Abstract
Ceramidases play a critical role in generating sphingosine-1-phosphate by hydrolyzing ceramide into sphingosine, a substrate for sphingosine kinase. In order to elucidate its transcriptional regulation, we identify here a putative promoter region in the 5’-UTR of the human neutral CDase (nCDase) gene. Using human genomic DNA, we cloned a 3000 base pair region upstream of the translational start site of the nCDase gene. Luciferase reporter analyses demonstrated that this 3000 bp region had promoter activity, with the strongest induction occurring within the first 200 bp. Computational analysis revealed the 200 bp essential promoter region contained several well-characterized promoter elements, lacked a conical TATA box, but did contain a reverse oriented CCAAT box, a feature common to housekeeping genes. Electrophoretic mobility shift assays demonstrated that the identified candidate transcriptional response elements (TRE) bind their respective transcription factors, including NF-Y, AP-2, Oct-1, and GATA. Mutagenic analyses of the TRE revealed that these sites regulated promoter activity and mutating an individual site decreased promoter reporter activity by up to 50%. Together, our findings suggest that regulation of nCDase expression involves coordinated TATA-less transcriptional activity.
Keywords: ceramidase, ceramide, sphingosine, promoter, transcriptional regulation, AP-1
1. Introduction
Ceramide (Cer) is the central molecule in most sphingolipid metabolic pathways [1–3]. Cer is also the substrate for several enzymes, including, but not limited to, ceramide kinase, glucosylceramide synthase, galactosylceramide synthase, and sphingomyelin synthase. Cer has been shown to function as lipid-bound second messengers inducing apoptosis or cell growth arrest [4]. Cer’s have also been linked to autophagy, differentiation, growth senescence, anti-inflammatory responses, and regulation of gene transcription [5–9]. Differential effects of Cer in different tissues may reflect unique molecular species of Cer, in terms of their fatty acid composition, altered metabolism of Cer species, or distinct subcellular localization of Cer species. Cer can be metabolized by ceramidases (CDases) to form sphingosine, which is subsequently phosphorylated by sphingosine kinase to form sphingosine-1-phosphate (So1P). The ceramide metabolite, So1P, promotes normal cell growth and differentiation [10] and inhibits the apoptotic effects of Cer [11]. Based on these and other findings, the rheostat model has been proposed in which the overall relative level of Cer and So1P to each other drives a cell towards apoptosis or normal growth, survival, and/or differentiation [10]. However, it is becoming apparent that this rheostat model is perhaps too simplified. In fact, increasing evidence, aided in part by mass spectrometry-based lipidomic strategies, have begun to show that changes in sphingolipid metabolism are not linear and often involve several sphingolipid metabolic pathways and differential regulation of fatty acid within a single class of sphingolipids [12–16]. Sphingosines also appear to be a critical metabolic branch point as they can also be phosphorylated, glycosylated, or re-acylated. In fact, CDase, is the predominant enzyme to generate sphingosine, as sphingosine is not a metabolite in de novo synthesis of sphingolipids and the role of sphingosine-1-phosphate phosphatases to generate sphingosine is still controversial [17]. Moreover, the exact biological role(s) and biochemical regulation of sphingosine metabolism have not been fully elucidated. Thus, a thorough understanding of the transcriptional regulation of the enzyme that predominantly regulates ceramide to sphingosine metabolism, is warranted.
Even though three families of distinct CDases have been cloned and classified based on their pH optima: acidic (human [18] and mouse [19]), neutral (human [20], mouse [21], rat [22], bacteria [23], and drosophila [24]), and alkaline (yeast [25], mouse [26]. and human [27], we have chosen to focus the present work on the human neutral ceramidase (nCDase) for several reasons. First, nCDases are integral membrane proteins, localizing predominantly to the plasma membrane [28], in contrast to alkaline CDases that have been shown to localize primarily within the endoplasmic reticulum or the golgi as well as acidic CDases, predominantly localized to lysosomes. Second, nCDases have been reported to be regulated by various cytokines and growth factors [29–32]. Third, recent studies support co-localization of nCDase enzyme, substrate, and signaling elements within defined membrane structures [22, 33], alluding to the importance of nCDase in modulating lipid-mediated signaling in response to various cytokine and growth factor stimuli.
To date, nCDases have only been characterized by tissue and species expression. The transcriptional mechanisms and unique response elements underlying this tissue and species specificity have not, as yet, been defined. As examples, characterization of mouse nCDase revealed that levels of nCDase mRNA were higher in liver than brain [21], indicative of tissue specific neutral CDase regulation. Furthermore, in rats, nCDase mRNA levels were highest in the kidney and brain, yet low in the liver [22]. Similarly, human nCDase mRNA is likely regulated in a tissue specific manner. Although mRNA was ubiquitously expressed in all tissues tested, skeletal muscle, heart, and kidney expressed high levels of neutral CDase mRNA, whereas the liver and brain expressed significantly lower mRNA levels [20]. Thus, to better comprehend the regulation of the human neutral CDase gene, we characterized the 5’-untranslated region for promoter function.
2. Materials and Methods
2.1 Cell Culture
Human embryonic kidney 293 (HEK 293) cells were obtained from American Type Culture Collection (Rockville, MD), and were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS).
2.2 Cloning of 5’-Flanking Region of Human NCDase Gene
To obtain the 5’-flanking sequence of the human nCDase gene, human GenomeWalker kit (Clontech, Mountain View, CA), containing four libraries of human genomic DNA, was used according to manufacturer’s instructions. Human nCDase gene specific primers GSwalk1 (5’-TTCAATGGTCCCACTGGTGATAAACAAG-3’) and GSwalk2 (5’-AGAAGGGCCACTGTGATGGCACTCAT-3’) were designed and synthesized by Integrated DNA Technologies (IDT, Coralville, IA). Primary polymerase chain reaction (PCR) was carried out using 1 uL of genomic DNA, gene specific primer GSwalk1 and adaptor primer AP1, and Advantage 2 Polymerase Mix (Clontech). The genomic library was amplified by PCR (94°C for 2 sec, then 72°C for 3 min for 7 cycles, followed by 32 cycles of 94°C for 2 sec, then 67°C for 3 min). A final extension step was done at 67°C for 4 min. The primary PCR products were diluted 1:50 with sterile water and used as a template for a second PCR reaction with the nested gene specific primer GSwalk2 and the nested adapter primer AP2. The secondary PCR was performed at the same parameters as the primary reaction with 5 and 20 cycles, respectively. The reaction was again held at 67°C for 4 min for final extension. The final PCR products were separated by agarose gel electrophoresis, cloned into pGEM-T Easy vector (Promega, Madison, WI), and sequenced in both directions using pUC/M13 forward and reverse primers for pGEM-T and internally designed primers.
2.3 Sequence Analysis
Putative transcription factor binding sites were predicted with Transcriptional Element Search Software (TESS) (http://cbil.upenn.edu/tess/). Sequencing comparisons between and alignments of human and mouse nCDase putative promoter regions were accomplished using ClustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
2.4 Construction of Reporter Genes
Specific sequences of the 5’-flanking regions of the human nCDase gene were amplified by PCR. The nucleotide range of each amplified region and the respective 5’forward primers used to amplify them are as follows: −3000/−1 TRE (CPF3-KpnI: 5’-GAGCGGTACCTATCAATACTCTTTAATCTCATAC-3’), −2500/−1 TRE (CPF4-KpnI: 5’-GAGCGGTACCATATAATTATTGGCTTAGCTCTAC-3’), −2000/−1 TRE (CPF5-KpnI: 5’-GAGCGGTACCAACTAGAGATACTGGGTTTGAAGT-3’), −1500/−1 TRE (CPF6-KpnI: 5’-GAGCGGTACCCAGTGTCATCATATTTTACGAATT-3’), −1000/−1 TRE (CPF7-KpnI: 5’-GAGCGGTACCATAAGATGATCATAATCATATTCC-3’), −500/−1 TRE (CPF8-KpnI: 5’-AGTCGGTACCAGTCCATGACCACAGAGTACTGTC-3’), −400/−1 TRE (CPF-400-KpnI: 5’-AGTCGGTACCAGCCCAGACCCATGTACAGATCCA-3’), −300/−1 TRE (CPF-300-KpnI: 5’-AGTCGGTACCTCAGTGGCCAATGCTAAATCATGA-3’), −200/−1 TRE (CPF-200-KpnI: 5’-AGTCGGTACCAATTGATTGGTCTTGTCTGCCATG-3’), −100/−1 TRE (CPF-100-KpnI: 5’-AGTCGGTACCAGATATCTTAACCTCGGTTGGCTT-3’), −50/−1 TRE (CPF-50-KpnI: 5’-AGTAGGTACCTTCTTCCTCTTCAGTATTTCTTCT-3’). The 3’ reverse primer used to amplify all regions was CPR1-XhoI (5’-TGACCTCGAGTTCTTCTCAGGTACAGCAGAGATG). The underlined bases are restriction digest sites for KpnI for the 5’ primers and XhoI for the 3’ primer. PCR’s were performed using the pGEM-T vector containing the 5’-flanking region as a template. PfuUltra High Fidelity DNA polymerase (Stratagene, La Jolla, CA) was used according to manufacturer’s instructions. The cycling parameters were 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min/kb, for 30 cycles. The DNA fragments generated were cloned into a TOPO vector using Zero Blunt TOPO PCR Cloning Kit (Invitrogen), propagated in Ecoli, and followed by plasmid isolation with Qiaprep Spin Mini Kit (Qiagen, Valencia, CA). The plasmids containing the 5’-flanking regions of DNA were digested with KpnI and XhoI, and the specific regions were purified by gel electrophoresis and cloned into the firefly luciferase expression vector pGL3-Basic vector (Promega) using the same restriction sites.
2.5 Construction of Site-Directed Mutational Reporter Plasmids
Site-directed mutagenesis or deletion of the putative transcription factor binding sites was done using Quikchange Site-Directed Mutagenesis Kit (Stratagene) according to manufacturer’s protocol. Primers used to alter the binding sites were designed with the help of Stratagene’s web-based primer design software program (http://labtools.stratagene.com/QC). Sense primers used are given in Table 2; antisense primers (not shown) are the reverse complement of the given sense primers. PCR was then performed with the appropriate primers using 25 ng of PGL-200 (pGL3-basic vector with −200 to −1 bp of CDase promoter) as a template. The cycling parameters were 95°C for 30 sec, 55°C for 1 min, and 68°C for 5 min, for 12 cycles. The parental DNA was then digested with 1 uL Dpn restriction enzyme for 1 h at 37°C. The remaining mutated vectors from the digested DNA were propagated by transformation of XL1-Blue supercompetent cells (Stratagene).
Table 2.
Primers for the site-directed mutagenesis at TF binding sites
| TF site | Original sequencea |
Primer sequence (sense strand)b,c,d |
|---|---|---|
| AP-1/CCAAT | TGATTGGTC | 5'-cagaacatttctctatcgataggtaccaat---------ttgtctgccatgggcttggtctatggaat-3’ |
| Ap-2 | GCCATGGG | 5'-ctatcgataggtaccaattgattggtcttgtctATTATGAGcttggtctatggaatgagtgg-3' |
| Rsrfc4/Oct-a | TATTAATATG | 5'-aagtgctctgtacttgtcgttcttggtatagtcTACCAGCATGaaagagctaagatatcttaac-3' |
| Gata | AGATATCT | 5'-ggtatagtctattaatatgaaagagctaACTTAAGTtaacctcggttggtttaaagcatttgca-3' |
| Oct-b | ATTTGCAT | 5'-gatatcttaacctcggttggtttaaagcGCTTACGTtttaatttgtggtttcttcctcttcag-3' |
Nucleotides underlined were altered.
Binding site shown in caps. Altered nucleotides are shown in bold.
Deleted nucleotides are represented by a dash, ‘-’
Antisense primers are the reverse complement of respective sense primers
2.6 Preparation of Nuclear Extract
Nuclear extracts were prepared using the Pierce Nuclear and Cytoplasmic Extraction Reagent Kit (NE-PER) (Pierce/Thermo Fisher Scientific, Rockford, IL) according to manufacturer’s instructions with slight modifications. Briefly, HEK 293 cells were grown to 90% confluency in a T175 flask, detached with trypsin, and isolated by centrifugation to yield a packed cell volume of approximately 300 uL. After the addition of 3 ml of CERI (cytoplasm extraction reagent I) buffer, the pellet was vortexed and incubated on ice for 10 min. CERII (165 uL) was then added followed by vortexing and an additional incubation on ice. The nuclear fraction was then separated by centrifugation and the supernatant (cytoplasmic extract) was pipetted off. The pellet containing the nuclei was then resuspended in 375 uL NER (Nuclear extraction reagent) buffer and incubated on ice for 40 min with periodic vortexing. The suspension was centrifuged a final time and the supernatant (nuclear extract) was removed and dialyzed in 4% glycerol, 10 mM Tris (pH 7.5) buffer for two hours to remove excess salt. The nuclear extract was stored at −80°C, and the protein concentration was measured using the Bio-Rad Protein Assay Kit.
2.7 Electrophoretic Mobility Shift Assays (EMSA)
All DNA oligonuceotides were synthesized by IDT (Table 1). The Gel Shift Assay System (Promega) was used according to manufacturer’s instructions. Synthetic complementary nucleotides were annealed, end-labeled with [γ-32P]ATP (7000Ci/mmol, American Radiolabeled Chemicals, St. Louis, MO) and T4 polynucleotide kinase, and purified with Sephadex G-25 Quick Spin Columns (Roche Diagnostics, Indianapolis, IN).
Table 1.
Sequence (5’-3’) of double-stranded oligonucleotides used in EMSA analyses
| Ccaat | sense | CTGTAATTGATTGGTCTTGTCT |
| antisense | AGACAAGACCAATCAATTACAG | |
| Ap2/Sp1 | sense | TTGTCTGCCATGGGCTTGGTCTATGG |
| antisense | CCATAGACCAAGCCCATGGCAGACAA | |
| Rsrfc4/Oct | sense | ATAGTCTATTAATATGAAAGAG |
| antisense | CTCTTTCATATTAATAGACTAT | |
| Gata | sense | AAGAGCTAAGATATCTTAACCT |
| antisense | AAGAGCTAAGATATCTTAACC | |
| Oct-b | sense | TTAAAGCATTTGCATTTTAATT |
| antisense | AATTAAAATGCAAATGCTTTAA | |
| NF-Y Consensus | sense | AGACCGTACGTGATTGGTTAATCTCTT |
| antisense | AAGAGATTAACCAATCACGTACGGTCT | |
| AP-2 Consensus | sense | GATCGAACTGACCGCCCGCGGCCCGT |
| antisense | ACGGGCCGCGGGCGGTCAGTTCGATC | |
| SP-1 Consensus | sense | ATTCGATCGGGGCGGGGCGAGC |
| antisense | GCTCGCCCCGCCCCGATCGAAT | |
| GATA Consensus | sense | CACTTGATAACAGAAAGTGATAACTCT |
| antisense | AGAGTTATCACTTTCTGTTATCAAGTG | |
| OCT-1 Consensus | sense | TGTCGAATGCAAATCACTAGAA |
| antisense | TTCTAGTGATTTGCATTCGACA | |
| C/EBP Consensus | sense | TGCAGATTGCGCAATCTGCA |
| antisense | TGCAGATTGCGCAATCTGCA |
Bold letters in oligonucleotide sequences indicate putative transcription factor binding sites.
EMSA assays were performed with 35–40 fmol of double-stranded end-labeled probes and 12–15 ug of HEK 293 nuclear protein for 20 minutes at room temperature. For competition assays, nuclear protein was preincubated with a 50-fold excess (except where noted) of unlabeled double-stranded EMSA probes for 10 min before the addition of labeled probe. Sequences of labeled probes are given in Table 1. Gel-loading buffer without dye was added before the protein/DNA complexes were resolved on Novex 6% DNA retardation gels (Invitrogen) using 0.5× TBE buffer (Sigma Aldrich, St. Louis, MO) at 200 V for 20 min. For supershift assays, nuclear protein was preincubated with rabbit or goat polyclonal antibodies against Oct-1 (sc-232×) and AP-2α (sc-184×) obtained from Santa Cruz (Santa Cruz, CA) for 1 h on ice. After electrophoresis, gels were dried and autoradiographed.
2.8 Transient Transfection and Luciferase Assays
Human embryonic kidney 293 (HEK 293) cells (40–60% confluent) were transfected with each reporter construct (250 ng) and the Renilla luciferase expression vector phRL-null (100 ng, Promega) using Lipofectamine 2000 (3:1, µL Lipofectamine: µg DNA) (Invitrogen) in 250µL Opti-MEM (GIBCO-Invitrogen) in 12 well plates. HEK 293 cells were grown in 1 ml DMEM supplemented with 10% FBS, and the media was not changed after the addition of the transfection reagents. Twenty-four hours after transfection, cells were lysed by the addition of 250 uL of Passive Lysis Buffer (Promega). For serum induced promoter activity, 30–40% confluent HEK 293 cells were transfected overnight and serum deprived with basal media the next morning for 24 hours. FBS was then added to a final concentration of 10% v/v, followed by lysing at the time points given in the results section. The luciferase activity in the cell lysates was determined using the Dual Luciferase Reporter System (Promega). Firefly luciferase activities of the human nCDase promoter and luciferase gene chimeras were normalized to that of Renilla luciferase (or to total protein where noted) and expressed relative to the activity of the pGL3-Basic plasmid. For overexpression of c-Jun and c-Fos, 500 ng of each vector was co-transfected with the reporter vectors, while maintaining a 3:1 ratio (µL: µg) of Lipofectamine 2000:DNA. For c-Jun knockdown, 10 pmol of c-Jun siRNA (JUN Stealth RNAi DuoPak, Invitrogen) was delivered with the reporter DNA in 3 µL of Lipofectamine 2000 per well of a 12-well plate.
2.9 Statistical Analysis
The results are expressed as mean ± standard error of at least three independent experiments. Probability (p) values ≤ 0.05 (Student’s t-test) were considered to indicate statistically significant differences.
3. Results
3.1 Cloning of human nCDase promoter
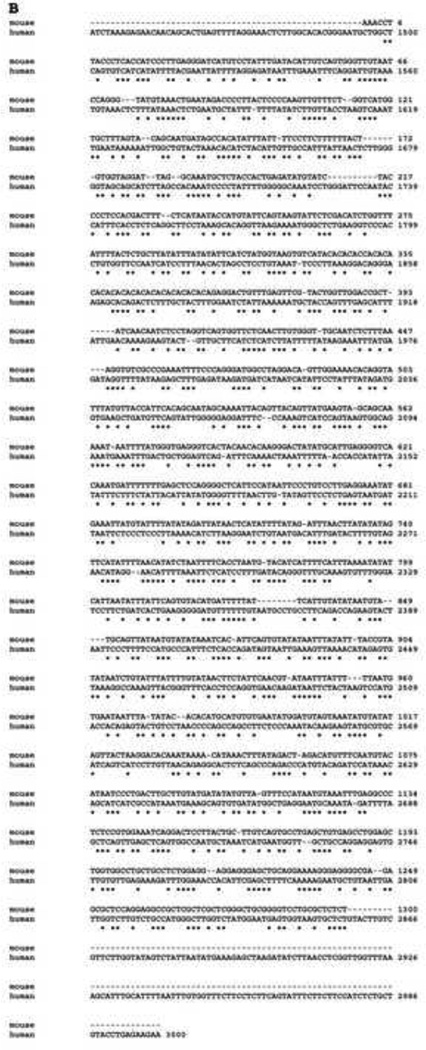
To identify the human nCDase promoter region, the DNA 5’ upstream of the open reading frame (ORF) of the nCDase gene was obtained via a PCR-based genomic walking method. Using human genomic DNA along with CDase specific primers, a 3.0 kb fragment of the 5’-flanking region of the human nCDase gene corresponding to the nucleotides −3000 to −1, relative to the translation start site (Fig. 1A), was cloned into a general cloning vector (pGEM-T easy). After aligning this cloned human nCDase promoter with the mouse nCDase promoter described by others [34] with ClustalW software, the first 1494 base pairs of the human nCDase promoter did not show any significant alignment with the mouse promoter. Starting from nucleotide −1495 of the human nCDase promoter, only a 46% nucleotide sequence identity with the mouse nCDase promoter was observed (Fig. 1B). In contrast, there is an 81% cDNA sequence identity for the ORF between the two species [35]. Furthermore, none of the identified putative transcriptional response elements were conserved between the two species. Hence, transcriptional regulation of nCDase appears to be less evolutionarily conserved than the enzyme itself; thus, it is likely that the nCDase transcriptional response elements may be different in humans compared to murine or other species.
Figure 1. The 5’-UTR region of the human nCDases gene aligned with mouse nCDase promoter.
A) The 3000 bp sequence from the human nCDase 5’-UTR cloned from genomic DNA is shown. B) Best fit alignment of 5’-UTR region of the human nCDases gene to the published sequence of mouse nCDase promoter. No conservation of potential transcription factor binding sites were detected between the two species.
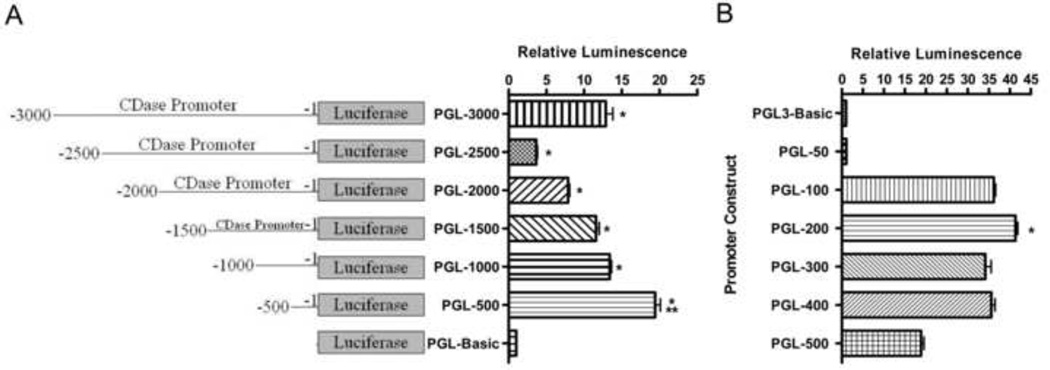
In order to determine if the unique initial 3000 bp of the 5’-UTR of human nCDase had promoter activity, we cloned this 5’-region into the 5’-end of pGL3-basic luciferase reporter vector to create PGL-3000. The luciferase activity from the HEK 293 cells transfected with PGL-3000 was about 15-fold higher than that from cells transfected with the pGL3-basic control vector (Fig. 2A, PGL-3000 vs. PGL-Basic), indicating that the 3000 bp region functions as a promoter for the human nCDase gene. To localize DNA elements responsible for this transcriptional activity, successive 500 bp deletions of the 5’-flanking region were generated by site-directed PCR amplification and inserted into the pGL3-basic vector to construct PGL-2500, PGL-2000, PGL-1500, PGL-1000, and PGL-500. These vectors were then transiently transfected into HEK 293 cells and assayed for luciferase activity. Luciferase activity of PGL-Basic was set as negative control with a value of 1, and all others activities were normalized to the control vector. Luciferase activity was highest from the reporter construct carrying a fragment from −500 to −1 (Fig. 2A, PGL-500). Successive increases in 5’-flanking fragment length resulted in a continued decrease in luciferase activity through 2500 bp, where the activity from PGL-2500 was decreased by >80% compared to PGL-500 (Fig. 2A). These results suggest that positive regulatory elements may exist between bp −500 and −1 and also upstream of bp − 2500, whereas negative regulatory elements are likely located between bp −2500 and −500. Most notably, the luciferase activity from the reporter construct containing the shortest fragment (PGL-500) was even higher than the activity from the full-length construct.
Figure 2. Deletion analysis of the 5’-UTR identified a 200 bp region as a serum-inducible, proximal promoter of the human neutral CDase gene.
Firefly luciferase (fLuc) expression vectors containing sequentially smaller regions of the nCDase promoter were cotransfected into HEK 293 cells with a plasmid (phRL-null) containing the renilla luciferase (rLuc) gene. After 24 h incubation, post-transfection, with DMEM containing FBS, fLuc and rLuc activities were measured. The fLuc/rLuc ratios were determined, and means ±S.E.M for a minimum of three transfections were calculated. Values are expressed as fold increase relative to the luminescence ratio observed in pGL3-Basic transfected cells (relative value = 1). A) Sequential 500 bp deletion analysis of the cloned 3000 base pair nCDase promoter region was used to identify regions likely to contain activator and/or repressor regions. Single asterisk represents significance compared to PGL-Basic. Double asterisks represent significance compared to PGL-3000. B) Sequential deletion analysis of bp −500 to −1 of the 5’-UTR identified the proximal promoter as the region containing the major positive regulatory elements of the human nCDase gene. Single asterisk represents significance compared to all data sets.
In order to further delineate where the promoter regulatory elements lie within this 500 bp region, successive deletions of the sequence were inserted into pGL3-basic to make PGL-400, PGL-300, PGL-200, PGL-100, and PGL-50. Luciferase activity was highest from the reporter construct carrying a fragment from −200 to −1 (Fig. 2B). In fact, activity from PGL-200 showed over 40-fold increased luciferase activity compared to the control pGL-basic vector, whereas the PGL-500 showed an approximate 20-fold increase. Deletion of bp −200 to −51 of the 200 bp 5’-flanking region completely abolished all luciferase activity (Fig. 2B). These results suggested that the major positive regulatory elements of the human nCDase gene reside within this 200 bp region of the 5’-flanking sequence, establishing this sequence as the proximal promoter of the human nCDase gene.
3.2 Identification of transcriptional response elements within the 200 bp human nCDase proximal promoter
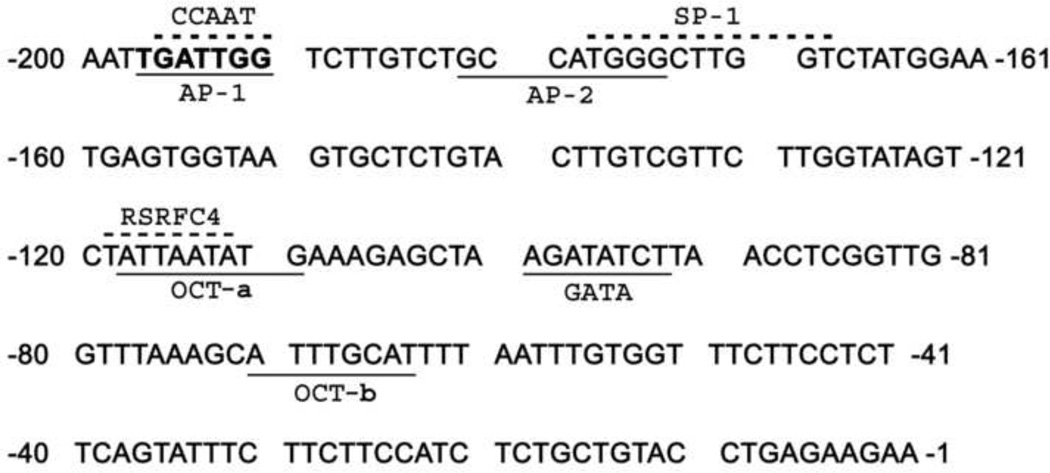
The above data indicate that the 200 bp proximal promoter region contains transcriptional response elements (TREs), which could possibly regulate nCDase transcription. To identify the TREs that may be responsible for nCDase transcriptional activity, the 200 bp region of human nCDase gene 5’-UTR was analyzed using Transcription Element Search Software (TESS). This software is designed to search nucleotide sequences for potential transcription factor binding sites using site or consensus strings and positional weight matrices from TRANSFAC 6.0. TESS analysis indicated that the proximal promoter contained multiple potential transcription factor binding sites. These include putative AP-1, AP-2, SP-1, Oct, RSRFC4 (related to serum response factor C4), and GATA sites, and also a CCAAT box (Fig. 3). Interestingly, the analyses did not detect a TATA box throughout the entire 5’-UTR of the human nCDase gene.
Figure 3. Several potential transcription factor sites are present within the 200 bp minimal essential promoter region.
Nucleotide sequence of the 5’-flanking region of the human nCDase gene was analyzed for consensus transcription factor-binding elements using TESS website. Putative cis-elements are underlined or over-scored in the case of overlapping sites. −1 indicates the first nucleotide of the 5’-UTR.
3.3 The 200 bp human nCDase proximal promoter contains a functional overlapping CCAAT binding site
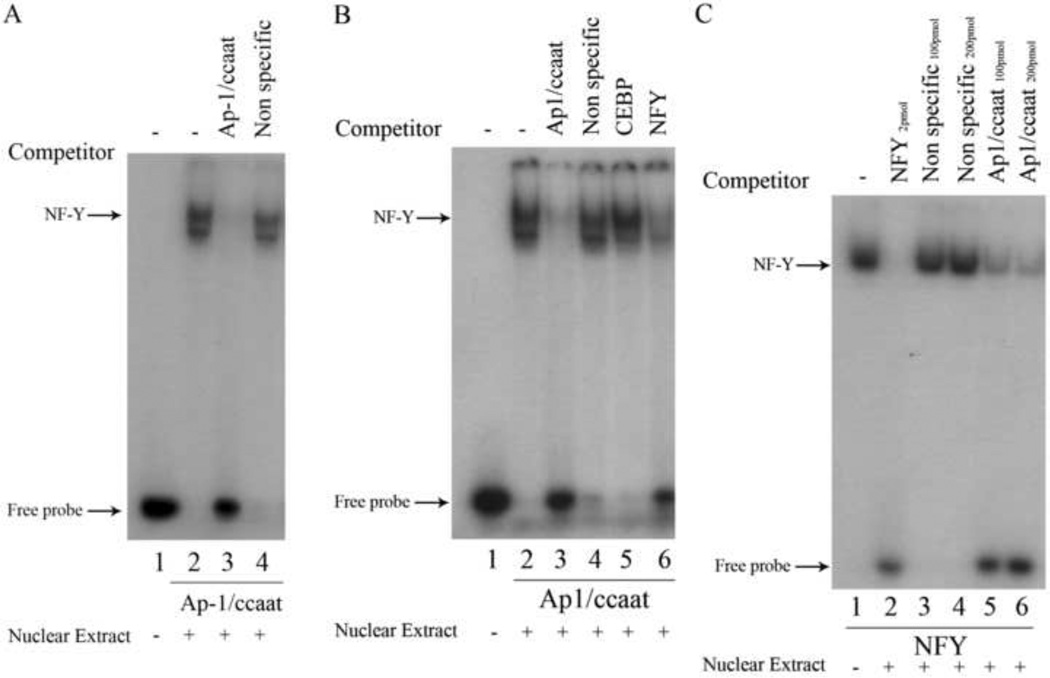
Given that the 200 bp proximal promoter contained multiple putative transcription factor binding sites, electrophoretic mobility shift assays (EMSAs) were performed to validate the functional role of these binding sites. We first examined the overlapping putative CCAAT box and AP-1 cis-elements because CCAAT boxes have been shown to have important roles in regulating TATA-less genes [36, 37]. To determine if the putative AP-1/CCAAT site of the human nCDase gene can functionally bind transcription factors, EMSAs were performed using nuclear extracts prepared from HEK 293 cells and a 32P-radiolabeled 22-nucleotide probe (bp −204 to −183) containing the putative AP-1/CCAAT site centrally positioned, designated “Ap1/ccaat” (see Table 1 for sequence). The results from EMSAs revealed two distinct shifted bands (Fig. 4A, lane 2) compared to the control labeled probe without nuclear extract (Fig. 4A, lane 1). Preincubation with 50-molar excess of unlabeled Ap-1/ccaat oligonucleotide completely inhibited the binding of nuclear proteins to the labeled probe (Fig. 4A, lane 3), indicating the specific interaction of potential transcription factors with AP-1/CCAAT DNA. To rule out nonspecific DNA-protein interactions, pre-incubation with 50-molar excess of an unlabeled, nonspecific oligonucleotide (bp −108 to −87 of the proximal promoter) did not affect the shifted bands (Fig. 4A, lane 4). These results indicated the putative AP-1/CCAAT binding site binds specifically to nuclear proteins from HEK 293 cells.
Figure 4. Competition EMSAs revealed the proximal promoter of the human nCDase gene contains overlapping Ap-1 and NF-Y binding sites with different nucleotide specificity for binding.
Nuclear extracts from HEK 293 cells were subjected to EMSAs with radiolabeled Ap1/ccaat probe (Table 1) containing nucleotides −204 to −183, or with radiolabeled NFY probe (Table 1). Lanes containing each labeled probe are underlined with the corresponding labeled probe marked below the underlined lanes. Oligonucleotides used as competitors are marked above the corresponding lanes. Unlabeled Ap1/ccaat, C/EBP consensus, NF-Y consensus, and non-specific oligonucleotides were used as competitors in a 50-fold excess in the corresponding lanes. A) EMSAs demonstrated that nuclear proteins specifically bind to the putative AP-1/CCAAT site. B) EMSAs demonstrated that unlabeled NF-Y consensus oligonucleotide is a competitor against binding of nuclear extract proteins to labeled probe containing the putative AP-1/CCAAT binding site (probe Ap1/ccaat). C) Conversely, EMSAs also demonstrated that unlabeled Ap1/ccaat oligonucleotide competitively inhibits binding of labeled NF-Y consensus probe to nuclear extract proteins. In lanes labeled with Ap1/ccaat 100pmol and Ap1/ccaat 200pmol, a 2500 and 5000 molar excess of unlabeled Ap1/ccaat probe was used to compete for NF-Y probe binding.
Several transcription factors have been shown to bind the DNA sequence CCAAT, including CCAAT/Enhancer Binding Protein (C/EBP) and Nuclear Factor Y (NF-Y) [37]. In order to determine the transcription factor(s) that directly bind to AP-1/CCAAT DNA, competition EMSAs were performed. We utilized oligonucleotides containing either NF-Y or C/EBP consensus-binding sequences (“NFY,” “CEBP,” Table 1) as competitors to the labeled Ap1/ccaat probe. Both bands were unaffected by preincubation with a 50-molar excess of unlabeled CEBP (Fig. 4B, lane 5). In contrast, an excess of unlabeled NFY oligonucleotide was able to significantly inhibit the formation of the upper band while minimally affecting the lower band (Fig. 4B, lane 6). This suggested that the upper band was the result of binding of NF-Y to the Ap1/ccaat probe. As a control for specific binding, EMSAs were performed using a reverse strategy whereby the oligonucleotide containing the NF-Y consensus sequence was used as a labeled probe. Labeled NFY consensus probe generated a band (Fig. 4C, lane 1) that was inhibited using unlabeled NFY oligonucleotide (Fig. 4C, lane 2), but was unaffected by unlabeled non-specific oligonucleotides (Fig. 4C, lanes 3 & 4). Most importantly, excess unlabeled Ap1/ccaat oligonucleotide drastically reduced the intensity of the band formed by binding of NF-Y protein to labeled NFY consensus probe (Fig. 4C, lanes 5 & 6). These results indicate that the transcription factor NF-Y binds to the AP-1/CCAAT sequence of the human nCDase gene.
Although we identified the upper band as an NF-Y/DNA complex, the identity of the trans-activator(s) responsible for forming the lower band was still unknown. We hypothesized that transcription factor AP-1 binding to the overlapping AP-1/CCAAT site would generate the lower band. Indeed, in our correlating paper submitted concurrently to Archives Biochemistry and Biophysics, AP-1 is shown to bind to the AP-1/CCAAT site and form the lower band that is seen in Figure 4 A and B. For these studies and further studies on the physiological significance of AP-1 binding and its serum- or growth factor-induced activation on nCDase gene transcription, please see this concomitant manuscript.
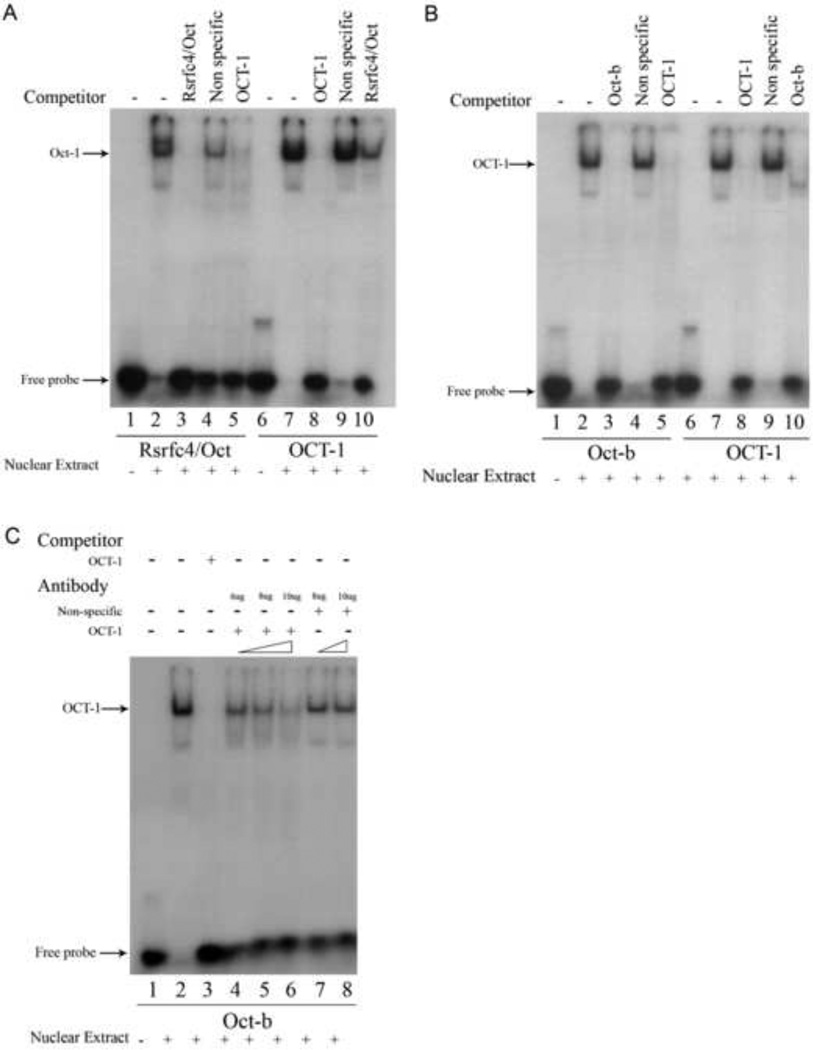
3.4 The human nCDase proximal promoter contains two functional Oct binding sites
In addition to the AP-1/CCAAT site, TESS analysis identified a putative binding site for another serum-regulated transcription factor, RSRFC4 (related to serum response factor-C4). We examined if the putative RSRFC4 site was able to function as an actual trans-activator binding site for nCDase. TESS analysis also revealed that the RSRFC4 site overlaps with an Oct (Octamer-binding) site, starting at nucleotide −118 (Fig. 3). We designed and labeled a double-stranded, 22 bp oligonucleotide probe, from bp −125 to −104 (Table 1, Rsrfc4/Oct), containing the putative sites to identify any transcription factors that may bind this overlapping TRE. The 32P-labeled Rsrfc4/Oct probe showed a band shift following incubation with the nuclear extract from HEK 293 cells (Fig. 5A, lane 2). Competition EMSAs with excess amounts of the identified unlabeled oligonucleotides (Fig. 5A, lanes 3&4) indicated that the lower band in Fig. 5 represents specific interactions between nuclear proteins and the Oct/Rsrfc4 probe and that the apparent upper band is the result of non-specific DNA-protein interactions. In Fig. 5, lane 4, we see that the upper band is competed out by an unlabeled random DNA oligo, i.e. the protein that binds to the Rsrfc4/Oct probe to form the upper band also binds an unrelated DNA sequence. This suggests that an unidentified transcription factor, or other DNA binding protein, with promiscuous DNA binding affinity may be responsible for the upper band in lane 2.
Figure 5. Competition EMSAs revealed the human nCDase proximal promoter contains two binding sites with varying affinity for Oct-1 at positions −118t to −110 and −71 to −64.
Nuclear extracts from HEK 293 cells were subjected to EMSAs with radiolabeled Rsrfc4/Oct probe (Table 1), Oct-b probe (Table 1), or with radiolabeled OCT-1 probe (Table 1) containing the Oct-1 consensus sequence. Lanes containing each labeled probe are underlined with the corresponding labeled probe marked below the underlined lanes. Unlabeled Rsrfc4/Oct, Oct-b, OCT-1 consensus, or non-specific oligonucleotides were used as competitors in a 50-fold excess to determine the specificity for Oct-1 binding. Oligonucleotides used as competitors are marked above the corresponding lanes. A) The putative Oct-1 site between nucleotides −118 and −110 bound to Oct-1, but with a weaker affinity than the Oct-1 consensus oligonucleotide. B) The putative Oct-1 site between nucleotides −71 and −64 bound to Oct-1 with comparable affinity as the Oct-1 consensus oligonucleotide. C) EMSAs with blocking antibodies confirmed the binding of Oct-1 to the putative Oct-1 cis-element at position −71 to −64. Additional EMSAs were performed using the radiolabeled Oct-b probe with the addition of antibodies to induce supershifts. Antibodies against Oct-1, antibodies against AP-2α (for control), and the amounts of antibodies used in each blocking assay are marked above their corresponding lane. Positive antibody blocking assays resulted in decreased labeled probe/protein complex (band) intensity.
To identify which nuclear protein(s) specifically bound the putative RSRFC4/Oct site, competitive EMSAs were performed with Oct-1 consensus oligonucleotide (Table 1, OCT1). An excess of unlabeled OCT1 completely prevented a band shift with labeled Oct/Rsrfc4 probe (Fig. 5A, lane 5). Furthermore, 32P-labeling the OCT1 probe revealed a specific DNA/Oct-1 complex induced band shift correlating with the aforementioned lower band (Fig. 5A, lane 7 vs. lane 2). This band was significantly attenuated by competition with a 50-molar excess of unlabeled Oct/Rsrfc4 oligonucleotide (Fig. 5A, lane 10). These results indicate that the putative Oct/RSRFC4 site in the human nCDase proximal promoter is a functional Oct binding site.
A second putative Oct site was also identified in the human nCDase proximal promoter starting at nucleotide −71 (Fig. 3). A 32P-labeled 22 nucleotide (bp −78 to −57) probe containing this binding site centrally positioned (Oct-b, Table 1) was used in EMSAs to determine its functionality. These EMSAs resulted in a distinct, specific DNA/transcription factor band shift (Fig. 5B, lanes 2–4). Competition assays with unlabeled OCT1 consensus oligonucleotide and labeled OCT1 probe confirmed the second putative Oct site bound to an Oct transcription factor (Fig. 5B, lane 5–10). To determine that the transcription factor binding the Oct-b probe is Oct-1, EMSAs with blocking antibodies were performed. Inclusion of an antibody against Oct-1 in the incubation stage of EMSAs continuously decreased the intensity of the shifted band with increasing concentrations of antibody (Fig. 5C, lanes 4–6). This decrease was specific to the anti-OCT-1 antibody, since increasing concentrations of an antibody directed against a different transcription factor did not affect the shifted band (Fig. 5C, lanes 7 and 8). The results from Figure 5B and the blocking antibody (Fig. 5C) clearly showed that Oct-1 transcription factor bound to the second putative Oct site within the human nCDase proximal promoter.
3.5 The human nCDase proximal promoter contains both functional AP-2 and GATA binding sites
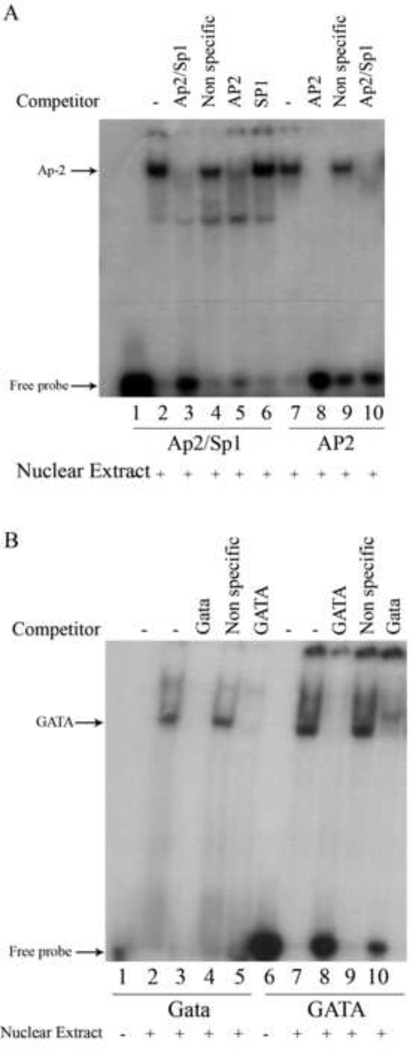
The proximal promoter of the human nCDase gene also contains a putative SP-1 binding site partially overlapping an AP-2 binding site (Fig. 3), which was of interest because cooperation between NF-Y and SP-1 in regulating promoters has been demonstrated for multiple genes [36], and SP-1 cis-elements have been found to be functionally important in many TATA-less promoters [38]. EMSAs using a 32P-labeled 26-nucleotide oligonucleotide (bp −188 to −163) containing the putative SP-1 and AP-2 sites centrally positioned (Table 1, Ap2/Sp1) resulted in one major band shift, indicating specific binding between transcription factors and the labeled probe (Figure 6A, lanes 1–4).
Figure 6. Competition EMSAs reveal the human nCDase proximal promoter contains binding sites for AP-2 and GATA factors.
Nuclear extracts from HEK 293 cells were subjected to competition EMSAs with the indicated radiolabeled probes. Lanes containing each labeled probe are underlined with the corresponding labeled probe marked below the underlined lanes. Oligonucleotides used as competitors are marked above the corresponding lanes. A) EMSAs with the Ap2/Sp1 probe (Table 1) containing the putative AP-2 site at nucleotides −182 to −175 or with radiolabeled AP2 probe (Table 1) containing the AP-2 consensus sequence. To rule out SP-1 binding, an unlabeled SP-1 oligonucleotide (Table 1) containing the SP-1 consensus sequence was also used, in addition to the AP-2 consensus sequence, as a competitor against labeled Ap2/Sp1 probe. B) EMSAs with the Gata probe (Table 1) containing the putative GATA site at nucleotides −100 to −93 or with radiolabeled GATA probe (Table 1) containing the GATA binding factor consensus sequence.
Since the Ap2/Sp1 probe formed only one major complex, while containing two putative binding sites, it was next determined which transcription factor, AP-2 or SP-1, preferentially interacts with the DNA. Competitive EMSAs with oligonucleotides containing either AP-2 consensus sequence (Table 1, AP2) or SP-1 consensus sequence (Table 1, SP1) demonstrated that AP-2, but not SP-1, likely bound to the Ap2/Sp1 probe (Fig. 6A, lane 5 vs. 6). Furthermore, when AP2 was 32P-labeled and used in EMSAs, it formed a shifted band that aligned with the band in lane 2 (Fig. 6A, lane 7) and that was inhibited by unlabeled Ap2/Sp1 oligonucleotide (Fig. 6A, lane 10). Taken together, these results indicated that the putative AP-2/SP-1 sequence in the human nCDase proximal promoter is able to bind the transcription factor AP-2, but not SP-1, in HEK 293 cells.
TESS analysis also revealed a possible GATA-factor binding site (Fig. 3). EMSAs using a 22 bp (bp −108 to −87), 32P-labeled probe containing this putative GATA site centrally positioned (Table 1, Gata) demonstrated there was specific binding of transcription factors to this oligonucleotide (Fig. 6B, lanes 1–4). Competition assays with unlabeled GATA consensus oligonucleotide suggested the putative GATA site bound to a GATA transcription factor (Fig. 6B, lanes 5). Furthermore, when GATA oligonucleotide was 32P-labeled and used in EMSAs, a shifted band that aligned with the band in lane 2 (Fig. 6B, lane 7) was observed and was inhibited by unlabeled Gata oligonucleotide (Fig. 6A, lane 10). Taken together, these results indicated that the putative GATA sequence in the human nCDase proximal promoter is able to bind a transcription factor from the GATA family.
3.6 Multiple cis-elements are functionally important for human nCDase proximal promoter activity
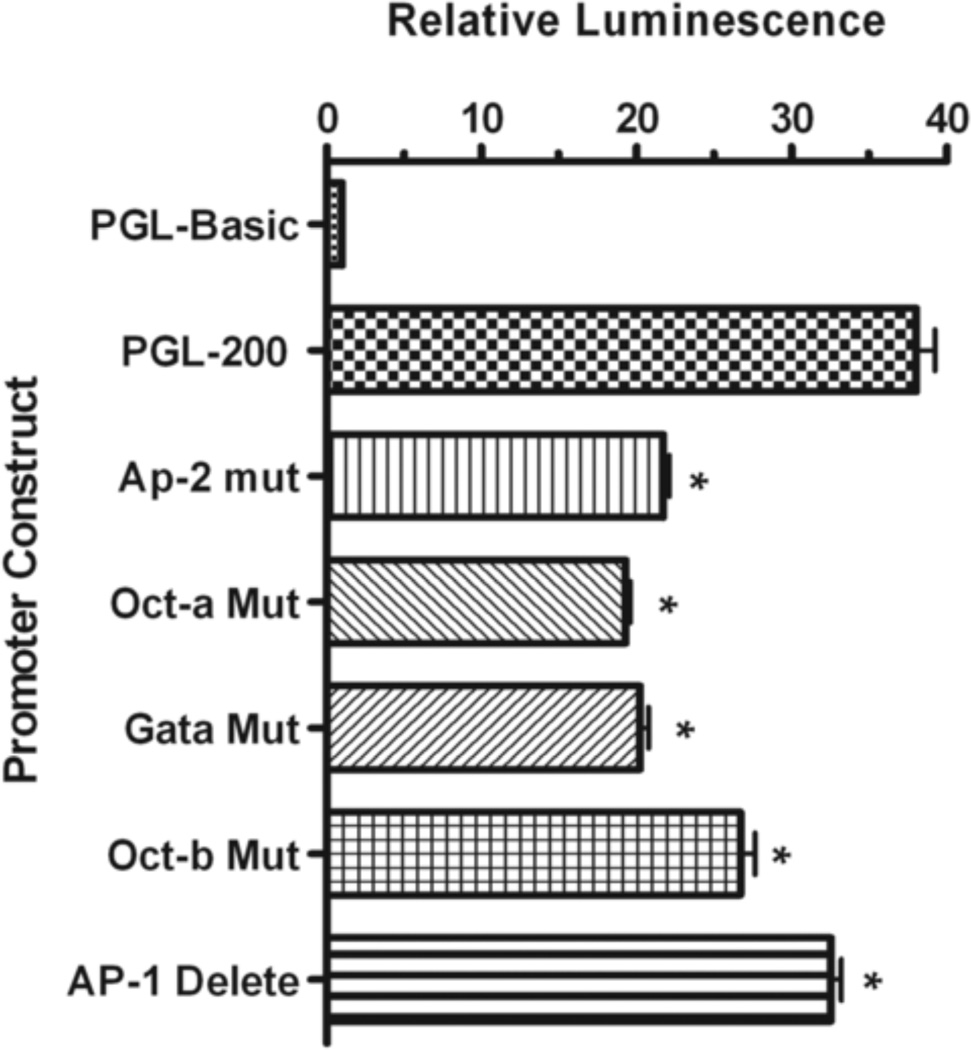
Several transcription factors that bind to cis-elements within the human nCDase proximal promoter have been identified. To explore the biological relevance of each of these transcription factor-binding sites, the sites were individually disrupted by introducing mutations into the proximal promoter/reporter vector as shown in Table 2. Mutation of the AP-2, GATA, Oct-a (bp −118 to −110), and Oct-b (−71 to −64) cis-elements down regulated luciferase activity 41%, 44%, 49%, and 28%, respectively, compared to the luciferase activity of HEK 293 cells transfected with PGL-200 (Fig. 7). To address the importance of the AP-1/CCAAT element to the overall activity of the proximal promoter, we introduced a 9 bp deletion into the PGL-200 reporter vector, eliminating the entire element (ΔAP-1/CCAAT). This deletion significantly, albeit less dramatically, reduced luciferase activity by 14% compared with the wild-type PGL-200 reporter (Fig. 7). As disruption of these individual binding elements was not sufficient to abrogate all luciferase activity, it implies that multiple and/or combinatorial transcription factor regulation is important to achieve maximal activity of the human nCDase proximal promoter. These data further suggest that the activity of the proximal promoter depends on concerted interactions of multiple transcription factors.
Figure 7. The identified cis-elements are each individually important for maximal activity of the human nCDase proximal promoter.
Mutations within the respective putative transcription factor binding sites (see Table 2) were introduced into the PGL-200 vector, and transfections were performed using HEK 293 cells. After 24 h incubation with DMEM containing FBS, firefly luciferase activity was measured and normalized to renilla luciferase activity. The binding sites for AP-2, GATA, and Oct-1 were each individually mutated to prevent binding. Additionally, the putative AP-1/NF-Y site was deleted from the PGL-200 vector. The results are represented as fold increase in activity from cells transfected with the pGL-3 basic control vector. Significantly different decreases in activity in the mutated vectors compared to PGL-200 are marked by an asterisk. Results represent the mean ± S.E.M of at least three individual experiments.
4. Discussion
The present study analyzes the transcription regulatory elements of human nCDase in order to further understand the central role of ceramide in sphingolipid metabolism and signaling. We isolated and identified the 5’-flanking region of the human nCDase gene. This sequence allowed us to analyze the human nCDase promoter region and identify cis-element regions involved in the regulation of the human nCDase gene. Luciferase reporter assays revealed a promoter region likely containing multiple activators and repressors, yet having maximal activity within the first 200 bp. Using EMSA, transfection, and mutational analyses, we were able to identify several promoter sequences within the minimal essential promoter region that are responsive to multiple trans-activating factors including NF-Y, AP-2, GATA factors, and Oct factors.
CDase’s have a critical role in sphingolipid metabolism through enzymatic hydrolysis of ceramide to form sphingosine. In fact, deficiencies in acidic CDase could cause Farber’s Disease in humans in which ceramide accumulates within lysosomes [39]. Neutral CDase can modulate lipid mediated signaling by changing the balance of sphingolipid metabolites in response to various cytokine and growth factor stimuli. With such a crucial role in lipid signaling, it is of interest that nCDase mRNA expression varies considerably between tissues and that this differential expression is different from species to species [20–22, 35]. Furthermore, the expression of human nCDase mRNA can be regulated in differentiation, as late-stage differentiated human keratinocytes express significantly less nCDase mRNA then undifferentiated, proliferating keratinocytes [40]. These observations are consistent with our identification of multiple cis-regulatory elements within the minimal essential promoter region.
Detailed analysis of the 5’-flanking region revealed the human nCDase gene lacks a canonical TATA box. The TATA box is an important sequence in the initiation of transcription, found in many, if not the majority, of eukaryotic genes. Although it lacked a TATA box, we observed a CCAAT box within the human nCDase promoter. CCAAT boxes have also been shown to associate with proteins within transcription initiation complex [41]. Promoters containing one or more CCAAT boxes, but no TATA box, are often observed in housekeeping genes [42, 43]. The promoter of murine nCDase gene also has typical features of housekeeping genes. Similar to human nCDase gene, it lacks a TATA box, but it also lacks any CCAAT box, instead containing a GC box [34]. Conservation of a housekeeping gene throughout species is expected, however the differences in the cis-elements between mouse and human is not surprising when there is only 40% homology between the 5’-flanking regions of the two genes. A few transcription factors have been shown to bind CCAAT elements. NF-Y is the major factor that binds the CCAAT box sequence [37], and it is a trimeric TF consisting of NF-YA, NF-YB, and NF-YC subunits, all of which are essential for DNA binding [44–46]. Our studies revealed that NF-Y binds the CCAAT box within the human nCDase promoter. Furthermore, mutational analysis revealed that the NF-Y binding site is necessary for maximum human nCDase promoter activity.
CCAAT boxes are invariantly flanked by at least one functionally important cis-element [37]. The mutual interactions between the respective trans-activating factor and NF-Y are essential for optimal regulation of transcription [37]. NF-Y has the capacity to enhance transactivation through direct protein-protein interactions and/or facilitate the positioning of transcription factors through DNA conformation changes. As briefly aforementioned, the CCAAT box in the nCDase promoter overlaps a functional AP-1 binding site. Our concurrent manuscript (submitted) investigates the cooperatively between NF-Y and AP-1 more thoroughly. Here we will focus on potential interactions of NF-Y with transcription factors that may bind other cis-elements identified in the nCDase promoter.
Interestingly, SP-1 and NF-Y cooperatively regulate many genes, and such interactions have been implicated in activation of TATA-less promoters [37, 47, 48]. Although the untranslated region of the human nCDase gene contains a putative SP-1 binding site flanking the CCAAT box, we were not able to detect SP-1 binding at this site using EMSA. However, interactions between other transcription factors and NF-Y have also been reported [49, 50]. Within the human nCDase gene, two putative Oct response elements bind Oct-1 transcription factors as demonstrated by EMSA. Oct-1 and NF-Y may concertedly activate the human nCDase promoter as they do with the cell cycle-arresting protein Gadd45 promoter [51]. Alternatively, Oct transcription factors may repress the human nCDase promoter by limiting NF-Y binding, similar to Oct-1 repression of HLA-DRA (a cell surface antigen binding protein subunit) transcription [52], though the former scenario is more consistent with mutation analysis data showing each Oct binding site is necessary for maximum human nCDase promoter activity. Furthermore, Oct-1 can interact with other transcription factors other than NF-Y, as Oct-1 synergizes with, or enhances the, action of AP-1 in several human genes including the H3.3B histone gene [53] and the IL-2 and IL-5 genes [54]. EMSA results also indicated GATA factors bind to cis-elements within the human nCDase gene, and GATA factors and NF-Y have been shown to directly interact with each other [55] or bind cooperatively [56].
In conclusion, our data indicate that the human nCDase gene is under control of a promoter region with multiple activator and repressor regions, however the major positive regulatory region resides within the first 200 bp of the 5’-flanking region. EMSA analyses demonstrated that putative cis-elements were able to bind several transcription factors, such as AP-1, NF-Y, AP-2, Oct, and GATA. Although each transcription factor binding site contributed to promoter activity, basal activity of the proximal promoter was not dependent on any individual transcription factor. Interestingly, we found that individually mutating several of the transcriptional response elements caused a greater reduction in promoter activity than deletion of the region from −101 to −200, which contains several of these elements (Fig. 2B vs. Fig. 7). A possible explanation for this observation could be as yet unidentified repressor transcription elements between base pairs −100 to −200. The complexity of transcriptional regulatory control is further exemplified by regions between −500 and −2500, which suppress transcriptional activity (Figure 2A), and were not analyzed in the present study, which focused solely on the proximal promoter of the hCDase gene. Regardless, these facts further raise questions about the complexity of the interplay of the multiple trans-activating factors in regulating human nCDase transcription. It is probable that several factors function in concert with each other to tightly regulate the promoter in response to multiple biological stimuli. The possible combinations of transcription factors that are necessary to activate the proximal promoter are numerous, and extensive research will be needed to delineate the role of each trans-factor and help unravel the intricacies of human nCDase transcription. However, these results provide the first insight into the regulatory mechanisms that control expression of the human nCDase gene and will form the foundation for understanding its tissue- and differentiation-specific regulation.
Research Highlights.
▶A 3000bp promoter region of human neutral ceramidase was cloned ▶ The strongest promoter activity is within the first 200bp of the transcriptional start site ▶ NF-Y, AP-2, Oct-1, and GATA transcription factors bind to their predicted transcriptional response elements ▶ Mutatation of these transcriptional response elements inhibits promoter activity.
Acknowledgments
NIH Grants HL66371 and HL76789 to MK supported this work. We would like to acknowledge the Molecular Genetics Core Facility at the Penn State College of Medicine for their assistance in DNA sequencing analyses. We would also like to thank Nicole Divittore for assistance with manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bartke N, Hannun YA. J. Lipid Res. 2009;50(Suppl):S91–S96. doi: 10.1194/jlr.R800080-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lahiri S, Futerman AH. Cell Mol. Life Sci. 2007;64:2270–2284. doi: 10.1007/s00018-007-7076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fyrst H, Saba JD. Nat. Chem. Biol. 2010;6:489–497. doi: 10.1038/nchembio.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolesnick R. J Clin Invest. 2002;110:3–8. doi: 10.1172/JCI16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grether-Beck S, Felsner I, Brenden H, Krutmann J. J. Biol. Chem. 2003;278:47498–47507. doi: 10.1074/jbc.M309511200. [DOI] [PubMed] [Google Scholar]
- 6.Zheng W, Kollmeyer J, Symolon H, Momin A, Munter E, Wang E, Kelly S, Allegood JC, Liu Y, Peng Q, Ramaraju H, Sullards MC, Cabot M, Merrill AH., Jr Biochim. Biophys. Acta. 2006;1758:1864–1884. doi: 10.1016/j.bbamem.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Kester M, Kolesnick R. Pharmacol. Res. 2003;47:365. doi: 10.1016/s1043-6618(03)00048-3. [DOI] [PubMed] [Google Scholar]
- 8.Grether-Beck S, Salahshour-Fard M, Timmer A, Brenden H, Felsner I, Walli R, Fullekrug J, Krutmann J. Oncogene. 2008;27:4768–4778. doi: 10.1038/onc.2008.116. [DOI] [PubMed] [Google Scholar]
- 9.Sun Y, Fox T, Adhikary G, Kester M, Pearlman E. J. Leukoc. Biol. 2008;83:1512–1521. doi: 10.1189/jlb.0108076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spiegel S, Kolesnick R. Leukemia. 2002;16:1596–1602. doi: 10.1038/sj.leu.2402611. [DOI] [PubMed] [Google Scholar]
- 11.Pyne S. Subcell Biochem. 2002;36:245–268. [PubMed] [Google Scholar]
- 12.Gangoiti P, Granado MH, Wang SW, Kong JY, Steinbrecher UP, Gomez-Munoz A. Cell Signal. 2008;20:726–736. doi: 10.1016/j.cellsig.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Munoz A, Kong JY, Parhar K, Wang SW, Gangoiti P, Gonzalez M, Eivemark S, Salh B, Duronio V, Steinbrecher UP. FEBS Lett. 2005;579:3744–3750. doi: 10.1016/j.febslet.2005.05.067. [DOI] [PubMed] [Google Scholar]
- 14.Fox TE, Kester M. Adv. Exp. Med. Biol. 2010;688:206–216. doi: 10.1007/978-1-4419-6741-1_14. [DOI] [PubMed] [Google Scholar]
- 15.Karahatay S, Thomas K, Koybasi S, Senkal CE, Elojeimy S, Liu X, Bielawski J, Day TA, Gillespie MB, Sinha D, Norris JS, Hannun YA, Ogretmen B. Cancer Lett. 2007;256:101–111. doi: 10.1016/j.canlet.2007.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Senkal CE, Ponnusamy S, Bielawski J, Hannun YA, Ogretmen B. FASEB J. 2010;24:296–308. doi: 10.1096/fj.09-135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 18.Koch J, Gartner S, Li CM, Quintern LE, Bernardo K, Levran O, Schnabel D, Desnick RJ, Schuchman EH, Sandhoff K. J Biol Chem. 1996;271:33110–33115. doi: 10.1074/jbc.271.51.33110. [DOI] [PubMed] [Google Scholar]
- 19.Li CM, Hong SB, Kopal G, He X, Linke T, Hou WS, Koch J, Gatt S, Sandhoff K, Schuchman EH. Genomics. 1998;50:267–274. doi: 10.1006/geno.1998.5334. [DOI] [PubMed] [Google Scholar]
- 20.El Bawab S, Roddy P, Qian T, Bielawska A, Lemasters JJ, Hannun YA. J Biol Chem. 2000;275:21508–21513. doi: 10.1074/jbc.M002522200. [DOI] [PubMed] [Google Scholar]
- 21.Tani M, Okino N, Mori K, Tanigawa T, Izu H, Ito M. J Biol Chem. 2000;275:11229–11234. doi: 10.1074/jbc.275.15.11229. [DOI] [PubMed] [Google Scholar]
- 22.Mitsutake S, Tani M, Okino N, Mori K, Ichinose S, Omori A, Iida H, Nakamura T, Ito M. J Biol Chem. 2001;276:26249–26259. doi: 10.1074/jbc.M102233200. [DOI] [PubMed] [Google Scholar]
- 23.Okino N, Ichinose S, Omori A, Imayama S, Nakamura T, Ito M. J Biol Chem. 1999;274:36616–36622. doi: 10.1074/jbc.274.51.36616. [DOI] [PubMed] [Google Scholar]
- 24.Yoshimura Y, Okino N, Tani M, Ito M. J Biochem. 2002;132:229–236. doi: 10.1093/oxfordjournals.jbchem.a003215. [DOI] [PubMed] [Google Scholar]
- 25.Mao C, Xu R, Bielawska A, Szulc ZM, Obeid LM. J Biol Chem. 2000;275:31369–31378. doi: 10.1074/jbc.M003683200. [DOI] [PubMed] [Google Scholar]
- 26.Mao C, Xu R, Szulc ZM, Bielawski J, Becker KP, Bielawska A, Galadari SH, Hu W, Obeid LM. J Biol Chem. 2003;278:31184–31191. doi: 10.1074/jbc.M303875200. [DOI] [PubMed] [Google Scholar]
- 27.Mao C, Xu R, Szulc ZM, Bielawska A, Galadari SH, Obeid LM. J Biol Chem. 2001;276:26577–26588. doi: 10.1074/jbc.M102818200. [DOI] [PubMed] [Google Scholar]
- 28.Tani M, Iida H, Ito M. J Biol Chem. 2003;278:10523–10530. doi: 10.1074/jbc.M207932200. [DOI] [PubMed] [Google Scholar]
- 29.Coroneos E, Martinez M, McKenna S, Kester M. J Biol Chem. 1995;270:23305. doi: 10.1074/jbc.270.40.23305. [DOI] [PubMed] [Google Scholar]
- 30.Nikolova-Karakashian M, Morgan ET, Alexander C, Liotta DC, A HM., Jr J Biol Chem. 1997;272:18718. doi: 10.1074/jbc.272.30.18718. [DOI] [PubMed] [Google Scholar]
- 31.Franzen R, Pautz A, Brautigam L, Geisslinger G, Pfeilschifter J, Huwiler A. J. Biol. Chem. 2001;276:35382. doi: 10.1074/jbc.M102153200. [DOI] [PubMed] [Google Scholar]
- 32.Franzen R, Fabbro D, Aschrafi A, Pfeilschifter J, Huwiler A. J Biol Chem. 2002;277:46184–46190. doi: 10.1074/jbc.M204034200. [DOI] [PubMed] [Google Scholar]
- 33.Stover T, Kester M. J Pharmacol Exp Ther. 2003;307:468–475. doi: 10.1124/jpet.103.054056. [DOI] [PubMed] [Google Scholar]
- 34.Okino N, Mori K, Ito M. Biochem Biophys Res Commun. 2002;299:160–166. doi: 10.1016/s0006-291x(02)02540-8. [DOI] [PubMed] [Google Scholar]
- 35.Choi MS, Anderson MA, Zhang Z, Zimonjic DB, Popescu N, Mukherjee AB. Gene. 2003;315:113–122. doi: 10.1016/s0378-1119(03)00721-2. [DOI] [PubMed] [Google Scholar]
- 36.Mantovani R. Nucleic Acids Res. 1998;26:1135. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantovani R. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 38.Blake MC, Jambou RC, Swick AG, Kahn JW, Azizkhan JC. Mol. Cell. Biol. 1990;10:6632. doi: 10.1128/mcb.10.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugita M, Dulaney JT, Moser HW. Science. 1972;178:1100–1102. doi: 10.1126/science.178.4065.1100. [DOI] [PubMed] [Google Scholar]
- 40.Houben E, Holleran WM, Yaginuma T, Mao C, Obeid LM, Rogiers V, Takagi Y, Elias PM, Uchida Y. J Lipid Res. 2006;47:1063–1070. doi: 10.1194/jlr.M600001-JLR200. [DOI] [PubMed] [Google Scholar]
- 41.Bellorini M, Lee DK, Dantonel JC, Zemzoumi K, Roeder RG, Tora L, Mantovani R. Nucleic Acids Res. 1997;25:2174. doi: 10.1093/nar/25.11.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dynan WS, Tjian R. Nature. 1985;316:774–778. doi: 10.1038/316774a0. [DOI] [PubMed] [Google Scholar]
- 43.Roy B, Lee AS. Mol Cell Biol. 1995;15:2263–2274. doi: 10.1128/mcb.15.4.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vuorio T, Maity SN, de Crombrugghe B. J Biol Chem. 1990;265:22480–22486. [PubMed] [Google Scholar]
- 45.Maity SN, Vuorio T, de Crombrugghe B. Proc Natl Acad Sci U S A. 1990;87:5378–5382. doi: 10.1073/pnas.87.14.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinha S, Maity SN, Lu J, de Crombrugghe B. Proc Natl Acad Sci U S A. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pugh BF, Tjian R. Genes Dev. 1991;5:1935–1945. doi: 10.1101/gad.5.11.1935. [DOI] [PubMed] [Google Scholar]
- 48.Liang F, Schaufele F, Gardner DG. J Biol Chem. 2001;276:1516–1522. doi: 10.1074/jbc.M006350200. [DOI] [PubMed] [Google Scholar]
- 49.Ueda A, Takeshita F, Yamashiro S, Yoshimura T. J Biol Chem. 1998;273:19339–19347. doi: 10.1074/jbc.273.30.19339. [DOI] [PubMed] [Google Scholar]
- 50.Jacobs SB, Coss D, McGillivray SM, Mellon PL. Mol Endocrinol. 2003;17:1470–1483. doi: 10.1210/me.2002-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirose T, Sowa Y, Takahashi S, Saito S, Yasuda C, Shindo N, Furuichi K, Sakai T. Oncogene. 2003;22:7762. doi: 10.1038/sj.onc.1207091. [DOI] [PubMed] [Google Scholar]
- 52.Osborne AR, Zhang H, Fejer G, Palubin KM, Niesen MI, Blanck G. J. Biol. Chem. 2004;279:28911. doi: 10.1074/jbc.M403118200. [DOI] [PubMed] [Google Scholar]
- 53.Witt O, Albig W, Doenecke D. FEBS Lett. 1997;408:255. doi: 10.1016/s0014-5793(97)00436-5. [DOI] [PubMed] [Google Scholar]
- 54.Thomas MA, Mordvinov VA, Sanderson CJ. Eur. J. Biochem. 1999;265:300. doi: 10.1046/j.1432-1327.1999.00732.x. [DOI] [PubMed] [Google Scholar]
- 55.Lui WY, Wong EW, Guan Y, Lee WM. J Cell Physiol. 2007;211:638–648. doi: 10.1002/jcp.20970. [DOI] [PubMed] [Google Scholar]
- 56.Gorshkova EV, Kaledin VI, Kobzev VF, Merkulova TI. Biochemistry (Mosc) 2005;70:1180–1184. doi: 10.1007/s10541-005-0244-7. [DOI] [PubMed] [Google Scholar]