Abstract
Noroviruses (NoVs) are increasingly being recognized as an important enteric pathogen. We investigated a nosocomial NoV outbreak at a university-based hospital that was originally attributed to Clostridium difficile infection (CDI). We describe the unique challenges and the important lessons learned in the identification of noroviruses as the true etiologic pathogen in an outbreak healthcare setting, where CDI is endemic.
Keywords: norovirus, Clostridium difficile, nosocomial diarrhea
Introduction
Noroviruses are the most common cause of foodborne diseases in the United States1, 2 and cause approximately 23 million cases of gastroenteritis each year in the US.3 With its low infectious dose, environmental stability, and resistance to most cleaning agents, NoVs are frequently associated with epidemic outbreaks.4, 5 Although a self-limited illness, NoV gastroenteritis causes a tremendous public health burden, leading to temporary closures of affected institutions, overwhelming of capabilities to manage infected persons, and substantial financial loss.6, 7, 8 We describe a challenging outbreak originally attributed to Clostridium difficile and the important lessons learned from this outbreak.
Methods
Setting
The Michael E. DeBakey Veterans Affairs Medical Center is a 357-bed university-affiliated teaching hospital in Houston, Texas. Psychiatry unit 1 has 22 patient beds, and adjacent psychiatry unit 2 has 28 beds.
Outbreak Investigation
We were asked to investigate a C. difficile (CD) outbreak that occurred March 8–24, 2008. Initial outbreak stool samples collected tested positive for CD toxin by ELISA (Premier C. Difficile Toxin A+B, Meridian Bioscience). However, due to the rapid dissemination of disease, failure to identify CDI in subsequent cases, lack of response to conventional CD therapy, and history of a previous NoV outbreak in psychiatry unit 1 in 2004, an investigation for NoV infection was also performed. Cases were defined as patients or healthcare workers (HCWs) in contact with the psychiatry or adjacent units or existing cases and suffering acute gastroenteritis manifesting as vomiting or diarrhea through March 31, 2008. Diarrhea was considered as ≥ 2 loose stools within a 24-hour period. A standardized questionnaire was used to collect information from cases including demographics, symptom characteristics, and epidemiological risk factors.
Microbiologic Evaluation
C. difficile studies
Stool samples were tested for CD toxins A and B by an ELISA assay (Premier C. Difficile Toxin A+B, Meridian Bioscience). All available stool samples were also cultured on cycloserine-cefoxitin-fructose agar (CCFA) for CD after alcohol shock treatment with 100% ethanol to kill vegetative cells. CD isolates were further characterized by testing for 16S rRNA, tcdA (toxin A), tcdB (toxin B), tcdC (putative negative regulator of toxin A&B production), cdtA and cdtB genes (binary toxin) as described previously.9 Evaluation for the presence of tcdC gene deletions, associated with the “epidemic” BI/NAP1/027 strains was performed using Microsatellites DNA technology.10, 11
NoV studies
Stool samples from prospective cases were sent to the Center for Infectious Diseases (CID) Laboratory at the University of Texas-Houston School of Public Health and to the Houston City Health Department for evaluation for noroviruses. The CID laboratory performed conventional reverse transcriptase polymerase chain reaction (RT-PCR) as previously described for NoV detection,10 while the City Health Department laboratory performed real-time RT-PCR. Sequence analysis of NoV-positive specimens was performed by previously described methods, with nucleotide sequences compared with sequences of prototype strains to determine the genotype.12
Environmental Investigation
Environmental sampling of the two psychiatry units was performed with sterile cotton swabs moistened in Luria-Bertani (LB) broth to evaluate for CD contamination. Surface swabs of toilets and other surfaces commonly contaminated with CD such as sinks, bathroom railings, bed railings, door knobs, window sills, floors, and chairs of case patients’ rooms and common living areas were collected.13, 14, 15 The swabs were cultured on CCFA for CD after alcohol shock treatment.
Results
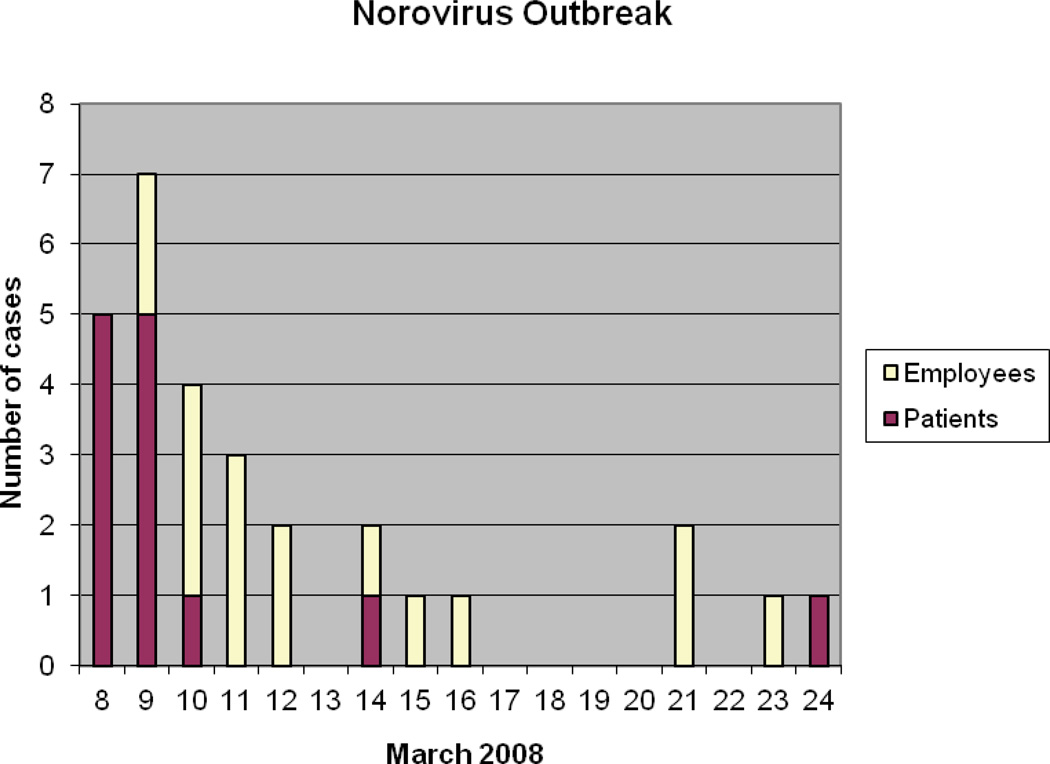
From March 8 – 24, 2008, 13 patients and 16 hospital employees met the case definition (Figure). Initially, there was confusion concerning the etiologic agent of the outbreak because 5 employee stools tested positive for CD toxin. However, the subsequent 12 case stools tested revealed no evidence of CDI. Stool specimens from the remaining 12 cases were unavailable for testing. At least one case with presumed CDI was given metronidazole without clinical response, but persisted to have nausea, vomiting, and diarrhea.
Figure.
Epidemic Curve of Norovirus Outbreak
Three of the 5 CD-positive stools eventually were shown to have NoV RNA. Five of 5 patient stool specimens and 7 of 12 employee stools tested revealed NoV gastroenteritis. Results from the City Health Department and our CID laboratory were completely consistent. The NoV genotype was determined for 4 of the 12 NoV strains detected in this outbreak. The four NoV strains had capsid nucleotide sequences 100% identical to the GII.4 NoV Hu/GII-4/C5-159/South Korea strain (Accession number EU003965).
Secondary transmission among the patients was believed to be facilitated by the close quarters, shared bathrooms and living areas, and encouragement to actively socialize with other patients. All ill HCWs either worked on the affected wards or interacted with the case patients. Once the outbreak on psychiatry unit 1 was recognized, new admissions were halted, but patients on this unit were allowed to have access to the adjacent psychiatry unit 2 for group therapy, where potentially four employees became infected.
Interestingly, CD was isolated on CCFA under anaerobic conditions from one CD toxin-positive employee stool and 2 CD toxin-negative patient stools. C. difficile 16S rRNA genes were detected in all 3 isolates. The presence of the tcdA, tcdB, and tcdC genes were demonstrated in only 1 patient CD isolate, but the stool tested negative by ELISA for toxin A and B production. The hypervirulent CD strain was not detected, as there was no evidence of the tcdC deletion or binary toxin production. Both patients with CD were infected with NoVs.
Environmental sampling for contamination of the psychiatric units with CD spores yielded no CD isolates. It should be noted, however, that cleaning and disinfection measures had already been initiated before sampling of these areas could be performed.
Rapid implementation of infection-control measures by the VA Hospital Infection Control team limited the extent and led to early termination of this outbreak. These interventions included closure of the psychiatry units to new admissions, instruction of ill HCWs to not report to work until at least 72 hours after symptom resolution, and careful surveillance for new exposures and cases. Disinfection of patients’ rooms involved first cleaning with an all-purpose cleaner (35% 2-butoxyethanol and 5–20% nonionic surfactant), followed by the application of a bleach-impregnated disposable cloth to environmental surfaces three times daily. Strict hand hygiene practice with soap and water was reinforced and monitored by Infection Control staff.
Discussion
During outbreaks of acute gastroenteritis at healthcare facilities, where patients are at risk for C. difficile infection, detection of CD can create confusion regarding the causative agent. Increased collection and testing of stool samples for CD, in addition to false positives associated with CD assays, may be reflected by increased rates of CD detection.16, 17,18 In this NoV outbreak, testing individuals who lacked traditional risk factors for CDI, such as employees, likely led to CD false positives. However, there was no evidence of increased CD testing during this outbreak compared to previous and following months. Asymptomatic CD colonization also contributed to the confusion in this outbreak hospital setting, where CD is endemic. Interactions between distinct enteric pathogens such as C. difficile and noroviruses have not been studied, and it is unknown whether CDI may somehow augment the pathogenesis of NoV infections, or vice versa. Distinguishing the true enteric pathogen responsible for an outbreak has important therapeutic implications, such as supportive treatment for NoVs and antibiotics for CD.
We believe that coincidental CD colonization and false positives were detected in this NoV outbreak due to fecal sampling to detect potential cases and a heightened awareness of diarrheal cases. Five of 7 cases with CD, detected by either ELISA or stool culture, were shown to be infected with NoVs. At least one of these cases with both enteropathogens was given appropriate antimicrobial therapy for C. difficile, but failed to improve clinically.
Two clusters of cases are apparent from the epidemic curve, with the second cluster occurring 5 days after the 25th case. Although the incubation period for NoV infection is typically 24 – 48 hours, NoV persistence in fecal shedding19 and on environmental surfaces20 may have led to the second group of cases. Prolonged NoV fecal shedding up to several weeks after infection has been well-described.19 Infected HCWs were allowed to return to work 72 hours after symptom resolution, in accordance with CDC recommendations.21 These recommendations regarding the resumption of duties by HCWs recovering from acute NoV gastroenteritis may need to be reevaluated in the future.
As recognition of NoVs as a cause of acute gastroenteritis increases with improvements and increased accessibility to molecular diagnostic testing, we expect the importance of this enteric pathogen to continue to become more apparent. A better understanding of the significance of detection of multiple potential pathogens, such as NoVs and C. difficile, in a single outbreak is critical to improve implementation of the optimal therapeutic and preventive strategies for patients.
Acknowledgments
Financial Support: National Institutes of Health (P01-AI-057788 to RLA).
Footnotes
Presented at the 48th Annual Interscience Conference on Antimicrobial Agents and Chemotherapy/Infectious Diseases Society of America 46th Annual Meeting, Washington, D.C., October 26, 2008.
References
- 1.Lynch M, Painter J, Woodruff R, et al. Surveillance for foodborne-disease outbreaks--United States, 1998–2002. MMWR Surveill Summ. 2006;55:1–42. [PubMed] [Google Scholar]
- 2.Bresee JS, Widdowson MA, Monroe SS, Glass RI. Foodborne viral gastroenteritis: challenges and opportunities. Clin Infect Dis. 2002;35:748–753. doi: 10.1086/342386. [DOI] [PubMed] [Google Scholar]
- 3.Mead PS, Slutsker L, Dietz V, et al. Food-Related Illness and Death in the United States. Emerg Infect Dis. 1999;5:841–842. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Guyader FS, Atmar RL. Binding and inactivation of viruses on and in food, with a focus on the role of the matrix. In: Koopmans M, Bosch A, Cliver D, editors. Food-borne Viruses: A Multidisciplinary Review of Progress and Challenges in Food-borne Viruses Research. Washington, DC: ASM Press; 2008. in press. [Google Scholar]
- 5.NIAID Biodefense Research. NIAID Category A, B & C Priority Pathogens. [Accessed January 10, 2008]; [online resource]. http://www.niaid.nih.gov/dmid/biodefense/bandc_priority.htm.
- 6.Johnston CP, Qiu H, Ticehurst JR, Dickson C, Rosenbaum P, Lawson P, Stokes AB, Lowenstein CJ, Kaminsky M, Cosgrove SE, Green KY, Perl TM. Outbreak management and implications of a nosocomial norovirus outbreak. Clin Infect Dis. 2007;45:534–540. doi: 10.1086/520666. [DOI] [PubMed] [Google Scholar]
- 7.White C. Scale of norovirus outbreak may have been exaggerated. BMJ. 2008;336:66. doi: 10.1136/bmj.39454.807234.4E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopman BA, Brown DW, Koopmans M. Human caliciviruses in Europe. J Clin Virol. 2002;24:137–160. doi: 10.1016/s1386-6532(01)00243-8. [DOI] [PubMed] [Google Scholar]
- 9.Borriello SP, Honour P. Simplified procedure for the routine isolation of Clostridium difficile from feces. J Clin Pathol. 1981;34:1124–1127. doi: 10.1136/jcp.34.10.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kassai-Jáger E, Ortutay C, Tóth G, Vellai T, Gáspári Z. Distribution and evolution of short tandem repeats in closely related bacterial genomes. Gene. 2008;410:18–25. doi: 10.1016/j.gene.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 11.Guo DC, Milewicz DM. Methodology for using a universal primer to label amplified DNA segments for molecular analysis. Biotechnol Lett. 2003;25:2079–2083. doi: 10.1023/b:bile.0000007075.24434.5e. [DOI] [PubMed] [Google Scholar]
- 12.Green KEA. Caliciviridae: the noroviruses. In: Knipe DEA, editor. Fields Virology. vol 5. Lippincott: Williams & Wilkins; 2007. pp. 949–979. [Google Scholar]
- 13.Verity P, Wilcox MH, Fawley W, Parnell P. Prospective evaluation of environmental contamination by Clostridium difficile in isolation side rooms. J Hosp Infect. 2001;49:204–209. doi: 10.1053/jhin.2001.1078. [DOI] [PubMed] [Google Scholar]
- 14.Wilcox MH, Fawley WN, Wigglesworth N, Parnell P, Verity P, Freeman J. Comparison of the effect of detergent versus hypochlorite cleaning on environmental contamination and incidence of Clostridium difficile infection. J Hosp Infect. 2003;54:109–114. doi: 10.1016/s0195-6701(02)00400-0. [DOI] [PubMed] [Google Scholar]
- 15.Dubberke ER, Reske KA, Noble-Wang J, Thompson A, Killgore G, Mayfield J, Camins B, Woeltje K, McDonald JR, McDonald LC, Fraser VJ. Prevalence of Clostridium difficile environmental contamination and strain variability in multiple health care facilities. Am J Infect Control. 2007;35:315–318. doi: 10.1016/j.ajic.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Martin AJ, Collins CJ, Ruddy R, Drudy D, Hannan MM, Kyne L. Simultaneous control of norovirus and Clostridium difficile outbreaks due to enhanced infection prevention and control measures. J Hosp Infect. 2008;68:180–181. doi: 10.1016/j.jhin.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 17.Bignardi GE, Staples K, Majmudar N. A case of norovirus and Clostridium difficile infection: casual or causal relationship? J Hosp Infect. 2007;67:198–200. doi: 10.1016/j.jhin.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Barrett SP, Holmes AH, Newsholme WA, Richards M. Increased detection of Clostridium difficile during a norovirus outbreak. J Hosp Infect. 2007;66:394–395. doi: 10.1016/j.jhin.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 19.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Graham DY. Norwalk virus shedding after experimental human infection. Emerg Infect Dis. 2008;14:1553–1557. doi: 10.3201/eid1410.080117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D'Souza DH, Sair A, Williams K, Papafragkou E, Jean J, Moore C, Jaykus L. Persistence of caliciviruses on environmental surfaces and their transfer to food. Int J Food Microbiol. 2006;108:84–91. doi: 10.1016/j.ijfoodmicro.2005.10.024. [DOI] [PubMed] [Google Scholar]
- 21.LeBaron CW, Furutan NP, Lew JF, Allen JR, Gouvea V, Moe C, Monroe SS. Viral agents of gastroenteritis. Public health importance and outbreak management. MMWR Recomm Rep. 1990;39(RR-5):1–24. [PubMed] [Google Scholar]