Abstract
Sex chromosome divergence, which follows the cessation of recombination and degeneration of the sex-limited chromosome, can cause a reduction in expression level for sex-linked genes in the heterozygous sex, unless some mechanisms of dosage compensation develops to counter the reduction in gene dose. Because large-scale perturbations in expression levels arising from changes in gene dose might have strong deleterious effects, the evolutionary response should be strong. However, in birds and in at least some other female heterogametic organisms, wholesale sex chromosome dosage compensation does not seem to occur. Using RNA-seq of multiple tissues and individuals, we investigated male and female expression levels of Z-linked and autosomal genes in the collared flycatcher, a bird for which a draft genome sequence recently has been reported. We found that male expression of Z-linked genes was on average 50% higher than female expression, although there was considerable variation in the male-to-female ratio among genes. The ratio for individual genes was well correlated among tissues and there was also a correlation in the extent of compensation between flycatcher and chicken orthologs. The relative excess of male expression was positively correlated with expression breadth, expression level, and number of interacting proteins (protein connectivity), and negatively correlated with variance in expression. These observations lead to a model of compensation occurring on a gene-by-gene basis, supported by an absence of clustering of genes on the Z chromosome with respect to the extent of compensation. Equal mean expression level of autosomal and Z-linked genes in males, and 50% higher expression of autosomal than Z-linked genes in females, is compatible with that partial compensation is achieved by hypertranscription from females’ single Z chromosome. A comparison with male-to-female expression ratios in orthologous Z-linked genes of ostriches, where Z–W recombination still occurs, suggests that male-biased expression of Z-linked genes is a derived trait after avian sex chromosome divergence.
Keywords: sex chromosomes, dosage compensation, sex-biased gene expression, expression variance, collared flycatcher
Introduction
It is generally considered that monosomy of individual genes can introduce severe problems for the organism due to dose dependence of expression levels and the associated risk for suboptimal interactions of the encoded proteins (i.e., haploinsufficiency [Veitia 2002; Deutschbauer et al. 2005]). When it comes to entire chromosomes, autosomal monosomy is typically lethal, at least in humans (Hassold and Hunt 2001). In birds, which is the taxonomic group in focus in this study, trisomies have been shown to be a source of embryo mortality (Forstmeier and Ellegren 2010), and the rare incidence of monosomies in screenings of early chicken embryos suggest that avian monosomies are effectively lethal (Lodge et al. 1973; Thorne et al. 1991). The deleterious effects of aneuploidy may apply to sex chromosome evolution. When recombination ceases between proto-sex chromosomes, the sex-limited chromosome (Y in organisms with male heterogamety and W in organisms with female heterogamety, like in birds) is bound to face the deleterious consequences associated with lack of recombination. For example, a lowered effective population size makes purifying selection less efficient and the accumulation of deleterious mutations may be further accentuated by the processes of Muller’s ratchet and selective sweeps (Charlesworth B and Charlesworth D 2000). This will lead to degeneration of the nonrecombining chromosome and is readily seen in many organisms by significant loss of genetic material causing chromosomal diminutivization and heterochromatization (Mank 2012). As a consequence, large parts of the recombining sex chromosome (X in organisms with male heterogamety and Z in organisms with female heterogamety) become monosomic in the heterogametic sex. This might result in an evolutionary response in some genes to regulate expression levels in the heterogametic sex in a way that the original stoichiometric balance of protein–protein interactions within biochemical pathways is maintained or re-established. Alternatively, buffering of gene dose might for some genes rather be a passive effect of the regulatory and transcriptional machinery (Birchler et al. 2001; Lundberg et al. 2012; Malone et al. 2012).
Sex chromosome dosage compensation has often been viewed as a means to equalize expression levels of sex-linked genes in the two sexes. However, it was earlier recognized that a more fundamental problem is the need for maintaining a balance (or more specifically, maintaining the original relationship) between expression levels of autosomal genes and sex-linked genes in the hemizygous sex (Ohno 1967). According to this point of view, selection should favor increased expression levels of sex-linked genes. However, unless this increase is sex and chromosome specific, rather than just chromosome specific, it generates the problem of overexpression of sex-linked genes (relative to autosomal genes) in the homogametic sex. This situation would thus introduce a selection pressure as a secondary response for reducing sex-linked gene expression specifically in the homogametic sex, one means of which would be X-chromosome inactivation (see further in Discussion).
In light of the earlier discussion, it came as a surprise when it was reported that chicken does not show a chromosome-wide regulation of Z-linked genes to reach equal expression levels in males and females, or equal levels of autosomal and Z-linked genes in females (Ellegren et al. 2007; Itoh et al. 2007). With sex chromosome dosage compensation previously seen as a norm, why and how can a bird do without? Moreover, the subsequent replication of incomplete dosage compensation in other bird species (Itoh et al. 2010; Wolf and Bryk 2011; Naurin et al. 2012) and in other organism groups with female heterogamety (Zha et al. 2009; Vicoso and Bachtrog 2011; Harrison et al. 2012) might now be taken to question the necessity of balanced expression of sex-linked genes in ZW systems. It has also led to the more general questioning of the ubiquity of dosage compensation across broad organism groups, including male heterogametic organisms (Mank et al. 2011; Julien et al. 2012).
Previous work in chicken has indicated that the mean male-to-female ratio in expression level of Z-linked genes is 1.5–1.7 (Ellegren et al. 2007; Itoh et al. 2007, 2010; Melamed and Arnold 2007; Mank et al. 2008; Mank and Ellegren 2009; Zhang et al. 2010; Julien et al. 2012), with some variation seen among tissues. This ratio is thus intermediate to chromosome wide and complete compensation (a ratio of 1) and a situation with no compensation and expression level solely reflecting gene dose (a ratio of 2), and is open to several possible explanations. These results were based on microarray hybridizations to measure expression levels in male and female chicken (but see Julien et al. 2012; Wright et al. 2012), and a debate has recently arisen about the suitability of microarrays to detect relatively subtle differences in gene expression in studies of dosage compensation (Castagne et al. 2011; Malone and Oliver 2011). In parallel, deep transcriptome sequencing (RNA-seq) has clearly emerged as state-of-the-art for large-scale quantification of gene expression levels (Garber et al. 2011; Malone and Oliver 2011; Ozsolak and Milos 2011) and has therefore been considered superior in studies of dosage compensation.
We have recently sequenced and assembled the 1.1. Gb genome of the collared flycatcher Ficedula albicollis (Ellegren et al. 2012), a songbird species of the order Passeriformes, which is an ecological model species in fields such as life history evolution and speciation research (Gustafsson and Sutherland 1988; Gustafsson and Part 1990; Gustafsson et al. 1995; Ellegren et al. 1996; Saetre et al. 1997; Qvarnström et al. 2000, 2006; Merila et al. 2001; Veen et al. 2001; Saether et al. 2007). As one of few avian genomes yet sequenced, this provides a useful resource for studying many aspects of molecular ecology and genome evolution. Here, we present the results of an extensive RNA-seq study conducted across multiple tissues of flycatcher aimed at addressing the case for dosage compensation in the avian genome. We do this by comparing expression levels of sex-linked and autosomal genes, and of sex-linked genes in male and female birds. We also study how dosage compensation is related to level, breadth, and variance in gene expression, and to protein connectivity.
Materials and Methods
Sample Collection
Ten unrelated adult birds (5 females and 5 males) were collected in 2009 on the Baltic Sea island Öland. Birds were killed by decapitation and immediately dissected in the field. Brain, kidney, liver, lung, muscle, skin, ovary, and testis tissues were isolated and stored in 1 ml of RNAlater (Qiagen) at −80 °C. In addition, eight eggs were collected shortly after laying, and before brooding started, and incubated until the age of seven days when embryos were sampled and stored in buffer. All sampling was conducted according to permissions and rules of the Swedish ethics committee for wild animals (2007/C319 – Uppsala Djurförsöksetiska nämnd).
RNA Extraction and Library Preparation
One hundred milligrams of kidney, muscle, ovary, testis, or embryo tissue was homogenized using TissueRuptor (Qiagen) for 30 s in 5 ml of TRIzol reagent (Invitrogen). Tissues rich in fat (brain, liver, lung, and skin) were homogenized in QIAzol lysis reagent (Qiagen). We used RNeasy kit or RNeasy lipid kit (fat-rich tissues) for total RNA extraction according to the manufacturer’s instructions (Qiagen). All RNA extractions yielded sufficient total concentrations of RNA (1–10 µg/µl) for preparation of sequencing libraries, with RNA integrity numbers higher than 8.
By the time libraries were prepared, there was no protocol available by Illumina for indexing of mRNA libraries available by Illumina. Therefore, we combined instructions from mRNA sample preparation kit, paired-end sample preparation guide, and multiplexing sample oligonucleotide kit (all Illumina) to produce 4 bp indexes for multiplexing of libraries. In short, mRNA was poly-A enriched, transcribed into cDNA, fragmented, end-repaired, adenylated, and adapters ligated. In the library enrichment step, we used 0.25 µM concentration of primer lnPE 2.0 instead of 0.5 µM and we used 25 amplification cycles instead of 10–15 as suggested by the paired-end sample preparation kit. In total, 156 libraries were then diluted to an equal concentration of 0.5 µM and sequenced using an Illumina Genome Analyzer IIx for 100 sequencing cycles. Eight to 12 libraries were pooled per Illumina flow cell lane.
Raw Read Processing, Transcriptome Assembly, Quantification, and Normalization
We checked libraries for duplicate reads and discarded all but three copies of pair-end reads. Reads were then trimmed for low-quality bases using ConDeTri (Smeds and Künstner 2011) with default settings. We retrieved 15,574 nonoverlapping protein-coding cDNA sequences for zebra finch from BioMart (ENSEMBL version 60) and, in the absence of an annotated flycatcher genome assembly, mapped flycatcher RNA-seq reads onto these genes using Stampy version 1.0.12 (Lunter and Goodson 2011). The substitution rate for mapping was relaxed to 0.06, which is still conservative compared with the observed divergence between zebra finch and flycatchers of 16% (Ellegren et al. 2012). We extracted flycatcher-specific gene models from these mappings and used those as templates for mapping all reads again using bwa. Mapped reads were summarized per gene and per library to get read counts per individual and tissue. Mapping pairs (from paired libraries) were counted only once. Raw read counts were normalized as FPKM (fragments per kilobase of exon per million fragments mapped) and log2 transformed, following Mortazavi et al. (2008).
To test the robustness of our results to normalization procedure, we tried out two additional normalization methods: calculation of TMM normalization factors according to Robinson and Oshlack (2010) and removal of any outlier genes that had greater or less log2 gene expression than 2.5 the distance between quartile and median compared with the median. None of these procedures changed our results significantly, see supplementary text, table S1, and figure S1, Supplementary Material online.
We measured breadth of gene expression as τ (Yanai et al. 2005), which ranges from 0 for equal expression in all tissues to 1 for expression limited to a single tissue (Yanai et al. 2005). We tested for the effect of expression breadth on the M:F ratio using an analysis of covariance (ANCOVA) framework with sex bias as response variable and expression breadth and expression level as explanatory covariates. Reported test statistics represent the F-test for expression breadth, corrected for confounding effects of expression level. Chromosomal location of genes in the flycatcher genome was taken from the collared flycatcher assembly version 1.5 (Ellegren et al. 2012).
Estimates of Expression Ratios
We compared expression levels between males and females as well as between autosomal and Z chromosomal genes in both sexes. Inactive genes (FPKM = 0) were excluded from the analyses and tissue-specific genes were thus only considered in tissues where they were actively expressed. Using a cut-off of FPKM >0 was motivated by that it gave data from a larger number of genes than at higher thresholds while not biasing the results (supplementary text and figs. S2 and S3, Supplementary Material online). Variance was calculated based on observed expression levels in the individuals of each sex (where FPKM#0) and for each tissue. There is a fundamental difference between estimating ZZ:Z (which should be understood as the male-to-female expression of Z-linked genes) ratios and AA:Z (or AA:ZZ) ratios. When calculating expression ratios between the sexes for Z-linked genes, one can divide male by female expression for each gene and then provide summary statistics. When calculating AA:(Z)Z ratios this is not possible, so we divided the autosomal expression median by the median expression of Z-linked genes and determined the median of the distribution. Confidence intervals of the AA:(Z)Z ratio were determined by 10,000 bias-corrected (calculated using the cumulative normal distribution) bootstrap replicates.
We tested for differences in variance between groups with different sex-bias using F-tests. In an ANCOVA framework, log2 variance was used as the response variable, sex-bias as a two-level factor, and expression level as a covariate. We report statistics for the effect of sex-bias alone, thus being corrected for confounding effects of expression level. We determined the strength of sex-biased gene expression by calculating log2-fold changes between groups. All analyses were performed using R 2.15.2. False discovery rate corrections were performed with the approach of Benjamini and Hochberg (1995); reported P values are q values according to this method.
Sex-Bias and Protein Interactions
We acquired information on protein–protein interactions (metabolic chain connections and protein complex involvement) for chicken from the FunCoup database (Alexeyenko et al. 2012); chicken is the only bird for which such information is available on a genome-wide basis. Data were extracted for 1:1 orthologous genes of chicken and flycatcher. As the distribution of the number of interactions per gene was highly skewed toward extreme values, we grouped genes into three categories for statistical analysis: 0, 1–6, or >6 interactions per gene. We tested for an effect of the number of interactions on expression variance and sex-bias of Z-linked genes using ANCOVAs. Log2 variance or sex-bias were used as response variable, number of interactions (in protein complexes or metabolic chains) as a three-level factor (0 interations, 1–6 interactions, and >6 interactions), and expression level as a covariate.
Results
Transcriptome sequencing yielded more than 1.1 billion 100 bp Illumina paired-end reads, on average 7.1 million reads per library (i.e., per tissue/individual combination). After removing duplicated reads and trimming of low-quality bases, 621 million reads and 58 billion bases were kept for analysis, which corresponds to 55.5% of the raw reads and 51.9% of the raw bases, respectively. Filtered reads were subsequently mapped to 15,574 flycatcher orthologs to annotated zebra finch genes. In total, 15,301 genes were found to be expressed in at least one of the nine tissues analyzed. The number of reads mapped to flycatcher orthologs per individual varied between tissues, ranging from on average 378,000 in brain to 982,000 in kidney. We note that these numbers may be considered low for accurate expression estimates for mid to lowly expressed genes. However, we believe that this is mitigated by that we average over large groups of genes and that estimates from individual genes are based on up to five replicates.
Relative Levels of Male and Female Expression of Genes on the Z Chromosome
The median male-to-female (M:F) Z chromosome expression ratio was in the range of 1.41–1.61 for the seven nonreproductive tissues analyzed (mean = 1.49 ± 0.07). The lowest ratio was seen in kidney and embryo (1.41), and the highest in lung (1.61) (table 1). The distribution of M:F ratios within each of these three tissues (supplementary fig. S4, Supplementary Material online) was significantly different from the distribution in all other tissues combined (kidney: P < 0.05; lung: P < 10−6; embryo: P < 0.01; Mann–Whitney U tests). Overall, however, the M:F ratio of individual genes was correlated among tissues, typically with Spearman’s ρ ≈ 0.31 in pairwise comparisons, with the exception of correlations between embryo and other tissues that had ρ ≈ 0.24. As notable exceptions, some genes showed considerable variation in the M:F ratio among tissues (supplementary fig. S5 and table S2, Supplementary Material online), indicating tissue-specific regulation of relative expression levels in males and females.
Table 1.
Number of Z-Linked Flycatcher Genes Expressed in Different Tissues, Their Median Male-to-Female (M:F) Expression Ratio, and Median Autosome-to-Z Chromosome Gene Expression Ratio in Females (AA:Z) and Males (AA:ZZ) Per Tissue
| Tissue | No. of Z-Linked Genes | Median M:F Ratio | Median AA:Z Ratioa | Median AA:ZZ Ratiob |
|---|---|---|---|---|
| Brain | 570 | 1.49 | 1.71* | 1.13 |
| Kidney | 592 | 1.41 | 1.54*** | 1.04 |
| Liver | 520 | 1.47 | 1.41*** | 0.96 |
| Lung | 588 | 1.61 | 1.37*** | 0.87 |
| Muscle | 524 | 1.50 | 1.44*** | 1.11 |
| Skin | 594 | 1.54 | 1.94 | 1.22** |
| Ovary | 610 | NA | 1.47** | NA |
| Testis | 611 | NA | NA | 0.86 |
| Embryo | 600 | 1.41 | 1.45*** | 0.97 |
aMann–Whitney U tests of the observed AA:Z ratios deviating from 2.
bMann–Whitney U tests of the observed AA:ZZ ratios deviating from 1.
*P < 0.05; **P < 0.01; ***P < 0.001; FDR corrected for multiple testing.
To analyze the evolutionary conservation of dosage compensation, we compared M:F ratios of orthologous Z-linked genes of flycatcher and chicken, two lineages representing the deepest divergence (≈80 Ma) among neognath birds. (Julien et al. 2012) obtained comparable RNA-seq data from chicken brain, liver, and kidney, although only a single male and a single female were sequenced. Despite the obvious limitations associated with that data set, we found statistically significant correlations between the M:F ratio of flycatcher and chicken for brain (ρ = 0.30, P < 10−6) and kidney (ρ = 0.14, P < 0.05), but not for liver (P = 0.58).
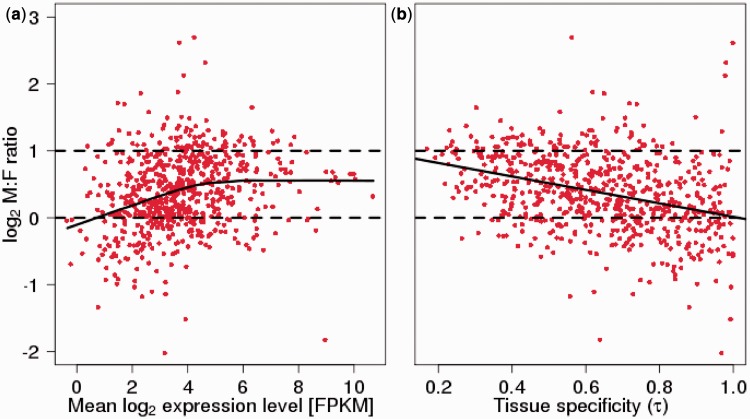
With expression data from multiple tissues at hand, we investigated the relationship between expression profile and dosage compensation. There was a negative correlation between tissue specificity (τ) and the M:F ratio, showing that narrowly expressed genes had more similar expression levels in the two sexes than ubiquitously expressed genes (P < 0.01 for all tissues except lung and embryo, F-test corrected for confounding effects by expression level; slope estimate β = −0.38 [brain] to −0.99 [liver]). On average, tissue-specific genes (τ ≈ 1) had about equal expression in males and females, whereas genes with close to flat expression profiles showed about two times higher expression in males than in females (fig. 1).
Fig. 1.—
(a) Relationship between log2 of the mean expression level (FPKM; across seven tissues) and log2 of the male-to-female (M:F) expression ratio of Z-linked flycatcher genes. The solid line represents a LOWESS regression, and the dashed lines indicate 2-fold excess of male expression (log2 = 1) and equal levels of expression between males and females (log2 = 0), respectively. For plots of individual tissues, see supplementary figure S6, Supplementary Material online. (b) Relationship between tissue specificity (τ) and log2 of the male-to-female (M:F) expression ratio (averaged over tissues with expression) of Z-linked flycatcher genes. The solid line is a linear regression for the correlation, and the dashed lines indicate 2-fold excess of male expression (log2 = 1) and equal levels of expression between males and females (log2 = 0), respectively.
We found significant positive correlations between expression level and the M:F ratio for all seven tissues investigated (Spearman’s rank correlation, ρ = 0.13 [brain] to 0.45 [liver]; P < 0.001 for all tissues except brain with P < 0.01). Graphical inspection indicated a nonlinear relationship, with a positive correlation only given for approximately the lower half of expression levels above which (at FPKM ≈ 24 = 16) the correlation seemed to level off (fig. 1; supplementary fig. S6, Supplementary Material online). Using only genes with FPKM >24 (corresponding to 39–54% of the genes, depending on tissue), there were weak negative correlations in two tissues (brain: ρ = −0.12; liver: ρ = −0.16; P < 0.05), a weak positive correlation in kidney (ρ = 0.14; P < 0.05), and nonsignificant correlations in the remaining four tissues (P > 0.05).
The variation in M:F ratio for Z-linked genes with most falling in the range of 1–2 has led to a model of dosage compensation in birds occurring on a per-gene rather than on a wholesale per-chromosome (cf. X chromosome inactivation) basis (Mank and Ellegren 2009). Although the results presented earlier demonstrate some correlates of the variation in M:F ratio, a model of dosage compensation on a per-gene basis could predict that variation in the M:F ratio is random with respect to the location of genes on the Z chromosome. On the other hand, a nonrandom distribution of M:F ratios along the Z chromosome would not necessarily be incompatible with a per-gene model if the distribution of genes itself is nonrandom with respect to their sensitivity to perturbation in dosage. Alternatively, a nonrandom distribution of M:F ratios would be expected if there are localized loci of dosage compensation, for example, originating from chromatin modification (Melamed and Arnold 2007). To test the hypothesis that dosage compensation is organized in clusters, that is, that genes localized close to each other are more likely to be either compensated or uncompensated, we obtained information on the location of genes on the flycatcher Z chromosome (supplementary fig. S7, Supplementary Material online). We then calculated the difference in the M:F ratio between neighboring genes and between distant genes, respectively. There was no significant difference between those groups in any of the tissues (P > 0.05, Mann–Whitney U test). Moreover, a sliding window analysis of the distribution of the M:F ratio of genes on the Z chromosome did not reveal any apparent clusters of similar M:F ratios consistent across tissues.
Relative Expression Levels of Genes on Autosomes and the Z Chromosome
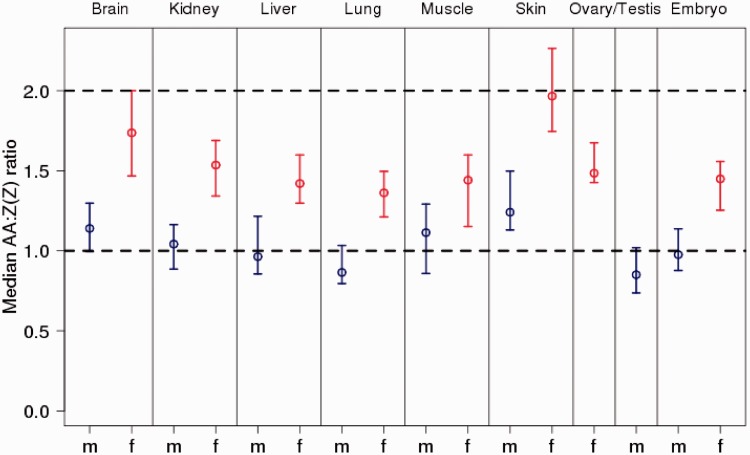
We next compared tissue-specific expression levels of genes on autosomes and the Z chromosomes for males and females separately, in this case also including data from ovary and testis. Median autosome to median Z chromosome expression level in females (AA:Z) was significantly different from 1 in all eight tissues (mean = 1.54 ± 0.19; range: 1.37–1.94; fig. 2 and table 1). In contrast, median autosome-to-Z chromosome expression level in males (AA:ZZ) was close to unity (mean = 1.02 ± 0.13; range: 0.87–1.22). Skin was an outlier in both female (1.94) and male (1.22) comparisons with a more pronounced excess of autosomal over Z-linked expression; excluding skin, AA:Z had a mean of 1.48 ± 0.11 (range: 1.37–1.71) and AA:ZZ of 0.99 ± 0.11 (range: 0.86–1.13). Together with the observation of the median M:F ratio for Z-linked genes being ≈1.5, these results indicate that expression levels of Z-linked genes are lowered in females compared with that of males, as well as compared with that of autosomal genes in both sexes.
Fig. 2.—
Median autosome-to-Z chromosome [AA:Z(Z)] gene expression ratio for seven nonreproductive and two reproductive tissues in flycatchers. Blue is male and red is female expression. Error bars indicate 95% confidence intervals for the median as determined by 10,000 bias-corrected bootstrap replicates. The dashed lines indicate 2-fold excess of autosomal expression and equal levels of expression between autosomes and Z chromosome, respectively.
Z-linked Genes That Are Dosage Compensated Show High Variance in Expression
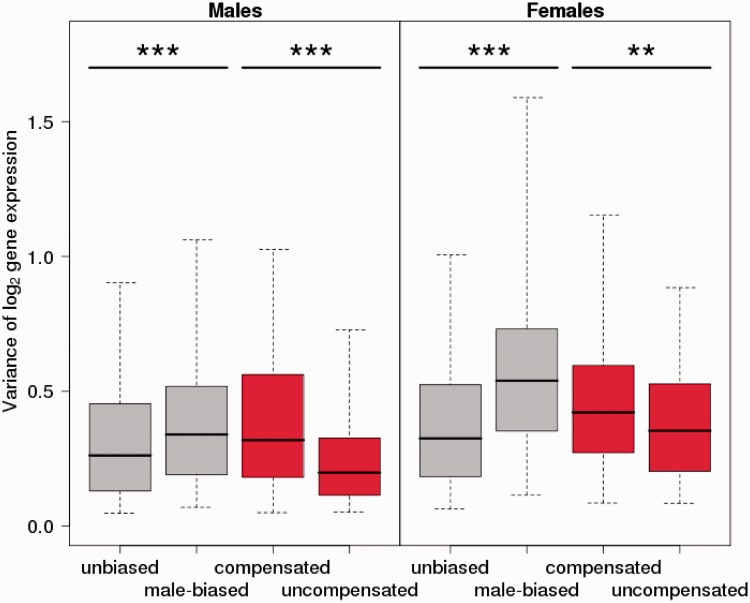
The gene-by-gene model of avian dosage compensation means that for some genes the precise expression level might be more critical than for others, for example, because their fine-tuned regulation is essential for function in networks or pathways. A dose-dependent reduction in female expression level of expression-sensitive genes during sex chromosome evolution would under this scenario be more deleterious than for genes where expression noise is more tolerable. As an evolutionary response to a potentially deleterious effect on female expression level, we should thus expect to see more of dosage compensation in the former category of genes than in the latter. To test this hypothesis, we used variance in male gene expression of Z-linked genes as an indication of tolerable variation in expression level. We divided the genes into two categories, those with M:F ratio in the range 0.5–1.5 and those in the range 1.5–2.5, respectively. However, contrary to the prediction, uncompensated Z-linked genes showed significantly lower variance in male expression than compensated genes, both when averaged over tissues (P < 10−11, F-test of log2 variance between compensated and uncompensated genes, corrected for correlation between expression mean and variance; fig. 3) and for all tissues analyzed separately (supplementary fig. S8, Supplementary Material online). Moreover, the same difference was seen when female expression was considered, with uncompensated genes showing lower variance (P < 0.01; fig. 3). So despite that this group of genes has not responded by compensating the difference in gene dose arising during sex chromosome evolution, their expression levels seem tightly regulated in both sexes.
Fig. 3.—
Variance in male (left panel) and female (right panel) expression of autosomal (gray) and Z-linked (red) flycatcher genes averaged over seven nonreproductive tissues. Genes are categorized as having an M:F ratio of 0.5–1.5 (unbiased or compensated) or 1.5–2.5 (male-biased or uncompensated). Error bars indicate 95% confidence intervals for the distribution.
As a comparison, we divided autosomal genes into the same two expression categories (and where it is more appropriate to refer to genes with M:F ratio of 0.5–1.5 as unbiased and genes with M:F ratio of 1.5–2.5 as male biased). Here, male-biased genes had significantly higher variance in male expression averaged over tissues than unbiased genes (P < 10−6, fig. 3; also significant in all tissues when analyzed separately, supplementary fig. S8, Supplementary Material online). The lower variance in male expression of Z-linked genes that are expressed at higher levels in males than in females (uncompensated) is thus not a general feature of male-biased genes in the flycatcher genome.
Level and Breadth of Z-linked Gene Expression in Relation to Involvement in Protein–Protein Interactions
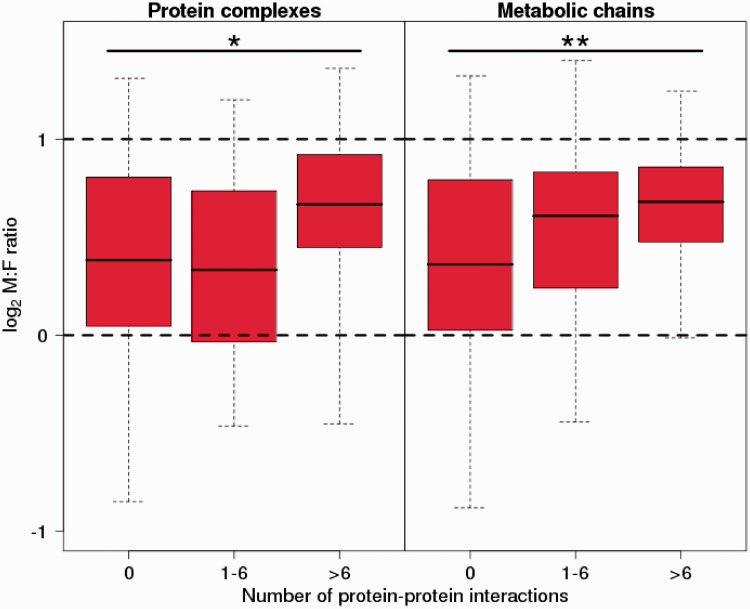
We acquired information on protein–protein interactions (in metabolic chains or protein complexes) from chicken and integrated this information with data on the M:F ratio and male expression variance for 1:1 Z-linked orthologs in flycatcher. We divided genes into three categories, with no evidence for interactions (N = 447 in protein complexes, N = 425 in metabolic chains), 1–6 interactions (N = 37 and N = 52), and more than 6 interactions (N = 28 and N = 35, respectively). We found that expression variance decreased with the number of interactions (protein complexes: P < 0.001 for 1–6 interactions, P < 0.05 for >6 interactions; metabolic chains: P < 0.001 for 1–6 interactions, P < 10−6 >6 interactions, t test), indicating that expression level is more constrained for genes involved in many interactions, which seems intuitive. The same was observed for male and female variances (in female protein complexes: P < 0.05 for 1–6 interactions, P < 0.01 for >6 interactions; metabolic chains: P < 0.001 for 1–6 interactions, P < 10−7 for >6 interactions, respectively). However, when controlling for expression level, the effect of interconnectivity on variance disappeared almost entirely (F-test, male protein complexes: P < 0.05 for 1–6 interactions, P > 0.5 for >6 interactions; metabolic chains: P > 0.05 for 1–6 interactions, P < 0.05 for >6 interactions; similar for females). Perhaps, more surprisingly, we found that the M:F ratio increased with the number of interactions, even when controlling for confounding effects through correlations with expression level (F-test, protein complexes: P > 0.05 for 1–6 interactions, P < 0.01 for >6 interactions; metabolic chains: P > 0.05 for 1–6 interactions, P < 0.001 for >6 interactions; fig. 4).
Fig. 4.—
Male-to-female expression ratio of Z-linked flycatcher genes averaged over seven nonreproductive tissues. Genes are divided into three categories according to their involvement in protein–protein interactions: 0, 1–6, and >6 interactions partners, respectively. Error bars show 95% confidence intervals for the distribution. The dashed lines indicate 2-fold excess of male expression (log2 = 1) and equal levels of expression between males and females (log2 = 0), respectively.
Dosage Compensation in the Course of Avian Sex Chromosome Evolution
Avian sex chromosome evolution has generally progressed to a stage where recombination only occurs in a small pseudoautosomal region and where the nonrecombining W chromosome is highly degenerated. However, paleognath birds make an exception. The deepest split among contemporary birds is that between Paleognathae and Neognathae 100–120 Ma (van Tuinen and Hedges 2001), with the former being represented by ratites and tinamous, whereas the latter contains >99% of all extant avian species. In ratites, sex chromosomes are either largely homomorphic or only show limited differentiation, with recombination occurring over most of the Z and the W chromosomes (Tsuda et al. 2007). As sex chromosomes are fully syntenic across Aves (Shetty et al. 1999; Nanda et al. 2008), this makes it possible to compare sex-specific expression levels of Z-linked genes prior to (Paleognathae) and subsequent to (Neognathae) cessation of recombination and sex chromosome divergence.
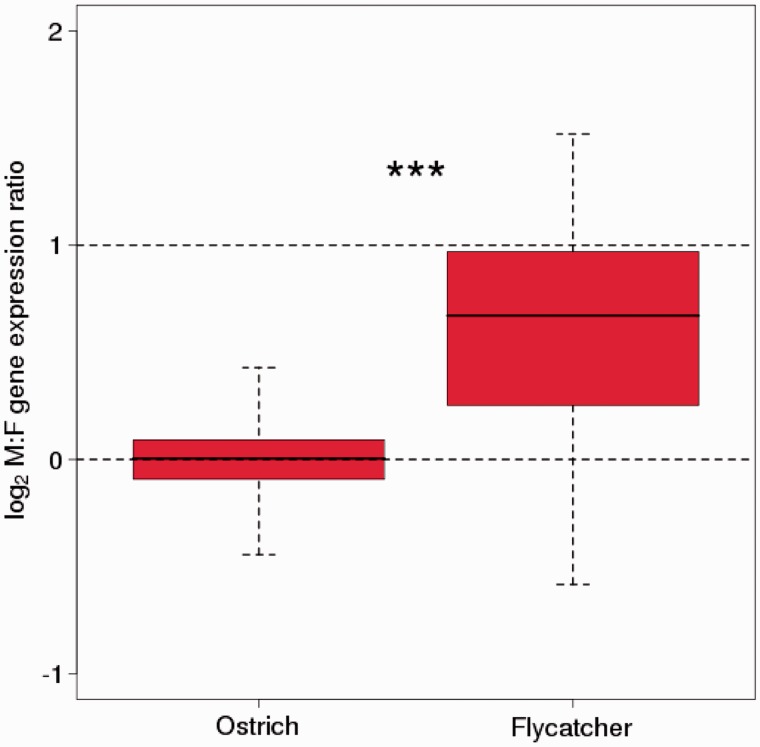
Using data on male and female expression level from brain and liver of 297 ostrich and flycatcher Z-linked orthologs, such a comparison shows two things. First, the M:F ratio of Z-linked genes has increased after the Z chromosome became hemizygous in females in the Neognathae lineage. The median M:F ratio of recombining Z-linked genes in ostriches was 1.00 (brain) and 1.09 (liver), whereas the median ratio of the orthologous genes in flycatcher was 1.51 (liver) and 1.59 (brain) (P < 10−10, Mann–Whitney U test; fig. 5). Second, we found nothing to suggest that uncompensated genes (M:F ratio 1.5–2.5) in flycatchers were more prone to be male biased in ostriches than compensated genes (M:F ratio 0.5–1.5) (median M:F ratio in ostrich of 0.042 and 0.098, respectively, P = 0.58). Moreover, the M:F ratios of flycatcher-ostrich Z-linked orthologs were not correlated (P = 0.81 for brain, P = 0.42 in liver, Spearman’s rank correlation). This would suggest that genes that have escaped dosage compensation after avian sex chromosome differentiation did not generally have an ancestral state of higher expression in males than in females. Male-biased expression of Z-linked genes after sex chromosome differentiation thus seems to represent a derived character state.
Fig. 5.—
Box-plot of log2 of the male-to-female (M:F) expression ratio in brain of orthologous Z-linked genes in ostrich and flycatcher. The included genes are from the region of the ostrich Z chromosome where Z-W recombination still occurs. Ostrich data are from Adolfsson and Ellegren (2013).
Discussion
Based on the results presented herein, we can suggest a model of gene expression during sex chromosome evolution in birds, and in particular in the lineage leading to flycatcher, as follows. 1) When genes on the avian proto-sex chromosomes ceased to recombine in females and W-linked gene copies degenerated and eventually became lost, there was a shift in the distribution of male-to-female expression ratio of Z-linked genes, from a median of no difference between sexes to a median of about 50% higher expression in males. This is supported by comparison of sex-specific expression levels of orthologous Z-linked genes when they recombine (ostrich) and when they do not recombine (flycatcher). 2) Male-biased expression of Z-linked genes is generally a consequence of lowered expression levels in hemizygous females. This is supported by that male expression levels of autosomal and Z-linked genes do not differ significantly, whereas female expression levels of Z-linked genes are significantly lower than of autosomal genes; the difference corresponds well to the difference between male and female expression of Z-linked genes. 3) Z-linked genes with similar expression levels in males and females differ in some respects from those are not compensated, including expression breadth, level, and variance. We stress that the model described earlier refers to the large-scale patterns of Z-linked genes treated as a group. This does not necessarily mean that individual genes conform to these patterns or exclude that other evolutionary forces either augment or counteract the effect of reduced gene dose during sex chromosome evolution.
Overall, our study confirms the conclusion (Ellegren et al. 2007; Itoh et al. 2007) that birds have no general mechanism for sex chromosome dosage compensation. However, observed male-to-female expression ratios of Z-linked genes do not generally correspond to the 2:1 ratio of the number of Z chromosomes in males and females, and there is considerable variation in the M:F ratio among genes. There is thus some compensation on a gene-by-gene basis, in this study of flycatchers resulting in a median male-to-female gene expression ratio of ≈1.5. This is similar to what has previously been observed in other bird species from the large group of Neognathae (Ellegren et al. 2007; Itoh et al. 2007, 2010; Wolf and Bryk 2011; Naurin et al. 2012). The similar levels of male expression from autosomal and Z-linked genes suggest that the compensation is achieved via upregulation of female expression (hyperexpression from the single Z chromosome) (Julien et al. 2012), rather than downregulation of male expression (by hypoexpression, or inactivation, of one or both of the Z chromosomes). This is also supported by the observation that still recombining genes on the ostrich sex chromosomes show similar expression levels as autosomal genes in both males and females (Adolfsson and Ellegren 2013). This observation is critical because it is not self-evident that overall expression levels of autosomal and sex-linked should be equal. In a recent study of emus (Vicoso et al. 2013), another ratite bird, late (but not early) embryonic expression of some recombining Z-linked genes showed slightly higher male than female expression, although the difference was not as pronounced as observed for nonrecombining Z-linked genes in flycatchers and birds.
In chicken, a male hypermethylated region on the Z chromosome, from which a long noncoding RNA is transcribed (Teranishi et al. 2001; Bisoni et al. 2005), has been suggested to constitute a focus for dosage compensation such that genes located in this region would be more compensated than others (Melamed and Arnold 2007; Itoh et al. 2010). In zebra finch, Itoh et al. (2010) failed to find a specialized region of greater dosage compensation along the Z chromosome. Similarly, we found no evidence for clustering of genes on the flycatcher Z chromosome with respect to the M:F ratio and no deviating patterns around the DMRT1 gene, which has been implicated in avian sex determination (Smith et al. 2009) (supplementary fig. S7, Supplementary Material online). This adds further support to the idea that partial dosage compensation has evolved on a per-gene basis.
It is reasonable to assume that some genes are particularly sensitive to variation in gene dose (being dosage-sensitive) because of the encoded proteins’ involvement in interactions with other proteins, that is, protein connectivity (the balance model) (Papp et al. 2003). This hypothesis is not specific to gene dose in the context of sex chromosome evolution but should also apply to, for example, gene duplication and expansion of gene or protein domain families (Veitia 2005). We found that uncompensated Z-linked genes (defined as having an M:F ratio of 1.5–2.5) generally had less interindividual variance in male as well as female gene expression than compensated genes (M:F ratio 0.5–1.5). At first glance, this might seem surprising since genes with the most confined expression levels could be thought of as being most sensitive to perturbations affecting gene expression, for example, changes in gene dose. By this way of reasoning, there should have been a strong impetus to compensate, via transcriptional control, changes in dose of such genes. However, there might be constraints to regulation of expression levels that hinder compensation to be achieved. If, for whatever reason, some genes are tightly regulated with limited variance in expression level, then such genes may also be resistant to sex-specific regulation. This could potentially explain why not only male but also female expression variance correlated negatively with the M:F ratio. From this perspective, the extent of dosage compensation of Z-linked genes in birds may be seen to represent a trade-off between the costs and benefits of restoring the original balance between male and female sex chromosome and autosome expression levels (discussed later).
Pessia et al. (2012) analyzed protein–protein interactions of X-linked human genes and found that levels of autosomal and X-linked expression were similar for genes encoding components of large complexes, thereby being considered dosage sensitive. However, and contrary to the observations for X-linked genes (Pessia et al. 2012), we found that Z-linked genes with many interacting partners had the highest M:F expression ratio. This is not easily perceived in the context of the balance model. It was also somewhat surprising to find that broadly expressed genes had the highest M:F ratios, while tissue-specific genes tended to have equal expression levels in males and females. There might be a category of broadly expressed genes (house-keeping genes?) that is less amendable to sex-specific regulation for reasons related to the lower variance in expression seen for genes with high M:F ratios.
Theoretical work predicts sex chromosomes to be enriched for genes involved with sexual antagonism (Rice 1984; Charlesworth et al. 1987), where dominance of new mutations and type of heterogamety will affect the expectations for accumulation of genes with differential fitness effects in the two sexes (Ellegren and Parsch 2007). For several male heterogametic organisms, the X chromosome has been reported to be demasculinized (Sturgill et al. 2007) while in chicken there is some evidence for an enrichment of male-beneficial genes (Ellegren 2011) and male-biased expression on the Z chromosome during avian evolution (Wright et al. 2012). This is in line with prediction for at least partly dominant genes; in birds, male-beneficial alleles/genes will be exposed to selection more often when Z-linked than autosomal. These theoretical underpinnings have fostered the idea that incomplete dosage compensation in birds would be an adaptive trait, to mediate male-biased expression of sexually antagonistic genes (Kaiser and Ellegren 2006; Storchová and Divina 2006; Wright et al. 2012), rather than reflecting constraint in, or necessity of, sex-specific regulation to restore the ancestral expression levels of Z-linked genes in males and females. We do not exclude an adaptive scenario and we also note that the two models should not need to be mutually exclusive. However, it remains to be revealed what might be the links between protein connectivity, variance in gene expression and expression breadth, and sexual antagonism. Wright et al. (2012) recently suggested that there has been a successive masculinization of the chicken Z chromosome during the course of sex chromosome evolution, resulting from male-specific selection and being manifested in a correlation between M:F ratios and time since recombination ceased between different regions of the Z chromosome and the W chromosome. This is an interesting model that warrants further investigation and replication across birds once the temporal dynamics of sex chromosome evolution in other avian lineages than chicken has been characterized.
Our results demonstrate a conservation of M:F ratios among tissues of flycatchers but also a conservation of tissue-specific ratios in the evolutionary distant comparison of flycatcher and chicken, two lineages that split ≈80 Ma. Interestingly, we found several examples of genes that displayed significant variation in the M:F ratio among tissues. This resembles the situation previously noted for sex-biased expression of autosomal genes in chicken (Mank et al. 2008, 2010) and mice (Yang et al. 2006). Genes showing strikingly different M:F ratios among tissues warrant further investigation as they may represent candidates for phenotypic differences in the physiology.
Conclusions
Studies of dosage compensation, like this, are faced with the general caveat associated with analyses of gene expression, namely that expression is measured at the intermediate level, at the stage of RNA. In theory, dosage compensation could occur at the level of regulation of translation or even at stage of post-translational processes. We emphasize that proteomic analyses of the amount of gene product encoded from the sex chromosomes in males and females constitute a conceptually important question for future research. In systems where deviations from what at least previously has been considered as norm in the context of dosage compensation, like in birds, it will be particularly be valuable to obtain independent support for incomplete compensation from analyses of protein abundance. It could be noted in this context that one of the first molecular reports indicating incomplete avian dosage compensation was actually based on analyses at the protein level, viz. by studies of enzymatic activity of Z-linked cytoplasmic aconitase (Baverstock et al. 1982). It is also noteworthy that for two of the three investigated species the observed excess of male enzyme activity (1.42 in sparrow and 1.59 chicken) was close to the average excess of male gene expression found in our and other RNA-based studies (though in the third species, pigeon, the excess was 2.4).
Supplementary Material
Supplementary text, figures S1–S8, and tables S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors thank Göran Arnqvist, Carina Mugal, Pall Olason, Holger Schielzeth, Linnéa Smeds, and Jochen Wolf for helpful discussion. This work was supported by the European Research Council (grant no.249976); a Knut and Alice Wallenberg Foundation Wallenberg Scholar Grant; and the Swedish Research Council (grant no. 2010-5650).
Literature Cited
- Adolfsson S, Ellegren H. Lack of dosage compensation accompanies the arrested stage of sex chromosome evolution in ostriches. Mol Biol Evol. 2013;30:806–810. doi: 10.1093/molbev/mst009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexeyenko A, et al. Comparative interactomics with Funcoup 2.0. Nucleic Acids Res. 2012;40:D821–D828. doi: 10.1093/nar/gkr1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baverstock PR, Adams M, Polkinghorne RW, Gelder M. A sex-linked enzyme in birds - Z-chromosome conservation but no dosage compensation. Nature. 1982;296:763–766. doi: 10.1038/296763a0. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- Bisoni L, Batlle-Morera L, Bird AP, Suzuki M, McQueen HA. Female-specific hyperacetylation of histone H4 in the chicken Z chromosome. Chromosome Res. 2005;13:205–214. doi: 10.1007/s10577-005-1505-4. [DOI] [PubMed] [Google Scholar]
- Castagne R, et al. The choice of the filtering method in microarrays affects the inference regarding dosage compensation of the active X-chromosome. PLoS One. 2011;6:e23956. doi: 10.1371/journal.pone.0023956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Phil Trans R Soc Lond Ser B. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton NH. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- Deutschbauer AM, et al. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Emergence of male-biased genes on the chicken Z-chromosome: sex-chromosome contrasts between male and female heterogametic systems. Genome Res. 2011;21:2082–2086. doi: 10.1101/gr.119065.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Gustafsson L, Sheldon BC. Sex ratio adjustment in relation to paternal attractiveness in a wild bird population. Proc Natl Acad Sci U S A. 1996;93:11723–11728. doi: 10.1073/pnas.93.21.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–698. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- Ellegren H, et al. Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol. 2007;5:40. doi: 10.1186/1741-7007-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H, et al. The genomic landscape of species divergence in Ficedula flycatchers. Nature. 2012;491:756–760. doi: 10.1038/nature11584. [DOI] [PubMed] [Google Scholar]
- Forstmeier W, Ellegren H. Trisomy and triploidy are sources of embryo mortality in the zebra finch. Proc Biol Sci. 2010;277:2655–2660. doi: 10.1098/rspb.2010.0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- Gustafsson L, Pärt T. Acceleration of senescence in the collared flycatcher Ficedula albicollis by reproductive costs. Nature. 1990;347:279–281. [Google Scholar]
- Gustafsson L, Qvarnstrom A, Sheldon BC. Trade-offs between life-history traits and a secondary sexual character in male collared flycatchers. Nature. 1995;375:311–313. [Google Scholar]
- Gustafsson L, Sutherland WJ. The costs of reproduction in the collared flycatcher Ficedula albicollis. Nature. 1988;335:813–815. [Google Scholar]
- Harrison PW, Mank JE, Wedell N. Incomplete sex chromosome dosage compensation in the Indian meal moth, Plodia interpunctella, based on de novo transcriptome assembly. Genome Biol Evol. 2012;4:1118–1126. doi: 10.1093/gbe/evs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2:280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- Itoh Y, et al. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, et al. Sex bias and dosage compensation in the zebra finch versus chicken genomes: general and specialized patterns among birds. Genome Res. 2010;20:512–518. doi: 10.1101/gr.102343.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, et al. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Ellegren H. Nonrandom distribution of genes with sex-biased expression in the chicken genome. Evolution. 2006;60:1945–1951. [PubMed] [Google Scholar]
- Lodge JR, Fechheimer NS, Miller RC. Deletion, monosomy, and multiple monosomy-trisomy mosaicism in chicken embryos. Poultry Sci. 1973;52:397–399. doi: 10.3382/ps.0520397. [DOI] [PubMed] [Google Scholar]
- Lundberg LE, Figueiredo MLA, Stenberg P, Larsson J. Buffering and proteolysis are induced by segmental monosomy in Drosophila melanogaster. Nucleic Acids Res. 2012;40:5926–5937. doi: 10.1093/nar/gks245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2011;21:936–939. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone J, et al. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012;13:R28. doi: 10.1186/gb-2012-13-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone JH, Oliver B. Microarrays, deep sequencing and the true measure of the transcriptome. BMC Biol. 2011;9:34. doi: 10.1186/1741-7007-9-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank J, Hultin-Rosenberg L, Webster M, Ellegren H. The unique genomic properties of sex-biased genes: insights from avian microarray data. BMC Genomics. 2008;9:148. doi: 10.1186/1471-2164-9-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. Small but mighty: the evolutionary dynamics of W and Y sex chromosomes. Chromosome Res. 2012;20:21–33. doi: 10.1007/s10577-011-9251-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity. 2009;102:312–320. doi: 10.1038/hdy.2008.116. [DOI] [PubMed] [Google Scholar]
- Mank JE, Hosken DJ, Wedell N. Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution. 2011;65:2133–2144. doi: 10.1111/j.1558-5646.2011.01316.x. [DOI] [PubMed] [Google Scholar]
- Mank JE, Nam K, Brunström B, Ellegren H. Ontogenetic complexity of sexual dimorphism and sex-specific selection. Mol Biol Evol. 2010;27:1570–1578. doi: 10.1093/molbev/msq042. [DOI] [PubMed] [Google Scholar]
- Melamed E, Arnold A. Regional differences in dosage compensation on the chicken Z chromosome. Genome Biol. 2007;8:R202. doi: 10.1186/gb-2007-8-9-r202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merila J, Kruuk LEB, Sheldon BC. Cryptic evolution in a wild bird population. Nature. 2001;412:76–79. doi: 10.1038/35083580. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Nanda I, Schlegelmilch K, Haaf T, Schartl M, Schmid M. Synteny conservation of the Z chromosome in 14 avian species (11 families) supports a role for Z dosage in avian sex determination. Cytogenet Genome Res. 2008;122:150–156. doi: 10.1159/000163092. [DOI] [PubMed] [Google Scholar]
- Naurin S, Hasselquist D, Bensch S, Hansson B. Sex-biased gene expression on the avian Z chromosome: highly expressed genes show higher male-biased expression. PLoS One. 2012;7:e46854. doi: 10.1371/journal.pone.0046854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex linked genes. Berlin (Germany): Springer Verlag; 1967. [Google Scholar]
- Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B, Pal C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvarnström A, Brommer JE, Gustafsson L. Testing the genetics underlying the co-evolution of mate choice and ornament in the wild. Nature. 2006;441:84–86. doi: 10.1038/nature04564. [DOI] [PubMed] [Google Scholar]
- Qvarnström A, Pärt T, Sheldon BC. Adaptive plasticity in mate preference linked to differences in reproductive effort. Nature. 2000;405:344–347. doi: 10.1038/35012605. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saether SA, et al. Sex chromosome-linked species recognition and evolution of reproductive isolation in flycatchers. Science. 2007;318:95–97. doi: 10.1126/science.1141506. [DOI] [PubMed] [Google Scholar]
- Saetre GP, et al. A sexually selected character displacement in flycatchers reinforces premating isolation. Nature. 1997;387:589–592. [Google Scholar]
- Shetty S, Griffin DK, Graves JAM. Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res. 1999;7:289–295. doi: 10.1023/a:1009278914829. [DOI] [PubMed] [Google Scholar]
- Smeds L, Künstner A. ConDeTri - A content dependent read trimmer for Illumina data. PLoS ONE. 2011;6:e26314. doi: 10.1371/journal.pone.0026314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CA, et al. The avian Z-linked gene DMRT1 is required for male sex determination in the chicken. Nature. 2009;461:267–271. doi: 10.1038/nature08298. [DOI] [PubMed] [Google Scholar]
- Storchová R, Divina P. Nonrandom representation of sex-biased genes on chicken Z chromosome. J Mol Evol. 2006;63:676–681. doi: 10.1007/s00239-006-0022-1. [DOI] [PubMed] [Google Scholar]
- Sturgill D, Zhang Y, Parisi M, Oliver B. Demasculinization of X chromosomes in the Drosophila genus. Nature. 2007;450:238–241. doi: 10.1038/nature06330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teranishi M, et al. Transcripts of the MHM region on the chicken Z chromosome accumulate as non-coding RNA in the nucleus of female cells adjacent to the DMRT1 locus. Chromosome Res. 2001;9:147–165. doi: 10.1023/a:1009235120741. [DOI] [PubMed] [Google Scholar]
- Thorne MH, Collins RK, Sheldon BL. Chromosome analysis of early embryonic mortality in layer and broiler-chickens. Br Poultry Sci. 1991;32:711–722. doi: 10.1080/00071669108417397. [DOI] [PubMed] [Google Scholar]
- Tsuda Y, Nishida-Umehara C, Ishijima J, Yamada K, Matsuda Y. Comparison of the Z and W sex chromosomal architectures in elegant crested tinamou (Eudromia elegans) and ostrich (Struthio camelus) and the process of sex chromosome differentiation in palaeognathous birds. Chromosoma. 2007;116:159–173. doi: 10.1007/s00412-006-0088-y. [DOI] [PubMed] [Google Scholar]
- van Tuinen M, Hedges SB. Calibration of avian molecular clocks. Mol Biol Evol. 2001;18:206–213. doi: 10.1093/oxfordjournals.molbev.a003794. [DOI] [PubMed] [Google Scholar]
- Veen T, et al. Hybridization and adaptive mate choice in flycatchers. Nature. 2001;411:45–50. doi: 10.1038/35075000. [DOI] [PubMed] [Google Scholar]
- Veitia RA. Exploring the etiology of haploinsufficiency. Bioessays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- Veitia RA. Gene dosage balance: deletions, duplications and dominance. Trends Genet. 2005;21:33–35. doi: 10.1016/j.tig.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. Lack of global dosage compensation in Schistosoma mansoni, a female-heterogametic parasite. Genome Biol Evol. 2011;3:230–235. doi: 10.1093/gbe/evr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Kaiser V, Bachtrog D. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc Natl Acad Sci U S A. 2013;110:6453–6458. doi: 10.1073/pnas.1217027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf J, Bryk J. General lack of global dosage compensation in ZZ/ZW systems? Broadening the perspective with RNA-seq. BMC Genomics. 2011;12:91. doi: 10.1186/1471-2164-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Moghadam HK, Mank JE. Trade-off between selection for dosage compensation and masculinization on the avian Z chromosome. Genetics. 2012;192:1433–1445. doi: 10.1534/genetics.112.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- Yang X, et al. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zha XF, et al. Dosage analysis of Z chromosome genes using microarray in silkworm, Bombyx mori. Insect Biochem Mol Biol. 2009;39:315–321. doi: 10.1016/j.ibmb.2008.12.003. [DOI] [PubMed] [Google Scholar]
- Zhang S, Mathur S, Hattem G, Tassy O, Pourquie O. Sex-dimorphic gene expression and ineffective dosage compensation of Z-linked genes in gastrulating chicken embryos. BMC Genomics. 2010;11:13. doi: 10.1186/1471-2164-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.