Abstract
The circadian system regulates daily rhythms in lipid metabolism and adipose tissue function. Although disruption of circadian clock function is associated with negative cardiometabolic end points, very little is known about interindividual variation in circadian-regulated metabolic pathways. Here, we used targeted lipidomics-based approaches to profile the time course of 263 lipids in blood plasma in 20 healthy individuals. Over a span of 28 h, blood was collected every 4 h and plasma lipids were analyzed by HPLC/MS. Across subjects, about 13% of lipid metabolites showed circadian variation. Rhythmicity spanned all metabolite classes examined, suggesting widespread circadian control of lipid-mediated energy storage, transport, and signaling. Intersubject agreement for lipids identified as rhythmic was only about 20%, however, and the timing of lipid rhythms ranged up to 12 h apart between individuals. Healthy subjects therefore showed substantial variation in the timing and strength of rhythms across different lipid species. Strong interindividual differences were also observed for rhythms of blood glucose and insulin, but not cortisol. Using consensus clustering with iterative feature selection, subjects clustered into different groups based on strength of rhythmicity for a subset of triglycerides and phosphatidylcholines, suggesting that there are different circadian metabolic phenotypes in the general population. These results have potential implications for lipid metabolism disorders linked to circadian clock disruption.
Keywords: metabolomics, chronobiology
The circadian clock in the suprachiasmatic nucleus (SCN) of the hypothalamus regulates daily rhythms in behavior and physiology, ensuring that metabolic pathways are temporally coordinated with 24-h cycles of rest–activity and feeding (1). The circadian system controls lipid and carbohydrate homeostasis, thus optimizing energy storage and utilization across the day (2). In humans, this is reflected in part by diurnal variation in glucose tolerance (3, 4), as well as circadian rhythms of glucose, insulin, triglycerides, and adipose-derived hormones in blood (5, 6). At the cellular level, circadian rhythms are generated by transcriptional and posttranscriptional feedback loops, with daily rhythms of cell biology driven by core clock genes and their protein products. The SCN neural rhythm is normally entrained by the 24-h solar cycle, whereas the phase of rhythms in peripheral tissues is thought to be determined primarily by daily rest–activity and/or feeding cycles (7).
Disruption of circadian rhythms has been implicated in metabolic syndrome and dyslipidemia (2), and chronic circadian misalignment is thought to contribute to increased risk of cardiovascular disease and obesity in shift workers (8, 9). Although the mechanisms have yet to be fully elucidated, consumption of meals at night is associated with higher postprandial triglyceride levels, compared with meals consumed during the daytime (10, 11). In nocturnal mice, the circadian timing of food intake contributes to weight gain, such that animals fed a high-fat diet during the inactive phase (daytime) gain weight at a faster rate than animals fed during the active phase (nighttime) (12). In addition, the amplitude of diurnal rhythms of behavior is reduced in mice fed a high-fat diet, and clock gene expression and function is altered in liver and adipose tissue (13). Together, these studies demonstrate that circadian timing and metabolic health are interrelated, and that disruption to one can impact the other.
Several studies have demonstrated circadian variation in the mammalian transcriptome across different tissues including liver, skeletal muscle, and brown and white adipose tissue, suggesting widespread circadian control of metabolic pathways (14–21). Similarly, studies of the metabolome in blood plasma and liver suggest that up to 20% of metabolites show circadian variation (22–25). To date, however, studies of the human circadian metabolome in plasma have been performed in small numbers of subjects or using pooled blood specimens. Previous research suggests an important role for the circadian clock in regulating energy homeostasis, but there is almost no information on interindividual variation in circadian-regulated metabolism. In the present study, we used targeted lipidomics profiling to examine individual differences in circadian control of lipids in plasma. Here, we report marked intersubject variability in the timing and strength of lipid metabolite rhythms, and we provide evidence for different circadian metabolic phenotypes.
Results
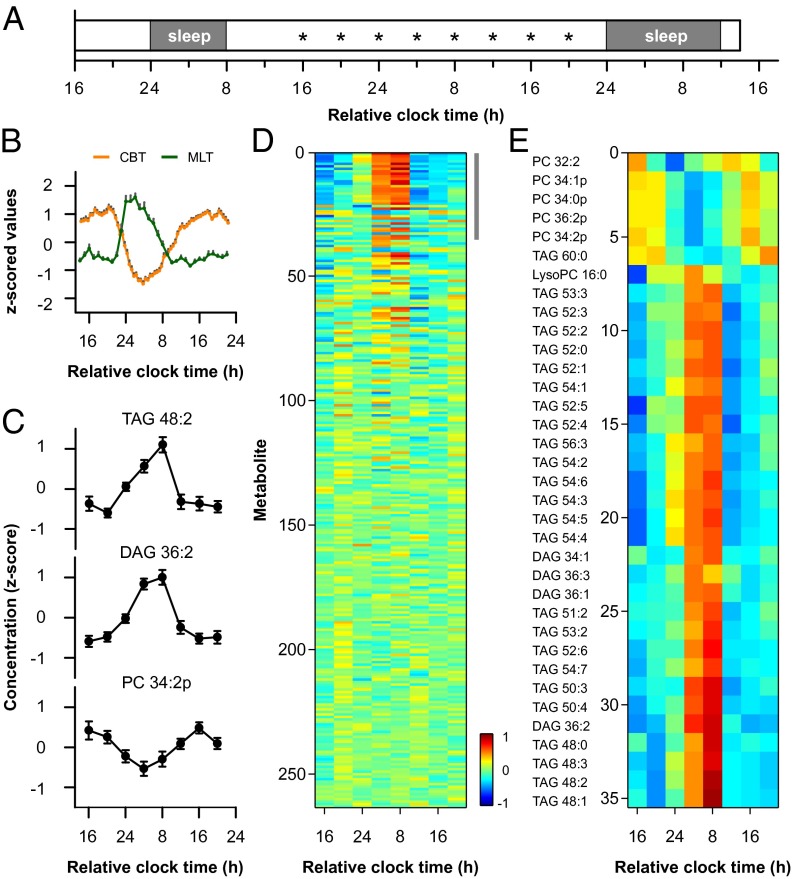
The time course of plasma lipids was examined in 20 healthy males aged 21–28 y. In a 4-d laboratory protocol, circadian rhythms were assessed under constant environmental conditions (Fig. 1A). Circadian timing of the core body temperature rhythm was similar across subjects, with the nadir occurring 4 h 33 min ± 1 h (SD) after habitual bedtime (Fig. 1B). Blood samples were drawn every 4 h over a span of 28 h, and plasma lipids were analyzed by HPLC/MS. Using targeted lipidomics-based approaches, we examined circadian regulation of glycerolipids (GLs; diacyl and triacyl forms, 76 species), glycerophospholipids (GPs, 142 species), and sphingolipids (SPs, 43 species) (Table S1; Fig. S1) (26, 27). We also measured free cholesterol and cholesterol esters using HPLC/MS (28), but did not measure other sterol lipids, fatty acids, or prenol lipids. In parallel analyses, we examined plasma cortisol, glucose, insulin, and low-density lipoprotein (LDL) cholesterol. Based on group-level analysis, 13.3% of lipids showed circadian variation (Fig. 1 C and D; Table S2). Of the 35 metabolites that were identified as group-rhythmic (Fig. S2), most were GLs consisting of triglycerides (TAGs) and diglycerides (DAGs). Whereas TAGs and DAGs exhibited peak levels in the morning (Fig. 1E), a small number of phosphatidylcholines (PCs), in particular the plasmalogen form, were also identified as group-rhythmic with peak concentrations in the evening. Plasma cortisol showed a strong circadian rhythm, with lowest levels at usual bedtime and highest levels at usual wake time (Fig. 2). In group-level analyses, we did not detect a circadian rhythm in blood glucose, but there was low-amplitude cycling of insulin levels. Plasma insulin was lowest at bedtime and increased during the usual hours of sleep, similar to the cortisol rhythm. LDL cholesterol did not show a clear circadian rhythm, but there was a trend for higher levels during the daytime.
Fig. 1.
Circadian regulation of plasma lipids. (A) Healthy subjects (n = 20) were studied in the laboratory across 4 calendar days. Asterisks indicate blood sample times. (B) Participants showed clear circadian variation in physiologic measures, as shown for core body temperature (CBT, orange) and melatonin (MLT, green) rhythms. (C) Representative lipid metabolites are shown that displayed a circadian rhythm. (D) Heat map shows that about 13% of lipids exhibited circadian rhythmicity when data were pooled across subjects. Colors show z-scored concentration levels averaged between subjects. Metabolites are ranked in order of strength of rhythmicity assessed using the JTK_CYCLE algorithm. (Upper Right) Gray bar shows metabolites identified as group-rhythmic. (E) Based on group-level analyses, 35 lipid metabolites were under circadian control, consisting primarily of GLs. From top to bottom, metabolites are arranged by the timing of their peak concentration levels. In B and C, the mean ± SEM is shown. The total number of acyl carbons and double bonds is indicated for each TAG and DAG species. Plasmalogen PCs are indicated by the suffix “p.”
Fig. 2.
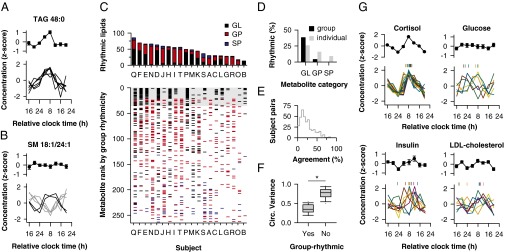
Interindividual differences in circadian control of lipids. (A) A representative TAG is shown that was identified as group-rhythmic (top), with rhythmicity driven by six participants with a similar 24-h profile. (B) A SM species is shown that was not rhythmic based on group-level analysis (top), but was nonetheless rhythmic in six subjects (bottom) with differently timed rhythms (gray and black traces). In the top of A and B, the mean ± SEM is shown (n = 20). (C) (Upper) Bar graph shows the number of rhythmic metabolites in each subject (labeled A–T) across different lipid categories. (Lower) In the scatter plot, metabolites are indicated that were rhythmic on a per-individual basis (GL, black; GP, red; SP, blue). From top to bottom, metabolites are arranged in order of group rhythmicity assessed using the JTK_CYCLE algorithm, as in Fig. 1D. Gray box shows the 35 lipid species identified as group-rhythmic. (D) Percentage of group-rhythmic versus individually rhythmic metabolites is shown across different lipid categories. (E) Distribution of agreement across circadian lipid profiles is shown for all pairwise comparisons between subjects. (F) Box plots show the distribution of circular variance in circadian phase for metabolites in which six or more participants displayed circadian rhythmicity (group-rhythmic, 30 lipid species; group-arrhythmic, 32 lipid species). Boxes show the interquartile range and whiskers indicate the 90th percentile of the distribution. (G) Black traces show the time course of plasma cortisol, glucose, insulin, and LDL cholesterol averaged across all subjects (mean ± SEM), and colored traces show individual profiles with circadian variation. For individual rhythms, the fitted peak is indicated at the top of each plot by vertical marks of the same color.
Next, we examined metabolite rhythms on a per-individual basis. For lipid species that were categorized as rhythmic in group analyses, the time course of individually rhythmic profiles was similar between subjects, as expected (Fig. 2A). We were surprised to find, however, that many lipid species that failed to show rhythmicity in group-level analyses (i.e., when data were pooled) nonetheless showed circadian variation at an individual level (Fig. 2B). Peak concentration levels occurred up to 12 h apart between subjects, suggesting large interindividual differences in metabolite rhythms. To assess this in greater detail, we systematically examined differences in circadian-regulated lipids between subjects (Fig. 2C; Table S3). The percentage of metabolites that exhibited circadian variation ranged from 5% to 33% across subjects (Fig. 2C) and included all lipid classes examined (Fig. 2D; Table S3). Across individuals, the set of lipid species identified as rhythmic differed substantially, however, with median agreement of 20% for all pairwise comparisons between subjects (Fig. 2E). Variance in circadian phase was greatest for metabolites categorized as arrhythmic in group-level analyses (Fig. 2F; Wilcoxon rank-sum test, U = 12, P < 0.001), which presumably explains why most GPs and SPs were not considered rhythmic when data were pooled across subjects. In contrast with these lipids, there was little variation in the plasma cortisol rhythm (Fig. 2G). In 19 out of 20 subjects, a high-amplitude circadian rhythm in cortisol was observed with peak levels near usual wake time, and the other participant (subject L) displayed a bimodal pattern with a secondary cortisol peak in the afternoon. Several participants showed circadian variation in blood glucose levels, but with substantial individual differences in timing of the rhythm (Fig. 2G). Some participants showed peak glucose levels close to habitual bedtime, whereas others exhibited peak levels in the morning. There were also strong interindividual differences in timing of insulin and LDL cholesterol rhythms. Most subjects who displayed circadian variation in insulin showed their highest levels between usual wake time and lunch time, whereas LDL cholesterol was typically low near bedtime and highest in the subjective daytime. The concentrations of total TAGs, glucose, and LDL cholesterol were within the normal range (≤120 mg dL−1, ≤118 mg dL−1, and ≤107 mg dL−1, respectively) across all subjects and time points, but note that our study conditions differed from those typically encountered in a clinical setting.
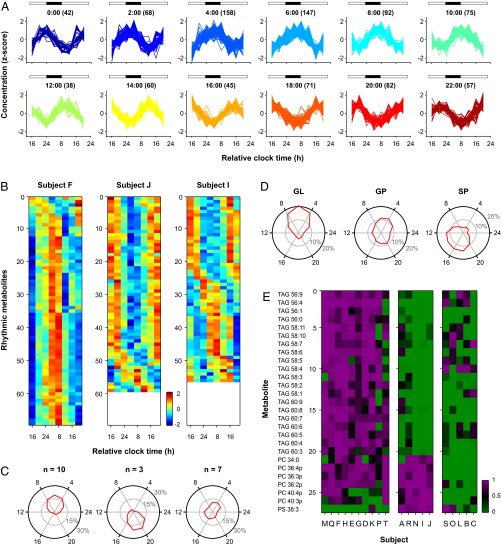
Across all participants, 17.8% of individually analyzed lipid profiles showed a significant circadian rhythm (Fig. S3; P < 0.05, 935 out of 5,260 metabolite profiles). The timing of metabolite rhythms was distributed broadly across the circadian cycle (Fig. 3 A and B), although the greatest number of metabolites reached their peak levels in the morning hours (Fig. S3). In half of participants (n = 10), more than twice as many lipid species exhibited peak concentrations in the morning relative to the evening (Fig. 3 B and C). Conversely, in a small group of subjects (n = 3), more than twice as many metabolites exhibited peak levels in the evening relative to the morning. In the remaining participants (n = 7), there was no clear circadian phase predominance. These differences in timing of lipid rhythms were not related to phase of the core body temperature rhythm, suggesting that timing of the SCN clock was similar across groups. In lipid “morning types,” the nadir of the body temperature rhythm occurred 4 h 40 min ± 62 min (SD) after habitual bedtime, which was similar to “evening types” (4 h 24 min ± 41 min) and subjects without a clear phase predominance in peak lipid concentrations (4 h 25 min ± 70 min; Kruskal–Wallis ANOVA, P = 0.79). Similarly, there was no difference between groups in chronotype assessed by Horne–Östberg morningness–eveningness questionnaire scores (49.6 ± 6.3, 54.3 ± 4.2, and 46.6 ± 4.9; Kruskal–Wallis ANOVA, P = 0.19), or in the average timing of breakfast, lunch, or dinner in the week before the laboratory study (Kruskal–Wallis ANOVA, P > 0.09 for all comparisons). Also, there did not appear to be any relationship between the timing of glucose and insulin rhythms and different lipid rhythm phenotypes. We found, however, that the distribution of peak concentration levels differed across lipid categories. GLs nearly always reached their peak levels in the late night or early morning, GPs showed no clear phase predominance, and SPs were more likely to peak in the afternoon and evening (Fig. 3D).
Fig. 3.
Characterization of different circadian metabolic phenotypes. (A) For lipids identified as rhythmic on a per-individual basis (935 traces), metabolites are arranged in 2-h bins by their peak concentration levels with time indicated at the top of each plot and the number of rhythmic metabolites in parentheses. Horizontal bars show the usual timing of wakefulness (white) and sleep (black). (B) Heat maps are shown for representative subjects whose rhythmic metabolites showed peak levels at different times of day (F, morning; J, evening; I, morning and evening). Colors show z-scored concentration levels. (C) Polar plots show the distribution of peak metabolite levels over time, as estimated by JTK_CYCLE analysis, for individuals whose lipid metabolites reached their highest concentration levels in the morning (Left), evening (Center), or with no phase predominance (Right). The radial axis shows the percentage of metabolites that reached their peak concentration, averaged across subjects in 2-h bins. (D) Distribution of peak levels over time is shown for GL, GP, and SP. (E) Subjects clustered into different groups based on rhythmicity for a subset of lipid metabolites. Colors show strength of rhythmicity (1 – P value), with magenta indicating metabolites identified as individually rhythmic. PS, phosphatidylserine.
Although there were large interindividual differences in lipid species that were identified as rhythmic versus arrhythmic, we hypothesized that participants might nonetheless cluster into groups based on similarities in their circadian lipid profiles. To assess this, we applied consensus clustering of participants based on strength of rhythmicity for each metabolite, i.e., using the P value determined from the Jonckheere-Terpstra-Kendall (JTK)_CYCLE algorithm (29, 30). We found that subjects clustered into three groups based primarily on rhythmicity for a subset of TAGs and plasmalogen-containing PCs (Fig. 3E and Fig. S4). In one cluster (n = 10), participants were rhythmic for TAGs, in particular those carrying long-chain polyunsaturated fatty acids (LC-PUFAs). In the second cluster (n = 5), subjects were arrhythmic for these TAGs, but were rhythmic for a subset of PCs. In the third cluster (n = 5), subjects were arrhythmic for most TAGs and PCs. These results suggest that healthy individuals can be classified into distinct circadian metabolic phenotypes based on differences in rhythmicity for a relatively small number of lipid species.
Discussion
In this study, we found substantial interindividual variation in the timing and strength of circadian rhythms across lipid species. The majority of lipids examined were rhythmic in at least a few subjects; however, for any given metabolite the number of subjects who showed circadian variation rarely exceeded half. Whereas the timing of TAG and DAG rhythms was similar in most subjects, there was high variability in circadian rhythms for GPs and SPs, with some participants exhibiting rhythms that were antiphase to one another. The circadian timing of glucose and insulin rhythms also differed substantially across individuals, whereas the plasma cortisol rhythm showed little intersubject variation. We observed morning and evening types based on the time of day that lipids reached their peak concentration levels, and participants clustered into different groups based on rhythmicity for a subset of TAG and PC species. These findings suggest that healthy individuals in the general population exhibit different circadian lipid phenotypes, which could have implications for metabolic disorders linked to circadian disruption.
In a previous study using nontargeted metabolomics profiling of pooled blood samples, it was estimated that about 15% of metabolites in plasma are under circadian control, with the highest proportion of rhythmic metabolites consisting of lipids (22). In that study, free fatty acids showed a strong circadian rhythm, most notably LC-PUFAs which showed peak levels near usual lunch time. By comparison, we found that many TAGs carrying LC-PUFAs reached their peak concentration levels near usual wake time. Together, these results could suggest that the circadian clock increases lipolysis of TAGs in the morning hours under normally entrained conditions, which would coincide with termination of fasting/sleep and increased energy expenditure associated with waking. Alternatively, the time course of TAG and fatty acid concentration levels could reflect circadian regulation of dietary lipid catabolism (see discussion below). Because we used targeted lipidomics profiling to monitor GLs, GPs, and SPs, we did not assess the time course of free fatty acids, and there was no overlap in lipid species examined between studies. Irrespective of methodological differences, both studies demonstrate that many lipids are under circadian control in human plasma. Here, ∼13% of lipids were identified as rhythmic. This might be a conservative estimate, however, as lipids that show low-amplitude circadian variation or a nonsinusoidal time course may have been overlooked using our methodology.
Recently, the circadian metabolome was examined in six individuals using nontargeted metabolomics profiling (24). About a dozen circadian-regulated lipid species were identified, including several metabolites monitored in the present study that showed substantial circadian variation at the level of individual subjects [e.g., sphingomyelin (SM) (d18:1/18:1), PC 32:2, PC 34:2, PC 36:4, PC 36:2, and LysoPC(16:0)]. Two of these lipid species were identified as rhythmic in our group-level analysis [LysoPC(16:0) and PC 32:2)], and the timing of peak concentration levels matched closely with that reported previously (i.e., <10 min apart). In their study, which focused on constructing a metabolite timetable, it was reported that rhythmic lipids showed high intersubject variability in their phase distribution. Our study confirms and extends these findings for hundreds of lipid species in a much larger group of subjects. Given that rhythms of core body temperature, salivary melatonin, and plasma cortisol were similar across subjects, we consider it unlikely that the broad distribution in lipid metabolite rhythms can be explained by differences in timing of the master clock in the SCN. For example, the SD of the fitted minimum of the core body temperature rhythm was 1.0 h across subjects, whereas the phase of individual lipid rhythms occurred up to 12 h apart. Also, because the timing and content of meals was the same for all subjects during the laboratory study, it is unlikely that food intake differentially reset the timing of peripheral clocks between subjects.
We found that the set of lipid species under circadian control was more different than similar for most pairs of subjects. Nonetheless, subjects clustered into different groups, with half of participants exhibiting rhythms for TAGs storing LC-PUFAs. Notably, LC-PUFAs are key components of neuronal membranes, and are important for neurodevelopment and psychiatric health (31). It is possible that daily rhythms in TAGs storing LC-PUFAs contribute to circadian control of neural function; however, we do not know if our findings in blood plasma parallel changes in brain lipid content. Participants also clustered into groups based on rhythmicity for a subset of GPs, especially plasmalogen-containing PCs, which could have implications for circadian regulation of lipoprotein homeostasis. Because we focused primarily on three categories of lipids (GLs, GPs, and SPs), but more than 500 lipids have been identified in human plasma (26), additional studies are needed to establish whether our results extend to other major categories of lipids including fatty acids, sterol lipids, and prenol lipids. Also, for each lipid we determined the number of fatty acyl carbons and double bonds, but we did not determine the composition of each fatty acyl group individually, e.g., PC34:2 includes both PC(18:1/16:1) and PC(18:2/16:0).
Although our study focused primarily on lipids, we also observed substantial individual differences in the timing of blood glucose and insulin rhythms, raising the possibility that there are also different circadian phenotypes for carbohydrate metabolism. In a study that used similar methodology as ours, blood glucose increased before usual bedtime and then decreased before usual wake time (5), whereas in another study glucose increased before wake time and then decreased in the afternoon (6). We observed glucose rhythms that were consistent with both of these studies when assessed at the level of individual subjects. At the group level, however, these rhythms appeared to cancel each other out, resulting in no detectable group rhythm.
An important limitation of our study is that we do not know the source of the lipids measured in blood plasma. The lipids could derive from catabolic breakdown of dietary fat or anabolic synthesis of lipids in liver or other tissues. Because we did not assess circadian regulation of lipids under fasting conditions, or in response to different types of meals, it is possible that some metabolites were derived from the granola snacks that were fed to subjects (Materials and Methods). In mice, the circadian metabolome is relatively stable across fed and fasted conditions (25), but it has not been tested whether these findings are generalizable to humans or to the lipids examined in the present study. Circadian regulation of lipid hydrolysis, absorption, and secretion from enterocytes could explain, in part, the time course of plasma lipid levels that we observed (32). For example, the increase in TAGs during the early morning hours could reflect increased efficiency in the digestion and absorption of lipids consumed in hourly snacks, or a circadian decrease in lipid tolerance during the biological night. Our findings for plasma lipids could also be explained by circadian regulation of liver and adipose tissue function, driven either by the SCN pacemaker or peripheral clocks. The liver plays a central role in temporally coordinating opposing processes of glycolysis and gluconeogenesis, as well as lipogenesis and fatty acid oxidation (33). Lipid homeostasis is determined, at least in part, by circadian regulation of nuclear receptors that sense fat-soluble hormones and dietary lipids (34), including the core clock protein REV-ERBα, which is important for orchestrating fatty acid and TAG biosynthesis. The circadian clock also regulates peroxisome proliferator-activated receptor α, which promotes hepatic fatty acid oxidation. Recent studies indicate that the circadian clock controls lipolysis and mobilization of free fatty acids in white adipose tissue (35), providing yet another pathway for circadian regulation of plasma lipids.
Although the sources of variation in the timing and strength of lipid rhythms remains to be determined, we hypothesize that different circadian metabolic phenotypes might associate with interindividual differences in susceptibility to the effects of circadian disruption. This would be especially important for shift workers who are at increased risk of metabolic syndrome and cardiovascular disease. Postprandial TAG levels are higher for meals taken during the night versus the day (10, 11), coinciding with a circadian increase in plasma TAG concentrations, as shown here and in other studies (5, 36). Given that postprandial lipemia increases risk of atherogenesis (37), and the occurrence of myocardial infarction is greatest near usual wake time (38), interindividual differences in circadian regulation of lipid metabolism could potentially contribute to differential disease risk and health outcomes in persons whose circadian clock is regularly misaligned with feeding and rest–activity cycles. As such, in future studies it will be important to link genetically determined differences in the lipidome with circadian control of energy homeostasis.
Materials and Methods
Subjects and Study Design.
Twenty healthy males (aged 21–28 y) enrolled in a 4-d laboratory study. After a baseline night of sleep with 8 h of time in bed, subjects underwent 40 h of sustained wakefulness under highly controlled environmental conditions. Participants were given a standardized diet consisting of hourly equicaloric snacks, consisting of a small portion of granola and cranberry juice. Starting in the afternoon, blood was drawn from the antecubital vein every 4 h, and plasma lipid concentrations were determined using HPLC/MS (LipidProfiles, National University of Singapore). Plasma cortisol and insulin were measured by ELISAs, and glucose and LDL cholesterol were measured by colorimetry assays (Duke–National University of Singapore Metabolomics Facility, Singapore). Core body temperature was measured continuously using an ingestible transmitter, and salivary specimens were collected hourly to assess the melatonin rhythm. Subjects were given 12 h of time in bed for recovery sleep before being discharged from the laboratory study. See SI Materials and Methods for details of study procedures.
HPLC/MS Identification of Lipids.
GPs and SPs were analyzed as described previously (27), and GLs (TAGs and DAGs) were analyzed using a modified version of reverse phase HPLC/electrospray ionization/MS (39). See SI Materials and Methods for details.
Analysis of Lipid Rhythmicity.
Circadian variation in lipid metabolite levels was assessed using the JTK_CYCLE algorithm (29). Consensus clustering (30) was used to determine subject grouping based on the strength of metabolite rhythmicity across different lipid species, as well as the minimal subset of lipids that determined the clustering. See SI Materials and Methods for details.
Supplementary Material
Acknowledgments
We thank Dr. Ivan Ho Mien, Eric Fang, Wen-Qi Tan, Jonathan Chua, Merryn Ang, Sze-Ching Lee, Esther Peh, and Hui-Ning Lim for their assistance in carrying out these studies; Dr. Karl Kornacker for helping us to implement the JTK_CYCLE algorithm; and Dr. Zheng-Deng Lei for his advice on biostatistics. We also thank Dr. Tal Burt for his contributions to the SingHealth Foundation grant. This work was supported by the Duke–National University of Singapore Signature Research Program funded by the Agency for Science, Technology and Research, Singapore, and the Ministry of Health, Singapore; National Medical Research Council (NMRC), Singapore under a New Investigator Grant (NIG) NMRC/NIG/1000/2009 (to J.J.G.); SingHealth Foundation (SHF), Singapore, under SHF/FG410P/2009 (to K.P.); and the National University of Singapore via the Life Sciences Institute and Competitive Research Program Award No. 2007-04 from the Singapore National Research Foundation (to M.R.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222647110/-/DCSupplemental.
References
- 1.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maury E, Ramsey KM, Bass J. Circadian rhythms and metabolic syndrome: From experimental genetics to human disease. Circ Res. 2010;106(3):447–462. doi: 10.1161/CIRCRESAHA.109.208355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aparicio NJ, et al. Circadian variation of the blood glucose, plasma insulin and human growth hormone levels in response to an oral glucose load in normal subjects. Diabetes. 1974;23(2):132–137. doi: 10.2337/diab.23.2.132. [DOI] [PubMed] [Google Scholar]
- 4.Carroll KF, Nestel PJ. Diurnal variation in glucose tolerance and in insulin secretion in man. Diabetes. 1973;22(5):333–348. doi: 10.2337/diab.22.5.333. [DOI] [PubMed] [Google Scholar]
- 5.Morgan L, et al. Effects of the endogenous clock and sleep time on melatonin, insulin, glucose and lipid metabolism. J Endocrinol. 1998;157(3):443–451. doi: 10.1677/joe.0.1570443. [DOI] [PubMed] [Google Scholar]
- 6.Shea SA, Hilton MF, Orlova C, Ayers RT, Mantzoros CS. Independent circadian and sleep/wake regulation of adipokines and glucose in humans. J Clin Endocrinol Metab. 2005;90(5):2537–2544. doi: 10.1210/jc.2004-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levi F, Schibler U (2007) Circadian rhythms: Mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol 47:593–628. [DOI] [PubMed]
- 8.Knutsson A. Health disorders of shift workers. Occup Med (Lond) 2003;53(2):103–108. doi: 10.1093/occmed/kqg048. [DOI] [PubMed] [Google Scholar]
- 9.Wang XS, Armstrong ME, Cairns BJ, Key TJ, Travis RC. Shift work and chronic disease: The epidemiological evidence. Occup Med (Lond) 2011;61(2):78–89. doi: 10.1093/occmed/kqr001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampton SM, et al. Postprandial hormone and metabolic responses in simulated shift work. J Endocrinol. 1996;151(2):259–267. doi: 10.1677/joe.0.1510259. [DOI] [PubMed] [Google Scholar]
- 11.Ribeiro DC, Hampton SM, Morgan L, Deacon S, Arendt J. Altered postprandial hormone and metabolic responses in a simulated shift work environment. J Endocrinol. 1998;158(3):305–310. doi: 10.1677/joe.0.1580305. [DOI] [PubMed] [Google Scholar]
- 12.Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17(11):2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kohsaka A, et al. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Akhtar RA, et al. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12(7):540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 15.Kita Y, et al. Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics. 2002;12(1):55–65. doi: 10.1097/00008571-200201000-00008. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy JJ, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31(1):86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panda S, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 18.Reddy AB, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16(11):1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 19.Storch KF, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417(6884):78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 20.Ueda HR, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418(6897):534–539. doi: 10.1038/nature00906. [DOI] [PubMed] [Google Scholar]
- 21.Zvonic S, et al. Characterization of peripheral circadian clocks in adipose tissues. Diabetes. 2006;55(4):962–970. doi: 10.2337/diabetes.55.04.06.db05-0873. [DOI] [PubMed] [Google Scholar]
- 22.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eckel-Mahan KL, et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc Natl Acad Sci USA. 2012;109(14):5541–5546. doi: 10.1073/pnas.1118726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kasukawa T, et al. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci USA. 2012;109(37):15036–15041. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Minami Y, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci USA. 2009;106(24):9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quehenberger O, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51(11):3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shui G, et al. Comparative plasma lipidome between human and cynomolgus monkey: Are plasma polar lipids good biomarkers for diabetic monkeys? PLoS ONE. 2011;6(5):e19731. doi: 10.1371/journal.pone.0019731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shui G, et al. Derivatization-independent cholesterol analysis in crude lipid extracts by liquid chromatography/mass spectrometry: Applications to a rabbit model for atherosclerosis. J Chromatogr A. 2011;1218(28):4357–4365. doi: 10.1016/j.chroma.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monti S, Tamayo P, Mesirov J, Golub T. Consensus clustering: A resampling-based method for class discovery and visualization of gene expression microarray data. Mach Learn. 2003;52(1–2):91–118. [Google Scholar]
- 31.Richardson AJ. Long-chain polyunsaturated fatty acids in childhood developmental and psychiatric disorders. Lipids. 2004;39(12):1215–1222. doi: 10.1007/s11745-004-1350-z. [DOI] [PubMed] [Google Scholar]
- 32.Douris N, et al. Nocturnin regulates circadian trafficking of dietary lipid in intestinal enterocytes. Curr Biol. 2011;21(16):1347–1355. doi: 10.1016/j.cub.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li MD, Li CM, Wang Z. The role of circadian clocks in metabolic disease. Yale J Biol Med. 2012;85(3):387–401. [PMC free article] [PubMed] [Google Scholar]
- 34.Yang X, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126(4):801–810. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 35.Shostak A, Meyer-Kovac J, Oster H. Circadian regulation of lipid mobilization in white adipose tissues. Diabetes. 2013;62(7):2195–2203. doi: 10.2337/db12-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rajaratnam SM, Arendt J. Health in a 24-h society. Lancet. 2001;358(9286):999–1005. doi: 10.1016/S0140-6736(01)06108-6. [DOI] [PubMed] [Google Scholar]
- 37.Jackson KG, Poppitt SD, Minihane AM. Postprandial lipemia and cardiovascular disease risk: Interrelationships between dietary, physiological and genetic determinants. Atherosclerosis. 2012;220(1):22–33. doi: 10.1016/j.atherosclerosis.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 38.Portaluppi F, Lemmer B. Chronobiology and chronotherapy of ischemic heart disease. Adv Drug Deliv Rev. 2007;59(9-10):952–965. doi: 10.1016/j.addr.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 39.Shui G, et al. Toward one step analysis of cellular lipidomes using liquid chromatography coupled with mass spectrometry: Application to Saccharomyces cerevisiae and Schizosaccharomyces pombe lipidomics. Mol Biosyst. 2010;6(6):1008–1017. doi: 10.1039/b913353d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.