheres than DOX-minus cells or cells from tumors expressing only HER2 (Fig. 4 A and B). The pan-PI3K inhibitor BKM120 (22) markedly decreased the ability of HER2+PIK3CA cells to form mammospheres, whereas trastuzumab had no effect on mammosphere formation (Fig. 4C), further suggesting that aberrant PI3K activity is associated with CSC characteristics.
Fig. 4.
Induction of PI3KH1047R promotes mammosphere formation. (A) Mammospheres were imaged using the 10× objective of an Olympus CK40 microscope. (Scale bar, 100 µm.) (B) Total mammosphere volume from each well was quantified using Gelcount software on day 13. (C) DOX-treated HER2+PIK3CA mammospheres were treated with 20 µg/mL trastuzumab, 1 µM BKM120, or the combination. Total mammosphere volume from each well was quantified using Gelcount software on day 12. Data represent the average ± SD of three replicate wells. **P < 0.01, Student t test.
Mutant PIK3CA Confers Innate Resistance to Trastuzumab and Combinations of Anti-HER2 Therapies.
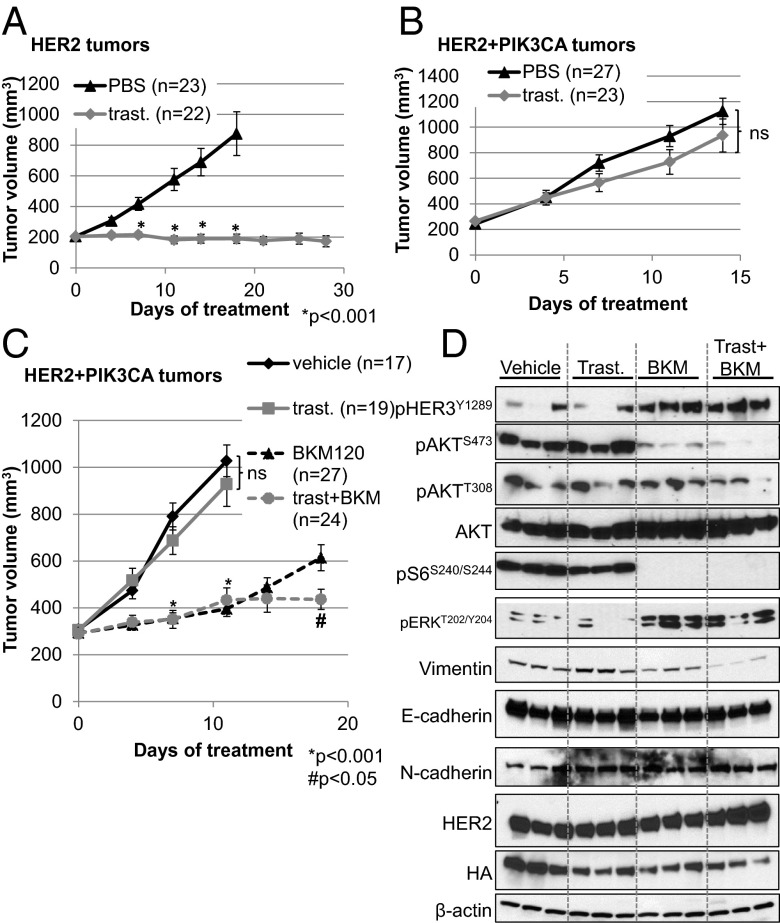
We transplanted tumors from HER2 or HER2+PIK3CA mice into recipient nude mice; once tumors reached a volume of ≥200 mm3, mice were treated with PBS or trastuzumab. Treatment with trastuzumab inhibited growth of HER2-induced but not HER2+PIK3CA tumor transplants (Fig. 5 A and B). We next determined whether these tumors responded to BKM120, which has currently completed phase I studies in patients with cancer (23). Treatment with BKM120 inhibited tumor growth but did not induce tumor regressions. The combination of trastuzumab and BKM120 was moderately superior to single-agent BKM120, suggesting that inhibition of both oncogenes is required to inhibit tumor growth in this bitransgenic model (Fig. 5C).
Fig. 5.
HER2+PIK3CA tumors are resistant to trastuzumab, but respond to the combination of trastuzumab + BKM120. (A and B) Nude mice were transplanted with HER2-expressing (A) or HER2+PIK3CA (B) cells harvested from transgenic tumors. When tumors reached ≥200 mm3, mice were treated with vehicle or trastuzumab twice per week. Tumors were measured with calipers twice weekly. (C) Mice transplanted with HER2+PIK3CA tumors were randomized to receive vehicle control, trastuzumab, BKM120, or the combination. Each data point represents the mean tumor volume in mm3 ± SEM. P values were calculated by Student t test. *, relative to vehicle control; #, relative to BKM120. (D) At the study endpoint, HER2+PIK3CA tumors from C were harvested and lysed in Nonidet P-40–containing lysis buffer. Lysates were probed with the indicated antibodies. β-Actin was used as a loading control.
We next examined pharmacodynamic biomarkers of target inactivation by Western blot analysis of tumor lysates and IHC of formalin-fixed, paraffin embedded (FFPE) tumor sections. BKM120 strongly inhibited p-AKTS473 and more modestly blocked p-AKTT308 following 18 d of treatment, whereas trastuzumab alone failed to inhibit p-AKT, p-S6, p-HER3, or p-ERK in HER2+PIK3CA tumors (Fig. 5D and Fig. S9). In addition, the combination of BKM120 and trastuzumab decreased vimentin expression, although E-cadherin and N-cadherin levels were unchanged (Fig. 5D). HER2+PIK3CA tumors treated with BKM120 displayed up-regulation of p-HER3Y1289 and p-ERK, consistent with relief of PI3K-driven negative repression of HER3 transcription/expression and ERK signaling (11–13, 24). However, the up-regulation of pERK did not rescue the inhibition of p-S6. Total HER2 and HA-PI3K levels remained unchanged with treatment.
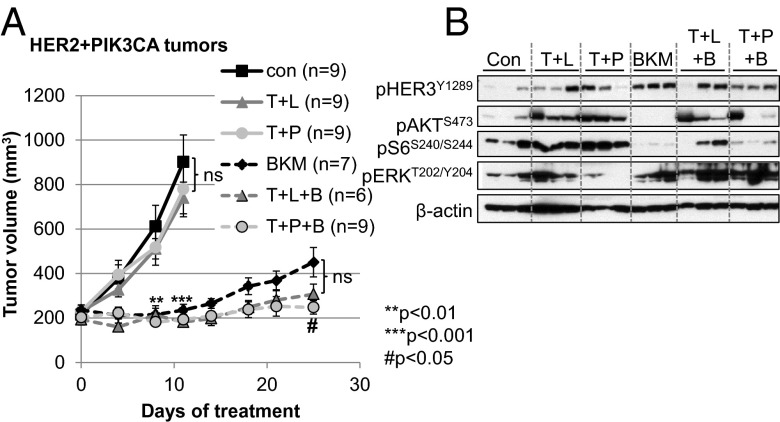
We finally examined whether mutant PIK3CA also conferred resistance to currently approved anti-HER2 combinations, specifically trastuzumab/lapatinib and trastuzumab/pertuzumab (25–27). HER2+PIK3CA tumors failed to respond to these combinations (Fig. 6A). However, the combination of trastuzumab + pertuzumab + BKM120 inhibited tumor growth better than BKM120 alone. There was also a trend toward better inhibition with trastuzumab + lapatinib + BKM120, but this was not statistically significant. Likewise, dual HER2 blockade also failed to inhibit p-AKT or p-S6 (Fig. 6B). The addition of pertuzumab reduced BKM120-induced up-regulation of p-HER3, but not p-ERK. Lapatinib + trastuzumab also failed to prevent BKM120-induced p-ERK, suggesting that dual HER2 blockade does not prevent feedback compensation upon inhibition of PI3K in these tumors. Whether the addition of MEK inhibitors will improve the efficacy of the BKM120-containing combination will require additional investigation.
Fig. 6.
HER2+PIK3CA tumors are resistant to dual blockade of HER2. (A) Mice with established HER2+PIK3CA tumor transplants were randomized to receive vehicle control, BKM120, trastuzumab + lapatinib ± BKM120 (T + L ± B), or trastuzumab + pertuzumab ± BKM120 (T + P ± B) as described in SI Materials and Methods. Each data point represents the mean tumor volume in mm3 ± SEM. P values were calculated by Student t test. ** or ***, relative to vehicle control; #, relative to BKM120. (B) Tumor lysates harvested at the study endpoint were probed with the indicated antibodies. β-Actin was used as a loading control.
Discussion
Resistance to current anti-HER2 therapies is a major hurdle in the eradication of HER2+ breast cancer. Several studies have suggested aberrant activation of the PI3K pathway as a mechanism of resistance to anti-HER2 therapies, although confirmation in the clinic is still unclear. Herein, we uniquely characterized a genetically engineered mouse mammary tumor model harboring both (human) HER2 and mutant PIK3CA. We showed that coexpression of both oncogenes in the mouse mammary gland accelerated tumor formation with an average tumor latency of just 2.5 mo following DOX induction. These tumors were histologically heterogeneous, exhibited features of EMT and stem-like cells, and displayed a high metastatic propensity. HER2+PIK3CA tumors also were resistant to trastuzumab as a single agent and in combination with lapatinib or with pertuzumab. Drug resistance was partially reversed by the PI3K inhibitor BKM120. We propose that this conditional mouse model will be a useful tool for testing novel drugs and combinations for the treatment of HER2+/PIK3CA-mutant breast cancer.
HER2 and PI3KH1047R cooperated to promote transformation of the mammary epithelium, cancer establishment, and metastasis. The mechanism by which these two oncogenes cooperate in this setting is unclear. Whereas it is possible that HER2 overexpression may further enhance the activity of mutant PI3K, we did not observe an increase in phosphorylation of AKT or downstream targets in the HER2+PIK3CA mammary glands or tumors compared with tumors expressing PIK3CA alone (Figs. 1B and 2B). HER2 is also capable of activating the MAPK pathway. Whereas p-ERK levels were low in HER2 tumors, they were elevated in HER2+PIK3CA tumors (Fig. 2B). However, p-ERK levels were also high in untransformed mammary glands, so the significance of elevated MAPK signaling in these tumors is unclear. Table S2 presents a list of genes that are differentially expressed in the HER2+PIK3CA tumors compared with the tumors expressing either oncogene alone; some of these genes may account for the increased tumorigenicity observed in this model.
The histopathological heterogeneity observed in the PIK3CA tumors is consistent with other mouse models of PI3K-induced breast cancer (28), including PIK3CAH1047R knock-in models (29, 30). This heterogeneity also reflects the fact that although PIK3CA mutations are enriched in the luminal A subtype, they are found across all subtypes of human breast cancer (3). Interestingly, ∼40% of the tumors in our PIK3CA mouse model were classified as metaplastic carcinomas, consistent with the high rate of PIK3CA mutations (∼50%) found in human metaplastic carcinomas (18). This suggests that PIK3CA-induced tumors may be useful models for the study of this rare form of breast cancer. The link with metaplastic breast carcinoma is further strengthened by the finding that the PIK3CA-driven tumors were associated with the claudin-low subtype of breast cancer, because claudin-low and metaplastic breast carcinomas exhibit similar gene expression patterns (16, 18). Furthermore, we found that the PIK3CA and HER2+PIK3CA tumors displayed high expression of EMT and stem cell markers and that induction of PIK3CA promoted mammosphere formation. These data are in agreement with in vitro studies that have implicated the PI3K pathway in EMT (31, 32) and stem cell maintenance in breast cancer (33–35). Therefore, the association between PIK3CA mutations and the claudin-low subtype of breast cancer, as well as EMT and stem-like features, suggests that HER2+ breast cancers harboring PIK3CA mutations will exhibit a more virulent behavior than HER2+ tumors with wild-type PIK3CA.
Several mechanisms may explain why mutational activation of PI3K promotes resistance to therapeutic inhibitors of HER2 in our model system. First, HER2 blockade did not affect p-AKT levels in the HER2+PIK3CA tumors (Fig. 6B), suggesting that it can no longer inhibit PI3K signaling in the presence of mutant PIK3CA. In agreement, a recent study showed that the PI3K H1047R mutation enhanced lipid binding, basal catalytic activity, and interaction with the plasma membrane (36) and hence may be insensitive to dual blockade of HER2 upstream. The receptor-independent strong catalytic activity of PI3K H1047R is further supported in our studies by the lack of response of the bigenic tumors to trastuzumab/lapatinib and transtuzumab/pertuzumab. Second, HER2+PIK3CA tumors displayed heterogeneous, nonoverlapping expression of HER2 and HA-PI3K transgenes (Fig. 1C); presumably trastuzumab would not inhibit HER2-negative, HA-PI3K–expressing cancer cells within these heterogeneous tumors. However, we do not believe that this contributes to trastuzumab resistance in our model, because treatment with trastuzumab did not affect HA-PI3K or HER2 expression (Fig. 5D). Third, both EMT and CSCs may play a role in trastuzumab resistance. The EMT transcription factors Snail and Slug, both transcriptionally up-regulated in our HER2+PIK3CA tumors (Fig. 3B), were recently shown to confer resistance to trastuzumab in HER2+ cells (37). CSCs may be less sensitive to trastuzumab than their more differentiated counterparts (38), and long-term trastuzumab treatment has been shown to enrich for CSCs that display an EMT phenotype (39). Indeed, mutant PIK3CA-containing tumors were associated with increased stem cell characteristics, such as the ability to form mammospheres in nonadherent conditions (Fig. 4); trastuzumab did not inhibit the mammosphere-forming capacity of HER2+PIK3CA cells (Fig. 4C). Finally, the elevated PI3K activity may counteract the trastuzumab-induced immune response required for an antitumor effect (39). One limitation of our mouse model is that it may not accurately recapitulate the genomic complexity of human tumors, so these studies should be complemented by future experiments using patient-derived xenografts. It is possible that a combination of multiple mechanisms contribute to the decreased sensitivity of HER2+/PIK3CA-mutant tumors to HER2 inhibitors.
In summary, HER2 and mutant PI3K cooperate to promote mammary tumor establishment and metastatic progression. These tumors are resistant to trastuzumab and combinations of direct inhibitors of HER2. Addition of a PI3K inhibitor reversed resistance in these tumors, suggesting that combining anti-HER2 therapies with PI3K inhibitors may be beneficial for the clinical treatment of HER2+/PIK3CA-mutant breast cancers. We speculate the addition of MEK inhibitors to this combination may prevent feedback compensation (Fig. 5D) and potentially eliminate these tumors. We propose that the mouse model presented here will be a valuable tool to investigate the underlying biology of HER2+/PI3K-mutant breast cancers and for preclinical testing of therapeutic strategies against this subtype of breast cancer.
Materials and Methods
MMTV-HER2 (human HER2), MMTV-rtTA, and TetOp-HA-PIK3CA-H1047R-luciferase mice were described previously (9, 10). All mice were housed in the Association for Assessment and Accreditation of Laboratory Animal Care International-approved facilities under Vanderbilt University's Institutional Animal Care and Use Committee guidelines in a pathogen-free environment. Detailed descriptions of animal experiments, drug treatments, IHC, IF, Western blot analysis, microarray analysis, and the mammosphere assay are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by Department of Defense Postdoctoral Fellowship Award BC103785 (to A.B.H.); Breast Cancer Specialized Program of Research Excellence Grants P50CA98131 to Vanderbilt University and P50CA58223 to University of North Carolina; Vanderbilt-Ingram Cancer Center Support Grant P30CA68485; Breast Cancer Research Foundation grants (to C.L.A. and C.M.P.); American Cancer Society Clinical Research Professorship Grant CRP-07-234 (to C.L.A.); the Lee Jeans Translational Breast Cancer Research Program (C.L.A.); Stand Up to Cancer Dream Team Translational Research Grant, a program of the Entertainment Industry Foundation (SU2C-AACR-DT0209) (to C.L.A.); Susan G. Komen for the Cure Foundation Grants CCR12225834 (to H.C.), PDF12229712 (to J.M.B.), SAC100013 (to C.L.A.), and KG100677 (to R.S.C.); and National Institutes of Health R01s CA80195 (to C.L.A.), CA143126 (to R.S.C.), CA134502 and CA172461-01 (to J.J.Z.), and CA138255 and CA148761 (to C.M.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the University of North Carolina Microarray Database (UMD, https://genome.unc.edu/pubsup/breastGEO/clinicalData.shtml) and Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE41118).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1303204110/-/DCSupplemental.
References
- 1.Ross JS, Fletcher JA. The HER-2/neu oncogene in breast cancer: Prognostic factor, predictive factor, and target for therapy. Stem Cells. 1998;16(6):413–428. doi: 10.1002/stem.160413. [DOI] [PubMed] [Google Scholar]
- 2.Engelman JA. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 3.Koboldt DC, et al. Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakrabarty A, et al. H1047R phosphatidylinositol 3-kinase mutant enhances HER2-mediated transformation by heregulin production and activation of HER3. Oncogene. 2010;29(37):5193–5203. doi: 10.1038/onc.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrett JT, Arteaga CL. Resistance to HER2-directed antibodies and tyrosine kinase inhibitors: Mechanisms and clinical implications. Cancer Biol Ther. 2011;11(9):793–800. doi: 10.4161/cbt.11.9.15045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 7.Esteva FJ, et al. PTEN, PIK3CA, p-AKT, and p-p70S6K status: Association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am J Pathol. 2010;177(4):1647–1656. doi: 10.2353/ajpath.2010.090885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandarlapaty S, et al. Frequent mutational activation of the PI3K-AKT pathway in trastuzumab-resistant breast cancer. Clin Cancer Res. 2012;18(24):6784–6791. doi: 10.1158/1078-0432.CCR-12-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finkle D, et al. HER2-targeted therapy reduces incidence and progression of midlife mammary tumors in female murine mammary tumor virus huHER2-transgenic mice. Clin Cancer Res. 2004;10(7):2499–2511. doi: 10.1158/1078-0432.ccr-03-0448. [DOI] [PubMed] [Google Scholar]
- 10.Liu P, et al. Oncogenic PIK3CA-driven mammary tumors frequently recur via PI3K pathway-dependent and PI3K pathway-independent mechanisms. Nat Med. 2011;17(9):1116–1120. doi: 10.1038/nm.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garrett JT, et al. Transcriptional and posttranslational up-regulation of HER3 (ErbB3) compensates for inhibition of the HER2 tyrosine kinase. Proc Natl Acad Sci USA. 2011;108(12):5021–5026. doi: 10.1073/pnas.1016140108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarty A, Sánchez V, Kuba MG, Rinehart C, Arteaga CL. Feedback upregulation of HER3 (ErbB3) expression and activity attenuates antitumor effect of PI3K inhibitors. Proc Natl Acad Sci USA. 2012;109(8):2718–2723. doi: 10.1073/pnas.1018001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chandarlapaty S, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19(1):58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herschkowitz JI, et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007;8(5):R76. doi: 10.1186/gb-2007-8-5-r76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herschkowitz JI, et al. Comparative oncogenomics identifies breast tumors enriched in functional tumor-initiating cells. Proc Natl Acad Sci USA. 2012;109(8):2778–2783. doi: 10.1073/pnas.1018862108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prat A, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrell JC, et al. Genomic analysis identifies unique signatures predictive of brain, lung, and liver relapse. Breast Cancer Res Treat. 2012;132(2):523–535. doi: 10.1007/s10549-011-1619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hennessy BT, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal transition and stem cell characteristics. Cancer Res. 2009;69(10):4116–4124. doi: 10.1158/0008-5472.CAN-08-3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shipitsin M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11(3):259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Liu S, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66(12):6063–6071. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shackleton M, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439(7072):84–88. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 22.Maira SM, et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther. 2012;11(2):317–328. doi: 10.1158/1535-7163.MCT-11-0474. [DOI] [PubMed] [Google Scholar]
- 23.Bendell JC, et al. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol. 2012;30(3):282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 24.Serra V, et al. PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene. 2011;30(22):2547–2557. doi: 10.1038/onc.2010.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baselga J, et al. NeoALTTO Study Team Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379(9816):633–640. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swain SM, et al. Pertuzumab, trastuzumab, and docetaxel for HER2-positive metastatic breast cancer (CLEOPATRA study): Overall survival results from a randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2013;14(6):461–471. doi: 10.1016/S1470-2045(13)70130-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gianni L, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): A randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 28.Koren S, Bentires-Alj M. Mouse models of PIK3CA mutations: One mutation initiates heterogeneous mammary tumors. FEBS J. 2013;280(12):2758–2765. doi: 10.1111/febs.12175. [DOI] [PubMed] [Google Scholar]
- 29.Yuan W, et al. Conditional activation of Pik3ca(H1047R) in a knock-in mouse model promotes mammary tumorigenesis and emergence of mutations. Oncogene. 2013;32(3):318–326. doi: 10.1038/onc.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tikoo A, et al. Physiological levels of Pik3ca(H1047R) mutation in the mouse mammary gland results in ductal hyperplasia and formation of ERα-positive tumors. PLoS ONE. 2012;7(5):e36924. doi: 10.1371/journal.pone.0036924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xue G, et al. (2012) Akt/PKB-mediated phosphorylation of Twist1 promotes tumor metastasis via mediating cross-talk between PI3K/Akt and TGF-beta signaling axes. Cancer Discov 2(3):248–259. [DOI] [PubMed]
- 32.Wallin JJ, et al. Active PI3K pathway causes an invasive phenotype which can be reversed or promoted by blocking the pathway at divergent nodes. PLoS ONE. 2012;7(5):e36402. doi: 10.1371/journal.pone.0036402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou J, et al. Activation of the PTEN/mTOR/STAT3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc Natl Acad Sci USA. 2007;104(41):16158–16163. doi: 10.1073/pnas.0702596104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korkaya H, et al. Regulation of mammary stem/progenitor cells by PTEN/Akt/beta-catenin signaling. PLoS Biol. 2009;7(6):e1000121. doi: 10.1371/journal.pbio.1000121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hardt O, et al. Highly sensitive profiling of CD44+/CD24- breast cancer stem cells by combining global mRNA amplification and next generation sequencing: Evidence for a hyperactive PI3K pathway. Cancer Lett. 2012;325(2):165–174. doi: 10.1016/j.canlet.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110α (PIK3CA) Proc Natl Acad Sci USA. 2012;109(38):15259–15264. doi: 10.1073/pnas.1205508109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliveras-Ferraros C, et al. Epithelial-to-mesenchymal transition (EMT) confers primary resistance to trastuzumab (Herceptin) Cell Cycle. 2012;11(21):4020–4032. doi: 10.4161/cc.22225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reim F, et al. Immunoselection of breast and ovarian cancer cells with trastuzumab and natural killer cells: Selective escape of CD44high/CD24low/HER2low breast cancer stem cells. Cancer Res. 2009;69(20):8058–8066. doi: 10.1158/0008-5472.CAN-09-0834. [DOI] [PubMed] [Google Scholar]
- 39.Korkaya H, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47(4):570–584. doi: 10.1016/j.molcel.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.