Abstract
Neuropathic pain is a chronic pain disorder caused by nervous system lesions as a direct consequence of a lesion or by disease of the portions of the nervous system that normally signal pain. The spinal nerve ligation (SNL) model in rats that reflect some components of clinical pain have played a crucial role in the understanding of neuropathic pain. To investigate the direct effects of gabapentin on differential gene expression in cultured dorsal root ganglion (DRG) cells of SNL model rats, we performed a differential display reverse transcription-polymerase chain reaction analysis with random priming approach using annealing control primer. Genes encoding metallothionein 1a, transforming growth factor-β1 and palmitoyl-protein thioesterase-2 were up-regulated in gabapentin-treated DRG cells of SNL model rats. The functional roles of these differentially expressed genes were previously suggested as neuroprotective genes. Further study of these genes is expected to reveal potential targets of gabapentin.
Keywords: Dorsal root ganglion, Neuropathic pain, Spinal nerve ligation, Gabapentin
INTRODUCTION
Neuropathic pain is a chronic pain disorder caused by nervous system lesions as a direct consequence of a lesion or by disease of the portions of the nervous system that normally signal pain (Baron, 2000). Pharmacological management remains the most important therapeutic option for chronic neuropathic pain. Clinically effective drugs such as gabapentin, pregabalin, and duloxetine have anti-allodynic effects in the rat L5 and L6 SNL model (Hunter et al., 1997; Iyengar et al., 2004). However, neuropathic pain is less responsive and sometimes refractory to conventional analgesic treatments, including non-steroidal anti-inflammatory drugs (NSAIDs) and opioid derivatives. NSAIDs show poor effects, while systemically administered morphine induces partial recovery from neuropathic pain, but only at higher than therapeutically relevant doses in the SNL model, which is consistent with the clinical results (Lee et al., 1995; Granados-Soto et al., 2004). Identifying improved medicine and pharmacological treatments for neuropathic pain is an important goal.
While the underlying spinal mechanisms for the development and maintenance of neuropathic pain characterized by spontaneous pain, hyperalgesia and allodynia are still not fully understood (Bridges et al., 2001; Cavenagh et al., 2006), the development of experimental models that reflect some components of clinical pain has accelerated the understanding of neuropathic pain. A rat model frequently used to study the molecular mechanisms of neuropathic pain is the L5 and L6 spinal nerve ligation (SNL) model originally established by Kim and Chung (Kim and Chung, 1992). The injury, which results in hyperalgesia, an enhanced response to mechanical stimuli, has a well-characterized time course.
An understanding of gene expression profiles associated with pharmacological treatments is important as these may open up new pathways for targeting neuropathic pain. While studies have described gene expression profiles following nerve transection or nerve ligation models in animals (Wang et al., 2002; Xiao et al., 2002; Valder et al., 2003; Aldrich et al., 2009), reports defining gene expression in studies that utilize culture of DRG cells of SNL model are scarce.
In this study, the differentially expressed genes induced directly by gabapentin, which is used as a first-line treatment for neuropathic pain, in cultured DRG cells of SNL model rats 2 weeks after SNL surgery were investigated. We performed differential display reverse transcription-polymerase chain reaction (DDRT-PCR) incorporating an annealing control primer (ACP), which has specificity to the template and allows only real products to be amplified (Hwang et al., 2003; Kim et al., 2004), for the identification of differentially expressed genes.
MATERIALS AND METHODS
Materials
Gabapentin was obtained from Sigma-Aldrich (St. Louis, MO, USA). It was dissolved in distilled water to a concentration of 100 mM and kept in a small vial at 4℃ until use.
Experimental animals
Adult male 6-week-old Sprague-Dawley rats were used for the experiments. The rats were housed under a 12/12 h reversed light-dark cycle (dark cycle: 8:00 A.M.-8:00 P.M.) for at least one week before beginning any experiments. All experiments were carried out in accordance with the National Institute of Health’s Guide for the Care and Use of Laboratory Animals and the animal use protocol was approved by the Institutional Animal Care and Use Committee.
Neuropathic pain surgery
SNL was conducted based on a previously described method (Kim and Chung, 1992; Kim et al., 2006). In brief, the left L5 and L6 spinal nerves were tightly ligated with 6~0 silk thread, and the wound was then sutured and coated with antibiotic ointment. During surgery, the animals were anesthetized using a constant flow of halothane/oxygen inhalation.
Measurement of mechanical allodynia
Prior to killing at 2 weeks post-surgery, the animals were assessed for mechanical allodynia using calibrated von Frey monofilaments. Briefly, animals were placed in individual plastic cages with wire mesh bottoms and allowed to acclimate for 20 min before testing. Withdrawal threshold was determined by increasing and decreasing stimulus intensity and the paw withdrawal threshold was determined as previously described (Chaplan et al., 1994).
Primary culture of DRG
One day after behavioral testing, allodynic animals (n=6) were killed and the L5 and L6 DRGs were dissected from the ipsilateral (injured) side of the vertebral column and placed in Dulbecco’s modified Eagles medium (DMEM; Hyclone, Logan, UT, USA) containing 1% collagenase for 30 min at 37℃. After 30 min, the DRG cells were incubated in 0.25% trypsin for 25 min at 37℃. After enzymatic digestion, the cells were dissociated by mechanical trituration through a fire-polished glass pipette. DRG cells were then rinsed with DMEM supplemented with 5% fetal bovine serum (FBS; Hyclone) three times, and seeded on 100 mm-diameter poly-L-lysine-coated dishes. The cells were cultured in DMEM supplemented with 10% FBS, 1% Penicillin-Streptomycin and 1% L-glutamine for 3 days in the presence or absence of 25 μM gabapentin.
ACP-based DDRT-PCR
Total RNAs extracted from untreated DRG cells and gabapentin-treated DRG cells were used for the synthesis of first-strand cDNAs. Reverse transcription was performed for 1.5 h at 42℃ in a reaction mixture containing 3 μg of the purified total RNA, reaction buffer, 2 mM dNTPs, dT-ACP1 (5'-CGTGAATGCTGCGACTACGATIIIIIT(18)-3'), RNase Inhibitor (Promega, Madison, WI, USA), and MMLV reverse transcriptase (Promega). Second-strand cDNA was synthesized in a reaction mixture containing first-strand cDNA, 1 μl of 10 μM dT-ACP2 (5'-CGTGAATGCTGCGACTACGATIIIIIT(15)-3'), 10 μM arbitrary ACP primers and 2×Master Mix (Seegene, Seoul, Korea). The PCR protocol for second-strand synthesis was one cycle at 94℃ for 1 min, followed by 50℃ for 3 min and 72℃ for 1 min. After second-strand DNA synthesis was completed, the PCR amplification protocol was 40 cycles of 94℃ for 40 s, followed by 65℃ for 40 s, 72℃ for 40 s and a 5-min final extension at 72℃. The amplified PCR products were separated in 2% agarose gel and stained with ethidium bromide. The differentially expressed bands were evaluated based on the band intensities measured with a ChemiDoc XRS (Bio-Rad, Hercules, CA, USA). The differentially expressed bands were extracted from the gel and directly cloned into a TOPO TA cloning vector (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. The cloned plasmids were sequenced and the DNA sequence of each gene was confirmed by comparison with sequences in GenBank (NIH, Bethesda, MD, USA).
Real-time reverse transcription-PCR (RT-PCR) confirmation
The mRNA levels for the selected genes in gabapentin-treated DRG cells were analyzed by quantitative real-time RT-PCR using a Rotor-Gene 6000 system (Corbett Research, Sydney, Australia). The reaction mixture (20 μl) consisted of cDNA, a 1:75,000 dilution of SYBR Green I (Molecular Probes, Eugene, OR, USA), 0.25 μM of each of primers, and 1 X AnyDirect Max qPCR Mix (BioQuest, Seoul, Korea). Amplification involved initial denaturation at 95℃ for 5 min followed by 50 cycles of denaturation at 95℃ for 40 s, annealing at 58℃ for 40 s, and extension at 72℃ for 40 s. Fluorescence data were acquired at 72℃ during each cycle. To determine the specificity of PCR reactions, melt curve analysis was carried out after amplification by slow heating from 72℃ to 95℃, with fluorescence acquisition at 1℃ intervals and a 5-s hold at each increment. Triplex real-time PCR was performed under the same conditions as above.
RESULTS
Measurement of mechanical allodynia
We first measured the mechanical withdrawal threshold in the rat L5 and L6 SNL model. Two weeks following SNL, animals showed significant mechanical allodynia as defined by responses to von Frey monofilament stimulation. Only rats with withdrawal thresholds less than 4 g on the plantar surface of the hind paw ipsilateral to the SNL injury were considered allodynic and used in this study. Specifically, responses of the injured paw of allodynic rats used in this study following SNL were 3.2 ± 0.5 g (mean ± SEM). These responses were significantly (p<0.001) lower than those for naive animals (14.2 ± 0.8 g).
Detection of differentially expressed genes
One day after behavioral testing, the DRG cells from six rats were removed and cultured for 3 days in the presence or absence of 25 μM of gabapentin. Fig. 1 shows the cultured DRG cells for 3 days in the absence and presence of gabapentin, which showed a characteristic neuronal morphology with processes extending from the condensing cell bodies. We then performed a differential expression analysis with random priming approach using an ACP-based DDRT-PCR to identify novel candidate genes that were specifically expressed in response to gabapentin. To do this, the total RNAs from gabapentin-treated cultured DRG cells and untreated cultured DRG cells were extracted and subjected to ACP-based DDRT-PCR analysis, and the PCR products were loaded onto 2% agarose gels. Up-regulated and down-regulated genes were identified by comparison of band intensities. Fig. 2 shows the results of the screening, which revealed 16 differentially expressed genes indicated by arrows including 13 up-regulated and 3 down-regulated genes in the samples of gabapentin-treated cultured DRG cells compared with untreated cultured DRG cells. These genes were purified, cloned and sequenced. BLAST searches in GenBank revealed that the differentially expressed genes displayed significant similarities with known genes. The sequence similarities of differentially expressed genes are summarized in Table 1.
Fig. 1. Morphology of cultured DRG cells for three days in the absence (A) and presence (B) of gabapentin. Magnification, ×100.
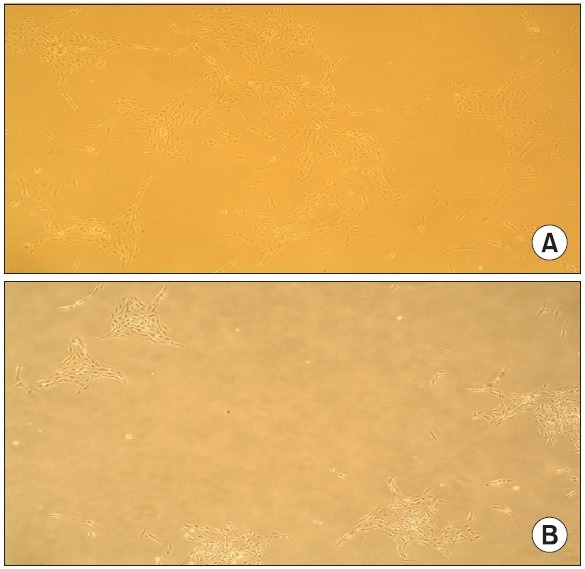
Fig. 2. Detection of differentially expressed genes in cultured DRG cells treated with gabapentin. Total RNA was extracted from DRG cells treated with gabapentin and subjected to ACP-based DDRT-PCR. Twenty arbitrary ACP primers (ACP1 to ACP20) were used to isolate the differentially expressed genes. Differential expression patterns were observed when the arbitrary ACP primer sets (indicated on the top) were used. The differential expression patterns were evaluated based on the band intensities. The arrows on the left-hand side indicate differential expressed bands between untreated DRG cells (U) and gabapentin-treated DRG cells (G). Bands were excised from the gel for sequencing. M, 100-bp DNA ladder.
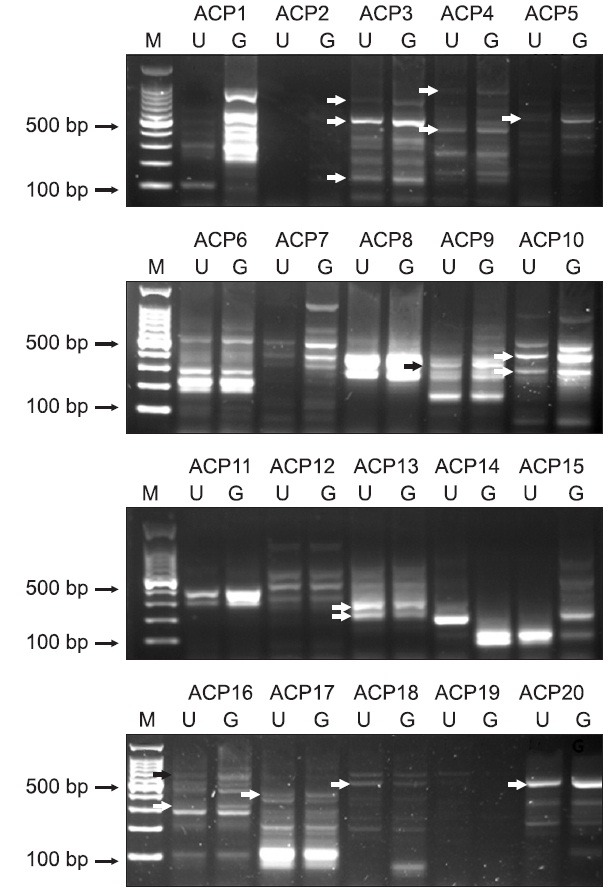
Table 1.
Sequence similarities of differentially expressed genes
| Clone name* | Identity | GeneBank accession No. |
|---|---|---|
| ACP3-1 (up) | Rattus norvegicus tropomyosin 2, beta (Tpm2) | NM_001024345 |
| ACP3-2 (up) | Rattus norvegicus spastic paraplegia 21 homolog (human) (Spg21) | NM_001006987 |
| ACP3-3 (up) | Rattus norvegicus strain Brown Norway clone CH230-206L9 | AC1469 |
| ACP4-1 (up) | Rattus norvegicus dipeptidylpeptidase 3, mRNA | BC107673 |
| ACP4-2 (up) | Rattus norvegicus TL0ABA40YJ05 mRNA | FQ209730 |
| ACP5 (up) | Rattus norvegicus metallothionein 1a, mRNA | BC058442 |
| ACP9 (up) | Rattus norvegicus growth arrest and DNA-damage-inducible, gamma (Gadd45g) | NM_001077640 |
| ACP10-1 (up) | Rattus norvegicus similar to LOC387763 protein (RGD1564664) | NM_001110055 |
| ACP10-2 (up) | Rattus norvegicus potassium channel modulatory factor 1 (Kcmf1), mRNA | NM_001128192 |
| ACP13-1 (dn) | Rattus norvegicus chromosome 20, major histocompatibility complex, assembled from 40 BACs, strain | BX883048 |
| ACP13-2 (dn) | Rattus norvegicus family with sequence similarity 96, member B (Fam96b) | NM_001144854 |
| ACP16-1 (up) | Mus musculus TGF-1 gene, exon 6 | L42461 |
| ACP16-2 (up) | Rattus norvegicus truncated palmitoyl-protein thioesterase (PPT-2) mRNA | AF067790 |
| ACP17 (up) | Rattus norvegicus TL0ACA52YI18 mRNA | FQ224907 |
| ACP18 (dn) | Rattus norvegicus TL0ACA31YH23 mRNA | FQ217242 |
| ACP20 (up) | Rattus norvegicus TL0AEA27YK05 mRNA | FQ226525 |
*Up or dn in parenthesis denotes up-regulated genes and down-regulated genes, respectively.
Confirmation of relative gene expression using real-time RT-PCR
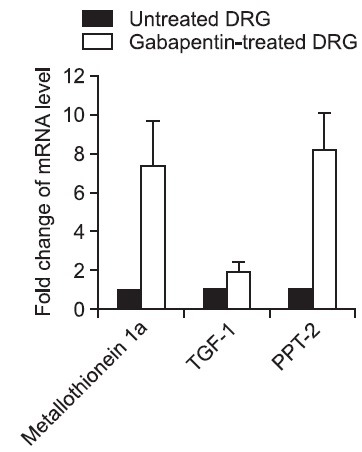
To confirm the efficacy of the ACP-based DDRT-PCR analysis, the differential expressions of three selected genes were confirmed by real-time RT-PCR using each gene specific primers. The sequences of the primers are shown in Table 2. We selected 3 up-regulated genes in the gabapentin-treated cultured DRG cells to validate the results of ACP-based DDRT-PCR analysis and to identify the relative expression of target sequences. The selected genes were metallothionein 1a, transforming growth factor-β1 (TGF-1) and palmitoyl-protein thioesterase-2 (PPT-2), because the functional roles of these genes were previously suggested as neuroprotective genes. Then, we compared the mRNA expression levels of these genes in the gabapentin-treated cultured DRG cells with untreated cultured DRG cells. To normalize the efficiency of real-time RT-PCR reaction, β-actin gene was used as an internal standard. The real-time RT-PCR assay revealed that, in accordance with the ACP-based DDRT-PCR analysis, metallothionein 1a, TGF-1 and PPT-2 were highly expressed in the gabapentin-treated DRG cells compared to the untreated control DRG cells (Fig. 3).
Table 2.
Primers used for real-time RT-PCR experiments
| Gene (clone name) | Primer sequence | Product size (bp) |
|---|---|---|
| β-actin | F*: 5’-TGCCCTGAGGCACTCTTC-3’ | 195 |
| R: 5’-GTGCCAGGGCAGTGATCT-3’ | ||
| Metallothionein 1a (ACP5) | F: 5’- ACCTCCTGCAAGAAGAGCTG -3’ | 190 |
| R: 5’- AAACTGGGTGGAGGTGTACG -3’ | ||
| TGF-1 (ACP16-1) | F: 5’-CAACAATTCCTGGCGTTACCTTGG-3’ | 128 |
| R: 5’-GAAAGCCCTGTATTCCGTCTCCTT-3’ | ||
| PPT-2 (ACP16-2) | F: 5’-CAGGCCACTCAGGACATTTT-3’ | 151 |
| R: 5’-CCTTGCAGGCCACAGATATT-3’ | ||
*F or R denotes forward primer and reverse primer, respectively.
Fig. 3. Confirmation by real-time RT-PCR of mRNA expression patterns of three selected genes. Real-time RT-PCR shows up-regulation of metallothionein 1a, TGF-1 and PPT-2 in cultured DRG cells treated with gabapentin. Data are shown as means +SD (bars) of samples conducted in triplicate determinations. To normalize the efficiency of real-time RT-PCR reaction, β-actin gene was used as an internal standard.
DISCUSSION
Gene expression profiling enables simultaneous analysis of the expression status of large number of genes that represent a comprehensive molecular signature of the tested samples and can be achieved by several approaches. Because of the large number of biochemical cascades and cellular reactions initiated after nerve injury, DNA microarrays have been used for broad analyses of gene transcription (Shalon, 1998). These arrays allow for scanning of large numbers of genes rapidly; however, these techniques have the relative disadvantage of being suitable only for analysis of a fixed number of predetermined gene sequences.
In this study, we used an ACP-based DDRT-PCR approach for differential expression profiling that was recently developed to allow direct visualization of the differentially expressed transcripts by agarose gel electrophoresis. This technology improves the specificity and sensitivity of PCR amplification so that it enables detection of only authentic PCR products. This approach has been previously adopted in differential expression studies of gastric cancer (Jang, 2004) and cervical cancer (Choi et al., 2007), as well as identifying developmental stage-specific genes in embryogenesis (Hwang et al., 2004).
Dorsal root ganglion (DRG) neurons have been the focus of much research to identify molecular targets of pain neurotransmission since they represent a primary site for pain processing. Previous studies using DRGs of animal pain models have measured mRNA levels by examining a targeted set of transcripts or through the use of global approaches such as microarray technology to study mRNA expression changes (Costigan et al., 2002; Xiao et al., 2002). A proteomic study demonstrated that 67 proteins are regulated in the SNL model (Komori et al., 2007).
We also used short-term cultured rat DRG cells instead of fresh tissue to study differential gene expression profiling, an approach taken for two reasons. Firstly, cultures were enriched in DRG neurons with very low non-neuronal gene contamination, thus biasing the data toward neuron specific gene expression changes associated with the vulnerability to painful stimuli triggered by inflammation or injury. Secondly, we wanted to see the direct effects of gabapentin on differential gene expression in cultured DRG cells of SNL model rats.
Since the complexity of the in vivo conditions makes it difficult to study the cellular and molecular mechanisms of pain, a useful alternative approach is an in vitro model of dissociated DRG neurons. Cultured DRG neurons are frequently used as model systems to study neuronal responses relating to pain, because the cultured DRG neurons can share characteristics with nociceptors in vivo (Passmore, 2005). A variety of chemical and environmental stimuli that might cause pain in vivo also cause stimulation on cultured DRG neurons (Senba et al., 2004). The use of cultured DRG neurons means that a defined mechanism can be more easily studied. The isolated neuron’s environment can easily be manipulated in vitro to try and reflect the in vivo environment or even the disease state. DRG cultures can be also used as a screening tool, because DRG neurons are very easy to harvest in adult animals and they survive well in culture.
Using the random priming ACP-based DDRT-PCR approach for differential expression profiling, we have identified 13 up-regulated and 3 down-regulated genes in the culture of DRG cells treated with gabapentin. The functional roles of all these genes that were identified were not fully characterized in terms of neuropathic pain. However, the functional roles of 3 up-regulated genes, metallothionein 1a, TGF-1 and PPT-2, were previously suggested as neuroprotective. Real-time RT-PCR was carried out to confirm the expression pattern of these three genes, and the data revealed that all of our target transcripts exhibited expression patterns consistent with the results of the ACP-based DDRT-PCR approach.
Metallothionein 1a is one of family of low-molecular-weight (6~7 kDa) intracellular metal-binding proteins. Metallothionein has been shown in a number of studies to exert a neuroprotective effect in the nervous system following injury or during disease and to stimulate regenerative growth (West et al., 2008).
The PPT-2 gene encodes a lysosomal thioesterase homologous to PPT-1, a lysosomal palmitoyl-protein thioesterase that cleaves thioester linkages in S-acylated proteins and removes palmitate residues facilitating the degradation of these proteins. PPT-1 is an essential lysosomal protein in the mammalian nervous system, defects result in a neurodegenerative disorder called infantile neuronal ceroid lipofuscinosis (Vesa et al., 1995). Although PPT2 deficiency has not been described in humans, PPT2 knock-out mice display many of the same neurodegenerative disorder phenotype and pathology (Gupta et al., 2003).
TGF-1, an injury-related cytokine, plays a crucial role in the regulation of neuronal survival (Krieglstein et al., 2002; Brionne et al., 2003). Its expression in the brain increases under conditions of neurodegenerative diseases and injuries. Previous studies have shown that within conditionally TGF-1-null mice, neuronal degeneration is widespread (Brionne et al., 2003), whereas TGF-1 over-expression results in neuroprotection from the mitochondrial toxin nitroproprionic acid (Ueberham et al., 2005). Recently, it has been reported that TGF-1 alleviates neuropathic pain in rats, and the suggested mechanism involves an inhibitory effect on the spinal cord inflammatory response to nerve injury and a neuroprotective effect on damaged neurons (Echeverry et al., 2009). Also, a recent study identified TGF-β family members as modulators of acute and chronic pain perception through the transcriptional regulation of genes encoding the endogenous opioids (Tramullas et al., 2010).
The detailed neuroprotective and pain related functions of these three genes and their products in neuropathic pain will require further study. But, this is a first attempt to analyze genes that are differentially expressed in cultured DRG cells of SNL model rats induced by gabapentin. Gabapentin interacts specifically with the α2δ-subunit of voltage-sensitive calcium channels (Gee et al., 1996). This property is believed to be of fundamental importance in mediating some, if not all, of the pharmacological effects of gabapentin. How this interaction contributes to the mechanism of action is not yet understood. Even in the absence of the altered subunit expression, gabapentin can exert anti-allodynic effects (Abe et al., 2002). Therefore, further studies of the presently identified genes are expected to reveal potential targets of gabapentin. Also, these genes and their products may be promising molecular targets for novel therapeutic agents for neuropathic pain management.
Acknowledgments
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2011-0023938).
References
- 1.Abe M., Kurihara T., Han W., Shinomiya K., Tanabe T. Changes in expression of voltage-dependent ion channel subunits in dorsal root ganglia of rats with radicular injury and pain. Spine (Phila. Pa 1976) (2002);27:1517–1524. doi: 10.1097/00007632-200207150-00007. [DOI] [PubMed] [Google Scholar]
- 2.Aldrich B. T., Frakes E. P., Kasuya J., Hammond D. L., Kitamoto T. Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience. (2009);164:711–723. doi: 10.1016/j.neuroscience.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baron R. Peripheral neuropathic pain: from mechanisms to symptoms. Clin. J. Pain. (2000);16:S12–20. doi: 10.1097/00002508-200006001-00004. [DOI] [PubMed] [Google Scholar]
- 4.Bridges D., Thompson S. W., Rice A. S. Mechanisms of neuropathic pain. Br. J. Anaesth. (2001);87:12–26. doi: 10.1093/bja/87.1.12. [DOI] [PubMed] [Google Scholar]
- 5.Brionne T. C., Tesseur I., Masliah E., Wyss-Coray T. Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron. (2003);40:1133–1145. doi: 10.1016/s0896-6273(03)00766-9. [DOI] [PubMed] [Google Scholar]
- 6.Cavenagh J., Good P., Ravenscroft P. Neuropathic pain: are we out of the woods yet? Intern. Med. J. (2006);36:251–255. doi: 10.1111/j.1445-5994.2006.01046.x. [DOI] [PubMed] [Google Scholar]
- 7.Chaplan S. R., Bach F. W., Pogrel J. W., Chung J. M., Yaksh T. L. Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods. (1994);53:55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 8.Choi Y. W., Kim Y. W., Bae S. M., Kwak S. Y., Chun H. J., Tong S. Y., Lee H. N., Shin J. C., Kim K. T., Kim Y. J., Ahn W. S. Identification of differentially expressed genes using annealing control primer-based GeneFishing in human squamous cell cervical carcinoma. Clin. Oncol. (2007);19:308–318. doi: 10.1016/j.clon.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Costigan M., Befort K., Karchewski L., Griffin R. S., D'Urso D., Allchorne A., Sitarski J., Mannion J. W., Pratt R. E., Woolf C. J. Replicate high-density rat genome oligonucleotide microarrays reveal hundreds of regulated genes in the dorsal root ganglion after peripheral nerve injury. BMC Neurosci. (2002);3:16. doi: 10.1186/1471-2202-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Echeverry S., Shi X. Q., Haw A., Liu H., Zhang Z. W., Zhang J. Transforming growth factor-beta1 impairs neuropathic pain through pleiotropic effects. Mol. Pain. (2009);5:16. doi: 10.1186/1744-8069-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee N. S., Brown J. P., Dissanayake V. U., Offord J., Thurlow R., Woodruff G. N. The novel anticonvulsant drug, gabapentin (Neurontin), binds to the alpha2delta subunit of a calcium channel. J. Biol. Chem. (1996);271:5768–5776. doi: 10.1074/jbc.271.10.5768. [DOI] [PubMed] [Google Scholar]
- 12.Granados-Soto V., Sánchez-Ramirez G., la Torre M. R., Caram-Salas N. L., Medina-Santillán R., Reyes-García G. Effect of diclofenac on the antiallodynic activity of vitamin B12 in a neuropathic pain model in the rat. Proc. West. Pharmacol. Soc. (2004);47:92–94. [PubMed] [Google Scholar]
- 13.Gupta P., Soyombo A. A., Shelton J. M., Wilkofsky I. G., Wisniewski K. E., Richardson J. A., Hofmann S. L. Disruption of PPT2 in mice causes an unusual lysosomal storage disorder with neurovisceral features. Proc. Natl. Acad. Sci. U.S.A. (2003);100:12325–12330. doi: 10.1073/pnas.2033229100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunter J. C., Gogas K. R., Hedley L. R., Jacobson L. O., Kassotakis L., Thompson J., Fontana D. J. The effect of novel anti-epileptic drugs in rat experimental models of acute and chronic pain. Eur. J. Pharmacol. (1997);324:153–160. doi: 10.1016/s0014-2999(97)00070-8. [DOI] [PubMed] [Google Scholar]
- 15.Hwang I. T., Kim Y. J., Kim S. H., Kwak C. I., Gu Y. Y., Chun J. Y. Annealing control primer system for improving specificity of PCR amplification. Biotechniques. (2003);35:1180–1184. doi: 10.2144/03356st03. [DOI] [PubMed] [Google Scholar]
- 16.Hwang K. C., Cui X. S., Park S. P., Shin M. R., Park S. Y., Kim E. Y., Kim N. H. Identification of differentially regulated genes in bovine blastocysts using an annealing control primer system. Mol. Reprod. Dev. (2004);69:43–51. doi: 10.1002/mrd.20156. [DOI] [PubMed] [Google Scholar]
- 17.Iyengar S., Webster A. A., Hemrick-Luecke S. K., Xu J. Y., Simmons R. M. Efficacy of duloxetine, a potent and balanced serotonin-norepinephrine reuptake inhibitor in persistent pain models in rats. J. Pharmacol. Exp. Ther. (2004);311:576–584. doi: 10.1124/jpet.104.070656. [DOI] [PubMed] [Google Scholar]
- 18.Jang J. H. FIGC, a novel FGF-induced ubiquitin-protein ligase in gastric cancers. FEBS Lett. (2004);578:21–25. doi: 10.1016/j.febslet.2004.10.071. [DOI] [PubMed] [Google Scholar]
- 19.Kim H. K., Kim J. H., Gao X., Zhou J. L., Lee I., Chung K., Chung J. M. Analgesic effect of vitamin E is mediated by reducing central sensitization in neuropathic pain. Pain. (2006);122:53–62. doi: 10.1016/j.pain.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Kim S. H., Chung J. M. An experimental model for peripheral neuropathy produced by segmental spinal nerve ligation in the rat. Pain. (1992);50:355–363. doi: 10.1016/0304-3959(92)90041-9. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y. J., Kwak C. I., Gu Y. Y., Hwang I. T., Chun J. Y. Annealing control primer system for identification of differentially expressed genes on agarose gels. Biotechniques. (2004);36:424–426. doi: 10.2144/04363ST02. [DOI] [PubMed] [Google Scholar]
- 22.Komori N., Takemori N., Kim H. K., Singh A., Hwang S. H., Foreman R. D., Chung K., Chung J. M., Matsumoto H. Proteomics study of neuropathic and nonneuropathic dorsal root ganglia: altered protein regulation following segmental spinal nerve ligation injury. Physiol. Genomics. (2007);29:215–230. doi: 10.1152/physiolgenomics.00255.2006. [DOI] [PubMed] [Google Scholar]
- 23.Krieglstein K., Strelau J., Schober A., Sullivan A., Unsicker K. TGF-beta and the regulation of neuron survival and death. J. Physiol. (Paris) (2002);96:25–30. doi: 10.1016/s0928-4257(01)00077-8. [DOI] [PubMed] [Google Scholar]
- 24.Lee Y. W., Chaplan S. R., Yaksh T. L. Systemic and supraspinal, but not spinal, opiates suppress allodynia in a rat neuropathic pain model. Neurosci. Lett. (1995);199:111–114. doi: 10.1016/0304-3940(95)12034-2. [DOI] [PubMed] [Google Scholar]
- 25.Passmore G. M. Dorsal root ganglion neurons in culture: a model system for identifying novel analgesic targets. J. Pharmacol. Toxicol. Methods. (2005);51:201–208. doi: 10.1016/j.vascn.2004.08.007. [DOI] [PubMed] [Google Scholar]
- 26.Senba E., Katanosaka K., Yajima H., Mizumura K. The immunosuppressant FK506 activates capsaicin- and bradykinin-sensitive DRG neurons and cutaneous C-fibers. Neurosci. Res. (2004);50:257–262. doi: 10.1016/j.neures.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 27.Shalon D. Gene expression micro-arrays: a new tool for genomic research. Pathol. Biol. (Paris) (1998);46:107–109. [PubMed] [Google Scholar]
- 28.Tramullas M., Lantero A., Díaz A., Morchón N., Merino D., Villar A., Buscher D., Merino R., Hurlé J. M., Izpisúa-Belmonte J. C., Hurlé M. A. BAMBI (bone morphogenetic protein and activin membrane-bound inhibitor) reveals the involvement of the transforming growth factor-beta family in pain modulation. J. Neurosci. (2010);30:1502–1511. doi: 10.1523/JNEUROSCI.2584-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueberham U., Ueberham E., Brückner M. K., Seeger G., Gärtner U., Gruschka H., Gebhardt R., Arendt T. Inducible neuronal expression of transgenic TGF-beta1 in vivo: dissection of short-term and long-term effects. Eur. J. Neurosci. (2005);22:50–64. doi: 10.1111/j.1460-9568.2005.04189.x. [DOI] [PubMed] [Google Scholar]
- 30.Valder C. R., Liu J. J., Song Y. H., Luo Z. D. Coupling gene chip analyses and rat genetic variances in identifying potential target genes that may contribute to neuropathic allodynia development. J. Neurochem. (2003);87:560–573. doi: 10.1046/j.1471-4159.2003.02016.x. [DOI] [PubMed] [Google Scholar]
- 31.Vesa J., Hellsten E., Verkruyse L. A., Camp L. A., Rapola J., Santavuori P., Hofmann S. L., Peltonen L. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. (1995);376:584–587. doi: 10.1038/376584a0. [DOI] [PubMed] [Google Scholar]
- 32.Wang H., Sun H., Della Penna K., Benz R. J., Xu J., Gerhold D. L., Holder D. J., Koblan K. S. Chronic neuropathic pain is accompanied by global changes in gene expression and shares pathobiology with neurodegenerative diseases. Neuroscience. (2002);114:529–546. doi: 10.1016/s0306-4522(02)00341-x. [DOI] [PubMed] [Google Scholar]
- 33.West A. K., Hidalgo J., Eddins D., Levin E. D., Aschner M. Metallothionein in the central nervous system: Roles in protection, regeneration and cognition. Neurotoxicology. (2008);29:489–503. doi: 10.1016/j.neuro.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao H. S., Huang Q. H., Zhang F. X., Bao L., Lu Y. J., Guo C., Yang L., Huang W. J., Fu G., Xu S. H., Cheng X. P., Yan Q., Zhu Z. D., Zhang X., Chen Z., Han Z. G., Zhang X. Identification of gene expression profile of dorsal root ganglion in the rat peripheral axotomy model of neuropathic pain. Proc. Natl. Acad. Sci. U.S.A. (2002);99:8360–8365. doi: 10.1073/pnas.122231899. [DOI] [PMC free article] [PubMed] [Google Scholar]


