Abstract
In eukaryotes, chromosome segregation during cell division is facilitated by the kinetochore, an assembly of proteins built on centromeric DNA. The kinetochore attaches chromosomes to spindle microtubules, modulates the stability of these attachments, and relays microtubule-binding status to the spindle assembly checkpoint, a cell cycle surveillance pathway that delays chromosome segregation in response to unattached kinetochores. Here, we discuss recent results that guide current thinking on how each of these kinetochore-centered processes is achieved, and how their integration ensures faithful chromosome segregation, focusing on the essential roles of kinase-phosphatase signaling and the microtubule-binding KMN protein network.
Introduction
The purpose of mitosis is to distribute chromosomes that have been duplicated in S phase to opposite ends of the cell prior to cytokinesis (Fig. 1a). This task is accomplished by the mitotic spindle, a bipolar, football-shaped structure, composed of microtubules, polymers of the cytoskeletal protein tubulin. To ensure that each daughter cell inherits an identical copy of the genome, the newly-replicated sister chromatids, which are held together by the cohesin complex, must bi-orient. That is, they must attach to microtubules emanating from opposite ends of the spindle. Once all chromosomes have attached to microtubules, cohesin is cleaved and the sister chromatids are irreversibly separated. Attachments between spindle microtubules and chromosomes depend on the kinetochore, a hierarchical protein assembly of nearly 100 proteins that links centromeric DNA to spindle microtubules to couple forces generated by microtubule dynamics to power chromosome movement. Core components of the kinetochore such as the Constitutive Centromere-Associated Network (CCAN)1 and the Knl1-Mis12 complex-Ndc80 complex (KMN) network2, which bind centromeric DNA and microtubules respectively, are conserved across eukaryotes, with additional contributions from species-specific auxiliary DNA- and microtubule-binding proteins.
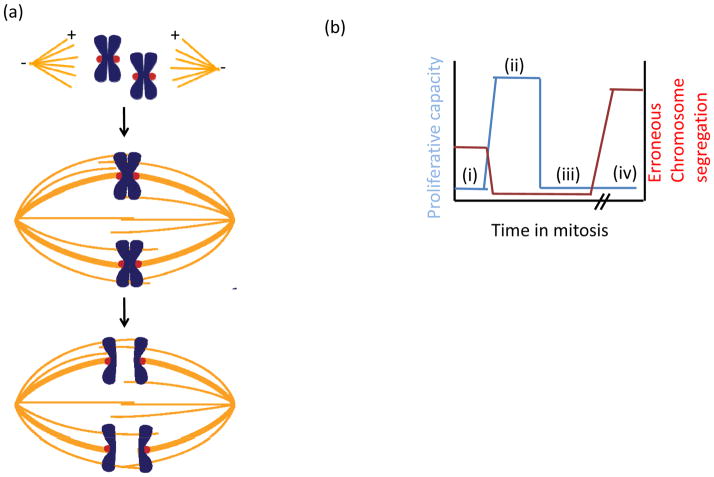
Figure 1. Speed and fidelity in cell division.
(a) Schematic of chromosome segregation. (top) At the start of mitosis, a bipolar spindle composed of microtubules (orange) assembles with plus-ends (+) oriented towards the spindle equator and minus-ends (−) clustered at the spindle pole. At the spindle poles, centrosomes (not shown) are a major site of microtubule nucleation. (middle) Pairs of replicated chromosomes (blue) attach to the spindle via kinetochores (red). Error-free chromosome segregation depends on each sister attaching to microtubules from opposite spindle poles (bi-orientation). (bottom) Once all chromosomes have bi-oriented, sister chromatids are segregated to opposite spindle poles. (b) Schematic plot of proliferative capacity after cell division (blue) and relative chromosome segregation errors (red) as a function of time spent in mitosis. (i) When the spindle assembly checkpoint (SAC) is inactive, exit from mitosis occurs rapidly, independent of kinetochore-microtubule attachment, increasing the frequency of erroneous chromosome segregation and hence decreasing proliferative capacity. (ii) In an unperturbed cell cycle, proper chromosome-spindle attachments are established quickly allowing for timely exit from mitosis. Meanwhile, the SAC ensures anaphase occurs only when all kinetochores have attached to the spindle. (iii) Modest increases in the time spent in mitosis, which can arise from SAC activity, decrease proliferative capacity. This can occur despite the absence of segregation defects, due to activation of stress-response pathways. (iv) When the SAC cannot be satisfied, for example, in the presence of anti-mitotic drugs, cells arrest in mitosis, followed by cell cycle arrest or apoptosis.
Sister chromatids that attach to the spindle but do not bi-orient, or chromatids that lack spindle attachments altogether, are at risk for mis-segregation. Errors in chromosome segregation are linked to the development of human cancers3 and occur more frequently in cancer cells that non-cancer cells4. Regulatory proteins at the kinetochore safeguard against erroneous segregation, and hence, increase the fidelity of mitosis, in two ways. First, attachments on bi-oriented kinetochore pairs are selectively stabilized, while erroneous attachment configurations are destabilized and eliminated, allowing for another opportunity for bi-orientation. Second, unattached kinetochores are the primary signal to launch the spindle assembly checkpoint (SAC), a cell cycle surveillance pathway that delays exit from mitosis. In the absence of the SAC, cohesin is cleaved and cells exit from mitosis irrespective of chromosome-spindle attachments, resulting in an increased frequency of chromosome mis-segregation events, decreased proliferative capacity, and lethality (Fig. 1b, i)5–8.
Juxtaposed with the need for fidelity in chromosome segregation is the need to transit through cell division rapidly, as core functionalities of the cell including protein trafficking, transcription, translation, and DNA repair are largely suppressed during mitosis. Consistent with this, protracted delays in mitosis can trigger apoptosis (Fig. 1b, iv), and are thought to underlie the efficacy of so-called ‘anti-mitotic’ drugs, including taxol and the vinca alkaloids, which are mainstays in cancer chemotherapy. These drugs bind tubulin and prevent proper kinetochore-microtubule attachments, resulting in sustained activation of the SAC (reviewed in9). While the proximal triggers of apoptosis that arise during mitotic arrest are not fully understood, accumulation of DNA damage is thought to contribute10, 11. While such delays are unlikely to be frequently encountered in nature, shorter delays may arise from aberrations found in cancers, including the presence of extra centrosomes12, or mis-regulation of the tumor suppressor protein phosphatase 2A (PP2A)13, each of which would be expected to delay bi-orientation, and hence, increase the time spent in mitosis. Interestingly, even modestly lengthening the time spent in mitosis can induce a cell cycle arrest in the following G1 phase14 (Fig. 1b, iii). This arrest occurs despite apparently normal chromosome segregation, suggesting cellular stress accumulates even when mitotic progression is transiently delayed.
The competing needs for speed and fidelity in chromosome segregation are integrated, in large part, at the kinetochore. This structure is essential for (i) SAC activation at unattached kinetochores, (ii) attachment to spindle microtubules, and (iii) SAC extinction at attached kinetochores. Here, we discuss each of these kinetochore-based activities, focusing on recent insights into requirements for kinase and phosphatase signaling. We focus our discussion around the KMN network, an essential and conserved regulator of microtubule binding and SAC signaling, focusing on other key players where appropriate to address concepts guiding current thinking on how the fidelity and timeliness of chromosome segregation are achieved.
What is the KMN network?
The KMN network is comprised of Kinetochore null 1 (Knl1), the four-subunit Mis-segregation 12 (Mis12) complex, and the four-subunit Nuclear division cycle 80 (Ndc80) complex (Fig. 2a). Across eukaryotes, the kinetochore-localized KMN network is essential for both microtubule binding and SAC signaling. It is worth emphasizing that such conservation was not necessarily expected, as other aspects of kinetochore assembly and function diverge significantly (e.g., centromeric DNA sequences are not conserved). Moreover, the functions of the KMN network were not immediately obvious, because although many kinetochore proteins are essential for chromosome segregation, few make direct contacts with microtubules. Rather, the central roles of the KMN network unfolded gradually as its individual components were first deemed important for chromosome segregation and, subsequently, found to assemble into a ‘super complex’15 (reviewed in16). These studies, together with reconstitution of the KMN network and a demonstration of its microtubule binding activity2 collectively established that the KMN network constitutes the core microtubule-binding site in eukaryotic chromosomes.
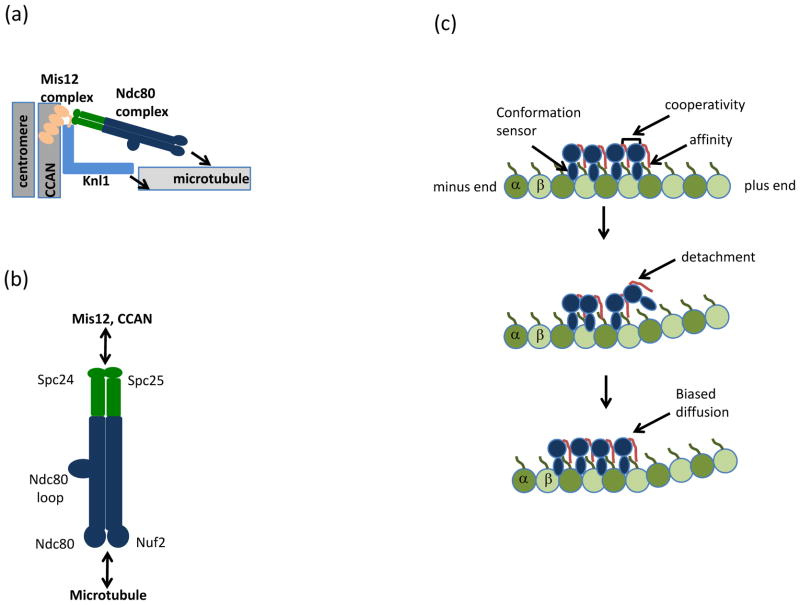
Figure 2. Architectural organization of the KMN network.
(a) Schematic of the Knl1-Mis12 complex-Ndc80 complex (KMN) network at the kinetochore. The four-subunit Mis-segregation 12 (Mis12) complex bridges Kinetochore null 1 (Knl1) and the Nuclear division cycle 80 (Ndc80) complex to the Conserved Centromere-Associated Network (CCAN) and underlying centromeric DNA. Arrows indicate microtubule binding activities in the Ndc80 complex and Knl1. (b) The Ndc80 complex, a dumbbell shaped heterotetramer, consists of a pair of microtubule-binding calponin-homology domains within the Ndc80 protein and Nuclear filamentous 2 (Nuf2) subunits (blue). The kinetochore domain is distal to the microtubule-binding domain and resides in the Spindle pole component 24 (Spc24) and Spc25 heterodimer (green). The subcomplexes interact via coiled-coil domains, which in the Ndc80 protein is interrupted by a loop that mediates protein-protein interactions. (c) Model for Ndc80 protein (blue) association with tubulin (green). Modified with permission from24. The Ndc80 protein has the unusual property of recognizing the region between α and β tubulin subunits, in contrast to most microtubule-binding proteins which bind microtubules once per tubulin dimer. Recognition of both intra- and inter-tubulin interfaces is thought to allow the Ndc80 complex to preferentially bind straight (top) versus the curled (middle) tubulin interfaces at disassembling microtubule tips. During detachment of the Ndc80 protein (middle) from curved microtubule structures, cooperativity between Ndc80 complexes (mediated by the Ndc80 tail, red) allows persistent microtubule and diffusion towards the microtubule minus-end (bottom). The positively charged Ndc80 tail also increases the affinity of the Ndc80 complex for microtubules via interaction with the negatively charged tubulin E-hook (green line) present in α and β tubulin subunits.
Recent analyses have provided important insights into the KMN network, in particular for the Ndc80 complex and its association with microtubules. The complex is a heterotetramer of the Ndc80 protein (also called Highly expressed in cancer protein 1 (Hec1) in humans), Nuclear filamentous 2 (Nuf2), Spindle pole component 24 (Spc24), and Spc25 (Fig. 2b). Heterodimers of Spc24-Spc25 and Ndc80-Nuf2 interact via coiled-coil domains, assembling into a dumbbell-like structure with distinct functionalities at either end17–20. The globular domains of the Ndc80-Nuf2 heterodimer each fold into a calponin-homology domain, which mediates microtubule binding2, 18, 21, while those of the Spc24-Spc25 heterodimer are essential for kinetochore targeting of the Ndc80 complex, directly binding to the Mis12 complex22 and/or Centromere protein (Cenp)-T23, a component of the CCAN.
To couple chromosome movement to microtubule dynamics, the Ndc80 complex must bind to and maintain persistent interactions with dynamic microtubules. A recent study has provided the first high-resolution picture of how this binding may occur24. The Ndc80 complex’s microtubule binding arises in at least two ways, both dependent on the Ndc80 protein (Fig. 2c, top). First, an electrostatic interaction between the basic N-terminal tails of the Ndc80 protein with the acidic E-hook of tubulin confers affinity 18, 21, 24. Second, the complex binds microtubules with a tubulin monomer repeat, recognizing both α and β tubulin at the inter- and intra-tubulin interfaces24. This striking property of the Ndc80 complex allows it to bind microtubules every 4nm (the spacing of tubulin monomers), in contrast to most microtubule-associated proteins, which bind every 8 nm (the spacing of tubulin dimers) and facilitates oligomerization of the Ndc80 complex on microtubules. Moreover, because the inter- and intra-tubulin dimer interfaces are disrupted by bending associated with depolymerizing filaments, this second mode of binding may allow the Ndc80 complex to function as a tubulin conformation ‘sensor’, causing detachment near depolymerizing microtubule tips (Fig. 2c, middle). The Ndc80 complex is nevertheless able to maintain processive association (Fig. 2c) because it oligomerizes. In this way, cooperative microtubule binding may facilitate retention of arrayed Ndc80 complexes, allowing persistent association with dynamic microtubules. Understanding how Nuf2’s calponin-homology domain, which was not observed to contact microtubules in the Ndc80 complex structure24 but nevertheless is required for high affinity Ndc80 complex binding to microtubules in vitro18, and for proper kinetochore-microtubule attachments in vivo25 will be important to address in future studies.
We currently lack high-resolution structures of the other KMN network components, Knl1 and the Mis12 complex. Knl1 has microtubule binding activity, which synergistically enhances KMN network association with microtubules in vitro2 whereas the heterotetrameric Mis12 complex, functions as an inter-complex scaffold, linking the KMN network to centromeric DNA via direct association with the CCAN protein Cenp-C 26, 27, as well as an intra-complex scaffold, bridging Knl1 and the Ndc80 complex at kinetochores22. Further work will be required to reveal precisely how these functions of Knl1 and the Mis12 complex are integrated with the Ndc80 complex to promote kinetochore function.
SAC activation at the kinetochore
In addition to constituting the site of chromosome-spindle attachments, the kinetochore plays an essential role in relaying microtubule-binding status to the SAC to delay exit from mitosis and chromosome segregation. In this section, we highlight recent progress on molecular mechanisms of SAC activation at the kinetochore.
The SAC and the kinetochore
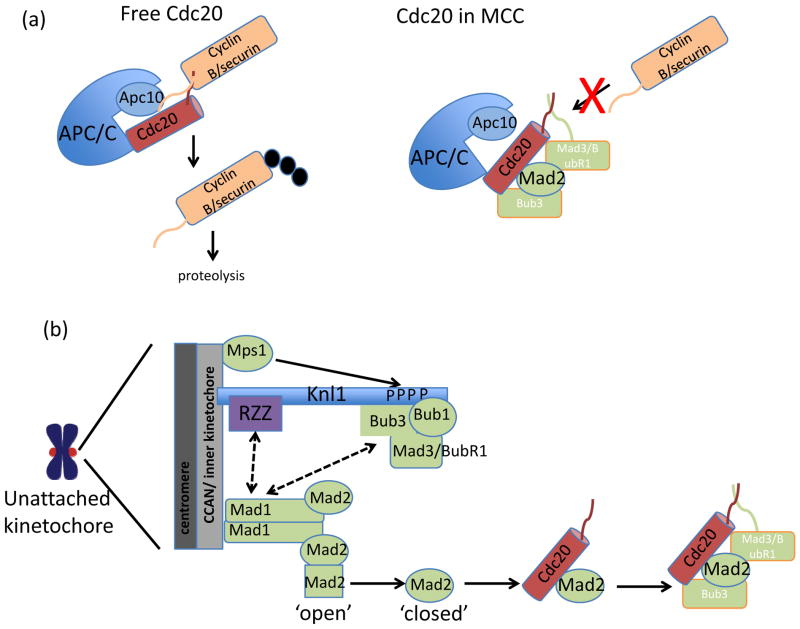
Conserved across eukaryotes, the SAC includes the serine/threonine kinases Monopolar spindle (Mps1) and Budding uninhibited by benomyl 1 (Bub1), as well as non-kinase components mitotic arrest deficient 1 (Mad1), Mad2, Bub3, and Bub1-related 1 (BubR1)/Mad328–30. Collectively, these proteins delay precocious chromosome segregation through the inactivation of Cell division cycle 20 (Cdc20)31, 32, a co-factor of an E3 ubiquitin ligase known as the anaphase promoting complex/cyclosome (APC/C)33, 34. The APC/C is a master regulator of cell division35, triggering both sister chromatid segregation and exit from mitosis via the ubiquitylation and subsequent proteasome-dependent destruction of cyclin B36, the master kinase regulating mitotic progression, and securin37, an inhibitor of separase, to allow cleavage of cohesion complexes38–42. The APC/C targets cyclin B and securin for ubiquitylation dependent on a destruction (D)-box36 sequence present in each of these proteins. APC/C co-factors, which, in addition to Cdc20 also includes Cdh143, 44, comprise the D-box recognition site, together with the APC/C subunit APC1045, 46. The SAC, in turn, catalyzes the formation of a Cdc20 inhibitory complex, referred to as the Mitotic Checkpoint Complex (MCC), which is a heterotetramer of Cdc20, Mad2, Mad3/BubR1 and Bub347.
Incorporation of Cdc20 into the MCC stabilizes securin and cyclin B in at least three ways. First, Mad3/BubR1 binding to Cdc20 blocks D-box recognition sites in Cdc2045, 48. Second, Cdc20 association with APC10 is disrupted45. Third, incorporation of Cdc20 into the MCC promotes APC/C-dependent autoubiquitylation of Cdc20, decreasing the levels of Cdc20 and hence, allowing for complete inhibition of remaining Cdc20 by the SAC49–51. In this way, SAC-catalyzed inhibition of securin and cyclin B proteolysis prevents precocious chromosome segregation and exit from mitosis.
During mitosis, unattached kinetochores play a central role in Cdc20 inhibition, and thus, SAC signaling. For example, in vertebrate tissue culture cells, a persistently unattached kinetochore can delay a cell in mitosis for several hours52 and, in yeast, mutations in centromeric DNA or kinetochore proteins result in a mitotic delay53, 54. Most SAC proteins, including Mps1, Bub1 Mad1, Mad2, Mad3/BubR1, and Bub3, as well as Cdc20, target to unattached kinetochores, and many are depleted from kinetochores upon microtubule attachment (reviewed in55). Consistent with essential roles of kinetochore signaling in maintaining mitotic arrest, concentration of Mps1 at the kinetochore is necessary for mitotic arrest56. Conversely, constitutive kinetochore-targeting of Mad1, which is normally lost from kinetochores upon microtubule attachment, is sufficient to sustain SAC signaling on attached kinetochores57. We note that kinetochores are not absolutely essential for the formation of the MCC, which is present during interphase in human cells (before recruitment of SAC proteins to kinetochores), and in yeast lacking functional kinetochores47, 58–60. However, in human cells, this kinetochore-independent pool of MCC is not sufficient to delay chromosome segregation in perturbed mitosis (e.g., continuous exposure to microtubule poisons). Rather, it sets a minimum length of mitosis, inhibiting the APC/C in prometaphase, when kinetochores are still assembling and recruiting SAC proteins61.
A key kinetochore-based reaction in SAC signaling is to promote the assembly of Mad2-Cdc20 complexes62. Mad2 exists in two states, a ‘closed’ conformer competent to bind Cdc20 and Mad1, the kinetochore receptor for Mad2, and an ‘open’ conformer that associates with neither binding partner63–66. Currently, a dominant model for the formation of kinetochore-catalyzed Mad2-Cdc20 complexes, the Mad2 ‘template’ model, posits that conversion of cytosolic Mad2 from an ‘open’ to a ‘closed’ state, and hence the ability of Mad2 to bind Cdc20, is catalyzed by a heterodimer of ‘closed’ Mad2 bound to Mad1 localized at the kinetochore62, 67 and reviewed in68, 69. Additional roles for the kinetochore in promoting the formation of Cdc20 inhibitory complexes, such as phosphorylation of Cdc2070, Mad271, or Mad3/BubR172, by kinetochore-localized SAC kinases may also contribute to robust assembly of MCC complexes.
SAC activation
Mechanistic and structural studies of activation of Mad2 at the kinetochore have provided key insights of what, in current models, are amongst the last stages kinetochore-based activation of the SAC (reviewed in55, 68, 69), but upstream events, in particular how the checkpoint machinery is targeted to unattached kinetochores, have only recently come into focus. Phospho-regulation clearly plays a role, with contributions from the Mps1 kinase, which has recently been shown to be required for recruitment of essentially all other SAC components to the kinetochore56, 73–75. However, a mechanistic understanding of how Mps1 contributes to SAC activation lagged, due in part to a difficulty in identifying essential Mps1 substrates.
Accumulating evidence now suggests that Knl1 plays a central and direct role in Mps1-dependent, kinetochore-based SAC activation. Disruption of the KMN network impairs recruitment of SAC proteins to the kinetochore and/or SAC activity76–80, and, very recently, Knl1 has recently been identified as a conserved substrate of Mps1 (Fig. 3a). Mps1 phosphorylates Knl1 at threonine residues within the conserved MELT repeats15 (Fig. 3c), plus additional sites near Knl1’s N-terminus81–83. Knl1 is a critical substrate of Mps1 at the kinetochore because mutation of all Mps1 phosphorylation sites in Knl1 abolishes SAC function in fission yeast83, with fewer mutations compromising SAC signaling82.
Figure 3. Molecular mechanism of kinetochore-based SAC activation.
(a) Chromosome segregation and exit from mitosis are triggered by the Anaphase Promoting Complex/Cyclosome (APC/C). Cell division cycle 20 (Cdc20), an activator of APC/C, forms a docking site with Apc10 that recognizes destruction box sequences in cyclin B and securin and promotes ubiquitylation (black circles) of these substrates. The SAC promotes the formation of a Cdc20 inhibitory complex known as Mitotic Checkpoint Complex (MCC). In the MCC, Cdc20 interaction with Apc10 is disrupted, and Mitotic arrest deficient 3 (Mad3)/Bub1-related 1 (BubR1) binds degron recognition sites in Cdc20, preventing ubiquitylation of cyclin B and securin. (b) Schematic of SAC activation at the kinetochore. On unattached kinetochores, Monopolar spindle 1 (Mps1) phosphorylates Knl1. Phosphorylated Knl1 binds Budding uninhibited by benomyl 1 (Bub1) and Bub3, and recruits Mad3/BubR1. In metazoans, Knl1 also recruits the Rod-ZW10-Zwilch (RZZ) complex. Together, these proteins recruit to the kinetochore heterodimer of Mad1 and ‘closed’ Mad2. The Mad1-Mad2 complex catalyzes the conversion of soluble ‘open’ Mad2 to ‘closed’ Mad2, which can associate with Cdc20 in the cytoplasm. Mad2-Cdc20 complexes are then bound by Mad3/BubR1 and Bub3 to form the MCC. (c) Knl1 schematic. Features of Knl1 that contribute to SAC signaling include the N-terminal basic patch that mediates microtubule binding (red) and protein phosphatase 1 (PP1) binding motif (pink), multiple MELT (M [D/E][I/L/V/M][S/T]) repeats (black lines), which are phosphorylated by Mps1, and coiled-coil domains (blue box) which mediate kinetochore targeting. The presence and/or number of these features in different systems are indicated below. Solid arrows indicate direct interactions; dashed arrows indicate unknown mechanism of recruitment.
Phosphorylation of Knl1 by Mps1 creates a docking site for the SAC kinase Bub1 and its binding partner Bub381–83 (Fig. 3b). Kinetochore-localized Bub1 is necessary and sufficient for localizing the SAC proteins Bub3 and Mad3/BubR184, 85. Bub1 also recruits Mad1, but the molecular mechanism is unclear (Fig. 3c). The requirement for Knl1 phosphorylation in SAC activation can be at least partially bypassed by artificially tethering Bub1 to the kinetochore and/or by introducing phospho-mimetic mutations at the sites of Mps1-dependent Knl1 phosphorylation82, 83. These studies are consistent with the notion that recruitment of Bub1 to kinetochores depends on Knl1 phosphorylation by Mps1, although the interdependencies of Bub1 and Bub3 kinetochore recruitment may vary between organisms85, 86.
A second Knl1-centered pathway of SAC activation operates in metazoans that depends on Zwint-1, which associates with both Knl1 and the three-subunit Rod-Zwilch-Zw10 (RZZ) complex, which, like Bub1, is required for Mad1 to kinetochores87, 88. Mps1 activity is required for kinetochore targeting of the RZZ complex56, 73, 75, however it is unclear whether this depends on phosphorylation of Knl1. The discovery of fast-acting chemical inhibitors to inactivate Mps1 kinase activity56, 73–75 should aid in the identification of substrates that promote RZZ complex association at the kinetochore. Finally, Bub1 and BubR1 directly bind human Knl1, at a site distinct from the MELT repeats89. Disrupting this interaction however, does not displace Bub1 and BubR1 from kinetochores90, suggesting it may be functionally redundant with Mps1 pathways, or, alternatively, that this interaction could contribute to SAC extinction.
Collectively, these results highlight the importance of the KMN network to integrate SAC signaling at the kinetochore and indicate Knl1 is a ‘landing pad’ for SAC activation. Interestingly, Mps1-dependent phosphorylation of Knl1 is also required for chromosome alignment 83. Whether this reflects a requirement for Bub1, Bub3 and/or BubR1, each of which contribute to bi-orientation85, 91–93, or whether additional regulators of microtubule attachment also recognize phosphorylated Knl1 is unclear. However, the recent advances described here provide a valuable framework that will guide the future studies that will unravel how precisely the SAC is activated.
Microtubule attachment stability
In human cells, interactions between kinetochores and spindle microtubules arise concurrent with spindle assembly. Thus, at the start of mitosis, all chromosomes lack spindle attachments. Initial interactions between chromosomes and spindle microtubules occur predominantly along the microtubule lattice, rather than the plus-ends94, but these interactions are eventually replaced by stable end-on attachments, which depend on the KMN network. In this section, we describe recent insights into how end-on kinetochore-microtubule attachments are established.
Phospho-regulation of attachments
Error-free chromosome segregation requires that microtubule binding be sufficiently dynamic such that erroneous attachments can be eliminated while proper attachments on bi-oriented chromosomes persist through anaphase. These two competing needs are balanced through reversible phosphorylation at the kinetochore (Fig. 4a), with essential contributions from the kinase Aurora B and the phosphatase B56-PP2A.
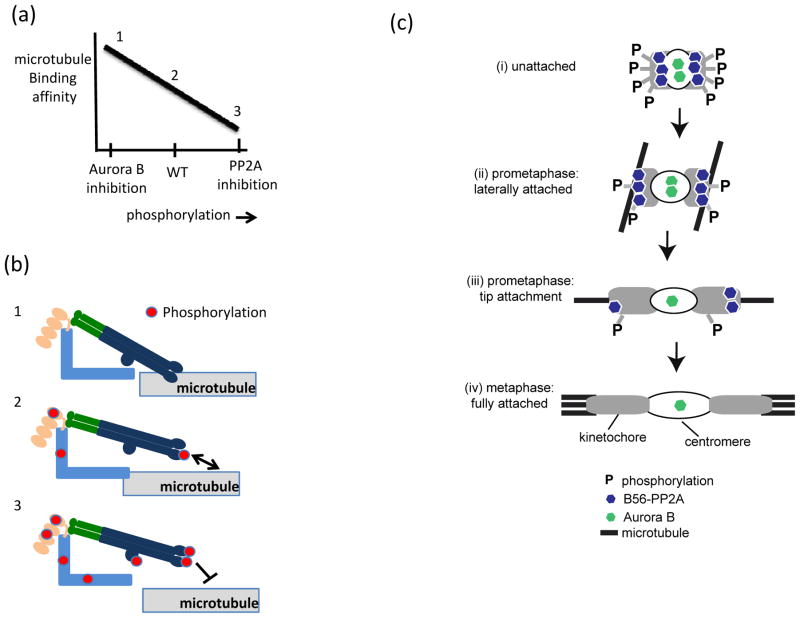
Figure 4. Molecular mechanisms of kinetochore-microtubule attachment.
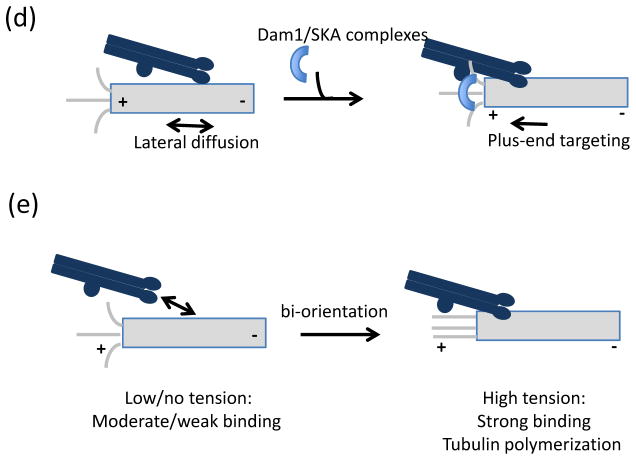
(a–b) Dynamic regulation of kinetochore-microtubule binding affinity by phospho-regulation. (a) Schematic plot of microtubule binding affinity versus phosphorylation at unattached kinetochores. (b) Molecular effects of altering the kinase/phosphatase balance on KMN network microtubule binding. KMN network diagrammed as in Fig. 2. (1) If Aurora B kinase is inhibited and phosphorylation is low, microtubule-binding affinity is high and erroneous attachments cannot be eliminated. (2) Partial phosphorylation allows attachments that are sufficiently dynamic to allow stabilization of attachments that result in bi-orientation and release of improper attachments. (3) If B56-protein phosphatase 2A (PP2A) is inhibited, phosphorylation marks generated by Aurora B kinase increase on the KMN network, preventing stable kinetochore-microtubule binding. (c) Speculative model of phospho-regulation of kinetochore (grey)-microtubule (black) binding through outer-kinetochore phosphorylation by Aurora B and B56-PP2A. (i) On unattached kinetochores, which lack tension, centromeric Aurora B kinase is close to its outer kinetochore substrates and phosphorylation is high, despite the localization of B56-PP2A phosphatase at kinetochores. (ii) During prometaphase, lateral interactions produce intermediate tension, increasing both inter- and intra-kinetochore stretching which increases the distance between Aurora B and outer kinetochore substrates, while B56-PP2A localization to relative to the outer kinetochore is unchanged. The net result in decreased phosphorylation and (iii) stabilization of initial microtubule tip interactions. (iv) At metaphase, full microtubule occupancy results in the loss of B56-PP2A from the kinetochore, and limited access of Aurora B to its substrates allowing for stable attachments. (d) Association of the Ndc80 complex with the Duo1 and Mps1 interacting factor 1 (Dam1) complex and the Spindle and Kinetochore-Associated (SKA) complex biases and strengthens Ndc80 complex localization to the microtubule plus-end, enabling the formation of persistent kinetochore-microtubule interactions. (d) Tension, arising from bi-orientation, stabilizes attachments both by increasing the lifetime of kinetochore-microtubule interactions and by promoting microtubule polymerization. As diagrammed here, force-dependent stabilization of kinetochore-microtubule interactions depends on the Ndc80 complex, but other microtubule-binding proteins at the kinetochore may also contribute.
Aurora B targets to centromeric DNA and has a conserved role in destabilizing, and hence, eliminating erroneous kinetochore-microtubule attachments95, 96, mediated in part through phosphorylation of the KMN network97. A large body of work suggests that phosphorylation of Aurora B substrates at the kinetochore reduces microtubule binding affinity, (reviewed in98 (Fig. 4a, b). In current models, this differential phosphorylation of Aurora B substrates arises because Aurora B is located closer to kinetochore substrates on erroneously attached kinetochores, where tension is low compared to bi-oriented kinetochores, where tension, and hence, the distance between the kinetochore and centromere, is high99. Additionally, Aurora B levels at centromeres are enriched on misaligned chromosomes100. The differential phosphorylation of Aurora B substrates at erroneously attached versus bi-oriented kinetochores can be explained through increased kinetochore substrates access. Paradoxically, however, Aurora B access to kinetochore substrates is expected to be highest in prometaphase when tension is low, and it is therefore unclear how stable attachments form at the start of mitosis.
A resolution to this paradox likely lies with two observations. First, although phosphorylation of KMN network proteins is indeed higher on kinetochores in prometaphase compared to metaphase, modification is not saturating. We know this because upon complete microtubule depolymerization, and hence, lack of tension, phosphorylation of KMN substrates is several-fold higher compared to prometaphase97(Fig, 4b, ii). Second, during prometaphase, these intermediate levels of Aurora B phosphorylation depend on the B56-PP2A phosphatase, which reduces phosphorylation of Aurora B substrates at the outer kinetochore, including the KMN network13 (see Box 1 for a discussion of serine/threonine phosphatases). Indeed, disrupting the kinase-phosphatase balance, through depletion of B56-PP2A, increases the phosphorylation of the KMN network, precluding stable kinetochore-microtubule interactions (Fig. 4a, b). Acute chemical inactivation of Aurora B kinase activity restores stable microtubule interactions in these cells suggesting that unopposed Aurora B activity accounts for the attachment defects. These findings also suggest that mis-regulation of B56-PP2A, commonly observed in human cancers, compromises the fidelity of chromosome segregation101, 102.
Box 1. Identifying substrates of serine/threonine phosphatases.
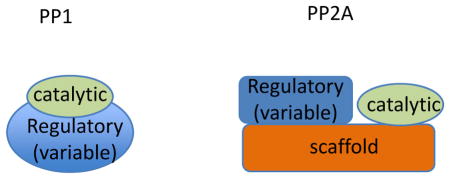
Dephosphorylation is essential for cellular function but linking specific serine/threonine phosphatases to signaling networks is challenging. This is largely because phosphorylation marks generated by the ~ 400 eukaryotic serine/threonine kinases are reversed by a handful of serine/threonine phosphatase catalytic subunit, with the majority of serine/threonine phosphatase activity coming from just two families of enzymes: protein phosphatase 1 (PP1) and protein phosphatase 2A (PP2A). PP1 and PP2A holoenzymes consist of a catalytic subunit, which is targeted to intracellular sites and/or substrates by a variable regulatory subunit. For PP2A, the catalytic and regulatory subunits assemble on a scaffolding subunit. Dozens of PP1 regulatory subunits have been identified, and most contain a short PP1-binding motif. For PP2A, four conserved families of regulatory subunits have been described with the total number of regulatory subunits varying between 3 (budding yeast) to at least 15 (humans).
Dissecting functions of regulatory subunits at specific stages of the cell cycle (e.g., mitosis) and/or at specific intracellular sites (e.g., the kinetochore) has been difficult, particularly because acute chemical inhibitors only target the common catalytic subunit and therefore can not inform on specific holoenzymes. Moreover, redundancy in regulatory subunit targeting has been observed. For example, in human cells, each of the five genes in the B56 (B′) regulatory subunit family can redundantly target PP2A catalytic activity to centromeres/kinetochores. Therefore, a requirement for B56-PP2A in stabilizing kinetochore-microtubule attachment was only revealed when levels all B56 proteins were simultaneously reduced by RNAi 13. In our view, linking phosphatases to specific signaling networks in vivo, in cell division and beyond, will require approaches that address the potential for redundancy in regulatory subunit targeting, such as combinatorial RNAi, analysis of localization dynamics 13, and phosphorylation sensors 154.
These results suggest the following model to describe how stable kinetochore-microtubule interactions are formed during prometaphase (Fig. 4c). First, on unattached kinetochores, at the start of mitosis or in the presence of microtubule poisons, Aurora B is close to its outer kinetochore substrates (e.g., the KMN network) and phosphorylation is maximal97. This occurs despite the localization of B56-PP2A, possibly because the close proximity of the kinase is sufficient for saturating levels of phosphorylation. Second, during prometaphase (Fig 4c, panel ii), kinetochores establish lateral interactions along microtubule walls94. Importantly, lateral attachments do not require the KMN network94, 103, and thus may be impervious to Aurora B and B56-PP2A signaling. Crucially, however, these lateral interactions produce tension94, increasing the distance between centromeric Aurora B and the KMN network. However, the proximity of B56-PP2A, which localizes to the outer kinetochore104, remains constant. As a result, phosphorylation of the KMN network proteins is decreased several-fold compared to unattached kinetochores97, facilitating the transition from side-on to tip attachments (Figure 4c, iii). Partial phosphorylation of the KMN network also creates an opportunity for combinatorial phosphorylation, permitting graded, rather than binary, changes in microtubule binding97. Finally, as microtubule occupancy increases (Fig. 4c iv), inter- and intra-kinetochore tension is established 105, 106 and the accessibility of the Aurora B kinase to the kinetochore is reduced while B56-PP2A is removed from the kinetochore. These two events, together with the targeting of the Protein Phosphatase 1 (PP1) to attached kinetochores107, 108, ensure phosphorylation remains low on bi-oriented kinetochore pairs. One weakness of this model is that it assumes that all substrates have equal affinities for both Aurora B and B56-PP2A. Therefore, in the future, it will be important to examine the enzymology of key kinetochore phospho-substrates in vitro.
Regulation of kinetochore-microtubule attachments depends on a network of additional proteins including the kinase Plk1100, 104, as well as SAC proteins, including Mps156, 75, BubR191, 104, Bub1, and Bub3 109 which perform double duties in both SAC activation and, in some instances, regulation of Aurora B and B56-PP2A. Thus, Aurora B targeting to centromeres depends in part on Bub1 phosphorylation of histone H2A110, whereas recruitment of B56-PP2A to the kinetochore depends on Plk1-dependent phosphorylation of BubR1104. B56-PP2A, in turn regulates phosphorylation of BubR1 and Plk1 kinetochore targeting13. Such interdependencies of kinase and phosphatase recruitment, combined with microtubule-attachment sensitive targeting, create the possibility for feedback mechanisms to rapidly respond to microtubule binding. A more complete understanding of the substrates and docking sites for Aurora B and B56-PP2A, and their kinetochore regulators, will be essential to understand how microtubule-dependent spatial rearrangements in the centromere and kinetochore are integrated with phospho-signaling networks to stabilize proper kinetochore-microtubule interactions.
Persistent Attachments
Proper chromosome segregation depends not only on establishing high-affinity interactions between the KMN network and microtubules (which can be tuned through phosphorylation) but also on maintaining persistent microtubule contacts as microtubules grow and shrink. Indeed, once kinetochores are bound to microtubule plus-ends, chromosome movement can be powered through polymer depolymerization111. Understanding how kinetochores remain persistently attached to dynamic microtubule plus-ends is therefore central to explain chromosome segregation.
Accumulating evidence suggests that persistent attachments between the KMN network and microtubules depend on additional microtubule-binding complexes, such as the Duo1 and Mps1 interacting protein 1 (Dam1) complex in yeast and the Spindle- and Kinetochore-Associated (SKA) complex in metazoans. Although structurally unrelated, the ten-subunit Dam1 complex and three-subunit SKA complex both bind processively to dynamic microtubules112–114, make processive attachments to dynamic microtubules113, 115–117 and have in common a striking feature: namely, the ability to retain Ndc80 complex at depolymerizing microtubule tips (Fig. 4d)118–120. Recombinant Ndc80 complexes maintain attachments to polymerizing and depolymerizing microtubules, but only as oligomers121. However, when associated with the Dam1 complex (yeast) or SKA complex (human), Ndc80 monomers can make processive, load-bearing attachments118–120. Consistent with this, proper chromosome spindle interactions in vivo depend on the Dam1 and SKA complexes in budding yeast122, 123 and humans113, 124–126, respectively. The SKA complex has also been proposed to regulate sister chromatid cohesion127, but further studies will be required to examine if cohesin loss after SKA depletion reflects a distinct biochemical activity of the SKA complex or whether this reflects an uncoordinated loss of cohesin arising from extended metaphase arrest128. Finally, both microtubule binding and Ndc80 complex association of the Dam1 and SKA complexes are negatively regulated by Aurora B-dependent phosphorylation118–120, 129, 130. Together, these studies suggest that establishment of processive microtubule binding may be yet another regulatory point to modulate chromosome-spindle interactions.
Additional proteins whose targeting to the Ndc80 complex may regulate kinetochore-microtubule interactions include the replication licensing factor Cdt1 in human cells131 and the microtubule polymerase, Dis1/XMAP215 in fission yeast132 which both associate with the Ndc80 loop, a short region on the Ndc80 protein that disrupts the coiled-coil domain. However, because these proteins bind microtubules (Dis1/XMAP215) or are required for chromosome duplication (Cdt1), analysis of contributions to kinetochore-microtubule binding will require in vitro analyses to establish direct binding to the Ndc80 complex as well as to identify biochemical effects of these protein-protein interactions on microtubule association.
(iii) Force stabilizes kinetochore-bound microtubules
Protein-protein interactions between kinetochores and microtubules lie at the heart of chromosome segregation. These associations must withstand force so that at anaphase sister chromatids are moved to opposite spindle poles. Prior to anaphase, when sister chromatids are held together by the cohesin complex, this force generates tension. Pioneering micromanipulation experiments in living cells revealed that tension stabilizes attachments133. However, it remained unclear tension might play a direct role in modulating the stability of kinetochore-microtubule attachments.
Further characterizing the role of tension required reconstitution of kinetochore-microtubule attachment in vitro, to allow for quantitative biochemical and biophysical measurements of microtubule binding as a function of force. This feat was recently accomplished using purified kinetochores from budding yeast134. These studies suggest a model in which force arising from tension on bi-oriented chromosomes increases the lifetime of kinetochore-microtubule attachments (Fig. 4e). These analyses indicate that tension suppresses microtubule catastrophes and increases the frequency of rescue, suggesting that under tension, kinetochore-bound microtubules spend more time in the assembling state, when kinetochore disassociation is less likely. Thus, in contrast to the vast majority of protein-protein interactions, which are destabilized by force, reconstituted kinetochore-microtubule interactions, which depend on the KMN network, become stronger under forces with magnitudes that are likely to be physiologically relevant. While clearly an important experimental and conceptual advance, future studies building on this work will require unraveling the molecular interactions that are stabilized by force, as well as how these interactions are integrated with dynamic phosphorylation at the kinetochore.
SAC extinction at the kinetochore
While SAC signaling is essential to ensure sufficient time for all chromosomes to attach to the spindle, equally important is that signaling be extinguished at each kinetochore upon microtubule binding. Indeed, kinetochore-based SAC signaling is intimately influenced by the binding of microtubules at the kinetochore. For example, several SAC proteins that are enriched at unattached kinetochores are depleted upon microtubule attachment, including Mad1, and Mad2 (reviewed in 55). Depletion of SAC proteins from kinetochores attached to spindle microtubules is likely to be a mechanistically relevant step in SAC extinction because constitutive targeting of Mad1 to the kinetochore is sufficient to sustain Mad2-dependent SAC signaling after chromosome bi-orientation57. Although the molecular mechanisms that couple microtubule binding to silencing of the SAC remain largely unclear, here we describe recent studies indicating that microtubule binding status is relayed through the KMN network to couple microtubule attachment to changes in SAC chemistry.
Considering that SAC activation at the kinetochore is dependent on phosphorylation, it is reasonable to suppose that a phosphatase might play a role in SAC extinction. Indeed, SAC extinction at the kinetochore has recently been shown to depend on recruitment of protein phosphatase 1 (PP1)135, 136 (Fig. 5). PP1 is targeted to the kinetochore via a PP1-binding motif present in Knl1 (Fig. 3c), indicating that the KMN network, and Knl1 in particular, contributes both to SAC activation and extinction. Deletion of Knl1’s PP1 interaction motif is lethal in budding yeast, due to failure to silence the SAC137. Viability can be rescued by fusing wild type, but not a catalytically dead mutant of PP1 directly to Knl1, arguing that dephosphorylation of kinetochore substrates contributes to SAC silencing. Similarly, in fission yeast and C. elegans, disruption of Knl1-dependent kinetochore targeting of PP1 compromises SAC inactivation138, 139.
Figure 5. Molecular mechanisms of SAC extinction at the kinetochore.
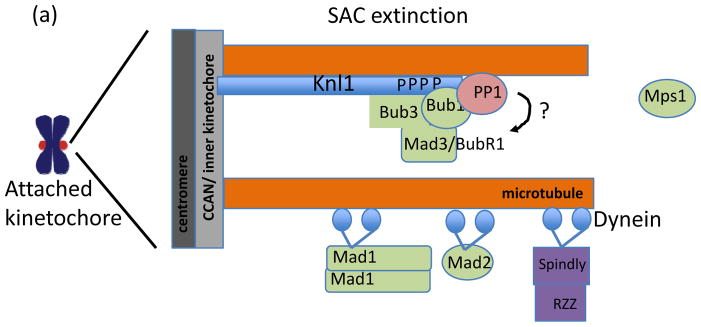
(a) SAC extinction at the kinetochore proceeds through PP1 targeting to Knl1, microtubule binding by Knl1, and, in metazoans, dynein-dependent stripping of kinetochore proteins including Mad1, Mad2, and Spindly, which binds to the RZZ complex. The substrates of PP1 at the kinetochore are not known but possible candidates include the MELT domains in Knl1 that are phosphorylated by Mps1. Together, these mechanisms lead to loss of SAC regulators from attached kinetochores but the interdependencies and molecular mechanisms of kinetochore eviction during SAC extinction remain largely unclear.
In C. elegans, a second Knl1-dependent mechanism of SAC silencing has recently been identified. At the extreme N-terminus of Knl1 resides a microtubule-binding domain2 (Fig. 3c). Unexpectedly, the short stretch of basic amino acids, adjacent to the PP1-binding motif, is not required for chromosome-spindle attachments in vivo, but rather contributes to SAC silencing138. Moreover, loss of both the microtubule-binding activity and PP1 targeting motif in Knl1 yields additive defects on SAC silencing, suggesting the two routes to SAC extinction are independent.
In human cells, the Knl1-PP1 interaction has thus far only been linked to stabilization of kinetochore-microtubule attachment107. A role for PP1 in SAC extinction may be obscured by redundant, Knl1-independent, mechanisms to silence kinetochore-based SAC signaling, in particular dynein-dependent removal of SAC regulators from the kinetochore140, 141. Dynein ‘strips’ proteins away from kinetochores towards microtubule minus ends, including Mad1, Mad2, and Spindly, a protein that promotes microtubule binding at the kinetochore140, 141. Of these, dynein-dependent removal of Spindly is important for SAC extinction at the kinetochore because removal of other cargo, such as Mad1 and Mad2, can also be achieved through dynein-independent, perhaps KMN network-dependent, mechanisms whereas Spindly removal can not140. It is not clear why the continued presence of Spindly at kinetochores interferes with SAC extinction. Spindly is not required for SAC signaling, and so its removal is unlikely to be directly related to SAC extinction. Rather, the removal of Spindly may be important for other activities at the kinetochore, such as the transition from lateral to tip-based microtubule attachments.
These recent insights are consistent with the notion that Knl1 is both a ‘scaffold’ for SAC activation and extinction (through recruitment of Bub1 and PP1, respectively) and a ‘sensor’, coupling microtubule binding to SAC extinction. How either extinction pathway is relayed to downstream SAC components remains unclear. Identifying substrates of PP1 at the kinetochore will be necessary to understand its role in SAC silencing, with phosphorylation sites in Knl1 as obvious candidates.
Finally, extinction of SAC signaling at the kinetochore must also be integrated with liberation of Cdc20 from its inhibitors Mad2, Bub3, and Mad3/BubR1 to activate the APC/C. APC/C-dependent ubiquitylation of Cdc20 plays a conserved role in disassembling MCC complexes50, 142–144, and, in metazoans, an additional protein, p31 (comet), a binding partner of ‘closed’ Mad2145, 146, promotes both disassembly of the MCC and ubiquitylation of Cdc20144, 147, 148. It will be important to examine whether microtubule capture at the kinetochore accelerates MCC disassembly, perhaps through phospho-Knl1, which, in addition to SAC activation, has also been linked to timely SAC silencing82. Understanding how the balance of SAC kinase and phosphatase activity at the kinetochore is altered upon microtubule binding to allow cell cycle progression is clearly an important area for future work.
Conclusions and perspective
We have outlined current thinking on how the major functions of the kinetochore, SAC activation, attachment to microtubules, and SAC silencing, are integrated to ensure accurate, yet timely chromosome segregation. Moving forward, one challenge is to understand how the precise balance of kinase and phosphatase activity contributes to kinetochore functions. In particular, this will require linking specific phosphatases, namely B56-PP2A and PP1, to kinetochore substrates. In contrast to serine/threonine kinases, many of which recognize substrates dependent on specific consensus sequences, PP1 and PP2A holoenzymes do not exhibit strong substrate selectivity, at least in vitro. This has implications for how these phosphatases can achieve specific functions at the kinetochore. In current models, kinetochore-targeted B56-PP2A dephosphorylates substrates at the kinetochore to stabilize attachment13, whereas kinetochore PP1 contributes to satisfaction of the SAC135, 136. This separation of function implies that the two phosphatases may dephosphorylate distinct substrates to ensure that, for example, enrichment of B56-PP2A on unattached kinetochores, does not extinguish SAC signaling. Such functional specialization could arise via differences in spatial targeting at the kinetochore between PP1 and PP2A. Nanometer-scale mapping149 of the relative positions of PP1 and PP2A, combined with quantitation of the relative number of molecules150 of kinases and phosphatases at the kinetochore would also shed light on how functional specialization may be achieved.
As our understanding of the molecular transactions at the kinetochore continues to expand, a long-term challenge, in our view, is to make equivalent progress towards understanding the micro-mechanical properties of the kinetochore. Chromosome segregation is a mechanical process and kinetochores experience active forces arising from microtubule-dependent interactions. Active forces, in turn, give rise to passive forces- elastic (e.g., centromere/kinetochore stretch) and/or friction. Forces at the kinetochore vary in magnitude (i.e., microtubule occupancy increasing from one to 25 microtubules bound151), timescale (brief initial movements along the microtubule lattice, versus persistent end-on attachment94), and orientation (towards the minus or plus end of microtubules). Throughout this process, intrinsic anisotropy across the centromere/kinetochore results in spatially distinct effects of force coupling. Finally, the molecular composition of the kinetochore/centromere is dynamic, with key proteins turning over on the seconds timescale, and/or depleted from the kinetochore in response to microtubule attachment.
A key focus for future work, therefore, is to identify how the kinetochore maintains functional stability while accommodating continuing fluctuations in forces and molecular composition. This will require (i) in vitro assays that couple molecular perturbations (e.g., substrate phosphorylation) to mechanical readouts (e.g., load-bearing microtubule attachments) and vice versa, at the level of single molecules and/or single kinetochores. The recent success of purifying functional kinetochores from budding yeast and reconstituting kinetochores in Xenopus egg extract are valuable steps towards this goal134, 152. (ii) in vivo measurements of forces at the kinetochore and the interplay between force and kinetochore composition. An important step towards this goal was recently reported, in which rearrangements in kinetochore architecture were correlated to microtubule polymerization status at the kinetochore153. Similar analyses will shed new light on the molecular effects of force at the kinetochore.
Mitosis is a vulnerable time for the cell. Mistakes in chromosome segregation lead to aneuploidy and/or DNA damage, and while the cell delays in mitosis to prevent such errors, essential metabolic transactions grind to a halt. While there remain many gaps in our understanding of this process, we now have a firm molecular grasp on some key aspects of the process. These advances will guide us towards a better understanding of pathways through which chromosome segregation is compromised in cancer cells, as well as how drugs that disrupt chromosome segregation lead to cell death.
Online Summary.
During mitosis, replicated sister chromatids attach to the mitotic spindle to facilitate their equal partitioning into two daughter cells.
Error-free chromosome segregation depends on establishing proper interactions between chromosomes and spindle microtubules, as well as delaying chromosome segregation until all chromosomes have attached to the spindle via activation of the spindle assembly checkpoint. Both of these processes depend on the kinetochore, an assembly of proteins that connects centromeric DNA to spindle microtubules.
The KMN protein network comprises an essential and conserved microtubule-binding activity at the kinetochore. Additionally, the KMN network is a ‘landing pad’ for recruitment of spindle assembly checkpoint proteins to the kinetochore, an essential step in activation of checkpoint signaling at the kinetochore.
Kinetochore-microtubule interactions are regulated by reversible protein phosphorylation, with essential contributions from kinases and phosphatases, as well as by protein-protein interactions to ensure that proper spindle attachments persist until anaphase while improper attachments are eliminated.
Phosphorylation-dependent protein-protein interactions between checkpoint proteins and the KMN network are important to coordinate the absence or presence of kinetochore-bound microtubules to activation or extinction of checkpoint signaling, respectively.
A complete understanding of how proper chromosome-spindle attachments are established and how their presence is relayed to the spindle assembly checkpoint to ensure error-free chromosome segregation will an integrated understanding of the molecular transactions at the kinetochore, together with its micro-mechanical properties, which function cooperatively to increase the fidelity of chromosome segregation.
Acknowledgments
T.M.K. is grateful to the NIH (GM98579). We apologize to colleagues whose work could not be discussed owing to space limitations.
Glossary
- Cytokinesis
The division of the cytoplasm to generate two daughter cells. It typically follows chromosome segregation
- Sister chromatids
A pair of identical DNA sequences formed as a result of DNA replication and held together by cohesin complexes
- Cohesin
A multi-protein complex that tethers replicated sister chromatids. Cohesin complexes enable sister chromatids to resist separation even when under microtubule-dependent pulling forces, possibly by encircling DNA. Proteolytic cleavage of cohesin by the enzyme separase allows chromosome segregation at anaphase
- Centromeric DNA
A specialized chromosomal locus, epigenetically defined by the presence of the histone H3 variant Cenp-A, which directs assembly of the kinetochore
- Microtubule plus-end
The end of the microtubule polymer with beta tubulin subunits exposed. It is more dynamic than the minus-end (in which alpha tubulin exposed). In cells, microtubule nucleation occurs only at the plus end
- CCAN
A conserved network of proteins assembled on centromeric DNA and required for recruitment of most kinetochore proteins
- KMN network
A conserved complex of proteins that constitutes the core microtubule-binding activity at the kinetochore and is a platform for spindle assembly checkpoint signaling
- Coiled-coil domain
A type of secondary structure composed of two or more alpha helices that entwine into a supercoil that, often mediating protein-protein interactions and oligomerization
- Calponin-homology domain
A protein module of ~110 amino acids formed found in many cytoskeletal and signal transduction proteins
- E-hook
The term for the C-terminal residues of alpha and beta tubulin, which are rich in acidic residues, glutamic acid (Glu, E) and aspartic acid (Asp, D)
- Cytoplasmic dynein
A multi-subunit, AAA+ ATPase, minus-end directed microtubule motor. During mitosis, it is essential for proper kinetochore-microtubule attachments and spindle pole organization
Biographies
Emily Foley is an Assistant Member in the Cell Biology program at Memorial Sloan-Kettering Cancer Center. She did her graduate work on understanding the basis of once-per-cell-cycle regulation of DNA replication with Johannes Walter at Harvard Medical School, followed by postdoctoral studies with Tarun Kapoor at The Rockefeller University, where she developed her interest in phospho-regulation of cell division.
Tarun Kapoor is Professor and Head of Laboratory of Chemistry and Cell Biology at the Rockefeller University. He received his Ph.D. in chemistry from Harvard University and did his postdoctoral research at Harvard Medical School. His laboratory develops and applies multi-disciplinary approaches to examine mechanisms that ensure error-free cell division.
Contributor Information
Emily A. Foley, Email: foleye1@mskcc.org, Cell Biology Program, Sloan-Kettering Institute, 1275 York Ave., New York, NY 10065
Tarun M. Kapoor, Email: Kapoor@rockefeller.edu, Laboratory of Chemistry and Cell Biology, Rockefeller University, 1230 York Ave, New York, NY 10065
References
- 1.McAinsh AD, Meraldi P. The CCAN complex: linking centromere specification to control of kinetochore-microtubule dynamics. Semin Cell Dev Biol. 2011;22:946–52. doi: 10.1016/j.semcdb.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A. The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell. 2006;127:983–97. doi: 10.1016/j.cell.2006.09.039. A seminal paper that identifies the KMN network as a conserved essential microtubule-binding activity at the kinetochore. [DOI] [PubMed] [Google Scholar]
- 3.Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- 4.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–9. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 5.Basu J, et al. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitagawa R, Rose AM. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat Cell Biol. 1999;1:514–21. doi: 10.1038/70309. [DOI] [PubMed] [Google Scholar]
- 7.Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 2000;101:635–45. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 8.Kalitsis P, Earle E, Fowler KJ, Choo KH. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 2000;14:2277–82. doi: 10.1101/gad.827500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gascoigne KE, Taylor SS. How do anti-mitotic drugs kill cancer cells? J Cell Sci. 2009;122:2579–85. doi: 10.1242/jcs.039719. [DOI] [PubMed] [Google Scholar]
- 10.Hayashi MT, Cesare AJ, Fitzpatrick JA, Lazzerini-Denchi E, Karlseder J. A telomere-dependent DNA damage checkpoint induced by prolonged mitotic arrest. Nat Struct Mol Biol. 2012;19:387–94. doi: 10.1038/nsmb.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Orth JD, Loewer A, Lahav G, Mitchison TJ. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol Biol Cell. 2012;23:567–76. doi: 10.1091/mbc.E11-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–82. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011 doi: 10.1038/ncb2327. Provides the first demonstration that a phosphatase is required to balance the activity of kinetochore kinases and promote the formation of proper kinetochore-microtubule interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–71. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheeseman IM, et al. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–68. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kline-Smith SL, Sandall S, Desai A. Kinetochore-spindle microtubule interactions during mitosis. Curr Opin Cell Biol. 2005;17:35–46. doi: 10.1016/j.ceb.2004.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciferri C, et al. Architecture of the human ndc80-hec1 complex, a critical constituent of the outer kinetochore. J Biol Chem. 2005;280:29088–95. doi: 10.1074/jbc.M504070200. [DOI] [PubMed] [Google Scholar]
- 18.Ciferri C, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–39. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei RR, et al. Structure of a central component of the yeast kinetochore: the Spc24p/Spc25p globular domain. Structure. 2006;14:1003–9. doi: 10.1016/j.str.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 20.Wei RR, Sorger PK, Harrison SC. Molecular organization of the Ndc80 complex, an essential kinetochore component. Proc Natl Acad Sci U S A. 2005;102:5363–7. doi: 10.1073/pnas.0501168102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–9. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- 22.Petrovic A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–52. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bock LJ, et al. Cnn1 inhibits the interactions between the KMN complexes of the yeast kinetochore. Nat Cell Biol. 2012 doi: 10.1038/ncb2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alushin GM, et al. The Ndc80 kinetochore complex forms oligomeric arrays along microtubules. Nature. 2010;467:805–10. doi: 10.1038/nature09423. This study provides a high-resolution view of the Ndc80 complex association with microtubules. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sundin LJ, Guimaraes GJ, Deluca JG. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol Biol Cell. 2011;22:759–68. doi: 10.1091/mbc.E10-08-0671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Przewloka MR, et al. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Screpanti E, et al. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–8. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–23. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–31. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 30.Hoyt MA, Totis L, Roberts BTS. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–17. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 31.Hwang LH, et al. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–4. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 32.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–7. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 33.Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–71. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 34.Kramer ER, Gieffers C, Holzl G, Hengstschlager M, Peters JM. Activation of the human anaphase-promoting complex by proteins of the CDC20/Fizzy family. Curr Biol. 1998;8:1207–10. doi: 10.1016/s0960-9822(07)00510-6. [DOI] [PubMed] [Google Scholar]
- 35.Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–56. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 36.Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–8. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto A, Guacci V, Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J Cell Biol. 1996;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–93. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- 39.Funabiki H, et al. Cut2 proteolysis required for sister-chromatid seperation in fission yeast. Nature. 1996;381:438–41. doi: 10.1038/381438a0. [DOI] [PubMed] [Google Scholar]
- 40.Holloway SL, Glotzer M, King RW, Murray AW. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993;73:1393–402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- 41.King RW, et al. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995;81:279–88. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- 42.Sudakin V, et al. The cyclosome, a large complex containing cyclin-selective ubiquitin ligase activity, targets cyclins for destruction at the end of mitosis. Mol Biol Cell. 1995;6:185–97. doi: 10.1091/mbc.6.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–93. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- 44.Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–3. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- 45.Chao WC, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–13. doi: 10.1038/nature10896. Structures of the APC/C reveal key insights into how the SAC inhibits APC/C activity towards mitotic substrates. [DOI] [PubMed] [Google Scholar]
- 46.da Fonseca PC, et al. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–8. doi: 10.1038/nature09625. Structures of the APC/C reveal key insights into how the SAC inhibits APC/C activity towards mitotic substrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–36. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–67. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foe IT, et al. Ubiquitination of Cdc20 by the APC occurs through an intramolecular mechanism. Curr Biol. 2011;21:1870–7. doi: 10.1016/j.cub.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Foster SA, Morgan DO. The APC/C Subunit Mnd2/Apc15 Promotes Cdc20 Autoubiquitination and Spindle Assembly Checkpoint Inactivation. Mol Cell. 2012;47:921–32. doi: 10.1016/j.molcel.2012.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–51. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–8. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spencer F, Hieter P. Centromere DNA mutations induce a mitotic delay in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1992;89:8908–12. doi: 10.1073/pnas.89.19.8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang Y, Burke DJ. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6838–44. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 56.Maciejowski J, et al. Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J Cell Biol. 2010;190:89–100. doi: 10.1083/jcb.201001050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol. 2011;13:475–82. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fraschini R, et al. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. EMBO J. 2001;20:6648–59. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim EM, Burke DJ. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008;4:e1000015. doi: 10.1371/journal.pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malureanu LA, et al. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev Cell. 2009;16:118–31. doi: 10.1016/j.devcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 62.De Antoni A, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–25. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 63.Luo X, et al. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–9. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 64.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 65.Luo X, et al. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–45. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 66.Sironi L, et al. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–43. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 68.Luo X, Yu H. Protein metamorphosis: the two-state behavior of Mad2. Structure. 2008;16:1616–25. doi: 10.1016/j.str.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mapelli M, Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr Opin Struct Biol. 2007;17:716–25. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 70.Tang Z, Shu H, Oncel D, Chen S, Yu H. Phosphorylation of Cdc20 by Bub1 provides a catalytic mechanism for APC/C inhibition by the spindle checkpoint. Mol Cell. 2004;16:387–97. doi: 10.1016/j.molcel.2004.09.031. [DOI] [PubMed] [Google Scholar]
- 71.Zich J, et al. Kinase activity of fission yeast Mph1 is required for Mad2 and Mad3 to stably bind the anaphase promoting complex. Curr Biol. 2012;22:296–301. doi: 10.1016/j.cub.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.King EM, Rachidi N, Morrice N, Hardwick KG, Stark MJ. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 2007;21:1163–8. doi: 10.1101/gad.431507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hewitt L, et al. Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J Cell Biol. 2010;190:25–34. doi: 10.1083/jcb.201002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kwiatkowski N, et al. Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function. Nat Chem Biol. 2010;6:359–68. doi: 10.1038/nchembio.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Santaguida S, Tighe A, D’Alise AM, Taylor SS, Musacchio A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J Cell Biol. 2010;190:73–87. doi: 10.1083/jcb.201001036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.He X, Rines DR, Espelin CW, Sorger PK. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 2001;106:195–206. doi: 10.1016/s0092-8674(01)00438-x. [DOI] [PubMed] [Google Scholar]
- 77.Janke C, et al. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 2001;20:777–91. doi: 10.1093/emboj/20.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Martin-Lluesma S, Stucke VM, Nigg EA. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 2002;297:2267–70. doi: 10.1126/science.1075596. [DOI] [PubMed] [Google Scholar]
- 79.McCleland ML, et al. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 2003;17:101–14. doi: 10.1101/gad.1040903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152:349–60. doi: 10.1083/jcb.152.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.London N, Ceto S, Ranish JA, Biggins S. Phosphoregulation of Spc105 by Mps1 and PP1 regulates Bub1 localization to kinetochores. Curr Biol. 2012;22:900–6. doi: 10.1016/j.cub.2012.03.052. These studies identify Knl1 as a critical substrate of Mps1 essential for SAC activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shepperd LA, et al. Phosphodependent Recruitment of Bub1 and Bub3 to Spc7/KNL1 by Mph1 Kinase Maintains the Spindle Checkpoint. Curr Biol. 2012 doi: 10.1016/j.cub.2012.03.051. These studies identify Knl1 as a critical substrate of Mps1 essential for SAC activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yamagishi Y, Yang CH, Tanno Y, Watanabe Y. MPS1/Mph1 phosphorylates the kinetochore protein KNL1/Spc7 to recruit SAC components. Nat Cell Biol. 2012;14:746–52. doi: 10.1038/ncb2515. These studies identify Knl1 as a critical substrate of Mps1 essential for SAC activity. [DOI] [PubMed] [Google Scholar]
- 84.Rischitor PE, May KM, Hardwick KG. Bub1 is a fission yeast kinetochore scaffold protein, and is sufficient to recruit other spindle checkpoint proteins to ectopic sites on chromosomes. PLoS One. 2007;2:e1342. doi: 10.1371/journal.pone.0001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vanoosthuyse V, Valsdottir R, Javerzat JP, Hardwick KG. Kinetochore targeting of fission yeast Mad and Bub proteins is essential for spindle checkpoint function but not for all chromosome segregation roles of Bub1p. Mol Cell Biol. 2004;24:9786–801. doi: 10.1128/MCB.24.22.9786-9801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson VL, Scott MI, Holt SV, Hussein D, Taylor SS. Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci. 2004;117:1577–89. doi: 10.1242/jcs.01006. [DOI] [PubMed] [Google Scholar]
- 87.Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–76. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 88.Kops GJ, et al. ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol. 2005;169:49–60. doi: 10.1083/jcb.200411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kiyomitsu T, Murakami H, Yanagida M. Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol Cell Biol. 2011;31:998–1011. doi: 10.1128/MCB.00815-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krenn V, Wehenkel A, Li X, Santaguida S, Musacchio A. Structural analysis reveals features of the spindle checkpoint kinase Bub1-kinetochore subunit Knl1 interaction. J Cell Biol. 2012;196:451–67. doi: 10.1083/jcb.201110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2005;7:93–8. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- 92.Meraldi P, Sorger PK. A dual role for Bub1 in the spindle checkpoint and chromosome congression. EMBO J. 2005;24:1621–33. doi: 10.1038/sj.emboj.7600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Windecker H, Langegger M, Heinrich S, Hauf S. Bub1 and Bub3 promote the conversion from monopolar to bipolar chromosome attachment independently of shugoshin. EMBO Rep. 2009;10:1022–8. doi: 10.1038/embor.2009.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Magidson V, et al. The spatial arrangement of chromosomes during prometaphase facilitates spindle assembly. Cell. 2011;146:555–67. doi: 10.1016/j.cell.2011.07.012. A must-read paper that combines high-resolution time-lapse imaging and electron microscopy reveal the predominance of lateral kinetochore-microtubule interactions in prometaphase. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–7. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- 96.Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- 97.Welburn JP, et al. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol Cell. 2010;38:383–92. doi: 10.1016/j.molcel.2010.02.034. This study identified phosphorylation sites in the KMN network, and revealed that combinatorial phosphorylation of the KMN network can produce graded changes in microtubule-binding activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lampson MA, Cheeseman IM. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2010 doi: 10.1016/j.tcb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liu D, Vader G, Vromans MJ, Lampson MA, Lens SM. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science. 2009;323:1350–3. doi: 10.1126/science.1167000. This study revealed that centromere tension results in a spatial separation between Aurora B and its kinetochore substrates, which contributes to proper stabilization of correct attachments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Salimian KJ, et al. Feedback control in sensing chromosome biorientation by the aurora B kinase. Curr Biol. 2011;21:1158–65. doi: 10.1016/j.cub.2011.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ruediger R, Ruiz J, Walter G. Human cancer-associated mutations in the Aalpha subunit of protein phosphatase 2A increase lung cancer incidence in Aalpha knock-in and knockout mice. Mol Cell Biol. 2011;31:3832–44. doi: 10.1128/MCB.05744-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Westermarck J, Hahn WC. Multiple pathways regulated by the tumor suppressor PP2A in transformation. Trends Mol Med. 2008;14:152–60. doi: 10.1016/j.molmed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 103.Cai S, O’Connell CB, Khodjakov A, Walczak CE. Chromosome congression in the absence of kinetochore fibres. Nat Cell Biol. 2009;11:832–8. doi: 10.1038/ncb1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Suijkerbuijk SJ, Vleugel M, Teixeira A, Kops GJ. Integration of Kinase and Phosphatase Activities by BUBR1 Ensures Formation of Stable Kinetochore-Microtubule Attachments. Dev Cell. 2012;23:745–55. doi: 10.1016/j.devcel.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 105.Maresca TJ, Salmon ED. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol. 2009;184:373–81. doi: 10.1083/jcb.200808130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Uchida KS, et al. Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol. 2009;184:383–90. doi: 10.1083/jcb.200811028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu D, et al. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–20. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Posch M, et al. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J Cell Biol. 2010;191:61–74. doi: 10.1083/jcb.200912046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Warren CD, et al. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol Biol Cell. 2002;13:3029–41. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–7. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 111.Coue M, Lombillo VA, McIntosh JR. Microtubule depolymerization promotes particle and chromosome movement in vitro. J Cell Biol. 1991;112:1165–75. doi: 10.1083/jcb.112.6.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–43. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- 113.Welburn JP, et al. The human kinetochore Ska1 complex facilitates microtubule depolymerization-coupled motility. Dev Cell. 2009;16:374–85. doi: 10.1016/j.devcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Westermann S, et al. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–90. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 115.Asbury CL, Gestaut DR, Powers AF, Franck AD, Davis TN. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc Natl Acad Sci U S A. 2006;103:9873–8. doi: 10.1073/pnas.0602249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Westermann S, et al. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–9. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- 117.Grishchuk EL, et al. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci U S A. 2008;105:6918–23. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–9. doi: 10.1083/jcb.200912021. These studies reveal that the Dam1 and SKA complexes target the Ndc80 complex to microtubule tips and enhance its ability to establish persistent contacts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Schmidt JC, et al. The Kinetochore-Bound Ska1 Complex Tracks Depolymerizing Microtubules and Binds to Curved Protofilaments. Dev Cell. 2012 doi: 10.1016/j.devcel.2012.09.012. These studies reveal that the Dam1 and SKA complexes target the Ndc80 complex to microtubule tips and enhance its ability to establish persistent contacts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tien JF, et al. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–23. doi: 10.1083/jcb.200910142. These studies reveal that the Dam1 and SKA complexes target the Ndc80 complex to microtubule tips and enhance its ability to establish persistent contacts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Powers AF, et al. The Ndc80 kinetochore complex forms load-bearing attachments to dynamic microtubule tips via biased diffusion. Cell. 2009;136:865–75. doi: 10.1016/j.cell.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cheeseman IM, Enquist-Newman M, Muller-Reichert T, Drubin DG, Barnes G. Mitotic spindle integrity and kinetochore function linked by the Duo1p/Dam1p complex. J Cell Biol. 2001;152:197–212. doi: 10.1083/jcb.152.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tanaka K, et al. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–94. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- 124.Gaitanos TN, et al. Stable kinetochore-microtubule interactions depend on the Ska complex and its new component Ska3/C13Orf3. EMBO J. 2009;28:1442–52. doi: 10.1038/emboj.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hanisch A, Sillje HH, Nigg EA. Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J. 2006;25:5504–15. doi: 10.1038/sj.emboj.7601426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Raaijmakers JA, Tanenbaum ME, Maia AF, Medema RH. RAMA1 is a novel kinetochore protein involved in kinetochore-microtubule attachment. J Cell Sci. 2009;122:2436–45. doi: 10.1242/jcs.051912. [DOI] [PubMed] [Google Scholar]
- 127.Daum JR, et al. Ska3 is required for spindle checkpoint silencing and the maintenance of chromosome cohesion in mitosis. Curr Biol. 2009;19:1467–72. doi: 10.1016/j.cub.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Stevens D, Gassmann R, Oegema K, Desai A. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS One. 2011;6:e22969. doi: 10.1371/journal.pone.0022969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chan YW, Jeyaprakash AA, Nigg EA, Santamaria A. Aurora B controls kinetochore-microtubule attachments by inhibiting Ska complex-KMN network interaction. J Cell Biol. 2012;196:563–71. doi: 10.1083/jcb.201109001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Gestaut DR, et al. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–14. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Varma D, et al. Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol. 2012;14:593–603. doi: 10.1038/ncb2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hsu KS, Toda T. Ndc80 internal loop interacts with Dis1/TOG to ensure proper kinetochore-spindle attachment in fission yeast. Curr Biol. 2011;21:214–20. doi: 10.1016/j.cub.2010.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nicklas RB, Koch CA. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J Cell Biol. 1969;43:40–50. doi: 10.1083/jcb.43.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Akiyoshi B, et al. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–9. doi: 10.1038/nature09594. A groundbreaking paper that reported the first reconstitution of kinetochore-microtubule attachment in vitro. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol. 2009;19:1176–81. doi: 10.1016/j.cub.2009.05.060. These studies identify PP1 association with Knl1 as a key step in checkpoint extinction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–7. doi: 10.1016/j.cub.2009.06.043. These studies identify PP1 association with Knl1 as a key step in checkpoint extinction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 Recruits PP1 to Silence the Spindle Assembly Checkpoint. Curr Biol. 2011;21:942–7. doi: 10.1016/j.cub.2011.04.011. These studies identify PP1 association with Knl1 as a key step in checkpoint extinction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Espeut J, Cheerambathur DK, Krenning L, Oegema K, Desai A. Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. J Cell Biol. 2012;196:469–82. doi: 10.1083/jcb.201111107. These studies identify PP1 association with Knl1 as a key step in checkpoint extinction. Also reveals that microtubule association of Knl1 functions together with PP1 recruitment to silence the checkpoint at kinetochores. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Meadows JC, et al. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev Cell. 2011;20:739–50. doi: 10.1016/j.devcel.2011.05.008. These studies identify PP1 association with Knl1 as a key step in checkpoint extinction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gassmann R, et al. Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev. 2010;24:957–71. doi: 10.1101/gad.1886810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Howell BJ, et al. Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol. 2001;155:1159–72. doi: 10.1083/jcb.200105093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13:1234–43. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Uzunova K, et al. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012 doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Varetti G, Guida C, Santaguida S, Chiroli E, Musacchio A. Homeostatic control of mitotic arrest. Mol Cell. 2011;44:710–20. doi: 10.1016/j.molcel.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 145.Xia G, et al. Conformation-specific binding of p31(comet) antagonizes the function of Mad2 in the spindle checkpoint. EMBO J. 2004;23:3133–43. doi: 10.1038/sj.emboj.7600322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mapelli M, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–84. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124:3905–16. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Teichner A, et al. p31comet Promotes disassembly of the mitotic checkpoint complex in an ATP-dependent process. Proc Natl Acad Sci U S A. 2011;108:3187–92. doi: 10.1073/pnas.1100023108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wan X, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–84. doi: 10.1016/j.cell.2009.03.035. These studies present technological breakthroughs in quantitative high-resolution imaging to give nanometer-scale resolution of the architecture of eukaryotic kinetochores. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Joglekar AP, Bloom K, Salmon ED. In vivo protein architecture of the eukaryotic kinetochore with nanometer scale accuracy. Curr Biol. 2009;19:694–9. doi: 10.1016/j.cub.2009.02.056. These studies present technological breakthroughs in quantitative high-resolution imaging to give nanometer-scale resolution of the architecture of eukaryotic kinetochores. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McEwen BF, Heagle AB, Cassels GO, Buttle KF, Rieder CL. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J Cell Biol. 1997;137:1567–80. doi: 10.1083/jcb.137.7.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. In vitro centromere and kinetochore assembly on defined chromatin templates. Nature. 2011;477:354–8. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Dumont S, Salmon ED, Mitchison TJ. Deformations within moving kinetochores reveal different sites of active and passive force generation. Science. 2012;337:355–8. doi: 10.1126/science.1221886. These studies present technological breakthroughs in quantitative high-resolution imaging to give nanometer-scale resolution of the architecture of eukaryotic kinetochores. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fuller BG, et al. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 2008;453:1132–6. doi: 10.1038/nature06923. [DOI] [PMC free article] [PubMed] [Google Scholar]