Significance
The three-dimensional fold and inherent structural dynamics of RNA are critical determinants of function. Although the hierarchical folding of RNA secondary structure followed by tertiary structure formation is well established, insights into how secondary structure units impact tertiary structure formation and dynamics remain elusive. Using single-molecule FRET imaging of the preQ1 class II bacterial riboswitch, we reveal that RNA employs stem–loop insertions into classical pseudoknots as a tool to tune ligand response and hence the outcome of natural gene-regulatory elements.
Keywords: SHAPE, single-molecule FRET, site-specific labeling, metabolite sensing RNAs, ligand recognition
Abstract
Structural and dynamic features of RNA folding landscapes represent critical aspects of RNA function in the cell and are particularly central to riboswitch-mediated control of gene expression. Here, using single-molecule fluorescence energy transfer imaging, we explore the folding dynamics of the preQ1 class II riboswitch, an upstream mRNA element that regulates downstream encoded modification enzymes of queuosine biosynthesis. For reasons that are not presently understood, the classical pseudoknot fold of this system harbors an extra stem–loop structure within its 3′-terminal region immediately upstream of the Shine–Dalgarno sequence that contributes to formation of the ligand-bound state. By imaging ligand-dependent preQ1 riboswitch folding from multiple structural perspectives, we reveal that the extra stem–loop strongly influences pseudoknot dynamics in a manner that decreases its propensity to spontaneously fold and increases its responsiveness to ligand binding. We conclude that the extra stem–loop sensitizes this RNA to broaden the dynamic range of the ON/OFF regulatory switch.
A variety of small metabolites have been found to regulate gene expression in bacteria, fungi, and plants via direct interactions with distinct mRNA folds (1–4). In this form of regulation, the target mRNA typically undergoes a structural change in response to metabolite binding (5–9). These mRNA elements have thus been termed “riboswitches” and generally include both a metabolite-sensitive aptamer subdomain and an expression platform. For riboswitches that regulate the process of translation, the expression platform minimally consists of a ribosomal recognition site [Shine–Dalgarno (SD)]. In the simplest form, the SD sequence overlaps with the metabolite-sensitive aptamer domain at its downstream end. Representative examples include the S-adenosylmethionine class II (SAM-II) (10) and the S-adenosylhomocysteine (SAH) riboswitches (11, 12), as well as prequeuosine class I (preQ1-I) and II (preQ1-II) riboswitches (13, 14). The secondary structures of these four short RNA families contain a pseudoknot fold that is central to their gene regulation capacity. Although the SAM-II and preQ1-I riboswitches fold into classical pseudoknots (15, 16), the conformations of the SAH (17) and preQ1-II counterparts are more complex and include a structural extension that contributes to the pseudoknot architecture (14). Importantly, the impact and evolutionary significance of these “extra” stem–loop elements on the function of the SAH and preQ1-II riboswitches remain unclear.
PreQ1 riboswitches interact with the bacterial metabolite 7-aminomethyl-7-deazaguanine (preQ1), a precursor molecule in the biosynthetic pathway of queuosine, a modified base encountered at the wobble position of some transfer RNAs (14). The general biological significance of studying the preQ1-II system stems from the fact that this gene-regulatory element is found almost exclusively in the Streptococcaceae bacterial family. Moreover, the preQ1 metabolite is not generated in humans and has to be acquired from the environment (14). Correspondingly, the preQ1-II riboswitch represents a putative target for antibiotic intervention. Although preQ1 class I (preQ1-I) riboswitches have been extensively investigated (18–28), preQ1 class II (preQ1-II) riboswitches have been largely overlooked despite the fact that a different mode of ligand binding has been postulated (14).
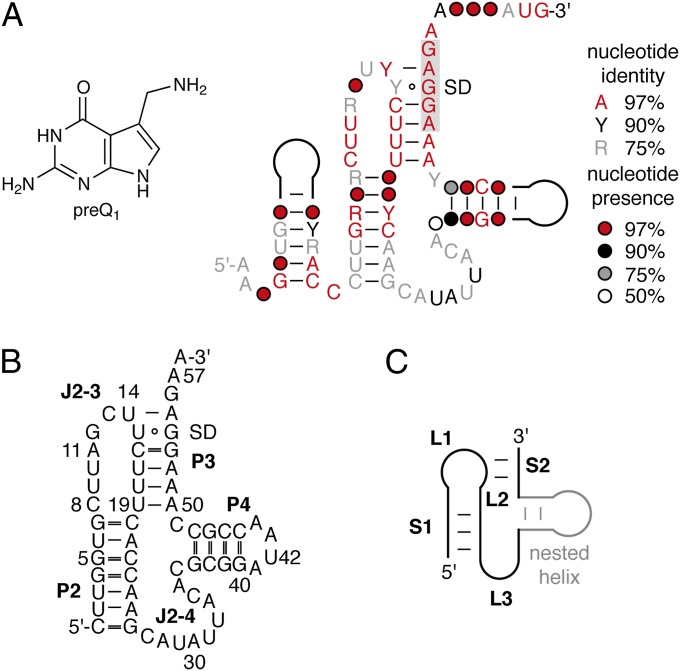
The consensus sequence and the secondary structure model for the preQ1-II motif (COG4708 RNA) (Fig. 1A) comprise ∼80–100 nt (14). The minimal Streptococcus pneumoniae R6 aptamer domain sequence binds preQ1 with submicromolar affinity and consists of an RNA segment forming two stem–loops, P2 and P4, and a pseudoknot P3 (Fig. 1B). In-line probing studies suggest that the putative SD box (AGGAGA; Fig. 1) is sequestered by pseudoknot formation, which results in translational-dependent gene regulation of the downstream gene (14).
Fig. 1.
PreQ1 class II riboswitch. (A) Chemical structure of 7-aminomethyl-7-deazaguanosine (preQ1); consensus sequence and secondary structure model for the COG4708 RNA motif (adapted from reference 14). Nucleoside presence and identity as indicated. (B) S. pneumoniae R6 preQ1-II RNA aptamer investigated in this study. (C) Schematics of an H-type pseudoknot with generally used nomenclature for comparison.
Here, we investigated folding and ligand recognition of the S. pneumoniae R6 preQ1-II riboswitch, using complementary chemical, biochemical, and biophysical methods including selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), mutational analysis experiments, 2-aminopurine fluorescence, and single-molecule fluorescence resonance energy transfer (smFRET) imaging. In so doing, we explored the structural and functional impact of the additional stem–loop element in the context of its otherwise “classical” H-type pseudoknot fold (29–32) (Fig. 1C). Our results reveal that the unique 3′-stem–loop element in the preQ1-II riboswitch contributes to the process of SD sequestration, and thus the regulation of gene expression, by modulating both its intrinsic dynamics and its responsiveness to ligand binding.
Results and Discussion
Temperature-Dependent SHAPE Indicates preQ1-II Pseudoknot Preorganization.
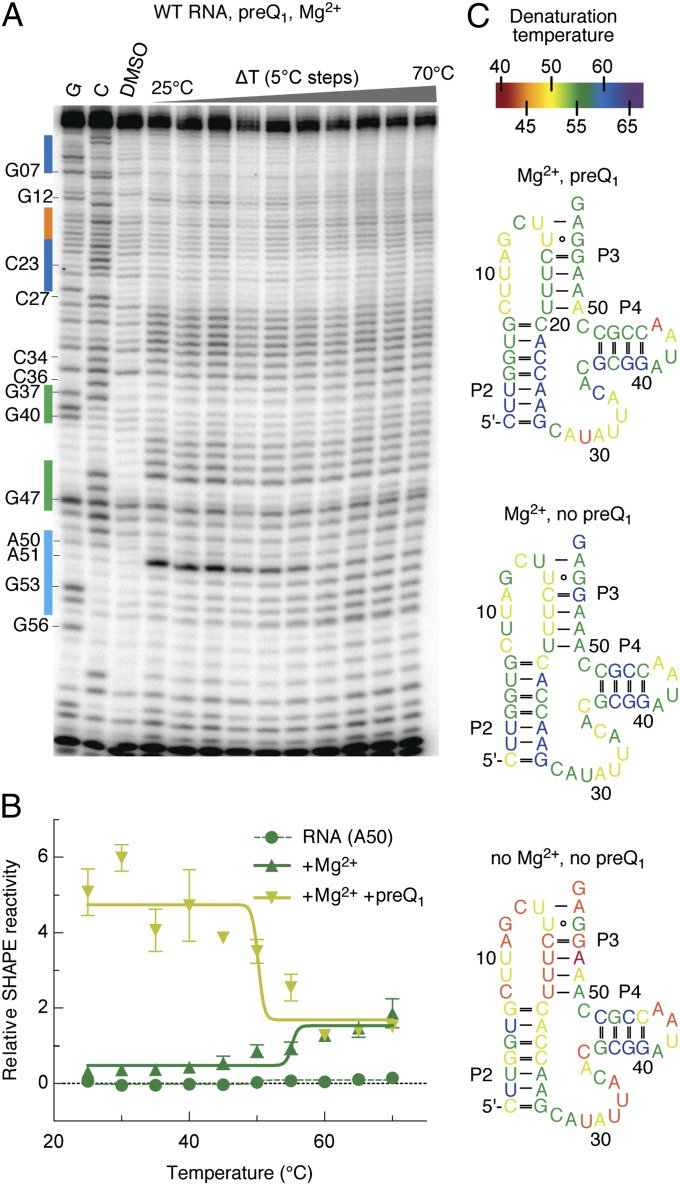
Previous bioinformatics and in-line probing studies on the preQ1-II riboswitch sequence from S. pneumonia R6 have yielded the putative secondary structure depicted in Fig. 1B (14). The characteristic pseudoknot (P3) interaction is defined by the pairing of the 3′-terminal sequence with junction J2–3, where the stem–loop P4 element is inserted immediately upstream of this interaction. To explore the structural and dynamic properties of this model, we used the chemical probing technique SHAPE (33) to probe the conformation of the preQ1-II riboswitch as a function of temperature. To do so, a transcribed riboswitch aptamer domain (85 nt in length; Materials and Methods and Fig. 2) was subjected to reaction with benzoyl cyanide (BzCN) at temperatures ranging from 25 to 70 °C in 5 °C increments. After reverse transcription, the discrete bands observed by gel electrophoresis were quantified using SAFA software (Fig. S1) (34). The relative 2′-OH reactivities were plotted as a function of temperature to evaluate the denaturation temperature of the RNA at the single-nucleotide level (Fig. 2 A and B, and Fig. S2). Here, band intensities represent the degree of 2′-hydroxyl acylation of the base identified by reverse transcription. This analysis was repeated for the preQ1-II RNA in the absence or presence of magnesium ions and the preQ1 ligand. The results from these experiments are presented on the proposed secondary structure and color-coded according to their denaturation temperature (Fig. 2C). The data obtained demonstrate that P2 and P4 are preformed in the absence of both Mg2+ and preQ1 ligands, whereas the pseudoknot nucleotides (U15–U19, A50–G54) are disordered and highly sensitive to thermal denaturation. The overall reactivity of the entire riboswitch decreases in the presence of magnesium over the full range of temperatures examined, especially the nucleotides of the pseudoknot and J2–3 regions (Fig. 2C). These residues were shown to possess an even higher denaturation temperature than neighboring pseudoknot nucleotides. With the addition of preQ1 ligand, a further increase in the denaturation temperature of the pseudoknot nucleotides was observed, with the exception of A50, which showed a 5 °C-lower denaturation temperature. This base exhibited enhanced SHAPE reactivity in the presence of preQ1 ligand (see ref. 35 and Fig. S3), suggesting that its solvent exposure increases as a consequence of preQ1 binding. Changes of this kind likely stem from disruption of the U19–A50 base pair in the ligand-bound complex. This conclusion is corroborated by the observation that a 2-aminopurine (2-Ap) nucleoside at position 50 exhibits an increase in fluorescence in the presence of Mg2+ and preQ1, trends that are in line with its movement from a stacked, or intrahelical, position to one that is extrahelical (35). Variations in denaturation temperature were also observed in junction J2–3 with certain nucleosides exhibiting greater temperature sensitivity (U9, G12, U14) than others (C13). These observations are consistent with conformational rearrangements in J2–3 upon formation of the preQ1-bound complex. Junction J2–4 nucleosides, close to P4 (C34 to C36), also become rigidified in the preQ1-bound complex, whereas those closer to P2 (A28, U29) become more flexible. Strikingly, only minor changes in denaturation temperatures were observed for the majority of nucleosides in the P4 stem–loop under the conditions tested, suggesting that this extra arm is highly stable relative to the rest of the structure.
Fig. 2.
Temperature-dependent SHAPE analysis of the preQ1-II RNA. (A) Representative gel for the SHAPE probing of the preQ1-II RNA structure with BzCN. Lanes from Left to Right: G and C base ladders, control in the absence of probing reagent, probing in the presence of 5 mM Mg2+ ions and 10 μM preQ1 across a temperature gradient from 25 to 70 °C in 5 °C increments. (B) Representative gel analysis for the estimation of denaturation temperatures Td of individual nucleotides (here A50) without ligands, with Mg2+ and with Mg2+ and preQ1. Td values were determined by the inflection point of the plot of relative reactivities over increasing temperature. (C) The apparent denaturation temperatures Td are colored as indicated onto the secondary structure for the three different conditions tested (free RNA, RNA with Mg2+, RNA with Mg2+ and preQ1).
Mutational Analysis of the preQ1-II Riboswitch.
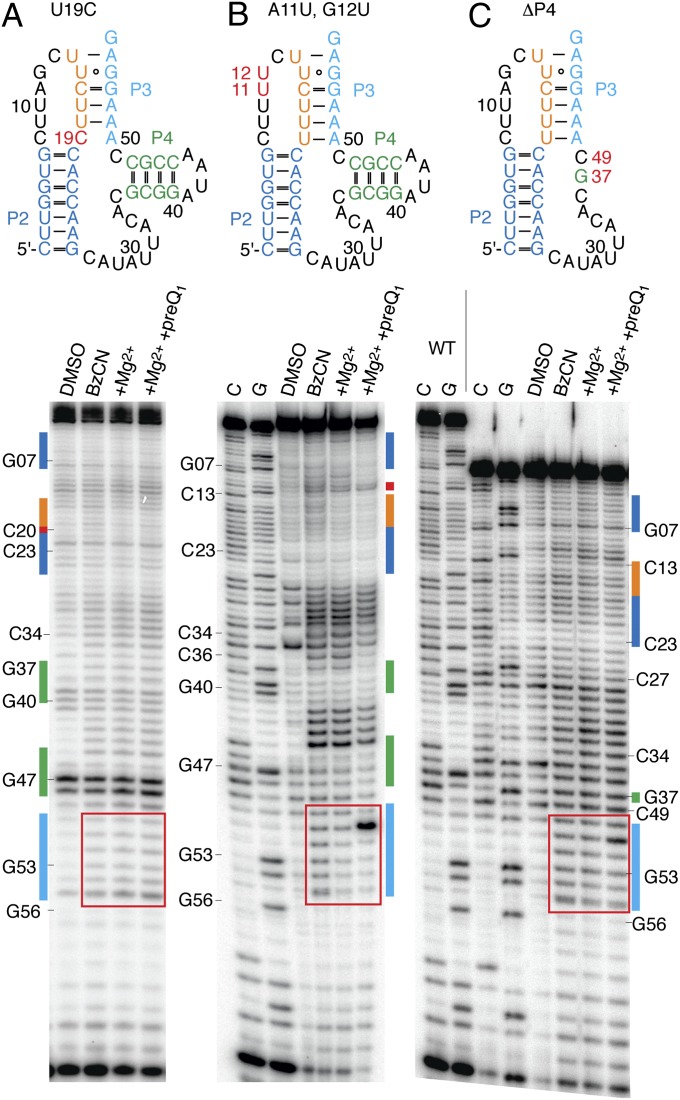
To assess the contribution of specific residues to pseudoknot formation and preQ1 binding, SHAPE analysis was performed on riboswitch constructs containing single point mutations. To test the role of U19 in preQ1 ligand recognition (discussed above), we first replaced this nucleotide with cytidine. Consistent with U19 being strictly required for pseudoknot preorganization and preQ1 binding, this mutant construct failed to exhibit any detectable changes in reactivity in the presence of magnesium and preQ1 (Fig. 3A). A similar analysis on constructs containing a C8-to-U mutation, a perturbation that causes an approximately two order of magnitude reduction in preQ1 affinity (14), also showed severely reduced binding (Fig. S3). We conclude that residues C8 and U19 either contribute to formation of the ligand binding pocket or directly interact with the preQ1 ligand in the bound complex. To evaluate whether nucleosides in the upper portion of junction J2–3 are also required for ligand recognition, residues A11 and G12 were mutated to uridines. These mutations were apparently tolerated, as SHAPE probing revealed a pattern of modification that was similar to the WT RNA (Fig. 3B and Fig. S3).
Fig. 3.
SHAPE analysis of preQ1-II RNA mutants. (A) Secondary structure and representative gel for the SHAPE probing of the preQ1-II C19U mutant with BzCN. Lanes from Left to Right: control in the absence of probing reagent (DMSO lane), probing in the absence of MgCl2 or ligand (BzCN) and presence of 5 mM Mg2+ ions, and 5 mM Mg2+ ions plus 10 μM preQ1. (B) Same as A for preQ1-II A11U/G12U mutant. (C) Same as A for a deletion mutant of preQ1-II RNA with ΔP4. The region that is highly indicative of preQ1 binding is highlighted by a red square. Secondary structures are color-coded to facilitate correlation of nucleoside positions with gel electrophoresis band shifts. Mutations and deletions highlighted by red lettering.
Consistent with the “extra” P4 stem–loop being critical for the preQ1-II riboswitch function, a double mutation (G39C/G40C) within this region was shown to abrogate interactions with preQ1 (14). Reasoning that the lack of binding may arise from the disruption of pseudoknot formation due to pairing of the G-rich sequence at the 3′ terminus (G53–G56) with the newly generated C track (C36–C40), a truncated aptamer was synthesized that entirely lacks the P4 stem–loop element (ΔP4; Fig. 3C). Here, a single guanosine (G37) was kept to directly bridge C36 of junction J2–4 and C49 of the aptamer 3′ terminal sequence. Strikingly, this construct retained the capacity to bind the preQ1 ligand (Fig. 3C). PreQ1-II RNAs with triple-uridine (ΔP4/U3) or triple-adenosine (ΔP4/A3) mutations of the bases surrounding the truncation (C36, G37, C49) also retained preQ1 binding, suggesting that the contribution of the J2-4 junction to preQ1 binding is largely sequence independent (Fig. S3 and Table S1).
To evaluate the relative ligand binding affinities of the WT and truncated aptamers in parallel, binding experiments were performed using riboswitch constructs containing a 2-Ap substitution at position A11 (Fig. S4). These experiments revealed that the truncated ΔP4 construct exhibited a Kd for preQ1 binding of 4.2 ± 0.1 μM, a roughly 10-fold decrease in affinity compared with the WT (A11Ap) aptamer construct (Kd = 0.43 ± 0.01 μM). Additional deletions in J2-4, which reduced the preQ1-II riboswitch motif to 41 nt, were also tolerated without further losses in preQ1 binding affinity (Table S1). We conclude that the P4 stem–loop is not essential but serves an important role of increasing the affinity of preQ1 binding.
To reveal the kinetics of the preQ1–aptamer interaction, pre–steady-state measurements were performed, where preQ1 binding was tracked by the increase in 2-Ap fluorescence over time. Here, the WT A11Ap system exhibited an apparent on rate, kon, of 2.9 ± 0.1 × 104 M−1⋅s−1 (Fig. S5). This rate, together with the estimated binding affinity, Kd, enabled us to estimate the dissociation rate of the preQ1 ligand, koff, as 1.2 × 10−2 s−1 (koff = Kd × kon). These estimates, which are based on local structural rearrangements of the RNA that report indirectly on the ligand binding process, are in line with the parameters reported previously for a number of other riboswitch aptamer domains (for comparison, see table S1 in ref. 22). Strikingly, the estimated kon for the truncated, ΔP4/A3 construct was ∼1.6 ± 0.1 × 106 M−1⋅s−1 (Fig. S5), more than an order of magnitude faster than the WT construct. Consequently, koff must also increase by an even greater extent (ca. 5 s−1). These findings show that the P4 helical element serves to modulate ligand binding affinity by altering both the rate of productive preQ1-riboswitch aptamer domain interactions as well as the stability of the ligand-bound state.
Dynamics of the preQ1-II Riboswitch Revealed by smFRET Imaging.
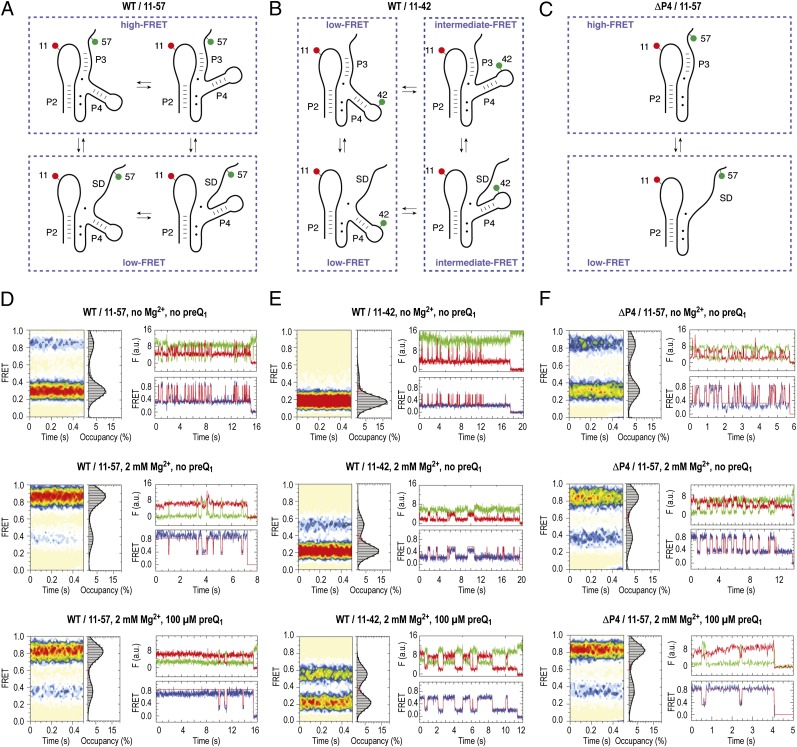
To elucidate dynamic features of the preQ1-II riboswitch RNA underpinning the folding and ligand recognition process, three distinct fluorescently labeled RNA constructs were created for smFRET imaging. These investigations focused on evaluating features of the regulatory interaction, namely the sequestration of the SD sequence through pairing with junction J2–3, as well as the role of the “extra” P4 stem–loop element. Two WT preQ1-II RNA constructs were synthesized carrying donor and acceptor fluorophores within junction J2–3 and very close to the SD sequence (Fig. 4A and Table S2), and within junction J2–3 and at the tip of P4, respectively (Fig. 4B and Table S2). To further examine the role of the P4 stem–loop element, a ΔP4 preQ1-II RNA construct was prepared, where donor and acceptor fluorophores were linked within the J2–3 and SD regions, respectively (Fig. 4C and Table S2). In each construct, the specific positions for fluorophore attachment were chosen based on our SHAPE probing data and mutational analysis, together with the 2-Ap data obtained here and elsewhere (35). We refer to these constructs as WT/11–57, WT/11–42, and ΔP4/11–57, where the acceptor fluorophore (Cy5) was linked at position 11 and the donor fluorophore (Cy3) at position 57 or 42. In each case, the dye linkage to the RNA was engineered via an extended linker to a 5-aminoallyluridine residue (Materials and Methods and Fig. S6). To enable surface immobilization of each construct for smFRET imaging, a 5′-biotin moiety was linked to the 5′-terminus. The final products were confirmed by liquid chromatography–electrospray ionization (LC-ESI) mass spectrometry (Fig. S6).
Fig. 4.
Dynamics of the preQ1 riboswitch aptamer analyzed by smFRET imaging and cartoon representations of hypothetic folding states predicted from secondary structure analysis and experimental FRET data. (A) Schematics of labeling pattern to sense pseudoknot formation. (B) Same as A but with labeling pattern to sense dynamics of the extra stem–loop P4. (C) Same as A but with labeling pattern to sense dynamics of pseudoknot formation of the ΔP4 deletion mutant. (D) Population FRET histograms showing the mean FRET values and percentage (%) occupancies of each state observed for the preQ1-II riboswitch (WT/11–57) in the absence of Mg2+ and preQ1 ligand, in the presence of 2 mM Mg2+ ions, and in the presence of 2 mM Mg2+ ions and 100 μM preQ1; corresponding fluorescence (green, Cy3; red, Cy5) and FRET (blue) trajectories of individual preQ1 aptamer molecules under the same conditions, where idealization of the data to a two-state Markov chain is shown in red. (E) Same as D but with WT/11–42 labeling scheme. (F) Same as D but with ΔP4 deletion.
Single-molecule imaging was performed using a wide-field total internal reflection fluorescence microscope as previously described (36). The dynamics of hundreds of individual surface-immobilized preQ1-II riboswitch molecules were tracked simultaneously over extended periods using an oxygen scavenging system in the presence of solution additives (37). Fluorescence resonance energy transfer efficiency (FRET) was calculated ratiometrically [FRET = ICy5/(ICy5 + ICy3)] to provide estimates of time-dependent changes in distance between donor and acceptor fluorophores (38, 39). Here, τFRET was ∼4.0 s, predominantly limited by photobleaching of the Cy5 fluorophore. The dynamic behaviors of individual molecules were assessed using hidden Markov modeling procedures (Materials and Methods), and ensemble information was obtained by combining FRET trajectories from individual molecules into population FRET histograms. Measurements of each construct were performed under three distinct conditions: (i) in the absence of Mg2+ and preQ1; (ii) in presence of 2 mM Mg2+ ions and absence of preQ1; and (iii) in presence of 2 mM Mg2+ ions and saturating concentrations of the preQ1 ligand (100 µM) (Fig. 4).
In the absence of both ligands, the WT/11–57 preQ1-II construct exhibited a dominant low-FRET (0.3) configuration (Fig. 4D, Top Left). This FRET value is consistent with a relatively open conformation in which the pseudoknot is not formed and the dyes are separated by ∼60–70 Å [assuming an R0 of ∼56 Å (40, 41)]. Under these conditions, this construct also exhibited roughly 20% high-FRET state occupancy (∼0.84 FRET) on a time-averaged basis, consistent with an interdye distance of <20 Å (Fig. 4D, Top Left). Inspection of individual FRET trajectories revealed that occupancy of the high-FRET state arose from frequent (∼4 s−1) excursions out of the predominant low-FRET configuration that were transient in nature (∼90-ms lifetime; Table S3 and Fig. 4D, Top Right). These data suggest that the WT preQ1-II riboswitch can spontaneously achieve a pseudoknot-like fold in the absence of Mg2+ and preQ1 ligands that is intrinsically unstable.
The high-FRET state was stabilized ∼10-fold in the presence of Mg2+ (2 mM), resulting in a 20:80 distribution of open (low-FRET) and compacted (high-FRET) riboswitch conformations (Fig. 4D, Middle Left). Visual inspection of individual smFRET trajectories revealed that this distribution could be attributed in part to a shift in the relative stability of the high-FRET state (ca. ∼1-s lifetime) and residual dynamics where the low-FRET, open configuration was transiently sampled (ca. ∼350-ms average lifetime). Notably, roughly two-thirds of the low-FRET population (∼15% of all molecules) appeared unaffected by the presence of Mg2+. The absolute value of the low-FRET state was observed to increase by 0.05 (whereas the high-FRET value remained unchanged), suggesting that Mg2+ binding promotes compaction or rigidification of the open conformation of the preQ1-II riboswitch fold. In the presence of Mg2+ and preQ1-II, the high-FRET state was further stabilized (approximately twofold), whereas the lifetime of the low FRET state and number of molecules apparently unaffected by the presence of ligands remained largely unchanged. Interestingly, the mean FRET value of the high-FRET state decreased by ∼0.05 to a value of 0.8. This change is consistent with the observations from SHAPE and 2-Ap measurements (see ref. 35 and Fig. S3), which suggested that the nucleobase at position 11 (to which Cy5 is attached), adopts an unstacked, extrahelical position upon preQ1 binding, a configuration that is expected to modestly increase the interdye distance. Taken together, these data suggest that the high-FRET, compacted state observed for this construct represents a fully folded riboswitch configuration.
Uncoupled Dynamic Behavior of P4 Positioning and Pseudoknot Folding.
We next set out to investigate the preQ1-II variant WT/11–42, whose labeling pattern directly reports on P4 movements relative to junction J2–3. Like the WT/11–57 construct, the dynamic behavior of this system is expected to be influenced by pseudoknot formation (Fig. 4B). In the absence of Mg2+ and preQ1, this construct exhibited a dominant low-FRET (∼0.18) configuration (Fig. 4E, Top Left). Stem P4, like P2, is folded under these conditions (see above), and therefore, the larger average distance between the two dyes in construct WT/11–42 (0.18 FRET) compared with WT/11–57 (0.3 FRET) can be rationalized by a conformation with stem P4 directed away from J2–3 (Fig. 4B, left-side cartoons). Under these conditions, an intermediate-FRET (around ∼0.5) configuration was also observed, albeit at very low occupancy (less than 10%) (Fig. 4E, Top). Inspection of individual FRET trajectories revealed that such conformations arise from frequent (∼2 s−1) excursions from the predominant, low-FRET configuration to transient (∼40-ms lifetime) intermediate-FRET states. As this lifetime approaches the imaging frame rate (15-ms integration time), the estimated lifetime of such configurations is likely an upper bound.
In the presence of Mg2+, intermediate-FRET (0.54) configurations exhibited a fivefold increase in average lifetime (∼190 ms; Table S3). Correspondingly, the intermediate-FRET configuration became significantly more populated on a time-averaged basis (∼20%) (Fig. 4E, Middle). Addition of saturating concentrations of preQ1 (100 µM) stabilized the intermediate-FRET configuration by an additional eightfold (∼1.6-s lifetime; Table S3). However, under these conditions, where our 2-Ap data (Fig. S4) suggest that the preQ1 binding site is saturated, the intermediate-FRET state exhibited only partial occupancy (roughly 50%) (Fig. 4E, Bottom). Although direct comparisons between these experiments must be interpreted with caution (e.g., different modifications, variations in Mg2+ concentrations, etc.), taken together with the distinct kinetic signatures of this construct relative to the WT/11–57 system, this observation suggests that the P4 element remains flexible in the context of an otherwise compacted riboswitch fold (Fig. 4 A and B). We speculate that the preQ1-II riboswitch undergoes conformational changes in the P4 stem–loop region that place the tip of P4 closer (∼55 Å) and further away (∼65 Å) from J2–3. Rearrangements in this region may conceivably arise if a rigid P4 stem undergoes a hinge-like motion relative to stem P2 (and the putatively coaxially stacked P3 stem) without making extensive tertiary contacts to the pseudoknot core. Such conformational plasticity is indeed consistent with our SHAPE analysis data (Figs. 2 and 3). In light of our mutational and 2-Ap data (Fig. 3 and Fig. S4), we conclude that these structural and dynamic features of the P4 stem contribute in some manner to preQ1 binding.
Truncation of P4-L4 Impacts the Dynamics and Stability of the Pseudoknot Fold.
Similar to the WT/11–57 system, in the absence of both Mg2+ and preQ1, the ΔP4/11–57 construct exhibited a two-state behavior, where low-FRET (∼0.3) and high-FRET (∼0.8) states were present at a ratio of 30:70 in favor of the lower-FRET, open riboswitch fold (Fig. 4F, Top). The modest increase in high-FRET state occupancy observed for this system (∼30% vs. ∼20% for the WT/11–57 construct) could be principally attributed to a roughly threefold increase in the high-FRET state lifetime relative to the WT system (270 ms vs. 90 ms). This finding suggests that the P4 stem–loop destabilizes the compacted pseudoknot configuration in the absence of ligand (Fig. 5 and Table S3). Collectively, these data suggest that truncation of the P4 stem–loop entropically favors pseudoknot formation.
Fig. 5.
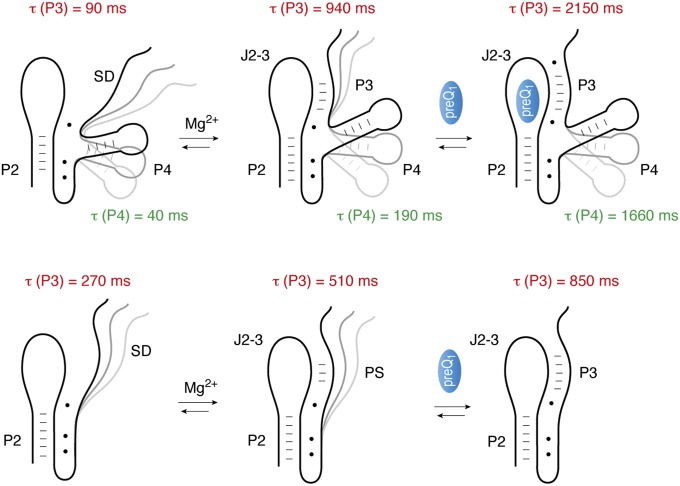
Model for ligand recognition and folding of the preQ1-II riboswitch. For direct comparison, lifetimes of closed pseudoknot conformations τ(P3) in the absence and presence of ligands (Mg2+, preQ1) are depicted (in red) next to each structure cartoon. Lifetimes of compacted P4/J2–3 conformations τ(P4) are indicated in green. The pseudoknot becomes prefolded in the presence of Mg2+ ions and further stabilized in the presence of preQ1. The extra stem–loop P4 behaves rather uncoupled from pseudoknot formation and retains higher dynamics throughout all three conditions (no ligands; Mg2+; Mg2+ and preQ1) investigated here, even in the fully folded, preQ1-bound state.
Similar to the WT system, the high-FRET configuration became dominant (75%) in the presence of Mg2+ (Fig. 4F, Middle). However, two distinctions from the WT system could be discerned. First, all molecules appeared to be Mg2+ responsive. Second, the lifetime of a high-FRET, compacted preQ1-II fold, was reduced nearly twofold compared with the WT system (510 ms for ΔP4/11–57 vs. 940 ms for WT/11–57; Table S3). This trend was even more pronounced in the presence of saturating (100 µM) concentrations of the preQ1 ligand (Fig. 4F, Bottom). The lifetime of the high-FRET pseudoknot configuration was approximately threefold lower for the ΔP4/11–57 construct compared with the WT/11–57 system (approximately ∼850 ms vs. 2.2 s) (Table S3). Similar to the WT construct, the absolute value of the high-FRET configuration decreased by about ∼0.05 to a value of 0.8 in the ligand-bound state, supporting the notion that WT and truncated versions behave comparably with respect to conformational changes toward the fully folded pseudoknot structure. The distinct dynamic behaviors of the ΔP4/11–57 construct shed light on the altered ligand binding affinity of this system. Here, the increased ligand on rate observed in bulk (Fig. S5) may relate to the increased propensity of this construct to adopt a pseudoknot fold, whereas the increased ligand dissociation rate may stem from the reduced stability of the ligand-bound state.
Model for Folding and Ligand Recognition of the preQ1-II Riboswitch.
Gene-regulating mRNA riboswitches often use pseudoknots as scaffolding for selective recognition of small molecules. The structural organization of a pseudoknot—namely, a stem–loop with a short sequence overhang that folds back onto the loop region—provides the platform for folding and biological function. Sequestration versus liberation of functional sequence elements within the pseudoknot thereby directly impacts transcriptional, translational or RNA processing machinery.
Recent high-resolution structures of pseudoknot-forming SAH, SAM-II, and preQ1-I riboswitch aptamers (15–17, 19, 20) have revealed how this fundamental RNA fold can create a high-affinity ligand binding pocket. Additionally, a detailed smFRET investigation of the SAM-II riboswitch has shed much-needed light on the principles of folding dynamics in a ligand-responsive pseudoknot that harbors the decision-making regulatory interaction (42). The SAM-II RNA pseudoknot employs a defined two-state behavior, where the closed, pseudoknot-like conformation is transiently sampled in the absence of ligand. Under physiological buffer conditions containing Mg2+ ions, the lifetime of the closed pseudoknot conformation increases to on the order of hundreds of milliseconds and becomes even more dramatically stabilized when the cognate SAM ligand is bound. The lifetime of the fully folded structure can reach the order of seconds or even tens of seconds (42).
Although the preQ1 class II riboswitch falls into a similar pseudoknot category as the preQ1-I and SAM-II riboswitches, it differs in that it contains an internal stem–loop extension immediately 5′ to the actual pseudoknot pairing interaction, the extra P4 stem–loop. When this second class of preQ1 riboswitches was discovered, this extension had been described as an essential element as mutations in this region abrogated ligand binding (14). By imaging this system from multiple structural perspectives using smFRET, the present investigation has shed important light on the functional role of this additional structural element.
The first unexpected observation was that pseudoknot conformations of the preQ1-II riboswitch were significantly more populated in the absence of the cognate ligand (∼85% occupancy in the presence of Mg2+ alone) compared with the SAM-II system (42). Nevertheless, like the SAM-II riboswitch, the preQ1-II system exhibited pronounced “breathing” with average lifetimes for the closed pseudoknot that were only increased roughly twofold compared with SAM-II. The second unexpected observation was that, although the pseudoknot interaction that is crucial to riboswitch function became stabilized upon preQ1 binding, persistent flexibility was observed in what should be the preQ1-bound state (200 times the apparent Kd). Such dynamics were particularly apparent in the position of the extra P4 stem–loop. Consistent with our SHAPE analysis, two configurations of P4 with roughly similar stability remained in dynamic exchange. This distinct dynamic signature suggests that pseudoknot dynamics and P4 stem–loop motions are structurally only loosely coupled.
Our investigations into the role of the P4 stem–loop extension using a truncated ΔP4/11–57 construct surprisingly revealed only a 10-fold decrease in preQ1 ligand affinity. This observation suggests that the truncated system may retain a significant degree of signaling functionality. However, the kinetic features of preQ1 binding of the truncated preQ1-II riboswitch were even more substantially altered: both the on- and off-rates were estimated to have increased by more than an orders of magnitude. Such disparities were manifested in the structural dynamics observed through smFRET, but the relationship was highly nonlinear. Here, the lifetimes of the pseudoknot-like conformation, both in the absence and presence of ligands, differed only modestly (Fig. 5) by comparison with the kinetic parameters of ligand binding deduced from our bulk studies (Fig. S5). Although a deeper understanding of the precise relationship between ligand binding and riboswitch conformation requires further investigation, we speculate based on our findings that the P4 stem–loop element serves to tune the structural stabilities of open and compacted configurations, and therefore the ligand responsiveness of the system, in a manner that is critical for proper regulation of the translation apparatus. Here, the extra stem–loop in preQ1-II indirectly affords submicromolar ligand affinity and tunes the dynamic features of the riboswitch folding landscape as well as the kinetic features underpinning ligand binding. By contrast, the extra P2/P2b stem–loop in the SAH riboswitch is essential because it directly contributes to ligand recognition and binding (17).
Large, Structurally Complex Riboswitches with Pseudoknots and Extensions.
Although the relatively small size of the preQ1-II, SAM-II, and SAH riboswitches (∼50 nt) may significantly reduce the complexity of the folding landscape, the present investigations demonstrating the impact of the P4 stem–loop element insertion on the kinetic and structural features of the preQ1-II riboswitch argue that substantial challenges remain toward gaining a complete understanding of even the most compact aptamer folds. Larger (100- to 200-nt) riboswitches such as SAM-I and B12 riboswitches, and the glmS riboswitch-ribozyme also contain pseudoknots as integral structural components (Table S4). In the case of the SAM-I riboswitch, a helical extension (P4) is located immediately 3′ to the pseudoknot and its deletion reduces ligand binding affinity (43). However, to the best of our knowledge, the effects of this extension on the dynamics of the SAM-I pseudoknot and the kinetic features of ligand binding have yet to be determined. In the glmS system (44), a large extension (P4-P4.1) impacts pseudoknot formation (P3.1) and is crucial for self-cleavage activity (45). In this system, the extension is involved in a series of tertiary interactions with the pseudoknot core and may be anticipated to affect both the stability of the overall fold as well the kinetics of ligand binding. For the B12 riboswitch, a large structural extension (P6 versus P6-P11) in the pseudoknot core aids in the ligand discrimination process (46, 47). Again, the influence of this extension on the dynamics of folding and ligand binding have yet to be explored.
Comparison with Other Riboswitches Analyzed by smFRET.
Detailed smFRET studies have been recently performed on a number of RNA systems, including the c-di-GMP, SAM-I, and lysine riboswitches, to elucidate the dynamics underlying the riboswitch folding process. In the case of the c-di-GMP riboswitch (48), such studies revealed that the free RNA occupies four distinct configurations that appeared to differ dramatically in their dynamic properties and apparent stabilities. Here, evidence was presented that all four states interconvert over very long (ca. minutes) timescales and that allosteric interactions accelerate ligand recognition by aiding preorganization of the RNA aptamer fold to favor rapid c-di-GMP binding. For the SAM-I riboswitch, a dynamic three-state exchange process was uncovered where the ligand-free state exhibited unfolding dynamics in the milliseconds regime and the most compacted fold was frozen in the presence of ligand (43). For the lysine riboswitch, smFRET was used to measure the opening/closing rate constants for the aptamer domain as well as the lysine dissociation constant (9). Although these studies have provided an important foundation for understanding the relationship between riboswitch dynamics, ligand binding and gene expression regulation, continued, in-depth investigations along these lines of pursuit are greatly needed. For instance, in most smFRET studies, only a single structural perspective on riboswitch dynamics has been obtained (e.g., from just one FRET pair). The importance of applying multiple structural perspectives in smFRET imaging experiments is exemplified by the present investigation, prior studies of the TPP riboswitch aptamer (49), as well as more complex systems such as DNA helicase (50) and the bacterial ribosome (51, 52). Such studies have the potential to uncover unexpected complexities in the structure and dynamics that would be difficult or impossible to discern through bulk techniques. This argument is particularly relevant to systems that exhibit asynchronous behaviors as well as static and dynamic heterogeneities where the relationship between structural dynamics and ligand binding is obscured. The present study on the preQ1-II riboswitch highlights this point given the nonlinear relationship between ligand binding kinetics and structural dynamics of the aptamer fold. Here, further clarification of the precise relationship between structure, dynamics and function would be greatly aided by multicolor smFRET studies where ligand binding and aptamer fold could be measured simultaneously. Ultimately, efforts of this kind are essential to enabling the rational design of artificial riboswitch systems as efficient tools for precise gene regulation in diverse cellular contexts.
Concluding Remarks.
During review and revision of the present investigation, the molecular details of the preQ1-RNA class II interaction were revealed at high-resolution for the first time (53). The results presented are entirely consistent with the structure presented in this study, which show the P4 helix organized roughly perpendicular to the long axis of aptamer pseudoknot.
Materials and Methods
Preparation of Cy3/Cy5-Labeled RNA.
Solid-phase RNA synthesis (54, 55) was performed as described in the SI Materials and Methods. (Sulfo-) Cy3 and (sulfo-) Cy5 NHS ester were purchased from GE Healthcare or Lumiprobe. DMSO was dried over activated molecular sieves. Dye-NHS ester (1 mg; ∼1,200 nmol) was dissolved in anhydrous DMSO (500 μL, dried over activated molecular sieves). Lyophilized RNA (20 nmol) containing a 5-aminoallyl-uridine modification was dissolved in labeling buffer (50–100 mM phosphate buffer, pH 8.0), and nanopure water was added to reach a fraction of 55% (vol/vol) (49 µL) of the intended final reaction volume (89 µL) with a final concentration of cRNA of 225 µM. The corresponding volume of the dye-NHS ester solution [45% (vol/vol)] (40 µL) was added to the RNA solution (to reach a concentration of cDye = 1,124 µM in the final reaction volume). The reaction mixture was gently tumbled on a shaker overnight at room temperature in the dark. The reaction was stopped by precipitation with absolute ethanol and sodium acetate for 30 min at –20 °C followed by centrifugation for 30 min at 4 °C at 13,000 rpm Eppendorf 5430R, rotor F-45-30-11. The colored pellets were dried, resuspended in water, and purified by anion-exchange chromatography on a Dionex DNAPac100 column (9 × 250 mm) at 60 °C. Flow rate was 2 mL/min; gradient was Δ12–22% B in A within 20 min; UV detection was at a wavelength λ of 260 nm (RNA), 548 nm (Cy3), and 646 (or 595) nm (Cy5). Fractions containing labeled oligonucleotide were loaded on a C18 SepPak cartridge (Waters/Millipore), washed with 0.1 M (Et3NH)+HCO3− and H2O, eluted with H2O/CH3CN (1/1), and lyophilized to dryness.
Enzymatic ligation.
PreQ1-II RNA aptamers containing 2-aminopurine or 5′-biotinylated, and Cy3/Cy5 labels were prepared by splinted enzymatic ligation of two chemically synthesized fragments (Fig. S6) using T4 DNA ligase (Fermentas) (56). Briefly, 10 μM of each RNA fragment, 15 μM of a 25-nt DNA splint oligonucleotide (IDT), and a final T4 DNA ligase (Fermentas) concentration of 0.5 U/μL in a total volume of 1 mL were incubated for 6–20 h at 37 °C in the provided buffer system. Analysis of the ligation reaction and purification of the ligation products were performed by anion exchange chromatography and the final product confirmed by LC-ESI mass spectrometry.
smFRET experiments.
smFRET data were acquired using a prism-based total internal reflection microscope, where the biotinylated preQ1 riboswitch was surface immobilized within PEG-passivated, streptavidin-coated quartz microfluidic devices (36). The Cy3 fluorophore was directly illuminated under 1.5 kW cm−2 intensity at 532 nm (Laser Quantum). Photons emitted from both Cy3 and Cy5 were collected using a 1.2 N.A. 60× Plan-APO water-immersion objective (Nikon), where optical treatments were used to spatially separate Cy3 and Cy5 frequencies onto two synchronized EMCCD devices (Evolve 512; Photometrics). Fluorescence data were acquired using MetaMorph acquisition software 13 (Universal Imaging Corporation) at a rate of 66.7 frames per second (15-ms integration). Fluorescence trajectories were selected from the movie files for analysis using automated image analysis software coded in MATLAB (The MathWorks). Fluorescence trajectories were selected on the basis of the following criteria: a single catastrophic photobleaching event, at least 6:1 signal-to-background noise ratio calculated from the total fluorescence intensity, and a FRET lifetime of at least 30 frames (450 ms) in any FRET state ≥0.15. smFRET trajectories were calculated from the acquired fluorescence data using the formula FRET = ICy5/(ICy3 + ICy5), where ICy3 and ICy5 represent the Cy3 and Cy5 fluorescence intensities, respectively. Equilibrium smFRET experiments were performed in 50 mM 3-(N-morpholino)propanesulfonate-KOH (KMOPS), 100 mM KCl, pH 7.5, buffer in the presence of an optimized oxygen scavenging and triplet state quenching mixture in the presence of an oxygen scavenging environment (1 unit protocatchuate-3,4-dioxygenase, 2 mM protocatechuic acid; 1 mM Trolox, 1 mM cyclooctatetraene, 1 mM nitrobenzyl-alcohol) (37). All smFRET experiments were performed at 25 °C. Concentrations of MgCl2 and preQ1 were as specified in the individual figure captions. FRET state occupancies and transition rates were estimated by idealization to a two-state Markov chain model using the segmental k-means algorithm implemented in QuB (57).
RNA Transcription for Chemical Probing.
PreQ1-II RNAs of 74–85 nt were synthesized using a pair of complementary oligonucleotides (IDT) including a T7 RNA promoter followed by the sequence of the RNA with flanking 5′ and 3′ linkers for reverse transcription. Following transcription at 37 °C for 2 h, phenol/chloroform extraction, and isopropanol precipitation, the RNA substrates were separated on a denaturing 8% (vol/vol) polyacrylamide gel (90 min, 28 W) and visualized by UV shadowing. The corresponding bands were excised and eluted from the gel by an overnight incubation in 0.1% SDS/0.5 M ammonium acetate. The RNAs were then precipitated with isopropanol, and the pellets were resuspended in nanopure water. The RNA substrates were then quantitated by spectrophotometry and stored at –20 °C.
RNA 2′-Hydroxyl Acylation by Benzoyl Cyanide.
Reaction mixtures containing T7-transcribed unlabeled RNA (5 pmol) with a 3′-end flanking sequence and 50 mM KMOPS, pH 7.5, 100 mM KCl, in the presence or absence of 5 mM MgCl2 and 10 µM preQ1 were heated at 65 °C for 2 min, cooled to 4 °C for 5 min, and incubated at 37 °C for 25 min in an Eppendorf Mastercycler (VWR). Following incubation, the control background reaction was treated with anhydrous DMSO, while the probing reagent benzoyl cyanide (BzCN), dissolved in DMSO, was added to the probing reaction mixtures for a final concentration of 55 mM. The RNA was recovered by ethanol precipitation with sodium acetate and glycogen. The RNA samples were resuspended in 8 μL of sterile water after centrifugation and stored at –20 °C.
Primer Extension.
DNA primers (18 nt) were 5′–end-labeled with [γ-32P]ATP (Hartmann Analytic) using T4 polynucleotide kinase (Fermentas) according to the manufacturer’s instructions. Three microliters of labeled DNA primer was added to 8 μL of RNA from BzCN 2′-hydroxyl acylation and allowed to anneal at 65 °C for 5 min, then incubated at 35 °C for 5 min, and cooled at 4 °C for 1 min in an Eppendorf Mastercycler. Eight microliters of a mix containing 4 μL of 5× first strand buffer (250 mM Tris⋅HCl, pH 8.3, 375 mM KCl, 15 mM MgCl2), 1 μL of 0.1 M DTT, 1 μL of 10 mM dNTPs mixture, and 2 μL of DMSO were then added to the reactions, followed by incubation at 61 °C for 1 min, addition of 0.4 μL of SuperScript III Reverse Transcriptase (Invitrogen), and further incubation at 61 °C for 10 min. Reactions were then stopped by addition of 1 μL of 4 M NaOH and incubation at 95 °C for 5 min. Radiolabeled cDNA strands were recovered by ethanol precipitation with sodium acetate and glycogen. The samples were resuspended in 8 μL of migration buffer [xylene cyanol, 97% (vol/vol) formamide, 10 mM EDTA] after centrifugation. Sequencing ladders were produced by adding 1 μL of 10 mM ddGTP or ddCTP in addition to the 8-μL reaction mixture of unmodified RNA samples, before incubation at 61 °C. Electrophoresis on a 10% (vol/vol) polyacrylamide gel for 95 min at 45 W was used to separate 300–500 cpm of the generated cDNA fragments. The gel was dried using a Vacuum Gel Dryer (VWR) at 75 °C for 45–60 min. Following overnight exposition on a 32P-sensitive phosphorscreen, the primer extension labeling was revealed by autoradiography. Band intensities visualized by gel electrophoresis were quantified using SAFA, version 1.1 [Semi-Automated Footprinting Analysis (34)]. Datasets were normalized for loading variations and RT efficiency by dividing all intensities by the intensity of the last bases of primer extension. Final results for graphical representation were obtained by subtracting the DMSO control background from the BzCN-probed reaction intensities.
Supplementary Material
Acknowledgments
This work was funded by Austrian Science Foundation (Fonds zur Förderung der Wissenschaftlichen Forschung) Projects I1040, P21641 (to R.M.), and M1449 (to M.F.S.), National Science Foundation Project 1223732 (to S.C.B.), and The Irma T. Hirschl/Monique Weill Caulier Trust (S.C.B. and R.B.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1304585110/-/DCSupplemental.
References
- 1.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43(6):867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garst AD, Edwards AL, Batey RT. Riboswitches: Structures and mechanisms. Cold Spring Harb Perspect Biol. 2011;3(6):a003533. doi: 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deigan KE, Ferré-D’Amaré AR. Riboswitches: Discovery of drugs that target bacterial gene-regulatory RNAs. Acc Chem Res. 2011;44(12):1329–1338. doi: 10.1021/ar200039b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serganov A, Patel DJ. Molecular recognition and function of riboswitches. Curr Opin Struct Biol. 2012;22(3):279–286. doi: 10.1016/j.sbi.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blouin S, Mulhbacher J, Penedo JC, Lafontaine DA. Riboswitches: Ancient and promising genetic regulators. ChemBioChem. 2009;10(3):400–416. doi: 10.1002/cbic.200800593. [DOI] [PubMed] [Google Scholar]
- 6.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29(1):11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 7.Schwalbe H, Buck J, Fürtig B, Noeske J, Wöhnert J. Structures of RNA switches: Insight into molecular recognition and tertiary structure. Angew Chem Int Ed Engl. 2007;46(8):1212–1219. doi: 10.1002/anie.200604163. [DOI] [PubMed] [Google Scholar]
- 8.Serganov A, Nudler E. A decade of riboswitches. Cell. 2013;152(1–2):17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fiegland LR, Garst AD, Batey RT, Nesbitt DJ. Single-molecule studies of the lysine riboswitch reveal effector-dependent conformational dynamics of the aptamer domain. Biochemistry. 2012;51(45):9223–9233. doi: 10.1021/bi3007753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corbino KA, et al. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 2005;6(8):R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang JX, Breaker RR. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol. 2008;86(2):157–168. doi: 10.1139/O08-008. [DOI] [PubMed] [Google Scholar]
- 12.Poiata E, Meyer MM, Ames TD, Breaker RR. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA. 2009;15(11):2046–2056. doi: 10.1261/rna.1824209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth A, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14(4):308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]
- 14.Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA. 2008;14(4):685–695. doi: 10.1261/rna.937308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilbert SD, Rambo RP, Van Tyne D, Batey RT. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol. 2008;15(2):177–182. doi: 10.1038/nsmb.1371. [DOI] [PubMed] [Google Scholar]
- 16.Klein DJ, Edwards TE, Ferré-D’Amaré AR. Cocrystal structure of a class I preQ1 riboswitch reveals a pseudoknot recognizing an essential hypermodified nucleobase. Nat Struct Mol Biol. 2009;16(3):343–344. doi: 10.1038/nsmb.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edwards AL, Reyes FE, Héroux A, Batey RT. Structural basis for recognition of S-adenosylhomocysteine by riboswitches. RNA. 2010;16(11):2144–2155. doi: 10.1261/rna.2341610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state. Wiley Interdiscip Rev RNA. 2012;3(3):369–384. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang M, Peterson R, Feigon J. Structural insights into riboswitch control of the biosynthesis of queuosine, a modified nucleotide found in the anticodon of tRNA. Mol Cell. 2009;33(6):784–790. doi: 10.1016/j.molcel.2009.02.019. [DOI] [PubMed] [Google Scholar]
- 20.Spitale RC, Torelli AT, Krucinska J, Bandarian V, Wedekind JE. The structural basis for recognition of the PreQ0 metabolite by an unusually small riboswitch aptamer domain. J Biol Chem. 2009;284(17):11012–11016. doi: 10.1074/jbc.C900024200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rieder U, Lang K, Kreutz C, Polacek N, Micura R. Evidence for pseudoknot formation of class I preQ1 riboswitch aptamers. ChemBioChem. 2009;10(7):1141–1144. doi: 10.1002/cbic.200900155. [DOI] [PubMed] [Google Scholar]
- 22.Rieder U, Kreutz C, Micura R. Folding of a transcriptionally acting preQ1 riboswitch. Proc Natl Acad Sci USA. 2010;107(24):10804–10809. doi: 10.1073/pnas.0914925107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng J, Walter NG, Brooks CL., 3rd Cooperative and directional folding of the preQ1 riboswitch aptamer domain. J Am Chem Soc. 2011;133(12):4196–4199. doi: 10.1021/ja110411m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Kang M, Peterson RD, Feigon J. Comparison of solution and crystal structures of preQ1 riboswitch reveals calcium-induced changes in conformation and dynamics. J Am Chem Soc. 2011;133(14):5190–5193. doi: 10.1021/ja111769g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eichhorn CD, et al. Unraveling the structural complexity in a single-stranded RNA tail: Implications for efficient ligand binding in the prequeuosine riboswitch. Nucleic Acids Res. 2012;40(3):1345–1355. doi: 10.1093/nar/gkr833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenkins JL, Krucinska J, McCarty RM, Bandarian V, Wedekind JE. Comparison of a preQ1 riboswitch aptamer in metabolite-bound and free states with implications for gene regulation. J Biol Chem. 2011;286(28):24626–24637. doi: 10.1074/jbc.M111.230375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santner T, Rieder U, Kreutz C, Micura R. Pseudoknot preorganization of the preQ1 class I riboswitch. J Am Chem Soc. 2012;134(29):11928–11931. doi: 10.1021/ja3049964. [DOI] [PubMed] [Google Scholar]
- 28.Yu C-H, Luo J, Iwata-Reuyl D, Olsthoorn RCL. Exploiting preQ1 riboswitches to regulate ribosomal frameshifting. ACS Chem Biol. 2013;8(4):733–740. doi: 10.1021/cb300629b. [DOI] [PubMed] [Google Scholar]
- 29.Han K, Byun Y. PSEUDOVIEWER2: Visualization of RNA pseudoknots of any type. Nucleic Acids Res. 2003;31(13):3432–3440. doi: 10.1093/nar/gkg539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aalberts DP, Hodas NO. Asymmetry in RNA pseudoknots: Observation and theory. Nucleic Acids Res. 2005;33(7):2210–2214. doi: 10.1093/nar/gki508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Batenburg FH, Gultyaev AP, Pleij CW, Ng J, Oliehoek J. PseudoBase: A database with RNA pseudoknots. Nucleic Acids Res. 2000;28(1):201–204. doi: 10.1093/nar/28.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hajdin CE, et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci USA. 2013;110(14):5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res. 2011;44(12):1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11(3):344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Soulière MF, Haller A, Rieder R, Micura R. A powerful approach for the selection of 2-aminopurine substitution sites to investigate RNA folding. J Am Chem Soc. 2011;133(40):16161–16167. doi: 10.1021/ja2063583. [DOI] [PubMed] [Google Scholar]
- 36.Munro JB, Altman RB, O’Connor N, Blanchard SC. Identification of two distinct hybrid state intermediates on the ribosome. Mol Cell. 2007;25(4):505–517. doi: 10.1016/j.molcel.2007.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dave R, Terry DS, Munro JB, Blanchard SC. Mitigating unwanted photophysical processes for improved single-molecule fluorescence imaging. Biophys J. 2009;96(6):2371–2381. doi: 10.1016/j.bpj.2008.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5(6):507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ditzler MA, Alemán EA, Rueda D, Walter NG. Focus on function: Single molecule RNA enzymology. Biopolymers. 2007;87(5–6):302–316. doi: 10.1002/bip.20819. [DOI] [PubMed] [Google Scholar]
- 40.Clegg RM. Fluorescence resonance energy transfer and nucleic acids. Methods Enzymol. 1992;211:353–388. doi: 10.1016/0076-6879(92)11020-j. [DOI] [PubMed] [Google Scholar]
- 41.Iqbal A, et al. Orientation dependence in fluorescent energy transfer between Cy3 and Cy5 terminally attached to double-stranded nucleic acids. Proc Natl Acad Sci USA. 2008;105(32):11176–11181. doi: 10.1073/pnas.0801707105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat Chem Biol. 2011;7(6):393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 43.Heppell B, et al. Molecular insights into the ligand-controlled organization of the SAM-I riboswitch. Nat Chem Biol. 2011;7(6):384–392. doi: 10.1038/nchembio.563. [DOI] [PubMed] [Google Scholar]
- 44.Klein DJ, Ferré-D’Amaré AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313(5794):1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 45.Wilkinson SR, Been MD. A pseudoknot in the 3′ non-core region of the glmS ribozyme enhances self-cleavage activity. RNA. 2005;11(12):1788–1794. doi: 10.1261/rna.2203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson JE, Jr, Reyes FE, Polaski JT, Batey RT. B12 cofactors directly stabilize an mRNA regulatory switch. Nature. 2012;492(7427):133–137. doi: 10.1038/nature11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peselis A, Serganov A. Structural insights into ligand binding and gene expression control by an adenosylcobalamin riboswitch. Nat Struct Mol Biol. 2012;19(11):1182–1184. doi: 10.1038/nsmb.2405. [DOI] [PubMed] [Google Scholar]
- 48.Wood S, Ferré-D’Amaré AR, Rueda D. Allosteric tertiary interactions preorganize the c-di-GMP riboswitch and accelerate ligand binding. ACS Chem Biol. 2012;7(5):920–927. doi: 10.1021/cb300014u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Haller A, Altman RB, Soulière MF, Blanchard SC, Micura R. Folding and ligand recognition of the TPP riboswitch aptamer at single-molecule resolution. Proc Natl Acad Sci USA. 2013;110(11):4188–4193. doi: 10.1073/pnas.1218062110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park J, et al. PcrA helicase dismantles RecA filaments by reeling in DNA in uniform steps. Cell. 2010;142(4):544–555. doi: 10.1016/j.cell.2010.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29(4):770–781. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Munro JB, et al. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci USA. 2010;107(2):709–714. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liberman JA, Salim M, Krucinska J, Wedekind JE. Structure of a class II preQ1 riboswitch reveals ligand recognition by a new fold. Nat Chem Biol. 2013;9(6):353–355. doi: 10.1038/nchembio.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pitsch S, Weiss PA, Jenny L, Stutz A, Wu X. Reliable chemical synthesis of oligoribonucleotides (RNA) with 2′-O-[(triisopropylsilyl)oxy]methyl (2′-O-tom)-protected phosphoramidites. Helv Chim Acta. 2001;84(12):3773–3795. [Google Scholar]
- 55.Micura R. Small interfering RNAs and their chemical synthesis. Angew Chem Int Ed Engl. 2002;41(13):2265–2269. doi: 10.1002/1521-3773(20020703)41:13<2265::AID-ANIE2265>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 56.Lang K, Micura R. The preparation of site-specifically modified riboswitch domains as an example for enzymatic ligation of chemically synthesized RNA fragments. Nat Protoc. 2008;3(9):1457–1466. doi: 10.1038/nprot.2008.135. [DOI] [PubMed] [Google Scholar]
- 57.Qin F, Li L. Model-based fitting of single-channel dwell-time distributions. Biophys J. 2004;87(3):1657–1671. doi: 10.1529/biophysj.103.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.