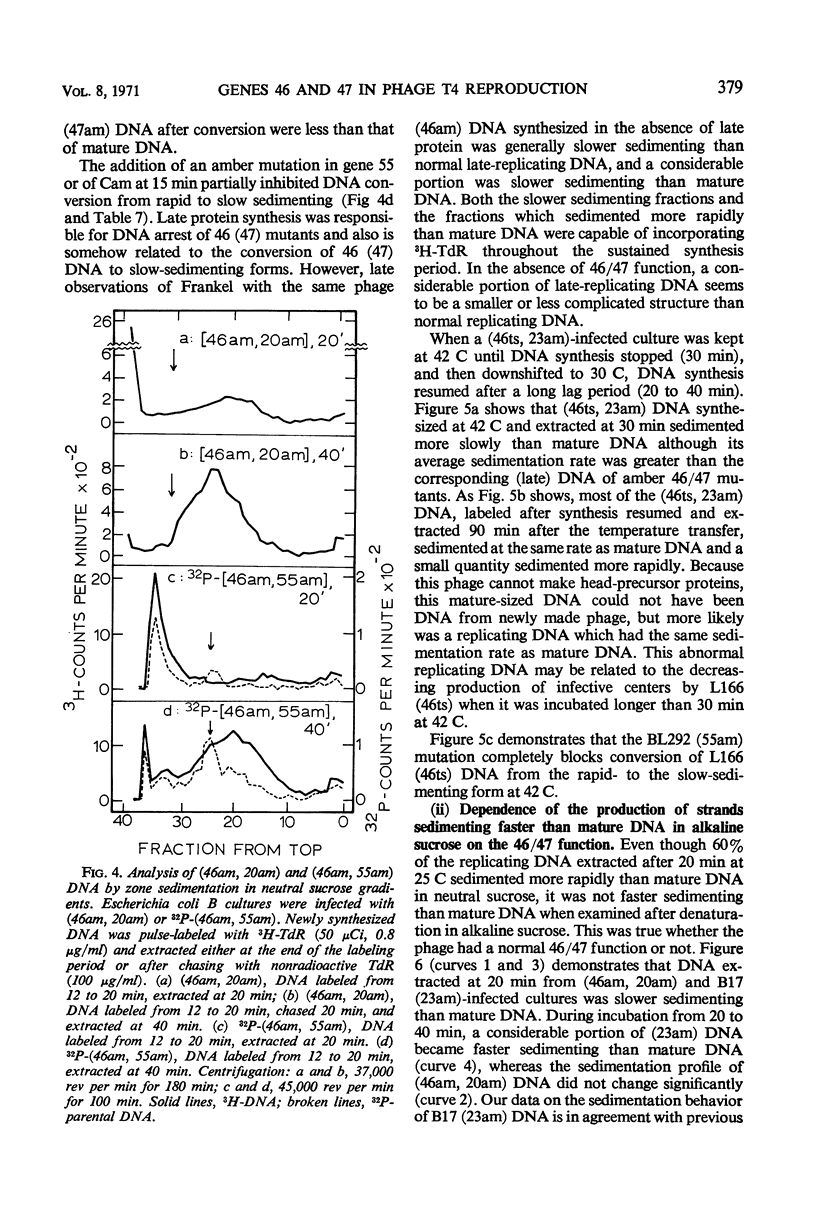
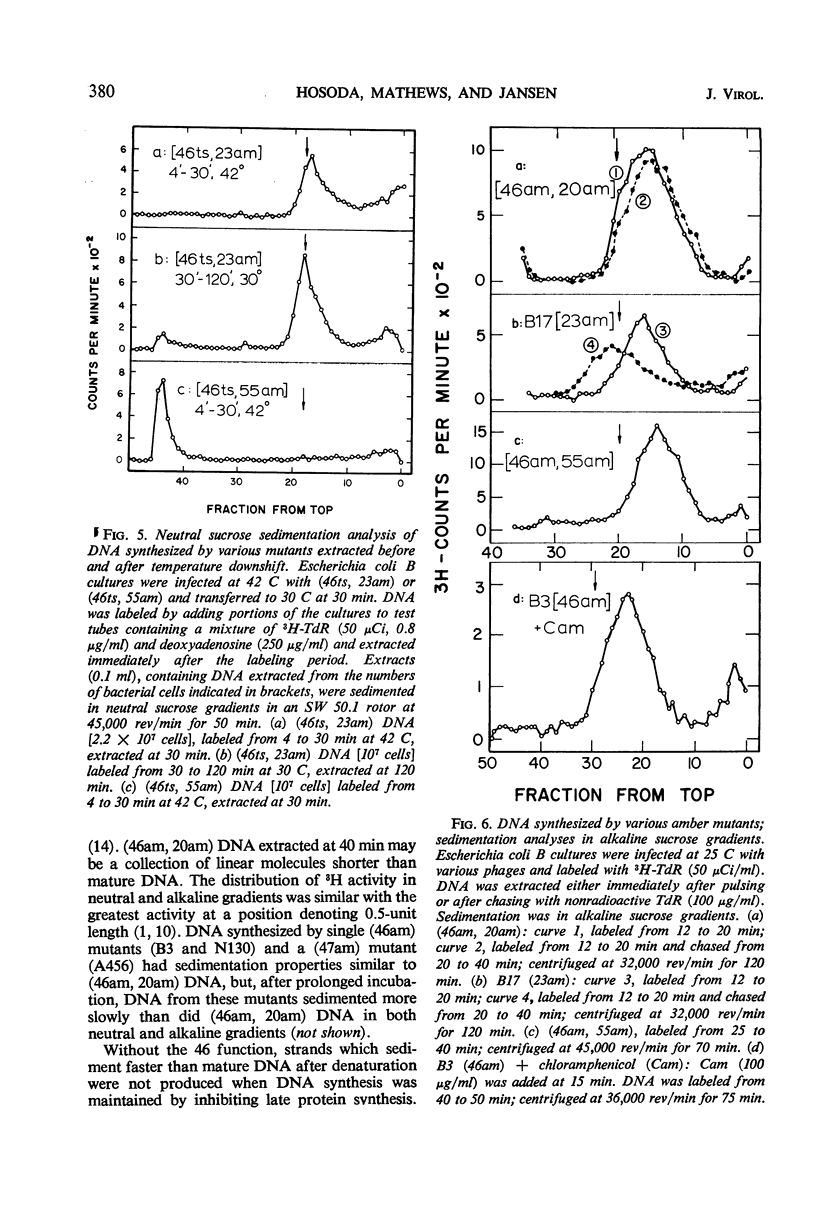
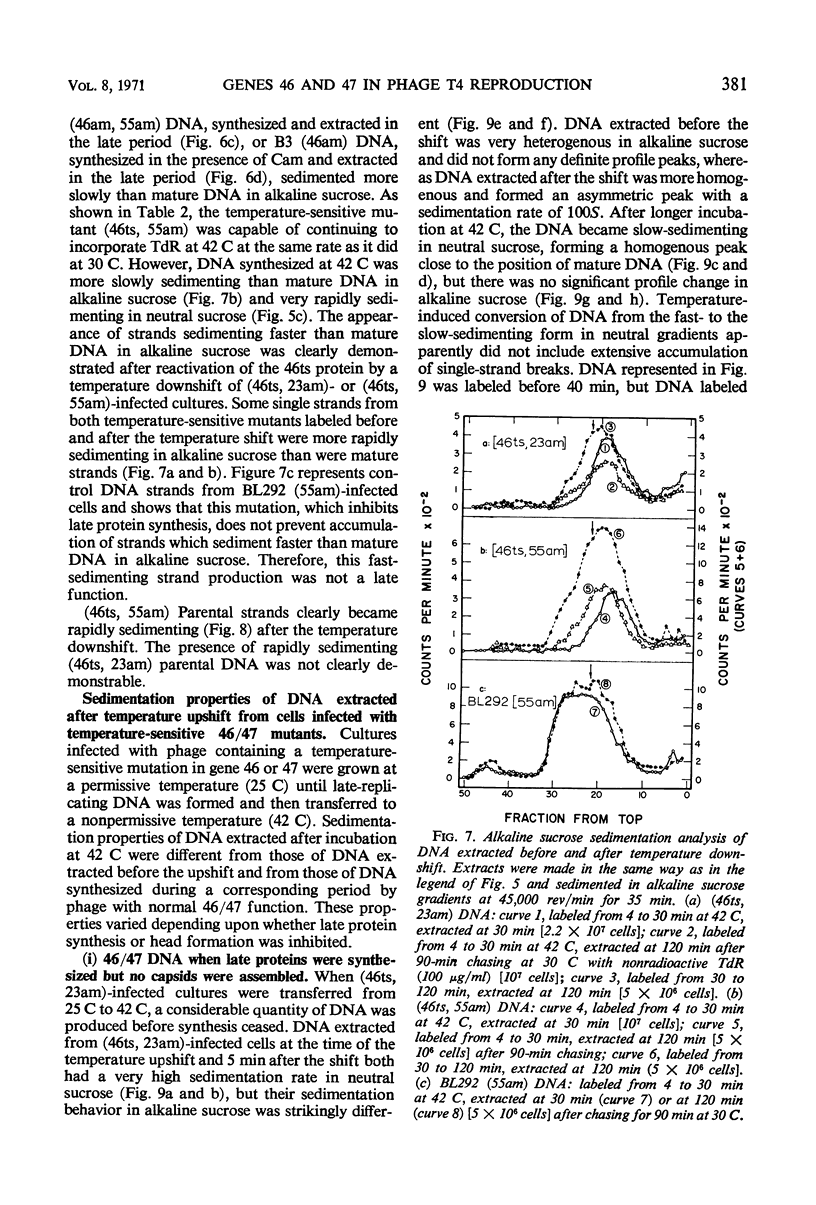
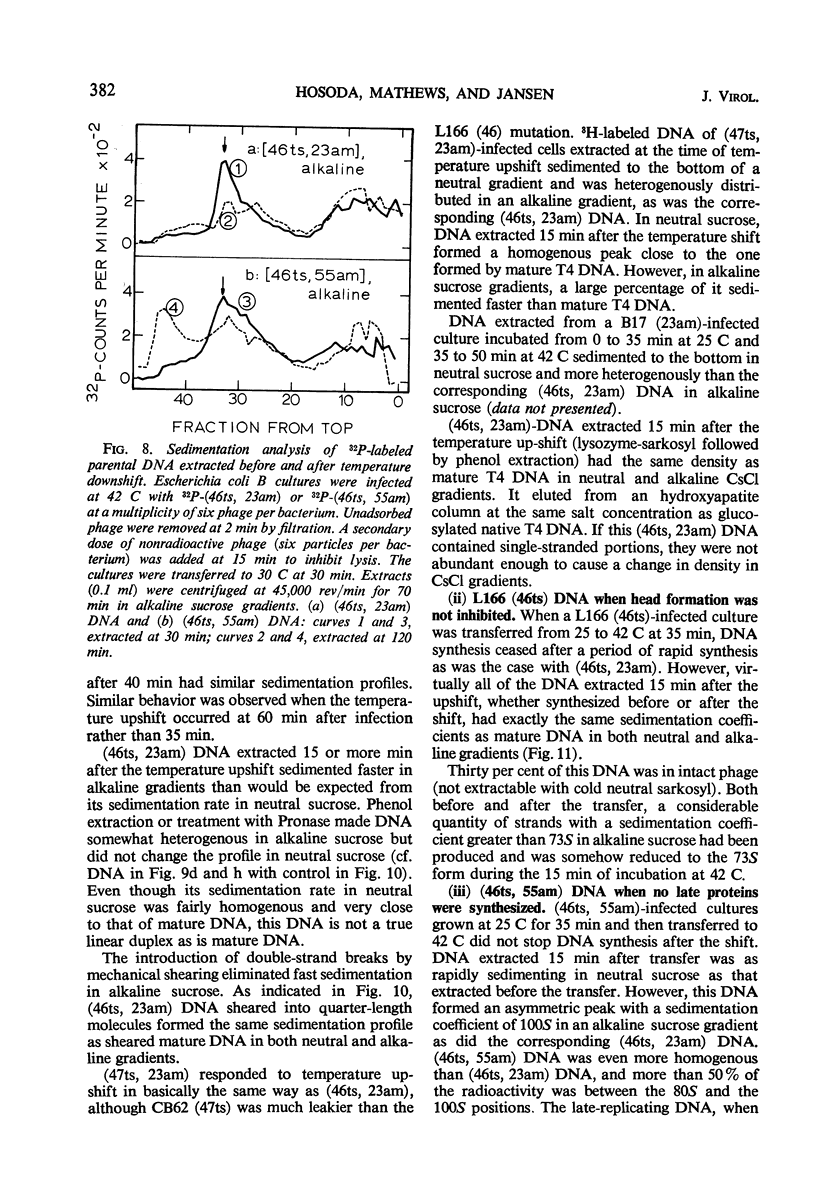
Abstract
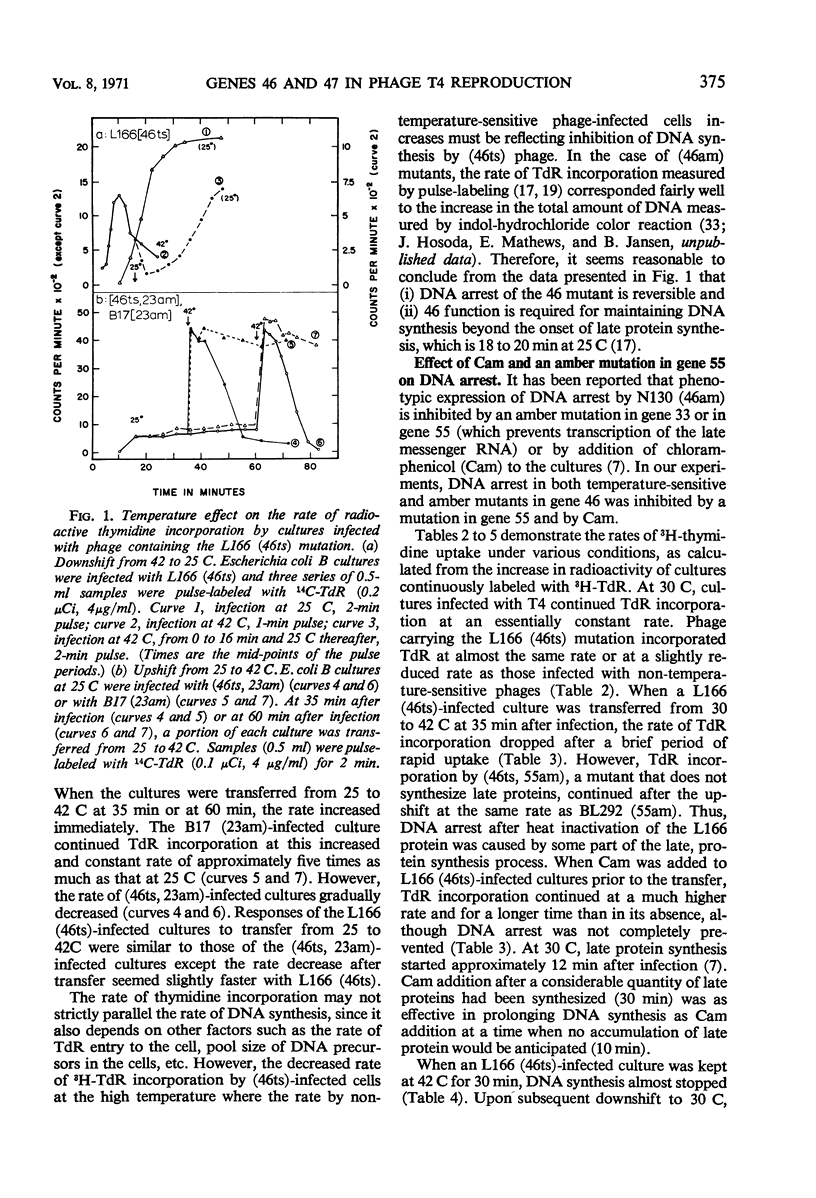
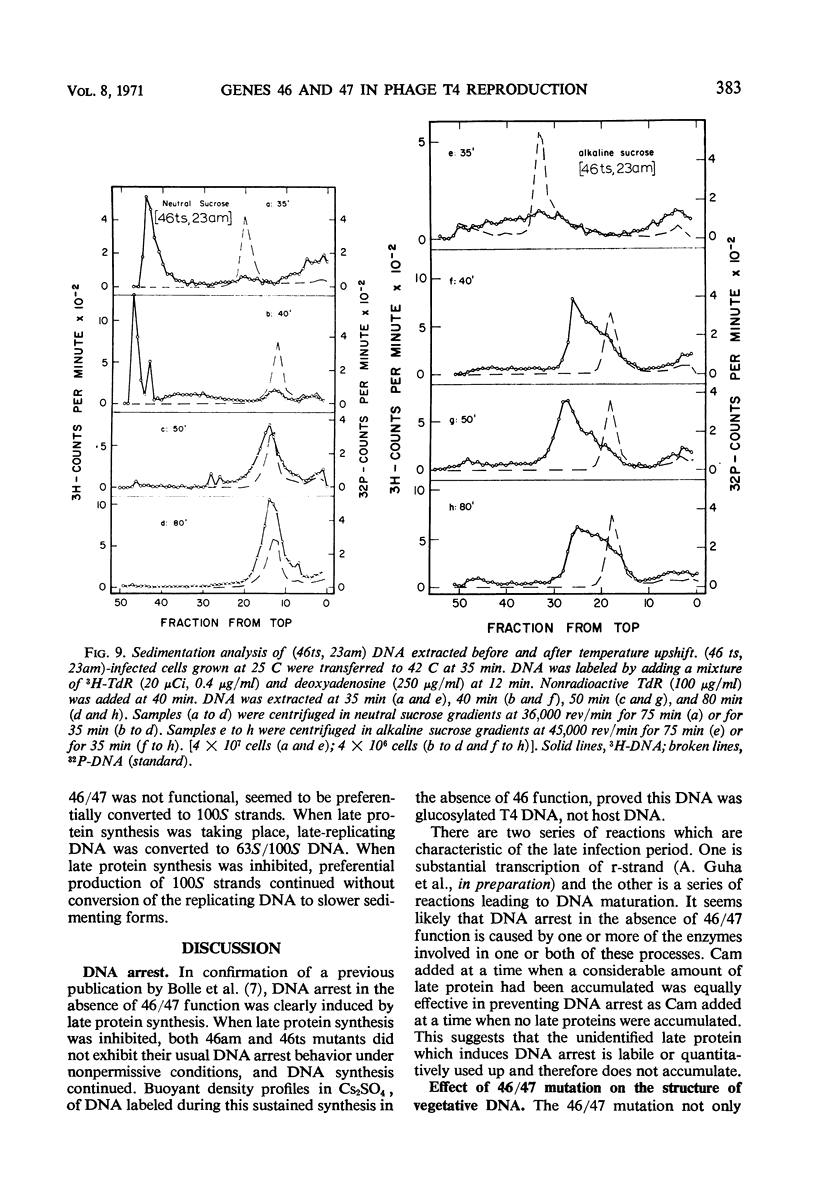
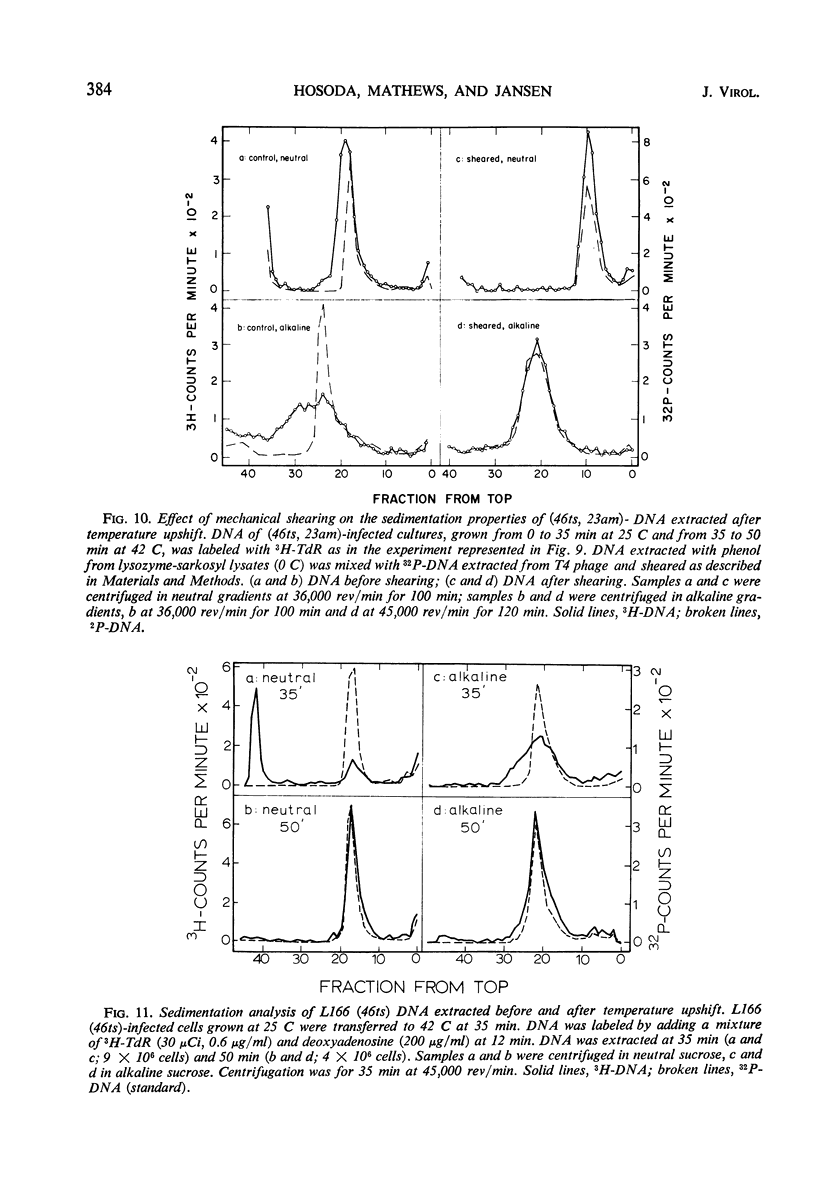
Functional proteins coded by genes 46 and 47 are required for (i) continuation of deoxyribonucleic acid (DNA) synthesis in the late period of T4 infection and (ii) production of normal, late replicating DNA which contains strands with a sedimentation coefficient in alkaline sucrose greater than that of mature DNA (73S). Continued DNA synthesis in the late period in the absence of functional genes 46 or 47 can be achieved by inhibiting late protein synthesis either by using bacterio-phage with a second mutation in gene 55 or by adding chloramphenicol to the culture before the decline in the rate of DNA synthesis. However, when functional 46/47 proteins are absent throughout infection, no strands with a sedimentation coefficient greater than 73S (in alkaline sucrose) are produced. This is the case even when DNA synthesis is allowed to continue. DNA arrest is accompanied by conversion of rapidly sedimenting, replicating DNA to slower sedimenting forms. When 46/47 is absent from the beginning of infection, the conversion product has a smaller sedimentation coefficient than mature DNA both in neutral and alkaline sucrose. When DNA arrest occurs midway in infection by heat-inactivating the ts46 enzyme, the conversion product has a sedimentation coefficient (i) the same as mature DNA in both neutral (63S) and alkaline sucrose if capsid assembly is allowed to take place and (ii) close to 63S in neutral sucrose but heterogenous and relatively greater (up to 100S) in alkaline sucrose if capsid assembly is inhibited. The structure of this DNA is unknown.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anraku N., Anraku Y., Lehman I. R. Enzymic joining of polynucleotides. 8. Structure of hybrids of parental T4 DNA molecules. J Mol Biol. 1969 Dec 28;46(3):481–492. doi: 10.1016/0022-2836(69)90191-0. [DOI] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. A relative molecular weight series derived from the nucleic acid of bacteriophage T2. J Mol Biol. 1961 Aug;3:458–472. doi: 10.1016/s0022-2836(61)80058-2. [DOI] [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylor M B, Hessler A Y, Baird J P. The Circular Linkage Map of Bacteriophage T2h. Genetics. 1965 Mar;51(3):351–361. doi: 10.1093/genetics/51.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger H., Warren A. J., Fry K. E. Variations in genetic recombination due to amber mutations in T4D bacteriophage. J Virol. 1969 Feb;3(2):171–175. doi: 10.1128/jvi.3.2.171-175.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H. Repair and recombination in phage T4. I. Genes affecting recombination. Cold Spring Harb Symp Quant Biol. 1968;33:325–331. doi: 10.1101/sqb.1968.033.01.037. [DOI] [PubMed] [Google Scholar]
- Bode V. C., Kaiser A. D. Changes in the structure and activity of lambda DNA in a superinfected immune bacterium. J Mol Biol. 1965 Dec;14(2):399–417. doi: 10.1016/s0022-2836(65)80190-5. [DOI] [PubMed] [Google Scholar]
- Bolle A., Epstein R. H., Salser W., Geiduschek E. P. Transcription during bacteriophage T4 development: requirements for late messenger synthesis. J Mol Biol. 1968 Apr 28;33(2):339–362. doi: 10.1016/0022-2836(68)90193-9. [DOI] [PubMed] [Google Scholar]
- Bose S. K., Warren R. J. Bacteriophage-induced inhibition of host functions. II. Evidence for multiple, sequential bacteriophage-induced deoxyribonucleases responsible for degradation of cellular deoxyribonucleic acid. J Virol. 1969 Jun;3(6):549–556. doi: 10.1128/jvi.3.6.549-556.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel F. R. DNA replication after T4 infection. Cold Spring Harb Symp Quant Biol. 1968;33:485–493. doi: 10.1101/sqb.1968.033.01.056. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. Evidence for long DNA strands in the replicating pool after T4 infection. Proc Natl Acad Sci U S A. 1968 Jan;59(1):131–138. doi: 10.1073/pnas.59.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel F. R. Studies on the nature of replicating DNA in T4-infected Escherichia coli. J Mol Biol. 1966 Jun;18(1):127–143. doi: 10.1016/s0022-2836(66)80081-5. [DOI] [PubMed] [Google Scholar]
- Hickson F. T., Roth T. F., Helinski D. R. Circular DNA forms of a bacterial sex factor. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1731–1738. doi: 10.1073/pnas.58.4.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda J., Levinthal C. Protein synthesis by Escherichia coli infected with bacteriophage T4D. Virology. 1968 Apr;34(4):709–727. doi: 10.1016/0042-6822(68)90092-5. [DOI] [PubMed] [Google Scholar]
- Hosoda J., Mathews E. DNA replication in vivo by a temperature-sensitive polynucleotide ligase mutant of T4. Proc Natl Acad Sci U S A. 1968 Nov;61(3):997–1004. doi: 10.1073/pnas.61.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoda J., Mathews E. DNA replication in vivo by polynucleotide-ligase defective mutants of T4. II. Effect of chloramphenicol and mutations in other genes. J Mol Biol. 1971 Jan 28;55(2):155–179. doi: 10.1016/0022-2836(71)90189-6. [DOI] [PubMed] [Google Scholar]
- Huberman J. A. Visualization of replicating mammalian and T4 bacteriophage DNA. Cold Spring Harb Symp Quant Biol. 1968;33:509–524. doi: 10.1101/sqb.1968.033.01.059. [DOI] [PubMed] [Google Scholar]
- King J. Assembly of the tail of bacteriophage T4. J Mol Biol. 1968 Mar 14;32(2):231–262. doi: 10.1016/0022-2836(68)90007-7. [DOI] [PubMed] [Google Scholar]
- Kozinski A. W., Kozinski P. B., James R. Molecular recombination in T4 bacteriophage deoxyribonucleic acid. I. Tertiary structure of early replicative and recombining deoxyribonucleic acid. J Virol. 1967 Aug;1(4):758–770. doi: 10.1128/jvi.1.4.758-770.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E. M., Wiberg J. S. Degradation of cytosin-containing bacterial and bacteriophage DNA after infection of Escherichia coli B with bacteriophage T4D wild type and with mutants defective in genes 46, 47 and 56. J Mol Biol. 1968 Dec;38(3):395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- MacHattie L. A., Ritchie D. A., Thomas C. A., Jr, Richardson C. C. Terminal repetition in permuted T2 bacteriophage DNA molecules. J Mol Biol. 1967 Feb 14;23(3):355–363. doi: 10.1016/s0022-2836(67)80110-4. [DOI] [PubMed] [Google Scholar]
- Oishi M. Studies of DNA replication in vivo. 3. Accumulation of a single-stranded isolation product of DNA replication by conditional mutant strains of T4. Proc Natl Acad Sci U S A. 1968 Jul;60(3):1000–1006. doi: 10.1073/pnas.60.3.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STREISINGER G., EDGAR R. S., DENHARDT G. H. CHROMOSOME STRUCTURE IN PHAGE T4. I. CIRCULARITY OF THE LINKAGE MAP. Proc Natl Acad Sci U S A. 1964 May;51:775–779. doi: 10.1073/pnas.51.5.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Shah D. B., Berger H. Replication of gene 46-47 amber mutants of bacteriophage T4D. J Mol Biol. 1971 Apr 14;57(1):17–34. doi: 10.1016/0022-2836(71)90117-3. [DOI] [PubMed] [Google Scholar]
- Shalitin C., Kahana S. Conversion of T4 gene 46 mutant deoxyribonucleic acid into nonviable bacteriophage particles. J Virol. 1970 Sep;6(3):353–362. doi: 10.1128/jvi.6.3.353-362.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalitin C., Naot Y. Role of gene 46 in bacteriophage T4 deoxyribonucleic acid synthesis. J Virol. 1971 Aug;8(2):142–153. doi: 10.1128/jvi.8.2.142-153.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G., Emrich J., Stahl M. M. Chromosome structure in phage t4, iii. Terminal redundancy and length determination. Proc Natl Acad Sci U S A. 1967 Feb;57(2):292–295. doi: 10.1073/pnas.57.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THOMAS C. A., Jr, RUBENSTEIN I. THE ARRANGEMENTS OF NUCLEOTIDE SEQUENCES IN T2 AND T5 BACTERIOPHAGE DNA MOLECULES. Biophys J. 1964 Mar;4:93–106. doi: 10.1016/s0006-3495(64)86771-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg J. S. Mutants of bacteriophage T4 unable to cause breakdown of host DNA. Proc Natl Acad Sci U S A. 1966 Mar;55(3):614–621. doi: 10.1073/pnas.55.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]
- Young E. T., 2nd, Sinsheimer R. L. Vegetative bacteriophage lambda-DNA. II. Physical characterization and replication. J Mol Biol. 1967 Nov 28;30(1):165–200. doi: 10.1016/0022-2836(67)90251-3. [DOI] [PubMed] [Google Scholar]


