Significance
Bacterial communication plays an important role in many population-based phenotypes and interspecies interactions, including those in host environments. Social interaction within bacterial communities, and particularly communication between pathogenic and commensal bacteria, is a subject of growing interest with relevance to ecology and human health. In this study, we show a case of interspecies communication where the intestinal pathogen Salmonella typhimurium increases its antibiotic tolerance in response to the bacterial signaling molecule indole, even though S. typhimurium does not natively produce indole. These results suggest that this intestinal pathogen can benefit from indole signaling produced by E. coli and other commensal bacteria.
Abstract
Bacterial communication plays an important role in many population-based phenotypes and interspecies interactions, including those in host environments. These interspecies interactions may prove critical to some infectious diseases, and it follows that communication between pathogenic bacteria and commensal bacteria is a subject of growing interest. Recent studies have shown that Escherichia coli uses the signaling molecule indole to increase antibiotic tolerance throughout its population. Here, we show that the intestinal pathogen Salmonella typhimurium increases its antibiotic tolerance in response to indole, even though S. typhimurium does not natively produce indole. Increased antibiotic tolerance can be induced in S. typhimurium by both exogenous indole added to clonal S. typhimurium populations and indole produced by E. coli in mixed-microbial communities. Our data show that indole-induced tolerance in S. typhimurium is mediated primarily by the oxidative stress response and, to a lesser extent, by the phage shock response, which were previously shown to mediate indole-induced tolerance in E. coli. Further, we find that indole signaling by E. coli induces S. typhimurium antibiotic tolerance in a Caenorhabditis elegans model for gastrointestinal infection. These results suggest that the intestinal pathogen S. typhimurium can intercept indole signaling from the commensal bacterium E. coli to enhance its antibiotic tolerance in the host intestine.
Rather than acting autonomously, bacterial cells communicate with one another to coordinate their efforts and relay vital information. Interspecies and intraspecies bacterial communication has been implicated in many community-dependent behaviors including virulence (1), biofilm formation (2), and antibiotic tolerance (3). Communication may therefore allow control of heterogeneity, which is important in determining fitness of microbial populations (4). Recently, we reported that bacterial communication through indole signaling induces persister formation in Escherichia coli (3). Persistence is an antibiotic-tolerant phenotype in which a dormant subpopulation of cells (persisters) survives antibiotic treatment without having genetically encoded resistance factors (5, 6). In E. coli, we found that indole signaling induced oxidative stress response and phage shock response pathways, thereby increasing the persister frequency within the population. This work suggested that bacteria can use intraspecies signaling to modify the antibiotic tolerance of their population in response to environmental conditions.
Indole signaling is used by bacteria in the distal intestine of humans and other mammals (7). In this environment, alkaline and low nutrient conditions induce expression of the indole-producing tryptophanase (tnaA) enzyme in commensal E. coli and related bacteria (8). Indole concentrations in the mammalian intestine (∼300 µM to 1 mM) (9, 10) can induce antibiotic tolerance in E. coli without adversely affecting growth (11). As the mammalian intestine contains a richly mixed microbial population (12), signaling molecules such as indole might be detected and used by both commensal and pathogenic bacteria. Although there is increasing interest in the roles that commensal bacteria play in mammalian health (13), the mechanisms by which commensal bacteria interact with invading pathogens are not yet well understood.
We hypothesized that pathogenic bacteria could use communication signals produced by commensal bacteria to sense and adjust their physiological state to the host environment. As indole induces antibiotic tolerance in E. coli, we hypothesized that it might also increase tolerance in related pathogens. Salmonella typhimurium is one such pathogen which, although it does not produce indole (14), has been shown to respond to signaling molecules produced by other bacteria (15). S. typhimurium is a common gastrointestinal pathogen and a major epidemiological threat, as it is a causative agent of gastroenteritis and sepsis. This pathogen can survive macrophage engulfment and persist within phagocytes, resulting in an asymptomatic but infectious carrier state (16) where antibiotic tolerance is a significant problem (17). We therefore sought to determine if indole signaling by E. coli might be exploited by S. typhimurium, leading to increased tolerance of the pathogen in a host intestinal environment.
Here we show that indole signaling can indeed increase the antibiotic tolerance of S. typhimurium. This tolerance can be induced by exogenous indole in Salmonella-only cultures or by indole produced by E. coli in a mixed-microbial population. Our data suggest that this tolerance is mediated, in part, by oxidative stress and phage shock response systems. Further, we find, using a Caernohabditis elegans infection model (18), that indole induces antibiotic tolerance of S. typhimurium in a mixed-microbial, intestinal environment.
Results
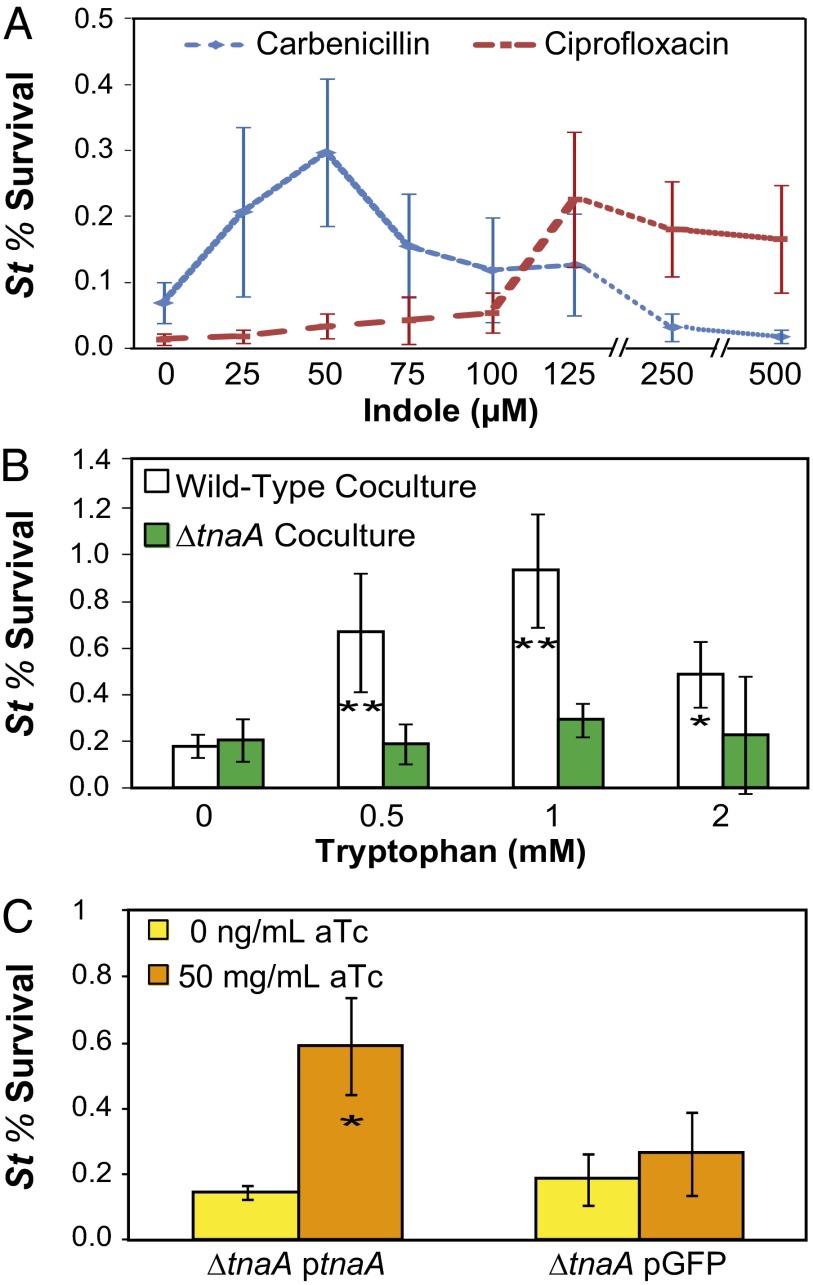
We first sought to test whether indole induced antibiotic tolerance in S. typhimurium (strain LT2). We treated exponential-phase S. typhimurium grown in tryptophan-free medium (M9CG [M9 + 0.2% Casamino acids + 0.4% (wt/vol) glucose]; Materials and Methods and SI Materials and Methods) with a range of indole concentrations to determine the effect on tolerance to carbenicillin and ciprofloxacin, antibiotics that are used in clinical treatment of enteric Salmonella infections (19) (Fig. 1A). We found that exogenous indole increased tolerance to both antibiotics by over threefold, demonstrating that indole enhances antibiotic tolerance despite the inability of S. typhimurium to produce this signal (14). Interestingly, the protective range of indole was different for the two antibiotics. Carbenicillin tolerance peaked in the presence of 50 μM indole and declined at higher indole concentrations, whereas ciprofloxacin tolerance was enhanced by higher concentrations of indole, peaking at 125 μM indole. The reason for this difference is unclear, but it may suggest that the protective processes induced by indole play differing roles against different antibiotics.
Fig. 1.
E. coli indole signaling induces antibiotic tolerance in Salmonella typhimurium. (A) Exponential-phase cultures of S. typhimurium (St, n ≥ 3) were incubated with indole for 1 h before treatment with carbenicillin (100 μg/mL) or ciprofloxacin (0.5 μg/mL). (B) Coculture of S. typhimurium with indole-producing cultures of E. coli (n ≥ 6). T tests were performed vs. the 0 mM tryptophan condition. (C) Expression of plasmid-borne tnaA in E. coli for induction of S. typhimurium tolerance in coculture (n ≥ 3). Cultures were grown in M9CG + 2 mM tryptophan with or without anhydrotetracycline (aTc) for induction of plasmid-borne genes. T tests were performed vs. the uninduced condition (0 ng/mL aTc). Assays in B and C were performed using ciprofloxacin (0.5 μg/mL). Error bars represent mean ± SD of biological replicates, and stars indicate significance level of two-sided two-sample t tests assuming unequal variance (*P ≤ 0.05; **P ≤ 0.01).
As addition of exogenous indole increased tolerance of S. typhimurium cultures, we postulated that indole produced by E. coli in a mixed-microbial environment would also induce S. typhimurium tolerance. To test this, we mixed an exponential-phase culture of S. typhimurium with a stationary-phase culture of E. coli [E. coli K-12 wild-type strain (EMG2) Pro, Table S1] grown in the presence of tryptophan to enable indole production. After a 1-h incubation, the mixed culture was treated with ciprofloxacin, as fluoroquinolone antibiotics are known to retain bactericidal activity in dense, starved cultures (20) such as those used in these assays. Antibiotic-treated cultures were plated on selective media to quantify the induced antibiotic tolerance in S. typhimurium (Materials and Methods and SI Materials and Methods). As expected, baseline tolerance of S. typhimurium was higher under dense coculture conditions than in exponential-phase culture (Fig. 1A) (21). We found that coculture with wild-type E. coli increased the tolerance of S. typhimurium to ciprofloxacin when cultures were grown in the presence of tryptophan (Fig. 1B and Fig. S1). Interestingly, the relationship between tolerance and tryptophan concentration was not strictly monotonic, with the benefit of tryptophan peaking at 1 mM. By measuring the E. coli-produced indole in cultures, we determined that 1 mM tryptophan corresponded to 125−300 µM of produced indole (Fig. S2), consistent with tolerance levels observed when indole was exogenously added (Fig. 1A). The conferred advantage for S. typhimurium of the mixed-microbe environment required indole production, as coculture with E. coli unable to produce indole (ΔtnaA strain) did not induce tolerance (Fig. 1B). Plasmid-based expression of tnaA, but not a gfp control, restored the coculture tolerance phenotype to the ∆tnaA knockout strain (Fig. 1C), further demonstrating the specific effect of indole on induced antibiotic tolerance in S. typhimurium. These results indicate that E. coli indole signaling can induce antibiotic tolerance in S. typhimurium.
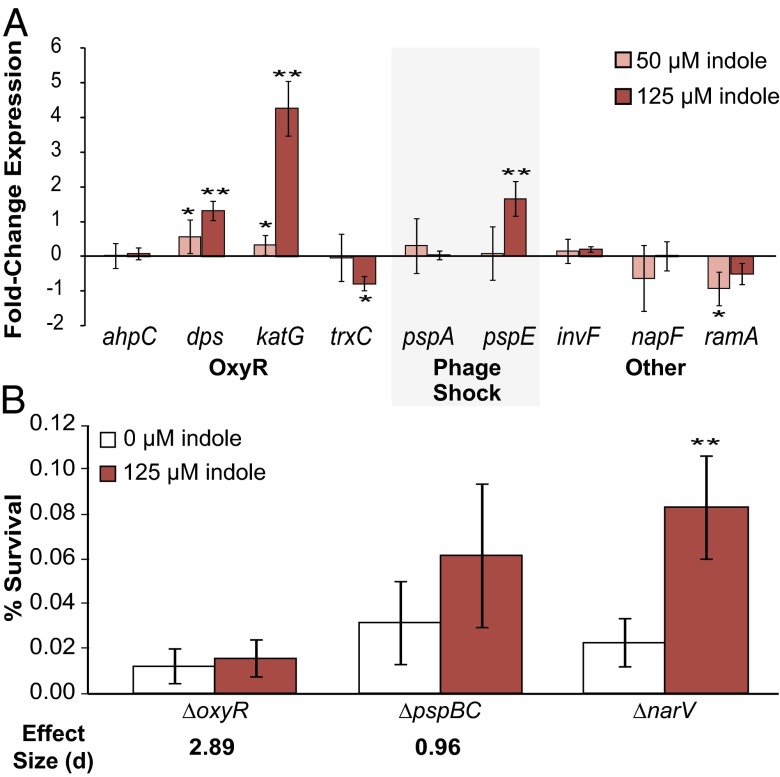
We previously determined that indole-induced tolerance in E. coli was mediated by the oxidative stress response (oxyR) and phage shock response (psp) (3). As these processes are largely conserved in S. typhimurium, with some differences in effectors and regulation (22, 23), we reasoned that indole-induced tolerance might function through a similar mechanism in S. typhimurium. To test this hypothesis, we performed qPCR on mRNA transcripts from several genes in these pathways from S. typhimurium cultures treated for 30 min with 50 μM and 125 μM indole, the concentrations that induced high carbenicillin and high ciprofloxacin tolerance, respectively. Our qPCR results (Fig. 2A) indicated that, as in E. coli, expression of the OxyR and phage shock regulons is induced in S. typhimurium by indole. We found that 125 μM indole strongly induced the expression of genes in the OxyR regulon [dps and hydroxyperoxidase I (katG)] and an effector of the phage shock pathway (pspE), although 50 μM indole produced weak induction of OxyR regulon genes and did not have a detectable effect on pspE (Fig. 2A). These results demonstrate that, as in E. coli, indole can stimulate in S. typhimurium expression of genes in the oxidative stress and phage shock pathways. Previous reports have indicated that higher levels of indole induced expression of genes involved in virulence and drug efflux pumps in S. typhimurium (24). We did not observe induction of these genes (presented here as “Other,” Fig. 2A) when cultures were treated with indole concentrations that induce increased antibiotic tolerance. Interestingly, indole treatment produced a small but consistent decrease in transcript abundance of a transcriptional regulator associated with multiple antibiotic resistance (ramA); this gene product has been shown to control expression of multidrug efflux pumps in S. typhimurium (24), suggesting that indole treatment (at the levels considered here) may actually decrease efflux in this system.
Fig. 2.
Response of OxyR and phage shock pathways to indole in S. typhimurium. (A) Transcriptional response of S. typhimurium to indole treatment as determined by qPCR. S. typhimurium cultures were treated with 0, 50 (pink bars) or 125 (red bars) μM indole (SI Materials and Methods). Results are shown as fold-change expression (−∆∆Ct) of indole-treated vs. untreated cultures. Error bars represent mean ± SD of three to four biological replicates. (B) Exponential-phase cultures of S. typhimurium ∆narV:CmR (control), ∆oxyR:CmR, and ∆pspBC:CmR were incubated with indole (0 or 125 μM) for 1 h before treatment with ciprofloxacin (0.5 μg/mL). Error bars represent mean ± SD of 8−13 biological replicates. Stars indicate significance level of two-sided two-sample t tests assuming unequal variance (*P ≤ 0.05; **P ≤ 0.01). Cohen's effect size (d) using pooled SD was calculated for the difference between indole-treated and untreated cultures compared with the difference observed in the ∆narV control; d > 0.8 suggests a large effect.
We next used genetic knockouts to determine whether the OxyR and phage shock pathways are involved in indole-induced antibiotic tolerance in S. typhimurium. The ∆oxyR and ∆pspBC mutants were constructed to allow inactivation of the OxyR and phage shock responses, respectively (SI Materials and Methods). We found that indole-induced tolerance was reduced in the ∆pspBC mutant and entirely eliminated in the ∆oxyR mutant relative to the control (Fig. 2B). These results suggest that both the OxyR and phage shock responses are involved in indole-induced antibiotic tolerance in S. typhimurium, as was previously observed in E. coli. Interestingly, although the mechanism of indole-induced antibiotic tolerance appears to be conserved between species, the role of the phage shock response is attenuated in S. typhimurium compared with E. coli.
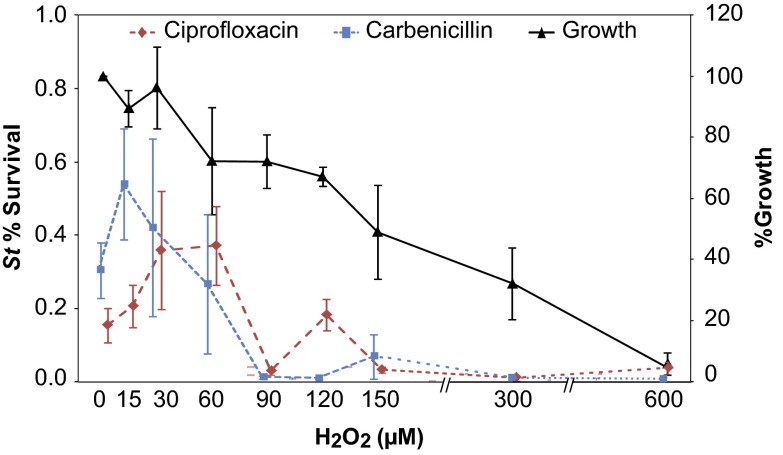
Given the high levels of induction in some aspects of the oxidative stress response (Fig. 2A), we hypothesized that the OxyR regulon might play a role in inducing antibiotic tolerance in S. typhimurium. To test this hypothesis, we treated S. typhimurium cultures with a range of H2O2 concentrations 1 h before treating cultures with antibiotic (Fig. 3). We found that low levels of oxidative stress (Fig. S3) could increase antibiotic tolerance of S. typhimurium to the same degree induced by indole, suggesting that indole may primarily act through the OxyR regulon to induce tolerance in S. typhimurium.
Fig. 3.
Hydrogen peroxide induces tolerance in S. typhimurium. Exponential-phase cultures of S. typhimurium LT2 in M9CG were incubated with H2O2 (0−600 μM) for 1 h before treatment with antibiotics. Black line indicates percent growth of LT2 cultures during incubation with hydrogen peroxide, where growth in the absence of hydrogen peroxide was used as reference (100%). Percent survival was determined after 4-h treatment with ciprofloxacin (0.5 μg/mL, dashed red line) or carbenicillin (100 μg/mL, dotted blue line). Error bars represent mean ± SD of at least six biological replicates.
The protective range of H2O2 was different for carbenicillin and ciprofloxacin, with better protection against carbenicillin at lower H2O2 concentrations and better protection against ciprofloxacin at higher H2O2 concentrations. Interestingly, this antibiotic-specific concentration dependence for H2O2 treatment (Fig. 3) resembles the concentration dependence observed during indole treatment (Fig. 1A). The qualitative similarities in the antibiotic tolerance curves suggest that the OxyR regulon plays a functional role in indole-induced antibiotic tolerance in S. typhimurium. These data may additionally suggest that contribution of reactive oxygen species (ROS) to antibiotic lethality differs between carbenicillin and ciprofloxacin, adding further nuance to a growing understanding of antibiotic-induced cell death (25–28).
Having demonstrated that S. typhimurium responds to E. coli-produced indole by inducing an oxidative stress response, thereby increasing antibiotic tolerance, we sought to determine whether this interspecies signaling occurred in an intestinal environment. We used a C. elegans model for S. typhimurium infection (18) to explore in vivo the relation between E. coli-produced indole and S. typhimurium tolerance. This model provides a tractable approach for investigating intestinal mixed-microbial interactions and has been used for determining in vivo antibiotic efficacy against pathogenic bacteria (29). Although clearly less complex than the mammalian intestine, the S. typhimurium intestine maintains many critical factors that pertain to microbes, including mixed, semiimmobilized bacterial populations with spatial distributions, innate antimicrobial defenses, and the requirement of pathogen-epithelium adhesion (30).
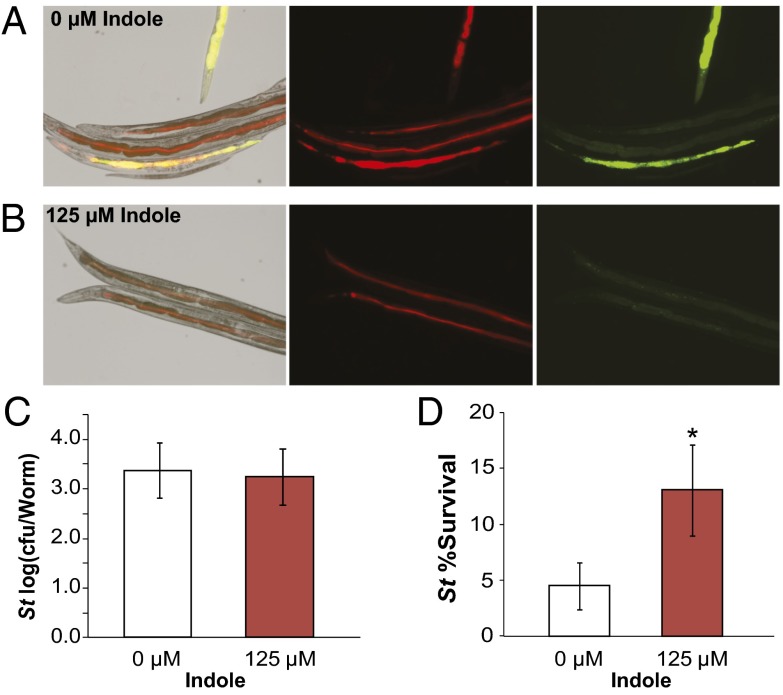
We used fluorescent microscopy to verify that E. coli and S. typhimurium established a mixed-microbial community in the C. elegans intestine (18). Synchronized cultures of adult C. elegans were fed on E. coli (mCherry) in tryptophan-free S-medium with or without 125 μM indole then incubated with S. typhimurium (GFP) to allow infection; indole was added exogenously to allow precise control of indole concentrations (Fig. 4 A and B; SI Materials and Methods). Worms generally showed observable fluorescence in both channels, indicating the simultaneous presence of E. coli and S. typhimurium in the intestine. Interestingly, indole-treated cultures (125 μM) showed low-intensity S. typhimurium infection with small adhered colonies in nearly all worms sampled (Fig. 4B), whereas untreated cultures contained a mix of heavily infected and uninfected worms (Fig. 4A). However, by measuring cfu/worm, we found that treatment with 125 µM indole did not affect the average pathogen count per worm (Fig. 4C), suggesting that indole altered heterogeneity of the S. typhimurium infection in C. elegans.
Fig. 4.
Indole increases antibiotic tolerance of S. typhimurium in the C. elegans intestine. All experiments were performed in S-Medium (SI Materials and Methods) at 25 °C to ensure sterility and survival of C. elegans AU37. Images were selected to represent the variation observable in each experimental condition. (A and B) Visualization of infection in the C. elegans intestine. C. elegans fed on E. coli EMG2 (mCherry) and infected with S. typhimurium (GFP) with (A) no indole or (B) 125 µM indole added to media were imaged 36 h after initial infection. Differential interference contrast (DIC) images are shown overlaid with red and green fluorescent channels, and fluorescent channels are shown in isolation (SI Materials and Methods). (C) Average intensity of infection by S. typhimurium (St). (D) Survival of S. typhimurium in the C. elegans intestine after treatment of worm cultures with ciprofloxacin (2 μg/mL). All error bars indicate mean ± SD of four biological replicates. Stars indicate significance level of one-sided two-sample t tests assuming unequal variance, comparing indole-treated and untreated cultures (*P ≤ 0.05; **P ≤ 0.01).
We next sought to determine if indole affected S. typhimurium antibiotic tolerance in the C. elegans model. Synchronized adult cultures of C. elegans in S-medium ± 125 μM indole were fed on E. coli (EMG2 ∆tnaA Pro; SI Materials and Methods). Infection with S. typhimurium [chloramphenicol resistance (CmR); SI Materials and Methods] was then facilitated as described above, and infected cultures were treated with ciprofloxacin to determine antibiotic tolerance. We observed that, although ciprofloxacin was effective in killing S. typhimurium in the C. elegans intestine, worms incubated with indole carried S. typhimurium with higher antibiotic tolerance. This finding suggests that indole increased antibiotic tolerance of S. typhimurium in the C. elegans host intestine (Fig. 4D and Fig. S4).
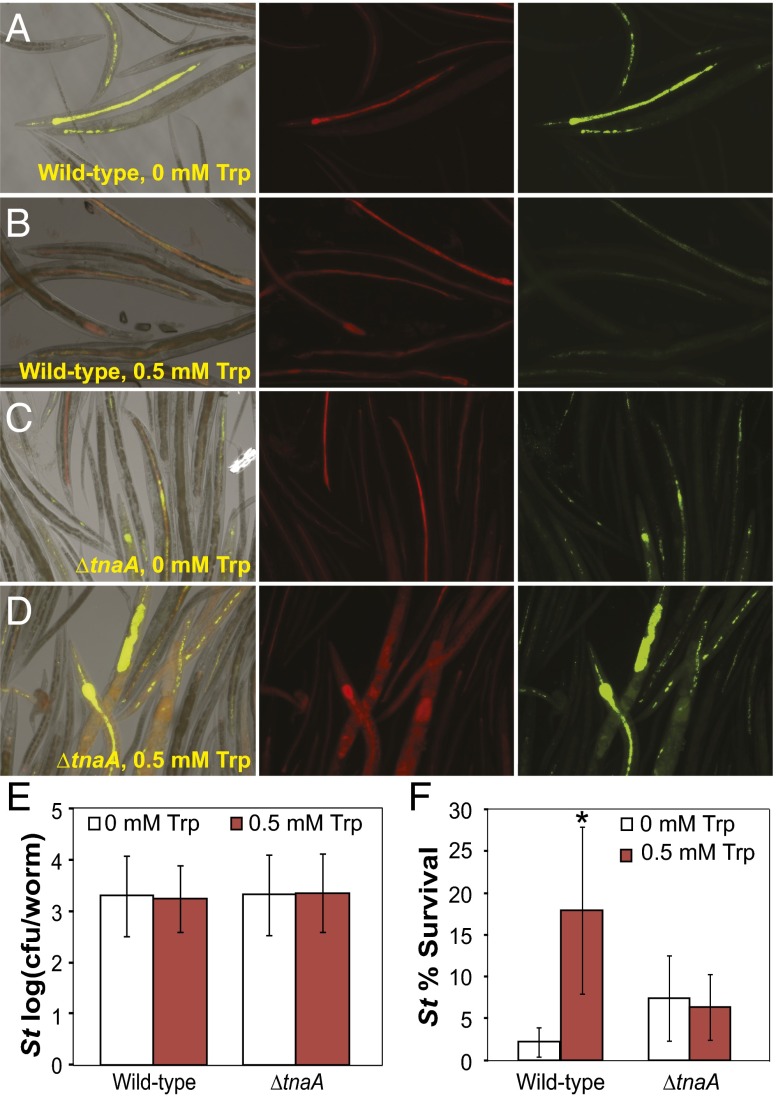
Having observed that indole could increase S. typhimurium tolerance in a host-commensal-pathogen model, we sought to test if indole produced by E. coli in this model was sufficient for increasing tolerance. We investigated this by incubating synchronized cultures of adult C. elegans in modified S-medium (pH∼7) with or without additional tryptophan using E. coli wild-type or ∆tnaA strains as the C. elegans food source (SI Materials and Methods). We found that addition of tryptophan to worm media encouraged the formation of small punctate intestinal colonies of S. typhimurium when worms were fed on wild-type E. coli but not when fed on the ∆tnaA E. coli strain (Fig. 5 A–D). Average pathogen count per worm was unchanged across all experimental conditions (Fig. 5E). We found that adding tryptophan to C. elegans culture media increased antibiotic tolerance of S. typhimurium when wild-type E. coli was present, but not in the presence of the ∆tnaA E. coli strain (Fig. 5F). These results suggest that S. typhimurium can intercept E. coli indole signaling in the C. elegans intestine, allowing the pathogen to increase its tolerance to antibiotics.
Fig. 5.
Indole produced by E. coli in a mixed-microbe population induces tolerance of S. typhimurium in the C. elegans intestine. All experiments were performed in modified S-Medium (SI Materials and Methods). Images were selected to represent the variation observable in each experimental condition. (A−D) Visualization of infection in the C. elegans intestine. C. elegans fed on (A and B) E. coli EMG2 (mCherry) or (C and D) ∆tnaA (mCherry) and infected with S. typhimurium (GFP) with (A and C) no tryptophan or (B and D) 0.5 mM tryptophan added to media were imaged 36 h after initial infection. DIC images are shown overlaid with red and green fluorescent channels, and fluorescent channels are shown in isolation. Images were autoscaled using software default settings (SI Materials and Methods). (E) Average intensity of infection by S typhimurium (St). (F) Survival of S. typhimurium in the C. elegans intestine after treatment of worm cultures with ciprofloxacin (2 μg/mL). Error bars indicate mean ± SD of four biological replicates. Stars indicate significance level of one-sided two-sample t tests assuming unequal variance, comparing tryptophan-treated and untreated cultures (*P ≤ 0.05; **P ≤ 0.01).
Discussion
Here, we report that physiologically relevant concentrations of indole, whether added exogenously or produced by E. coli in coculture, enhance the antibiotic tolerance of S. typhimurium. Indole-induced tolerance was observed consistently in S. typhimurium despite naturally occurring variation in baseline tolerance measurements (31). Physiological responses to indole have been observed in non-indole-producing bacteria (32), suggesting that indole can function as an interspecies signal (2, 33–35). The data presented here implicate indole signaling by commensal bacteria and exploitation of this signal by pathogenic bacteria as a potential factor in establishing antibiotic tolerance of pathogens.
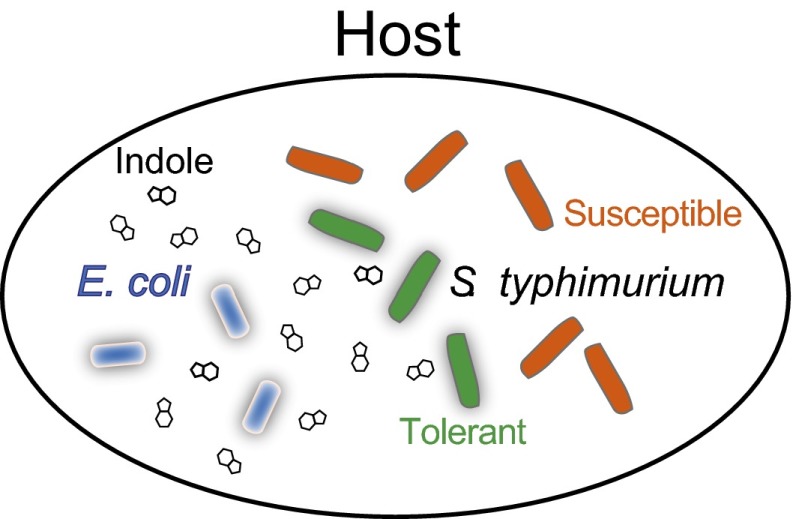
We also showed that indole signaling by E. coli enhances S. typhimurium tolerance in a C. elegans host intestinal model. Despite the relative simplicity of the C. elegans model, it has been suggested as a useful model for S. typhimurium infection of mammals based on the observation that strains attenuated in virulence in mammals were also attenuated in C. elegans (29). The C. elegans infection model was used here to create a spatially nonhomogenous mixed-bacterial culture with adhesion of bacteria to an epithelium. Our results with this system indicate that pathogenic S. typhimurium are able to “eavesdrop” on commensal bacterial communication by E. coli to enhance their antibiotic tolerance within the host intestine (Fig. 6).
Fig. 6.
Bacterial communication, signal interception, and antibiotic tolerance in the host environment. Within the C. elegans host intestine, pathogenic S. typhimurium encounters indole-producing E. coli (7) and is able to intercept indole signaling to enhance its antibiotic tolerance.
We found that indole-induced tolerance in S. typhimurium is mediated, in part, by the oxidative stress response and the phage shock response, as was seen previously in E. coli (3). Antioxidant capability has previously been linked to antibiotic tolerance in bacteria (36, 37), and in this case, induction of the oxidative stress response appears to be largely responsible for the indole-induced antibiotic-tolerant phenotype. We observed increased transcript levels for katG, which acts to protect the cell from damage under conditions of increased oxidative stress (38, 39), but not alkyl hydroperoxide reductase C (ahpC), which has been implicated in scavenging of low-level, endogenously produced H2O2 (40), suggesting that indole response prepares these bacteria for survival in stressful conditions.
We found some differences in indole-induced changes in transcript abundance between E. coli and S. typhimurium. For example, while both species show an indole-responsive increase in pspE transcript, indole-treated E. coli show increased levels of pspA transcript, whereas indole-treated S. typhimurium do not (3). Although stress response genes are largely conserved and highly homologous between E. coli and S. typhimurium, there are important functional and regulatory differences between these species (22, 23, 41), and it is therefore expected that indole-based induction of individual genes within the relevant stress response pathways will vary somewhat between species. However, it is notable that the same pathways are observed to respond to indole in both species, suggesting that the mechanism of indole-induced tolerance is conserved. It remains to be tested whether this cross-species protection extends to other bacterial pathogens, including Gram-positives.
Indole signaling may affect stress responses that are important for survival in a host and establishment of a chronic infection. Previous work has shown that survival in macrophages is critical for Salmonella virulence (42, 43) and that peroxide catalase activity plays an essential role in survival (39). OxyR-mediated processes may protect Salmonella intracellularly by inducing tolerance to the oxidative bursts that immune cells use to kill bacteria. The importance of oxidative stress in establishment and recurrence of Salmonella infection is demonstrated by chronic granulomatous disease, a hereditary immunodeficiency in which macrophage cannot generate the oxidative burst; patients with this disease are highly susceptible to bacteremic infection by nontyphoidal Salmonella and to recurrences of these infections (44). We found that indole signaling strongly induced expression of OxyR regulon genes in S. typhimurium, suggesting that the ability to detect commensal indole signaling may provide the pathogen with a method of altering its physiology to tolerate the stresses of the immune system and antibiotics and thereby establish a persistent infection.
More broadly, our results suggest that persistence of pathogens in the host environment may be induced by the interception of nonnatively produced bacterial-signaling molecules. There is a growing body of work exploring the complex relationship between the host, the innate microbiota, and gastrointestinal pathogens (45–49), and it is increasingly evident that the interplay between the established microbiota and introduced organisms in the intestine may be critical in determining the progress and resolution of infections and the aftermath of disease. Here, we observe a case where a bacterial pathogen has lost the capacity for production of a signal but has retained the ability to respond to signaling produced by commensals in the shared intestinal environment. Further, we show that interception of and response to nonnative signaling produces an antibiotic-tolerant phenotype in a bacterial pathogen, suggesting that interactions between the commensal microbiota and invading pathogens may in some cases improve stress tolerance in pathogens, thereby increasing recalcitrance of bacterial infections.
Materials and Methods
Bacterial Strains and Strain Construction.
All experiments were performed using laboratory strains of E. coli and Salmonella enterica serovar Typhimurium. Ancestral wild-type E. coli K-12 EMG2 +fertility factor plasmid (F+) obtained from Yale E. coli Genetic Stock Center (ECGC 4401) was the reference wild-type E. coli strain used in all experiments, and S. typhimurium LT2 [American Type Culture Collection (ATCC) 700720] was the reference strain of nontyphoidal Salmonella. C. elegans strains were provided by the C. elegans Genetic Stock Center, which is funded by National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). Bacterial strains and primers used in this study are presented in Table S1. Details of strain construction are presented in SI Materials and Methods.
Antibiotics and Chemicals.
The following concentrations of antibiotics were used in this study: 100 µg/mL carbenicillin, 10–60 µg/mL kanamycin, 0.5–2 µg/mL ciprofloxacin, and 1–5 µg/mL ofloxacin. Strains containing kanamycin-resistance plasmids were grown with 30–60 µg/mL kanamycin for selection, and strains containing spectinomycin-resistance cassettes were grown with 50 µg/mL spectinomycin. For induction of plasmid-borne genes, 25–50 ng/mL anhydrotetracycline (aTc) was added after 2–4 h of growth. Otherwise, antibiotic treatments were ≥10× minimum inhibitory concentration (MIC) to ensure killing of sensitive cells.
Growth and Tolerance Assays.
All experiments were performed in accordance with standard protocols unless otherwise stated. Briefly, bacterial cultures were grown in light-insulated shakers at 37 °C with shaking at 300 rpm (14 mL-Falcon tubes, Fisher Scientific) or 900 rpm (96-well, clear, flat-bottom culture plates, Fisher Scientific; with Breathe-Easy adhesive gas-permeable membrane, USA Scientific). Cultures were grown in tryptophan-free rich media (M9 + 0.2% casamino acids + 0.4% glucose, M9CG, pH >7.2) or in complete rich media (Luria-Bertani, LB).
Antibiotic tolerance was assessed by incubating cultures for at least 4 h with antibiotic to allow full killing of sensitive cells. Serial dilution and plating were used to determine cfu/mL before and after treatment (SI Materials and Methods).
For coculture experiments, E. coli K-12 EMG2 was grown to stationary phase in M9CG + 0–2 mM tryptophan to allow indole production. The ∆tnaA knockout was used to prevent indole production. Cultures of E. coli ∆tnaA pZA21-tnaA or pZA21-GFP were grown in M9CG + 2 mM tryptophan, with or without 50 ng/mL anhydrotetracycline (aTc) for induction of plasmid-borne genes. Twenty percent of culture volume was replaced with exponential-phase culture of S. typhimurium in M9CG, and cultures were incubated 1 h before treatment with ciprofloxacin.
Indole Quantification.
Indole quantification was performed via HPLC. Details of sample preparation and chromatography are presented in SI Materials and Methods.
Quantitative PCR.
RNA was collected from indole-treated and untreated cultures of S. typhimurium LT2. Cultures were inoculated 1:500 from overnight LB cultures into 1 mL of M9CG in 14-mL Falcon tubes and incubated 3.5 h before treatment with indole. After 30-min incubation with indole (0, 50, or 125 µM), cultures were stabilized with RNAprotect Bacteria Reagent (Qiagen) according to the manufacturer’s protocol. Details of RNA extraction, cDNA synthesis, and qPCR are presented in SI Materials and Methods.
Induction of OxyR.
Cultures were inoculated 1:200 (E. coli) or 1:500 (S. typhimurium) from overnight LB cultures into M9CG (1 mL in 14-mL Falcon tubes or 150 µL in 96-well plates) and allowed to grow to exponential (3.5 h) or stationary phase (24 h) at 37 °C. Cultures were incubated 1 h with hydrogen peroxide (0–600 µM) before treatment with antibiotics. Serial dilution plating was performed before and after 1 h of hydrogen peroxide incubation and after 4 h of ofloxacin treatment to determine survival at each stage.
C. elegans Intestinal Model for S. typhimurium Infection.
The temperature-sensitive germ line proliferation (glp) mitogen-activated protein kinase kinase (sek) mutant strain AU37 [glp-4 (bn2) I; sek-1 (km4) X] of C. elegans (Caenorhabditis Genetics Center) was used as a model organism for Salmonella pathogenesis. E. coli OP50 was used as a food source in maintenance cultures. E. coli EMG2 and ∆tnaA Pro were used as food sources during experiments, and S. typhimurium nitrate reductase 2 mutant (∆narV:CmR) was used as a pathogen. For fluorescent microscopy, worms were fed on E. coli EMG2 pZS4-mCherry or E. coli ∆tnaA pZS4-mCherry, and S. typhimurium pZA21-GFP was used as the infectious agent. Details of C. elegans culture and experimental conditions are given in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Ari E. Friedland for C. elegans experimental advice and Daniel J. Dwyer and D. Ewen Cameron for helpful suggestions on the manuscript. This work was supported by the US National Science Foundation, the National Institutes of Health Director’s Pioneer Award Program, and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308085110/-/DCSupplemental.
References
- 1. Rutherford ST, Bassler BL (2012) Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med 2(11):a012427. [DOI] [PMC free article] [PubMed]
- 2.Martino PD, Fursy R, Bret L, Sundararaju B, Phillips RS. Indole can act as an extracellular signal to regulate biofilm formation of Escherichia coli and other indole-producing bacteria. Can J Microbiol. 2003;49(7):443–449. doi: 10.1139/w03-056. [DOI] [PubMed] [Google Scholar]
- 3.Vega NM, Allison KR, Khalil AS, Collins JJ. Signaling-mediated bacterial persister formation. Nat Chem Biol. 2012;8(5):431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nevozhay D, Adams RM, Van Itallie E, Bennett MR, Balázsi G. Mapping the environmental fitness landscape of a synthetic gene circuit. PLOS Comput Biol. 2012;8(4):e1002480. doi: 10.1371/journal.pcbi.1002480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rotem E, et al. Regulation of phenotypic variability by a threshold-based mechanism underlies bacterial persistence. Proc Natl Acad Sci USA. 2010;107(28):12541–12546. doi: 10.1073/pnas.1004333107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allison KR, Brynildsen MP, Collins JJ. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473(7346):216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Botsford JL, Demoss RD. Escherichia coli tryptophanase in the enteric environment. J Bacteriol. 1972;109(1):74–80. doi: 10.1128/jb.109.1.74-80.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han TH, Lee JH, Cho MH, Wood TK, Lee J. Environmental factors affecting indole production in Escherichia coli. Res Microbiol. 2011;162(2):108–116. doi: 10.1016/j.resmic.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol. 1985;109(2):135–141. doi: 10.1007/BF00391888. [DOI] [PubMed] [Google Scholar]
- 10.Zuccato E, et al. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci. 1993;38(3):514–519. doi: 10.1007/BF01316508. [DOI] [PubMed] [Google Scholar]
- 11.Li G, Young KD. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology. 2013;159(Pt 2):402–410. doi: 10.1099/mic.0.064139-0. [DOI] [PubMed] [Google Scholar]
- 12.Hughes DT, et al. Chemical sensing in mammalian host-bacterial commensal associations. Proc Natl Acad Sci USA. 2010;107(21):9831–9836. doi: 10.1073/pnas.1002551107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blaser M, Bork P, Fraser C, Knight R, Wang J. The microbiome explored: Recent insights and future challenges. Nat Rev Microbiol. 2013;11(3):213–217. doi: 10.1038/nrmicro2973. [DOI] [PubMed] [Google Scholar]
- 14. Bergey DH, Breed RS (1957) Bergey's Manual of Determinative Bacteriology (Williams & Wilkins, American Society for Microbiology, Baltimore), 7th Ed.
- 15.Sperandio V. SdiA bridges chemical signaling between Salmonella enterica serovar Typhimurium and Yersinia enterocolitica in mice. J Bacteriol. 2010;192(1):21–22. doi: 10.1128/JB.01359-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruby T, McLaughlin L, Gopinath S, Monack D. Salmonella’s long-term relationship with its host. FEMS Microbiol Rev. 2012;36(3):600–615. doi: 10.1111/j.1574-6976.2012.00332.x. [DOI] [PubMed] [Google Scholar]
- 17.Sirinavin S, et al. Norfloxacin and azithromycin for treatment of nontyphoidal salmonella carriers. Clin Infect Dis. 2003;37(5):685–691. doi: 10.1086/377273. [DOI] [PubMed] [Google Scholar]
- 18.Aballay A, Yorgey P, Ausubel FM. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol. 2000;10(23):1539–1542. doi: 10.1016/s0960-9822(00)00830-7. [DOI] [PubMed] [Google Scholar]
- 19.Chiu C-H, Lin T-Y, Ou JT. In vitro evaluation of intracellular activity of antibiotics against non-typhoid Salmonella. Int J Antimicrob Agents. 1999;12(1):47–52. doi: 10.1016/s0924-8579(99)00038-2. [DOI] [PubMed] [Google Scholar]
- 20.Eng RH, Padberg FT, Smith SM, Tan EN, Cherubin CE. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob Agents Chemother. 1991;35(9):1824–1828. doi: 10.1128/aac.35.9.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levin BR, Rozen DE. Non-inherited antibiotic resistance. Nat Rev Microbiol. 2006;4(7):556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- 22.Farr SB, Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991;55(4):561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joly N, et al. Managing membrane stress: The phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol Rev. 2010;34(5):797–827. doi: 10.1111/j.1574-6976.2010.00240.x. [DOI] [PubMed] [Google Scholar]
- 24.Nikaido E, Shirosaka I, Yamaguchi A, Nishino K. Regulation of the AcrAB multidrug efflux pump in Salmonella enterica serovar Typhimurium in response to indole and paraquat. Microbiology. 2011;157(Pt 3):648–655. doi: 10.1099/mic.0.045757-0. [DOI] [PubMed] [Google Scholar]
- 25.Dwyer DJ, Kohanski MA, Collins JJ. Role of reactive oxygen species in antibiotic action and resistance. Curr Opin Microbiol. 2009;12(5):482–489. doi: 10.1016/j.mib.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130(5):797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 28.Brynildsen MP, Winkler JA, Spina CS, MacDonald IC, Collins JJ. Potentiating antibacterial activity by predictably enhancing endogenous microbial ROS production. Nat Biotechnol. 2013;31(2):160–165. doi: 10.1038/nbt.2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol. 2000;10(23):1543–1545. doi: 10.1016/s0960-9822(00)00833-2. [DOI] [PubMed] [Google Scholar]
- 30.Alegado RA, Tan M-W. Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell Microbiol. 2008;10(6):1259–1273. doi: 10.1111/j.1462-5822.2008.01124.x. [DOI] [PubMed] [Google Scholar]
- 31.Jõers A, Kaldalu N, Tenson T. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. J Bacteriol. 2010;192(13):3379–3384. doi: 10.1128/JB.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JH, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev. 2010;34(4):426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 33.Hamilton S, et al. The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genomics. 2009;10:599. doi: 10.1186/1471-2164-10-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Attila C, Cirillo SLG, Cirillo JD, Wood TK. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol. 2009;2(1):75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chu W, et al. Indole production promotes Escherichia coli mixed-culture growth with Pseudomonas aeruginosa by inhibiting quorum signaling. Appl Environ Microbiol. 2012;78(2):411–419. doi: 10.1128/AEM.06396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grant SS, Kaufmann BB, Chand NS, Haseley N, Hung DT. Eradication of bacterial persisters with antibiotic-generated hydroxyl radicals. Proc Natl Acad Sci USA. 2012;109(30):12147–12152. doi: 10.1073/pnas.1203735109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakamoto Y, et al. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339(6115):91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- 38.González-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179(2):382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hébrard M, Viala JPM, Méresse S, Barras F, Aussel L. Redundant hydrogen peroxide scavengers contribute to Salmonella virulence and oxidative stress resistance. J Bacteriol. 2009;191(14):4605–4614. doi: 10.1128/JB.00144-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183(24):7173–7181. doi: 10.1128/JB.183.24.7173-7181.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Storz G, et al. An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: Genetic characterization and cloning of ahp. J Bacteriol. 1989;171(4):2049–2055. doi: 10.1128/jb.171.4.2049-2055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monack DM, Bouley DM, Falkow S. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J Exp Med. 2004;199(2):231–241. doi: 10.1084/jem.20031319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fields PI, Swanson RV, Haidaris CG, Heffron F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc Natl Acad Sci USA. 1986;83(14):5189–5193. doi: 10.1073/pnas.83.14.5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56(6):413–422. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 45.Thiennimitr P, Winter SE, Bäumler AJ. Salmonella, the host and its microbiota. Curr Opin Microbiol. 2012;15(1):108–114. doi: 10.1016/j.mib.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahmer BM, Gunn JS. Interaction of Salmonella spp. with the intestinal microbiota. Front Microbiol. 2011;2:101. doi: 10.3389/fmicb.2011.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stecher B, et al. Like will to like: Abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6(1):e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parker CT, Sperandio V. Cell-to-cell signalling during pathogenesis. Cell Microbiol. 2009;11(3):363–369. doi: 10.1111/j.1462-5822.2008.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Srikanth CV, McCormick BA. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: A three-way crosstalk. Interdiscip Perspect Infect Dis. 2008;2008:626827. doi: 10.1155/2008/626827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.