Abstract
Protein modification by one or more ubiquitin chains serves a critical signalling function across a wide range of cellular processes. Specificity within this system is conferred by ubiquitin E3 ligases which target the substrates. Their activity is balanced by deubiquitylating enzymes which remove ubiquitin from both substrates and ligases. The RING-CH ligases were initially identified as viral immunoevasins involved in the downregulation of immunoreceptors. Their cellular orthologues, the Membrane Associated RING-CH (MARCH) family represent a subgroup of the classical RING genes. Unlike their viral counterparts the cellular RING-CH proteins appear highly regulated, and one of these in particular, MARCH7 was of interest due to a potential role in neuronal development and lymphocyte proliferation. Difficulties in detection and expression of this orphan ligase lead us to search for cellular co-factors involved in MARCH7 stability. Here we show that MARCH7 readily undergoes autoubiquitylation and associates with two deubiquitylating enzymes – USP9X in the cytosol and USP7 in the nucleus. Exogenous expression and siRNA depletion experiments demonstrate that MARCH7 can be stabilised by both USP9X and USP7, which deubiquitylate MARCH7 in the cytosol and nucleus respectively. We therefore demonstrate compartment-specific regulation of this E3 ligase through recruitment of site-specific deubiquitylating enzymes.
Keywords: Ubiquitin E3 ligase, MARCH7, Deubiquitylation, USP9X, USP7
Introduction
Protein modification by ubiquitin serves a critical signalling function across a diverse range of cellular processes. The combinatorial diversity within the ubiquitin pathway suggests it is the most complex regulatory system of the eukaryotic cell. Ubiquitylation principally involves three steps: (i) ubiquitin activation via an E1 enzyme, (ii) transfer of activated ubiquitin to an E2 conjugating enzyme and (iii) targeting of ubiquitin to the lysine residue of the substrate protein (1, 2). This last reaction is mediated by ubiquitin E3 ligases which therefore confer specificity to the ubiquitylation reaction. Two major families of E3 ligases are recognised. The HECT family forms an essential thiol ester intermediate during ubiquitin chain catalysis, while the RING family has no intrinsic catalytic activity, but acts as a scaffold to recruit the ubiquitin-charged E2 enzyme and promote ubiquitin transfer to the substrate (3).
The classical RING finger (RING-HC) consists of a series of interspersed cysteine and histidine residues in a C3HC4 configuration. The cysteine-histidine spacing allows binding of two zinc ions which stabilise the formation of a cross-brace structure (3). The recently described RING-CH E3 ligases are variants of the classical RING and, as their name describes, have an unusual C4HC3 (CH) cysteine/histidine configuration (4). Their founding members, mK3, K3 and K5 were originally identified as viral immunoevasins from gamma herpesviruses, and form a family of viral transmembrane E3 ligases which downregulate critical cell surface receptors, including MHC class I molecules (5-10). The unusual C4HC3 RING configuration initially led these proteins to be mistaken for PHD domains, but bioinformatic analysis (11, 12) and the solution structure of the K3 zinc binding motif confirmed them as true RINGs (13). The identification of additional RING-CH viral E3 ligases as evolutionarily distant as the herpes and pox viruses made it likely that this family was appropriated from the vertebrate host. This led to the subsequent description of a family of cellular RING-CH ligases, named the MARCH (Membrane Associated RING-CH) proteins, which are the orthologues of the viral RING-CH proteins (14, 15).
The MARCH family comprises eleven cellular gene products (5, 14-16). Recent studies have investigated their intracellular trafficking (17-20) and identified them as potential regulators of cell surface immunoreceptors. Exogenous expression of several of the MARCH genes results in the downregulation of MHC Class I, CD4, CD86, Fas and the transferrin receptor (5, 14, 15, 21). MARCH9 downregulates cell surface MHC Class I (14) and ICAM-1 (22) in a ubiquitin dependent manner. MHC Class II surface expression is also regulated by ubiquitylation (23) and MARCH1 and MARCH8 (c-MIR) were recently identified as key facilitators in MHC Class II downregulation. Both MARCH1 and MARCH8 ubiquitylate and downregulate cell surface MHC Class II molecules (15, 24-26) and depletion of MARCH1 results in high MHC Class II expression at the cell surface (25, 26).
MARCH7 (also known as Axotrophin or Axot) together with MARCH10 are conspicuous members of the cellular ‘MARCH’ family, as they encode a RING-CH domain but no predicted transmembrane domains. MARCH7 is highly expressed in stem cells, neurones and lymphocytes, suggesting a possible function in development as well as the immune system (27, 28). This is of particular interest as disruption of the MARCH7 locus using a random gene trap generated mice with reported abnormalities in both neurological development and the immune system (29, 30). Splenocytes from MARCH7 deficient mice were reported to show a MARCH7 gene-dose dependent increase in lymphocyte proliferation and Leukaemia Inhibitory Factor (LIF) secretion after stimulation with a T cell mitogen, as well as increased IL-2 secretion (31). In addition, MARCH7 has been implicated in the control of expression of the regulatory T lymphocyte transcription factor, Foxp3 and the suppressor of cytokine signalling 3 gene (SOCS3) (32, 33).
While viral members of the RING-CH family show stable expression, regulation of cellular E3 ligases represents a fine balance between ubiquitylation and deubiquitylation. Indeed, a key feature of all RING E3 ligases is their ability to autoubiquitylate. This provides an important mechanism for self-regulation and promotes targeting of the E3 ligase for proteasome-mediated degradation, (34, 35). The removal of ubiquitin chains by deubiquitylating enzymes (DUBs) plays a critical role in regulating the half-life and activity of many cellular ligases (36-38).
Our ongoing interest in the RING-CH proteins, together with the potential importance of MARCH7 in the immune and nervous system led to a further examination of the MARCH7 polypeptide. Despite high levels of transcriptional expression in many tissues, we were unable to detect endogenous MARCH7 protein. Furthermore, detection of exogenously expressed MARCH7 was only possible in a limited number of cell lines, suggesting MARCH7 is highly regulated.
In this study we show that MARCH7 readily undergoes autoubiquitylation in vivo, and is targeted for proteasome-mediated degradation. MARCH7 is found to interact independently with two compartment-specific DUBs - USP9X (also known as FAM) in the cytosol and USP7 (also known as HAUSP) in the nucleus. These DUBs are individually able to deubiquitylate and therefore stabilise MARCH7. We show that differential localisation of the DUBs provides a mechanism for compartment-specific stabilisation of this E3 ligase.
Results
MARCH7 is preferentially expressed in cells of neuronal origin and is stabilised by functional mutations within the RING-CH domain
The MARCH7 gene predicts a protein of 693 amino acids with a single recognised functional motif, the RING-CH domain, close to the C-terminus (Figure 1A). The large N-terminus is serine/proline rich and predicts a highly disordered structure. Unlike other MARCH family proteins there are no predicted transmembrane domains. MARCH7 is highly conserved in bony vertebrates and especially in mammals (39), with 85% homology between human and mouse MARCH7 and an identical RING-CH domain. In keeping with this high homology, gene expression profiles for MARCH7 between human and mouse are similar, showing MARCH7 to be highly expressed in immune, neuronal and embryonic stem (ES) cells (27, 28).
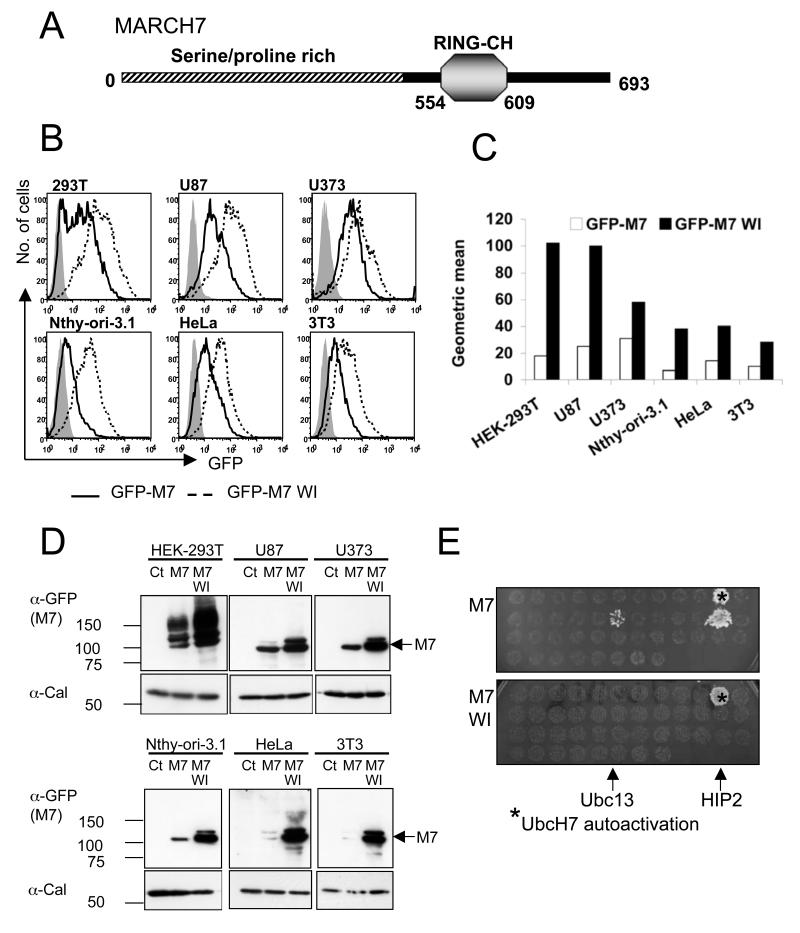
Figure 1. MARCH7 is preferentially expressed in cells of neuronal origin and is stabilised by functional mutations within the RING-CH domain.
A) Schematic of MARCH7, showing the RING-CH and serine/proline rich N-terminus (hatched area). B-D) The indicated cell lines were transduced with lentivirus encoding GFP-MARCH7, GFP-MARCH7 WI or a control virus encoding GFP alone and after 3 days analysed by flow cytometry (B) or western blotting (D). The multiplicity of infection (MOI) was estimated for each cell line by titration of the control GFP virus, and kept constant for all cell lines. The mean fluorescent intensity (MFI) for GFP-MARCH7 (open bars) and GFP-MARCH7 WI (closed bars) are shown in (C). The transduced cell lines were lysed in RIPA buffer and immunoblotted with an anti-GFP antibody for MARCH7 (D). Twice as much lysate was loaded for the non-neuronal cell lines (Nthy-ori-3.1, HeLa and 3T3) compared to the neuronal cell lines (HEK-293T, U87 and U373) to allow detection of GFP-MARCH7 protein. E) MARCH7 recruits the E2 conjugating enzymes HIP2 and Ubc13. Full length human MARCH7 and MARCH7 WI were expressed in a yeast two hybrid assay against a library of human E2 conjugating enzymes. The yeast colonies were selected on histidine treated plates. Wildtype MARCH7 associated with HIP2 and Ubc13 (top panel) whilst the MARCH7 WI mutant is unable to bind the E2 conjugating enzymes Ubc13 and HIP2 (bottom panel). UbcH7 autoactivates on histidine treated plates. M7 MARCH7, M7 WI MARCH7 WI, Cal calreticulin.
Despite apparently high transcriptional MARCH7 expression in ES cells, protein detection proved difficult. A MARCH7 gene-trapped ES cell line, expressing β-galactosidase (β-gal) under the endogenous MARCH7 promoter, verified high expression by β-gal staining, but no endogenous protein could be detected in wildtype ES cells (data not shown). Even overexpression of MARCH7 proved difficult. The only cells which allowed exogenous MARCH7 expression were HEK-293T cells. Although originally assumed to be of embryonic kidney origin, transcriptional profiles suggest a likely neuronal lineage for these cells (40), which led us to determine whether MARCH7 expression can be supported in other cell lines of neuronal origin. We therefore used flow cytometry to assess MARCH7 expression in six different cell lines transduced with lentivirus encoding a GFP-tagged MARCH7 (Figure 1B). Three cell lines were of neuronal origin (HEK-293Ts, the U87 glioblastoma and U373 glioblastoma cell lines). The other cell lines were epithelial (HeLa), thyroid epithelial (Nthy-ori-3.1) and fibroblasts (3T3). A control virus containing only GFP showed equivalent transduction efficiency for all six cell lines. While the GFP-MARCH7 protein is detected by western blotting in all cell lines (Figure 1D) higher levels of MARCH7 expression is seen in neuronal lineage cells (U373, U87 and HEK-293T cells) compared to other cell lines (HeLa, Nthy-Ori3.1 and 3T3 cells) (Figure 1B). The mean fluorescent intensity (MFI) for GFP-MARCH7 was at least 2 fold higher in cells of neuronal origin compared to non-neuronal cells (Figure 1C).
To determine whether an active RING contributes to MARCH7 instability, we mutated two residues in the MARCH7 RING, W589A and I556A (MARCH7 WI). These are well recognized mutants (41) which were previously shown to prevent recruitment of the E2 conjugating enzymes by the RING-CH domain (9), without affecting its structural integrity. The loss of function of this mutant was confirmed in a yeast two hybrid analysis using MARCH7 as bait against an E2 library as prey (Figure 1E). Wildtype MARCH7 binds two E2 conjugating enzymes, Huntingtin interacting protein 2 (HIP2) and Ubc13. These interactions were lost with the MARCH7 WI mutant which did not bind any E2 conjugating enzymes (Figure 1E).
It was therefore of interest to find that the GFP-MARCH7 WI mutant showed consistently higher expression levels than wildtype GFP-MARCH7 (Figure 1B, C and D). In all cell lines tested, expression of the GFP-MARCH7 WI mutant is two to four-fold higher than wildtype GFP-MARCH7, with significantly higher levels seen in neuronal as compared with non-neuronal cell lines (Figure 1B, C). This increased stability of the MARCH7 mutant was also seen on western blot analysis of MARCH7 lysates (Figure 1D). These data show that an active RING is required for MARCH7 regulation, suggesting a role for autoubiquitylation in the rapid turnover of MARCH7.
MARCH7 is ubiquitylated in vivo
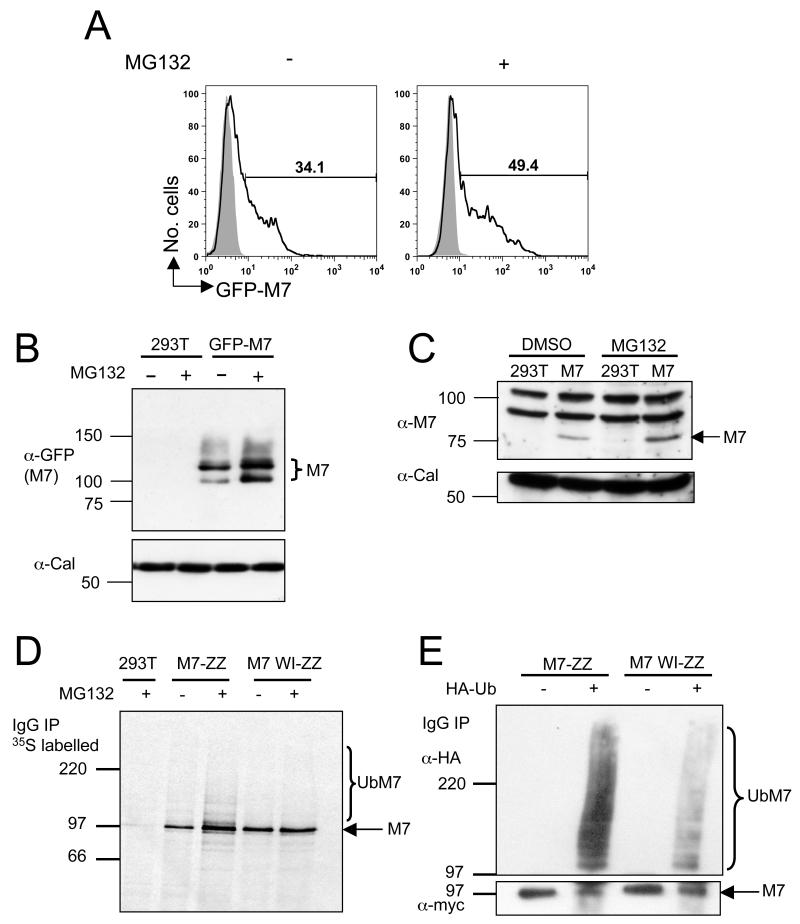
We first used flow cytometry to examine the effect of proteasome inhibitors on MARCH7 expression. In the presence of the proteasome and caspase inhibitor MG132, GFP-MARCH7 expression in transfected HEK-293T cells increased from 34.1% to 49.4% (Figure 2A), consistent with autoubiquitylation promoting proteasomal degradation. This increase in cellular MARCH7 expression following MG132 treatment is also evident by western blot analysis of untagged and GFP conjugated MARCH7 lysates (Figure 2B, C).
Figure 2. MARCH7 is ubiquitylated in vivo.
A-C) MARCH7 is stabilised with MG132 treatment. HEK-293T cells were transfected with MARCH7 or GFP-MARCH7 and incubated with 7.5μM MG132 or DMSO for 18 hours. GFP-MARCH7 expression was measured by flow cytometry and western blotting (A, B). Untagged MARCH7 expression was measured by immunoblotting using the chicken MARCH7 antisera (C). Calreticulin (Cal) was immunoblotted as a loading control. D, E) MARCH7 ubiquitylation. Radiolabeled MARCH7 detects higher molecular weight species consistent with ubiquitylation (D). HEK-293T cells were transfected with MARCH7-ZZ or MARCH7 WI-ZZ, radiolabelled with [35S] methionine and [35S] cysteine for 15 minutes and chased for 90 minutes. Cells were pretreated with 20μM MG132 or DMSO control from the starve until the end of the chase (150 minutes), lysed in RIPA buffer, immunoprecipitated on IgG sepharose beads and after SDS-PAGE separation, analysed by phosphoimager. E) HEK-293T cells co-transfected with or without HA tagged ubiquitin (HA-Ub) and either MARCH7-ZZ or MARCH7 WI-ZZ were lysed in RIPA buffer and MARCH7 immunoprecipitated by IgG pull down. Samples were separated by SDS-PAGE and immunoblotted for MARCH7 (c-myc antibody) and ubiquitin (HA antibody).
To determine whether MARCH7 is ubiquitylated we initially radiolabelled HEK-293Ts, transfected with ZZ-myc tagged MARCH7 or MARCH7 WI (MARCH7-ZZ and MARCH7 WI-ZZ), in the presence or absence of MG132 (Figure 2D). Following incubation with MG132, MARCH7 is not only stabilised, but an additional ladder of higher molecular weight MARCH7 species appear, consistent with ubiquitylation. To verify the nature of these bands, cell lysates from HEK-293T cells transfected with wildtype or mutant MARCH7-ZZ and HA-tagged ubiquitin, were immunoprecipitated for MARCH7 with IgG sepharose beads and immunoblotted for ubiquitin. Ubiquitylated MARCH7 species are readily detected (Figure 2E), with a marked decrease in ubiquitylation in the presence of the MARCH7 WI mutant. We believe that autoubiquitylation is likely to be responsible for MARCH7 ubiquitylation, though the presence of a very small amount of ubiquitylation with the MARCH7 WI mutant raises the possibility of an additional E3 ligase being involved.
MARCH7 associates with the DUBs USP9X and USP7
As MARCH7 is readily ubiquitylated we wanted to determine whether additional cellular cofactors involved in MARCH7 regulation could be identified. Large scale IgG-sepharose pull downs of lysates from ZZ-myc tagged MARCH7 and MARCH7 WI transfected HEK-293T cells were performed and protein bands detected were analysed by mass spectrometry. Two potentially regulatory MARCH7 binding partners were identified - the DUBs USP9X and USP7 (Supplementary Figure 1). While deubiquitylation is known to regulate the stability of several ubiquitin E3 ligases (36-38), the identification of these two DUBs was of particular interest as USP9X and USP7 show differential subcellular localisation - USP7 in the nucleus (42, 43) and USP9X in the cytosol (37, 44). We therefore hypothesised that MARCH7 may be regulated by deubiquitylation in distinct cellular compartments.
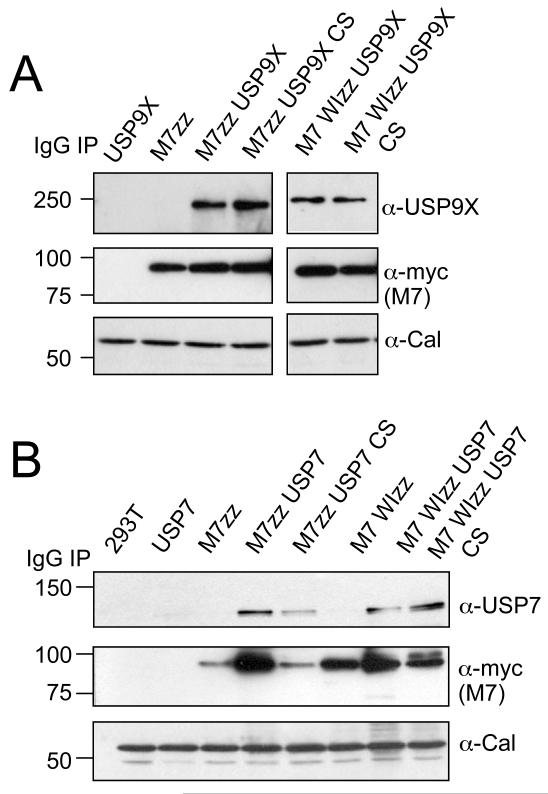
The interactions between MARCH7 and USP9X and USP7 could be confirmed in standard immunoprecipitations for MARCH7-ZZ and MARCH7 WI-ZZ and subsequent immunoblotting for USP9X and USP7 (Figure 3A and B). These interactions are dependent on neither the enzymatic activity of the DUBs, nor a functional RING, as MARCH7 and MARCH7 WI both associate with catalytically inactive (CS) mutant DUBs.
Figure 3. MARCH7 associates with the DUBs USP9X and USP7.
A, B) MARCH7 co-immunoprecipitates with USP7 and USP9X. HEK-293T cells were co-transfected with MARCH7-ZZ/MARCH7 WI-ZZ and (A) USP9X/USP9X CS or (B) USP7/USP7 CS, lysed in 1% Triton X-100 and MARCH7 immunoprecipitated on IgG sepharose beads. 1×106 cells equivalents were loaded per lane, separated by SDS-PAGE and immunoblotted for USP9X, USP7 and MARCH7 (c-myc antibody). 1×105 cells/lane of lysate were loaded and immunoblotted for calreticulin (Cal) as a loading control.
MARCH7 localises to the cytosol, nucleus and plasma membrane
The identification of two MARCH7-associated DUBs was of particular interest when we examined the subcellular localisation of MARCH7. In U87 glioblastoma cells, transduced with GFP-tagged MARCH7, localisation is predominantly nuclear with some cytosolic MARCH7 (Figure 4A). A very different pattern is seen with the MARCH7 WI mutant, which shows a punctate distribution at the plasma membrane, together with some nuclear staining. This pattern of MARCH7 subcellular localisation is consistent across different cell lines, including the mouse fibroblast cell line 3T3 (Supplementary Figure 2). We wanted to confirm that this same pattern is seen with untagged MARCH7. As our MARCH7 specific antibody did not work well on microscopy, cell fractionation experiments were performed to confirm the MARCH7 subcellular distribution. Cellular fractions isolated from HEK-293T cells transfected with MARCH7 or MARCH7 WI, identified MARCH7 and MARCH7 WI in the cytosolic, membrane and nuclear fractions (Figure 4B). Epitope tagged MARCH7 showed the same distribution in the cellular fractions of HEK-293T cells (data not shown). Thus wildtype MARCH7 is predominantly nuclear, while the MARCH7 WI mutant accumulates at the plasma membrane, with less protein seen in the nucleus.
Figure 4. MARCH7 localises to the cytosol, nucleus and plasma membrane.
A) MARCH7 and MARCH7 WI subcellular localisation. The U87 glioblastoma cell line was transduced with lentivirus encoding GFP-MARCH7 or GFP-MARCH7 WI (green). Cells were plated on coverslips and fixed with 4% paraformaldehyde after 3 days. DAPI was used as a nuclear stain (blue). B) MARCH7 is present in cytosolic, nuclear and membrane cellular fractions. HEK-293T cells were transfected with MARCH7 or MARCH7 WI and specific cellular fractions isolated after 48 hours, acetone precipitated, separated by SDS-PAGE and visualised with the chicken MARCH7 antisera. Five times more cell lysate was loaded in the MARCH7 transfected cells compared with the MARCH7 WI cells to allow comparison. C, D) MARCH7 co-localises with USP7 and USP9X. U87 cells were transduced with GFP-MARCH7 or GFP-MARCH7 WI lentivirus (green) and fixed on coverslips after 3 days. The coverslips were probed for USP7 (C) or USP9X (D) and detected using an anti-rabbit Alexafluor 568 antibody (red). DAPI (blue) was used as a nuclear counter stain. Bar=10μm Cy cytosolic fraction, Mb membrane fraction, Nu nuclear fraction
We examined the co-localisation of GFP-MARCH7 with USP9X and USP7. U87 cells transduced with GFP-MARCH7 or GFP-MARCH7 WI were stained with antibody for endogenous DUBs and analysed by confocal microscopy (Figure 4C and D). Both MARCH7 and MARCH7 WI co-localise with USP7 in the nucleus (Figure 4C). Endogenous USP9X is seen diffusely throughout the cytosol. While some localisation between the MARCH7 WI mutant and USP9X is present in the cyotsol there is no clear enrichment of USP9X at the plasma membrane (Figure 4D).
MARCH7 is stabilised by USP9X and USP7
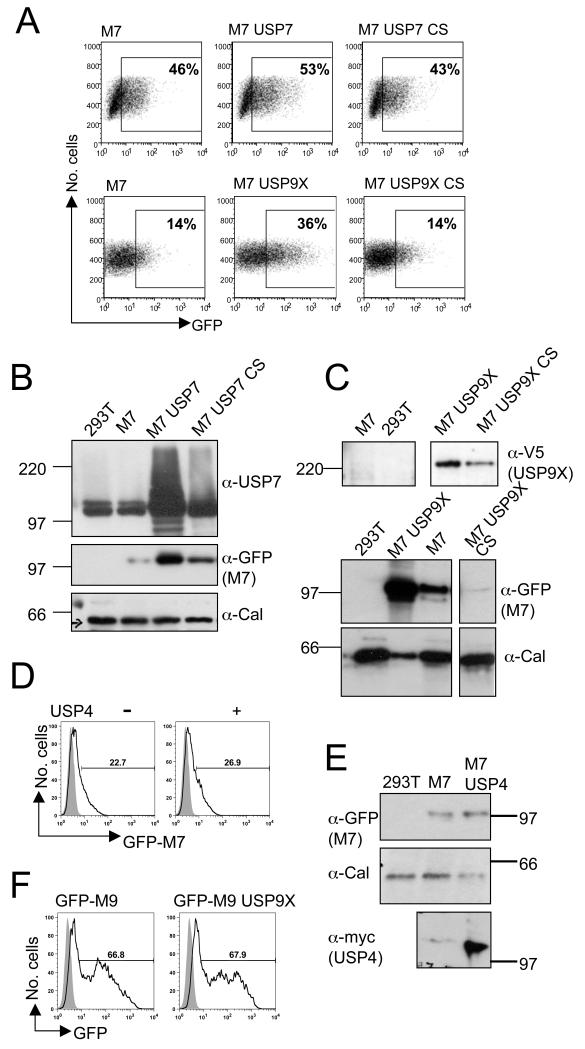
We used flow cytometry to determine how exogenous expression of USP7 and USP9X affect GFP-MARCH7 levels in HEK-293T cells. Both USP7 and V5-USP9X increase GFP-MARCH7 expression (Figure 5A). USP7 increased the percentage of GFP-MARCH7 cells from 46% to 53% cells with a much higher percentage of GFP-MARCH7high cells, while USP9X increases GFP-MARCH7 expression from 14% to 36% (Figure 5A). This increased MARCH7 expression is dependent on an enzymatically active DUB as the catalytically inactive CS mutant DUBs had no effect on GFP-MARCH7 expression. Stabilisation of GFP-MARCH7 by the DUBs could be visualised by immunoblotting (Figure 5B and C). Similar findings were observed in at least three independent experiments, showing that DUB activity stabilises MARCH7. The specificity of the USP9X and USP7 interaction was examined using unrelated DUBs, including USP4 (Figure 5D and E) and USP8 (data not shown), neither of which affected MARCH7 levels. Conversely, USP9X had no effect on expression of the related MARCH9 protein (Figure 5F).
Figure 5. MARCH7 is stabilised by USP7 and USP9X.
A-C) HEK-293T cells were co-transfected with GFP-MARCH7 and USP7 or USP7 CS (A, B) or GFP-MARCH7 and USP9X or USP9X CS (A, C). After 48 hours cells were harvested and analysed by flow cytometry (A) or western blotting (B, C). The percent GFP positive cells in the gated population out of the total number of live cells is indicated. Cell lysates were prepared using RIPA buffer and 1x105 cell equivalents loaded (B, C). Lysates were probed for MARCH7 (anti-GFP mAb.) (B and C, middle panels), USP7 (B, top panel) and USP9X (anti-V5 mAb.) (C, top panel). Calreticulin was blotted as control (B and C, bottom panels). D, E) USP4 has no effect on MARCH7 levels. HEK-293T cells were transfected with GFP-MARCH7 in the presence of myc tagged USP4 and analysed by flow cytometry (D). The shaded histogram shows the non-transfected population of cells. RIPA extracts were probed for MARCH7 (anti-GFP), USP4 (myc antibody - 9E10) and calreticulin (E). F) USP9X does not stabilise MARCH9. HEK-293T cells were co-transfected with GFP-MARCH9 and USP9X, and after 48 hours analysed by flow cytometry. The shaded histogram refers to the non-transfected control cells. Percentages refer to the gated GFP positive population.
SiRNA mediated depletion of USP9X and USP7 affects the stability of MARCH7
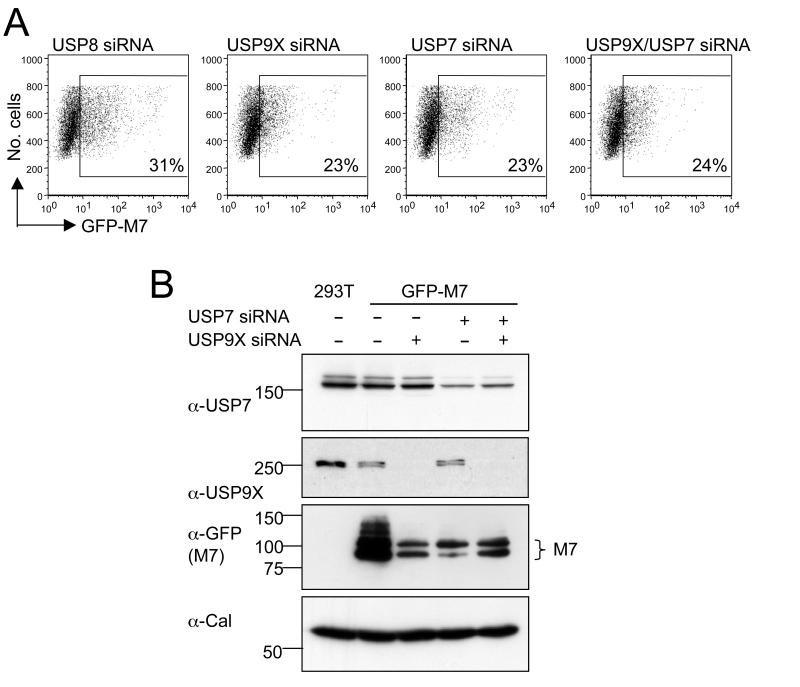
Having demonstrated MARCH7 stabilisation by exogenous DUB expression we investigated the effect of short interfering RNA (siRNA)-mediated depletion of endogenous USP9X and USP7. USP8 has no effect on MARCH7 expression and was therefore used as a control DUB. Individual siRNA-mediated depletions of either USP9X or USP7 (Figure 6B) reduced GFP-MARCH7 expression from 31% to 23% (Figure 6A). Depletion of USP9X and USP7 together also reduced GFP-MARCH7 levels to 24%. Therefore overexpression of USP9X and USP7 stabilises MARCH7 expression, while depletion of USP9X and USP7 destabilises MARCH7.
Figure 6. SiRNA mediated depletion of USP9X and USP7 affects the stability of MARCH7.
A, B) HEK-293T cells were transfected with siRNA specific for USP7, USP9X or USP8 (as a control). After 24 hours cells were transfected with GFP-MARCH7. Analysis by (A) flow cytometry or (B) immunoblotting was performed after a further 48 hours. The percentages refer to the geometric mean of GFP positive cells from the total live cell population. Cell lysates were prepared in RIPA buffer and blotted for endogenous USP7, USP9X and MARCH7 (GFP antibody). Calreticulin was used as a loading control.
MARCH7 is deubiquitylated by USP9X and USP7 in vivo
To confirm that the stabilisation of MARCH7 was dependent on the deubiquitylating activity of USP9X and USP7, we examined MARCH7 ubiquitylation in the presence and absence of exogenously expressed DUBs. The high level of MARCH7 ubiquitylation seen in 293T cells transfected with MARCH7-ZZ and HA-ubiquitin is markedly decreased in the presence of USP9X (Figure 7 lanes 3 versus 5) and completely abolished by USP7 (Figure 7 lane 4). Furthermore, the very low level of MARCH7 WI ubiquitylation (Figure 7 lane 9) can be further reduced in the presence of USP7 and USP9X (Figure 7 lanes 7 and 8). These data suggest that deubiquitylation is responsible for the USP9X and USP7 mediated stabilisation of MARCH7.
Figure 7. MARCH7 is deubiquitylated by USP7 and USP9X.
HEK-293T cells were co-transfected with ZZ-myc tagged MARCH7 or MARCH7 WI, HA tagged ubiquitin and either USP7 or USP9X. After 48 hours cells were lysed in RIPA buffer and MARCH7 immunoprecipitated on IgG sepharose beads. Membranes were probed for ubiquitin (HA antibody) and MARCH7 (myc antibody).
SiRNA mediated depletion of USP9X and USP7 alters the subcellular localisation of MARCH7
The subcellular distribution of USP9X and USP7 suggests their ability to deubiquitylate MARCH7 is compartment specific. To determine whether this was correct we examined the subcellular distribution of GFP-MARCH7 following siRNA-mediated depletion of endogenous USP9X and USP7 from U87 cells. USP7 depletion caused a decrease in nuclear GFP-MARCH7 (Figure 8A and B). This is further emphasised in the MARCH7 WI mutant where USP7 depletion leads to an almost complete loss of nuclear staining but GFP-MARCH7 WI remains prominent within the cytosol (Figure 8A). Conversely, siRNA mediated depletion of USP9X reduces the total levels of GFP-MARCH7, while no GFP-MARCH7 WI expression can be identified at the cytosol or plasma membrane (Figure 8C and D). Quantitative analysis of multiple images of GFP-MARCH7 localisation following siRNA depletion of USP7 and USP9X demonstrate similar results (Figure 8E), suggesting that USP9X and USP7 govern the stability of MARCH7 in a cell compartment-specific manner.
Figure 8. SiRNA mediated depletion of USP9X AND USP7 alters the subcellular localisation of MARCH7.
A-D) U87 cells were transduced with GFP-MARCH7 or GFP-MARCH7 WI lentivirus and after 24 hours transfected with siRNA specific for USP7 or USP9X. Cells were analysed by confocal microscopy (A, C) and immunoblotting (B, D) after a further 72 hours. For microscopy, the U87 cells were plated onto coverslips, fixed and stained for USP7 (A) or USP9X (C) as above. RIPA extracted cell lysates were probed for endogenous USP7 (B) or USP9X (D). Calreticulin was used as a loading control. (E) Quantitative analysis of GFP-MARCH7 expression following siRNA mediated depletion of USP7 and USP9X demonstrates compartment-specific regulation of MARCH7. U87 cells were transduced with GFP-MARCH7 or GFP-MARCH7 WI and transfected with siRNA specific for USP8, USP7 and USP9X as described previously. 10 image fields (containing between 1-6 cells) for each siRNA mediated depletion were analysed for GFP-MARCH7 intensity levels in the nucleus and cytosol using Volocity software. The results are presented as the mean GFP intensity (arbitrary units), with the error bars indicating the standard error of the mean. Bar=10μm.
Discussion
The balance between ubiquitylation and deubiquitylation provides an intrinsic mechanism for regulating cellular proteins (45) and E3 ligases in particular (34-37). In this study we show that the RING-CH MARCH7 E3 ligase is highly regulated and undergoes RING-dependent autoubiquitylation and proteasome-mediated degradation. MARCH7 associates with the cytosolic USP9X DUB as well as the nuclear USP7 DUB. Each of these DUBs can stabilise MARCH7 by deubiquitylation. USP9X appears to predominantly regulate MARCH7 in the cytosol, while USP7 regulates MARCH7 in the nucleus, demonstrating compartment specific regulation of this E3 ligase.
USP9X and USP7 are ubiquitin specific proteases (USPs), the largest family of the five classifications of DUBs (46). Members of the USP family are typically large proteins (60-300kDa) with a conserved catalytic domain allowing cleavage of ubiquitin and polyubiquitin conjugates by their cysteine peptidase activity (47, 48). USP7 was first identified through its interaction with the herpes simplex virus type 1 immediate early protein (ICP0 or Vmw110) (49). ICP0 binds USP7 and this interaction is required for ICP0 to alter gene expression (50). Not surprisingly, as a predominantly nuclear expressed DUB, USP7 has been shown to be important in regulation of a number of transcription factors. Specifically, deubiquitylation by USP7 modulates the activity of the tumour suppressor gene, p53 (51) and the forkhead transcription factor, FOXO4 (43).
USP9X is the mammalian homologue of the Drosophila fat-facets gene which regulates cell determination and normal embryonic growth in the fly (52, 53). While clearly of importance for normal mammalian development (54-56), USP9X specifically deubiquitylates critical molecules required for maintenance of cell adhesion and polarity. It interacts and stabilises AF-6 and the β-catenin E-cadherin complex, preserving the integrity of adherens junctions (44, 57-59). USP9X has also been implicated in neurological synapse formation where it co-localises with the clathrin coat adaptor Epsin 1 (56, 60, 61).
MARCH7 is highly expressed in stem cells, the brain, lung and the immune system (27, 28) and consistent with the DUBs ability to stabilise MARCH7, both USP7 and USP9X are expressed in tissues where high MARCH7 expression has been detected. Northern blot analysis of mouse tissues show USP9X expression is most abundant in the brain, heart, lung and thymus (37), and USP7 expression is highest in the brain, lung, testis and thymus (62). USP9X expression has also been identified in stem cells (27).
Both USP7 and USP9X are reported to stabilise other E3 ligases through deubiquitylation – USP9X the HECT E3 ligase Itch (37) and USP7 the RING type E3 ligase Mdm2 (38). However, the association of an E3 ligase with two or more DUBs is unusual and has only been reported in the context of highly regulated cellular processes. Mdm2, the critical regulator of the tumour suppressor gene p53 is deubiquitylated by USP7 (38) and the predominantly cytosolic USP2a (63, 64), providing intricate control of p53 activity (65). TRAF6, a RING E3 ligase and core component of the TNF-α complex, is deubiquitylated by the DUBs A20 (66) and CYLD (67), allowing the rapid termination NF-κB signalling. The stabilization of MARCH7 by two DUBs in different locations allows compartment-specific regulation of this ligase and provides an additional layer for control of MARCH7. The activity of MARCH7 will be affected by signalling pathways which differentially regulate expression of USP9X and USP7 within the cell, implying a dynamic role for the regulation of MARCH7 by deubiquitylation.
Whether additional post-translational modifications are involved in MARCH7 regulation have not been determined. The discrepancy between high MARCH7 mRNA expression and low protein levels suggests this may indeed be the case, and would include regulation by other E3 ligases or phosphorylation. Cross-talk between phosphorylation and ubiquitylation is increasingly recognised and phosphorylation may influence E3 ligase activity in several different ways, including substrate recognition, E3 ligase activity and subcellular localisation (68). At present we do not have clear evidence for MARCH7 phosphorylation, but the presence of a MARCH7 doublet on some gels suggests this modification may occur. Whether phosphorylation is required for MARCH7 trafficking between nucleus and cytosol, as reported for Mdm2, whose nuclear localisation is initiated by Akt phosphorylation (69), remains to be determined. Further studies are therefore required to discern whether the interplay between phosphorylation and ubiquitylation adds a further layer of complexity to MARCH7 regulation.
The interactions between MARCH7 and USP9X or USP7 are clearly not dependent on MARCH7’s E3 ligase activity or the DUBs cysteine protease function. Locating the binding sites within MARCH7 remains difficult due to its large size and disordered structure. Domain and structural analysis of USP7 has identified binding motifs for EBNA1 (70, 71), p53 (71-73) and Mdm2 (72, 73). However this consensus P/AXXS USP7 motif (72) is located at 18 possible conserved positions within MARCH7. No interaction domain within USP9X has been identified and detailed structural modelling are required to further elucidate these associations.
USP9X depletion decreases total MARCH7 levels, whereas USP7 appears to stabilise MARCH7 in the nucleus. The discrepancy between the effect of siRNA mediated-depletion of USP9X and USP7 on total MARCH7 levels suggests that MARCH7 may require stabilisation in the cytosol by USP9X before translocation to the nucleus. The absence of classical nuclear localisation signals in MARCH7 or USP7 (42) is consistent with this finding, implying that additional protein interactions may be required to allow MARCH7 to access the nucleus.
Distribution of MARCH7 at the plasma membrane only became apparent following expression of the MARCH7 WI mutant. Since this mutant has little ubiquitylation activity it is likely to bind, but no longer ubiquitylate its target(s), as we have found for other members of the RING-CH family (9). Similar findings have recently been observed for another member of the RING-CH family, MARCH2. Exogenous expression of MARCH2 demonstrates perinuclear distribution but the MARCH2 W mutant localises to points of cell-cell contact, where it identifies its substrate DLG1 (74). Since the MARCH7 WI mutant is likely to bind, but no longer ubiquitylate its target(s), it therefore suggests the plasma membrane as the site where MARCH7 recruits its substrates. In the absence of any clear target we can only speculate on why MARCH7 WI accumulates at the plasma membrane, but it may be involved in dynamic trafficking and signalling pathways between the plasma membrane and nucleus. Indeed, a number of proteins traffic from the plasma membrane to the nucleus. The plasma membrane EGF receptor is located in the nucleus of highly proliferating cells where it is thought to alter gene expression (75) and Notch and other gamma-secretase substrates requires proteolytic cleavage at the plasma membrane to release its intracellular domain, which translocates to the nucleus to from a transcriptional complex (76), some of which are thought to require ubiquitylation (77).
The DUBs themselves may share common substrates with the E3 ligase, as demonstrated by USP7, which deubiquitylates Mdm2 as well as p53 (38, 78) providing an elegant negative feedback loop for p53 regulation. It will be of interest to determine whether MARCH7 shares common substrates with USP7 and USP9X.
In summary, the localisation of MARCH7 in the nucleus and cytosol, and its association with site-specific DUBS suggest a novel form of compartment-specific regulation for these proteins.
Materials and Methods
Materials and antibodies
The following antibodies were used: Chicken antisera to MARCH7 (Abcam); mAb to GFP (Abcam); 9E10, mouse monoclonal to cmyc (Santa Cruz); mouse monoclonal to the V5 epitope (Invitrogen); rabbit polyclonal to USP7 (Bethyl Laboratories); PA3-900, rabbit polyclonal to calreticulin; 16B12 rabbit polyclonal to the HA epitope (Covance). The rabbit USP9X (FAM) antisera was generated as described previously (44, 57). The fluorescent secondary antibody was anti-rabbit Alexa Fluor 568 (Invitrogen). MG132 (Calbiochem) was used at the indicated concentrations.
Plasmids and mutagenesis
The full length cDNA IMAGE clone for mouse MARCH7 was purchased from RZPD (pCMV-Sport6, IMAGE ID 4935124). The open reading frame was amplified by PCR (Phusion DNA polymerase, New England Biolabs) and sequenced verified in pCR4Blunt-TOPO (Invitrogen) before final ligation into the mammalian expression construct. Untagged an MARCH7 constructs were generated in the pHRSIN (kind gift from Y. Ikeda) lentiviral construct. GFP tagged constructs were generated by amplification of EGFP by PCR from pEGFP-C1 (Clontech) and ligation into the pHRSIN backbone to generate the pHRSIN-GFP vector. MARCH7 was then ligated into pHRSIN-GFP, resulting in a N-terminal GFP tag on MARCH7. The MARCH7-ZZ construct was generated by ligation of MARCH7 into the pCANT-ZZ backbone (kind gift from J. Trowsdale). This construct encodes MARCH7 with a c-myc tag followed by a TEV cleavage site and then the ZZ tag (protein A). MARCH7-ZZ was then amplified by PCR into the pHRSIN lentiviral backbone. The IMAGE cDNA clone for human MARCH7 was purchased from RZPD (pCMV-Sport6, IMAGE ID 6169288). The open reading frame was amplified by PCR and cloned into yeast two hybrid bait and prey vectors pGAD-C1 and pGBDU-C1 (79). The following oligonucleotide primers were used: BglII EGFPfor AGATCTCCACCATGGTGAGCAAGGGCGAGGAGCTGT, BamHI EGFPrev GGATCCCTTGTACAGCTCGTCCATGCCGA, BamHI MARCH7for CGGGATCCACCATGGAGTCTAAACCTTCCAGGATT, NotI MARCH7rev TTGCGGCCGCCCATCTTCTTCAGAAATTCATCCTC, AgeI MARCH7rev ACCGGTGAATTCATCCTCTGAAGTTTCGA, BglII pZZfor AGATCTACCATGGTGACCCACGATGA, NotI pZZrev GCGGCCGCGATCACTAATTCGCGTCTACTTTC, ClaI hMARCH7for TCCCCCGGGGAGTCTAAACCTTCAAGGATTCC and SmaI hMARCH7rev CCATCGATTGAAGTTAGGCAATATCAAATGTC. The MARCH7 WI mutants were generated by site-directed mutagenesis using the QuickChange kit (Stratagene). The W589A and I556A mutations were made in the RING-CH domain of mouse MARCH7 using the primers: MARCH7 W589Afor GAGTGTATGAAAAAGGCGTTACAAGCCAAAATTAATTCTGG, MARCH7 W589Arev CCAGAATTAATTTTGGCTTGTAACGCCTTTTTCATACACTC, MARCH7 I556Afor GAAGGGGACTTATGTAGAGCTGTCAGATGGCAGCAGCG, and MARCH7 I556Arev CGCTGCTGCCATCTGACAAGCTCTACATAAGTCCCCTTC. The human MARCH7 W587A and I554A mutations were generated with the primers: MARCH7 W587Afor CAAGACTGTATGAAAAAGGCGTTACAGGCCAAAATTAAC, MARCH7 W587Arev GTTAATTTTGGCCTGTAACGCCTTTTTCATACAGTCTTG, MARCH7 I554Afor GAAGAAGGTGACTTATGTAGAGCTTGTCAAATGGCAGC and MARCH7 I554Arev GCTGCCATTTGACAAGCTCTACATAAGTCACCTTCTTC. The mouse MARCH7 WI construct was then ligated into the pZZ-CANT, pHRSIN and pHRSIN-GFP vectors. Human MARCH7 WI was ligated into the pGAD-C1 and pGBDU-C1 vectors. The mouse V5 tagged USP9X constructs, pEF-DEST51-USP9X and pEF-DEST51-USP9X C1556S, were generated as described previously (44). The USP7 vectors, pCI-USP7 and pCI-USP7 C223S, were donated by RD Everett (35, 49). The pCDNA3 HA-ubiquitin construct was a gift from I Dikic (80) and the myc tagged USP4 from K Lindsten.
Cell culture and transient transfections
U87 cells were grown in MEM (Sigma) supplemented with 10% foetal calf serum (FCS), penicillin and streptomycin (Sigma). HEK-293T cells and all other cell lines were grown in supplemented RPMI (Sigma). The MARCH7 gene trapped ES cell line (clone AE0078) was from WC Skarnes. Cells were transfected using TransIT 293 reagent (Mirus) in six well plates and harvested after 48 hours. The DNA molar ratios were kept constant for all co-transfections and when required, the total concentration of DNA was kept at 2μg using pCDNA3.1 as a filler.
Lentivirus preparation and transductions
The lentiviral preparation was performed as described for the pHRSIN Ub/Em vector with some modifications (81). HEK-293T cells were transfected with the pHRSIN vector, the VSV-G envelope (pMDG) and the pCMV 8.9 (plasmid containing the GAG and POL adaptor genes) in a T75 flask. The viral supernatant was harvested at 48 and 72 hours, 0.2μm filtered and stored at −80°C. Ultracentrifugation (30 000rpm, 4°C for 90 minutes) was performed to concentrate the virus using an SW40Ti rotor (Beckman-Coulter). The pellet was resuspended in the residual media and 25μl aliquots were stored at −80°C. Viral titration was performed by ten fold serial dilutions onto 104 cells in 30μl media in a 96 well plate. The plate was centrifuged at 700rpm for 20 minutes at 37°C and the total volume increased to 200μl with pre-warmed media. Transduction efficiency was assessed by GFP expression using flow cytometry and the titre calculated. The multiplicity of infection (MOI) for the cell line was then estimated from the viral titre. Transductions for experiments were performed by centrifugation (700rpm, 20 minutes) of 2×105 cells with virus in 100μl medium, using equivalent viral MOIs for each cell line. The cells were transferred into six well plates and analysed at the indicated time point.
SiRNA-mediated depletion
siRNA SMARTpool oligonucleotides for USP7, USP9X and USP8 were purchased from Dharmacon. The individual oligonucleotides for USP7 and USP9X were evaluated and all produced similar depletions. The siRNA oligonucleotides for USP7 were AAGCGUCCCUUUAGCAUUAUU, GCAUAGUGAUAAACCUGUAUU, UAAGGACCCUGCAAAUUAUUU, and GUAAAGAAGUAGACUAUCGUU. The siRNA oligonucleotides for USP9X were AGAAAUCGUGGUAUAAAUUU, ACACGAUGCUUUAGAAUUUUU, GUACGACGAUGUAUUCUCAUU and GAAAUAACUUCCUACCGAAUU. The siRNA were transfected using DharmaFECT 1 transfection reagent (Dharmacon) at a final concentration of 50nM. Cells were analysed after 72 hours after transfection.
Immunoprecipitations and immunoblotting
Cell lysates were extracted using RIPA buffer (150mM NaCl, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS with 50mM Tris pH 8.0) or 1% Triton X-100 in Tris buffered saline (TBS, pH 8.0) supplemented with 2mM ZnCl2, 0.5mM phenylmethylsulphonyl fluoride (PMSF), 10mM N-ethylmaleimide (NEM), 1mM iodoacetamide (IAA) and Complete Protease Inhibitor Cocktail (Roche). Extracts were incubated for 30 minutes at 4°C and after a brief centrifugation (13000rpm for 15 minutes) the supernatant was collected. For cell fractionation experiments the lysates were prepared using the Qproteome Cell Compartment Kit (Qiagen) according to the manufacturers recommendations. Samples were then heated at 70°C for 10minutes, separated by SDS-PAGE and detected by chemiluminescense. All immunoprecipitations were performed at 4°C and were initially pre-cleared on sepharose beads (Sigma). ZZ immunoprecipitations were performed by incubation with IgG-sepharose beads (GE Healthcare). GFP was immunoprecipitated using the mAb to GFP on protein A-sepharose beads (Sigma). The immunoprecipitates underwent four washes with 0.2% Triton X-100 and were heated at 70°C for ten minutes, prior to separation by SDS-PAGE and detection by chemiluminescense.
Mass spectrometry
Large scale pull-downs for mass spectrometry analysis were conducted as for the ZZ immunoprecipitation with some modifications. 108 HEK-293T cells were transfected using 15cm dishes. IgG-sepharose beads were washed with 0.5M acetic acid and 1% Triton-100 in TBS prior to use. Following the IgG pull down the beads were washed with four large volume (15ml) 0.2% Triton TBS washes and loaded onto a column and washed a further three times. A final ammonium acetate wash was used to remove salt and detergent and proteins were eluted from the beads with 0.5M acetic acid. The sample was then snap frozen and lyophilised. Pull downs were performed in a modified RIPA lysis buffer (150mM NaCl, 0.5% NP40, 0.25% sodium deoxycholate. Protein samples were separated by SDS-PAGE, visualised with Coomassie Brilliant Blue and then digested with trypsin to perform mass spectrometry analysis by LC-MS/MS on LTQ-Orbitrap (Thermo) or by MALDI-TOF on the ABI 4700 Proteomics Analyzer (Applied Biosystems). Peptides were identified by Pep-Miner and Sequest software against the human, mouse, rat bovine and Rabbit part of the non-redundant National Center for Biotechnology Information (NCBI) database. A peptide was considered as high quality if its Pep-Miner identification score was greater than 80 and the Sequest Xcore of 1.5 for singly charged peptides, 2.5 for doubly charged peptides and 3 for triply charged peptides.
Metabolic[35S]radiolabelling
Cells were starved in methionine and cysteine free medium for 45 minutes, labelled with [35S]methionine and [35S]cysteine (Promix) for 15 minutes and chased in media containing excess methionine and cysteine for the indicated time periods. Immunoprecipitations were performed as described with one modification. The IgG-sepharose beads were pre-incubated with HEK-293T whole cell lysate to minimise the effects of non-specific protein binding. Samples were separated by SDS-PAGE and detected by autoradiography or by phospho-imager.
Immunofluorescence and confocal microscopy
Cells were plated on coverslips, fixed with 4% paraformaldehyde and permeabilised with 0.2% Triton X-100 in phosphate buffered saline (PBS). Cells were then blocked with 0.2% bovine serum albumin (BSA) in PBS and probed with the desired antibody. The coverslips were washed with 0.2% BSA in PBS prior to mounting on slides. Slides were visualised with the LSM510 META Confocal Microscope (Ziess) and analysed using AxioVision software (Zeiss). Quantitative analysis of the subcellular localisation of MARCH7 was performed by measuring the intensity levels of GFP-MARCH7 and GFP-MARCH7 WI in the nucleus and cytosol using Volocity software (Improvision).
Flow cytometry
Cells were washed with PBS and GFP expression was measured using a FACSCalibur analyser (Becton-Dickinson). Analysis was performed with FloJo software (Tree Star).
Yeast two hybrid analysis
The pGAD-hMARCH7/hMARCH7 WI and pGBDU-hMARCH7/hMARCH7 WI constructs were used for a yeast two hybrid matrix screen against a library of all human E2 conjugating enzymes as described previously (13, 82).
Supplementary Material
Supplementary Figure 1: Identification of USP9X and USP7 by mass spectrometry analysis of MARCH7 pull downs. A) MARCH7 associates with USP9X. ZZ tagged MARCH7 and MARCH7 WI were transfected in HEK-293T cells. After 48 hours the cells were lysed in 1% Triton-X 100 TBS and immunoprecipitated on IgG sepharose beads. The beads were washed and the protein eluted in 0.5M acetic acid. The sample was frozen, lyophilised and then separated by SDS-PAGE. The gel was then analysed by MALDI-TOF mass spectrometry. USP9X (1) was identified in the MARCH7 and MARCH7 WI samples but not the control. MARCH7 (2) is also shown. B) MARCH7 associates with USP7. The pull down for MARCH7 was as indicated for (A) with some modifications. The cells were lysed in a modified RIPA buffer, as described in the materials and methods. The gel was analysed for mass spectrometry by LC-MS/MS. USP7 (1) was identified in the MARCH7 and MARCH7 WI samples but not the control HEK-293T sample. MARCH7 (2) is also shown.
Supplementary Figure 2: MARCH7 subcellular localisation in the 3T3 mouse fibroblast cell line. GFP-MARCH7 or GFP-MARCH7 WI encoding lentivirus was transduced into mouse 3T3 cells, which were then plated onto coverslips after three days. Cells were fixed and counter stained with the DAPI nuclear stain (blue). MARCH7 localisation is shown by GFP (green). Bar=10μm.
Acknowledgments
We thank Roger D Everett (MRC Virology Unit, Glasgow) for the USP7 constructs, Ivan Dikic (Goethe University Medical School) for the ubiquitin construct, Kristina Lindsten (Karolinska Institute) for the USP4 vector, William C Skarnes (Wellcome Trust Sanger Institute) for the MARCH7 gene trapped cell line, and Mike Harbour (Cambridge Insititute for Medical Research) for his contribution to the mass spectrometry analysis. This work was supported by grants from the Wellcome Trust (PJL, JAN), Medical Research Council (CMS, GM), Raymond and Beverly Sacker Foundation (JAN) and the Addenbrookes Trust Fund (SS). PJL holds a Lister Institute Research Prize.
References
- 1.Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–79. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–33. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 4.Swanson R, Locher M, Hochstrasser M. A conserved ubiquitin ligase of the nuclear envelope/endoplasmic reticulum that functions in both ER-associated and Matalpha2 repressor degradation. Genes Dev. 2001;15:2660–74. doi: 10.1101/gad.933301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lehner PJ, Hoer S, Dodd R, Duncan LM. Downregulation of cell surface receptors by the K3 family of viral and cellular ubiquitin E3 ligases. Immunol Rev. 2005;207:112–25. doi: 10.1111/j.0105-2896.2005.00314.x. [DOI] [PubMed] [Google Scholar]
- 6.Boname JM, de Lima BD, Lehner PJ, Stevenson PG. Viral degradation of the MHC class I peptide loading complex. Immunity. 2004;20:305–17. doi: 10.1016/s1074-7613(04)00047-0. [DOI] [PubMed] [Google Scholar]
- 7.Coscoy L, Ganem D. Kaposi’s sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci U S A. 2000;97:8051–6. doi: 10.1073/pnas.140129797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coscoy L, Sanchez DJ, Ganem D. A novel class of herpesvirus-encoded membrane-bound E3 ubiquitin ligases regulates endocytosis of proteins involved in immune recognition. J Cell Biol. 2001;155:1265–73. doi: 10.1083/jcb.200111010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hewitt EW, Duncan L, Mufti D, Baker J, Stevenson PG, Lehner PJ. Ubiquitylation of MHC class I by the K3 viral protein signals internalization and TSG101-dependent degradation. Embo J. 2002;21:2418–29. doi: 10.1093/emboj/21.10.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevenson PG, Efstathiou S, Doherty PC, Lehner PJ. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc Natl Acad Sci U S A. 2000;97:8455–60. doi: 10.1073/pnas.150240097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aravind L, Iyer LM, Koonin EV. Scores of RINGS but no PHDs in ubiquitin signaling. Cell Cycle. 2003;2:123–6. doi: 10.4161/cc.2.2.335. [DOI] [PubMed] [Google Scholar]
- 12.Scheel H, Hofmann K. No evidence for PHD fingers as ubiquitin ligases. Trends Cell Biol. 2003;13:285–7. doi: 10.1016/s0962-8924(03)00102-8. author reply 287-8. [DOI] [PubMed] [Google Scholar]
- 13.Dodd RB, Allen MD, Brown SE, Sanderson CM, Duncan LM, Lehner PJ, Bycroft M, Read RJ. Solution structure of the Kaposi’s sarcoma-associated herpesvirus K3 N-terminal domain reveals a Novel E2-binding C4HC3-type RING domain. J Biol Chem. 2004;279:53840–7. doi: 10.1074/jbc.M409662200. [DOI] [PubMed] [Google Scholar]
- 14.Bartee E, Mansouri M, Hovey Nerenberg BT, Gouveia K, Fruh K. Downregulation of major histocompatibility complex class I by human ubiquitin ligases related to viral immune evasion proteins. J Virol. 2004;78:1109–20. doi: 10.1128/JVI.78.3.1109-1120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohmura-Hoshino M, Goto E, Matsuki Y, Aoki M, Mito M, Uematsu M, Hotta H, Ishido S. A novel family of membrane-bound E3 ubiquitin ligases. J Biochem (Tokyo) 2006;140:147–54. doi: 10.1093/jb/mvj160. [DOI] [PubMed] [Google Scholar]
- 16.Morokuma Y, Nakamura N, Kato A, Notoya M, Yamamoto Y, Sakai Y, Fukuda H, Yamashina S, Hirata Y, Hirose S. MARCH-XI, a novel transmembrane ubiquitin ligase implicated in ubiquitin-dependent protein sorting in developing spermatids. J Biol Chem. 2007;282:24806–15. doi: 10.1074/jbc.M700414200. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura N, Fukuda H, Kato A, Hirose S. MARCH-II is a syntaxin-6-binding protein involved in endosomal trafficking. Mol Biol Cell. 2005;16:1696–710. doi: 10.1091/mbc.E04-03-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda H, Nakamura N, Hirose S. MARCH-III Is a novel component of endosomes with properties similar to those of MARCH-II. J Biochem. 2006;139:137–45. doi: 10.1093/jb/mvj012. [DOI] [PubMed] [Google Scholar]
- 19.Karbowski M, Neutzner A, Youle RJ. The mitochondrial E3 ubiquitin ligase MARCH5 is required for Drp1 dependent mitochondrial division. J Cell Biol. 2007;178:71–84. doi: 10.1083/jcb.200611064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakamura N, Kimura Y, Tokuda M, Honda S, Hirose S. MARCH-V is a novel mitofusin 2- and Drp1-binding protein able to change mitochondrial morphology. EMBO Rep. 2006;7:1019–22. doi: 10.1038/sj.embor.7400790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goto E, Ishido S, Sato Y, Ohgimoto S, Ohgimoto K, Nagano-Fujii M, Hotta H. c-MIR, a human E3 ubiquitin ligase, is a functional homolog of herpesvirus proteins MIR1 and MIR2 and has similar activity. J Biol Chem. 2003;278:14657–68. doi: 10.1074/jbc.M211285200. [DOI] [PubMed] [Google Scholar]
- 22.Hoer S, Smith L, Lehner PJ. MARCH-IX mediates ubiquitination and downregulation of ICAM-1. FEBS Lett. 2007;581:45–51. doi: 10.1016/j.febslet.2006.11.075. [DOI] [PubMed] [Google Scholar]
- 23.Shin JS, Ebersold M, Pypaert M, Delamarre L, Hartley A, Mellman I. Surface expression of MHC class II in dendritic cells is controlled by regulated ubiquitination. Nature. 2006;444:115–8. doi: 10.1038/nature05261. [DOI] [PubMed] [Google Scholar]
- 24.Ohmura-Hoshino M, Matsuki Y, Aoki M, Goto E, Mito M, Uematsu M, Kakiuchi T, Hotta H, Ishido S. Inhibition of MHC class II expression and immune responses by c-MIR. J Immunol. 2006;177:341–54. doi: 10.4049/jimmunol.177.1.341. [DOI] [PubMed] [Google Scholar]
- 25.Matsuki Y, Ohmura-Hoshino M, Goto E, Aoki M, Mito-Yoshida M, Uematsu M, Hasegawa T, Koseki H, Ohara O, Nakayama M, Toyooka K, Matsuoka K, Hotta H, Yamamoto A, Ishido S. Novel regulation of MHC class II function in B cells. Embo J. 2007;26:846–54. doi: 10.1038/sj.emboj.7601556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Gassart A, Camosseto V, Thibodeau J, Ceppi M, Catalan N, Pierre P, Gatti E. MHC class II stabilization at the surface of human dendritic cells is the result of maturation-dependent MARCH I down-regulation. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0708874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- 28.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, Patapoutian A, Hampton GM, Schultz PG, Hogenesch JB. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–70. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baker RK, Haendel MA, Swanson BJ, Shambaugh JC, Micales BK, Lyons GE. In vitro preselection of gene-trapped embryonic stem cell clones for characterizing novel developmentally regulated genes in the mouse. Dev Biol. 1997;185:201–14. doi: 10.1006/dbio.1997.8541. [DOI] [PubMed] [Google Scholar]
- 30.Metcalfe SM. Axotrophin and leukaemia inhibitory factor (LIF) in transplantation tolerance. Philos Trans R Soc Lond B Biol Sci. 2005;360:1687–94. doi: 10.1098/rstb.2005.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Metcalfe SM, Muthukumarana PA, Thompson HL, Haendel MA, Lyons GE. Leukaemia inhibitory factor (LIF) is functionally linked to axotrophin and both LIF and axotrophin are linked to regulatory immune tolerance. FEBS Lett. 2005;579:609–14. doi: 10.1016/j.febslet.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 32.Muthukumarana P, Chae WJ, Maher S, Rosengard BR, Bothwell AL, Metcalfe SM. Regulatory transplantation tolerance and “stemness”: evidence that Foxp3 may play a regulatory role in SOCS-3 gene transcription. Transplantation. 2007;84:S6–11. doi: 10.1097/01.tp.0000269116.06510.db. [DOI] [PubMed] [Google Scholar]
- 33.Muthukumarana PA, Lyons GE, Miura Y, Thompson LH, Watson T, Green CJ, Shurey S, Hess AD, Rosengard BR, Metcalfe SM. Evidence for functional inter-relationships between FOXP3, leukaemia inhibitory factor, and axotrophin/MARCH-7 in transplantation tolerance. Int Immunopharmacol. 2006;6:1993–2001. doi: 10.1016/j.intimp.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 34.Nuber U, Schwarz SE, Scheffner M. The ubiquitin-protein ligase E6-associated protein (E6-AP) serves as its own substrate. Eur J Biochem. 1998;254:643–9. doi: 10.1046/j.1432-1327.1998.2540643.x. [DOI] [PubMed] [Google Scholar]
- 35.Canning M, Boutell C, Parkinson J, Everett RD. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J Biol Chem. 2004;279:38160–8. doi: 10.1074/jbc.M402885200. [DOI] [PubMed] [Google Scholar]
- 36.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., 3rd Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol Cell Biol. 2004;24:7748–57. doi: 10.1128/MCB.24.17.7748-7757.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, Angers A. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 2006;281:38738–47. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- 38.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–86. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, Wong GK, Zheng W, Dehal P, Wang J, Durbin R. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–80. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaw G, Morse S, Ararat M, Graham FL. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. Faseb J. 2002;16:869–71. doi: 10.1096/fj.01-0995fje. [DOI] [PubMed] [Google Scholar]
- 41.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–9. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 42.Fernandez-Montalvan A, Bouwmeester T, Joberty G, Mader R, Mahnke M, Pierrat B, Schlaeppi JM, Worpenberg S, Gerhartz B. Biochemical characterization of USP7 reveals post-translational modification sites and structural requirements for substrate processing and subcellular localization. Febs J. 2007;274:4256–70. doi: 10.1111/j.1742-4658.2007.05952.x. [DOI] [PubMed] [Google Scholar]
- 43.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–73. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 44.Murray RZ, Jolly LA, Wood SA. The FAM deubiquitylating enzyme localizes to multiple points of protein trafficking in epithelia, where it associates with E-cadherin and beta-catenin. Mol Biol Cell. 2004;15:1591–9. doi: 10.1091/mbc.E03-08-0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Clague MJ, Urbe S. Endocytosis: the DUB version. Trends Cell Biol. 2006;16:551–9. doi: 10.1016/j.tcb.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–86. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 47.Pickart CM, Rose IA. Mechanism of ubiquitin carboxyl-terminal hydrolase. Borohydride and hydroxylamine inactivate in the presence of ubiquitin. J Biol Chem. 1986;261:10210–7. [PubMed] [Google Scholar]
- 48.Pickart CM, Rose IA. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J Biol Chem. 1985;260:7903–10. [PubMed] [Google Scholar]
- 49.Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. Embo J. 1997;16:1519–30. doi: 10.1093/emboj/16.7.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Everett RD, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–26. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–53. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y, Fischer-Vize JA. Undifferentiated cells in the developing Drosophila eye influence facet assembly and require the Fat facets ubiquitin-specific protease. Development. 1996;122:3207–16. doi: 10.1242/dev.122.10.3207. [DOI] [PubMed] [Google Scholar]
- 53.Huang Y, Baker RT, Fischer-Vize JA. Control of cell fate by a deubiquitinating enzyme encoded by the fat facets gene. Science. 1995;270:1828–31. doi: 10.1126/science.270.5243.1828. [DOI] [PubMed] [Google Scholar]
- 54.Pantaleon M, Kanai-Azuma M, Mattick JS, Kaibuchi K, Kaye PL, Wood SA. FAM deubiquitylating enzyme is essential for preimplantation mouse embryo development. Mech Dev. 2001;109:151–60. doi: 10.1016/s0925-4773(01)00551-2. [DOI] [PubMed] [Google Scholar]
- 55.Noma T, Kanai Y, Kanai-Azuma M, Ishii M, Fujisawa M, Kurohmaru M, Kawakami H, Wood SA, Hayashi Y. Stage- and sex-dependent expressions of Usp9x, an X-linked mouse ortholog of Drosophila Fat facets, during gonadal development and oogenesis in mice. Gene Expr Patterns. 2002;2:87–91. doi: 10.1016/s0925-4773(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Taya S, Kaibuchi K, Arnold AP. Spatially and temporally specific expression in mouse hippocampus of Usp9x, a ubiquitin-specific protease involved in synaptic development. J Neurosci Res. 2005;80:47–55. doi: 10.1002/jnr.20429. [DOI] [PubMed] [Google Scholar]
- 57.Kanai-Azuma M, Mattick JS, Kaibuchi K, Wood SA. Co-localization of FAM and AF-6, the mammalian homologues of Drosophila faf and canoe, in mouse eye development. Mech Dev. 2000;91:383–6. doi: 10.1016/s0925-4773(99)00294-4. [DOI] [PubMed] [Google Scholar]
- 58.Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K. The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 1999;4:757–67. doi: 10.1046/j.1365-2443.1999.00297.x. [DOI] [PubMed] [Google Scholar]
- 59.Taya S, Yamamoto T, Kano K, Kawano Y, Iwamatsu A, Tsuchiya T, Tanaka K, Kanai-Azuma M, Wood SA, Mattick JS, Kaibuchi K. The Ras target AF-6 is a substrate of the fam deubiquitinating enzyme. J Cell Biol. 1998;142:1053–62. doi: 10.1083/jcb.142.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cheng F, Wang HW, Cuenca A, Huang M, Ghansah T, Brayer J, Kerr WG, Takeda K, Akira S, Schoenberger SP, Yu H, Jove R, Sotomayor EM. A critical role for Stat3 signaling in immune tolerance. Immunity. 2003;19:425–36. doi: 10.1016/s1074-7613(03)00232-2. [DOI] [PubMed] [Google Scholar]
- 61.Cadavid AL, Ginzel A, Fischer JA. The function of the Drosophila fat facets deubiquitinating enzyme in limiting photoreceptor cell number is intimately associated with endocytosis. Development. 2000;127:1727–36. doi: 10.1242/dev.127.8.1727. [DOI] [PubMed] [Google Scholar]
- 62.Lim SK, Shin JM, Kim YS, Baek KH. Identification and characterization of murine mHAUSP encoding a deubiquitinating enzyme that regulates the status of p53 ubiquitination. Int J Oncol. 2004;24:357–64. [PubMed] [Google Scholar]
- 63.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. Embo J. 2007;26:976–86. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gousseva N, Baker RT. Gene structure, alternate splicing, tissue distribution, cellular localization, and developmental expression pattern of mouse deubiquitinating enzyme isoforms Usp2-45 and Usp2-69. Gene Expr. 2003;11:163–79. doi: 10.3727/000000003108749053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cummins JM, Vogelstein B. HAUSP is required for p53 destabilization. Cell Cycle. 2004;3:689–92. [PubMed] [Google Scholar]
- 66.Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, Ma A, Koonin EV, Dixit VM. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–9. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 67.Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424:801–5. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- 68.Hunter T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 2007;28:730–8. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 69.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci U S A. 2001;98:11598–603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J Biol Chem. 2003;278:47753–61. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- 71.Saridakis V, Sheng Y, Sarkari F, Holowaty MN, Shire K, Nguyen T, Zhang RG, Liao J, Lee W, Edwards AM, Arrowsmith CH, Frappier L. Structure of the p53 binding domain of HAUSP/USP7 bound to Epstein-Barr nuclear antigen 1 implications for EBV-mediated immortalization. Mol Cell. 2005;18:25–36. doi: 10.1016/j.molcel.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 72.Sheng Y, Saridakis V, Sarkari F, Duan S, Wu T, Arrowsmith CH, Frappier L. Molecular recognition of p53 and MDM2 by USP7/HAUSP. Nat Struct Mol Biol. 2006;13:285–91. doi: 10.1038/nsmb1067. [DOI] [PubMed] [Google Scholar]
- 73.Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cao Z, Huett A, Kuballa P, Giallourakis C, Xavier RJ. DLG1 is an anchor for the E3 ligase MARCH2 at sites of cell-cell contact. Cell Signal. 2008;20:73–82. doi: 10.1016/j.cellsig.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–8. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- 76.Kopan R, Goate A. A common enzyme connects notch signaling and Alzheimer’s disease. Genes Dev. 2000;14:2799–806. doi: 10.1101/gad.836900. [DOI] [PubMed] [Google Scholar]
- 77.Gupta-Rossi N, Six E, LeBail O, Logeat F, Chastagner P, Olry A, Israel A, Brou C. Monoubiquitination and endocytosis direct gamma-secretase cleavage of activated Notch receptor. J Cell Biol. 2004;166:73–83. doi: 10.1083/jcb.200310098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. following 486. [DOI] [PubMed] [Google Scholar]
- 79.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Multiple monoubiquitination of RTKs is sufficient for their endocytosis and degradation. Nat Cell Biol. 2003;5:461–6. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 81.Demaison C, Parsley K, Brouns G, Scherr M, Battmer K, Kinnon C, Grez M, Thrasher AJ. High-level transduction and gene expression in hematopoietic repopulating cells using a human immunodeficiency [correction of imunodeficiency] virus type 1-based lentiviral vector containing an internal spleen focus forming virus promoter. Hum Gene Ther. 2002;13:803–13. doi: 10.1089/10430340252898984. [DOI] [PubMed] [Google Scholar]
- 82.Estevez AM, Lehner B, Sanderson CM, Ruppert T, Clayton C. The roles of intersubunit interactions in exosome stability. J Biol Chem. 2003;278:34943–51. doi: 10.1074/jbc.M305333200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Identification of USP9X and USP7 by mass spectrometry analysis of MARCH7 pull downs. A) MARCH7 associates with USP9X. ZZ tagged MARCH7 and MARCH7 WI were transfected in HEK-293T cells. After 48 hours the cells were lysed in 1% Triton-X 100 TBS and immunoprecipitated on IgG sepharose beads. The beads were washed and the protein eluted in 0.5M acetic acid. The sample was frozen, lyophilised and then separated by SDS-PAGE. The gel was then analysed by MALDI-TOF mass spectrometry. USP9X (1) was identified in the MARCH7 and MARCH7 WI samples but not the control. MARCH7 (2) is also shown. B) MARCH7 associates with USP7. The pull down for MARCH7 was as indicated for (A) with some modifications. The cells were lysed in a modified RIPA buffer, as described in the materials and methods. The gel was analysed for mass spectrometry by LC-MS/MS. USP7 (1) was identified in the MARCH7 and MARCH7 WI samples but not the control HEK-293T sample. MARCH7 (2) is also shown.
Supplementary Figure 2: MARCH7 subcellular localisation in the 3T3 mouse fibroblast cell line. GFP-MARCH7 or GFP-MARCH7 WI encoding lentivirus was transduced into mouse 3T3 cells, which were then plated onto coverslips after three days. Cells were fixed and counter stained with the DAPI nuclear stain (blue). MARCH7 localisation is shown by GFP (green). Bar=10μm.