Abstract
Catalytic antibody 14D9 catalyzes the enantioselective protonation of prochiral enol ethers with high enantioselectivity (>99% ee) and a practical turnover (kcat = 0.4 s-1), allowing for preparative scale applications. This antibody represents one of the rare examples of catalytic antibodies promoting acid-catalyzed processes. Antibody 14D9 was cloned and expressed as a chimeric Fab fragment in Escherichia coli. Crystal structures of Fab 14D9 as apo form and of its close analog 19C9 in complex with the transition state analog were determined at 2.8-Å resolution. A series of site-directed mutagenesis experiments was carried out to probe the role of individual active-site amino acids. Proton transfer to carbon is catalyzed by a hydrogen bond network formed by the side chains of AspH101 and TyrL36 with a water molecule serving as a relay. The intermediate oxocarbonium ion formed during the protonation step is trapped by the same water molecule, resulting in an overall syn-addition of water to the enol ether's double bond. The enantioselectivity is caused by steric crowding at the active site, mainly because of the side chain of PheH84. The 20-fold lower activity of 19C9 compared with 14D9 was traced down to residue ThrL46, which forms a nonproductive hydrogen bond with the catalytic residue AspH101, which competes with the critical AspH101—TyrL36 hydrogen bond and therefore reduces catalytic efficiency. The catalytic activity of 19C9 was restored to that of 14D9 by using either site-directed mutagenesis (ThrL46Ala) or chain shuffling.
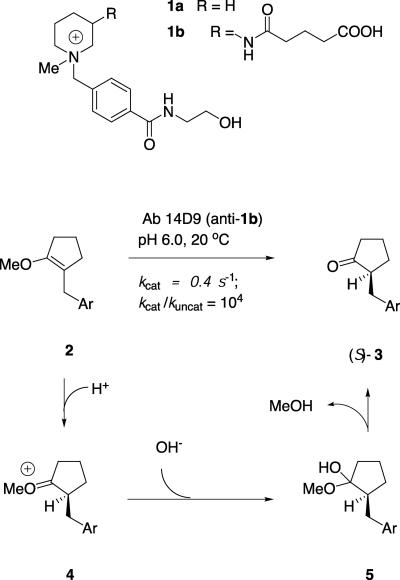
Proton transfer to or from carbon is a fundamental process in enzyme catalysis (e.g., occurring in mandelate racemase and triose phosphate isomerase) (1–3). Carbon deprotonation processes have been studied in a number of catalytic antibodies (4), including β-fluoride and aldol elimination (5–8), intramolecular aldol condensation (9), and the deprotonation of Kemp's benzisoxazole (10). These antibodies generally operate by means of a glutamate residue acting as a general base for proton abstraction (11). By contrast, only very few antibodies have been described to catalyze carbon protonation processes, including enol ester hydrolysis (12), and the protonation of enol ethers by antibody 14D9 (13). This antibody catalyzes the highly enantioselective (>99% ee) protonation of enol ether 2 to give ketone (S)-3 (Fig. 1). The reaction has a practical turnover of kcat = 0.4 s-1 and provided the first examples of an enantioselective gram-scale synthesis with a catalytic antibody (14). This antibody and its close relative 19C9, were obtained from an immunization against the piperidinium hapten 1b (15). Herein we report the x-ray crystal structure determinations of 14D9 in the apo form and 19C9 as a complex with hapten 1a. The antibodies were sequenced, cloned, and expressed as chimeric humanized Fab fragments in Escherichia coli. Structural information combined with site-directed mutagenesis and chain shuffling allow us to formulate the detailed reaction mechanism of carbon protonation and shed light on the origin of the enantioselectivity of these antibodies.
Fig. 1.
Enantioselective protonation reaction catalyzed by antibody 14D9.
Materials and Methods
Affinity Chromatography Resin Preparation. A hapten affinity resin based on DEAE-cellulose was prepared in the following procedure. Ten milliliters of water-saturated DEAE-Sephacel gel (1.1–1.6 mmol amine function) was washed extensively with acetone. A solution of N-hydroxyethyl-4-iodomethyl-benzamide (1.0 g/2.5 mmol) obtained by a standard Finkelstein procedure from the corresponding chloromethyl derivative in 5 ml of acetone was then added, and the reaction mixture was kept in the dark at 20°C for 12 h while being shaken. The unreacted ligand was then washed away with acetone extensively, and the coupled resin was resaturated with water and PBS.
Cloning of Antibody 14D9 and 19C9 Fab Gene Fragments. Total RNA was isolated from the hybridoma cell producing antibody 14D9 or 19C9 with RNeasy kit (Qiagen). The cDNA library of each antibody was constructed with the Omniscript kit (Qiagen). The Fd and κ chain gene fragments of 14D9 or 19C9 were amplified by PCR with degenerated primers as described (16). Amplified PCR products were digested with XhoI–SpeI and SacI–XbaI (for the Fd and κ chain gene fragments, respectively) and ligated into phage display vector pcomb3H to give their corresponding combinatorial libraries (17). The final clones, pcombIIIH-14D9 or pcombIIIH-19C9, were obtained after selection with phage display panning methods against the hapten-BSA conjugate (18). All PCRs were carried out with VentR polymerase (New England Biolabs) to ensure high fidelity.
Construction of Chimeric Fab Fragment. The variable light (VL) and variable heavy (VH) genes of antibodies 14D9 and 19C9 were amplified with their respective vector as template with the following primers: 14D9-VL sense, GCGACTGAGCTCGATGTTTTGTTGACCCAGAC (the SacI restriction site is underlined); VL antisense, GCATCAAAGCTTTGTGCCTCCACCGAACGT (HindIII); VH sense, GCACCACTCGAGCAGTCTGGACCTGAACTG (XhoI); 14D9-VH antisense, GCA ACTGGTGACCGTGGTGCCT TGGCC (BstEII); 19C9-VL sense, CCAGATGTGAGCTCGTCATGACCCAGTCTCCA (SacI); and 19C9-VH antisense, GGAGACGGTGACCGTGGTGCCTGGGCCCCAG (BstEII). The amplified fragments were cloned into appropriate sites in the vector p4xH-M13 (19) to give a plasmid in which the VL and VH segments of the antibody 14D9 or 19C9 are fused to human Cκ and Cγ CH1 regions. pBAD-14D9 and pBAD-19C9 were constructed by subcloning the chimeric Fab gene fragment into the pBAD vector (20) by using restriction sites SacI and SalI. Vectors for the recombinant Fab mutant expression is constructed by further PCR amplification with VL sense and CH1 antisense, CACCGCCGGTCGACTCAGTGGTGGTGGTGGTGGTGTGTGTGAGTTTTGTCAC (SalI), and subcloning of the chimeric Fab fragment with the SacI and SalI restriction sites. A hexa-His tag was added at the C terminus of the chimeric Fd. The resulting expression plasmids pBAD-14D9his and pBAD-19C9his allow expression of hexa-His-tagged chimeric Fab 14D9 or 19C9.
Production and Purification of Chimeric Fab Fragments for Crystallization. The TOP10 E. coli strain (Invitrogen) was transformed with pBAD-14D9 or pBAD-19C9. Preculture (1/40 volume) was inoculated into LB medium containing 100 μg/ml ampicillin at 20°C, and overexpression was performed after induction with 0.1% l-(+)-arabinose at OD600 of 1.5. One-tenth culture volume of Terrific Broth was added to implement nutrition for overexpression. After induction for 14 h, the cells were harvested and sonicated in 20 mM Tris·HCl, pH 7.8/500 mM NaCl, and the supernatant of lysis was loaded into the hapten affinity resin. The chimeric Fabs 14D9 and 19C9 were eluted with 0.2 M Gly-HCl (pH 2.8) and neutralized with 1 M Tris·HCl (pH 9.0) Concentrations of purified antibody were determined by measuring A280 and A260 (21).
Site-Directed Mutagenesis and Purification of Mutants. Site-directed mutagenesis was accomplished by a QuickChange site-directed mutagenesis protocol (Stratagene) with minor modifications and including silent restriction sites. pBAD-14D9his or pBAD-19C9his are used as template. All PCRs were performed with the High Fidelity System (Roche, Gipf-Oberfrick, Switzerland). Mutant codons were generally chosen to exploit the most frequently used codon for a particular amino acid among highly expressed genes in E. coli. Oligonucleotides were custom-synthesized and purified by Microsynth (Balgach, Switzerland) as follows.
For all primers, mutagenized positions are denoted in lower-case. The silent restriction sites are in italics. DH101A forward, GTA ACT TCT T TGcgTACTGGGGCCA AGGtACCACTCTCAC (KpnI); DH101A reverse, GTGAGAGTGGtACCTTGGCCCCAGTAcgCAAAGAAGTTAC (KpnI); DH101N forward, GT-AACTTCTTTGaaTACTGGGGCCAAGGtACCACTCTCAC (KpnI); DH101N reverse, GTGAGAGTGGtACC-TTGGCCCCAGTAttCAAAGAAGTTAC (KpnI); DH101E forward, G-TA ACT TCT T TGagTACTGGGGCCA AGGtACCACTCTCAC (KpnI); DH101E reverse, GTGAGAGTGGtACCTTGGCCCCAGTActCAAAGAAGTTAC (KpnI); DH101H forward, G-TAACTTCTTTcACTACTGGGGCCAAGGtACCACTCTCAC (KpnI); DH101H reverse, GTGAGAGTGGtACCT TGGCCCCAGTAGTgA A AGA AGT TAC (KpnI); FH98A forward, GTGCCATCgcaTATGGTAACTTCTTTGAC (NdeI); FH98A reverse, GTTACCATAtgcGATGGCACAGTAATAGACTGC (NdeI); FH98D forward, CTGTGCgATCgacTATGGTAACTTCTTTGACTAC (PvuI); FH98D reverse, GTTACCATAgtcGATcGCACAGTAATAGACTGCAGAG (PvuI); YH35F forward, GACTTCACaATATtCTGGGTGAAACAGAGCCATGG (SspI); YH35F re-verse, CACCCAGaATATtGTGAAGTCAGTGATTGAGTAAGC (SspI); LA46T forward, CCTAAAaccCTGATTTACTCGACtagtTACCGGTACAGTG (SpeI); LA46T reverse, CGAGTA A ATCAGggtT T TAGGAGGT TGCCCTGG; LT46A forward, CTCCAAAAgCttTGATTTACTCGGCATCCTAC (HindIII); LT46A reverse, GTA A ATCAaaGcT TT TGGAGATTGCCCTGG (HindIII); QL89D forward, CCGAATaTTTCTGTGACCAATATAACATCTATCCTGTG (SspI); QL89D reverse, GTTATATTGGTCACAGAAAtATTCGGCCAAGTCTTCAGAC (SspI).
All mutants were expressed by using the same procedure as before. After sonication in lysis buffer (20 mM Tris·HCl, pH 8.0/10 mM imidazole/300 mM NaCl), the supernatant was applied to a metal-chelate affinity column (Ni2+-NTA; Qiagen) and washed with lysis buffer containing 40 mM imidazole. Fab mutants were eluted with a buffer containing 20 mM Tris, pH 8.0, 300 mM NaCl, and 250 mM imidazole and were dialyzed afterward against PBS (pH 7.4).
Catalytic Activity Assay. Catalytic reactions were carried out by mixing 0.6 μl of a 10 mM stock solution of substrate 2 in a 1:1 acetonitrile/water solution (Fig. 1) with 19.2 μl of protein solutions at a concentration of 0.5 mg/ml in PBS (pH 7.4) at 22°C (300 μM final concentration). An identically prepared solution with 0.2 μl of 5 mM inhibitor 1a (50 μM final concentration) was used to check specific inhibition. All reactions were monitored by HPLC. The substrate and product were separated on an analytical RP-C18 column (218TP54, 22 cm × 0.45 cm, 300-Å pore size; Vydac, Hesperia, CA) after a 30-min incubation. Retention times (tR) were as follows (eluent, 1.5 ml/min; H2O/acetonitrile = 75:25; detection by UV at 230 nm): tR (ketone 3) = 3.3 min, tR (substrate 2) = 15.7 min. The percentage conversion indicating the catalytic activity was calculated from peak integration. All percentages shown in Table 2 refer to the hexa-His-tagged chimeric Fab 14D9 wild type.
Table 2. Catalytic activity and enantioselectivity of 14D9 and 19C9 wild type and mutants.
| Antibody | Product conversion, %* | Enantioselectivity, %† |
|---|---|---|
| MAb 14D9 | 92 | 97 |
| Fab 14D9-His | 100 | 98 |
| Fab 19C9-His | 4 | 92 |
| 14D9L-19C9H | 86 | 95 |
| 19C9L-14D9H | 3 | 91 |
| AspH101Ala | 0 | — |
| AspH101Asn | 0 | — |
| AspH101Glu | 60 | 95 |
| AspH101His | 25 | 93 |
| TyrL36Phe | 0 | — |
| TyrH35Phe | 30 | 94 |
| GlnL89Glu | 0 | — |
| GlnL89Asp | 0 | — |
| PheH98Ala | 10 | 89 |
| PheH98Asp | 0 | 0 |
| AlaL46Thr | 4 | 90 |
| 19C9-ThrL46Ala | 80 | 95 |
The activity was determined by hydrolysis of enol ether 3 and monitored with RP-HPLC
The purified product of ketone 5 was analyzed by employing an achiral HPLC system, and the enantioselectivity was calculated from the peak areas of each enantiomer of ketone 5
Enantioselectivity Assay. Substrate 2 was incubated with the antibody samples as described above on a 200-μl scale. The reaction mixture was separated by RP-C18-HPLC, and the peak of ketone 3 was collected, lyophilized, redissolved in 1:1 hexane/isopropanol solution, and analyzed on a 22 cm × 0.45 cm chiral-phase column (Chiralpak OD; Daicel Chemical Industries, Osaka) with a 7:3 hexane/isopropanol eluent at 1 ml/min {tR[(R)- ketone 3] = 8.6 min, tR[(S)-ketone 3] = 12.4 min} (14). The absolute configuration of the products has been established earlier by chemical correlation (22).
Crystallization. All crystallization experiments were performed by using the vapor diffusion method at 20°C. The crystals of the chimeric Fab fragment 14D9 were grown by using the sitting-drop procedure with reservoirs containing 80 μl of 8% polyethylene glycol 3350/50 mM citric acid, pH 3.0. Drops were set up by mixing 2 μl of a protein solution (20 mg/ml Fab in 5 mM NaCl) with 1 μl of the reservoir solution. The purified chimeric Fab 19C9 could be cocrystallized with its hapten 1a by using the hanging-drop procedure and wells containing 500 μl of 45% (vol/vol) 2-methyl-2,4-pentanediol, 0.2 M mono ammonium dihydrogen phosphate, and 0.1 M Tris·HCl (final pH 8.5). Three microliters of a protein solution (16 mg/ml Fab in 5 mM NaCl/0.6 mM hapten 1a) was mixed with 2 μl of the well solution. An additional 0.5 μl of 1 M NaI was added to the drop only to modify the crystal growth.
X-Ray Data Collection and Structure Determination. Crystals of chimeric Fab 14D9 were flash-cooled to 110 K by using 30% PEG 400 as cryoprotectant. For the 19C9–hapten 1a (19C9–1a) complex, the crystallization buffer was used as cryoprotectant directly. Data were processed with mosflm 6.0 and scala of the ccp4 program suite (23). The structure of the Fab 14D9 was solved by molecular replacement with the program cns 1.05; the search models used were the Fv (variable region Fab fragment) domain of germline Fab AZ-28 (Protein Data Bank ID 1D5I) and its constant domain (24). The atomic model was refined by alternating cycles of model reconstruction with the program o (25) and refinement with cns and refmac (26) (Table 1). The structure of the 19C9 complex was solved by using the program molrep with the 14D9 apo model as search probe.
Table 1. Data collection and refinement statistics.
| Fab 14D9 | Fab 19C9-1a | |
|---|---|---|
| Data collection | ||
| Space group | C2 | P21 |
| Unit cell | a = 110.04 Å, b = 142.70 Å, c = 125.42 Å, β = 104.87° three molecules per a.s.u. | a = 68.47 Å, b = 97.92 Å, c = 70.88 Å, β = 99.28° two molecules per a.s.u. |
| Resolution range, Å* | 20–2.67 (2.83–2.67) | 20–2.79 (2.96–2.79) |
| Observation | 157,522 | 85,877 |
| Unique reflections | 49,857 | 22,991 |
| Completeness, %* | 93.8 (77.6) | 99.4 (97.2) |
| Mean I/σ(I)* | 22.4 (5.4) | 9.0 (3.6) |
| Rsym* | 0.035 (0.172) | 0.137 (0.375) |
| Refinement statistics | ||
| No. of atoms/water molecules | 9,703/25 | 6,500/92 |
| Rcryst, % | 21.6 | 19.7 |
| Rfree, % | 27.0 | 28.0 |
| rms bond length, Å | 0.006 | 0.008 |
Rsym = Σhkl Σi |(hkl;i) – 〈I(hkl)〉|/Σhkl Σi I(hkl;i). Rcryst; Rfree = Σhkl | Fobs(hkl) – Fcalc(hkl)|/Σhkl Fobs(hkl) whereby the summation runs over the working set and test set reflections, respectively. a.s.u., asymmetric unit.
Values in parentheses are for the outmost resolution shell
Results and Discussion
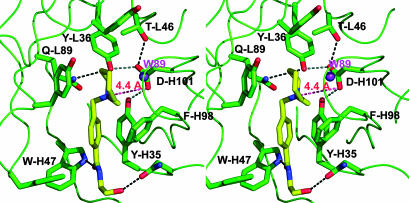
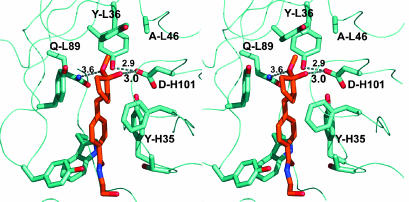
Ligand Binding of 19C9 and 14D9. In the structure of the complex Fab 19C9–1a, the ligand is clearly visible in the 2Fo-Fc electron density map, allowing unequivocal identification of the catalytic pocket as the cleft formed by TyrL36, ThrL47, TyrL49, TyrL55, TyrL91, GlnL89, ValL96, PheL98, TyrH35, PheH98, AsnH100A and AspH101. This pocket is open and exposes half of the hapten 1a to the solvent (Fig. 2). The ethanolamide side chain of hapten 1a is immobilized by two hydrogen bonds, one between the terminal OH group and the main-chain nitrogen of AspH52 and another between the carboxamide oxygen atom and the side-chain N atom of TrpH47. The hapten's aromatic group interacts with PheH98. Ligand recognition also involves multiple van der Waals contacts and a weak electrostatic interaction between the hapten's ammonium group and AspH101, which will be shown to be the critical catalytic carboxyl group (see below).
Fig. 2.
Stereoview of the binding pocket of antibody 19C9 with bound hapten. Hydrogen-bonding distances are denoted by black dashed lines. The red line indicates the distance (4.4 Å) between the catalytic AspH101 and the quaternary ammonium group of the hapten. Water W89 is shown as magenta spheres.
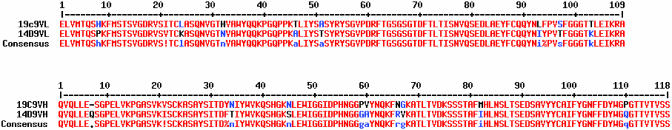
The structure of Fab 14D9 apo form is almost superimposable with that of Fab 19C9, which is not surprising given their high degree of sequence identity (light chain, 92.7%; heavy chain, 92.4%) (Fig. 3). A notable difference is found at position L46, where an Ala in 14D9 replaces the Thr present in 19C9. The variability of this residue influences the hydrogen-bonding interactions of the critical AspH101. In antibody 19C9, the AspH101 carboxylate interacts with the hydroxyl groups of ThrL46 and TyrH35. In 14D9, by contrast, the hydrogen-bonding OH group of ThrL46 is absent, and TyrL36 replaces ThrL46 as the second hydrogen-bonding partner of the AspH101 carboxylate.
Fig. 3.
Fv domain gene sequence alignment of Ab 14D9 and 19C9. The identical residues are shown in red; different residues are shown in blue or black.
Proton Transfer Mechanism. The recombinant humanized Fab 14D9 and Fab 19C9 catalyze the enantioselective protonation of enol ether 2 to form ketone (S)-3 with high enantioselectivity (>95% ee), with antibody 19C9 being ≈20-fold less active than 14D9, in agreement with the catalytic activities of the parent hybridoma-derived immunoglobulins (27). A possible role of AspH101 in catalysis is suggested by the spatial proximity of this group to the hapten's ammonium cation. This hypothesis was confirmed by mutating each of the three acidic residues found within the complementarity-determining region regions to Ala or Asn. Ala replacements at position H31 and H52 resulted in a 10% and 30% reduction in catalytic activity. By contrast, there was a complete loss of catalysis in the AspH101Ala and AspH101Asn mutants, clearly identifying AspH101 as the key catalytic residue.
The chemical reaction mechanism for the hydrolysis of enol ether 2 is well established (28). The rate-limiting step is the initial general-acid-catalyzed proton transfer from a catalytic acid to the β-carbon of the enol ether to form intermediate 4, which is trapped by water to give hemiacetal 5. The hemiacetal is unstable and collapses to the carbonyl reaction product and methanol. Kinetic studies with antibody 14D9 suggest that the antibody-catalyzed reaction follows the same mechanism, with the stereogenerating protonation step being catalyzed by an ionizable residue of pKa = 4.5.
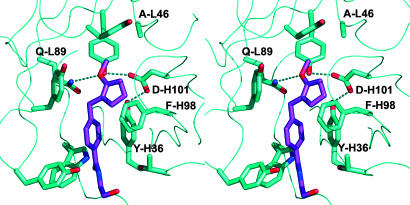
To gain a structural insight into the reaction mechanism, we placed enol ether 2 in the antibody-binding pocket by overlaying its aromatic benzamide side chain with that of hapten 1a as observed in the 19C9–1a complex (Fig. 4). This arrangement is supported by earlier structure–activity relationship data measured with 11 different reactive substrates and 23 inhibitory hapten analogs (27, 28). Despite steric crowding, especially between the methoxycyclopentene group and PheH98, the substrate could be arranged with its reactive double bond pointing toward the catalytic residue AspH101. The position of the aromatic ring of PheH98 is ill defined in two of the three crystallographically independent molecules, indicating a high degree of flexibility. Hence, it is reasonable to assume that it could adopt the conformation observed in the 19C9 complex, avoiding steric clashes.
Fig. 4.
Stereoview of the 14D9 binding pocket with docked substrate. No energy minimization was performed.
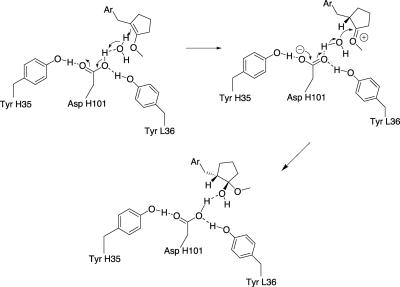
The distance between the enol ether's β-carbon and the carboxyl group is too long (≈4.4 Å) to be compatible with direct proton transfer from that group and suggests that a water molecule transiently hydrogen-bonded to AspH101 might serve as a relay. This water molecule would also be optimally placed to trap the intermediate carbocation 4 to form hemiacetal 5, resulting in an overall syn-addition of water to the enol ether double bond (Fig. 5). Indeed, water molecule W89, which was found in the 19C9 complex structure and is hydrogen-bonded to the phenol group of TyrL55 and the main-chain carbonyl oxygens of residues H100 and H100C, is located in a position that is favorable for the attack of the protonated carbocation. On the other hand, its position is not particularly suitable to transfer a proton to the β-carbon of the reactive double bond. In a docking study, hemiacetal 5 is readily placed in the binding pocket with its hydroxyl group interacting in hydrogen-bonding fashion with AspH101 and being close to water W89 (Fig. 6). The critical role of TyrL36 was evidenced by the fact that the TyrL36Phe mutant was catalytically inactive, whereas the structurally similar TyrH35Phe mutant retained 30% of catalytic power and a high enantioselectivity as the wild type. Remarkably, the AspH101Glu and AspH101His mutants retained good catalytic activity with unaltered enantioselectivity. In these mutants, a direct proton transfer from the catalytic group at position H101 probably takes place with subsequent addition of a water molecule from the solvent, resulting in an overall trans-addition of water.
Fig. 5.
Proposed mechanism of protonation reaction catalyzed by antibody 14D9. The rate-limiting step is a proton transfer from AspH101 to Re-face of enol ether by means of a water molecule. This water molecule then attacks the oxocarbonium intermediate 4 to form hemiketal 5 as the primary reaction product.
Fig. 6.
Stereoview of the 14D9 binding pocket with docked hemiketal 5.
Origin of the Enantioselectivity. The enantioselectivity of the protonation reaction is explained by the fact that enol ether 2 can only be placed within the binding pocket with the Si-face of enol ether pointing toward the catalytic residues. In this conformation, the methoxy group is in hydrogen-bonding distance to the side-chain of GlnL89, which is very important for catalysis as shown by mutagenesis (Table 2). The conformation exposing the Re-face is not accessible because of a steric clash between the methoxy group and PheH98. Furthermore, to expose the Re-face to AspH101, a stereochemically unfavorable conformation has to be occupied, where the methoxy group lies on top of the aromatic ring. Similarly, only the (S)-configured oxocarbonium intermediate 4 can be properly positioned within the antibody-binding pocket while the enantiomeric (R)-configured intermediate again clashes its methoxy group against PheH98.
Two mutagenesis experiments were carried out in an attempt to alter the enantioselectivity of antibody 14D9. First, the PheH98Ala mutant was prepared to release the steric clash observed by docking. However, this mutant lost 90% activity relative to the wild type but still retained the same (S)-enantioselectivity. A second attempt was directed toward residue GlnL89, which is positioned on the opposite side of the binding pocket relative to AspH101 and is similarly hydrogen-bonded to TyrL36. Considering that the hydrogen-bonded pair AspH101—TyrL36 was essential for activity, the double mutant AspH101Asn GlnL89Glu was prepared as a possible catalyst with inverted enantioselectivity. This mutant showed no catalytic activity, although the distance of 4.5 Å from the ammonium center of the hapten to the carboxamide group of GlnL89 is similar to that to the carboxylate of AspH101. The single mutant GlnL89Glu also showed no activity.
Remote Control of Activity by Hydrogen Bonding. The differential hydrogen-bonding pattern of AspH101 in 19C9 and 14D9 pointed to the possibility that residue L46 alone might explain the 20-fold activity difference between the two antibodies. This was indeed confirmed by mutagenesis experiments. Thus, the AlaL46Thr mutant of 14D9 showed a reduced activity close to that of antibody 19C9. Conversely, the ThrL46Ala mutant of 19C9 showed a catalytic activity comparable to that of antibody 14D9. The observation was further confirmed by chain shuffling (29, 30) between 14D9 and 19C9. The Fab 14D9L-19C9H retained 80% activity compared with Fab 14D9, and 19C9L-14D9H showed a similar activity to Fab 19C9. These clearly showed that the 19C9 heavy-chain was catalytically competent when combined with the proper 14D9-light-chain bearing the nonhydrogen-bonding Ala residue at position L46.
Conclusion
The structural and mutagenesis experiments above demonstrate the molecular mechanism of the 14D9-catalyzed enantioselective protonation reaction. The role played by the hydrogen-bonding network surrounding the catalytic residue AspH101, in particular the control of activity by the “noncatalytic” residues TyrL36 and ThrL46, is remarkable. These observations disprove the notion that catalytic antibodies are mere binding pockets equipped with a single catalytic residue, and they illustrate the necessity for subtle interactions to obtain catalytic activity. It must be pointed out that most of the interactions leading to catalysis were not designed. Indeed the immunogenic hapten 1 merely ensured the selection of antibodies capable of binding the substrates at the aromatic benzamide group and possessing a binding pocket for the reactive functional group favoring the formation of a positive charge. Whether the occurrence of catalysis was purely serendipitous or whether it was made possible by a natural tendency of antibodies to acquire catalytic activity remains an open question. Additional modeling experiments should rationalize (on the basis of the present crystal structure) other hydrolytic reactions that are related to the hydrolysis of enol ether 2 and are also catalyzed, albeit more weakly, by antibody 14D9 (31–35).
Acknowledgments
We thank Dr. Carlos Barbas (The Scripps Research Institute, La Jolla, CA) for the kind gift of pComb3H-myc vector, Martine T. Reymond for preparing the hybridoma cells of 14D9 and 19C9 and for assistance in antibody purification, and the staff of beamline ID14-4 at the European Synchrotron Radiation Facility (Grenoble, France) for help with data collection on the 14D9 Fab. This work was supported by the Swiss National Science Foundation.
Abbreviations: VL, variable light; VH, variable heavy.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PBD ID codes 1UWE and 1UWG for 14D9 and 19C9, respectively).
References
- 1.Kemp, D. S. (1995) Nature 373, 196-197. [DOI] [PubMed] [Google Scholar]
- 2.Mitra, B., Kallarakal, A. T., Kozarich, J. W., Gerlt, J. A., Clifton, J. G., Petsko, G. A. & Kenyon, G. L. (1995) Biochemistry 34, 2777-2787. [DOI] [PubMed] [Google Scholar]
- 3.Davenport, R. C., Bash, P. A., Seaton, B. A., Karplus, M., Petsko, G. A. & Ringe, D. (1991) Biochemistry 30, 5821-5826. [DOI] [PubMed] [Google Scholar]
- 4.Stevenson, J. D. & Thomas, N. R. (2000) Nat. Prod. Rep. 17, 535-537. [DOI] [PubMed] [Google Scholar]
- 5.Shokat, K. M., Leumann, C. J., Sugasawara, R. & Schultz, P. G. (1989) Nature 338, 269-271. [DOI] [PubMed] [Google Scholar]
- 6.Uno, T. & Schultz, P. G. (1992) J. Am. Chem. Soc. 114, 6573-6574. [Google Scholar]
- 7.Shokat, K., Uno, T. & Schultz, P. G. (1994) J. Am. Chem. Soc. 116, 2261-2262. [Google Scholar]
- 8.Cravatt, B. F., Ashley, J. A., Janda, K. D., Boger, D. L. & Lerner R. A. (1994) J. Am. Chem. Soc. 116, 6013-6014. [Google Scholar]
- 9.Koch, T., Reymond, J.-L. & Lerner, R. A. (1995) J. Am. Chem. Soc. 117, 9383-9387. [Google Scholar]
- 10.Thorn, S. N., Daniels, R. G. & Auditor, M.-T. M. Hilvert, D. (1995) Nature 373, 228-230. [DOI] [PubMed] [Google Scholar]
- 11.Gigant, B., Charbonnier, J. B., Eshhar, Z., Green, B. S. & Knossow, M. (1997) Proc. Natl. Acad. Sci. USA 94, 7857-7861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujii, I., Lerner, R. A. & Janda, K. D. (1991) J. Am. Chem. Soc. 113, 9528-8529. [Google Scholar]
- 13.Reymond, J.-L., Janda, K. D. & Lerner, R. A., (1992) J. Am. Chem. Soc. 114, 2257-2258. [Google Scholar]
- 14.Reymond, J.-L., Reber, R. A. & Lerner, R. A. (1994) Angew. Chem. Int. Ed. 33, 475-478. [Google Scholar]
- 15.Reymond, J.-L., Janda, K. D. & Lerner, R. A. (1991) Angew. Chem. Int. Ed. 30, 1711-1714. [Google Scholar]
- 16.Huse, W. D., Sastry, L., Iverson, S. A., Kang, A. S., Alting-Mees, M., Burton, D. R., Benkovic, S. J. & Lerner, R. A. (1989) Science 246, 1250-1251. [DOI] [PubMed] [Google Scholar]
- 17.Barbas, C. F., Kang, A. S., Lerner, R. A. & Benkovic, S. J. (1991) Proc. Natl. Acad. Sci. USA 88, 7978-7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang, A. S., Barbas, C. F., Janda, K. D., Benkovic, S. J. & Lerner, R. A. (1991) Proc. Natl. Acad. Sci. USA 88, 4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulrich, H. D., Patten, P. A., Yang, P. L., Romesberg, F. E. & Schultz, P. G. (1995) Proc. Natl. Acad. Sci. USA 92, 11907-11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin, J., Mundorff, E. C., Yang, P. L., Wendt, K. U., Hanway, D., Stevens, R. C. & Schultz, P. G. (2001) Biochemistry 40, 10764-10773. [DOI] [PubMed] [Google Scholar]
- 21.Harlow, E. & Lane, D. (1988) Antibodies: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY)
- 22.Sinha, S. C. & Keinan, E. (1995) J. Am. Chem. Soc. 117, 3653-3654. [Google Scholar]
- 23.Collaborative Computational Project Number 4. (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 24.Mundorff, E. C., Hanson, M. A., Varvak, A., Ulrich, H., Schultz, P. G. & Stevens, R. C. (2000) Biochemistry 39, 627-632. [DOI] [PubMed] [Google Scholar]
- 25.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 26.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240-255. [DOI] [PubMed] [Google Scholar]
- 27.Jahangiri, G. K., Reymond, J.-L. & Lerner, R. A. (1994) J. Am. Chem. Soc. 116, 11264-11274. [Google Scholar]
- 28.Reymond, J.-L., Jahanghiri, G. K., Stoudt, C. & Lerner, R. A. (1993) J. Am. Chem. Soc. 115, 3909-3917. [Google Scholar]
- 29.Kang A. S., Jones T. M. & Burton D. R. (1991) Proc. Natl. Acad. Sci. USA 88, 11120-11123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, G. P., Posner, B. A. & Benkovic, S. J. (1997) Bioorg. Med. Chem. 5, 581-590. [DOI] [PubMed] [Google Scholar]
- 31.Sinha, S. C., Keinan, E. & Reymond, J.-L., (1993) J. Am. Chem. Soc. 115, 4893-4894. [Google Scholar]
- 32.Shabat D., Itzhaky, H., Reymond, J.-L. & Keinan, E. (1995) Nature 374, 143-145. [DOI] [PubMed] [Google Scholar]
- 33.Sinha, S. C., Keinan, E. & Reymond, J.-L. (1993) Proc. Natl. Acad. Sci. USA 90, 11910-11913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shabat, D., Sinha, S. C., Reymond, J.-L. & Keinan, E. (1996) Angew. Chem. Int. Ed. 35, 2628-2631. [Google Scholar]
- 35.Shabat, D., Shulman, H., Itzhaky, H., Reymond, J.-L. & Keinan, E. (1998) Chem. Commun. 1759-1760.