SUMMARY
During development of the embryonic neocortex, tightly regulated expansion of neural stem cells (NSCs) and their transition to intermediate progenitors (IPs) are critical for normal cortical formation and function. Molecular mechanisms that regulate NSC expansion and transition remain unclear. Here, we demonstrate that the microRNA (miRNA) miR-17-92 cluster is required for maintaining proper populations of cortical radial glial cells (RGCs) and IPs through repression of Pten and Tbr2 protein. Knockout of miR-17-92 and its paralogs specifically in the developing neocortex restricts NSC proliferation, suppresses RGC expansion, and promotes transition of RGCs to IPs. Moreover, Pten and Tbr2 protectors specifically block silencing activities of endogenous miR-17-92 and control proper numbers of RGCs and IPs in vivo. Our results demonstrate a critical role for miRNAs in promoting NSC proliferation and modulating the cell-fate decision of generating distinct neural progenitors in the developing neocortex.
INTRODUCTION
In the developing neocortex, self-renewal of neural stem cells (NSCs), proliferation of neural progenitors, and subsequent differentiation are regulated by conserved complex interactions of multiple genes (Guillemot, 2005; Kriegstein et al., 2006; Merkle and Alvarez-Buylla, 2006; Molyneaux et al., 2007; Shen et al., 2006). Radial glial cells (RGCs), the primary cortical neural progenitors, are transformed from neuroepithelial cells/NSCs and reside in the ventricular zone (VZ). Intermediate progenitors (IPs) or basal progenitors are transited from RGCs and populate mostly the subventricular zone (SVZ) (Chenn and McConnell, 1995; Englund et al., 2005; Götz and Huttner, 2005; Haubensak et al., 2004; Noctor et al., 2001; Rakic, 2003). Expansion of NSCs is regulated by both positive and negative factors. For example, ablation of the tumor suppressor gene Pten results in a larger cortex and an expansion of NSCs and neural progenitors (Groszer et al., 2001; Zheng et al., 2008), and transcription factor Tbr2 promotes expansion of IPs and elevates IP transition from RGCs (Arnold et al., 2008; Englund et al., 2005; Sessa et al., 2008, 2010). However, the accurate modulation of expression levels of positive and negative regulators that control proper expansion of NSCs and RGCs and transition to IPs is not well understood.
Emerging evidence has shown that microRNAs (miRNAs) play an important role in cortical development. Global depletion of miRNA functions using Dicer ablation results in a smaller cortex and affects NSCs survival and differentiation (Andersson et al., 2010; De Pietri Tonelli et al., 2008;Kawase-Koga et al., 2009, 2010; Nowakowski et al., 2011). miRNAs are frequently transcribed together as polycistronic primary transcripts that are processed into multiple individual mature miRNAs. An important miRNA polycistron is the miR-17-92 cluster and its paralogs miR-106a-363 and miR-106b-25 (Mendell, 2008). Knockout mice of the miR-17-92 cluster and paralogs display embryonic lethality, indicating a critical role in mouse development (Ventura et al., 2008). miR-17-92 has been shown to be oncogenic and promotes tumorigenesis (Mavrakis et al., 2010; Mu et al., 2009; Olive et al., 2009). Considering the conserved targets of the miR-17-92 family in different tissues, miR-17-92 likely plays a general role in cell proliferation and survival during normal development and under tumorigenesis.
We here show that knockout of the miR-17-92 cluster restricts expansion of NSCs and RGCs and promotes transition to IPs. miR-17-92 balances the proper RGC and IP populations by suppressing Pten and Tbr2. Pten and Tbr2 mRNA protectors can specifically block endogenous miR-17-92 silencing activities and regulate RGC and IP numbers. Our results demonstrate that the miR-17-92 cluster is an important regulator controlling distinct neural progenitor populations by balancing proper protein output.
RESULTS
The miR-17-92 Family Promotes Expansion of RGCs and NSCs
The miR-17-92 cluster and paralogs yield 15 mature miRNAs that can be categorized into miR-17, miR-18, miR-19, and miR-92 subfamilies according to their conserved seed sequences (Figure 1A). Due to the function of the miR-17-92 family in regulating cancer cell proliferation, we predicted that miR-17-92 may be essential for neural progenitor expansion in the embryonic cortex. We first examined expression levels of miR-17-92 using the northern blotting assay. All miR-17-92 subfamilies were expressed in developing mouse cortices, with high expression levels in embryonic day 12.5 (E12.5) cortices and low levels in postnatal day 0 (P0) cortices (Figure S1). Moreover, miR-17, 19a, 92a, and 106a displayed strong expression in the VZ in E12.5 cortices detected by in situ hybridization (Figure S1). The high expression of the miR-17-92 family in the VZ suggests their role in cortical NSC and neural progenitor development.
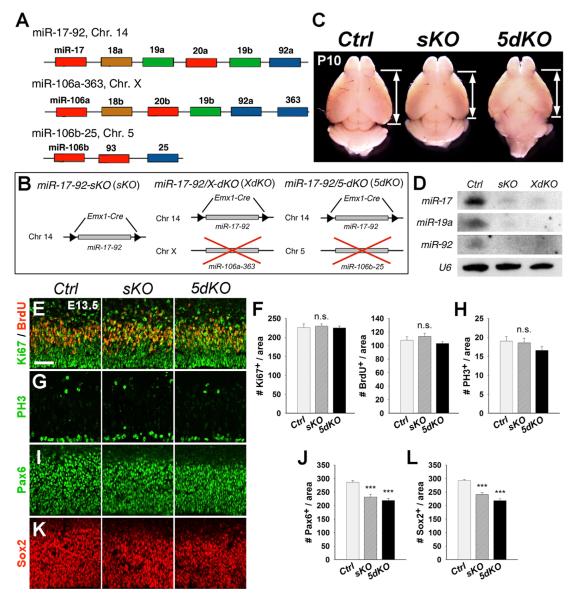
Figure 1. Deletion of the miR-17-92 Cluster and Its Paralogs Causes Reduced Number of Radial Glial Cells (RGCs) but Not Overall Neural Progenitors in E13.5 Cortices.
(A) The schematic genomic organization of the miR-17-92 cluster and its paralogs miR-106a-363 and miR-106b-25 on the mouse chromosome 14 (Chr. 14), Chr. X, and Chr. 5, respectively.
(B) The strategy of generating miR-17-92 single- and double-knockout mice using the Emx1-Cre line.
(C) Dorsal view of control (Ctrl), miR-17-92 sKO, and 5dKO brains at P10. Reduced cortical size was found in miR-17-92 KO mice.
(D) Reduced expression levels of representative miRNAs of the miR-17-92 cluster in miR-17-92 KO cortices detected by northern blots. U6 was used as a loading control.
(E and F) Numbers of BrdU+ and Ki67+ progenitors were unaffected in miR-17-92 KO cortices compared to controls.
(G and H) Numbers of PH3+ cells did not show significant changes in miR-17-92 KO cortices.
(I–L) Numbers of Pax6- and Sox2-expressing RGCs were decreased in miR-17-92 KO cortices.
Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (***p < 0.001). n.s., not significant. See also Figures S1 and S2.
To determine the function of the miR-17-92 family in the developing cortex, we generated mouse models in which miR-17-92 and its paralogs were genetically ablated by applying the Creloxp system using Emx1-Cre mice due to their specific activity in the cortex (Gorski et al., 2002). Emx1-Cre mice were bred with floxed miR-17-92 transgenic mice to generate miR-17-92flox/flox;Emx1-Cre mice, called miR-17-92 single-knockout (KO) mice (miR-17-92 sKO, sKO) (Figure 1B). We also bred miR-17-92 sKO mice with miR-106a-363 or miR-106b-25 knockout mice, called XKO and 5KO, respectively, to generate double-KO (dKO) mice, called miR-17-92/X-dKO, XdKO, and miR-17-92/5-dKO, 5dKO, respectively (Figure 1B). miR-17-92 KO mice displayed slightly reduced cortical thickness at P1, and the reduction was more pronounced at P10 (Figures 1C and S1). Moreover, expression levels of representative miRNAs were decreased in P1 KO cortices detected using the northern blotting assay (Figure 1D). miRNAs in miR-106a-363 and miR-106b-25 paralogs were not upregulated in miR-17-92 sKO cortices as detected by quantitative real-time RT-PCR (qRT-PCR) (data not shown). Our results indicate a successful cortical depletion of miR-17-92 and no significant compensation by the other two paralogs after miR-17-92 deletion.
We next examined miR-17-92 function in neural progenitor and NSC development in miR-17-92 KO mice. Neural progenitors can be detected by labeling cells in the S phase with a 30 min pulse of bromodeoxyuridine (BrdU), in the G1, S, G2, and M phase with Ki67, and in the M phase with phospho-histone H3 (PH3). Numbers of BrdU+, Ki67+, and PH3+ cells were not changed in E13.5 miR-17-92 sKO and 5dKO cortices compared to wild-type controls (Figures 1E–1H). However, the numbers and percentages of Pax6+ and Sox2+ RGCs were significantly reduced in E13.5 and E15.5 miR-17-92 KO cortices, suggesting a continuous loss of RGCs (Figures 1I–1L; data not shown). There was no significant increase in apoptotic cells in E13.5, E15.5, or P1 miR-17-92 KO cortices as detected by a TUNEL assay (data not shown).
We then cultured NSCs collected from E13.5 cortices as neurospheres. After two passages, compared to controls, the numbers and sizes of the neurospheres derived from miR-17-92 sKO cortices were reduced (Figure S2). miR-17-92-deficient neurospheres properly expressed NSC markers such as Nestin and SSEA-1. However, after a 6 hr pulse of BrdU labeling on attached NSCs, 40% fewer miR-17-92-deficient NSCs had incorporated BrdU compared to controls (Figure S2). Our results indicate that, under culture conditions with FGF2 that maintains self-renewal of NSCs and RGCs (Kang et al., 2009), the miR-17-92 cluster enhances growth of cortical NSCs by promoting proliferation.
The miR-17-92 Family Suppresses Transition from RGCs to IPs
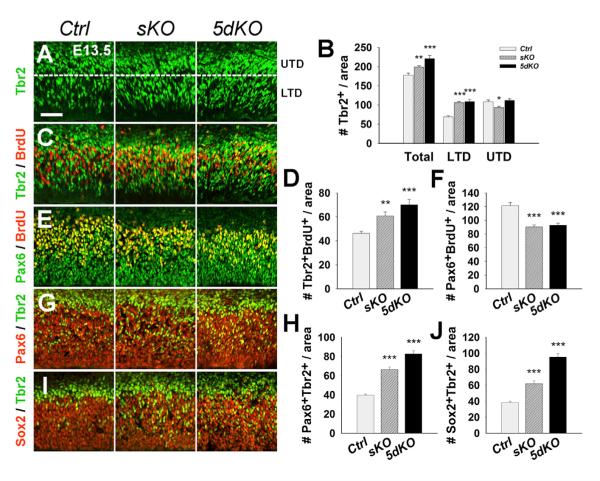
We next questioned why the overall number of neural progenitors was not changed while the number of RGCs was reduced in miR-17-92 KO cortices. We speculated that the RGC reduction might be caused by abnormal transition to IPs. We divided the Tbr2-expressing area into two subdomains: the “lower Tbr2 domain” (LTD), where IPs are just derived from RGCs in the VZ, and the “upper Tbr2 domain” (UTD), where IPs differentiate into neurons in the SVZ (Munji et al., 2011). We found a significant increase of Tbr2+ IPs in the LTD of E13.5 miR-17-92 KO cortices compared with controls, but the number of Tbr2+ cells in the UTD was unchanged or slightly reduced (Figures 2A and 2B). While the number of Tbr2+/BrdU+ was increased, the number of Pax6+/BrdU+ was decreased in miR-17-92 KO cortices, indicating that miR-17-92 depletion promotes transition into proliferative IPs from dividing RGCs (Figures 2C–2F).
Figure 2. The miR-17-92 Cluster Suppresses Transition of Cortical Intermediate Progenitors.
(A) Increased numbers of IPs labeled with Tbr2 in E13.5 miR-17-92 sKO and 5dKO cortices as compared with controls (Ctrl). The expression region of Tbr2 was divided into the “lower Tbr2 domain” (LTD) and the “upper Tbr2 domain” (UTD).
(B) Quantification of the total number of IPs and IPs within the UTD and LTD.
(C and D) Numbers of Tbr2+/BrdU+ IPs were significantly higher in miR-17-92 KO cortices than controls.
(E and F) Numbers of Pax6+/BrdU+ RGCs in miR-17-92 KO cortices were significantly less compared to controls.
(G–J) Increased transition to IPs from RGCs as detected by Tbr2+/Pax6+ and Tbr2+/Sox2+ cells in miR-17-92 KO cortices compared to controls. Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*p < 0.03, **p < 0.007, ***p < 0.001). See also Figures S3, S4, and S5.
We next examined RGC to IP transition by colabeling Tbr2 with Pax6 and Sox2. A significant increase in the numbers of Tbr2+/Pax6+ and Tbr2+/Sox2+ cells was detected in miR-17-92 KO cortices, suggesting that more RGCs were in the process of converting into IPs (Figures 2G–2J). The increased transition to IPs from RGCs was maintained in E15.5 miR-17-92 KO cortices (Figure S3). However, in E17.5 miR-17-92 KO cortices, decreased numbers of Pax6+ RGCs and Tbr2+ IPs were detected, suggesting a reduction of the IP population after E15.5, likely due to progressive loss of RGCs (Figure S4). Furthermore, while miR-17-92 sKO cortices showed no changes in the ratio of Tbr2+/BrdU+ cells to Tbr2+ cells throughout development, 5dKO cortices revealed a transient increase of proliferative Tbr2+ cells at E15.5 followed by a decrease at E17.5 (Figure S4). Our results indicate that the transient increase in total Tbr2+ cells in miR-17-92 KO cortices is largely caused by an elevated generation of IPs from RGCs.
Cortical Ablation of the miR-17-92 Family Affects Neuronal Production
To determine whether neuronal production was affected, we examined early-born and late-born neurons. The numbers of Tbr1+ cells were temporarily increased in E13.5 cortices but subsequently decreased in E15.5 and P1 cortices of miR-17-92 KO mice (Figures S5A–5C). A transient increase of Satb2+ and Cux1+ neurons was detected in E15.5 cortices, and a decrease of those cells was observed in E17.5 and P1 cortices of miR-17-92 KO mice (Figures S5D–S5F; data not shown). We then normalized relative numbers of RGCs and IPs at E13.5, E15.5, and E17.5 to those of E13.5 controls. Compared to E13.5 controls, a consistent reduction of RGCs was detected at all three stages in miR-17-92 KO cortices, while an increase of IPs was detected at E13.5 and E15.5, and a decrease of IPs was observed at E17.5 (Figure S5G). Moreover, we normalized relative numbers of Tbr1+ and Satb2+ neurons to those of E13.5 and E15.5 controls, respectively. These neurons were temporarily increased in miR-17-92 KO cortices at E13.5 and E15.5, which is consistent with the ectopic transition from RGCs to IPs at these stages (Figure S5H). However, a reduction of both Tbr1+ and Satb2+ neurons was detected after E15.5 and E17.5, respectively, when the numbers of RGCs and IPs were decreased in miR-17-92 KO cortices. Our results suggest that cortical miR-17-92 deletion suppresses RGC expansion and promotes RGC transition to IPs, which results in a transient increase of neuronal production in early developmental stages and a reduction of neurogenesis in late stages, likely due to exhaustion of the RGC pool (Figure S5I).
miR-17-92 Regulates RGC Expansion by Targeting Pten
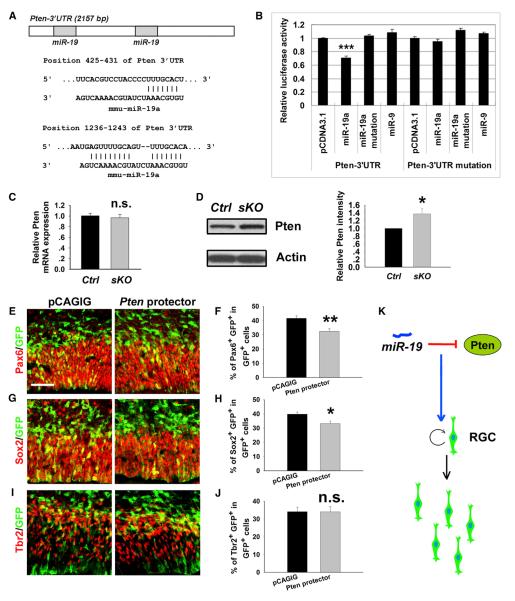
miRNAs function by silencing target genes. Since the miR-17-92 family promotes RGC expansion and suppresses IP transition, we speculated that their target genes likely negatively regulate RGC expansion and positively direct IP transition. We found that the 3′ untranslated region (3′ UTR) of Pten contains two targeting sites for miR-19a (Figure 3A). To validate the targeting effect, we performed a luciferase assay. The 3′ UTR sequence of Pten was cloned into a luciferase vector and cotransfected with miR-19a. While luciferase activities in constructs containing the 3′ UTR of Pten were not affected by the mutated miR-19a or a control miRNA miR-9, they were significantly reduced by miR-19a (Figure 3B). Furthermore, Pten 3′ UTR containing mutations in the seed sequence of miR-19a binding sites abolished miR-19a-targeting effects, proving Pten as a specific target of miR-19a (Figure 3B).
Figure 3. Pten Is a Putative Target Gene of miR-19a in Regulating Cortical RGC Numbers.
(A) Predicted targeting sites of miR-19a on the 3′ untranslated region (3′ UTR) of Pten.
(B) Luciferase assays of miR-19a targeting effects on the Pten 3′ UTR. miR-19a but not the mutation of miR-19a and control miRNA miR-9 recognized the 3′ UTR sequence of Pten and reduced luciferase activities. Pten 3′ UTR mutation for the seed sequences of miR-19a binding sites abolished miR-19a targeting effects.
(C) Pten mRNA level was not affected in E15.5 miR-17-92 sKO cortices compared to controls (Ctrl), as detected by qRT-PCR.
(D) The protein level of Pten was increased in the E15.5 miR-17-92 sKO cortex, detected by western blotting assays.
(E–H) Pten mRNA protectors specifically blocked silencing effects of miR-19 in the Pten 3′ UTR in vivo. Ectopic expression of Pten protectors in E13.5 mouse cortices, analyzed at E14.5, decreased the number of RGCs colabeled with GFP and Pax6 or Sox2.
(I and J) Ectopic expression of Pten protectors had no effect on the number of IPs colabeled with GFP and Tbr2.
(K) A summary of miR-19 function on expanding RGCs by repressing Pten.
Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*p < 0.03, **p < 0.004, ***p < 0.001). n.s., not significant. See also Figure S6.
We then tested whether the miR-17-92 silencing effect on Pten is due to degradation of messenger RNA (mRNA) or blockage of translation. Total RNA and protein were extracted from E15.5 control and miR-17-92 sKO cortices. Pten mRNA levels were not significantly altered between the control and miR-17-92 sKO cortices as detected by qRT-PCR (Figure 3C). Conversely, protein levels of Pten were increased in miR-17-92 sKO cortices detected by the western blotting assay, indicating a posttranscriptional regulation of Pten by miR-17-92 (Figure 3D).
To validate the specificity of the miR-19 targeting effect on the Pten 3′ UTR, we designed Pten mRNA protectors, which are complementary sequences that bind to the Pten 3′ UTR and block access to the miR-19 targeting sites (Zhang et al., 2012) (Figure S6A). Pten protectors showed a blocking effect on miR-19-silencing activities in the Pten 3′ UTR in luciferase assays (Figure S6B). We next introduced Pten protectors in E13.5 cortices using in utero electroporation, which should partially block endogenous silencing activities of miR-19 and result in phenotypes similar to the upregulation of Pten. The numbers of Pax6+ and Sox2+ RGCs were significantly decreased, while the number of Tbr2+ cells was not greatly changed, after Pten protectors were electroporated (Figures 3E–3J). Our results indicate a specific targeting regulation of miR-19 on Pten activity in expanding RGCs (Figure 3K).
miR-17-92 Balances Tbr2 Activity on Transition of RGCs to IPs
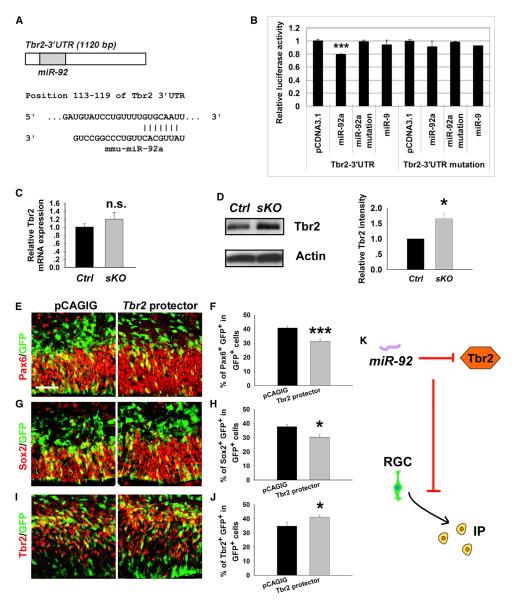
Our results have shown that knockout of miR-17-92 increases generation of IPs in the embryonic neocortex. Because Tbr2 has been shown to promote expansion of IPs, we examined the Tbr2 3′ UTR and found a binding site for miR-92a (Figure 4A). We next performed luciferase assays. Luciferase activities in the construct containing the 3′ UTR of Tbr2 were reduced by miR-92a, but not by the mutated miR-92a or miR-9 (Figure 4B). Tbr2 3′ UTR containing mutations in the seed sequence of miR-92a binding sites abolished miR-92a targeting effects, indicating Tbr2 as a specific target of miR-92 (Figure 4B). Moreover, similar to Pten, Tbr2 was regulated at a posttranscriptional level by miR-92 (Figures 4C and 4D).
Figure 4. miR-92a Regulates Cortical IP Numbers by Targeting Tbr2.
(A) A predicted targeting site of miR-92a on the 3′ UTR of Tbr2.
(B) Luciferase activities of the Tbr2 3′ UTR construct were greatly reduced by miR-92a, but not by miR-92a mutation and miR-9. Tbr2 3′ UTR mutation for the seed sequence of miR-92a abolished miR-92a targeting effects.
(C) Tbr2 mRNA level was not affected in E15.5 miR-17-92 sKO cortices compared to controls (Ctrl), as detected by qRT-PCR.
(D) The protein level of Tbr2 was increased in the E15.5 miR-17-92 sKO cortex, detected by western blotting assays.
(E–H) Ectopic expression of Tbr2 protectors in E13.5 mouse cortices, analyzed at E14.5, decreased the number of RGCs colabeled with GFP and Pax6 or Sox2.
(I and J) Ectopic expression of Tbr2 protectors increased the number of IPs colabeled with GFP and Tbr2.
(K) A summary of miR-92 function on the transition to IPs from RGCs by repressing Tbr2.
Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*p < 0.04, ***p < 0.005). See also Figure S6.
We next examined the specific targeting effect of miR-92 on Tbr2 expression in neural progenitor development using Tbr2 mRNA protectors (Figure S6). In E14.5 cortices electroporated with Tbr2 protectors at E13.5, the numbers of Pax6+ and Sox+ cells were significantly decreased, while the number of Tbr2+ cells was increased, indicating a specific blockade of silencing activity of the endogenous miR-92 on the Tbr2 3′ UTR in regulating IP transition from RGCs (Figures 4E–4K).
DISCUSSION
Maintaining proper populations of distinct neural progenitors is essential for controlling the precise architectural assembly of a functional neocortex. In this study, we have demonstrated an essential role of the miR-17-92 cluster in controlling expansion of NSCs and RGCs, and transition to IPs. Our results reveal an important regulatory mechanism of miR-17-92 in balancing expression levels of target gene Pten, which negatively controls RGC production, and Tbr2, which positively promotes IP generation.
During early neurogenesis, RGCs divide asymmetrically to generate one RGC and one postmitotic neuron (PN) (Molyneaux et al., 2007). As neurogenesis proceeds, RGCs give rise to IPs that normally undergo symmetric division to directly produce PNs (Noctor et al., 2007). Emerging studies have shown that, like coding genes, miRNAs play critical roles in cortical development (Bian and Sun, 2011; Fineberg et al., 2009; Lang and Shi, 2012; Qureshi and Mehler, 2012). We here show that even though the overall numbers of neural progenitors were not changed, a significant reduction of RGCs and an increase of IPs were detected in miR-17-92 KO cortices. The elevated transition to IPs from RGCs in miR-17-92 KO cortices is likely because more RGCs acquire IP cell fate due to upregulated expression of Tbr2, a putative target of miR-92. Therefore, the normal regulatory function of the miR-17-92 family is to promote the expansion of RGCs and to suppress transition to IPs.
The altered proportions of the RGC and IP populations in the miR-17-92 KO cortices ultimately perturb proper neurogenesis. While neuronal production is temporarily increased in miR-17-92 KO cortices at E13.5 and E15.5, it is eventually reduced after E17.5 when numbers of both RGCs and IPs are decreased. These phenotypes are likely caused by the following factors. First, the continuous reduction of RGCs may have a more profound impact on neurogenesis than the transient increase of IPs. Second, increased conversion to IPs may progressively exhaust the RGC pool, which eventually results in a reduced IP population. Thus, reduced neurogenesis at perinatal stages is a result of the overall disruption of proportional RGC and IP development when miR-17-92 is ablated.
Most functional studies of the miR-17-92 cluster have been focused on tumorigenesis, in which miR-17-92 promotes proliferation and survival of tumor cells (Mavrakis et al., 2010; Mu et al., 2009; Olive et al., 2009). In our study, we have validated Pten as a target for miR-19 during cortical development, suggesting a conserved role of miR-17-92 in regulating progenitor expansion. Furthermore, we have identified a target gene of the miR-17-92 cluster—Tbr2, which normally diverts and suppresses RGC development (Arnold et al., 2008; Englund et al., 2005; Sessa et al., 2008). An important role of miR-17-92 is likely to ensure proper protein output of factors such as Pten and Tbr2 in neural progenitors, which, in turn, controls accurate progenitor development and specification, and modulates subsequent neuronal production. Even though the changes of Pten and Tbr2 protein levels by miR-17-92 are relatively moderate, the outcome of progenitor alteration is significant. These results further support the hypothesis that fine-tuning gene regulation by miRNAs plays a critical role in development (Hobert, 2007;Karres et al., 2007).
Because there are four subfamilies with distinct seed sequences within the miR-17-92 family, it is expected that perhaps there are more genes directly affected by the miR-17-92 family in the cortex other than Pten and Tbr2. The next step will be to identify more target genes and establish an interaction network of miR-17-92 and its targets, which collaborate to modulate the expansion and conversion of NSCs and different types of progenitors. Nonetheless, in the current scope of our work, we have demonstrated two major players, Pten and Tbr2, which respond to miR-17-92 silencing activity in cortical NSC and neural progenitor development. Our work has revealed a mechanism of miRNAs controlling distinct progenitor populations in the developing neocortex.
EXPERIMENTAL PROCEDURES
Transgenic Mouse Lines
To knock out the miR-17-92 cluster in the cortex, Emx1-Cre mice (The Jackson Laboratory) were bred with floxed miR-17-92 mice to generate miR-17-92flox/flox;Emx1-Cre, called here miR-17-92 single-knockout (KO) mice. miR-106a-363 and miR-106b-25 knockout mice were maintained as homozygote (Ventura et al., 2008). To generate double-knockout mice, miR-17-92flox/flox mice were first bred with miR-106a-363 or miR-106b-25 knockout mice, and then bred with miR-17-92flox/+;Emx1-Cre mice.
For staging of embryos, midday of the day of vaginal-plug formation was considered E0.5; the first 24 hr after birth were defined as P0. Animal use was overseen by the Animal Facility at Weill Cornell Medical College.
mRNA Protectors for Pten and Tbr2
The mRNA protectors for mouse Pten and Tbr2 were designed as 60 bp complementary sequences covering the miRNA binding sites in their 3′ UTRs. After annealing, they were inserted into the pCAGIG vector for electroporation, and into the pCDNA3.1 vector for the luciferase assay.
Statistics
For the neurosphere-formation assay, three independent experiments were performed. For electroporated mouse sections, at least three brains from each group were analyzed. Statistical comparison was made by an analysis of variance (unpaired Student’s t test).
For further details, please refer to Extended Experimental Procedures.
Supplementary Material
Figure S1. The miR-17-92 Family Is Expressed in Neural Progenitors in the Mouse Embryonic Neocortex, Related to Figure 1
(A) Expression levels of representative subfamilies of the miR-17-92 cluster in cortices of embryonic day 12.5 (E12.5), E15.5, and postnatal day 0 (P0) mice as detected by Northern blots. Ubiquitously expressed small RNA U6 was used as a loading control.
(B) Expression patterns of representative subfamilies of the miR-17-92 cluster in E12.5 mouse cortices, as detected by in situ hybridization. miR-17, miR-19a, miR-92a and miR-106b were highly expressed in neural progenitors (arrowheads) in the ventricular zone of the E12.5 cortex.
(C) Coronal sections of P10 brains with Nissl staining of control (Ctrl), miR-17-92 sKO and 5dKO mice. Marginal zone (mz) and layer II-VI are labeled.
(D) Quantification of the relative thickness of the cortical wall in P10 Ctrl, miR-17-92 sKO and 5dKO brains. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (***: p < 0.0003).
Figure S2. The miR-17-92 Cluster Promotes Proliferation of Cortical Neural Stem Cells In Vitro, Related to Figure 1
(A–C) The size and number of neurospheres derived from the E13.5 miR-17-92 knockout (sKO) cortices were decreased. Neurospheres > 2472 (B), neurospheres > 435 (C).
(D and E) Neurospheres derived from miR-17-92 sKO cortices expressed NSC markers such as Nestin and SSEA-1 similarly to control (Ctrl) neurospheres.
(F and G) The numbers of BrdU-labeled cortical neural stem cells, after 6 hr BrdU pulse, were decreased in miR-17-92 sKO mice. Cell number > 1500. Data are presented as mean ± SEM; n = 3 independent experiments carried out; p values in relation to control (*: p < 0.04, **: p < 0.008, ***: p < 0.001).
Figure S3. The miR-17-92 Cluster Suppresses Transition of Intermediate Progenitors at E15.5, Related to Figure 2
(A) Increased numbers of IPs labeled with Tbr2 in E15.5 miR-17-92 sKO and 5dKO cortices as compared with controls (Ctrl).
(B) Quantification of the total number of IPs as well as IPs within the UTD and LTD among E15.5 control and miR-17-92 KO cortices.
(C and D) The numbers of Tbr2+/BrdU+ IPs were significantly higher in the miR-17-92 KO cortices than controls.
(E and F) The numbers of Pax6+/BrdU+ radial glial cells (RGCs) in the miR-17-92 KO cortices were significantly less compared to controls.
(G) The transitioning RGCs to IPs were labeled with Pax6 and Tbr2.
(H) Increased conversion of IPs as detected by Tbr2+/Pax6+ cells in miR-17-92 KO cortices compared to controls.
(I and J) Increased conversion of IPs as detected by Tbr2+/Sox2+ cells in miR-17-92 KO cortices compared to controls.
Scale bar: 100 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*: p < 0.05, **: p < 0.004, ***: p < 0.001).
Figure S4. Decreased Radial Glial Cells and Intermediate Progenitors in E17.5 Cortices of miR-17-92 sKO and 5dKO Mice, Related to Figure 2
(A–C) The numbers of Pax6+ RGCs and Tbr2+ IPs were decreased in E17.5 miR-17-92 sKO and 5dKO cortices compared to controls (Ctrl).
(D) Ratios of Tbr2+/BrdU+ cells to Tbr2+ cells. The ratio was increased in E15.5 miR-17-92 5dKO cortices at E15.5 and decreased at E17.5 compared to controls (Ctrl).
Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*: p < 0.04, **: p < 0.004, ***: p < 0.001). n.s.: not significant.
Figure S5. Neurogenesis Is Affected in miR-17-92 KO Cortices, Related to Figure 2
(A–C) The numbers of early born neurons labeled with Tbr1 were increased in miR-17-92 KO cortices at E13.5 but were decreased at E15.5 and P1, compared to controls (Ctrl).
(D–F) The numbers of late born neurons labeled with Satb2 were increased in miR-17-92 KO cortices at E15.5 but were decreased at E17.5 and P1.
(G) Normalized quantification of Pax6+ and Tbr2+ cell numbers per area at E13.5, E15.5 and P1 relative to wild-type controls at E13.5.
(H) Normalized quantification of Tbr1+ cell numbers per area at E13.5, E15.5 and P1 and normalized quantification of Satb2+ cell numbers at E15.5, E17.5 and P1 relative to wild-type controls at E13.5 and E15.5, respectively.
(I) An illustrative summary of altered neural progenitor populations and neurogenesis in miR-17-92 KO cortices at different developmental stages.
Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*: p < 0.03, **: p < 0.008, ***: p < 0.001.). n.s.: not significant.
Figure S6. Pten and Tbr2 mRNA Protectors Specifically Block Silencing Effects of miR-19a in the Pten 3′ UTR and miR-92a in the Tbr2 3′ UTR, Respectively, Related to Figures 3 and 4
(A and B) Design of Pten and Tbr2 protectors that can block binding of miR-19a and miR-92a to their 3′ UTR, respectively.
(C and D) Pten and Tbr2 protectors rescued the reduction of luciferase activities caused by the miR-19a and miR-92a targeting effect on the 3′ UTR of Pten and Tbr2, respectively.
Data are presented as mean ± SEM; n ≥ 3; p values in relation to control (*: p < 0.05).
ACKNOWLEDGMENTS
We thank Dr. Sally Temple for insightful discussions and Drs. John Cave and Harriet Baker for critical reading of the manuscript. We appreciate Drs. Andrea Ventura and Tyler Jacks at MIT for providing mice and Drs. David Price and Tom Nowakowski for discussing unpublished data. This work was supported by the Hirschl/Weill-Caulier Trust (T.S.), an NPRP grant from the Qatar National Research Fund (T.S.), and an R01-MH083680 grant from the NIH/NIMH (T.S.).
Footnotes
SUPPLEMENTAL INFORMATION Supplemental Information includes six figures and Extended Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2013.03.037.
REFERENCES
- Andersson T, Rahman S, Sansom SN, Alsiö JM, Kaneda M, Smith J, O’Carroll D, Tarakhovsky A, Livesey FJ. Reversible block of mouse neural stem cell differentiation in the absence of dicer and microRNAs. PLoS ONE. 2010;5:e13453. doi: 10.1371/journal.pone.0013453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, Molnár Z, Robertson EJ, Groszer M. The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev. 2008;22:2479–2484. doi: 10.1101/gad.475408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian S, Sun T. Functions of noncoding RNAs in neural development and neurological diseases. Mol. Neurobiol. 2011;44:359–373. doi: 10.1007/s12035-011-8211-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–641. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englund C, Fink A, Lau C, Pham D, Daza RA, Bulfone A, Kowalczyk T, Hevner RF. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J. Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineberg SK, Kosik KS, Davidson BL. MicroRNAs potentiate neural development. Neuron. 2009;64:303–309. doi: 10.1016/j.neuron.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz M, Huttner WB. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Groszer M, Erickson R, Scripture-Adams DD, Lesche R, Trumpp A, Zack JA, Kornblum HI, Liu X, Wu H. Negative regulation of neural stem/progenitor cell proliferation by the Pten tumor suppressor gene in vivo. Science. 2001;294:2186–2189. doi: 10.1126/science.1065518. [DOI] [PubMed] [Google Scholar]
- Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr. Opin. Cell Biol. 2005;17:639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Haubensak W, Attardo A, Denk W, Huttner WB. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:3196–3201. doi: 10.1073/pnas.0308600100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobert O. miRNAs play a tune. Cell. 2007;131:22–24. doi: 10.1016/j.cell.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Kang W, Wong LC, Shi SH, Hebert JM. The transition from radial glial to intermediate progenitor cell is inhibited by FGF signaling during corticogenesis. J. Neurosci. 2009;29:14571–14580. doi: 10.1523/JNEUROSCI.3844-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karres JS, Hilgers V, Carrera I, Treisman J, Cohen SM. The conserved microRNA miR-8 tunes atrophin levels to prevent neuro-degeneration in Drosophila. Cell. 2007;131:136–145. doi: 10.1016/j.cell.2007.09.020. [DOI] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev. Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Low R, Otaegi G, Pollock A, Deng H, Eisenhaber F, Maurer-Stroh S, Sun T. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J. Cell Sci. 2010;123:586–594. doi: 10.1242/jcs.059659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegstein A, Noctor S, Martínez-Cerdeño V. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat. Rev. Neurosci. 2006;7:883–890. doi: 10.1038/nrn2008. [DOI] [PubMed] [Google Scholar]
- Lang MF, Shi Y. Dynamic roles of microRNAs in neurogenesis. Front. Neurosci. 2012;6:71. doi: 10.3389/fnins.2012.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS, et al. Genome-wide RNA-mediated interference screen identifies miR-19 targets in Notch-induced T-cell acute lymphoblastic leukaemia. Nat. Cell Biol. 2010;12:372–379. doi: 10.1038/ncb2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle FT, Alvarez-Buylla A. Neural stem cells in mammalian development. Curr. Opin. Cell Biol. 2006;18:704–709. doi: 10.1016/j.ceb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Menezes JR, Macklis JD. Neuronal subtype specification in the cerebral cortex. Nat. Rev. Neurosci. 2007;8:427–437. doi: 10.1038/nrn2151. [DOI] [PubMed] [Google Scholar]
- Mu P, Han YC, Betel D, Yao E, Squatrito M, Ogrodowski P, de Stanchina E, D’Andrea A, Sander C, Ventura A. Genetic dissection of the miR-17~92 cluster of microRNAs in Myc-induced B-cell lymphomas. Genes Dev. 2009;23:2806–2811. doi: 10.1101/gad.1872909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munji RN, Choe Y, Li G, Siegenthaler JA, Pleasure SJ. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J. Neurosci. 2011;31:1676–1687. doi: 10.1523/JNEUROSCI.5404-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Neural stem and progenitor cells in cortical development. Novartis Found. Symp. 2007;288:59–73. discussion 73–58, 96–58. [PubMed] [Google Scholar]
- Nowakowski TJ, Mysiak KS, Pratt T, Price DJ. Functional dicer is necessary for appropriate specification of radial glia during early development of mouse telencephalon. PLoS ONE. 2011;6:e23013. doi: 10.1371/journal.pone.0023013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive V, Bennett MJ, Walker JC, Ma C, Jiang I, Cordon-Cardo C, Li QJ, Lowe SW, Hannon GJ, He L. miR-19 is a key oncogenic component of mir-17-92. Genes Dev. 2009;23:2839–2849. doi: 10.1101/gad.1861409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nat. Rev. Neurosci. 2012;13:528–541. doi: 10.1038/nrn3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P. Developmental and evolutionary adaptations of cortical radial glia. Cereb. Cortex. 2003;13:541–549. doi: 10.1093/cercor/13.6.541. [DOI] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, Broccoli V. Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron. 2008;60:56–69. doi: 10.1016/j.neuron.2008.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Colasante G, Nini A, Klein WH, Broccoli V. Tbr2-positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev. 2010;24:1816–1826. doi: 10.1101/gad.575410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Wang Y, Dimos JT, Fasano CA, Phoenix TN, Lemischka IR, Ivanova NB, Stifani S, Morrisey EE, Temple S. The timing of cortical neurogenesis is encoded within lineages of individual progenitor cells. Nat. Neurosci. 2006;9:743–751. doi: 10.1038/nn1694. [DOI] [PubMed] [Google Scholar]
- Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Shykind B, Sun T. Approaches to manipulating micro-RNAs in neurogenesis. Front. Neurosci. 2012;6:196. doi: 10.3389/fnins.2012.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Ying H, Yan H, Kimmelman AC, Hiller DJ, Chen AJ, Perry SR, Tonon G, Chu GC, Ding Z, et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. The miR-17-92 Family Is Expressed in Neural Progenitors in the Mouse Embryonic Neocortex, Related to Figure 1
(A) Expression levels of representative subfamilies of the miR-17-92 cluster in cortices of embryonic day 12.5 (E12.5), E15.5, and postnatal day 0 (P0) mice as detected by Northern blots. Ubiquitously expressed small RNA U6 was used as a loading control.
(B) Expression patterns of representative subfamilies of the miR-17-92 cluster in E12.5 mouse cortices, as detected by in situ hybridization. miR-17, miR-19a, miR-92a and miR-106b were highly expressed in neural progenitors (arrowheads) in the ventricular zone of the E12.5 cortex.
(C) Coronal sections of P10 brains with Nissl staining of control (Ctrl), miR-17-92 sKO and 5dKO mice. Marginal zone (mz) and layer II-VI are labeled.
(D) Quantification of the relative thickness of the cortical wall in P10 Ctrl, miR-17-92 sKO and 5dKO brains. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (***: p < 0.0003).
Figure S2. The miR-17-92 Cluster Promotes Proliferation of Cortical Neural Stem Cells In Vitro, Related to Figure 1
(A–C) The size and number of neurospheres derived from the E13.5 miR-17-92 knockout (sKO) cortices were decreased. Neurospheres > 2472 (B), neurospheres > 435 (C).
(D and E) Neurospheres derived from miR-17-92 sKO cortices expressed NSC markers such as Nestin and SSEA-1 similarly to control (Ctrl) neurospheres.
(F and G) The numbers of BrdU-labeled cortical neural stem cells, after 6 hr BrdU pulse, were decreased in miR-17-92 sKO mice. Cell number > 1500. Data are presented as mean ± SEM; n = 3 independent experiments carried out; p values in relation to control (*: p < 0.04, **: p < 0.008, ***: p < 0.001).
Figure S3. The miR-17-92 Cluster Suppresses Transition of Intermediate Progenitors at E15.5, Related to Figure 2
(A) Increased numbers of IPs labeled with Tbr2 in E15.5 miR-17-92 sKO and 5dKO cortices as compared with controls (Ctrl).
(B) Quantification of the total number of IPs as well as IPs within the UTD and LTD among E15.5 control and miR-17-92 KO cortices.
(C and D) The numbers of Tbr2+/BrdU+ IPs were significantly higher in the miR-17-92 KO cortices than controls.
(E and F) The numbers of Pax6+/BrdU+ radial glial cells (RGCs) in the miR-17-92 KO cortices were significantly less compared to controls.
(G) The transitioning RGCs to IPs were labeled with Pax6 and Tbr2.
(H) Increased conversion of IPs as detected by Tbr2+/Pax6+ cells in miR-17-92 KO cortices compared to controls.
(I and J) Increased conversion of IPs as detected by Tbr2+/Sox2+ cells in miR-17-92 KO cortices compared to controls.
Scale bar: 100 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*: p < 0.05, **: p < 0.004, ***: p < 0.001).
Figure S4. Decreased Radial Glial Cells and Intermediate Progenitors in E17.5 Cortices of miR-17-92 sKO and 5dKO Mice, Related to Figure 2
(A–C) The numbers of Pax6+ RGCs and Tbr2+ IPs were decreased in E17.5 miR-17-92 sKO and 5dKO cortices compared to controls (Ctrl).
(D) Ratios of Tbr2+/BrdU+ cells to Tbr2+ cells. The ratio was increased in E15.5 miR-17-92 5dKO cortices at E15.5 and decreased at E17.5 compared to controls (Ctrl).
Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*: p < 0.04, **: p < 0.004, ***: p < 0.001). n.s.: not significant.
Figure S5. Neurogenesis Is Affected in miR-17-92 KO Cortices, Related to Figure 2
(A–C) The numbers of early born neurons labeled with Tbr1 were increased in miR-17-92 KO cortices at E13.5 but were decreased at E15.5 and P1, compared to controls (Ctrl).
(D–F) The numbers of late born neurons labeled with Satb2 were increased in miR-17-92 KO cortices at E15.5 but were decreased at E17.5 and P1.
(G) Normalized quantification of Pax6+ and Tbr2+ cell numbers per area at E13.5, E15.5 and P1 relative to wild-type controls at E13.5.
(H) Normalized quantification of Tbr1+ cell numbers per area at E13.5, E15.5 and P1 and normalized quantification of Satb2+ cell numbers at E15.5, E17.5 and P1 relative to wild-type controls at E13.5 and E15.5, respectively.
(I) An illustrative summary of altered neural progenitor populations and neurogenesis in miR-17-92 KO cortices at different developmental stages.
Scale bar: 50 μm. Data are presented as mean ± SEM; n ≥ 3 in all genotypes; p values in relation to control (*: p < 0.03, **: p < 0.008, ***: p < 0.001.). n.s.: not significant.
Figure S6. Pten and Tbr2 mRNA Protectors Specifically Block Silencing Effects of miR-19a in the Pten 3′ UTR and miR-92a in the Tbr2 3′ UTR, Respectively, Related to Figures 3 and 4
(A and B) Design of Pten and Tbr2 protectors that can block binding of miR-19a and miR-92a to their 3′ UTR, respectively.
(C and D) Pten and Tbr2 protectors rescued the reduction of luciferase activities caused by the miR-19a and miR-92a targeting effect on the 3′ UTR of Pten and Tbr2, respectively.
Data are presented as mean ± SEM; n ≥ 3; p values in relation to control (*: p < 0.05).