Abstract
Although significant effort is expended on identifying transcripts/proteins that are up-regulated in cancer, there are few reports on systematic elucidation of transcriptional mechanisms underlying such druggable cancer-specific targets. The mesothelin (MSLN) gene offers a promising subject, being expressed in a restricted pattern normally, yet highly overexpressed in almost one-third of human malignancies and a target of cancer immunotherapeutic trials. CanScript, a cis promoter element, appears to control MSLN cancer-specific expression; its related genomic sequences may up-regulate other cancer markers. CanScript is a 20-nt bipartite element consisting of an SP1-like motif and a consensus MCAT sequence. The latter recruits TEAD (TEA domain) family members, which are universally expressed. Exploration of the active CanScript element, especially the proteins binding to the SP1-like motif, thus could reveal cancer-specific features having diagnostic or therapeutic interest. The effcient identification of sequence-specific DNA-binding proteins at a given locus, however, has lagged in biomarker explorations. We used two orthogonal proteomics approaches— unbiased SILAC (stable isotope labeling by amino acids in cell culture)/DNA affnity-capture/mass spectrometry survey (SD-MS) and a large transcription factor protein microarray (TFM)—and functional validation to explore systematically the CanScript interactome. SD-MS produced nine candidates, and TFM, 18. The screens agreed in confirming binding by TEAD proteins and by newly identified NAB1 and NFATc. Among other identified candidates, we found functional roles for ZNF24, NAB1 and RFX1 in MSLN expression by cancer cells. Combined interactome screens yield an effcient, reproducible, sensitive, and unbiased approach to identify sequence-specific DNA-binding proteins and other participants in disease-specific DNA elements.
Keywords: mesothelin, SILAC, protein microarray, ZNF24, NAB1
INTRODUCTION
Cancer-specific proteins serve as starting points for studies of carcinogenesis and for developing diagnostics and therapeutics. Exploring their transcriptional regulation could unveil cancer-specific processes. There have been, however, relatively few successes in such transcriptional studies; for example, the most recent report exploring the detailed mechanism of transcriptional regulating a familiar cancer marker, CEA (CEACAM5), was published in 1994.1 Technical challenges, such as a lack of comprehensive unbiased screening methods, previously hindered the identification of locus-specific DNA-binding proteins. Here, we revisited this challenge using several technologies to determine the interactome of the MSLN cancer-specific element (the CanScript sequence) and then assayed the in vivo relevance of some of these interactions.
Historically, protein libraries were used to screen for unknown proteins binding to known DNA sequences. For example, phage display of cDNA libraries successfully uncovered several zinc finger proteins2,3 (including Can-Script-regulating candidate KLF6), by using specific transcriptionally active DNA elements as probes to generate qualitative results. The yeast one-hybrid screening of cDNA libraries was the in vivo analog of such screens and provided a quantitative output. Limitations have included studying libraries of inadequate complexity, poor modeling of native protein structure, omission of relevant binding partners, and blindness to the indirect interactions as would occur in assembled protein complexes.
Protein microarrays and yeast screens of custom polypeptide libraries are practical for screening DNA-binding proteins, carry similar limitations, but offer greater reproducibility (~80% hits reproducible between two experiments)4 and investigator control than natural libraries. DNA–protein interaction databases were established from their results,4,5 in which thousands of purified and arrayed proteins directly bound in vitro to applied DNA probes of interest.
Bioinformatics tools offer predictions of DNA-binding proteins.6,7 Their computational power rests on databases generated from hundreds of known DNA–protein interactions. The predicted results are limited by database size and accuracy, sampling biases, disparate informational sources, omission of cooperative binding patterns, etc. We used such tools in prior explorations.8
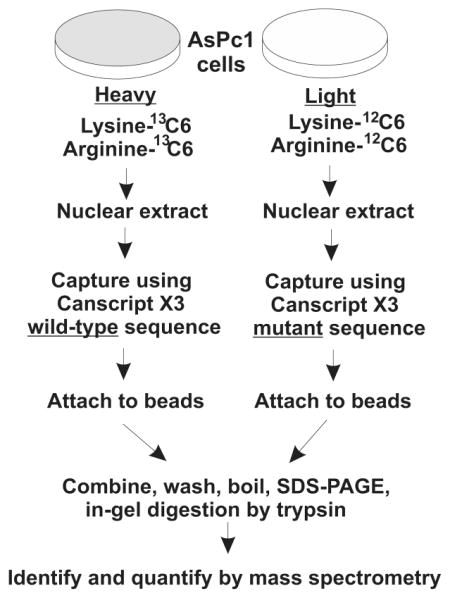
Tandem mass spectrometry allows identification and quantitation of native protein interactomes affnity-captured by defined DNA sequences directly from cell lysates.9,10 One of the major limitations of this method is that specific binding proteins are usually obscured by a background of nonspecific protein binding, even when incorporating stringent controls. To overcome this limitation, protein samples can be isotope labeled to distinguish the binding proteins in control and test samples to discern sequence-specific DNA-binding proteins. This is because mass spectrometry allows one to compare the intensities of the different isotopic forms in the MS spectrrum.9–12 We used the SILAC method for labeling proteins that were then subjected to affnity capture by target or control biotinylated oligonucleotides, mixed and analyzed together (Figure 1A). The protein samples were digested with trypsin, and each resulting peptide generates a quantitative ratio (heavy/light) of the paired peptides, which greatly increases the chance to identify specific binding proteins.10,11
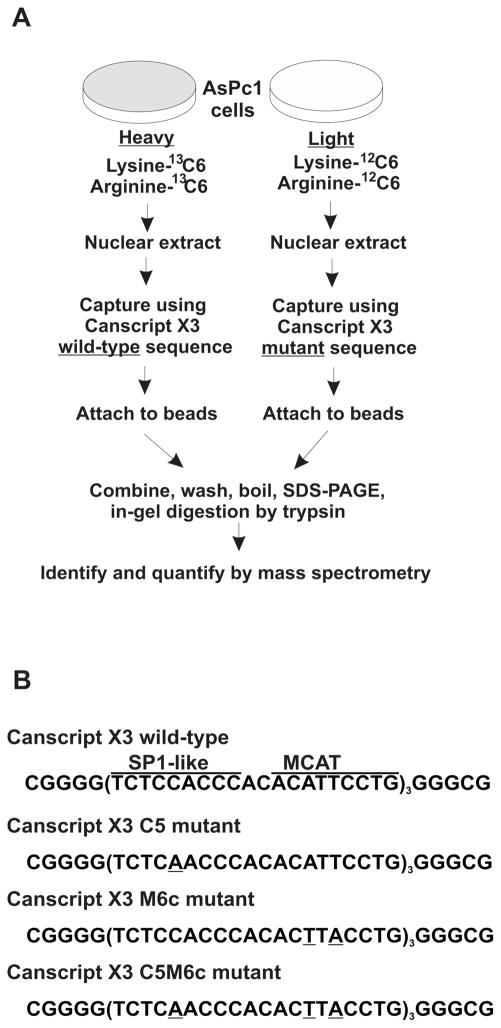
Figure 1.
Strategy: combining SILAC with DNA affnity capture mass spectrometry (SD-MS). (A) Schematic of SD-MS screen for wild-type CanScript-specific binding proteins; ~108 AsPc1 cells were cultured in parallel with either normal growth medium or lysine, arginine-depleted medium but supplemented with 13C6 labeled lysine and arginine. Crude nuclear extracts were prepared from strictly equal weights (~0.5 g) of labeled or unlabeled cell pellets. Nuclear extracts were incubated with wild-type and mutant 5′ biotinylated CanScript ×3 probes, respectively. DNA–protein complexes were captured by neutroavidin sepharose followed by brief washes. After SDS-PAGE and trypsin digestion, differentially labeled peptides were detected by mass spectrometry. Peptides having high heavy/light ratios were subjected to further MS/MS analysis to reveal their identities. (B) Wild-type and mutant CanScript ×3 sequences. SP1-like and MCAT motifs are overscored.
Normally, MSLN is expressed only in a thin layer of mesothelial cells lining body cavities. For reasons that are currently unknown, MSLN expression is strongly activated in many cancers, especially the vast majority of ovarian and ductal pancreatic adenocarcinomas13,14 and is the molecular target for antigen-directed experimental cancer immunotherapeutics.15,16 We previously ascribed MSLN cancer-overexpression to CanScript, a 20-nt sequence with promoter-like activity in MSLN-positive cancers.8,17 CanScript is bipartite, comprising an MCAT motif and an SP1-like motif separated by a short linker (Figure 1B). The MCAT motif recruits a complex of TEAD1 and YAP1 (yes-associated protein1),18 genes whose expression is almost universal. TEAD1 and YAP1 are necessary, but not sufficient, for MSLN gene expression and CanScript reporter activity.8 Interestingly, the 5′ flanking sequence of MCAT motif often plays a determining role in tissue-specific expression of genes, such as cardiac troponin T in striated muscle.19 We postulated that the SP1-like motif and its associated unknown transcription factors may lend the cancer-specificity. In previous study, we found KLF6 as a candidate associated with the SP1-like motif, but it did not fully explain the features of CanScript’s activity.8
Here, we sought to identify CanScript-binding proteins by SD-MS and TFM by comparing the binding of the wild-type CanScript sequence to a transcriptional inactive mutant sequence.8,17
EXPERIMENTAL PROCEDURES
Cell Lines and Culture
AsPc1, HeLa, RKO and HEK293 cell lines were obtained from the American Type Culture Collection. Cells were cultured in RPMI (AsPc1 only) or DMEM medium, supplemented with 10% FBS and 1% penicillin/streptomycin. The cultures were monitored for characteristic patterns of MSLN expression and morphology during passage.
CanScript Sequences Used for DNA Affinity-capture
The CanScript 3× wild-type sequence was CGGGG-(TCTCCACCCACACATTCCTG)3GGGCG; the mutant C5 sequence was CGGGG(TCTCAACCCACACATTCCTG)3-GGGCG; the mutant M6c sequence was CGGGG(TCTCCA-CCCACACTTACCTG)3GGGCG, the double mutant C5M6c sequence was CGGGG(TCTCAACCCACACTTACCTG)3-GGGCG. Biotin with a 15-atom spacer arm was covalently linked to the 5′ end of these polynucleotides. These were annealed with their complementary nonmodified sequence (both from IDT) to make double-stranded DNA.
Plasmids
The pRL-SV40 plasmids were obtained from Promega (E2231). pGL3-Can3 was described previously.8 TEAD1, YAP1 and ZNF24 sequences were inserted into pcDNA3.1-FLAG vector (Invitrogen) between BamHI and NotI to create pcDNA3.1-FLAG-TEAD1, pcDNA3.1-FLAG-YAP1, and pcDNA3.1-FLAG-ZNF24, TEAD1 was also inserted into the pRK5-HA vector (BD Pharmingen) between SalI and NotI to create pRK5-HA-TEAD1.
Nuclear Extraction
Cells were lysed by suspension using 1 mL hypotonic detergent (10 mM Tris pH 8.0, 10 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA, 0.5 mM DTT, 0.75% NP-40 with protease inhibitors, Roche 1183617001) per 0.2 g cell pellet for 15 min. The lysate was agitated followed by 10 min centrifugation at 3500× g for 30 s. The supernatant was collected as the cytoplasmic fraction. The nuclear pellet was resuspended in 1 mL isotonic solution A (20 mM Tris pH 7.5, 100 mM KCl, 50 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA with protease inhibitors) per 0.2 g cell pellet. The nuclear suspension was sonicated for ten seconds and clarified by 10 min centrifugation at 13000× g. This supernatant was collected as the nuclear extract. Separation of cytoplasmic and nuclear proteins was confirmed by immunoblot to detect alpha-tubulin and lamin B, respectively.
DNA Affinity-capture Assay
For immunoblot analysis of captured proteins, nuclear extracts from ~107 AsPc1 cells were incubated with 0.5 μM biotinylated CanScript ×3 probes and 50 μg/mL dI-dC for 1 h, followed by addition of 1/20 volume NeutrAvidin beads (Pierce, 29200). For DS-MS, cell numbers and solution volumes were increased 10-fold excepting the beads, which were doubled. After 30 min binding, beads were washed three times in solution A. Bound proteins were released by boiling beads in 1× loading buffer. Released proteins were separated by SDS-PAGE before immunoblot or mass spectrometric analysis.
Immunoblot
For analysis of unseparated proteins, cells were washed in PBS and lysed by RIPA buffer (containing protease inhibitors), followed by sonication. Protein concentrations were determined by DC protein assay (Bio-Rad 500–011). Samples were boiled with loading buffer and proteins separated by SDS-PAGE. For all immunoblots, proteins transferred onto a PVDF membrane were detected using one or more primary and secondary antibodies: anti-MSLN (Abcam, ab-3362); anti-TEAD1 (Santa Cruz, sc-81396); anti-TEAD2 (Santa Cruz, sc-32427); anti-YAP1 (Santa Cruz, sc-101199); anti-SP1 (Santa Cruz, sc-420); anti-KLF6 (Santa Cruz sc-7158); anti-ZNF24 (Santa Cruz, sc-101079); anti-RFX1 (Santa Cruz sc-10650); anti-NAB1 (Santa Cruz, sc-81565); anti-NFATC1 (Santa Cruz, sc-7294); anti-NFATC2 (Santa Cruz, sc-7296); anti-CEA/CEACAM5 (Santa Cruz, sc-23928); anti-IKZF1 (Santa Cruz, sc-13039); anti-HBP1 (Santa Cruz, sc-25390); anti-PRDM14 (Santa Cruz, sc-133923); antialpha-tubulin (Santa Cruz, sc-8035); antilamin B (Santa Cruz, sc-6216); anti-GRSF (Novus Biologicals, NBP1–57316); anti-HA (Santa Cruz, sc-7392 HRP); anti-FLAG (Sigma A8592). HRP-linked secondary antibodies were from Santa Cruz (antimouse sc-2005; antirabbit sc-2004; antigoat sc-2020). Membranes were developed using chemiluminescence substrate (Millipore WBKLS0500) and exposed to film.
SILAC Labeling Procedure
AsPc1 cells were cultured in lysine and arginine-free RPMI (Cambridge, RPMI-500) supplied with 13C6 L-Lysine-2HCl (Pierce, 89988), 13C6 L-Arginine-HCl (Pierce, 88210) and 10% dialyzed FBS (Invitrogen, 26400036) for at least five cell generations for the complete incorporation of heavy stable isotope-labeled amino acids.
Mass Spectrometry
In-gel digestion of affnity-purified proteins was done prior to protein identification and quantification. Colloidal Coomassie blue-stained gel bands were excised and destained. Gel bands were subjected to reduction (5 mM DTT) and alkylation (20 mM iodoacetamide) and subsequently proteolyzed using sequencing-grade modified porcine trypsin (Promega, V5111) as described.20 Dried extracted peptides were resuspended in 0.1% formic acid and loaded for liquid chromatography–tandem mass spectrometry (LC–MS/MS) analysis. We carried out SD-MS experiments for three replicate, distinct experiments. Proteins identified twice to have wild-type/mutant (i.e., heavy/light) ratios of at least 10.0 were arbitrarily chosen as high-confidence interactors.
Protein identification by LC–MS/MS analysis of peptides was performed using the LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) interfaced with a 2D nanoLC system (Eksigent) and Agilent 1100 autosampler (Meadows Instrumentation Inc.). Peptides were loaded on a 75 μm × 2.5 cm trap packed with Magic AQ C18, 5 μm 100 Å reversed-phase material, then separated by reversed-phase HPLC on the same material using an acetonitrile/0.1% formic acid gradient of 60 min with a flow rate of 300 nL/min.
Identification and quantitation of proteins were carried out using Proteome Discoverer 1.3 software with the Mascot and SEQUEST search algorithms and a human protein database (NCBI RefSeq, Reference Sequence collection, Release 46). Program parameter settings were: trypsin as the digestion enzyme (missed cleavage 1 allowed), peptide tolerance at 10 ppm, fragment tolerance at 0.6 Da (carbamidomethylation on Cys, deamidation of asparagine and glutamine, oxidation at methionine), lysine (13C6) and arginine (13C6) as variable modifications. Proteins were considered positive when their peptides were identified with a cutoff of <1% false discovery rate (FDR). SILAC-based quantitation was carried out using the Proteome Discoverer Event Detector (mass precision of 2 ppm) and Precursor Ions Quantifier algorithms to measure the area under extracted ion chromatograms. The data associated with this manuscript may be downloaded from Proteome-Commons.org Tranche using the following hash: 9jrzamhW-qUF+MRkk6cUzXJ0z+xt9yZWv+T5LZGXbYO2np +BQ8p6pRoFro2HS4bMti7q0TRun/OJedyz4qFK6clYVm-b0AAAAAAAAL/A==
Transcription Factor Protein Microarray
The fluorophore Cy5 was covalently linked to the 5′ end of wild-type or C5M6c mutant CanScript ×3 sequences, which were then annealed to their complementary nonmodified sequences to form double-stranded DNA probes. Wild-type and mutant probes were incubated individually with identical TFM chips as described.21 The chips included 4191 human GST-tagged proteins purified from yeast. The software of GenePix@ Pro 7 was used to read spots’ intensities on the protein microarray. The raw intensity of the spot was defined as the ratio (in logarithmic scale, base 2) of foreground median to local background median. We adopted quantile normalization to modify the signals across microarrays under different conditions (wild-type and mutant in the study). Intensities of less than zero were treated as a half of the background (noise) data and its symmetric part (>0) was generated. Both parts formed the whole of the background noise, which approximately fitted to a normal distribution with the mean value of zero. Then, each spot’s signal was standardized according to
where Z is the Z-score of each spot, I is the intensity of the spot after quantile normalization, and σ is the standard deviation of the background noise. Since all proteins had duplicated spots on each microarray, only proteins having both spots’ Z-scores greater than three were considered as positive for DNA probe-binding activity. Those positive proteins for the wild-type probe (Z > 3), but not the mutant probe, were considered having wild-type binding specificity. We listed only those with wild-type/mutant ratio at least 10.0 in Table 2.
Table 2.
Proteins Binding Canscript Sequence as Identified by TFM
| gene ID | ratio (WT/MT)a | identified protein full name |
|---|---|---|
| NFATc1b,c | 33.5 | nuclear factor of activated T cells, cytoplasmic 1 |
| DHX36 | 29.3 | deah box polypeptide 36 |
| RBM12 | 29.3 | RNA-binding motif protein 12 |
| SUMO4 | 26 | small ubiquitin-like modifier 4 |
| CAMK2D | 22.3 | calcium/calmodulin-dependent protein kinase II-delta |
| IKZF1c | 18.3 | IKAROS family zinc finger 1 |
| TEAD2b | 15.9 | transcriptional enhancer factor 4 |
| HBP1c | 14.8 | HMG box-containing protein 1 |
| PRDM14c | 13 | PR domain-containing protein 14 |
| NAB1b,c | 12 | NFFI-A-binding protein 1 |
| BZW1 | 11.6 | basic leucine zipper and W2 domains 1 |
| ING2 | 11.3 | inhibitor of growth 2 |
| ACTL6B | 10.6 | actin-like 6B |
| RBM9 | 10.6 | RNA-binding motif protein 9 |
| YY1 | 10.5 | transcription factor YY1 |
| CRAT | 10.4 | carnitine acetyltransferase |
| Tcf15 | 10.2 | transcription factor 15 |
| CD80 | 10.1 | B-lymphocyte activation antigen B7-1 |
Candidates having a WT/MT ratio of at least 10 are listed. WT, Canscript wild-type; MT, Canscript C5M6C mutant.
Also identified this protein or a member of this protein family by SD-MS.
Included among the proteins examined by immunoblot after DNA affnity-capture.
Transfection
We primarily used HeLa cells for transient transfections, owing to low transfection effciency in AsPc1 cells. We seeded 1.5 × 105 cells per well of a 6-well plate one day before transfection by Lipofectamine (Invitrogen 18324–012, manufacturer’s instructions). pGL3-Can3 firefly luciferase vector (0.2 μg) and pRL-SV40 Renilla luciferase control vector (20 ng) were transfected per well. Transfection medium was replaced by growth medium at 6 h.
Purification of FLAG-tagged Protein
2.5 million HEK293 cells were seeded in a 10-cm dish followed by transfection with 12 μg pcDNA3.1-FLAG vector. Cells were harvested after 48 h and sonicated in 300 μL PBS buffer. After 10 min centrifuge at 10,000 × g, supernatant was collected and incubated with 20 μL anti-FLAG M2 magnetic beads (Sigma, M8823) at 4 °C for 1 h. The beads were washed and protein eluted using 150 ng/μL 3 × FLAG peptide (Sigma, F4799). The yield was ~1 μg/dish.
Luciferase Reporter Assays
The dual luciferase reporter assay system (Promega E1910) was used. Cells were harvested 24 h after cotransfection with pGL3-Can3 and pRL-SV40 vector. Cells were lysed by the kit-provided lysis solution (300 μL/well). Each sample was measured using 20 μL of cell lysate mixed with luciferase reagent II (LARII, 100 μL per well). After gathering the firefly luminescent signal by photometer, Stop and Glo reagent was added (100 μL per well), followed by the Renilla luminescence reading. Firefly luciferase readings were normalized using the Renilla readings of the same wells. The average relative luciferase activity (RLA) was obtained from the triplicate wells in each experiment (the differences among three wells were usually less than 5%). Two independent experiments performed on different days had high agreement; a representative experiment was presented (Supporting Information Figure S3).
siRNA sequences and transfection. siRNA oligonucleotides were synthesized by Ambion. An effcient siRNA from an irrelevant gene (FANCD2, Irr) was systematically used as a negative control. Except for the previously reported TEAD1 and TEAD2 genes,17 three nonoverlapping siRNA sequences were evaluated for each gene (total five siRNA sequences were tried on RFX1 and only three could effectively reduce RFX1 protein level). Immunoblot assays were used to verify siRNA effciencies of gene depletion. The sense sequences were: FANCD2 (Irr) si:CCAUGUCCUUAGUAGCCGATT; TEAD1 si: GCCCUGUUUCUAAUUGUGGTT; ZNF24 si1: GGCACUGUGAUGAUGAUGGTT; ZNF24 si2: GUUC-CUGGCACUCUCAAUATT; ZNF24 si3: GCAUUCAGCC-GAAGUUCCATT; RXF1 si1: GAGUACAUGUACUACCU-GATT; RXF1 si2: GAGAGAUCUGUGGUCCAGGTT; RXF1 si3: GAUGGAAGGCAUGACCAACTT; NAB1 si1: GGA-GUUCCUUUGCAACCAATT; NAB1 si2: GUAGCAUACC-CAUCUAUAATT. NAB1 si3: GAGUGAAGAACUUGCAG-CUTT. siRNA transfections were done in HeLa cells using Oligofectamine (Invitrogen 12252011). The protocol was similar to our transient plasmid transfection except that 0.2 nM siRNA and 4 μL Oligofectamine were added into each well.
RESULTS
DNA Affinity-capture
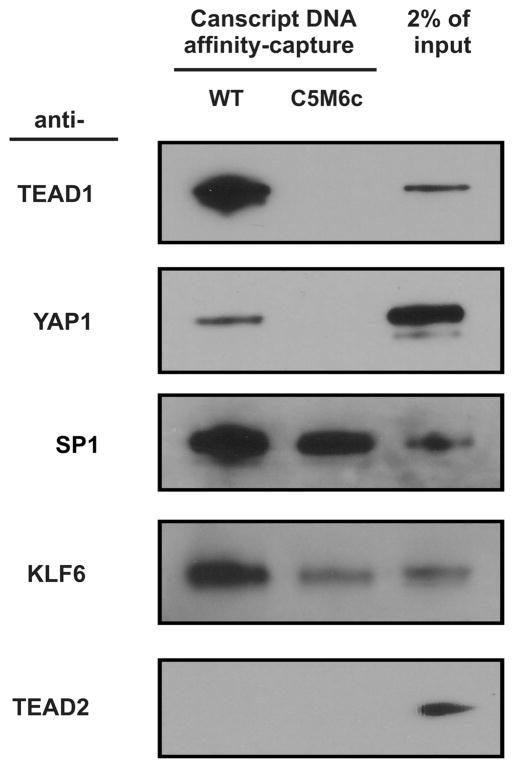
Using an immunoblot assay following DNA affnity-capture (Figure 1A), the positive control TEAD1 was retained at greater than 2% of the input amount using the wild-type CanScript sequence, but not retained by the known inactive C5M6c mutant (Figure 1B)8,17 (Figure 2). KLF6, a reported candidate,8 had a similar although weaker pattern. TEAD2, which shared DNA-binding characteristics with TEAD1, yet in our previous study was not recruited to CanScript in vivo,17 was not detected (Figure 2). The negative control SP1 bound to CanScript wild-type and mutant indistinguishably (Figure 2), confirming our findings that SP1 was not a regulator of CanScript activity.8 SP1 transcriptional function is not expected to tolerate the central adenine in the Canscript sequence. The TEAD1-binding partner YAP1 was specifically effciently captured by wild-type CanScript, reflecting indirect capture of a member of the complex (as contrasted with exclusively reflecting direct DNA-binding).
Figure 2.
Confirmation of known CanScript-binding candidates. TEAD1, YAP1, SP1, KLF6 and TEAD2 were examined for their binding to DNA (wild-type or transcriptionally inactive C5Mc6 mutant CanScript ×3) by immunoblot analysis after DNA affnity-capture. For each blot, one-third of the final captured sample was resolved by SDS-PAGE. 2% of the nuclear extract was used as the input reference. WT, wild-type.
Screening by SD-MS
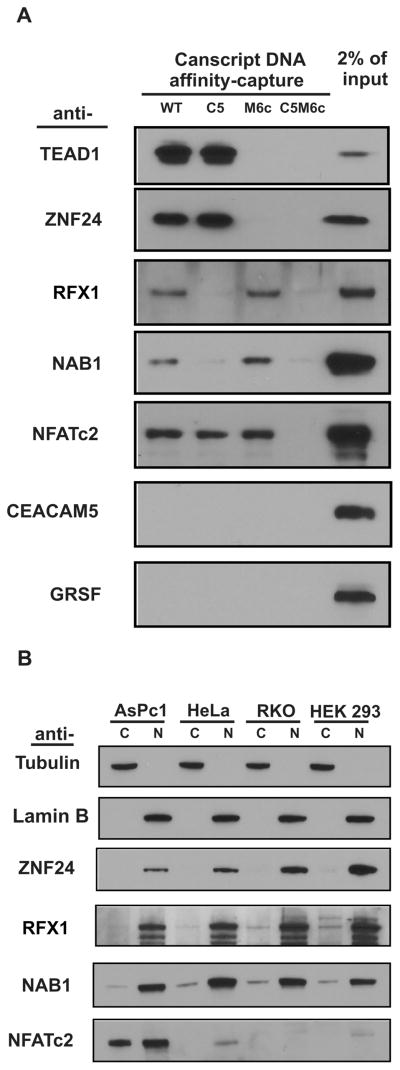
We attempted to visually select “probe-specific” protein bands appearing after DNA affnity-capture (Supporting Information Figure S1). To be complete, we excised and analyzed several of the visible differential protein bands even in the mutant bound lane, but they seemed nonrelevant (such as universally expressed ribosome-binding proteins). We then adopted the SILAC approach coupled to the DNA-affnity capture. A total of 850 proteins, based on 7,700 unique peptides, were identified from three replicate SD-MS experiments. Only 15 proteins preferentially bound to wild-type probe (wild-type/mutant ratio more than 10) whereas 83 proteins preferred C5M6c mutant probe (ratio less than 0.1). Nine hits were identified using the predefined criterion (10 as threshold ratio) (Table 1). TEAD1 (Figure 3A), an interactor reported previously by us (and thus the positive control), was one of the two with the highest ratios. TEA family members TEAD3 and TEAD4 were the first and the fourth in the order, respectively. TEAD2 was not identified by SD-MS. The discovery of a structural redundancy of TEAD family members (TEAD1–3) might indicate a functional redundancy, also. These results reflected those found using immunoblots (above) in place of mass spectrometry. We subsequently chose seven out of the nine hits (excluding TEAD3 and TEAD4 as being biochemically similar) to examine their cognate binding motifs (MCAT or SP1-like) by immunoblot analysis to measure DNA capture (Figure 4A). ZNF24, a TCAT repeat-binding protein,22 was identified as specifically recruited by the MCAT motif (Figure 3B). ZNF24 was reported as a transcriptional repressor for vascular endothelial growth factor.23,24 The knockout of ZNF24 in mice resulted in early embryo lethality,25 attributed to inhibition of proliferation of neural progenitor cells.26 In this study we demonstrated that ZNF24 might function as a transcriptional activator for MSLN expression. In contrast, RFX1 (regulatory factor X, 1) and NAB1 (NGF1A-binding protein 1) were specifically captured by the SP1-like motif. RFX1 is a transactivator for expression of MHC class II,27 proliferating cell nuclear antigen (PCNA),28 and certain viral proteins such as HBsAg.29 It binds as a monomer to the consensus sequence RGYAAC and as a dimer to palindromic DNA29,30 with a preference for methylated sequence.31 NFATc2 (cytoplasmic nuclear factor of activated T cells) bound wild-type CanScript and tolerated each of the tested single mutants (C5 and M6c), but not the double mutant, C5M6c. The NFATc consensus-binding sequence WGGAAANH,32 however, was not present in CanScript. NFATc family members NFATc1–4 are long known to play pivotal and redundant roles in activation of immune system.32 The DNA binding core sequences RAHYETEG is conserved in all NFATc family members.32 Binding to CanScript was not confirmed by immunoblots for CEACAM5 and GRSF; the highly overexpressed membrane protein CEACAM533 was considered a false positive due to all membrane components in the nuclear extract.
Table 1.
Proteins Binding Canscript Sequence as Identified by SD-MS
| accession | gene IDa | # of peptides identified | times identifiedb | ratio (heavy/light, WT/MT)c |
|---|---|---|---|---|
| NP_003205.2 | TEAD3d | 10 | 3/3 | ≥100 |
| NP_068780.2 | TEAD1d,e | 12 | 3/3 | ≥93 |
| NP_008896.2 | ZNF24e | 7 | 3/3 | ≥41 |
| NP_003204.2 | TEAD4d | 9 | 2/3 | ≥100 |
| NP_004354.2 | CEACAM5e | 2 | 2/3 | ≥100 |
| NP_002909.4 | RFX1e | 2 | 2/3 | ≥54 |
| NP_001129493.1 | NFATC2d,e | 3 | 2/3 | ≥49 |
| NP_005957.2 | NAB1d,e | 2 | 1/3 | ≥40 |
| NP_001091947.1 | GRSF1e | 2 | 2/3 | ≥15 |
Full names of identified proteins are provided in Supporting Information Table S1.
SD-MS was replicated three times using independent extracts.
Candidates having a WT/MT ratio of at least 10 are listed. WT, Canscript wild-type; MT, Canscript C5M6C mutant.
Also identified this protein or a member of this protein family by TFM.
Included among the proteins examined by immunoblot after DNA affnity-capture.
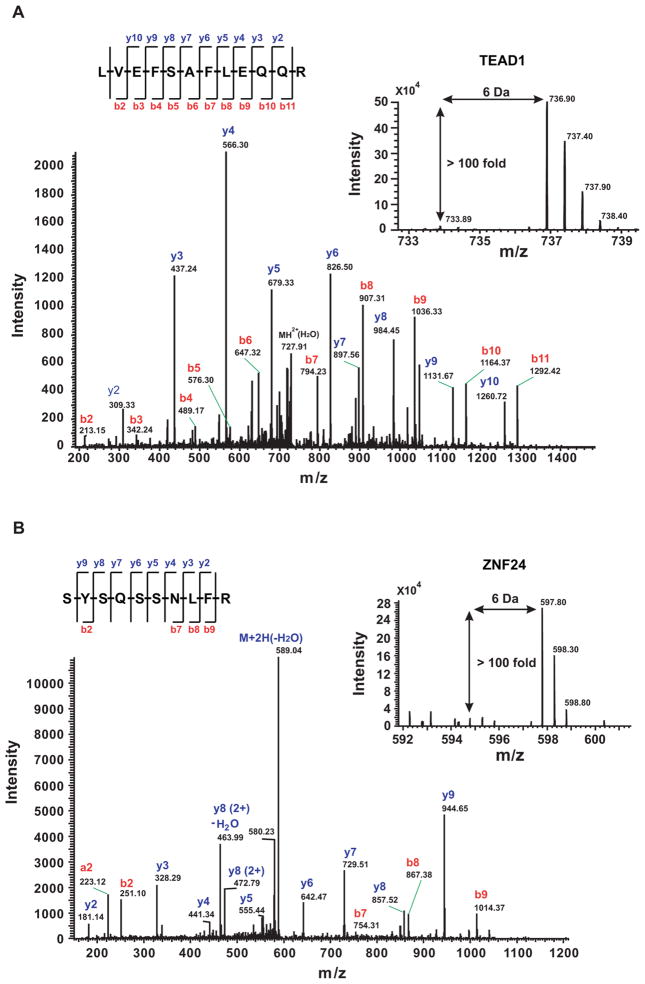
Figure 3.
Representative MS and MS/MS spectra of peptides from selected proteins specifically bound to wild-type Canscript ×3 probe. (A) LVEFSAFLEQQR from TEAD1 and (B) SYSQSSNLFR from ZNF24. (Inset) Heavy stable isotope-labeled peptide precursor ion and corresponding position of the lighter version. A list of y and b fragment ions are indicated in the MS/MS spectra.
Figure 4.
Verification of hits identified by SD-MS. (A) Seven of nine hits listed in Table 1 were examined by immunoblot following DNA affnity-capture. Single mutant (C5 and M6c) CanScript ×3 probes were used in parallel to attribute which binding motif was responsible for selected hits. The protocol was as described in Figure 2. (B) Expression pattern and subcellular distribution of ZNF24, RFX1, NAB1 and NFATc2 in MSLN-positive (AsPc1 and HeLa) and -negative (RKO and HEK293) cell lines. Alpha-tubulin was used as a cytoplasmic marker (C) and lamin B as a nuclear marker (N).
We next examined whether the four immunoblot-confirmed hits (ZNF24, RFX1, NAB1 and NFATc2) might correlate to MSLN expression patterns among various cell lines (Figure 4B). This was not the general pattern, for ZNF24, RFX1 and NAB1 were expressed in both MSLN-positive (AsPc1 and HeLa) and -negative cells (RKO and HEK 293). NFATc2 had high expression in AsPc1 cells, yet a very low level in HeLa and HEK293 cells and was not detected in RKO cells. Not surprisingly, these proteins identified as transcriptionally active interactors were predominantly distributed in the nuclear fraction (Figure 4B). This is interesting for NFATc2 because calcium influx is required for NFAT nuclear translocation in T cells.34 It is unclear whether NFATc2 nuclear localization in cancer cells is calcium influx-dependent.
Screening by TFM
We probed Cy5-labeled double-stranded wild-type or C5M6c mutant CanScript ×3 sequences individually on our TFM chips. Eighteen hits that bound specifically to the wild-type sequence were identified at a threshold ratio of 10 (wild-type/mutant signal intensity, logarithmic transformed) (Table 2). Due to fluorescent signals being a more sensitive reading (i.e., producing data for a greater proportion of the analytes), we expected and attained diversity in the ratios from this screen. Three of the TFM hits shared gene family members with SD-MS hits: NAB1 was a hit in both; NFATc1 and TEAD2 from the TFM hits shared identical DNA-binding protein sequences with NFATc2 and TEAD1, respectively, identified by SD-MS. It was not surprising to identify TEAD2 by TFM because the 72-amino acid DNA-binding domain is identical between TEAD1 and TEAD2,35 and a report had demonstrated that in vitro-synthesized TEAD2 could bind to the MCAT motif36 (note that TFM is an in vitro screen). Similarly, recombinant NFATc1 and NFATc2 were known to have share cognate DNA-binding sequences.37 Identification of NAB1 by TFM indicated that NAB1 could directly bind to CanScript. NAB1 might be an unconventional DNA-binding protein as described previously,4 because it has not been reported to have direct DNA-binding activity. NAB1 was reported as a zinc finger protein EGR1 (Early Growth Response factor 1) binding partner,38 which is implicated in promoting growth and progression of prostate cancer.39 ERG1 itself was not a hit on either of our screens. Interestingly, HMG box transcription factor 1 (HBP1)40 shared the binding consensus sequence TCAT repeat with ZNF24, identified by SD-MS (Table 1).
We next evaluated further an arbitrarily chosen five of the ten top hits (Table 2) by immunoblot assay after DNA-affnity capture (Supporting Information Figure S2). NAB1 and NFATc1 were specifically captured by wild-type CanScript. Immunoblotting did not confirm the binding of HBP1 and PRDM14 with CanScript, despite being expressed in the tested cells (AsPc1). In contrast, IKZF1 had CanScript C5M6c mutant-specific binding.
Functional Studies
To validate some of these newly identified candidates, we asked whether they played roles in MSLN expression or CanScript reporter activity. We chose to examine ZNF24, NAB1 and RFX1 because: (1) ZNF24 was one of the two CanScript-binding candidates whose captured fraction was greater than 2% of the input (Figure 4A); (2) NAB1 and RFX1 were the only two proteins specifically recruited by the wild-type SP1-like motif, thought to be the likely cancer-specific component;8 (3) NAB1 was an unconventional DNA-binding protein and had been identified with both orthogonal proteomics approaches.
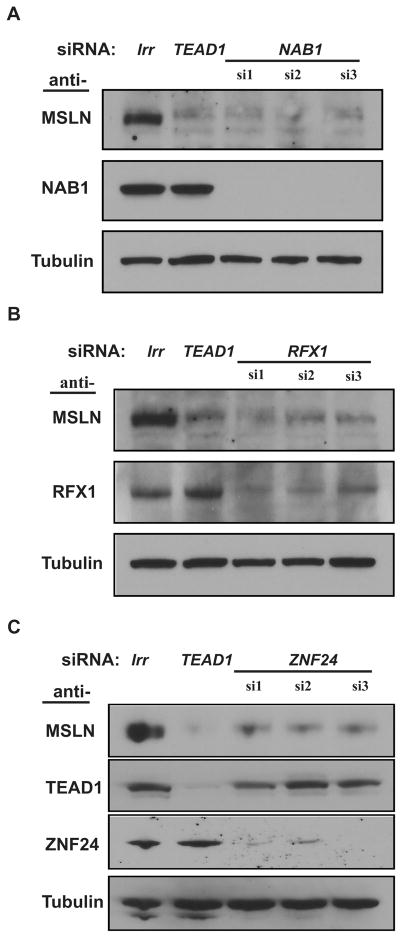
For functional studies, we used siRNA to individually knock down ZNF24, NAB1 and RFX1 in HeLa cells. Testing three sequences per gene, the expression levels of all three genes were effectively reduced by their siRNAs (Figure 5A–C). NAB1 and RFX1 siRNA reduced most MSLN expression with efficacies similar to our positive control, the TEAD1 siRNA (Figure 5A and B), whereas MSLN protein was moderately reduced by ZNF24 siRNAs in HeLa cells (Figure 5C). TEAD1 expression was slightly reduced by two of three ZNF24 siRNAs (Figure 5C, si1 and si3), but not by NAB1 or RFX1 siRNAs (Supporting Information Figure S4). We did not observe prominent cell loss or cell death induced by any siRNA (the reported proliferation inhibition of ZNF24 RNAi could be cell line-specific).25 Similarly, two of three ZNF24 and NAB1 siRNAs reduced CanScript ×3 reporter activity (Supporting Information Figure S3) with one of the ZNF24 siRNAs being as effective as TEAD1 siRNA. In contrast, two RFX1 siRNAs increased CanScript reporter activity; one reached almost 3-fold induction. The inconsistent effects of RFX1 manipulation on MSLN expression and CanScript reporter activity, are as yet unexplained but might represent a complex regulatory effect or a plasmid-dependent artifact.8,41–44
Figure 5.
Dependence of MSLN expression on expression of three interactome genes. siRNA treatments of (A) NAB1, (B) RFX1 and (C) ZNF24 were examined for knockdown efficacy and for effects on MSLN expression in HeLa cells. Each gene was tested with three nonoverlapping siRNA sequences. Irrelevant target FANCD2 (Irr) and TEAD1 siRNA were used as negative and positive control, respectively. Alpha-tubulin was used as a comparison for sample loading.
Overexpression of ectopic wild-type ZNF24, NAB1 or RFX1 neither switched on MSLN expression nor significantly augmented CanScript reporter activity in MSLN-negative cell lines (data not shown). The overall results suggested that the two transcription factors and NAB1 might be necessary or contributory, but not sufficient, for MSLN overexpression.
The association of ZNF24 with the MCAT motif raised a question, whether ZNF24 participated in a complex with TEAD1-YAP1 or bound to CanScript independently. ZNF24 was reported to bind to DNA only through its heterodimers,45,46 its homodimerization being weak when compared to its heterodimerization, as when it was paired with the leucine-rich SCAN domain-containing partners ZNF202 or SDP1,45 which were not identified in our screens. We did not observe the ZNF24-TEAD1 interaction when we attempted to coimmunoprecipitate ZNF24 using anti-TEAD1 antibody in transiently transcfected HeLa cells, yet the YAP1-TEAD1 interaction was confirmed (data not shown). We also failed to observe direct CanScript interaction with purified ZNF24 by EMSA (data not shown). These negative results were thus inconclusive.
DISCUSSION
We combined two orthogonal proteomic screens, SD-MS and TFM, systematically exploring the interactome at a structurally novel cancer-specific promoter driving the expression of a therapeutically and diagnostically relevant cancer marker. The combination provided an effcient, reproducible; and unbiased approach for identification of interactors having DNA-binding specificity. The results agreed well with each other: eight of 27 (9 + 18) hits overlapped at the level of protein families. The two approaches were practically complementary to each other as well, SD-MS presumably scanning all relevant native nuclear proteins specifically in relevant cells, and TFM providing evidence of direct DNA-binding.
Multiple advantages attend SD-MS as an approach for identifying DNA sequence-specific interactomes. Mass spectrometry is quite sensitive, requiring only tens of nanograms of captured protein for identification. Also, SILAC provides a facile and numerical prioritization among captured proteins. SD-MS is an unbiased screen, owing to all native proteins of the nuclear extract being interrogated by the DNA probe, and includes analysis of protein complexes that might mimic the in vivo functional binding as well or better than could purified proteins.
The major challenge of SD-MS is that the DNA-binding activities of native proteins captured and resolved in vitro are subject to arbitrary experimental conditions. For example, using a conventional nuclear extract protocol (high salt, ~ 0.4 M NaCl) solution, we found the TEAD1-Canscript interaction greatly impaired (data not shown). Our modified nuclear extract also did not fully exclude contamination from membrane proteins (CEACAM5, Table 1).
TFM has a unique advantage of detecting only direct interactions with DNA. The use of individual purified proteins on a chip also provides a clean background free of the nonspecific interferences expected from cell lysates. Thus, TFM could discover weak or rare DNA binding proteins that are at a competitive disadvantage in the SD-MS screen. TFM’s disadvantages, however, include: (1) purified recombinant proteins (here, provided in yeast) might not mimic the in vivo functional conformations; (2) DNA-binding proteins dependent on protein cooperation (such as ZNF24) might evade discovery.
We identified nine hits from SD-MS and 18 from TFM comparing the Canscript sequence with a transcriptionally inactive C5Mc6 mutant CanScript matched control and with stringent cutoffs. The screens confirmed the previous candidate TEAD1, but also reflected the other TEAD family members, as an expected redundancy. We selected eleven hits (TEAD1, ZNF24, RFX1, NFATc1, NFATc2, NAB1, CEACAM5, GRSF1, IKZF1, HBP1 and PRDM14) and confirmed the binding activities of six (TEAD1, ZNF24, RFX1, NFATc1, NFATc2 and NAB1) by immunoblot following DNA affnity-capture.
We chose MCAT motif-bound ZNF24 and SP1-like motif-bound NAB1 and RFX1 for further functional analysis. Knockdown of any of these three transcription factors could effectively reduce MSLN expression in MSLN-positive cells (Figure 5), suggesting they were necessary for MSLN expression. In a test of sufficiency, their overexpression did not switch on MSLN expression in MSLN-negative cells; this was not surprising, owing to their being nonspecifically expressed in MLSN-positive and -negative cells (Figure 4B). In addition, CanScript itself is likely necessary but not sufficient for MSLN expression; the reporter activity of the pGL3-CanScript ×1 plasmid, when tested exclusive of MSLN flanking sequences, was almost at the basal level of an empty reporter construct (data not shown),8,17 in contrast to the MSLN cancer-specific promoter region (CanScript is located at the very 5′ end) containing ~160 bp, which had greater than a 100-fold increase of transcriptional activity over the basic construct. Additional cis factors in the MSLN promoter region presumably cooperate with CanScript to fully activate MSLN cancer-specific overexpression.
Four additional annotated transcription factors would be considered had we chose 2.0 rather than 10.0 as our criterion in SD-MS (Table 1): ZFP64, MAFG, MAFF and MAFK. The MAF proteins belong to the AP-1/JUN superfamily of basic leucine zipper proteins. MAF homo- and heterodimers are reported to recognize a palindromic consensus DNA sequence TMARE (TGCTGAG/CTCAGCA),47 which is not present in CanScript, yet their possible roles related to MSLN expression may worth investigation.
Interestingly, 9 of the 15 (60%) detected wild-type Canscript-specific binding proteins were known transcription factors (Supporting Information Table S1), whereas only 12 of 83 (14%) mutant Canscript-specific binding proteins were known transcription factors (Supporting Information Table S2). We can not speculate what roles these mutant Canscript-binding transcription factors might play in vivo; the human genome lacks exact copies of that mutant sequence.
The study of DNA–protein interactions is a key bridge connecting genetics and proteomics. Combinatorial and unbiased approaches, such as those explored here, could effciently explore transcriptional controls driving cancer- or disease-specific, transcriptionally active, DNA elements known to be distributed among diverse genome loci in cancer and in other diseases. Better knowledge of such interactions could suggest novel targets for pharmacologic disruption, or draw fresh attention to overlooked influences in the recurring biochemical signatures of disease.
Supplementary Material
Acknowledgments
This work has been supported by NIH grant CA62924, NIH Technology Centers for Networks and Pathways grant U54 RR020839 and the Everett and Marjorie Kovler Professorship in Pancreas Cancer Research.
ABBREVIATIONS
- MS
mass spectrometry
- EDTA
ethylenediaminetetraacetic acid
- DTT
dithiothreitol
- PAGE
polyacrylamide gel electrophoresis
- HA
hemagglutinin
Footnotes
The authors declare no competing financial interest.
Supplementary figures and methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hauck W, Nedellec P, Turbide C, Stanners CP, Barnett TR, Beauchemin N. Transcriptional control of the human biliary glycoprotein gene, a CEA gene family member down-regulated in colorectal carcinomas. Eur J Biochem. 1994;223(2):529–41. doi: 10.1111/j.1432-1033.1994.tb19022.x. [DOI] [PubMed] [Google Scholar]
- 2.Koritschoner NP, Bocco JL, Panzetta-Dutari GM, Dumur CI, Flury A, Patrito LC. A novel human zinc finger protein that interacts with the core promoter element of a TATA box-less gene. J Biol Chem. 1997;272(14):9573–80. doi: 10.1074/jbc.272.14.9573. [DOI] [PubMed] [Google Scholar]
- 3.Thiagalingam A, De Bustros A, Borges M, Jasti R, Compton D, Diamond L, Mabry M, Ball DW, Baylin SB, Nelkin BD. RREB-1, a novel zinc finger protein, is involved in the differentiation response to Ras in human medullary thyroid carcinomas. Mol Cell Biol. 1996;16(10):5335–45. doi: 10.1128/mcb.16.10.5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu S, Xie Z, Onishi A, Yu X, Jiang L, Lin J, Rho HS, Woodard C, Wang H, Jeong JS, Long S, He X, Wade H, Blackshaw S, Qian J, Zhu H. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139(3):610–22. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37(Database issue):D77–82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kumar M, Gromiha MM, Raghava GP. Identification of DNA-binding proteins using support vector machines and evolutionary profiles. BMC Bioinform. 2007;8:463. doi: 10.1186/1471-2105-8-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan C, Terribilini M, Wu F, Jernigan RL, Dobbs D, Honavar V. Predicting DNA-binding sites of proteins from amino acid sequence. BMC Bioinform. 2006;7:262. doi: 10.1186/1471-2105-7-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ren YR, Patel K, Paun BC, Kern SE. Structural analysis of the cancer-specific promoter in mesothelin and in other genes overexpressed in cancers. J Biol Chem. 2011;286(14):11960–9. doi: 10.1074/jbc.M110.193458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ranish JA, Hahn S, Lu Y, Yi EC, Li XJ, Eng J, Aebersold R. Identification of TFB5, a new component of general transcription and DNA repair factor IIH. Nat Genet. 2004;36(7):707–13. doi: 10.1038/ng1385. [DOI] [PubMed] [Google Scholar]
- 10.Mittler G, Butter F, Mann M. SILAC-based, A DNA protein interaction screen that identifies candidate binding proteins to functional DNA elements. Genome Res. 2009;19(2):284–93. doi: 10.1101/gr.081711.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butter F, Kappei D, Buchholz F, Vermeulen M, Mann M. A domesticated transposon mediates the effects of a single-nucleotide polymorphism responsible for enhanced muscle growth. EMBO Rep. 2010;11(4):305–11. doi: 10.1038/embor.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11(1):73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 13.Hassan R, Ebel W, Routhier EL, Patel R, Kline JB, Zhang J, Chao Q, Jacob S, Turchin H, Gibbs L, Phillips MD, Mudali S, Iacobuzio-Donahue C, Jaffee EM, Moreno M, Pastan I, Sass PM, Nicolaides NC, Grasso L. Preclinical evaluation of MORAb-009, a chimeric antibody targeting tumor-associated mesothelin. Cancer Immun. 2007;7:20. [PMC free article] [PubMed] [Google Scholar]
- 14.Argani P, Iacobuzio-Donahue C, Ryu B, Rosty C, Goggins M, Wilentz RE, Murugesan SR, Leach SD, Jaffee E, Yeo CJ, Cameron JL, Kern SE, Hruban RH. Mesothelin is overexpressed in the vast majority of ductal adenocarcinomas of the pancreas: identification of a new pancreatic cancer marker by serial analysis of gene expression (SAGE) Clin Cancer Res. 2001;7(12):3862–8. [PubMed] [Google Scholar]
- 15.Hassan R, Cohen SJ, Phillips M, Pastan I, Sharon E, Kelly RJ, Schweizer C, Weil S, Laheru D. Phase I clinical trial of the chimeric anti-mesothelin monoclonal antibody MORAb-009 in patients with mesothelin-expressing cancers. Clin Cancer Res. 2010;16(24):6132–8. doi: 10.1158/1078-0432.CCR-10-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kreitman RJ, Hassan R, Fitzgerald DJ, Pastan I. Phase I trial of continuous infusion anti-mesothelin recombinant immunotoxin SS1P. Clin Cancer Res. 2009;15(16):5274–9. doi: 10.1158/1078-0432.CCR-09-0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hucl T, Brody JR, Gallmeier E, Iacobuzio-Donahue CA, Farrance IK, Kern SE. High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor dependent MCAT motif. Cancer Res. 2007;67(19):9055–65. doi: 10.1158/0008-5472.CAN-07-0474. [DOI] [PubMed] [Google Scholar]
- 18.Mahoney WM, Jr, Hong JH, Yaffe MB, Farrance IK. The transcriptional co-activator TAZ interacts differentially with transcriptional enhancer factor-1 (TEF-1) family members. Biochem J. 2005;388(Pt 1):217–25. doi: 10.1042/BJ20041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larkin SB, Farrance IK, Ordahl CP. Flanking sequences modulate the cell specificity of M-CAT elements. Mol Cell Biol. 1996;16(7):3742–55. doi: 10.1128/mcb.16.7.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68(5):850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 21.Hu S, Xie Z, Blackshaw S, Qian J, Zhu H. Characterization of protein-DNA interactions using protein microarrays. Cold Spring Harb Protoc. 2011;2011(5):1. doi: 10.1101/pdb.prot5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Albanese V, Biguet NF, Kiefer H, Bayard E, Mallet J, Meloni R. Quantitative effects on gene silencing by allelic variation at a tetranucleotide microsatellite. Hum Mol Genet. 2001;10(17):1785–92. doi: 10.1093/hmg/10.17.1785. [DOI] [PubMed] [Google Scholar]
- 23.Harper J, Yan L, Loureiro RM, Wu I, Fang J, D’Amore PA, Moses MA. Repression of vascular endothelial growth factor expression by the zinc finger transcription factor ZNF24. Cancer Res. 2007;67(18):8736–41. doi: 10.1158/0008-5472.CAN-07-1617. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Chen X, Liu Y, Ding L, Qiu L, Hu Z, Zhang J. The transcriptional repression of platelet-derived growth factor receptor-beta by the zinc finger transcription factor ZNF24. Biochem Biophys Res Commun. 2012;397(2):318–22. doi: 10.1016/j.bbrc.2010.05.110. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Chen X, Yang H, Wang S, Guo B, Yu L, Wang Z, Fu J. The zinc finger transcription factor 191 is required for early embryonic development and cell proliferation. Exp Cell Res. 2006;312(20):3990–8. doi: 10.1016/j.yexcr.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 26.Khalfallah O, Ravassard P, Lagache CS, Fligny C, Serre A, Bayard E, Faucon-Biguet N, Mallet J, Meloni R, Nardelli J. Zinc finger protein 191 (ZNF191/Zfp191) is necessary to maintain neural cells as cycling progenitors. Stem Cells. 2009;27(7):1643–53. doi: 10.1002/stem.88. [DOI] [PubMed] [Google Scholar]
- 27.Pugliatti L, Derre J, Berger R, Ucla C, Reith W, Mach B. The genes for MHC class II regulatory factors RFX1 and RFX2 are located on the short arm of chromosome 19. Genomics. 1992;13(4):1307–10. doi: 10.1016/0888-7543(92)90052-t. [DOI] [PubMed] [Google Scholar]
- 28.Liu M, Lee BH, Mathews MB. Involvement of RFX1 protein in the regulation of the human proliferating cell nuclear antigen promoter. J Biol Chem. 1999;274(22):15433–9. doi: 10.1074/jbc.274.22.15433. [DOI] [PubMed] [Google Scholar]
- 29.Siegrist CA, Durand B, Emery P, David E, Hearing P, Mach B, Reith W. RFX1 is identical to enhancer factor C and functions as a transactivator of the hepatitis B virus enhancer. Mol Cell Biol. 1993;13(10):6375–84. doi: 10.1128/mcb.13.10.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emery P, Strubin M, Hofmann K, Bucher P, Mach B, Reith W. A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Mol Cell Biol. 1996;16(8):4486–94. doi: 10.1128/mcb.16.8.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garcia AD, Ostapchuk P, Hearing P. Methylation-dependent and -independent DNA binding of nuclear factor EF-C. Virology. 1991;182(2):857–60. doi: 10.1016/0042-6822(91)90629-p. [DOI] [PubMed] [Google Scholar]
- 32.Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–47. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 33.Klavins JV. Tumor markers of pancreatic carcinoma. Cancer. 1981;47(6 Suppl):1597–601. doi: 10.1002/1097-0142(19810315)47:6+<1597::aid-cncr2820471423>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 34.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFA. Genes Dev. 2003;17(18):2205–32. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 35.Kaneko KJ, DePamphilis ML. Regulation of gene expression at the beginning of mammalian development and the TEAD family of transcription factors. Dev Genet. 1998;22(1):43–55. doi: 10.1002/(SICI)1520-6408(1998)22:1<43::AID-DVG5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 36.Yasunami M, Suzuki K, Houtani T, Sugimoto T, Ohkubo H. Molecular characterization of cDNA encoding a novel protein related to transcriptional enhancer factor-1 from neural precursor cells. J Biol Chem. 1995;270(31):18649–54. doi: 10.1074/jbc.270.31.18649. [DOI] [PubMed] [Google Scholar]
- 37.Hoey T, Sun YL, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2(5):461–72. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 38.Russo MW, Sevetson BR, Milbrandt J. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc Natl Acad Sci USA. 1995;92(15):6873–7. doi: 10.1073/pnas.92.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adamson E, de Belle I, Mittal S, Wang Y, Hayakawa J, Korkmaz K, O’Hagan D, McClelland M, Mercola D. Egr1 signaling in prostate cancer. Cancer Biol Ther. 2003;2(6):617–22. [PubMed] [Google Scholar]
- 40.Zhuma T, Tyrrell R, Sekkali B, Skavdis G, Saveliev A, Tolaini M, Roderick K, Norton T, Smerdon S, Sedgwick S, Festenstein R, Kioussis D. Human HMG box transcription factor HBP1: a role in hCD2 LCR function. EMBO J. 1999;18(22):6396–406. doi: 10.1093/emboj/18.22.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Q, Suzuki K, Hirschler-Laszkiewicz I, Rothblum LI. Paradoxical effect of eukaryotic expression vectors on reporters. Biotechniques. 2002;33(1):74, 76, 78. doi: 10.2144/02331st04. passim. [DOI] [PubMed] [Google Scholar]
- 42.Hong SJ, Chae H, Kim KS. Promoterless luciferase reporter gene is transactivated by basic helix-loop-helix transcription factors. Biotechniques. 2002;33(6):1236–8. 40. doi: 10.2144/02336bm11. [DOI] [PubMed] [Google Scholar]
- 43.Osborne SA, Tonissen KF. pRL-TK induction can cause misinterpretation of gene promoter activity. Biotechniques. 2002;33(6):1240–2. doi: 10.2144/02336bm12. [DOI] [PubMed] [Google Scholar]
- 44.Jensen LE, Whitehead AS. ELAM-1/E-selectin promoter contains an inducible AP-1/CREB site and is not NF-kappa B-specific. Biotechniques. 2003;35:54–6. 58. doi: 10.2144/03351bm05. [DOI] [PubMed] [Google Scholar]
- 45.Schumacher C, Wang H, Honer C, Ding W, Koehn J, Lawrence Q, Coulis CM, Wang LL, Ballinger D, Bowen BR, Wagner S. The SCAN domain mediates selective oligomerization. J Biol Chem. 2000;275(22):17173–9. doi: 10.1074/jbc.M000119200. [DOI] [PubMed] [Google Scholar]
- 46.Nam K, Honer C, Schumacher C. Structural components of SCAN-domain dimerizations. Proteins. 2004;56(4):685–92. doi: 10.1002/prot.20170. [DOI] [PubMed] [Google Scholar]
- 47.Kurokawa H, Motohashi H, Sueno S, Kimura M, Takagawa H, Kanno Y, Yamamoto M, Tanaka T. Structural basis of alternative DNA recognition by Maf transcription factors. Mol Cell Biol. 2009;29(23):6232–44. doi: 10.1128/MCB.00708-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.