Significance
A global reduction of microRNAs (miRNAs) in growth plate chondrocytes results in defects in both proliferation and differentiation; however, specific miRNAs that regulate these processes have not been identified. In this study, we provide evidence that let-7 miRNAs and microRNA-140 (miR-140), among other miRNAs, play major roles in endochondral bone development. We found that suppression of let-7 miRNAs in chondrocytes reduced proliferation, whereas the chondrocyte-specific miR-140 was necessary for differentiation into proliferating chondrocytes. Deficiency of both let-7 and miR-140 caused a dramatic growth defect, synergistically reducing proliferating chondrocyte mass. Thus, these miRNAs coordinately regulate distinct steps of endochondral bone development.
Keywords: chondrocyte differentiation, chondrocyte proliferation, mouse
Abstract
MicroRNAs (miRNAs) play critical roles in multiple processes of skeletal development. A global reduction of miRNAs in growth plate chondrocytes results in defects in both proliferation and differentiation; however, specific microRNAs responsible for these defects have not been identified. In this study, we provide evidence that let-7 miRNAs and microRNA-140 (miR-140), among other miRNAs expressed in chondrocytes, play major roles in endochondral bone development. We overexpressed lin-28 homolog A (Lin28a) to inhibit let-7 miRNA biogenesis in growth plate chondrocytes. Lin28a overexpression efficiently and specifically reduced let-7 miRNAs and up-regulated let-7 target genes. However, unlike the previous notion that let-7 miRNAs inhibit proliferation and growth, suppression of let-7 miRNAs via Lin28a overexpression decreased proliferation in growth plate chondrocytes, likely through up-regulation of the let-7 target cell cycle regulators cell division cycle 34 (Cdc34) and E2F transcription factor 5 (E2F5). Deficiency of the chondrocyte-specific miRNA, miR-140, causes a differentiation defect in growth plate chondrocytes. Although either Lin28a overexpression or miR-140 deficiency alone caused only mild growth impairment, mice with both miR-140 deficiency and Lin28a overexpression in chondrocytes showed a dramatic growth defect. Deregulation of distinct processes in the absence of these miRNAs synergistically decreased the proliferating chondrocyte mass; miR-140 deficiency reduced differentiation into proliferating chondrocytes, whereas Lin28a overexpression decreased proliferation per se.
Skeletal growth is primarily driven by the growth plate (1). Differentiation and proliferation of growth plate chondrocytes are tightly coordinated to achieve normal skeletal growth. Growth plate chondrocytes are divided into roughly three groups based on their differentiation and proliferation statuses. Resting chondrocytes, located at the most epiphyseal end, proliferate infrequently and differentiate into columnar proliferating chondrocytes that vigorously proliferate while forming orderly columns. Columnar proliferating chondrocytes then further differentiate into postmitotic hypertrophic chondrocytes. Thus, alterations of chondrocyte proliferation and/or differentiation influence the number and size of hypertrophic chondrocytes, which are the major determinant of the shape and growth speed of long bones.
MicroRNAs (miRNAs) regulate gene expression mainly at the posttranscriptional level. Direct binding of miRNAs to their target RNAs usually suppresses gene expression and facilitates RNA degradation (2, 3). miRNAs have been shown to regulate important biological functions in diverse organisms, including mice (4). We have previously shown that global miRNA deficiency in growth plate chondrocytes via conditional ablation of Dicer, a gene encoding an RNase III that catalyzes miRNA maturation, results in reduced chondrocyte proliferation and accelerated chondrocyte differentiation (5). However, roles of specific miRNAs in skeletal development remain largely unknown.
MicroRNA-140 (miR-140) and miR-140*, generated from the Mir140 gene, are abundantly and relatively specifically expressed in chondrocytes. We and others have found that loss of the Mir140 gene causes a mild skeletal growth defect (6, 7). Mir140 deficiency causes a defect in chondrocyte differentiation at multiple steps, whereas it does not affect proliferation (7). Because Dicer deficiency causes a dramatic proliferation defect, this finding suggests that miRNAs species, other than miR-140 or miR-140*, play an important role in regulating chondrocyte proliferation.
Chondrocytes express a few hundred detectable miRNAs, and let-7 family miRNAs collectively constitute the largest miRNA species in chondrocytes (5). The murine let-7 family is composed of 12 members expressed from eight genomic loci (let-7a-1, let-7a-2, let-7b, let-7c-1, let-7c-2, let-7d, let-7e, let-7f-1, let-7f-2, let-7g, let-7i, and miR-98). The let-7 miRNAs are ubiquitously expressed in most somatic cells, but their expression is suppressed in ES cells and cancer stem cells. The suppression of let-7 is crucial for maintenance of the undifferentiated state and for self-renewal of stem cells (8). In contrast, ectopic let-7 induction rescues differentiation defects of miRNA-deficient ES cells, demonstrating that let-7 miRNAs facilitate stem cell differentiation (9). The let-7 miRNAs are also considered to be tumor suppressor miRNAs. The down-regulation of let-7 is associated with poor prognosis in patients who have lung cancer (10). The causal role of let-7 down-regulation in tumorigenesis has been established recently (11–13). The let-7 miRNAs generally suppress cell proliferation through down-regulation of a series of oncogenic molecules, including RAS and high mobility group AT-hook 2 (HMGA2) (14–16).
Although the importance of let-7 suppression in stem cells and cancer cells has been relatively well established, specific roles of let-7 miRNAs in many types of somatic cells, including skeletal cells, have been poorly defined. This is mainly due to the technical difficulty in applying the conventional gene targeting method to let-7 miRNA genes for loss-of-function studies, because let-7 miRNAs are encoded in eight different genomic loci. However, a recent finding that the RNA binding proteins lin-28 homolog A (LIN28A) and LIN28B, abundantly expressed in stem cells, specifically inhibit let-7 biogenesis has provided a novel means to suppress endogenous let-7 miRNAs. LIN28 proteins bind to the loop region of pri– and pre–let-7 miRNA transcripts to inhibit processing into mature let-7 miRNAs and to facilitate degradation (17). Transgenic mice ubiquitously expressing LIN28 proteins have been generated; a low-level ubiquitous expression of LIN28 proteins causes overgrowth, delayed puberty, and insulin hypersensitivity in mice (18, 19), whereas ubiquitous let-7 overexpression causes a mild growth defect and glucose intolerance (19, 20). Although these mice show altered skeletal growth, it is not known whether their skeletal phenotypes are a direct consequence of LIN28 or let-7 overexpression in skeletal cells.
In this study, we investigate the role of the two abundantly expressed miRNA species in chondrocytes, let-7 miRNAs and miR-140/140*, in skeletal development. We show that Lin28a overexpression reduces let-7 miRNAs, decreases proliferation, and increases cell death in growth plate chondrocytes, which is due, at least in part, to the reduction of let-7 miRNAs and up-regulation of let-7 target genes. We also demonstrate that simultaneous Mir140 loss and Lin28a overexpression in chondrocytes cause a substantial growth defect resembling that of mice missing Dicer in chondrocytes, providing evidence that these two species of miRNAs, let-7 miRNAs and miR-140/140*, among other miRNAs, play important roles in skeletal development by regulating different processes of endochondral bone growth.
Results
Generation of Cre-Inducible Lin28a Transgenic Mice.
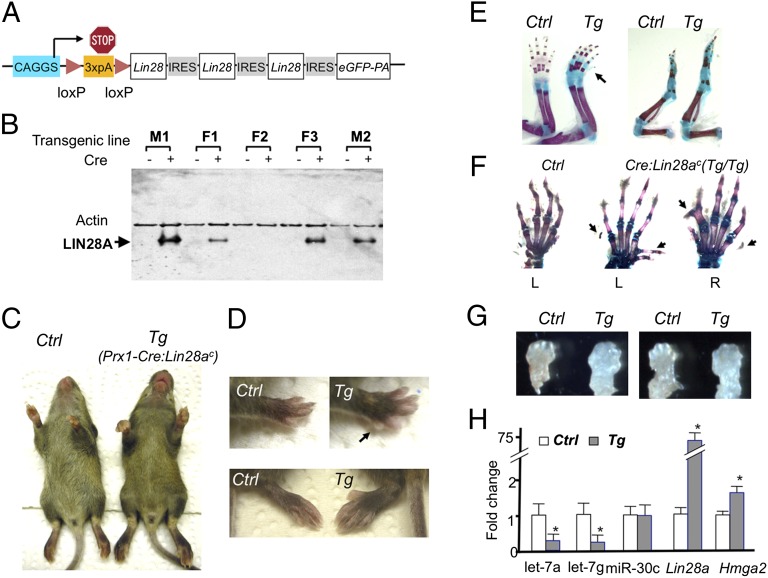
To suppress let-7 miRNAs in vivo, we generated a Cre-dependent binary system to overexpress Lin28a. In this system, Lin28a is expressed from the ubiquitous promoter, CAGGS, after Cre recombinase removes a loxP-flanked stop sequence (Fig. 1A). Five transgenic mouse lines carrying the Cre-dependent Lin28a transgene construct (Lin28ac) were established. Cre-dependent LIN28A induction was confirmed in four lines (Fig. 1B). Two lines (M1 and M2) were used for this study. Both lines gave similar results.
Fig. 1.
Lin28a overexpression in limb mesenchymal cells causes overgrowth and polydactyly. (A) Cre-inducible Lin28a expression construct (Lin28ac). Three copies of the Lin28a coding sequence along with an EGFP cDNA, connected by IRES, were placed downstream of the ubiquitous promoter, CAGGS, followed by a loxP-flanked “stop cassette” composed of three copies of a poly(A) (3×pA) signal sequence. (B) LIN28A is induced in tail fibroblasts from four transgenic lines on adenovirus-mediated Cre expression. (C) Limb size is greater in 3-wk-old Prx1-Cre:Lin28ac (Tg) mice than in Cre-negative control (Ctrl) littermates. (D) Overgrowth of forelimbs (Upper) and hind limbs (Lower) in Tg mice. The arrow indicates an additional digit at the posterior side. Polydactyly is usually absent in hind limbs, but it can be observed in homozygous Tg mice. (E) Skeletal preparation of forelimbs (Left) and hind limbs (Right). The skeletal size is increased in Tg mice compared with Ctrl. The arrow indicates an additional digit. (F) Homozygous Tg mice show diverse digit abnormalities. Left (L) and right (R) forelimbs are shown. (G) Increased limb bud size of forelimbs (Left) and hind limbs (Right) of E12.5 Tg embryos. (H) Gene expression of E13.5 limbs. Prx1-Cre:Lin28ac transgenic limbs show decreased expression of let-7a and let-7g, and increased expression of the let-7 target gene, Hmga2. Expression levels are shown relative to Ctrl levels (n = 4; *P < 0.05 vs. Ctrl).
Lin28a Overexpression in the Limb Mesenchyme Causes Overgrowth and Polydactyly.
Lin28a up-regulation is associated with various cancers (21), and a low-level ubiquitous expression of Lin28a causes overgrowth, including that of skeletal tissues (18), suggesting that Lin28a promotes cellular proliferation. To examine the effect of Lin28a overexpression in skeletal progenitor cells, we first overexpressed Lin28a in limb mesenchymal cells using Prx1-Cre transgenic mice (22). LIN28A overexpression in embryo day (E) 12.5 limb buds was confirmed in Prx1-Cre:Lin28ac doubly transgenic mice (Fig. S1A). Prx1-Cre:Lin28ac doubly transgenic mice developed limb overgrowth and postaxial polydactyly in forelimbs (Fig. 1 C–E). When Lin28a was overexpressed from both alleles (homozygous Lin28ac) in the limb mesenchyme, polydactyly and syndactyly were found also in the anterior forelimbs and occasionally in hind limbs (Fig. 1F). This phenotype is likely a consequence of overgrowth of the limb mesenchyme during development (Fig. 1G), because various genetic conditions in mice and humans that cause limb overgrowth, such as GPC3 mutation (23), mutations in the PI3K pathway (24, 25), and Cdkn1c mutations (26), are often accompanied by polydactyly. It is also possible that Lin28a overexpression affects signaling systems that govern digit patterning, although we did not find evidence that suggested alterations in Shh or BMP signaling that controls digit patterning (Fig. S1C). The precise mechanism for the overgrowth is not clear because we were unable to detect a significant increase in cell proliferation in E11.5 and E12.5 limb buds (Fig. S1 D and E). Given the magnitude of overgrowth, it is possible that the change of proliferation at a given time point is modest but its accumulative effect over time can be appreciable. Analysis of limb bud RNA demonstrated that Lin28a overexpression significantly down-regulated let-7 miRNAs and up-regulated the well-known let-7 target gene, Hmga2 (Fig. 1H). This overgrowth phenotype is in line with a previous report in which ubiquitous low-level expression of Lin28a in mice increased the size of the animals (18). Because the overgrowth in Prx1-Cre:Lin28ac mice is limited to the Prx1-expressing tissue, this result demonstrates that Lin28a-induced overgrowth is caused in a cell-autonomous fashion.
Lin28a Overexpression in Chondrocytes Reduces Proliferation and Increases Cell Death.
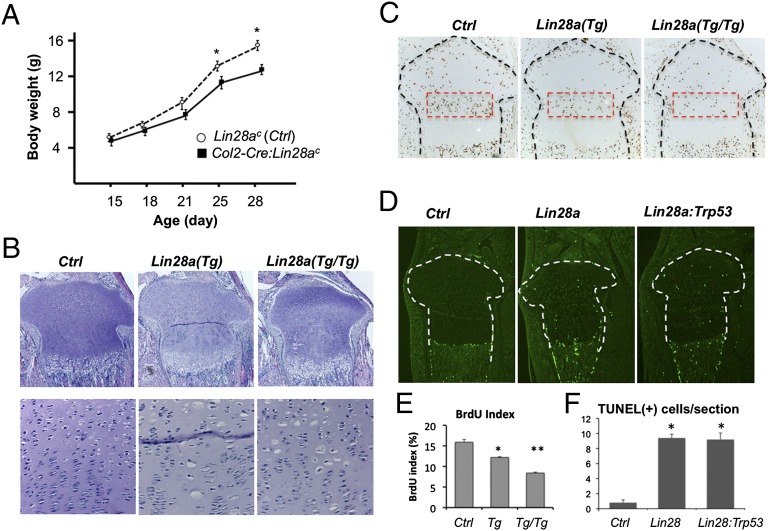
To investigate the effect of Lin28a overexpression in growth plate chondrocytes, we overexpressed Lin28a using Col2-Cre transgenic mice (27). In this system, Lin28a is overexpressed after differentiation of mesenchymal cells into the chondrocytic lineage. Unlike the Prx1-Cre:Lin28ac mice, which showed overgrowth, Col2-Cre:Lin28ac mice showed a mild growth defect (Fig. 2A). Histological analysis revealed that Lin28a overexpression caused an enlargement of cell size in some proliferating chondrocytes, although maintaining a relatively normal growth plate structure (Fig. 2B). These enlarged chondrocytes are not aberrantly differentiated hypertrophic chondrocytes because they were negative for Col10a1, a specific marker for hypertrophic chondrocytes (Fig. S2). Lin28a overexpression decreased proliferation of columnar proliferating chondrocytes in a dose-dependent manner (Fig. 2 C and E). We also found an increase in cell death in the growth plate overexpressing Lin28a (Fig. 2 D and F). The increase in cell death was caused by a mechanism independent of TRP53 (p53), a tumor suppressor molecule that is activated on aberrant oncogenic or genotoxic stimuli to mediate cell cycle arrest and apoptosis, because simultaneous ablation of the Trp53 gene was not able to rescue this phenotype (Fig. 2D). These growth plate phenotypes are also observed in Prx1-Cre:Lin28ac mice (Fig. S3).
Fig. 2.
Lin28a overexpression decreases proliferation and increases cell death in chondrocytes. (A) Growth of Col2-Cre:Lin28ac mice (black squares), assessed by body weight, is mildly impaired compared with that of Cre-negative Ctrl (open circles) (n = 5; *P < 0.05 vs. Ctrl). Error bars depict mean ± SEM. (B) Growth plate morphology of the proximal tibia of postnatal 4.5-d-old mice. Col2-Cre:Lin28ac mice with one allele (heterozygote) and two alleles (homozygote) of the Lin28ac transgene are indicated by Lin28a (Tg) and Lin28a (Tg/Tg), respectively. (Lower) Cell density of the growth plate is decreased in the columnar proliferating zone of Lin28a transgenic mice. Transgenic mice show a somewhat disorganized columnar structure with the appearance of enlarged “hypertrophic-like” chondrocytes. (Original magnification, Top, ×40; Bottom, ×200.) (C) BrdU labeling index is reduced in postnatal 4.5-d-old heterozygous (Tg) and homozygous (Tg/Tg) Lin28a transgenic mice. (Magnification, ×40.) (D) Number of TUNEL-positive cells was increased in the growth plate of the transgenic tibia of postnatal 2.5-d-old Lin28a mice. Cell death was not rescued by simultaneous ablation of Trp53 (Col2-Cre:Lin28ac:TrP53fl/fl, labeled as Lin28a:Trp53). (Magnification, ×40.) (E) Quantification of the BrdU labeling index (n = 3; *P < 0.05 vs. Ctrl; **P < 0.05 vs. Tg). (F) Quantification of TUNEL-positive cells (*P < 0.05 vs. Ctrl). There is no significant difference between Lin28a transgenic mice and Lin28a:Trp53 doubly mutant mice.
Lin28a Overexpression Decreases let-7 miRNAs and Increases let-7 Target mRNA in Chondrocytes.
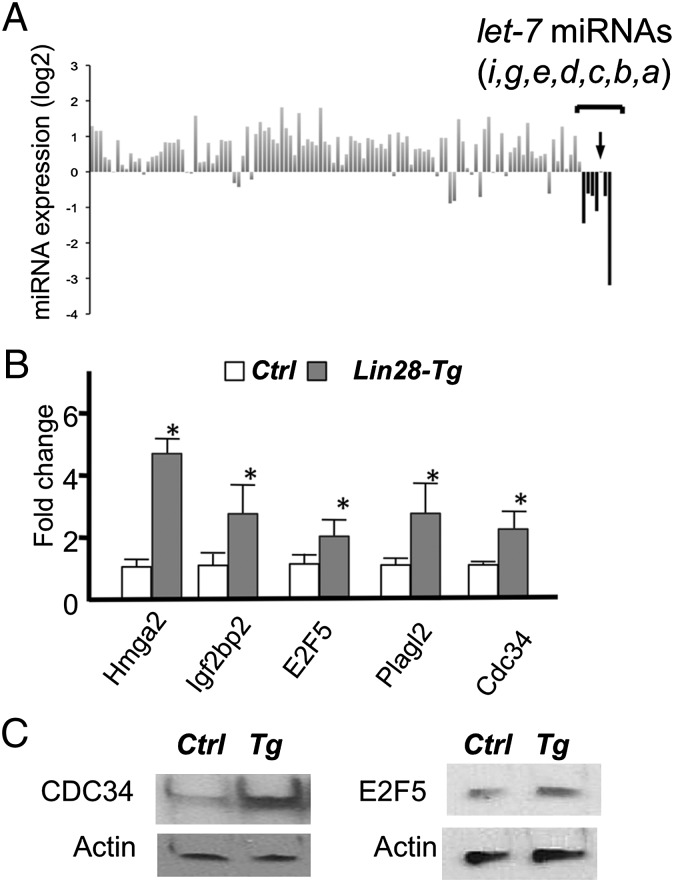
The best-known role of LIN28A is inhibition of biogenesis of let-7 miRNAs. Recently, it was found that LIN28A also binds to other RNA species and is suggested to have other functions, such as regulation of RNA splicing and translation of certain RNAs (28–32). To examine the effect of Lin28a overexpression in chondrocytes, we performed miRNA and RNA profiling. RNA was extracted from primary rib chondrocytes. Lin28a overexpression relatively specifically suppressed expression of most let-7 miRNAs, although it had a marginal effect on let-7c (Fig. 3A). This is presumably due to the fact that pre–let-7c miRNA has a less optimal binding sequence for LIN28 proteins (28, 32). We performed microarray analysis to compare expression profiles in primary rib chondrocytes between Col2-Cre:Lin28ac and control mice. Among those whose signal intensity was greater than 200 (30,725 probes of a total 35,512 probes, with an average intensity of 2,838), we found that expression of 56 genes was up-regulated by more than 40% in chondrocytes overexpressing Lin28a (Table S1). Among the 56 genes, 10 (17.8%) were predicted to be let-7 target genes by the computational prediction program, TargetScan (33). Because TargetScan predicted 928 conserved let-7 target genes among 18,615 genes in the genome (4.9%), predicted let-7 target genes were significantly enriched in genes up-regulated in Lin28a-overexpressing chondrocytes (P < 0.001, χ2 test). The predicted let-7 target genes that were up-regulated in Lin 28a transgenic mice included potential cell proliferation regulators, such as Hmga2, E2F transcription factor 5 (E2F5), cell division cycle 34 (Cdc34), and pleiomorphic adenoma gene-like 2 (Plagl2) (Fig. 3 B–C). These results demonstrate that Lin28a overexpression specifically reduces let-7 miRNAs and up-regulates let-7 target genes in chondrocytes.
Fig. 3.
Lin28a overexpression suppresses let-7 expression and derepresses let-7 target genes in chondrocytes. (A) miRNA expression profiles in primary rib chondrocytes of Cre-negative Ctrl and Col2-Cre:Lin28ac (Lin28-Tg) mice. Expression levels of 165 miRNAs detected in chondrocytes were normalized by that of the U6 level. Expression levels of miRNAs in Lin28-Tg chondrocytes are shown as fold differences relative to those of controls. The let-7 miRNAs were relatively specifically down-regulated in Lin28-Tg, except for let-7c (arrow). (B) Target let-7 genes were up-regulated in Lin28-Tg rib chondrocytes. Some of microarray results (Table S1) were verified by qRT-PCR (n = 3; *P < 0.05 vs. Ctrl). (C) Up-regulation of CDC34 and E2F5 proteins in primary rib chondrocytes of Lin28-Tg mice was verified.
Up-regulation of the let-7 Target Gene Hmga2 Does Not Play a Causal Role in Lin28a Transgenic Phenotype Development.
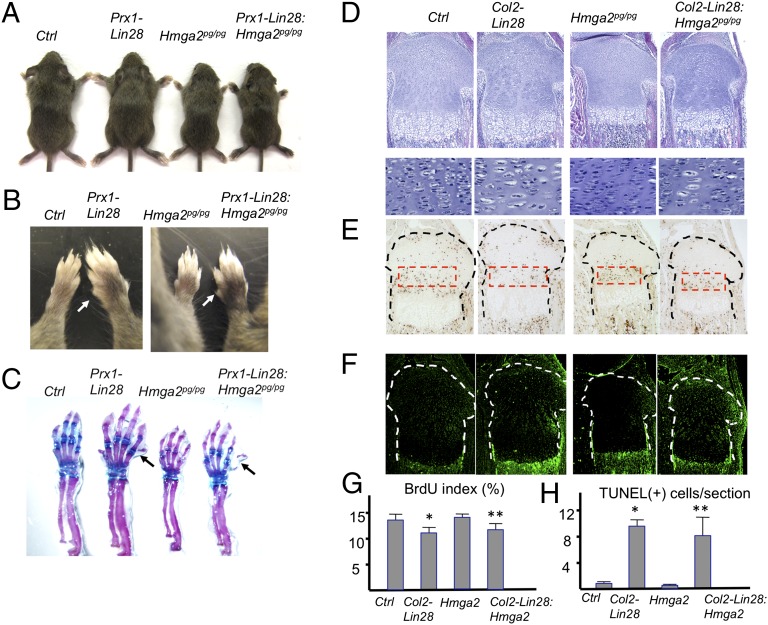
The DNA architectural factor and protooncogene Hmga2 is often overexpressed in tumor cells. HMGA2 directly interacts with chromatin and facilitates RNA transcription (34). The Hmaga2 gene, predicted to have six conserved let-7 binding sites, is one of the best-characterized let-7 target genes. Loss of let-7 binding to Hmga2 transcripts due to genomic rearrangements causes Hmga2 overexpression in tumor cells, which likely plays a causal role in tumorigenesis (15, 16). We hypothesized that Hmga2 up-regulation might contribute to skeletal phenotypes of Lin28a transgenic mice. To test this hypothesis, we crossed spontaneous Hmga2-null mutant (pygmy) mice (35) to Lin28a transgenic mice to generate compound mutant mice. Homozygous Hmga2-null mice are proportionally smaller than controls (Fig. 4A). However, the absence of Hmga2 did not rescue the overgrowth or polydactyly in Prx1-Cre:Lin28ac transgenic mice (Fig. 4 A–C). Likewise, Hmga2 loss had no effect on the morphological changes of chondrocytes (Fig. 4D), proliferation defects (Fig. 4 E and G), or cell death (Fig. 4 F and H) observed in Col2-Cre:Lin28ac chondrocytes. These results demonstrate that Hmga2 up-regulation in Lin28a transgenic mice does not play a causal role in their skeletal phenotypes.
Fig. 4.
Hmga2 up-regulation does not contribute to skeletal phenotypes of Lin28a transgenic mice. (A) Limb overgrowth of Prx1-Cre:Lin28ac (Prx1-Lin28) mice is present in the absence of Hmga2 . An Hmga2-null allele [pygmy (pg)] was bred into Prx1-Cre:Lin28ac mice to generate compound mutant mice, Prx1-Cre:Lin28ac:Hmga2pg/pg (Prx1-Lin28:Hmga2pg/pg). Three-wk-old littermates are shown. (B) Polydactyly and overgrowth of forelimbs are present in Lin28:Hmga2pg/pg mice. (C) Skeletal preparation demonstrates bone overgrowth and polydactyly in Lin28:Hmga2pg/pg mice. (D) Growth plate abnormalities [hypocellularity and appearance of large chondrocytes in Col2-Cre:Lin28ac (Col2-Lin28)] are present in the absence of Hmga2. (Original magnifications, Top, ×40; Bottom, ×200.) (E) BrdU labeling assay. The proliferation defect is still present in Lin28:Hmga2pg/pg mice. (Magnification, ×40.) (F) TUNEL assay. Chondrocyte cell death caused by Lin28a overexpression is not rescued by the absence of Hmga2. (Magnification, ×40.) (G) Quantification of E (n = 3; *P < 0.05 vs. Ctrl; **P < 0.05 vs. Ctrl; not significant vs. Lin28). (H) Quantification of F (n = 3; *P < 0.05 vs. Ctrl; **P < 0.05 vs. Ctrl; not significant vs. Col2-Lin28).
Overexpression of the let-7 Target Genes Cdc34 and E2F5 Suppresses Chondrocyte Proliferation.
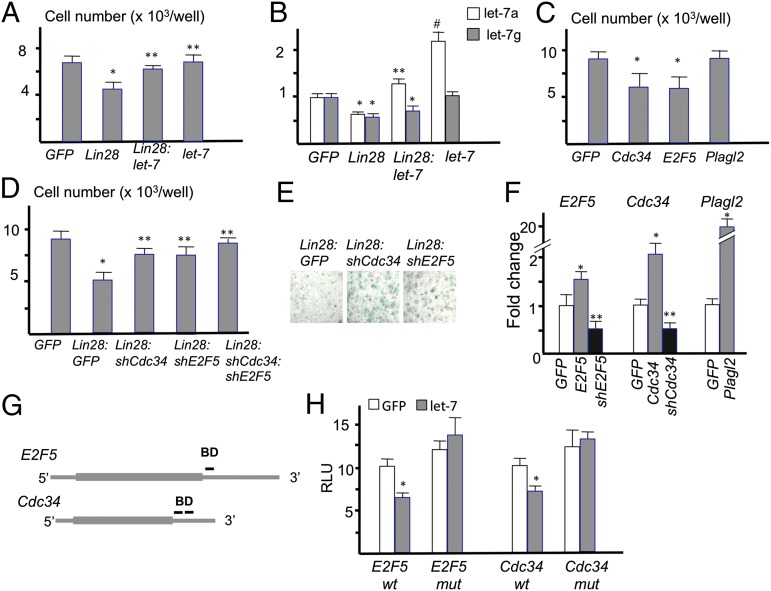
In addition to Hmga2, three of the genes up-regulated in the Lin-28a–overexpressing chondrocytes Cdc34, E2F5, and Plagl2 were predicted to be let-7 target genes and were also implicated in the regulation of cell cycle progression (Fig. 3 B and C). To investigate the roles of let-7 miRNAs and their target genes in chondrocyte proliferation, we first established a primary chondrocyte culture system. Primary rib chondrocytes were isolated from mice at postnatal day 1.5 and infected with retroviruses. In this system, we typically achieved 60–80% infection rates as determined by GFP coexpressed from the virus vector. Lin28a overexpression significantly reduced cell number, whereas co-overexpression of let-7 partially rescued the proliferation defect (Fig. 5A). The decrease or increase of let-7 levels after Lin28a or let-7 virus infection was confirmed (Fig. 5B). These results suggest that the proliferation defect in Lin28a-overexpressing chondrocytes is caused, at least in part, through let-7 suppression. Next, we overexpressed Cdc34, E2F5, or Plagl2. We found that overexpression of Cdc34 or E2F5 significantly reduced the chondrocyte cell number, whereas Plagl2 overexpression had no effect (Fig. 5C). In contrast, knockdown of E2F5 and/or Cdc34 using shRNA viruses rescued the cell number decrease caused by Lin28a overexpression (Fig. 5D) and subsequently increased the number of Alcian blue-positive cartilaginous nodules (Fig. 5E). The knockdown efficiency of these shRNA viruses was confirmed (Fig. 5F). These results suggest that the up-regulation of E2F5 and Cdc34 plays a causal role in the proliferation defect observed in chondrocytes overexpressing Lin28a. Because knockdown of E2F5 or Cdc34 increased Alcian blue positivity, it is also possible that these genes negatively regulate proteoglycan synthesis and contribute to the cartilage phenotype of Lin28a transgenic mice. Multiple computational programs predicted conserved let-7 binding sites in the 3′-UTR of the E2F5 and Cdc34 genes (Fig. 5G). To determine whether let-7 miRNAs regulate gene expression through these sequences, DNA containing the predicted let-7 binding site(s) was cloned into a luciferase expression construct in the 3′-UTR region of the luciferase gene. After transfection with luciferase vectors carrying these let-7 binding sequences, luciferase activity was significantly suppressed by let-7 coexpression, whereas luciferase activity after transfection with constructs carrying mutated let-7 binding sequences was not suppressed by let-7 cotransfection (Fig. 5H). These results suggest that let-7 miRNAs suppress E2F5 and Cdc34 expression through these predicted binding sequences.
Fig. 5.
Overexpression of let-7 target genes decreases chondrocyte proliferation. (A) Primary rib chondrocytes are infected with MSCV-GFP viruses with no insert (GFP), Lin28a (Lin28), and/or let-71a-f1 (let-7). The cell number was counted 48 h after infection (n = 5; *P < 0.01 vs. GFP; **P < 0.05 vs. Lin28). (B) Expression levels of let-7a and let-7g. The let-7 virus contains let-7a and let-7f but not let-7g (n = 5; *P < 0.05 vs. GFP; **P < 0.05 vs. Lin28; #P < 0.01 vs. GFP). (C) Cell numbers of primary rib chondrocytes 48 h after infection of indicated viruses (n = 5; *P < 0.05 vs. GFP control). (D) Cell numbers of primary rib chondrocytes 72 h after infection with indicated viruses (n = 5; *P < vs. GFP; **P < 0.05 vs. Lin28). (E) Alcian blue staining of primary rib chondrocytes isolated from Col2-Cre:Lin28ac mice. Cartilaginous nodules were stained 4 d after infection with the indicated viruses. (Original magnification, ×40.) (F) Validation of gene knockdown and overexpression by qRT-PCR. E2F5, Cdc34, and Plagl2 levels were determined after infection with viruses with empty (GFP), overexpression (gray bars), or knockdown (black bars) constructs of indicated genes (n = 4; *P < 0.05 vs. GFP; **P < 0.05 vs. GFP). (G) Schematic representation of let-7 binding sites in the E2F5 and Cdc34 mRNAs. DNA containing the predicted let-7 binding site(s) (BD) is indicated by bars in 3′-untranslated regions. (H) Luciferase activities after transfection with luciferase reporter constructs carrying WT (wt) or mutated (mut) BD sequences with control (GFP) or a let-7a1-f1 expression construct (let-7) (n = 5; *P < 0.05 vs. control).
Combination of Lin28a Overexpression and Mir140 Loss Substantially Impairs Skeletal Growth.
We have previously shown that a global miRNA reduction through conditional ablation of Dicer in chondrocytes causes a severe growth defect due to a dramatic reduction in chondrocyte proliferation and altered chondrocyte differentiation (5). In this study, we have demonstrated that let-7 deficiency by means of Lin28a overexpression reduces chondrocyte proliferation. Nevertheless, unlike Dicer deficiency, it results in only a relatively mild skeletal growth defect. These findings suggest that in addition to let-7 miRNAs, other miRNAs play important roles in skeletal growth. The miR-140 and miR-140* are encoded by the Mir140 gene, and they are abundantly and relatively specifically expressed in chondrocytes. We and others have previously reported that loss of the Mir140 gene causes a mild skeletal growth defect (6, 7). Mir140 deficiency causes a defect in chondrocyte differentiation at multiple steps (7). Mir140 deficiency also accelerates chondrocyte differentiation into postmitotic hypertrophic chondrocytes and inhibits differentiation of resting chondrocytes into columnar proliferating chondrocytes, whereas it does not affect chondrocyte proliferation. Because the major growth plate phenotypes of Dicer-deficient growth plates are defects in both proliferation and differentiation, we hypothesized that the two miRNA species, let-7 and Mir140 miRNAs, might play major roles in skeletal development by regulating chondrocyte proliferation and differentiation, respectively; thus, simultaneous deficiency of both miRNA species might result in skeletal phenotypes similar to those observed in mice missing Dicer in chondrocytes.
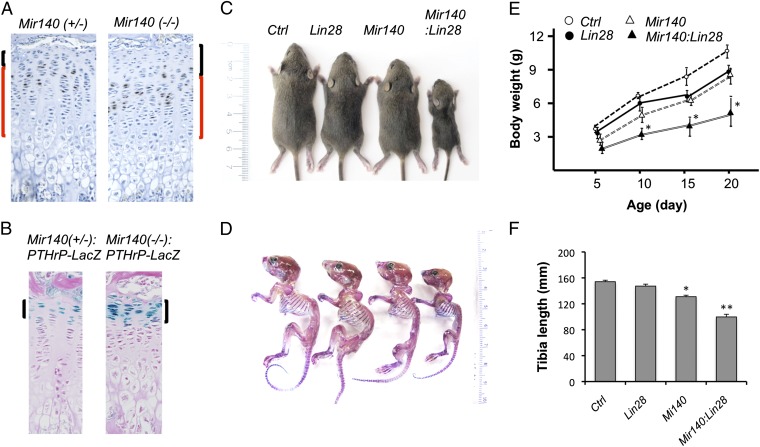
Genetic ablation of the Mir140 gene causes an expansion of the resting zone, the area mostly devoid of BrdU-positive cells and marked by PTHrP expression, and a mild reduction in the proliferating zone (Fig. 6 A and B). To generate mice deficient for both let-7 and Mir140 miRNAs, we crossed Col2-Cre:Lin28ac transgenic mice and Mir140-null mice. Single-mutant mice for either Col2-Cre:Lin28ac or Mir140−/− showed mild growth defects (10–20% reduction in body weight), whereas compound-mutant mice (i.e., Col2-Cre:Lin28ac:Mir140−/−) were substantially smaller than single-mutant mice (50–60% reduction in body weight), suggesting a synergistic effect of Mir140 deficiency and Lin28a overexpression (Fig. 6 C–E). Col2-Cre:Lin28ac:Mir140−/− mice showed a severe impairment in maxillary growth, occasionally leading to underbite, a phenotype observed in mice missing Dicer in chondrocytes (Fig. S4A). To test whether the synergistic effect of miR140 deficiency and Lin28a overexpression was conserved in other animal species, we manipulated miR-140 and Lin28a in zebrafish. Knockdown of miR-140 causes a mild elongation of the palatal cartilage in zebrafish (36). The body size of fish with a single manipulation, either miR-140 knockdown or Lin28a overexpression, was only marginally smaller than that of controls; however, simultaneous manipulation of miR-140 knockdown and Lin28a overexpression resulted in a significant decrease in the size of the body and head (Fig. S4B), a phenotype reminiscent of Dicer knockdown fish (37). In addition, we found palatal defects only in doubly manipulated fish (Fig. S4 C and D). These results suggest that miR-140 and let-7 miRNAs together also play an important role in skeletal development in zebrafish, thus further suggesting the conserved roles of these miRNAs in skeletogenesis in vertebrates.
Fig. 6.
Simultaneous manipulation of Lin28a and Mir140 causes a substantial growth defect. (A) BrdU staining depicts an expansion of the resting zone and mild shortening of the proliferating columnar zone of the tibial growth plate of 3-wk-old Mir140-null mice. The resting zone, composed of mostly BrdU-negative cells, is indicated by black brackets. The columnar proliferating zone flanked by the resting zone and hypertrophic zone is indicated by red brackets. (Original magnification, ×200.) (B) Expansion of the resting zone is demonstrated by the increase in cells expressing LacZ from the endogenous PTHrP gene promoter (brackets). PTHrP is expressed exclusively by the resting zone chondrocytes in the growth plate. (Magnification, ×200.) (C) Doubly mutant mice, Col2-Cre:Lin28ac:Mir140−/− (Mir140:Lin28), are significantly smaller than single-mutant mice, Col2-Cre:Lin28a (Lin28) or Mir140−/− (Mir140), at 3 wk of age. (D) Skeletal preparation of C. (E) Growth of single- and double-mutant mice (n ≥ 5; *P < 0.05 vs. Lin28 or Mir140 single-mutant mice). (F) Tibia of double-mutant mice is significantly shorter than that of Ctrl or single-mutant mice (n ≥ 3, *P < 0.05 vs. Ctrl; **P < 0.05 vs. Lin28 or Mir140 single-mutant mice).
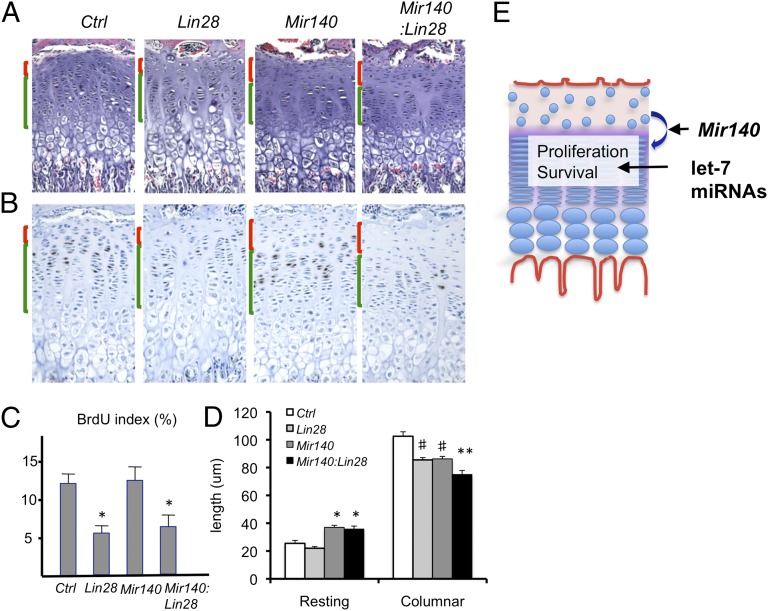
Histological analysis of Col2-Cre:Lin28ac:Mir140−/− mice showed the presence of both features of Mir140−/− and Col2-Cre:Lin28ac growth plate abnormalities: an expansion of the resting zone and a reduction in the proliferating zone observed in Mir140−/− mice and reduced cell proliferation found in Col2-Cre:Lin28ac mice (Fig. 7 A–D). The decrease in proliferating chondrocytes, caused by suppressed differentiation of resting chondrocytes into columnar proliferating chondrocytes missing Mir140, appears to be exacerbated further by the proliferation defect caused by Lin28a overexpression.
Fig. 7.
miR-140 and let-7 regulate distinct processes of growth plate development. (A) Histological appearances of Ctrl, Col2-Cre:Lin28ac (Lin28), Mir140−/− (Mir140), and Col2-Cre:Lin28ac:Mir140−/− (Mir140:Lin28) mice. The resting zone and columnar proliferating zone are indicated by red and green brackets, respectively. An expansion of the resting zone and shortening of the columnar proliferating zone are observed in Mir140:Lin28 mice. (Original magnification, ×100.) (B) BrdU staining shows an expansion of the resting zone and a reduction in the proliferation of columnar proliferating chondrocytes in Mir140:Lin28 mice. (Magnification, ×100.) (C) BrdU index in the columnar proliferating region is reduced in Lin28 and Mir140:Lin28 transgenic growth plates compared with Ctrl or Mir140 mice (n = 3 each group; *P < 0.05 vs. Ctrl or Mir140 mice). (D) Length of the resting and columnar regions of growth plates. The resting is zone is expanded in Mir140 and Mir140:Lin28 mice, whereas the columnar region is reduced in both Lin28 and Mir140 mice. Mir140:Lin28 mice show a further reduction in the length of the columnar region (n ≥ 3; *P < 0.05 vs. Ctrl or Lin28 mice; **P < 0.05 vs. Ctrl, Lin28, or Mir140 mice; #P < 0.05 vs. Ctrl). (E) Proposed model. Mir140 miRNAs are necessary for normal differentiation of resting zone chondrocytes into columnar proliferating chondrocytes. Proper regulation of this process is necessary to maintain an adequate number of proliferating chondrocytes, whereas let-7 miRNAs are necessary for normal proliferation and cell survival of columnar proliferating chondrocytes. Pathways regulated by Mir140 and let-7 miRNAs thus control the mass of proliferating chondrocytes through distinct mechanisms.
Discussion
The let-7 miRNAs comprise the most abundant miRNA species in chondrocytes. In mammals, the roles of let-7 miRNAs have been investigated mainly in the context of cancer and stem cell biology. Despite their abundance in most somatic cells, little is known about their physiological roles in normal tissues, including skeletal tissues. Because let-7 miRNAs are encoded in eight different genomic loci, the conventional gene targeting strategy is difficult to apply to a loss-of-function study in mice. An alternative is to use a sponge vector that expresses a pseudotarget RNA containing let-7 binding sites to sequester endogenous let-7 miRNAs (38, 39). We generated Cre-dependent let-7 sponge transgenic mice; although let-7 sponge transgenic mice develop skeletal abnormalities, including increased cell death and reduced proliferation of growth plate chondrocytes after crossing with Col2-Cre transgenic mice, we failed to confirm reductions in let-7 miRNAs, up-regulation of let-7 target genes, or let-7 luciferase reporter activities. Therefore, sponge expression, at least in this context, was not able to suppress let-7 functions but caused “nonspecific” abnormalities. Another strategy we took in this study was based on the recent discovery that LIN28A and LIN28B inhibit let-7 miRNA biogenesis. This finding provided a genetic means to suppress endogenous let-7 miRNAs efficiently.
A low-level ubiquitous expression of Lin28a causes overgrowth in mice, including the skeletal system (18). The mechanism for the Lin28a-induced overgrowth is not known but is speculated to occur through up-regulation of let-7 target genes. In the present study, we demonstrate that Lin28a overexpression in developing limb mesenchymal cells causes overgrowth in limbs. This finding demonstrates that the overgrowth of Lin28a overexpression is caused in a tissue-autonomous fashion. Unlike in limb mesenchymal cells, Lin28a overexpression in chondrocytes caused a growth defect due to reduced chondrocyte proliferation, suggesting that suppression of let-7 miRNAs results in different consequences dependent on cell type. It is also possible that let-7–independent functions of LIN28A contributed to the differences of phenotypes between limb mesenchymal cells and chondrocytes; it has been reported that LIN28 proteins bind to various mRNAs and regulate their translation, although the physiological significance of these findings remains to be determined (28, 30, 31).
Lin28a overexpression reduced chondrocyte proliferation, whereas it did not overtly affect chondrocyte differentiation. The Lin28a-induced proliferation defect was partly rescued by let-7 overexpression in vitro, suggesting that the reduced let-7 miRNAs are responsible for the proliferation defect of Lin28a transgenic chondrocytes. A major growth plate abnormality observed in mice missing Dicer in chondrocytes is a proliferation defect. Therefore, it is possible that the proliferation defect in Dicer-deficient chondrocytes is mainly mediated by the reduction in let-7 miRNAs.
Lin28a overexpression efficiently reduced let-7 miRNAs and up-regulated let-7 target genes both in chondrocytes and limb mesenchymal cells. Hmga2 is a well-characterized let-7 target gene, and it was one of the most robustly up-regulated genes in Lin28a-overexpressing chondrocytes in this study. Hmga2 up-regulation, due to mutations that disrupt let-7 binding to Hmga2 transcripts, is often observed in multiple types of cancers and is considered to contribute to tumorigenesis (15, 16). However, in this study, we found that Hmga2 deletion had no effects on skeletal phenotypes caused by Lin28a overexpression. This result suggests that Hmga2 is not a physiologically important let-7 target in skeletal cells.
We were able to show that other let-7 target genes, E2F5 and Cdc34, influence the chondrocyte cell number using an in vitro primary chondrocyte culture system. The observation that knockdown of these genes rescued the cell number reduction caused by Lin28a overexpression suggests that the up-regulation of E2F5 and Cdc34 plays a causal role in the proliferation defect of the Lin28a transgenic growth plate. E2F5 directly interacts with pocket proteins and inhibits cell cycle progression (40). On the other hand, CDC34, an E2 ubiquitin-conjugating enzyme, is known to promote cell cycle progression through ubiquitination of cell cycle regulators, such as CDKN1B (P27kip1) (41) and Wee1 (42), in nonchondrocytic cells. Our study suggests that CDC34 suppresses cell proliferation in chondrocytes. The role of CDC34 has not been studied in skeletal cells; thus, the mechanism by which CDC34 overexpression suppresses chondrocyte proliferation is presently unclear.
Mir140 regulates chondrocyte differentiation. Mir140-deficient growth plates show an expansion of the resting zone and shortening of the columnar proliferating zone, possibly due to deregulation of miR-140 target genes, such as Dnpep, whereas Mir140 loss does not affect chondrocyte proliferation (7). Singly mutant mice, either Mir140−/− or Col2Cre:Lin28ac, showed relatively mild skeletal growth defects, even though each modification caused clear alterations in either chondrocyte differentiation or proliferation. This finding suggests that a proliferation or differentiation defect alone caused by these mutations can be compensated for to some extent. In contrast, Col2Cre:Lin28ac:Mir140−/− doubly mutant mice showed a substantially more severe skeletal growth defect, a phenotype reminiscent of mice with chondrocytes missing Dicer. Because Col2Cre:Lin28ac:Mir140−/− doubly mutant chondrocytes are deficient for both let-7 miRNAs and miR-140/miR-140*, this observation suggests that these miRNAs, among other chondrocytic miRNAs, play a major role in regulating skeletal growth. The histology of compound mutant mice revealed that both of the abnormalities observed in Mir140−/− and Col2Cre:Lin28ac mice (i.e., a differentiation defect of resting chondrocytes and a proliferation defect of columnar proliferating chondrocytes, respectively) were present. This result provides evidence that these two abundantly expressed miRNA species in chondrocytes regulate different processes of growth plate development and that defects in these regulations result in failure to maintain an adequate number of columnar proliferating chondrocytes (Fig. 7E). Mir140 miRNAs facilitate differentiation of resting chondrocytes and inhibit hypertrophic differentiation to increase columnar proliferating chondrocytes, whose normal proliferation and survival require let-7 miRNAs.
Materials and Methods
Mice.
A Cre-inducible Lin28a expression transgene (Lin28ac) was constructed by inserting a mouse Lin28a coding sequence into an expression cassette containing the ubiquitous promoter CAGGS, followed by a loxP-flanked stop sequence and an internal ribosomal entry site (IRES) and EGFP cDNA (Fig. S1A). This expression cassette is identical to the one previously described (43), except that it carries EGFP instead of nLacZ. A 1-kb-long coding sequence of Lin28a was PCR-amplified using primers 5′-ACGGGCTCAGCAGACGACCATG-3′ and 5′-CCGTCTGGCAAGGGAAATATCACA-3′. Three copies of the amplified cDNA were connected in tandem with IRES sequences and inserted into the above expression cassette between the floxed stop sequence and the IRES-EGFP sequence (Fig. 1A). The vector sequence and the puromycin resistance gene contained in the vector were removed by digesting the construct with SphI and AscI. The purified DNA fragment was subjected to pronuclear injection. Five transgenic lines were established. Transgenic mice were screened for LIN28A and EGFP expression in tail primary fibroblasts after infection with adenoviruses expressing a Cre recombinase (Fig. 1B). Two lines that showed Cre-dependent LIN28A induction were selected and further characterized. For genotyping, the Lin28ac transgene was detected by PCR using primers tpA-5, 5′-ATCTTATCATGTCTGGATCCCC-3′, and Lin28AS, 5′-CGCAGTTGTAGCACCTGTCTC-3′.
The spontaneous Hmga2-null mutant mice (pygmy) were previously described (44). We performed genome walking and found that insertion of an LTR-like sequence deleted the first and second exons of the Hmga2 gene in the pygmy allele. The wild-type Hmga2 allele was detected using primers Hmga2-L, 5′-TGGGAATCATCACACACACA-3′, and Hmga2-R, 5′-CCATTAGGACTCCAGCGTACA-3′. The pygmy allele was detected by primers Hmga2pg-F, 5′-AGCCCAGAAGATCAGTCTTTG-3′, and Hmga2pg-R, 5′-TTTGATGGCGGTGTCATGTAG-3′. Mir140 KO mice (7), Col2-Cre (27) and Prx1-Cre (22) transgenic mice, and PTHrP-LacZ knock-in mice (45) were previously described. Mice were in a mixed genetic background. Comparison was always made between littermates. The animal experiments were approved by the Institutional Animal Care and Use Committee of the Massachusetts General Hospital and performed in accordance with its regulations and guidelines.
Skeletal Preparation and Histology.
Alizarin red staining and Alcian blue staining were performed using a modified method of McLeod (46). For histological analysis, mice were dissected; fixed in 10% (vol/vol) formalin; decalcified in 10% (wt/vol) EDTA; paraffin-processed; cut; and subjected to H&E staining, in situ hybridization, and BrdU staining.
Proliferation and Cell Death Assays.
For BrdU labeling, 50 μg of BrdU per gram of body weight was injected in mice i.p. 2 h before euthanasia. BrdU was detected using the BrdU in situ staining kit (Invitrogen). The BrdU labeling index was calculated as the ratio of BrdU-positive nuclei over total nuclei in columnar proliferating chondrocytes of the growth plate. Cell death was evaluated on sections using an in situ cell death detection kit (Roche) according to the manufacturer’s instructions.
Primary Chondrocyte Isolation and Culture.
Isolation and culture of primary rib chondrocytes were performed as previously described (7). After overnight culture, cells were trypsinized and replated at a concentration of 5 × 105 cells per milliliter in the DMEM containing 10% (vol/vol) FCS. Cell numbers were counted using a C-Chip hemocytometer (Incyto) after trypsinization.
Retrovirus Production and Infection.
Retroviruses were constructed using a modified murine stem cell virus vector (pMSCV; Clontech) in which the puromycin resistance cassette is replaced by EGFP. The full-length cDNA for E2F5 was purchased from Openbiosystems (clone no. 3482306). A SalI/EcoRI fragment containing the coding sequence was subcloned into the XhoI/EcoRI site of the pMSCV-EGFP retrovirus vector, in which the puromycin resistance gene in pMSCV-puro (Clontech) is replaced by EGFP (47). Coding sequences for Plagl2 and Cdc34 were PCR-amplified using primers Cdc34-5Xho1, TTCTCGAGCCATGGCCCGGCCGCTGG; Cdc34-3EcoR1, CCGAATTCAGGACTCTTCGGTGCCAG; Plag2l-5Xho1, GTCTCGAGCCTTGCCATGACCACATTT; and Plag2l-3EcoR1, CTGAATTCTACTGGAATGCTTGGTGGA (restriction enzyme recognition sites are underlined). Small hairpin constructs were synthesized and subcloned into pMSCV-EGFP. The cloning site of this vector is flanked by sequences of the stem region of the pri–miR-30 gene to facilitate generation of shRNAs through the miRNA processing pathway (48). For shRNA constructs for E2F5 (shE2F5) and Cdc34 (shCdc34), the following sequence was synthesized and subcloned into pMSCV-EGFP: shE2F5, 5′-gtCTCGAGGGCCTATCCATGTGCTACTTATAAATTCAAGAGATTTATAAGTAGCACATGGATAGGCCGAATTCTG -3′, and Cdc34, 5′-GTCTCGAGGGCCGCTGGTGCCAAGCTCGCAGAATTGAAGACATTCTGCGAGCTTGGCACCAGCGGCCGAATTCTG -3′ (restriction enzyme sites are underlined). pMSCV-EGFP with an insert was cotransfected with Ecopack (Clontech) into 293 cells to produce viruses. Viral particles were concentrated by standard PEG precipitation. Virus infection of primary chondrocytes was performed in the presence of 4 μg/mL Polybrene (American Bioanalytical). Infection efficiency was determined by EGFP expression.
A let-7 expression vector was constructed by cloning a genomic sequence containing the let-7a-1 and let-7f-1 genes into pMSCV-GFP at the XhoI and EcoRI sites. The genomic fragment was amplified using primers let-7-X, 5′GGCTCGAGTAAGCTTCAGTATTAGACAGTGGT-3′ (the underlined XhoI site was added for cloning purposes), and let-7-S, 5′-GCGGTCGAGTGCCCATTTACCTGTGACATCTTT-3′ (the underlined SalI site was added for cloning purposes). The PCR fragment was digested with XhoI and EcoRI, which are endogenously present in the amplified region. The resultant 770-bp-long fragment was cloned into the pMSCV-EGFP vector. A Lin28a expression vector was constructed by cloning PCR-amplified Lin28a cDNA, using primers Lin28-5′, GCAATCGATCTCGAGACGGGCTCAGCAGACGACCATG, and Lin28-3′, GCGAATTCCGTCTGGCAAGGGAAATATCACA (additional XhoI and EcoRI sites are underlined), into the pMSCV-EGFP vector.
Transfection and Luciferase Assay.
A 130-bp-long sequence that contains the predicted let-7 binding sequence (underlined) of E2F5 (E2F5-Wt, 5′-ATTCCATGGAAACTTGGGACTATTATCTACCTCTATAACATTTTAGAATTCTTTAATAACCTAAGTATTTAAAATTATGAATGTAACACCTTTTTAGTTCACTGATTCTGAAGTGTTCTTCCCTAACATT-3′) was PCR-amplified and cloned into the pMirReport luciferase vector (Ambion) using SpeI and HindIII sites. The same sequence containing a mutated binding site (changed from 5′-CTACCTC-3′′ to 5′-GATGGAG-3′) was synthesized and similarly subcloned into the same vector. An 86-bp-long sequence containing two predicted let-7 binding sites (underlined) of Cdc34 (Cdc34-Wt, 5′-CCCTGAAATAAACTTACAAATTTTACCTCAGCAGGACCCGTGTGGGCCTCAGGCGACAGACTACCTCACCGAGGTTCCATGTGGGC-3′) was PCR-amplified. A mutant sequence containing mutated binding sites (underlined) (Cdc34-mut, 5′-CCCTGAAATAAACTTACAAATTTAGGAGTGCAGGACCCGTGTGGGCCTCAGGCGACAGAGATGGAGTCCGAGGTTCCATGTGGGC-3′) was synthesized and cloned into the pMirReport luciferase vector. DNA containing 0.02 μg of a luciferase construct, 0.02 μg of Renilla luciferase control vector, and 0.06 μg of the let-7 expression vector or control vector (GFP alone) was transfected into chondrogenic ATDC5 cells using the Fugene HD transfection reagent (Promega). Luciferase and Renilla activities were measured 48 h after transfection using the Dual-Luciferase Reporter Assay System (Promega).
Microarray Analysis and miRNA Profiling.
RNA was extracted from primary rib chondrocytes from newborn mice. Gene expression profiling was performed using the Affymetrix Mouse Gene 1.0 ST. miRNA profiling was performed using the TaqMan array microRNA 384-well card (Life Science).
Quantitative RT-PCR Analysis.
cDNA synthesis was performed using random hexamers and the DyNAmo cDNA Synthesis Kit (Finnzymes). Quantitative PCR analysis was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems) and Eva Green quantitative RT-PCR (qRT-PCR) mix (Solix BioDyne). Primer sequences are as follows: Actb-L, 5′-GCACTGTGTTGGCATAGAGG-3′, and Actb-R, 5′-GTTCCGATGCCCTGAGGCTCTT-3′; Lin28a-L, 5′-CTTTTGCCAAAGCATCAACC-3′, and Lin28a-R, 5′-GGGCTGTGGATCTCTTCCTC-3′; Hmga2-L, 5′-ATCCAACTTTCTCCCCGTTC-3′, and Hmga2-R, 5′-AGGTATTGCCACAAGCAAGC-3′; Igf2bp2-L, 5′-CCACTCCGGATACTTCTCCA-3′, and Igf2bp2-R, 5′-AGCGAGCTGTTTGATGTGTG-3′; Cdc34-L, 5′-TGGCACCCAAACATCTATGA-3′, and Cdc34-R, 5′-AAGGTGTTGGGCTCATTCAG-3′; Plagl2-L, 5′-GGCTTTTGCCTCCAAGTACA-3′, and Plagl2-R, 5′-ATGGTCCTTTCGGTGAAACA-3′; and E2F5-L, 5′-AGAAGCCAGACTTTCCAGCA-3′, and E2F5-R, 5′-CATCTGCTGGGGTAGGAGAA-3′. Expression of let-7a, let-7g, and miR-30c was determined using the mirVana qRT-PCR miRNA Detection Kit (Ambion).
Western Blot Analysis.
Anti-LIN28A antibody (no. 3978) was purchased from Cell Signaling Technology. Anti-Cdc34 antibody (GTX105013) was purchased from GeneTex. Anti-E2F5 antibody (sc-999) and anti-actin antibody (I-19) were purchased from Santa Cruz Biotechnology. Western blot analysis was performed according to the standard procedure.
Supplementary Material
Acknowledgments
We thank Dr. Andy McMahon for the Cre-dependent expression construct pBGSApBpACAGftIGn. We thank Dr. Arthur Broadus for PTHrP-LacZ knock-in mice. We thank the Massachusetts General Hospital Transgenic Core Facility for pronuclear injection and the Beth Israel Deaconess Medical Center Genomics and Proteomics Center for microarray analysis. This work was supported by National Institutes of Health Grants AR054500 and AR056645 (to T.K.) and American Society for Bone and Mineral Research Grant CEA0811 (to T.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302797110/-/DCSupplemental.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423(6937):332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134(9):1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park CY, Choi YS, McManus MT. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19(R2):R169–R175. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi T, et al. Dicer-dependent pathways regulate chondrocyte proliferation and differentiation. Proc Natl Acad Sci USA. 2008;105(6):1949–1954. doi: 10.1073/pnas.0707900105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyaki S, et al. MicroRNA-140 plays dual roles in both cartilage development and homeostasis. Genes Dev. 2010;24(11):1173–1185. doi: 10.1101/gad.1915510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamura Y, Inloes JB, Katagiri T, Kobayashi T. Chondrocyte-specific microRNA-140 regulates endochondral bone development and targets Dnpep to modulate bone morphogenetic protein signaling. Mol Cell Biol. 2011;31(14):3019–3028. doi: 10.1128/MCB.05178-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li MA, He L. microRNAs as novel regulators of stem cell pluripotency and somatic cell reprogramming. Bioessays. 2012;34(8):670–680. doi: 10.1002/bies.201200019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463(7281):621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osada H, Takahashi T. let-7 and miR-17-92: Small-sized major players in lung cancer development. Cancer Sci. 2011;102(1):9–17. doi: 10.1111/j.1349-7006.2010.01707.x. [DOI] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7(6):759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 12.Kumar MS, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci USA. 2008;105(10):3903–3908. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trang P, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29(11):1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 15.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21(9):1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315(5818):1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22(9):474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu H, et al. Lin28a transgenic mice manifest size and puberty phenotypes identified in human genetic association studies. Nat Genet. 2010;42(7):626–630. doi: 10.1038/ng.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu H, et al. DIAGRAM Consortium MAGIC Investigators The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frost RJ, Olson EN. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proc Natl Acad Sci USA. 2011;108(52):21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Viswanathan SR, et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat Genet. 2009;41(7):843–848. doi: 10.1038/ng.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Logan M, et al. Expression of Cre Recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33(2):77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 23.Chiao E, et al. Overgrowth of a mouse model of the Simpson-Golabi-Behmel syndrome is independent of IGF signaling. Dev Biol. 2002;243(1):185–206. doi: 10.1006/dbio.2001.0554. [DOI] [PubMed] [Google Scholar]
- 24.Delatycki MB, Danks A, Churchyard A, Zhou XP, Eng C. De novo germline PTEN mutation in a man with Lhermitte-Duclos disease which arose on the paternal chromosome and was transmitted to his child with polydactyly and Wormian bones. J Med Genet. 2003;40(8):e92. doi: 10.1136/jmg.40.8.e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivière JB, et al. Finding of Rare Disease Genes (FORGE) Canada Consortium De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet. 2012;44(8):934–940. doi: 10.1038/ng.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romanelli V, et al. CDKN1C (p57(Kip2)) analysis in Beckwith-Wiedemann syndrome (BWS) patients: Genotype-phenotype correlations, novel mutations, and polymorphisms. Am J Med Genet A. 2010;152A(6):1390–1397. doi: 10.1002/ajmg.a.33453. [DOI] [PubMed] [Google Scholar]
- 27.Ovchinnikov DA, Deng JM, Ogunrinu G, Behringer RR. Col2a1-directed expression of Cre recombinase in differentiating chondrocytes in transgenic mice. Genesis. 2000;26(2):145–146. [PubMed] [Google Scholar]
- 28.Cho J, et al. LIN28A is a suppressor of ER-associated translation in embryonic stem cells. Cell. 2012;151(4):765–777. doi: 10.1016/j.cell.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Ma W, et al. Lin28 regulates BMP4 and functions with Oct4 to affect ovarian tumor microenvironment. Cell Cycle. 2013;12(1):88–97. doi: 10.4161/cc.23028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng S, et al. Genome-wide studies reveal that Lin28 enhances the translation of genes important for growth and survival of human embryonic stem cells. Stem Cells. 2011;29(3):496–504. doi: 10.1002/stem.591. [DOI] [PubMed] [Google Scholar]
- 31.Polesskaya A, et al. Lin-28 binds IGF-2 mRNA and participates in skeletal myogenesis by increasing translation efficiency. Genes Dev. 2007;21(9):1125–1138. doi: 10.1101/gad.415007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilbert ML, et al. LIN28 binds messenger RNAs at GGAGA motifs and regulates splicing factor abundance. Mol Cell. 2012;48(2):195–206. doi: 10.1016/j.molcel.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia DM, et al. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat Struct Mol Biol. 2011;18(10):1139–1146. doi: 10.1038/nsmb.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashar HR, Chouinard RA, Jr, Dokur M, Chada K. In vivo modulation of HMGA2 expression. Biochim Biophys Acta. 2010;1799(1-2):55–61. doi: 10.1016/j.bbagrm.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 35.Xiang X, Benson KF, Chada K. Mini-mouse: Disruption of the pygmy locus in a transgenic insertional mutant. Science. 1990;247(4945):967–969. doi: 10.1126/science.2305264. [DOI] [PubMed] [Google Scholar]
- 36.Eberhart JK, et al. MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nat Genet. 2008;40(3):290–298. doi: 10.1038/ng.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35(3):217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 38.Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4(9):721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rybak A, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008;10(8):987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 40.Gaubatz S, et al. E2F4 and E2F5 play an essential role in pocket protein-mediated G1 control. Mol Cell. 2000;6(3):729–735. doi: 10.1016/s1097-2765(00)00071-x. [DOI] [PubMed] [Google Scholar]
- 41.Tam SW, Theodoras AM, Pagano M. Kip1 degradation via the ubiquitin-proteasome pathway. Leukemia. 1997;11(Suppl 3):363–366. [PubMed] [Google Scholar]
- 42.Michael WM, Newport J. Coupling of mitosis to the completion of S phase through Cdc34-mediated degradation of Wee1. Science. 1998;282(5395):1886–1889. doi: 10.1126/science.282.5395.1886. [DOI] [PubMed] [Google Scholar]
- 43.Stenman JM, et al. Canonical Wnt signaling regulates organ-specific assembly and differentiation of CNS vasculature. Science. 2008;322(5905):1247–1250. doi: 10.1126/science.1164594. [DOI] [PubMed] [Google Scholar]
- 44.Zhou X, Benson KF, Ashar HR, Chada K. Mutation responsible for the mouse pygmy phenotype in the developmentally regulated factor HMGI-C. Nature. 1995;376(6543):771–774. doi: 10.1038/376771a0. [DOI] [PubMed] [Google Scholar]
- 45.Chen X, et al. Initial characterization of PTH-related protein gene-driven lacZ expression in the mouse. J Bone Miner Res. 2006;21(1):113–123. doi: 10.1359/JBMR.051005. [DOI] [PubMed] [Google Scholar]
- 46.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22(3):299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 47.Lu J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14(6):843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silva JM, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37(11):1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.