Abstract
Although the objective of any ‘omic science is broad measurement of its constituents, such coverage has been challenging in metabolomics because the metabolome is comprised of a chemically diverse set of small molecules with variable physical properties. While extensive studies have been performed to identify metabolite isolation and separation methods, these strategies introduce bias toward lipophilic or water-soluble metabolites depending on whether reversed-phase (RP) or hydrophilic interaction liquid chromatography (HILIC) is used respectively. Here we extend our consideration of metabolome isolation and separation procedures to integrate RPLC/MS and HILIC/MS profiling. An aminopropyl-based HILIC/MS method was optimized on the basis of mobile-phase additives and pH, followed by evaluation of reproducibility. When applied to the untargeted study of perturbed bacterial metabolomes, the HILIC method enabled the accurate assessment of key, dysregulated metabolites in central carbon pathways (e.g., amino acids, organic acids, phosphorylated sugars, energy currency metabolites), which could not be retained by RPLC. To demonstrate the value of the integrative approach, bacterial cells, human plasma, and cancer cells were analyzed by combined RPLC/HILIC separation coupled to ESI positive/negative MS detection. The combined approach resulted in the observation of metabolites associated with lipid and central carbon metabolism from a single biological extract, using 80 % organic solvent (ACN:MeOH:H2O 2:2:1). It enabled the detection of more than 30,000 features from each sample type, with the highest number of uniquely detected features by RPLC in ESI positive mode and by HILIC in ESI negative mode. Therefore, we conclude that when time and sample are limited, the maximum amount of biological information related to lipid and central carbon metabolism can be acquired by combining RPLC ESI positive and HILIC ESI negative mode analysis.
Keywords: Untargeted metabolomics, Global metabolite profiling, Metabolome, Liquid chromatography Electrospray-Mass Spectrometry, Hydrophilic interaction chromatography, Reversed phase chromatography, Central carbon metabolism
INTRODUCTION
Metabolites are a direct readout of biochemical activity and, as the end products of genomic, transcriptomic, and proteomic expression, provide a representation of cellular state.1–3 In contrast to genes, transcripts, and proteins, which are composed of a defined set of building blocks, metabolites contain a high level of chemical diversity.3–5 Given the wide range of physical properties characteristic of the metabolome, measuring the complete set of metabolites present in complex biological matrices represents a major challenge for untargeted, mass spectrometry-based metabolomics.1, 3–4, 6 Examining a broad range of chemically diverse metabolites, however, is important when studying biological systems. In diabetes, for example, in addition to perturbations in carbohydrate metabolism there are alterations in the lipid profile that also contribute to disease pathogenesis.7–8
To study the chemically complex metabolome, liquid chromatography-mass spectrometry (LC/MS) equipped with an electrospray ionization (ESI) source has become a key analytical tool given the number of analytes that can be simultaneously measured.4, 9–11 The number of metabolite features detected by LC/MS analyses, defined by unique m/z and LC retention times, has previously been used as a relevant measure of the overall coverage of a metabolome.6, 12 The steps in LC/MS-based untargeted metabolomics that most directly influence the number and intensity of detected metabolite features, include metabolite extraction, LC separation, and MS detection.4, 6 Metabolite extraction in particular, has been studied extensively for different types of biological samples (biofluids, tissues, cells).13–17 Although there is no consensus for a universal extraction method, minimum sample handling was reported as essential for reproducibility in large-scale metabolomic studies.16–17
Furthermore, efficient chromatographic separation prior to MS is of importance for the analysis of complex metabolite-rich samples to reduce ion suppression at the MS source and improve signal sensitivity.4, 18 Historically, reversed-phase liquid chromatography (RPLC) using C18 or C8 columns has dominated most LC/MS profiling in untargeted metabolomic studies. This is due to its high versatility, stability, and ability to cover a large set of physiologically important metabolites. However, water soluble, highly polar, and ionic metabolites are typically not effectively retained in RPLC making their separation, relative quantification, and tandem MS/MS identification unreliable.4, 6 Ion-pairing agents such as tetrabutylamine or decyl sulfate in the RPLC mobile phase have improved results for the analysis of polar compounds (e.g., sugar phosphates, nucleotides, carboxylic acids, peptides, amino acids),4, 9 however, they contaminate the LC/MS instrumentation and are therefore generally not preferable.
Hydrophilic interaction chromatography (HILIC) has become increasingly popular as an alternative to RPLC for the analysis of polar metabolites.18–19 Indeed, key cellular metabolites involved in central carbon metabolism pathways have been successfully analyzed by using HILIC.5, 20–23 Specifically, an amino-column based HILIC chromatography at high pH (9.45) pioneered by Rabinowitz and colleagues has been effectively used to separate a diverse set of central carbon metabolites5, and has been successfully applied to targeted metabolomic studies of bacteria and cancer cell metabolism.4, 20–21, 24–26 For untargeted metabolic profiling, however, the ability to analyze polar metabolites is in the process of maturation, where generating reproducible and robust data still represents a significant challenge.18, 27 The reports on HILIC/MS application for global metabolic profiling are mainly limited to studies of urine.27–30
The development of a reproducible HILIC method as a complement to standard RPLC is required to improve the measurement accuracy of water-soluble metabolites and to make untargeted metabolomics more comprehensive. This is also crucial for the integration of metabolomics into multi-‘omic’ systems biology (genomics, transcriptomics and proteomics) to provide functional assignment of genes and proteins.2, 23, 31–32 This can be achieved by deciphering enzymatic activity, metabolic pathway regulation33 and by the identification of yet unknown, altered pathways in perturbed systems (disease, environmental stress, etc.).34–35 Significant effort has been invested to minimize the bias related to sample preparation and extraction,15, 17 whereas systematic evaluation of LC/MS analytical techniques has been examined less frequently in untargeted studies.28, 36 Moreover, the extraction protocols have mostly been considered in the scope of one column type (e.g., RP6, 16, 37 or HILIC5, etc.), rarely assuming that an appropriately designed single extraction method can be adapted for comparative, multiple platform profiling.15
Here, a comprehensive study of the HILIC method was initially evaluated to expand the metabolome coverage to hydrophilic metabolites in a reliable and reproducible manner for the purpose of large-scale untargeted profiling. A study of the bacterial (Shewanella oneidensis) metabolic response to nitric oxide stress underlined the importance of gaining insight into central carbon metabolism from an untargeted perspective. To proceed toward ‘omic scale comprehensive analysis in metabolomic experiments, two complementary separation techniques (HILIC and RPLC) together with ESI positive and ESI negative were integrated into a single extraction-dual separation LC/MS workflow. Metabolome coverage was evaluated on three different types of biological samples: bacterial cells, human cancer cells, and human plasma.
EXPERIMENTAL SECTION
Chemical materials
Chemical standards were purchased from Sigma Aldrich (St. Louis, MO, USA) and Cayman Chemical (Ann Arbor, MI, USA). For each of the hydrophilic and hydrophobic standard mixtures, 50 standards (Table S1 and S2) were prepared at a final concentration of 20 µg/mL. Ammonium acetate (NH4Ac), ammonium hydroxide (NH4OH), ammonium fluoride (NH4F), ammonium carbonate ((NH4)2CO3) and ammonium formate (NH4HCOO) were purchased from Sigma Aldrich. LC/MS grade 0.1 % formic acid (FA) in acetonitrile (ACN), 0.1 % FA in water (H2O), and methanol (MeOH) were purchased from Honeywell (Muskegon, MI, USA). Acetonitrile was purchased from Fisher Scientific (Morris Plains, NJ, USA) and water was purchased from J.T. Baker (Phillipsburg, NJ, USA). Standard plasma samples (P9523) were purchased from Sigma Aldrich (St. Louis, MO, USA).
Metabolite extraction and reconstitution
Detailed preparation of bacteria (Shewanella oneidensis MR-1 and Escherichia coli W3110), human cancer cells (Raji Burkitt’s lymphoma), and human plasma samples is included in the Supporting Information. Bacteria cell pellets (~109 cells) and human cancer cell pellets (~107 cells) were extracted using a MeOH:ACN:H2O (2:2:1, v/v) solvent mixture. A volume of 1 mL of cold solvent was added to each pellet, vortexed for 30 s, and incubated in liquid nitrogen for 1 min. The samples were then allowed to thaw at room temperature and sonicated for 10 min. This cycle of cell lysis in liquid nitrogen combined with sonication was repeated three times. To precipitate proteins, the samples were incubated for 1 h at −20 °C, followed by 15 min centrifugation at 13,000 rpm and 4 °C. The resulting supernatant was removed and evaporated to dryness in a vacuum concentrator. The dry extracts were then reconstituted in 100 µL of ACN:H2O (1:1, v/v), sonicated for 10 min and centrifuged 15 min at 13000 rpm and 4 °C to remove insoluble debris. The supernatants were transferred to HPLC vials and stored at −80 °C prior to LC/MS analysis.
Standard human plasma samples (200 µL) were extracted with 800 µL of cold MeOH:ACN (1:1, v/v) to keep the same MeOH:ACN:H2O (2:2:1, v/v) ratio. The samples were then vortexed for 30 s, and sonicated for 10 min. The rest of the procedure was the same as described for bacteria and cancer cell pellets.
LC/MS analysis
Analyses were performed using an HPLC system (1200 series, Agilent Technologies) coupled to a 6538 UHD Q-TOF (Agilent Technologies). Samples were analyzed using a XBridge C18, 3.5 µm, 150 mm × 1.0 mm I.D. column (Waters) for RPLC/MS analysis, and Luna Aminopropyl, 3 µm, 150 mm × 1.0 mm I.D. column (Phenomenex) for HILIC/MS analysis. The standard mobile phase, A = 0.1% formic acid in water and B = 0.1% formic acid in acetonitrile, was used for RPLC in ESI positive mode. To optimize the analysis conditions for RPLC in ESI negative mode, as well as for HILIC in both ESI positive and negative modes, different additives in the mobile phase were tested as listed in Table S3. The final mobile phase for RPLC in ESI negative mode was composed of A = 0.5 mM ammonium fluoride in water and B = 100% acetonitrile. For HILIC, in both ESI positive and negative modes, the final mobile phase was composed of A = 10 mM ammonium acetate and 10 mM ammonium hydroxide in 95% water and B = 95% acetonitrile. The linear gradient elution from 100 % B (0–5 min) to 100 % A (50–55min) was applied in HILIC (A = 95% H2O, B = 95% ACN, with appropriate additives) and from 100 % A (0–5 min) to 100% B (50–55min) in RPLC (A = 100% H2O, B = 100% ACN, with appropriate additives). The 10 minutes post-run was applied for HILIC, to insure the column re-equilibration and maintain the reproducibility. The flow rate was 50 µL/min and the sample injection volume was 8 µL.
ESI source conditions were set as followings: gas temperature 325 °C, drying gas 5 L/min, nebulizer 15 psi, fragmentor 120 V, skimmer 65 V, and capillary voltage 4000 V or −4000V in ESI positive or ESI negative modes, respectively. The instrument was set to acquire over the m/z range 60–1000, with the MS acquisition rate of 1.67 spectra/s. For the MS/MS of selected precursors the default isolation width was set as medium (4 Da), with a MS acquisition rate at 1.67 spectra/s and MS/MS acquisition at 1.67 spectra/s. The collision energy was fixed at 20 V.
Bacteria cells, human cancer cells, and human plasma samples, together with blank samples, were run in triplicate using both HILIC and RPLC coupled to both ESI positive and ESI negative modes, for a total of four different LC/MS conditions. Blank samples represented the reconstitution solution prior to injection, ACN/H2O (1:1). Experimentally perturbed bacteria (Shewanella oneidensis, “control” vs. “treated with nitric oxide”) were used to test the large-scale reproducibility and separation power of the optimized HILIC method. Three different, common types of samples, bacteria cells (Escherichia coli), human plasma, and human cancer cells (Raji cells) were used to evaluate the metabolome coverage of the combined HILIC/RPLC/MS approach.
Data analysis
A comparison of the different mobile phase conditions under different LC/MS modes was made based on the intensity of detected metabolites from standard mixtures. The intensities of EICs (Extracted Ion Chromatograms) matching the metabolites from the standard mixtures (Table S1 and S2) were extracted by using XCMS.12, 38 Data were then normalized by scaling the intensities for the same metabolite at different conditions to its maximal intensity value observed. The scaled values for different metabolites at the same analysis condition were averaged to obtain the overall scores for each chromatographic and ion-mode combination tested.
The raw LC/MS data were converted to mzXML files using ProteoWizard MS Convert version 3.0.4146.39 The mzXML files were processed using XCMS12, 38, 40 for peak detection, alignment, and isotope annotation. The parameters in XCMS were set as follows: centWave settings for feature detection (Δ m/z = 15 ppm, minimum peak width = 10 seconds and maximum peak width = 120 seconds); obiwarp settings for retention time correction (profStep = 1); and other parameters including mzwid = 0.015, minfrac = 0.5 and bw = 5 for chromatogram alignment. Overlap between different chromatographic modes was performed by using exact mass value defined by Δ m/z = 0.01 Da. The relative quantification of metabolite features was based on the integrated EIC peak areas.
Quality control was performed with pooled samples, representative of the entire sample batch, to monitor the reproducibility of the HILIC/MS method. Coefficients of variation (CV) of peak area and peak height were calculated based on annotated metabolite features with clear isotopic patterns (XCMS12, 38 and CAMERA41 software) across the entire run.
RESULTS AND DISCUSSION
Optimization of HILIC/MS Method
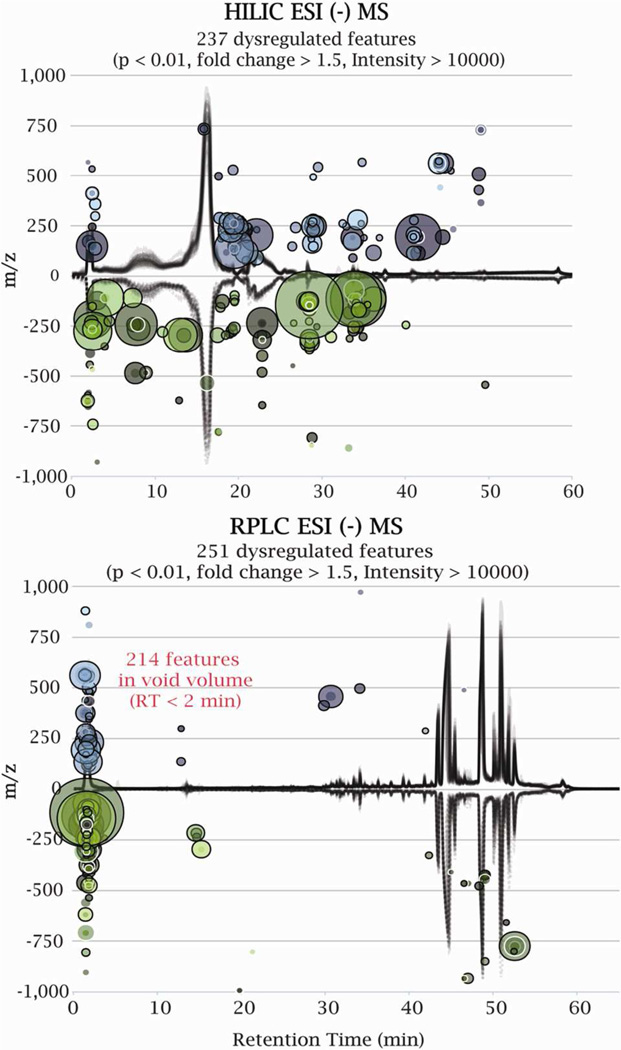
The reversed-phase LC/MS untargeted metabolic profiling of a bacterial (Shewanella oneidensis) response to nitric oxide stress revealed that the majority (~ 85 %, Figure 1) of the abundant, significantly dysregulated features were observed in the void volume or with minimal retention times (RT < 2 min). Dysregulated features represent peaks whose MS signal (peak area and intensity) has been significantly altered in a perturbed system, thus reflecting a disruption in biochemical activity. The abundant dysregulated features were filtered according to the following criteria: p-value < 0.01, a fold change > 1.5 and MS peak intensity > 10000 ion counts, representing the threshold required to generate high-quality MS/MS spectra on an Agilent Q-TOF instrument.6 These results suggest that metabolic changes induced by nitric oxide treatment occur mainly in the highly polar cellular metabolome, most likely affecting the core network of central carbon metabolism.
Figure 1.
Cloud plots showing dysregulated metabolite features (represented by “bubbles”) in a bacterial response to nitric oxide treatment analyzed by HILIC/MS and RPLC/MS. The dysregulated features (up-regulated in blue, down-regulated in green) in comparison to “control” were filtered according to statistical thresholds shown in plots. The ion intensity was used as radius scale of each bubble.
A HILIC/MS approach was optimized to chromatographically resolve the dysregulated polar metabolites, thus reducing the ionization suppression and enabling the accurate, relative quantification of metabolite changes. Since untargeted metabolic profiling has been mainly based on RPLC methods,28, 30 the optimization and evaluation of reproducibility of our HILIC/MS method was required for combining with RPLC/MS in large-scale, global profiling studies. For this purpose, the aminopropyl column from Phenomenex was chosen due to its good selectivity, high reproducibility, and broad coverage of hydrophilic metabolites.5, 26, 30
The additives in the mobile phase have a significant impact on metabolite separation and MS ionization efficiency6, 10, 19, 42 as they determine mobile phase pH and regulate stationary phase selectivity, thus influencing metabolite retention and separation.43 Here, a range of pH conditions and salts were tested for metabolite separation and MS sensitivity using a hydrophilic standard mixture composed of 50 common endogenous metabolites (Table S1, Figure S1). Among the three different pH conditions examined (pH 4.0, 6.9, and 9.8), a basic pH 9.8 yielded the best overall scores (sum of normalized ion intensities) and the highest number of resolved compounds in both ESI positive and negative ionization modes. This result was consistent with previous research performed on aminopropyl-based HILIC columns.5, 26 Specific examples include the polycarboxylic acids and phosphorylated compounds (succinic acid, malic acid, citric acid, sugar phosphates, and nucleotides) that were detected and resolved only in basic mobile phase conditions. The improved performance at basic pH conditions compared to neutral and acidic pH conditions is most likely due to weaker electrostatic interactions between the aminopropyl stationary phase (pKa of primary amine functional group = 9.8) and the negatively charged metabolites. Moreover, basic pH conditions are also known to improve the deprotonation of acidic analytes and enhance MS detection sensitivity.5, 42
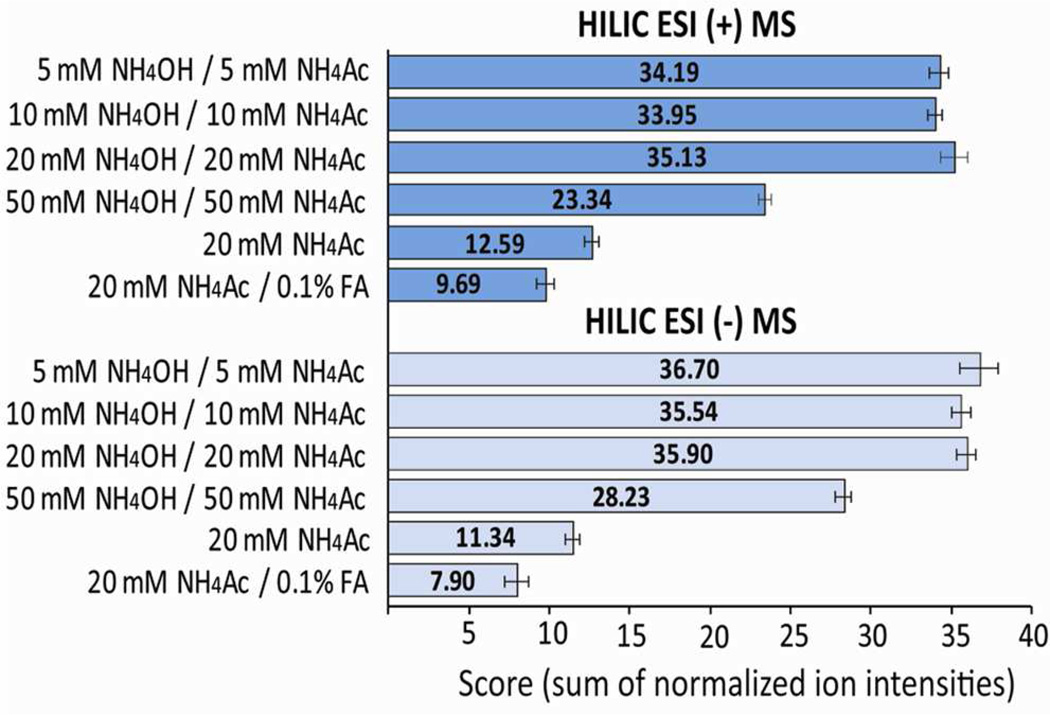
In addition to the mobile phase pH, the salt concentration determines the ionic strength of the mobile phase and enhances the elution of charged metabolites by mediating their electrostatic interaction with the stationary phase.19 Initially, a range of concentrations (5, 10, 20, and 50 mM) of ammonium acetate and ammonium hydroxide mixture were tested. It was found that 5, 10, and 20 mM salt concentrations compared to no addition of buffer enhanced the ESI sensitivity in HILIC/MS. This enhancement occurred in both positive and negative ion modes, with no significant difference between the three concentrations (Figure 2). The addition of 50 mM buffer led to a significant decrease in overall sensitivity likely due to ionization suppression. Further examination of different salt additives (e.g., NH4F, (NH4)2CO3, NH4HCOO) did not result in increased HILIC/MS sensitivity (Figure S1 and S2).
Figure 2.
Optimization of HILIC/MS analysis conditions. Each tested condition is presented by its overall score, calculated as a sum of normalized intensities of 50 metabolites from the hydrophilic standard mixture (Table S1, Table S3).
Based on these results, the optimum, high pH mobile phase conditions for HILIC/MS analyses using aminopropyl stationary phase were comprised of A = 10 mM NH4Ac and 10 mM NH4OH in 95 % water and B = 95 % acetonitrile, in both ESI positive and ESI negative modes.
Reproducibility and Separation Capabilities of Optimized HILIC/MS Method
Minimum retention time variation and high reproducibility of MS signal are crucial for high-throughput LC/MS analysis, especially in untargeted studies where it plays an important role in chromatogram alignment, peak matching across different samples, and downstream statistical analysis. To evaluate the reproducibility of our optimized HILIC method and to adequately measure the bacterial metabolic response to nitric oxide, the optimized method was applied to the same batch of bacteria samples that were previously analyzed by RPLC/MS (Figure 1). The analyses included 36 bacteria samples, 9 injections of pooled samples for quality control, and 12 blank injections for a total of over 60 hours of analysis time. Although a common challenge encountered with HILIC technologies is chromatographic reproducibility, the alignment of sample profiles acquired by the optimized HILIC method demonstrated that the deviation of retention time across all runs did not vary significantly in comparison to the deviation of retention time using RPLC (Figure 3). The observed shift in retention time across all these runs was approximately 1 min in a 60-min separation run (XCMS Online using nonlinear obiwarp method 38, 41), a typical deviation during high-throughput analysis that did not affect the sample alignment. Overlay of aligned total ion chromatograms (TICs) and an extracted ion chromatogram (EIC) are shown after retention time correction (Figure 3). The reproducibility assessment of MS signal was also evaluated using 9 injections of pooled samples across the 60 hours of analysis. The metabolite features, naturally present in the sample and annotated for their clear isotopic pattern (CAMERA software44), were used to monitor the reproducibility of peak area and peak height measurements. The average coefficient of variation (CV) was 19.1 ± 6.9 % and 21.7 ± 8.2 % based on peak height and peak area, respectively. More precisely, 84 % and 70 % of features satisfied the criterion of CV ≤ 25 % in terms of the peak intensity and peak area, respectively, and compared favorably to previous reports of metabolic profiling by HILIC/MS.28, 45 The minimal retention time shift, alignment accuracy, and reproducibility of the MS signal demonstrate the overall robustness of the optimized HILIC/MS method. It is important to note, however, that HILIC aminopropyl columns have shorter lifetimes (~300–500 injections depending on the biological matrix) than RPLC C18 columns (~1000 injections) and that the symmetry and sharpness of the peak shape from a HILIC column is more variable when compared to stable RPLC columns. Moreover, HILIC column conditioning, by blank and biological matrix runs, is recommended before starting the analysis.
Figure 3.
Reproducibility of optimized HILIC/MS and RPLC/MS in ESI negative mode. Curve plots represent the retention time deviation over a 60 minute run across 36 samples of Shewanella oneidensis extracts (“control” and “treated with nitric oxide”). Each sample is presented by a differentially colored curve. An overlay of total ion chromatograms (TICs) and one “zoomed in” example of an extracted ion chromatogram (EIC) are given below.
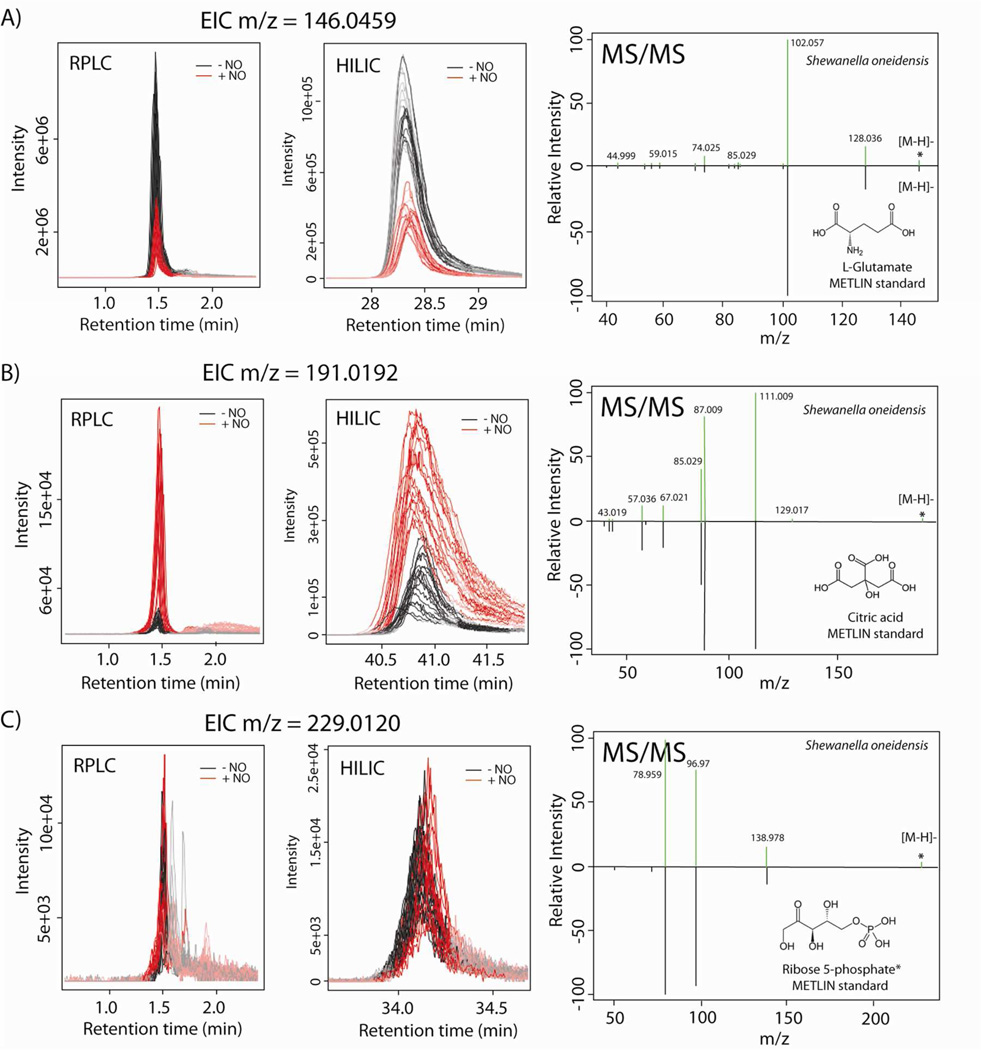
Importantly, the efficient chromatographic separation of dysregulated polar metabolites by HILIC (Figure 1) reduced the ion suppression and mass spectral complexity, thus facilitating the unbiased relative quantification as well as subsequent MS/MS analysis for identification (Figure 4). The alteration of three identified metabolites, covering a wide range of intensity (from 2×104 to 6×106 ion counts) and that eluted in the void volume by RPLC, but were well resolved by HILIC, is demonstrated by EICs (Figure 4). For highly abundant, glutamate, the relative quantification by RPLC matched closely the quantification by HILIC (see Figure 4A, p = 6.6×10−11, fold change = 2.9 in RPLC vs. p = 1.4×10−11, fold change = 3 in HILIC). However, for less abundant features, whose signals were significantly suppressed by matrix effects in the RPLC void volume, the relative quantification was biased (see Figure 4B, p = 4.06×10−9, fold change = 8.8 in RPLC vs. p = 0.0008, fold change = 4.6 in HILIC). Moreover, some low-abundant features (e.g., intensity < 20,000) eluting in the void volume were designated as significantly dysregulated by RPLC, whereas their relative quantification in HILIC indicated that they were not dysregulated (see Figure 4C, m/z 229.012 putatively identified as ribose 5-phosphate, p = 0.005, fold change = 1.5 in RPLC vs. p = 0.5, fold change = 1.1 in HILIC). Metabolite identification was performed by matching acquired MS/MS data against MS/MS data recorded for standard compounds in the METLIN database.
Figure 4.
Extracted ion chromatograms (from RPLC/MS and HILIC/MS) and matching MS/MS spectra for three identified dysregulated metabolites in bacteria exposed to nitric oxide stress. Each metabolite dysregulation is defined by the peak area, fold change, and independent t-test p-value. A) RPLC: p = 6.6×10−11, fold change = 2.9 vs. HILIC: p = 1.4×10−11, fold change = 3, B) RPLC: p = 4.06×10−9, fold change = 8.8 vs. HILIC: p = 0.0008, fold change = 4.6 and C) RPLC: p = 0.005, fold change = 1.5 vs. HILIC: p = 0.5, fold change = 1.1, *Note that the metabolite shown in panel C has been putatively identified as ribose 5-phosphate or ribulose 5-phosphate.
Additional experiments are required to identify other dysregulated metabolites in this study and place the observed changes into biological context. Yet even with the current results, the significant up-regulation of citric acid can be attributed to aconitase inactivation, caused by NO-mediated destruction of the aconitase Fe-S cluster46. Previous enzyme activity studies demonstrated that aconitases are cellular targets of nitric oxide toxicity.47 Further examination of dysregulated metabolites observed with HILIC/MS will likely reveal other, yet unknown changes in central biochemical pathways that are associated with nitric oxide toxicity. After the identification of metabolic changes by untargeted analysis, follow up with targeted analysis is required for absolute quantification of altered metabolites of interest.
Combining HILIC/RPLC to Expand the Metabolome Coverage: Single Extraction-Dual LC/MS Approach
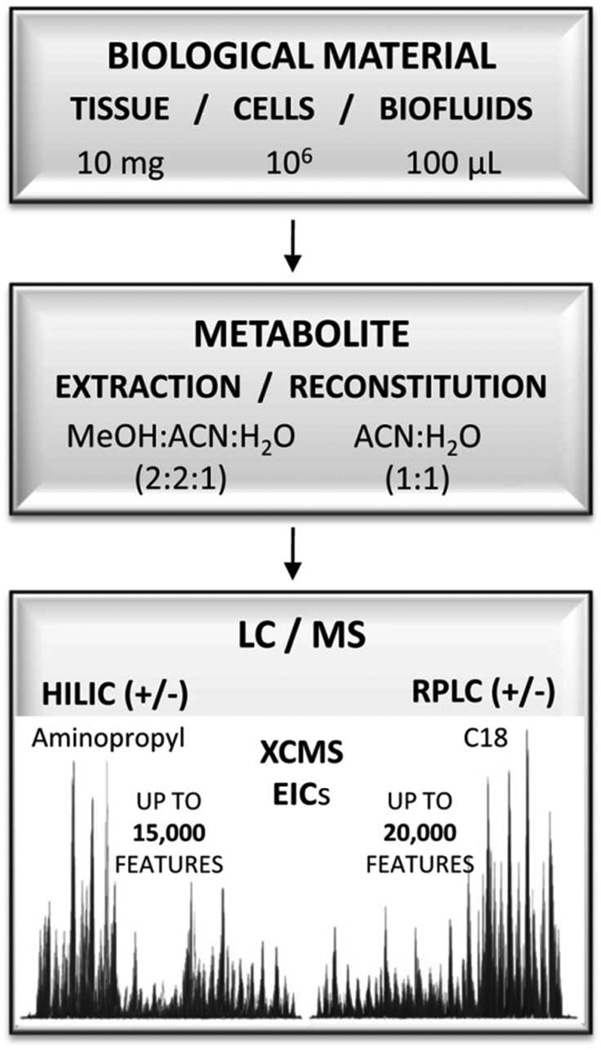
Following the high resolution and reproducibility of large-scale untargeted profiling by the selected HILIC/MS method, an analytical strategy combining HILIC and conventional RPLC separation was developed to maximize the metabolome coverage in untargeted metabolomic studies (Figure 5). The strategy consists of a single extraction and reconstitution method followed by dual LC separation by using HILIC and RPLC (in independent runs) combined with mass spectrometry analysis performed in both ESI positive and negative modes. A minimal sample handling, single-extraction method was chosen to enable the systematic and comparative analysis of the metabolome, profiled by HILIC/MS and by RPLC/MS. The extraction method composed of an 80% organic solvent (MeOH:ACN:H2O (2:2:1 (v/v/v)) mixture efficiently precipitated proteins while extracting a wide range of metabolites from highly hydrophilic polycarboxylic acids and phosphorylated compounds to hydrophobic phospholipids and fatty acids / amides over a variety of biological samples, like bacterial species (Figure S3), human plasma, and cancer cell lines. The ACN:H2O (1:1 (v/v)) reconstitution solution was chosen to re-suspend the majority of both polar and non-polar extracted metabolites, and to ensure analyte retention with both HILIC and RPLC. For HILIC and RPLC separations, small 1 mm diameter columns, low flow rates (50 µL/min), and 60 minute gradients were chosen to minimize matrix effects and maximize the separation and MS-sensitivity of metabolites in the complex biological matrices. HILIC and RPLC analysis were performed by using optimal mobile phase conditions as specified in the Experimental section.
Figure 5.
The integrated LC/MS workflow presenting the standard amount of material used, single extraction, and reconstitution protocol enabling comparative analysis by HILIC/MS and RPLC/MS in ESI positive and ESI negative ionization mode. The average number of features typically observed by each chromatographic / ionization mode is indicated as the number of EICs extracted by XCMS. Note that theoverall number of cells refers to human cells (~106). For significantly smaller size bacterial cells, the overall number was ~109 cells (10 mL of culture at OD~0.3).
The metabolome coverage of three distinct biological samples, bacterial cell pellets (Escherichia coli), human plasma, and human cancer cells (Raji Burkitt’s lymphoma) was analyzed and evaluated with the single extraction - dual chromatography - dual ionization mode strategy. The LC/MS data acquired for these samples were analyzed with the XCMS Online software for feature detection. The total number of features for each sample was obtained after comparing against blank samples to subtract the “background” (fold change < 1.5, p < 0.05) or noise artifacts (Table 1). As seen in Table 1, the total number of features summed across all four LC/MS conditions was above 30,000 for E. coli (56,421), plasma (40,836), and human cancer cells (33,654). The lower number for cancer cells may be due to a limited amount of biological material (Figure 5).
Table 1.
Metabolome coverage by HILIC and RPLC in both ESI positive and ESI negative modes expressed as a number of total aligned metabolite features detected by XCMS in different types of samples. Only dysregulated features compared to blank (to subtract the background) are displayed. The intensity threshold of 10,000 ion counts was used to filter out the abundant features.
| Escherichia coli | Human plasma | Human cancer cells | |||||
|---|---|---|---|---|---|---|---|
| LC mode | MS mode | Total features |
Abundant features |
Total features |
Abundant features |
Total features |
Abundant features |
| HILIC | ESI (+) | 15726 | 2865 | 9709 | 2250 | 11238 | 1903 |
| ESI (−) | 12208 | 2580 | 8122 | 2445 | 8448 | 1643 | |
| RPLC | ESI (+) | 20322 | 3055 | 15263 | 2071 | 8078 | 1077 |
| ESI (−) | 8165 | 2076 | 7742 | 1899 | 5890 | 1034 | |
Although the total number of features detected in ESI positive mode was highest in all analyzed samples compared to ESI negative mode, in both HILIC and RPLC, the difference was not significant (paired t-test, p > 0.01 for HILIC and RPLC), due to the variability observed between different types of samples (Table 1). Moreover, when only the number of highly abundant features was taken into account, the difference between the number of features detected in ESI positive and ESI negative was even less pronounced. The number of features was 2339 ± 487 for HILIC ESI positive mode, 2222 ± 507 for HILIC ESI negative mode, 2068 ± 989 for RPLC ESI positive mode, and 1670 ± 558 for RPLC ESI negative mode (Table 1). In addition to representing similar proportions of high-quality features (with intensities > 10,000 ion counts to provide high-quality MS/MS data) for HILIC/MS and RPLC/MS, these data also suggest that a greater number of low-abundant features is observed in positive ionization mode, most likely due to inherently more efficient ionization.6
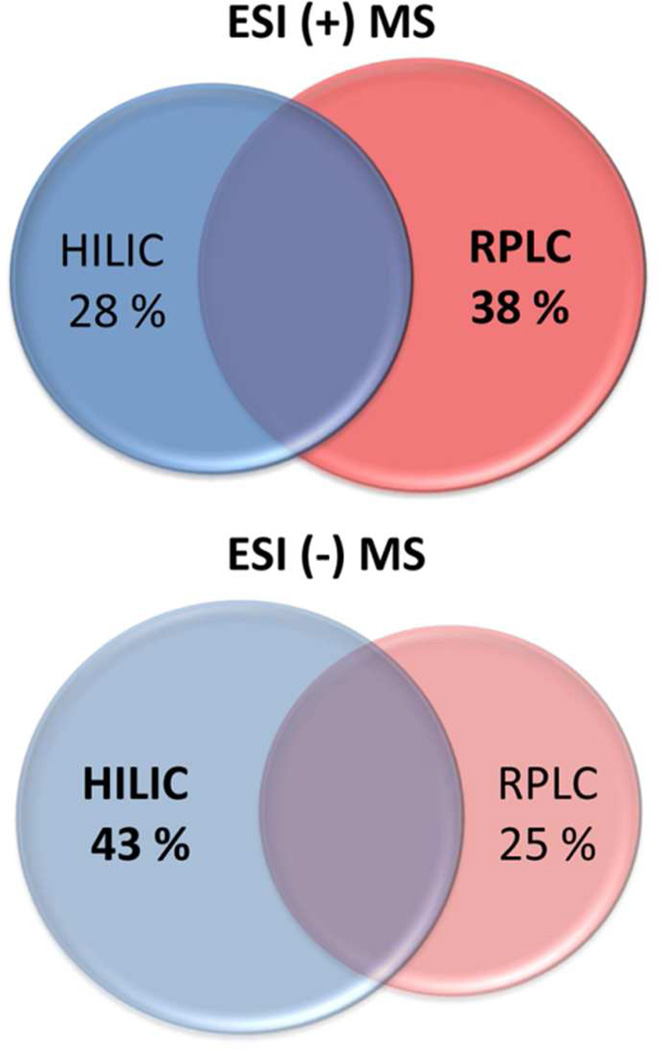
The metabolome coverage between HILIC and RPLC chromatographic modes was evaluated by using the unique ion features defined by their exact mass (Δ m/z = 0.01 Da) corresponding to the [M+H]+ and/or [M-H]− ions. These analyses revealed that the highest number of unique features was detected by RPLC in ESI positive mode and by HILIC in ESI negative mode, an average of 38 ± 9 % and 43 ± 7 %, respectively (Figure 6). The results were based on the sum of features detected in ESI positive or in ESI negative mode by HILIC and by RPLC mode. They highlight the importance of HILIC/MS profiling and the importance of integrating both chromatographic/ionization modes to get a more complete picture of the metabolome. The single-extraction method allows for the integration of HILIC/MS and RPLC/MS profiling and significantly extends the metabolome coverage, thereby improving untargeted profiling of highly polar, central carbon metabolites. To analyze the diversity of small molecules present in complex biological matrices, here we show the value of combining complementary analytical strategies (e.g., different chromatography/mass spectrometry platforms and modes) without multiplexing extraction methods.
Figure 6.
Metabolome coverage by HILIC and RPLC chromatographic mode in ESI positive and ESI negative mode (based on accurate mass only). Features uniquely identified by HILIC and by RPLC as well as the overlap were estimated as a percentage of the total number of metabolite features identified in ESI positive and in ESI negative mode, in three different types of samples (Table 1). The overlap between HILIC and RPLC is estimated at 34 ± 10 % and 32 ± 6 % in ESI positive mode and ESI negative mode, respectively.
Based on these results, it is suggested that when time and sample amount are limited for comprehensive analyses, performing RPLC ESI positive and HILIC ESI negative mode analysis provides the maximum amount of biological information related to lipid and central carbon metabolism (Figure S2). The choice of RPLC positive ionization mode and HILIC negative ionization mode analysis are supported by the number of unique assessed features largely representing positively charged lipid metabolites in RPLC and negatively charged intermediates and end products of central carbon pathways in HILIC.
CONCLUSIONS
The chemical heterogeneity of metabolites presents a significant challenge for obtaining comprehensive coverage of the metabolome. Here we introduce a single extraction –dual LC/MS approach combining RPLC/HILIC/MS to facilitate the accurate measurement of hydrophobic, lipid, hydrophilic, and central carbon metabolites in untargeted metabolic profiling. For this purpose, an aminopropyl-based HILIC/MS method was developed for global-scale analyses. Besides offering good resolution of highly hydrophilic metabolites, the optimized HILIC/MS method demonstrated high reproducibility with minimal retention time deviations over more than 60 hours of analysis, which is essential for interpretation of untargeted results. The value of the approach was highlighted by the analysis of bacterial cell, human cancer cell, and human plasma metabolomes where the combined HILIC/RPLC/MS approach generated over 30,000 metabolite features in each biological sample, with the highest number of unique features identified by RPLC in ESI positive mode and by HILIC in ESI negative mode.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the California Institute of Regenerative Medicine no. TR1-01219 (G.S.), the US National Institutes of Health nos. R01 CA170737 (G.S.), R24 EY017540 (G.S.), P30 MH062261 (G.S.), RC1 HL101034(G.S.), P01 DA026146 (G.S.), and 1R01 ES022181-01 (G.J.P.) and the US National Institutes of Health-National Institute on Aging no. L30 AG0 038036 (G.J.P.). Financial support was also received from the US Department of Energy grant nos. FG02-07ER64325 and DE-AC0205CH11231 (G.S.).
Footnotes
Supporting Information. Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Patti GJ, Yanes O, Siuzdak G. Nat. Rev. Mol. Cell Biol. 2012;13:263–269. doi: 10.1038/nrm3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weckwerth W. Anal. Bioanal. Chem. 2011;400:1967–1978. doi: 10.1007/s00216-011-4948-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker M. Nat. Methods. 2011;8:117–121. doi: 10.1038/nmeth.1722. [DOI] [PubMed] [Google Scholar]
- 4.Patti GJ. J. Sep. Sci. 2011;34:3460–9. doi: 10.1002/jssc.201100532. [DOI] [PubMed] [Google Scholar]
- 5.Bajad SU, Lu W, Kimball EH, Yuan J, Peterson C, Rabinowitz JD. J. of Chromatography A. 2006;1125:76–88. doi: 10.1016/j.chroma.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 6.Yanes O, Tautenhahn R, Patti GJ, Siuzdak G. Anal. Chem. 2011;83:2152–2161. doi: 10.1021/ac102981k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McGarry JD. Science. 1992;258:766–70. doi: 10.1126/science.1439783. [DOI] [PubMed] [Google Scholar]
- 8.Su X, Han X, Mancuso DJ, Abendschein DR, Gross RW. Biochemistry (Mosc.) 2005;44:5234–45. doi: 10.1021/bi047773a. [DOI] [PubMed] [Google Scholar]
- 9.Lu W, Bennett BD, Rabinowitz JD. J. Chromatogr. B. 2008;871:236–242. doi: 10.1016/j.jchromb.2008.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X, Clausen MR, Zhao X, Zheng H, Bertram HC. Anal. Chem. 2012;84:7785–7792. doi: 10.1021/ac3013835. [DOI] [PubMed] [Google Scholar]
- 11.Tautenhahn R, Cho K, Uritboonthai W, Zhu Z, Patti GJ, Siuzdak G. Nat. Biotechnol. 2012;30:826–828. doi: 10.1038/nbt.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rabinowitz JD, Kimball E. Anal. Chem. 2007;79:6167–6173. doi: 10.1021/ac070470c. [DOI] [PubMed] [Google Scholar]
- 14.Álvarez-Sánchez B, Priego-Capote F, Castro MDLd. Trends Anal. Chem. 2010;29:120–127. [Google Scholar]
- 15.Saric J, Want EJ, Duthaler U, Lewis M, Keiser J, Shockcor JP, Ross GA, Nicholson JK, Holmes E, Tavares MF. Anal. Chem. 2012;84:6963–72. doi: 10.1021/ac300586m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tulipani S, Llorach R, Urpi-Sarda M, Andres-Lacueva C. Anal. Chem. 2012;85:341–348. doi: 10.1021/ac302919t. [DOI] [PubMed] [Google Scholar]
- 17.Vuckovic D. Anal. Bioanal. Chem. 2012;403:1523–48. doi: 10.1007/s00216-012-6039-y. [DOI] [PubMed] [Google Scholar]
- 18.Spagou K, Tsoukali H, Raikos N, Gika H, Wilson ID, Theodoridis G. J. Sep. Sci. 2010;33:716–727. doi: 10.1002/jssc.200900803. [DOI] [PubMed] [Google Scholar]
- 19.Buszewski B, Noga S. Anal. Bioanal. Chem. 2012;402:231–47. doi: 10.1007/s00216-011-5308-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vander Heiden MG, Locasale JW, Swanson KD, Sharfi H, Heffron GJ, Amador-Noguez D, Christofk HR, Wagner G, Rabinowitz JD, Asara JM, Cantley LC. Science. 2010;329:1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang J-k, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiefer P, Portais J-C, Vorholt JA. Anal. Biochem. 2008;382:94–100. doi: 10.1016/j.ab.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Baran R, Bowen BP, Price MN, Arkin AP, Deutschbauer AM, Northen TR. ACS Chem. Biol. 2012;8:189–199. doi: 10.1021/cb300477w. [DOI] [PubMed] [Google Scholar]
- 24.Yi Caroline H, Pan H, Seebacher J, Jang I-H, Hyberts Sven G, Heffron Gregory J, Vander Heiden Matthew G, Yang R, Li F, Locasale Jason W, Sharfi H, Zhai B, Rodriguez-Mias R, Luithardt H, Cantley Lewis C, Daley George Q, Asara John M, Gygi Steven P, Wagner G, Liu C-F, Yuan J. Cell. 2011;146:607–620. doi: 10.1016/j.cell.2011.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Locasale J, Melman T, Song S, Yang X, Swanson K, Cantley L, Wong E, Asara J. Mol. Cell. Proteomics. 2012 doi: 10.1074/mcp.M111.014688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Min Y, Susanne BB, Xuemei Y, John MA. Nat. Protoc. 2012;7:872–881. [Google Scholar]
- 27.Cubbon S, Antonio C, Wilson J, Thomas-Oates J. Mass Spectrom. Rev. 2010;29:671–684. doi: 10.1002/mas.20252. [DOI] [PubMed] [Google Scholar]
- 28.Spagou K, Wilson ID, Masson P, Theodoridis G, Raikos N, Coen M, Holmes E, Lindon JC, Plumb RS, Nicholson JK, Want EJ. Anal. Chem. 2010;83:382–390. doi: 10.1021/ac102523q. [DOI] [PubMed] [Google Scholar]
- 29.Kind T, Tolstikov V, Fiehn O, Weiss RH. Anal. Biochem. 2007;363:185–195. doi: 10.1016/j.ab.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 30.Kloos DP, Lingeman H, Niessen WM, Deelder AM, Giera M, Mayboroda OA. J. Chromatogr. B. 2013 doi: 10.1016/j.jchromb.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Biochemistry (Mosc.) 2004;43:14332–14339. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 32.de Carvalho LPS, Zhao H, Dickinson CE, Arango NM, Lima CD, Fischer SM, Ouerfelli O, Nathan C, Rhee KY. Chem. Biol. 2010;17:323–332. doi: 10.1016/j.chembiol.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Gianoulis TA, Yip KY, Gerstein M, Snyder M. Cell. 2010;143:639–650. doi: 10.1016/j.cell.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patti GJ, Yanes O, Shriver LP, Courade J-P, Tautenhahn R, Manchester M, Siuzdak G. Nat. Chem. Biol. 2012;8:232–234. doi: 10.1038/nchembio.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang T, Creek DJ, Barrett MP, Blackburn G, Watson DG. Anal. Chem. 2012;84:1994–2001. doi: 10.1021/ac2030738. [DOI] [PubMed] [Google Scholar]
- 37.Masson P, Alves AC, Ebbels TMD, Nicholson JK, Want EJ. Anal. Chem. 2010;82:7779–7786. doi: 10.1021/ac101722e. [DOI] [PubMed] [Google Scholar]
- 38.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. Anal. Chem. 2006;78:779–87. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 39.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patti GJ, Tautenhahn R, Siuzdak G. Nat. Protocols. 2012;7:508–516. doi: 10.1038/nprot.2011.454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prince JT, Marcotte EM. Anal. Chem. 2006;78:6140–6152. doi: 10.1021/ac0605344. [DOI] [PubMed] [Google Scholar]
- 42.Kostiainen R, Kauppila TJ. J. Chromatogr. A. 2009;1216:685–699. doi: 10.1016/j.chroma.2008.08.095. [DOI] [PubMed] [Google Scholar]
- 43.Guo Y, Gaiki S. J. Chromatogr. A. 2011;1218:5920–5938. doi: 10.1016/j.chroma.2011.06.052. [DOI] [PubMed] [Google Scholar]
- 44.Kuhl C, Tautenhahn R, Böttcher C, Larson TR, Neumann S. Anal. Chem. 2011;84:283–289. doi: 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Want EJ, Masson P, Michopoulos F, Wilson ID, Theodoridis G, Plumb RS, Shockcor J, Loftus N, Holmes E, Nicholson JK. Nat. Protocols. 2013;8:17–32. doi: 10.1038/nprot.2012.135. [DOI] [PubMed] [Google Scholar]
- 46.Gupta KJ, Shah JK, Brotman Y, Jahnke K, Willmitzer L, Kaiser WM, Bauwe H, Igamberdiev AU. J. Exp. Bot. 2012;63:1773–84. doi: 10.1093/jxb/ers053. [DOI] [PubMed] [Google Scholar]
- 47.Gardner PR, Costantino G, Szabó C, Salzman AL. J. Biol. Chem. 1997;272:25071–25076. doi: 10.1074/jbc.272.40.25071. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.