Abstract
Transport of large viral nucleocapsids from replication centers to assembly sites requires contributions from the host cytoskeleton via cellular adaptor and motor proteins. For the Marburg and Ebola viruses, related viruses that cause severe hemorrhagic fevers, the mechanism of nucleocapsid transport remains poorly understood. Here we developed and used live-cell imaging of fluorescently labeled viral and host proteins to characterize the dynamics and molecular requirements of nucleocapsid transport in Marburg virus-infected cells under biosafety level 4 conditions. The study showed a complex actin-based transport of nucleocapsids over long distances from the viral replication centers to the budding sites. Only after the nucleocapsids had associated with the matrix viral protein VP40 at the plasma membrane were they recruited into filopodia and cotransported with host motor myosin 10 toward the budding sites at the tip or side of the long cellular protrusions. Three different transport modes and velocities were identified: (i) Along actin filaments in the cytosol, nucleocapsids were transported at ∼200 nm/s; (ii) nucleocapsids migrated from one actin filament to another at ∼400 nm/s; and (iii) VP40-associated nucleocapsids moved inside filopodia at 100 nm/s. Unique insights into the spatiotemporal dynamics of nucleocapsids and their interaction with the cytoskeleton and motor proteins can lead to novel classes of antivirals that interfere with the trafficking and subsequent release of the Marburg virus from infected cells.
Keywords: dual-color imaging, reverse genetics, viral inclusion bodies
The filoviruses Marburg (MARV) and Ebola (EBOV) cause severe hemorrhagic fever with high–case-fatality rates in humans and nonhuman primates (1–3). Although the interplay of filoviral proteins leading to the transcription and replication of the viral genome and the formation of the viral nucleocapsids (NCs) is rather well understood, we are only just beginning to unravel the complex interactions between the viral and cellular proteins that are necessary to transport the NCs from the sites of their formation to the budding sites. The central protein within the NC is the nucleoprotein NP, which forms complexes with VP35, VP30, and VP24 (4, 5). The helical MARV NC is composed of the RNA-dependent RNA polymerase (L), the polymerase cofactor VP35, the viral proteins VP30 and VP24, and NP, which encapsidates the viral genome (5–7). Within the viral particle, the NC is surrounded by a regular lattice of the matrix protein VP40 (5, 8, 9). The outside of the VP40 lattice contacts the viral envelope, in which the glycoprotein GP is inserted (10).
MARV virogenesis begins with the formation of perinuclear inclusions which, analogous to the EBOV, are considered to be sites of viral replication and the assembly of new NCs (8, 9, 11). Later in the replication cycle, NCs are detected in the cytosol, at the plasma membrane, and in filopodia, the preferred sites of MARV budding (12, 13). The glycoprotein GP reaches the plasma membrane via vesicular secretory membrane traffic and is recruited to sites where viral protein VP40 accumulates (14, 15). The multifunctional VP40 is a peripheral membrane-associated protein and the major player that orchestrates virogenesis. It (i) down-regulates viral replication and transcription, (ii) is the only viral protein that is recruited to sites of budding via the late retrograde endosomal transport pathway independently of other viral proteins, (iii) recruits NCs and GP to the sites of budding, and (iv) induces budding and release of filamentous particles (12–14, 16, 17). VP40 self-assembles into well-defined oligomers and polymerizes to form the regular lattice that is located underneath the plasma membrane of infected cells and beneath the viral envelope (18–20). In infected cells, VP40 is distributed to the plasma membrane and late endosomal compartments and is also detected in the viral inclusions (19). To date, it is unclear where the association between VP40 and NCs occurs, although this association is a prerequisite for the formation of infectious viral particles.
To investigate the spatiotemporal dynamics of NCs, live-cell imaging is considered to be the method of choice. Although reverse genetic systems for filoviruses expressing fluorescent proteins are available (3, 21), the efficient fluorescent labeling of filoviral NCs remains a problem. Although exchanging the gene encoding the EBOV polymerase with a gene encoding a fluorescently labeled version of the enzyme does not impair viral growth and enables the visualization of viral inclusions, its fluorescent intensity is not sufficient to observe individual EBOV NCs, most likely due to the few L particles associated with NCs (11). The filamentous MARV NCs of 892 times 91 nm could be large enough for visualization by fluorescence microscopy if the number of fluorescent molecules per NC is enhanced (6, 9). We hypothesized that labeling VP30, one of the abundant structural NC components, would allow us to visualize the NCs in living cells. In addition, we wanted to address where and when NCs become associated with the matrix protein VP40. Because VP40 suppresses filovirus replication and transcription as well as cellular transcription, we generated a fluorescently labeled VP40 that was expressed under the control of the MARV promoter. Regulation by the viral promoter should mitigate any disturbances of viral and cellular RNA synthesis due to the expression of labeled VP40 (18, 22). Here we established a dual-color live-cell imaging assay that enabled the simultaneous monitoring of NCs and VP40 in MARV-infected cells under biosafety level 4 (BSL-4) conditions. This approach demonstrated that NCs underwent actin-dependent long-distance transport from the viral inclusions to the plasma membrane, where they became associated with VP40. Only VP40-associated NCs were then recruited into filopodia and transported along actin filaments to the budding site at the tip or sides of the filopodia with the help of the motor protein myosin 10. This live-cell imaging approach enables an in-depth analysis into the dynamics of the cellular life cycle of MARV and the required virus–host interactions, and can be harnessed for potential antiviral therapies that interfere with the assembly and dissemination of infectious MARV.
Results
Development of a Dual-Color Live-Cell Imaging Assay to Monitor the Intracellular Transport of Nucleocapsids and VP40.
To investigate the intracellular dynamics of NCs in living cells, we labeled the NC-associated VP30 by attaching a green fluorescent protein (GFP) to its C terminus (VP30-GFP). As expected, upon transient coexpression with NP, VP30-GFP was recruited into NP-induced inclusions (Fig. S1A) (23) and, similarly, transiently expressed VP30-GFP was colocalized with inclusions in MARV-infected cells (Fig. S1B). In addition, those MARV-infected cells contained VP30-GFP–labeled NP-positive filamentous structures, which had an average length of 962 ± 100 nm (n = 50) (Fig. S1B, Upper Inset). The length of the filamentous structures and their NP content indicated that they were NCs (6, 24), which was supported by the observation that fluorescently labeled released MARV particles had similar dimensions (Fig. S1C). Finally, VP30-GFP was incorporated into released MARV particles, as demonstrated by immunoblot (Fig. S1D). These results indicate that VP30-GFP was targeted to all sites that contain NCs in MARV-infected cells, and that it was therefore suitable for tracking the dynamics of NCs in living cells.
We also wanted to address where and when NCs meet the viral matrix protein using live-cell imaging. We therefore constructed RFP-VP40, a fluorescent version of VP40 with RFP fused to its N terminus. RFP-VP40 behaved similarly to VP40 in terms of membrane association, induction of particle morphogenesis, and release (Fig. S2 A–C). Upon coexpression with VP40 and all other MARV proteins in an infectious virus-like particle (iVLP) assay, RFP-VP40 also supported the formation of fluorescent VLPs that were infectious for new target cells (Fig. S2D) (22). Thus, coexpression of RFP-VP40 and VP40 is a suitable approach to investigate the intracellular dynamics of VP40 by live-cell imaging.
Cloning, Rescue, Growth Kinetics, and Phenotype of Recombinant MARVRFP-VP40.
The gene encoding RFP-VP40 was inserted as an additional ORF into a unique AvrII restriction site of the plasmid encoding the full-length MARV genome (pFLMARV) upstream of the VP40 gene, thereby creating pFLMARVRFP-VP40 (Fig. S3A) (14, 21, 25). pFLMARVRFP-VP40 was transfected into cells together with helper plasmids encoding NP, VP35, VP30, L, and T7 polymerase (25). At 12 d posttransfection (p.t.), cells showed red fluorescence signals, indicating replication of the recombinant virus expressing RFP-VP40 (rMARVRFP-VP40). Supernatant from pFLMARVRFP-VP40–transfected cells was used to generate virus stock for subsequent experiments.
VeroE6 cells infected with rMARVRFP-VP40 expressed VP40 and RFP-VP40; both were incorporated into released MARV particles (Fig. S3B, lanes 3 and 4). Control cells, infected with recombinant MARV with the wild-type genome (rMARVwt), did not contain RFP-VP40 (Fig. S3B, lane 1). The growth kinetics of rMARVRFP-VP40 was comparable to that of rMARVwt during the first 72 h, suggesting that the presence of RFP-VP40 did not impair viral replication. At later stages of infection [4–7 d postinfection (p.i.)], 50% tissue culture infective dose (TCID50) titers of rMARVRFP-VP40 were 1 log lower than rMARVwt (Fig. S3C). All further experiments were therefore performed within 48 h p.i.
To determine the subcellular localization of RFP-VP40, we infected Huh-7 cells with rMARVRFP-VP40 expressing VP40 and RFP-VP40 (Fig. S3D). At 6 h p.i., the first RFP-VP40 signals were detected. At 12 h p.i., some of the RFP-VP40–positive structures resembled the shape of MARV-induced inclusion bodies (Fig. S3D, 12 h, arrows) (5, 8, 9). At 24 h p.i., RFP-VP40 was located also at the cell margins and in individual NCs that had accumulated primarily in the cell periphery and in cell protrusions (Fig. S3D, 24 h, arrows).
To investigate the distribution of RFP-VP40 with respect to the other viral proteins, especially the nucleoprotein NP, a colocalization study was performed by simultaneously labeling RFP-VP40, VP40, RFP, and NP in rMARVRFP-VP40–infected Huh-7 cells fixed at 24 h p.i. (Fig. S3E). RFP-VP40 colocalized completely with both the signals of anti-VP40 and anti-RFP antibodies, indicating that the intracellular distribution of the two proteins was completely overlapping. In combination with results gained from the plasmid-based expression of VP40 and RFP-VP40 in the iVLP system (Fig. S2D), these findings confirm that RFP-VP40 had the same subcellular localization as the untagged VP40, and both proteins are most likely oligomerized into mixed functional complexes. RFP-VP40 also colocalized with NP in inclusion bodies and partially within NCs. Therefore, RFP-VP40 was considered to be suitable as a marker to follow the fate of VP40 in rMARVRFP-VP40–infected cells.
Nucleocapsids Leaving Viral Inclusions Are Not Associated with Detectable Amounts of VP40.
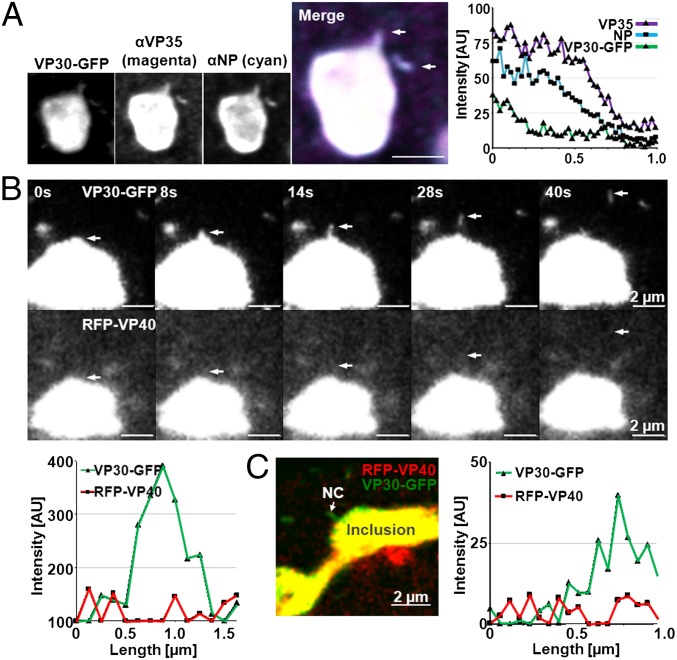
MARV NCs are considered to be formed in the virus-induced inclusions and then transported to the plasma membrane, where budding takes place (6, 9, 26). However, so far it has not been possible to trace individual NCs as they leave the inclusions on their way to the cell periphery. We therefore analyzed inclusions of cells infected with MARV and transfected with a plasmid encoding VP30-GFP by confocal immunofluorescence microscopy. Particles with the size of NCs were detected (962 ± 100 nm, n = 50), which were obviously in the process of leaving the inclusions. These structures were positive for the nucleocapsid-associated proteins NP, VP35, and VP30-GFP, confirming that those indeed represented NCs (Fig. 1A). This finding was confirmed by live-cell microscopy of rMARVRFP-VP40–infected cells expressing RFP-VP40 from the viral genome and VP30-GFP from a plasmid. We were able to detect NCs that protruded from the surface of the inclusion, which were later completely detached (Fig. 1B). This result supported the hypothesis that the inclusions are indeed the source of NCs, which are subsequently transported to the plasma membrane. It was then of interest whether VP40 was also associated with the emanating NCs, as RFP-VP40 had also been targeted to the inclusions. However, analysis of the fluorescence intensity profile over the entire length detected VP30-GFP but no RFP-VP40 in the emanating NCs (Fig. 1B, diagram). This finding was confirmed by confocal microscopy of fixed MARV-infected cells showing NCs in the process of separating from the inclusion (Fig. 1C). Taken together, these results indicated that newly formed NCs do not contain significant amounts of VP40.
Fig. 1.
NCs in the process of leaving viral inclusions do not contain VP40. (A) Huh-7 cells transiently expressing VP30-GFP were infected with MARV, fixed at 24 h p.i., and analyzed by confocal microscopy using monoclonal α-NP and guinea pig α-VP35 followed by goat α-mouse antibodies coupled to Alexa Fluor 647 and goat α-guinea pig antibodies coupled to Alexa Fluor 594. In addition, autofluorescence of VP30-GFP was recorded. A fluorescence intensity profile along the length axis of the emanating NC is displayed (diagram). (Scale bar, 2 μm.) (B) Huh-7 cells transiently expressing VP30-GFP were infected with rMARVRFP-VP40, and fluorescence signals were analyzed by time-lapse microscopy at 29 h p.i. Time between each frame: 3 s. The time in seconds is displayed in the upper left corner of each panel. Released NC is indicated by arrows. A fluorescence intensity profile along the length axis of the emanating NC in the panel (40 s) is displayed (diagram). (C) Huh-7 cells were treated as in B but fixed at 24 h p.i., and autofluorescence was analyzed by confocal microscopy. A fluorescence intensity profile along the length axis of the emanating NC is displayed (diagram).
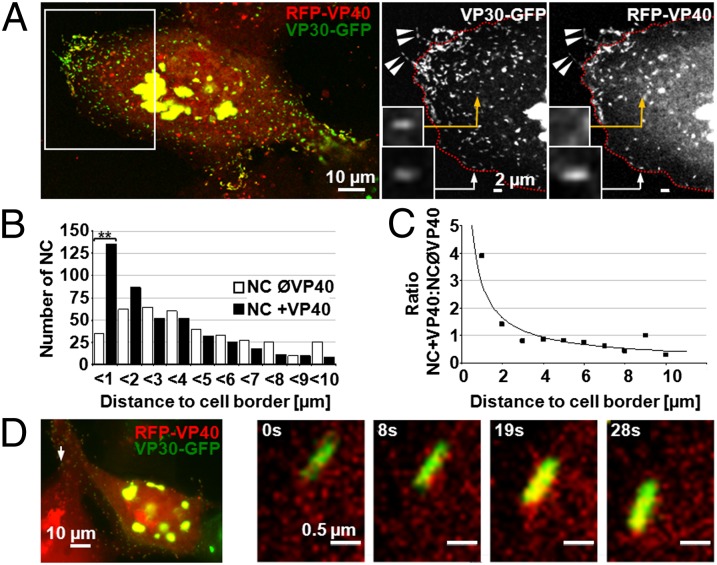
Association of RFP-VP40 with Nuclearcapsids Occurs Close to the Plasma Membrane.
Nevertheless, RFP-VP40–associated NCs were located close to the cell boundary and inside filopodia (Fig. 2A). We calculated the total number of VP30-GFP–labeled NCs and those that additionally were labeled with RFP-VP40 and measured their respective distance to the next cell border. The ratio between RFP-VP40–associated NCs and RFP-VP40–free NCs was 4 within 1 μm from the cell border, dropped to approximately 1 in the corridor between 1 and 2 μm from the cell border, and was reduced further to less than 1 in areas close to the inclusions (Figs. 2B and 2C). It was therefore of interest whether VP40 association occurred preferentially in the cell periphery or in the cell center close to the nucleus, and whether the VP40-associated NCs were then rapidly transported to the plasma membrane. Live-cell imaging analyses revealed that NCs only became associated with RFP-VP40 in close proximity to the plasma membrane but not in the cell center (Fig. 2D).
Fig. 2.
NCs become associated with VP40 close to the plasma membrane. Huh-7 cells transiently expressing VP30-GFP were infected with rMARVRFP-VP40 and analyzed by confocal microscopy (A–C) or time-lapse microscopy (D). (A) NCs associated with VP40 located in filopodia are indicated by arrowheads. VP40-positive NCs, not yet recruited into filopodia (Insets), are indicated by arrows. NC without RFP-VP40 is indicated by yellow arrows. Magnified images of the boxed region (Left) are depicted in the black and white pictures. The red dotted lines indicate the cell border; fluorescence signals outside the cell border reflect filopodia-associated NCs. (B) VP40-associated NCs are more frequently detected close to the cell border than in the cell body. NCs (VP30-GFP signals, n > 400) were analyzed for their association with RFP-VP40 and distance to the cell border at 24 h p.i. **P ≤ 0.001. (C) Quantitative analysis of the ratio of VP40-associated NCs (RFP-VP40 and VP30-GFP signals colocalized) and free NCs (only VP30-GFP signal) with respect to their distance to the cell border. (D) NC in the process of becoming associated with VP40. Time-lapse microscopy of Huh-7 cells infected with rMARVRFP-VP40 at 22 h p.i. The arrow points to the NC at the cell margins, which is magnified in the four pictures to the right.
Actin-Dependent Movement of Nucleocapsids.
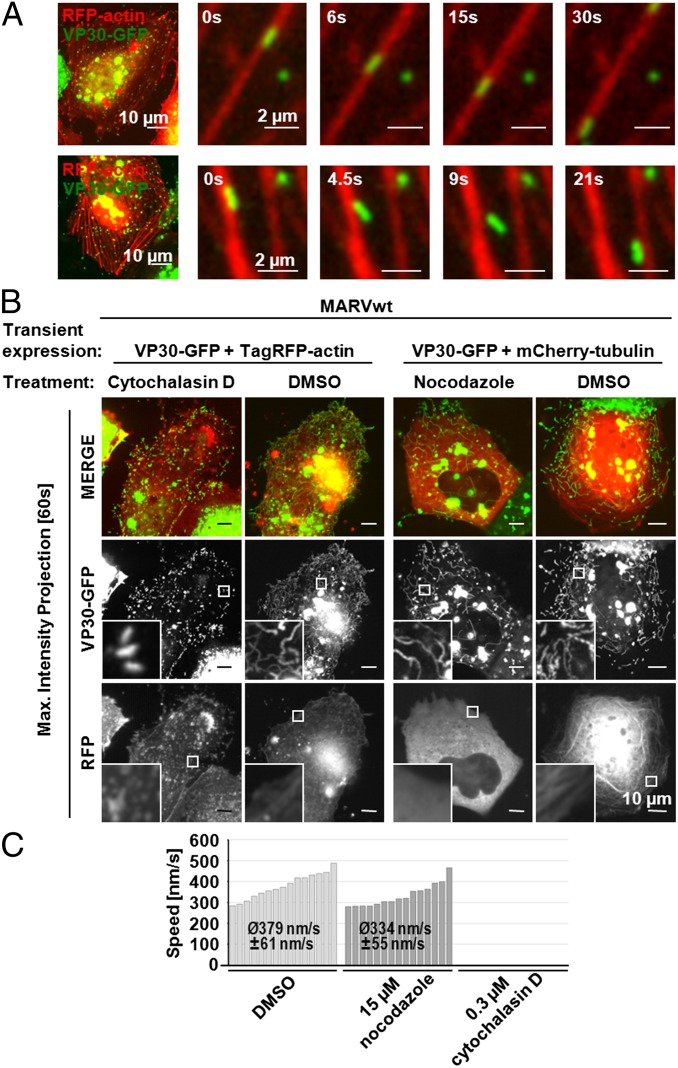
Time-lapse microscopy was used to measure the velocity of NCs in rMARVRFP-VP40–infected cells transiently expressing VP30-GFP (Movie S1). The speed of cytosolic NCs varied considerably between 200 and 500 nm/s; also, stop-and-go movement was observed (Fig. 3 and Movies S2 and S3). Interestingly, most of the VP40-associated NCs were considerably slower and moved primarily close to the cell border in parallel with the plasma membrane (100–200 nm/s; Fig. 3 and Movie S4). To determine whether NCs used actin- or microtubule-dependent transport, we investigated their migration pattern in rMARVwt-infected cells expressing VP30-GFP and TagRFP-actin or VP30-GFP and mCherry-tubulin. NCs were observed moving along actin filaments (Fig. 4A, Upper) and microtubules with an average speed of 193 ± 79 nm/s (n = 26). In infected cells expressing VP30-GFP and TagRFP-actin it was observed that NCs also moved across areas without detectable actin filaments (Fig. 4A, Lower and Movie S5). During those periods, NCs had an average speed of 411 ± 87 nm/s (n = 15). To investigate whether actin or microtubules or both were responsible for the active trafficking of NCs, rMARVwt-infected cells that expressed VP30-GFP and TagRFP-actin or VP30-GFP and mCherry-tubulin were treated with the actin-depolymerizing cytochalasin D or the microtubule-depolymerizing nocodazole. In nocodazole-treated cells, the microtubules had been depolymerized, but movement patterns of NCs were unaltered and their velocities were only slightly reduced (Fig. 4 B and C, nocodazole). Indeed, NCs still covered a distance of 14 ± 5.4 µm on average (n = 15), which is unusually long for actin-dependent transport. In contrast, treatment with cytochalasin D completely stopped the motion of NCs in the cell body as well as in the periphery, and only random movements of NCs were detected (Fig. 4 B and C, cytochalasin D and Inset and Movie S6). These experiments indicated that transport of NCs was primarily actin-based.
Fig. 3.
NCs migrate with higher velocity in the cell body than in the cortex or filopodia. Huh-7 cells transiently expressing VP30-GFP were infected with rMARVwt, and the velocity of NCs was analyzed by time-lapse microscopy. NCs (n = 30) were tracked over at least four frames.
Fig. 4.
Transport of NCs is dependent on actin. (A) NCs move along actin filaments and can change to neighboring filaments. Huh-7 cells transiently expressing VP30-GFP and TagRFP-actin were infected with rMARVwt, and fluorescence signals were monitored by time-lapse microscopy. (B) Cells transiently expressing VP30-GFP and either TagRFP-actin or mCherry-tubulin were infected with rMARVwt and treated with cytoskeleton-modulating drugs. TagRFP-actin–expressing cells were incubated with 0.3 µM cytochalasin D; mCherry-tubulin–expressing cells were incubated with 15 µM nocodazole. Forty frames (one frame per 1.5 s, reflecting 60 s) of time-lapse microscopy are displayed as maximal intensity projection. Magnified pictures of the boxed regions are shown (Insets). (C) Determination of NC velocities in cells treated with cytoskeleton-modulating drugs. NCs (n = 15) were analyzed and tracked as described in Fig. 3. No directed movements of NCs were determined in cells treated with cytochalasin D.
RFP-VP40–Associated Nucleocapsids Are Recruited into Filopodia.
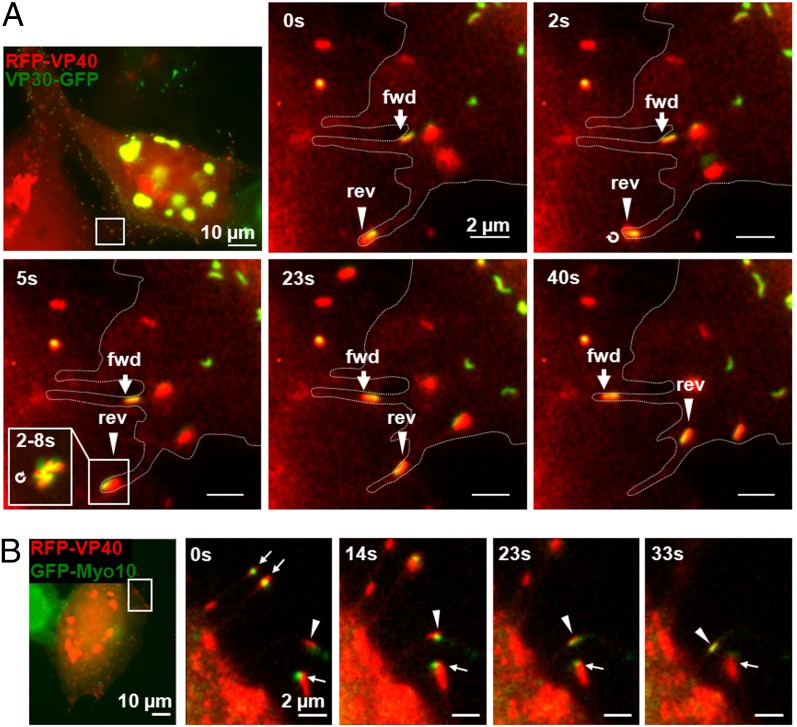
MARV budding takes place either at the side or tip of filopodia (12, 26). In cells infected with rMARVRFP-VP40, filopodia contained exclusively RFP-VP40–associated NCs, suggesting that this association was a prerequisite for the NCs to enter filopodia (Fig. 5A). NCs moved toward the tip of filopodia and also back to the cell body at ∼100 nm/s (Fig. 5A and Movie S7). To change direction, NCs turned around and moved back, thus indicating the polarity of the NCs, which supports previous observations (Fig. 5A, Lower Left, Inset) (6). Some NCs remained inside filopodia for hours, whereas others moved back to the cell body at different times after entering. NCs moving in opposite directions were also able to bypass each other within filopodia.
Fig. 5.
Movement of VP40-associated NCs inside filopodia. (A) Forward and backward NC movement inside filopodia. Huh-7 cells expressing VP30-GFP were infected with rMARVRFP-VP40 and analyzed at 29 h p.i. by time-lapse microscopy. Forward movement is indicated by arrows; backward movement is indicated by arrowheads. Cell border is indicated by dotted lines. Only VP40-associated NCs (yellow) were detected in filopodia and moved bidirectionally (velocity 100 nm/s). Backward movement requires a turn of the NCs (between 2 and 5 s). (Lower Left, Inset) (5 s) Maximal intensity projection reflects 6 s. (B) Cotransport of Myo10 with NCs inside filopodia. Huh-7 cells transiently expressing GFP-Myo10 were infected with rMARVRFP-VP40. NCs at the tip of a filopodium are indicated by arrows. Colocalization of NCs with GFP-Myo10 is indicated by arrowheads.
Intrafilopodial VP40-Associated Nucleocapsids Are Cotransported with Myosin 10.
Previous data indicated that the actin-dependent molecular motor protein myosin 10 (Myo10) is necessary for the transport of VP40 inside filopodia (12, 27). To analyze whether Myo10 is also involved in the transport of VP40-associated NCs, we transiently expressed GFP-tagged Myo10 (GFP-Myo10) in rMARVRFP-VP40–infected cells and analyzed its subcellular localization. GFP-Myo10 colocalized with RFP-VP40–associated NCs at the tips of filopodia (Fig. 5B, arrows). In addition, live-cell imaging demonstrated that GFP-Myo10 was cotransported with NCs inside filopodia at the same speed (Fig. 5B, arrowheads). These results suggest an important role of Myo10 in the VP40-induced release of NCs (12).
Taken together, we set up an assay that allowed investigating intracellular dynamics and interactions of MARV NCs and VP40. The transport of NCs across the cell body took place along actin filaments at ∼200 nm/s, whereas NCs close to the plasma membrane and within filopodia moved at 100 nm/s. The treatment of infected cells with nocodazole or cytochalasin D established that the transport of NCs was mediated by actin. Furthermore, we showed that NCs only became associated with VP40 in close proximity to the plasma membrane, and that only VP40-associated NCs entered filopodia, where they moved in association with the motor protein Myo10.
Discussion
In the present study, we constructed rMARVRFP-VP40 that expressed VP40 tagged by RFP and used this virus in combination with transient expression of VP30 tagged by GFP to establish dual-color live-cell microscopy in MARV-infected cells.
This enabled us to uncover the subcellular trafficking of NCs and VP40, their assembly, and further transport in filopodia. We could therefore analyze the spatiotemporal dynamics of NC transport and virion formation in MARV-infected cells. We tracked filoviral NCs in the process of leaving virus-induced inclusions, thus proving a long-held hypothesis that those inclusions were indeed the source of individual NCs in the cytoplasm. Although both RFP-VP40 and VP30-GFP were targeted to the inclusions, the emanating NC contained only VP30-GFP, whereas RFP-VP40 became associated with NCs only in close proximity to the plasma membrane. Upon association with VP40, NCs were then recruited into filopodia and further transported to the budding sites.
A study limited to the analysis of the intracellular localization of EBOV NCs in cells fixed at different times during infection led to two hypotheses concerning the interaction of NCs with VP40 (28): Either EBOV NCs may be considered to start their transport from the inclusions without significant amounts of VP40, or NCs and VP40 may be cotransported along the secretory pathway. Our data clearly support the first scenario, in which NCs highlighted by VP30-GFP were transported to the plasma membrane in the absence of detectable amounts of VP40. The interaction of matrix proteins with NCs is organized differently within the order of Mononegavirales. In cells infected with measles or Sendai virus, matrix protein and NCs seem to be cotransported to the plasma membrane (29, 30). Conversely, the NCs and matrix protein of vesicular stomatitis virus meet only at the plasma membrane, resulting in the condensation of the NCs (31). Although the separate transport of NCs and matrix proteins of vesicular stomatitis virus was similar to the mechanism operating in MARV-infected cells, MARV NCs obtained their final appearance already upon leaving the inclusions and, in contrast to vesicular stomatitis virus, their morphology did not change upon contact with VP40 at the plasma membrane (26).
Many viruses usurp microtubule-based mechanisms to mediate intracellular transport of NCs (32). Examples from the order Mononegavirales are vesicular stomatitis virus and Sendai virus (33–35). For vaccinia virus and African swine fever virus (DNA viruses), both microtubule-dependent transport and actin-based motility are used to drive viral particles (36–38). Intracellular vaccinia virus particles are transported along microtubules with speeds of 500–750 nm/s (35, 39). The actin-based propelling of extracellular vaccinia virus particles takes place with velocities of ∼170 nm/s (36). Intracellular transport of baculovirus in insect cells is only actin-based with velocities of ∼230 nm/s (40). Our study indicates that the trafficking of MARV NCs to the budding site was exclusively mediated by the actin cytoskeleton. Nevertheless, NCs were transported with different velocities in different regions of the cell. Along actin filaments, NCs moved at either 200 nm/s in the cell body or 100 nm/s in filopodia. Additionally, another transport velocity, 411 nm/s, was recorded for NCs switching between actin filaments. The evidence that the transport of NCs depended on actin fits with previous studies reporting a crucial role of polymerized actin for the release of MARV particles, whereas microtubules were not required (12).
So far, it is unclear which motor, adaptor, and signaling proteins mediate the transport of NCs along actin filaments in the cytosol, or whether actin filaments nucleated by viral proteins propel the NCs through the cytosol. The three different transport velocities of the NCs with on average 100, 200, or 400 nm/s could be caused by different sets or a differential regulation of actin-based motor proteins. The faster movement could be facilitated by plus end-directed myosin 5, for which speeds ranging from 200 to 1,000 nm/s have been reported (41). Alternatively, minus end-directed myosin 6 may be involved, as it transports cargo at speeds of 300–400 nm/s (42). The slower motion of NCs within the filopodia is in accordance with a Myo10-based transport (84 ± 36 nm/s; see below) (27). The use of different motors might explain why MARV NCs display such a variety of migration patterns, velocities, stop-and-go movements, and changes of transport direction as well as switches between actin filaments (43, 44).
It has long been assumed that microtubules are the major long-distance transport highways of the cell (43). However, Schuh recently described the actin-dependent transport of cellular Rab11-containing vesicles toward the plasma membrane that proceeded for more than 20 µm (45). In this case, an interdependent network of actin-linked vesicles moved collectively. Whereas the Rab11-positive vesicles had several actin filaments emanating from their surfaces, thus creating a communicating network, MARV NCs seem to either migrate along prominent bundles of actin filaments or to switch from one to another. It is possible that a combination of motor protein-dependent movement and actin propelling is responsible for the complex migration patterns reported here. To our knowledge, this is a unique example of viral NCs that are transported in mammalian cells over long distances exclusively by actin.
For their final step in intracellular transport, the NCs use filopodia. Filopodia are slender and highly dynamic cellular protrusions of up to several micrometers in length that are involved in mechanisms such as mechano-probing and cell attachment. Filopodia contain bundles of long parallel actin filaments that are characterized by the presence of the actin cross-linking protein fascin (46, 47). Previous experiments have demonstrated that MARV uses filopodia for its release (6, 12). Additionally, Myo10, an actin-based motor protein that can catalyze intrafilopodial movements, has been implicated in the intrafilopodial transport of VP40 (12, 27, 48–50). Our live-cell microscopy study now demonstrates a cotransport of Myo10 with RFP-VP40–associated NCs. Its velocity of 100 nm/s is consistent with another report on Myo10-dependent transport (27). Because Myo10 was necessary to transport VP40 inside filopodia (12) and because only VP40-associated NCs were targeted to filopodia, the association with VP40 might actually be the prerequisite for the recruitment and intrafilopodial transport of NCs (12). Several viruses, especially retroviruses, exploit filopodia for efficient egress from infected cells and spread to other cells (51, 52). Whereas initial publications reported that mouse leukemia retrovirus particles “surf” along the outside of filopodia, HIV type 1 particles have recently been described to bud from the tips of filopodia of infected dendritic cells (52, 53). MARV NCs are transported inside filopodia and bud either at the side or tip of the protrusions (6, 12, 26). This budding mode is advantageous for the virus because filopodia often make connections to neighboring cells, thus facilitating access to the next target cell. On the other hand, the release of budding virions directly to the target cells minimizes the appearance of virions in the extracellular space, which avoids recognition by the hostile environment (e.g., the immune system and host proteases).
Taken together, the present study describes the construction, characterization, and use of a recombinant MARV that expresses fluorescently labeled VP40. Transient coexpression of the NC-associated GFP-labeled VP30 in rMARVRFP-VP40–infected cells allowed us to describe the spatiotemporal dynamics of NCs. We showed that NCs use the actin cytoskeleton for long-distance transport to the plasma membrane, where NCs become associated with VP40 and are recruited into filopodia. Within filopodia, NCs are cotransported with Myo10 along actin filaments and are finally released close to neighboring cells.
Experimental Procedures
Experimental procedures for cell culture and virus infection as well as conventional microscopy have been performed as described (25). Molecular cloning and biochemical assays have been reported earlier (4, 14, 22). More detailed information on the experimental procedures including confocal laser scanning microscopy, virological methods, and all reagents is provided in SI Experimental Procedures.
For live-cell imaging, Huh-7 cells were seeded onto 35-mm µ-dishes (Ibidi) 24 h before infection. Cells were infected in 400 µL Opti-MEM without phenol red (Life Technologies) for 1 h, inoculum was removed, and, if needed, cells were transfected in a 500-µL final volume of CO2-independent Leibovitz’s medium (Life Technologies). Live-cell time-lapse experiments were recorded with a Leica DMI6000B using a 63× oil objective equipped with a remote-control device to operate the microscope from outside the BSL-4 facility. Confocal images of 4% (wt/vol) paraformaldehyde fixed and immunolabeled samples were acquired on a DMI6000B TCS SP5 laser scanning microscope using a 63× oil objective (Leica Microsystems), 488-nm argon laser, DPSS 561-nm laser, or helium 633-nm laser. Pictures and movie sequences were processed with the Leica LAS AF software package or Nikon NIS Elements 3.1, respectively.
Supplementary Material
Acknowledgments
We thank Astrid Herwig and Katharina Kowalski for excellent technical assistance, Walther Mothes for reading the manuscript and for helpful discussions, and Tobias Kleinhenz (Leica Microsystems) for excellent support in setting up live-cell imaging under BSL-4 conditions. BSL-4 work would not have been possible without the supervision of Markus Eickmann and technical support from Michael Schmidt and Gotthard Ludwig. The work was supported by a fellowship of the Leibniz Graduate School for Emerging Infectious Diseases (to G.S.) and by the German Research Foundation (Deutsche Forschungsgemeinschaft) through a Collaborative Research Center (Sonderforschungsbereich SFB 593, TP B12) and the Priority Program (Schwerpunktprogramm 1175).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307681110/-/DCSupplemental.
References
- 1.Colebunders R, et al. International Scientific and Technical Committee “DRC Watsa/Durba 1999 Marburg Outbreak Investigation Group” Marburg hemorrhagic fever in Durba and Watsa, Democratic Republic of the Congo: Clinical documentation, features of illness, and treatment. J Infect Dis. 2007;196(Suppl 2):S148–S153. doi: 10.1086/520543. [DOI] [PubMed] [Google Scholar]
- 2.Siegert R, Shu HL, Slenczka W, Peters D, Müller G. [On the etiology of an unknown human infection originating from monkeys.] Dtsch Med Wochenschr. 1967;92(51):2341–2343. doi: 10.1055/s-0028-1106144. [DOI] [PubMed] [Google Scholar]
- 3.Towner JS, et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J Virol. 2006;80(13):6497–6516. doi: 10.1128/JVI.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bamberg S, Kolesnikova L, Möller P, Klenk HD, Becker S. VP24 of Marburg virus influences formation of infectious particles. J Virol. 2005;79(21):13421–13433. doi: 10.1128/JVI.79.21.13421-13433.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker S, Rinne C, Hofsäss U, Klenk HD, Mühlberger E. Interactions of Marburg virus nucleocapsid proteins. Virology. 1998;249(2):406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 6.Bharat TA, et al. Cryo-electron tomography of Marburg virus particles and their morphogenesis within infected cells. PLoS Biol. 2011;9(11):e1001196. doi: 10.1371/journal.pbio.1001196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kiley MP, et al. Physicochemical properties of Marburg virus: Evidence for three distinct virus strains and their relationship to Ebola virus. J Gen Virol. 1988;69(Pt 8):1957–1967. doi: 10.1099/0022-1317-69-8-1957. [DOI] [PubMed] [Google Scholar]
- 8.Geisbert TW, Jahrling PB. Differentiation of filoviruses by electron microscopy. Virus Res. 1995;39(2-3):129–150. doi: 10.1016/0168-1702(95)00080-1. [DOI] [PubMed] [Google Scholar]
- 9.Kolesnikova L, Mühlberger E, Ryabchikova E, Becker S. Ultrastructural organization of recombinant Marburg virus nucleoprotein: Comparison with Marburg virus inclusions. J Virol. 2000;74(8):3899–3904. doi: 10.1128/jvi.74.8.3899-3904.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feldmann H, Will C, Schikore M, Slenczka W, Klenk HD. Glycosylation and oligomerization of the spike protein of Marburg virus. Virology. 1991;182(1):353–356. doi: 10.1016/0042-6822(91)90680-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoenen T, et al. Inclusion bodies are a site of ebolavirus replication. J Virol. 2012;86(21):11779–11788. doi: 10.1128/JVI.01525-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolesnikova L, Bohil AB, Cheney RE, Becker S. Budding of Marburgvirus is associated with filopodia. Cell Microbiol. 2007;9(4):939–951. doi: 10.1111/j.1462-5822.2006.00842.x. [DOI] [PubMed] [Google Scholar]
- 13.Dolnik O, Kolesnikova L, Becker S. Filoviruses: Interactions with the host cell. Cell Mol Life Sci. 2008;65(5):756–776. doi: 10.1007/s00018-007-7406-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittler E, Kolesnikova L, Herwig A, Dolnik O, Becker S. Assembly of the Marburg virus envelope. Cell Microbiol. 2013;15(2):270–284. doi: 10.1111/cmi.12076. [DOI] [PubMed] [Google Scholar]
- 15.Becker S, Klenk HD, Mühlberger E. Intracellular transport and processing of the Marburg virus surface protein in vertebrate and insect cells. Virology. 1996;225(1):145–155. doi: 10.1006/viro.1996.0582. [DOI] [PubMed] [Google Scholar]
- 16.Kolesnikova L, Bamberg S, Berghöfer B, Becker S. The matrix protein of Marburg virus is transported to the plasma membrane along cellular membranes: Exploiting the retrograde late endosomal pathway. J Virol. 2004;78(5):2382–2393. doi: 10.1128/JVI.78.5.2382-2393.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolesnikova L, Berghöfer B, Bamberg S, Becker S. Multivesicular bodies as a platform for formation of the Marburg virus envelope. J Virol. 2004;78(22):12277–12287. doi: 10.1128/JVI.78.22.12277-12287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoenen T, Jung S, Herwig A, Groseth A, Becker S. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology. 2010;403(1):56–66. doi: 10.1016/j.virol.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 19.Kolesnikova L, Bugany H, Klenk HD, Becker S. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J Virol. 2002;76(4):1825–1838. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weissenhorn W, Carfí A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998;2(5):605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 21.Schmidt KM, Schümann M, Olejnik J, Krähling V, Mühlberger E. Recombinant Marburg virus expressing EGFP allows rapid screening of virus growth and real-time visualization of virus spread. J Infect Dis. 2011;204(Suppl 3):S861–S870. doi: 10.1093/infdis/jir308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wenigenrath J, Kolesnikova L, Hoenen T, Mittler E, Becker S. Establishment and application of an infectious virus-like particle system for Marburg virus. J Gen Virol. 2010;91(Pt 5):1325–1334. doi: 10.1099/vir.0.018226-0. [DOI] [PubMed] [Google Scholar]
- 23.Modrof J, Mühlberger E, Klenk HD, Becker S. Phosphorylation of VP30 impairs ebola virus transcription. J Biol Chem. 2002;277(36):33099–33104. doi: 10.1074/jbc.M203775200. [DOI] [PubMed] [Google Scholar]
- 24.Bailey B, Farkas DL, Taylor DL, Lanni F. Enhancement of axial resolution in fluorescence microscopy by standing-wave excitation. Nature. 1993;366(6450):44–48. doi: 10.1038/366044a0. [DOI] [PubMed] [Google Scholar]
- 25.Krähling V, et al. Establishment of fruit bat cells (Rousettus aegyptiacus) as a model system for the investigation of filoviral infection. PLoS Negl Trop Dis. 2010;4(8):e802. doi: 10.1371/journal.pntd.0000802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Welsch S, et al. Electron tomography reveals the steps in filovirus budding. PLoS Pathog. 2010;6(4):e1000875. doi: 10.1371/journal.ppat.1000875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg JS, Cheney RE. Myosin-X is an unconventional myosin that undergoes intrafilopodial motility. Nat Cell Biol. 2002;4(3):246–250. doi: 10.1038/ncb762. [DOI] [PubMed] [Google Scholar]
- 28.Nanbo A, Watanabe S, Halfmann P, Kawaoka Y. The spatio-temporal distribution dynamics of Ebola virus proteins and RNA in infected cells. Sci Rep. 2013;3:1206. doi: 10.1038/srep01206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Runkler N, Pohl C, Schneider-Schaulies S, Klenk HD, Maisner A. Measles virus nucleocapsid transport to the plasma membrane requires stable expression and surface accumulation of the viral matrix protein. Cell Microbiol. 2007;9(5):1203–1214. doi: 10.1111/j.1462-5822.2006.00860.x. [DOI] [PubMed] [Google Scholar]
- 30.Stricker R, Mottet G, Roux L. The Sendai virus matrix protein appears to be recruited in the cytoplasm by the viral nucleocapsid to function in viral assembly and budding. J Gen Virol. 1994;75(Pt 5):1031–1042. doi: 10.1099/0022-1317-75-5-1031. [DOI] [PubMed] [Google Scholar]
- 31.Dancho B, McKenzie MO, Connor JH, Lyles DS. Vesicular stomatitis virus matrix protein mutations that affect association with host membranes and viral nucleocapsids. J Biol Chem. 2009;284(7):4500–4509. doi: 10.1074/jbc.M808136200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greber UF, Way M. A superhighway to virus infection. Cell. 2006;124(4):741–754. doi: 10.1016/j.cell.2006.02.018. [DOI] [PubMed] [Google Scholar]
- 33.Chambers R, Takimoto T. Trafficking of Sendai virus nucleocapsids is mediated by intracellular vesicles. PLoS ONE. 2010;5(6):e10994. doi: 10.1371/journal.pone.0010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das SC, Nayak D, Zhou Y, Pattnaik AK. Visualization of intracellular transport of vesicular stomatitis virus nucleocapsids in living cells. J Virol. 2006;80(13):6368–6377. doi: 10.1128/JVI.00211-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rietdorf J, et al. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat Cell Biol. 2001;3(11):992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 36.Dodding MP, Way M. Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe. 2009;6(6):536–550. doi: 10.1016/j.chom.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 37.Hollinshead M, et al. Vaccinia virus utilizes microtubules for movement to the cell surface. J Cell Biol. 2001;154(2):389–402. doi: 10.1083/jcb.200104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jouvenet N, et al. African swine fever virus induces filopodia-like projections at the plasma membrane. Cell Microbiol. 2006;8(11):1803–1811. doi: 10.1111/j.1462-5822.2006.00750.x. [DOI] [PubMed] [Google Scholar]
- 39.Herrero-Martínez E, Roberts KL, Hollinshead M, Smith GL. Vaccinia virus intracellular enveloped virions move to the cell periphery on microtubules in the absence of the A36R protein. J Gen Virol. 2005;86(Pt 11):2961–2968. doi: 10.1099/vir.0.81260-0. [DOI] [PubMed] [Google Scholar]
- 40.Ohkawa T, Volkman LE, Welch MD. Actin-based motility drives baculovirus transit to the nucleus and cell surface. J Cell Biol. 2010;190(2):187–195. doi: 10.1083/jcb.201001162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheney RE, et al. Brain myosin-V is a two-headed unconventional myosin with motor activity. Cell. 1993;75(1):13–23. doi: 10.1016/S0092-8674(05)80080-7. [DOI] [PubMed] [Google Scholar]
- 42.O’Connell CB, Tyska MJ, Mooseker MS. Myosin at work: Motor adaptations for a variety of cellular functions. Biochim Biophys Acta. 2007;1773(5):615–630. doi: 10.1016/j.bbamcr.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 43.Radtke K, Döhner K, Sodeik B. Viral interactions with the cytoskeleton: A hitchhiker’s guide to the cell. Cell Microbiol. 2006;8(3):387–400. doi: 10.1111/j.1462-5822.2005.00679.x. [DOI] [PubMed] [Google Scholar]
- 44.Akhmanova A, Hammer JA., III Linking molecular motors to membrane cargo. Curr Opin Cell Biol. 2010;22(4):479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuh M. An actin-dependent mechanism for long-range vesicle transport. Nat Cell Biol. 2011;13(12):1431–1436. doi: 10.1038/ncb2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Faix J, Rottner K. The making of filopodia. Curr Opin Cell Biol. 2006;18(1):18–25. doi: 10.1016/j.ceb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Yang C, Svitkina T. Filopodia initiation: Focus on the Arp2/3 complex and formins. Cell Adhes Migr. 2011;5(5):402–408. doi: 10.4161/cam.5.5.16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bohil AB, Robertson BW, Cheney RE. Myosin-X is a molecular motor that functions in filopodia formation. Proc Natl Acad Sci USA. 2006;103(33):12411–12416. doi: 10.1073/pnas.0602443103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nambiar R, McConnell RE, Tyska MJ. Myosin motor function: The ins and outs of actin-based membrane protrusions. Cell Mol Life Sci. 2010;67(8):1239–1254. doi: 10.1007/s00018-009-0254-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hirano Y, et al. Structural basis of cargo recognition by the myosin-X MyTH4-FERM domain. EMBO J. 2011;30(13):2734–2747. doi: 10.1038/emboj.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lehmann MJ, Sherer NM, Marks CB, Pypaert M, Mothes W. Actin- and myosin-driven movement of viruses along filopodia precedes their entry into cells. J Cell Biol. 2005;170(2):317–325. doi: 10.1083/jcb.200503059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sherer NM, et al. Retroviruses can establish filopodial bridges for efficient cell-to-cell transmission. Nat Cell Biol. 2007;9(3):310–315. doi: 10.1038/ncb1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aggarwal A, et al. Mobilization of HIV spread by diaphanous 2 dependent filopodia in infected dendritic cells. PLoS Pathog. 2012;8(6):e1002762. doi: 10.1371/journal.ppat.1002762. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.