Abstract
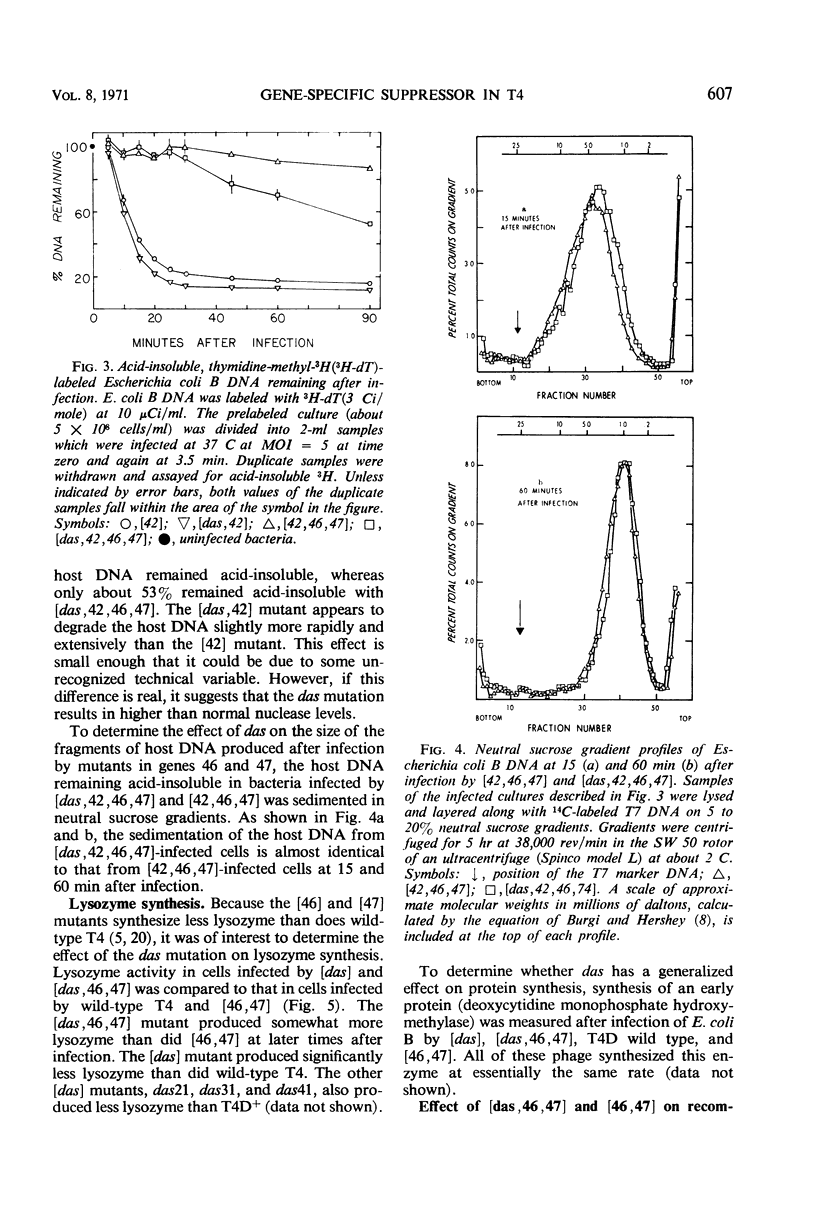
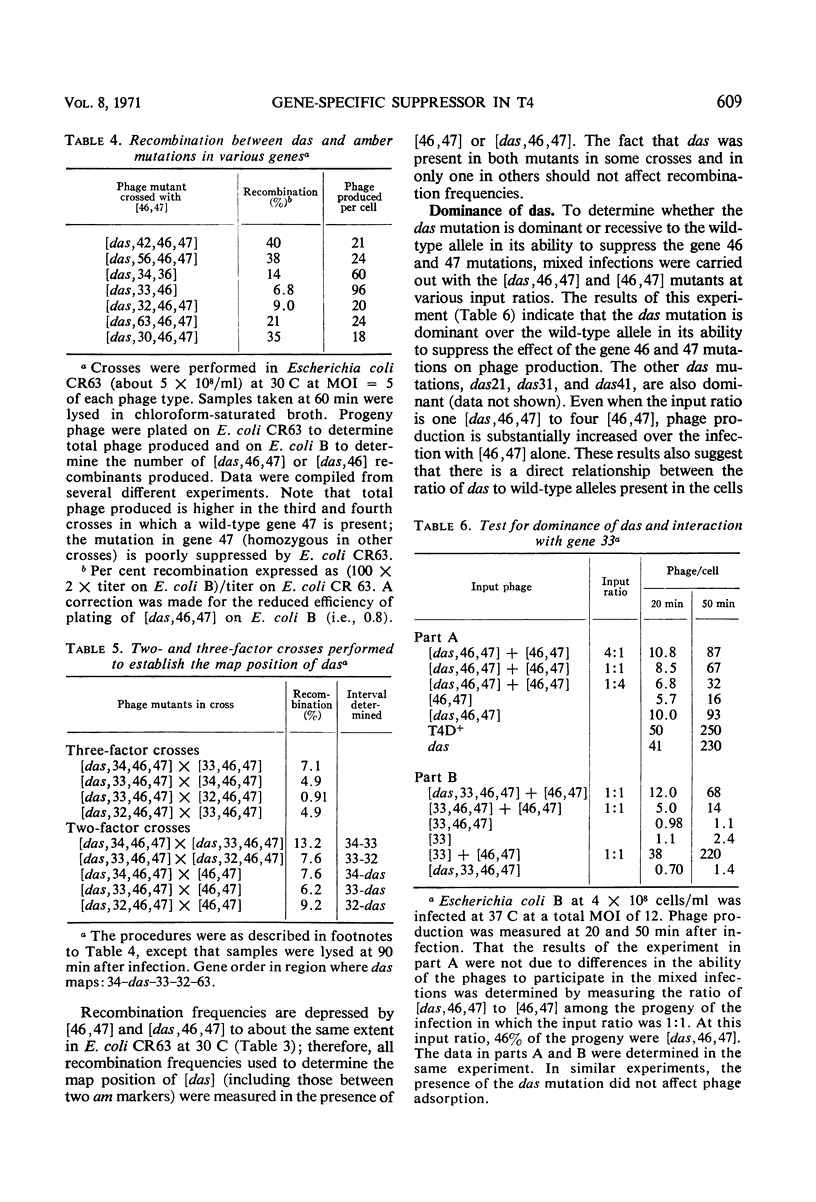
Mutants in T4 genes 46 and 47 exhibit early cessation of deoxyribonucleic acid (DNA) synthesis (“DNA arrest”) and decreased synthesis of late proteins and phage. In addition, mutants in genes 46 and 47 fail to degrade host DNA to acidsoluble products. It is shown here that this complex phenotype can be partially suppressed by mutation of a T4 gene external to genes 46 and 47 which has been named das for “DNA arrest suppressor.” The das mutations were discovered as third-site mutations in spontaneous pseudorevertants of [46, 47] mutants; the pseudorevertants make small plaques on Escherichia coli B, whereas [46, 47] mutants make none. The [das, 46, 47] triple mutant exhibits increased DNA, late protein, and viable phage production compared to the double mutant [46, 47]. The [das, 46, 47] mutant also degrades more of the host DNA to acid-soluble products than does the [46, 47] mutant. The suppressor effect of the das mutation appears to be gene-specific: it suppresses both amber and temperature-sensitive mutations in genes 46 and 47 and does not suppress amber mutations in any of the other genes tested. The [das] single mutants make normal-sized plaques on E. coli B and exhibit nearly normal host DNA degradation, DNA synthesis, late protein synthesis, and viable phage production. The das mutations either define a new gene between genes 33 and 34 or are special mutations within gene 33.
Full text
PDF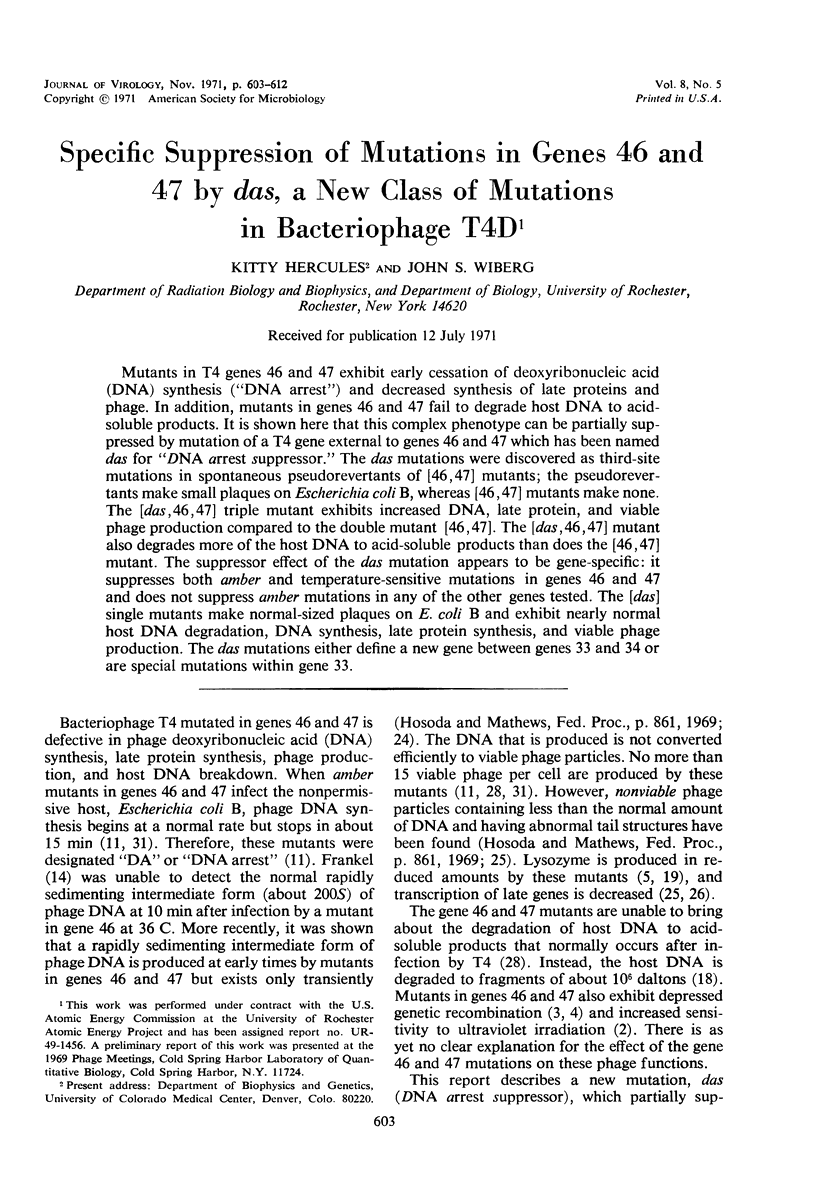







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alberts B. M., Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970 Sep 26;227(5265):1313–1318. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- BOLLUM F. J. Thermal conversion of nonpriming deoxyribonucleic acid to primer. J Biol Chem. 1959 Oct;234:2733–2734. [PubMed] [Google Scholar]
- BURGI E., HERSHEY A. D. Sedimentation rate as a measure of molecular weight of DNA. Biophys J. 1963 Jul;3:309–321. doi: 10.1016/s0006-3495(63)86823-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldy M. W. Repair and recombination in phage T4. II. Genes affecting UV sensitivity. Cold Spring Harb Symp Quant Biol. 1968;33:333–338. doi: 10.1101/sqb.1968.033.01.038. [DOI] [PubMed] [Google Scholar]
- Berger H., Warren A. J., Fry K. E. Variations in genetic recombination due to amber mutations in T4D bacteriophage. J Virol. 1969 Feb;3(2):171–175. doi: 10.1128/jvi.3.2.171-175.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein H. Repair and recombination in phage T4. I. Genes affecting recombination. Cold Spring Harb Symp Quant Biol. 1968;33:325–331. doi: 10.1101/sqb.1968.033.01.037. [DOI] [PubMed] [Google Scholar]
- Bolle A., Epstein R. H., Salser W., Geiduschek E. P. Transcription during bacteriophage T4 development: requirements for late messenger synthesis. J Mol Biol. 1968 Apr 28;33(2):339–362. doi: 10.1016/0022-2836(68)90193-9. [DOI] [PubMed] [Google Scholar]
- Botstein D. Synthesis and maturation of phage P22 DNA. I. Identification of intermediates. J Mol Biol. 1968 Jun 28;34(3):621–641. doi: 10.1016/0022-2836(68)90185-x. [DOI] [PubMed] [Google Scholar]
- CERIOTTI G. A microchemical determination of desoxyribonucleic acid. J Biol Chem. 1952 Sep;198(1):297–303. [PubMed] [Google Scholar]
- DIRKSEN M. L., HUTSON J. C., BUCHANAN J. M. HOST-DEPENDENT SYNTHESIS OF ALTERED DEOXYCYTIDYLATE HYDROXYMETHYLASE AFTER INFECTION OF ESCHERICHIA COLI WITH CERTAIN AMBER MUTANTS OF BACTERIOPHAGE T4. Proc Natl Acad Sci U S A. 1963 Sep;50:507–513. doi: 10.1073/pnas.50.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRASER D., JERREL E. A. The amino acid composition of T3 bacteriophage. J Biol Chem. 1953 Nov;205(1):291–295. [PubMed] [Google Scholar]
- Fareed G. C., Richardson C. C. Enzymatic breakage and joining of deoxyribonucleic acid. II. The structural gene for polynucleotide ligase in bacteriophage T4. Proc Natl Acad Sci U S A. 1967 Aug;58(2):665–672. doi: 10.1073/pnas.58.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel F. R. Studies on the nature of replicating DNA in T4-infected Escherichia coli. J Mol Biol. 1966 Jun;18(1):127–143. doi: 10.1016/s0022-2836(66)80081-5. [DOI] [PubMed] [Google Scholar]
- Frankel F. R. Studies on the nature of the replicating DNA in Escherichia coli infected with certain amber mutants of phage T4. J Mol Biol. 1966 Jun;18(1):144–155. doi: 10.1016/s0022-2836(66)80082-7. [DOI] [PubMed] [Google Scholar]
- Hercules K., Munro J. L., Mendelsohn S., Wiberg J. S. Mutants in a nonessential gene of bacteriophage T4 which are defective in the degradation of Escherichia coli deoxyribonucleic acid. J Virol. 1971 Jan;7(1):95–105. doi: 10.1128/jvi.7.1.95-105.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov V. N., Plotnikova T. G. A suppressor in the genome of phage T4 inhibiting phenotypic expression of mutations in genes 46 and 47. Genetics. 1971 Mar;67(3):319–326. doi: 10.1093/genetics/67.3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E. M., Wiberg J. S. Biological effects of substituting cytosine for 5-hydroxymethylcytosine in the deoxyribonucleic acid of bacteriophage T4. J Virol. 1969 Oct;4(4):439–453. doi: 10.1128/jvi.4.4.439-453.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutter E. M., Wiberg J. S. Degradation of cytosin-containing bacterial and bacteriophage DNA after infection of Escherichia coli B with bacteriophage T4D wild type and with mutants defective in genes 46, 47 and 56. J Mol Biol. 1968 Dec;38(3):395–411. doi: 10.1016/0022-2836(68)90394-x. [DOI] [PubMed] [Google Scholar]
- McClain W. H. UAG suppressor coded by bacteriophage T4. FEBS Lett. 1970 Jan 26;6(2):99–101. doi: 10.1016/0014-5793(70)80011-4. [DOI] [PubMed] [Google Scholar]
- Munro J. L., Wieberg J. S. Evidence that gene 56 of bacteriophage T4 is a structural gene for deoxycytidine triphosphatase. Virology. 1968 Nov;36(3):442–446. doi: 10.1016/0042-6822(68)90169-4. [DOI] [PubMed] [Google Scholar]
- Price A. R., Warner H. R. A structural gene for bacteriophage T4-induced deoxycytidine triphosphate-deoxyuridine triphosphage nucleotidohydrolase. Virology. 1968 Nov;36(3):523–526. doi: 10.1016/0042-6822(68)90183-9. [DOI] [PubMed] [Google Scholar]
- SEKIGUCHI M., COHEN S. S. THE SYNTHESIS OF MESSENGER RNA WITHOUT PROTEIN SYNTHESIS. II. SYNTHESIS OF PHAGE-INDUCED RNA AND SEQUENTIAL ENZYME PRODUCTION. J Mol Biol. 1964 May;8:638–659. doi: 10.1016/s0022-2836(64)80114-5. [DOI] [PubMed] [Google Scholar]
- Shah D. B., Berger H. Replication of gene 46-47 amber mutants of bacteriophage T4D. J Mol Biol. 1971 Apr 14;57(1):17–34. doi: 10.1016/0022-2836(71)90117-3. [DOI] [PubMed] [Google Scholar]
- Shalitin C., Kahana S. Conversion of T4 gene 46 mutant deoxyribonucleic acid into nonviable bacteriophage particles. J Virol. 1970 Sep;6(3):353–362. doi: 10.1128/jvi.6.3.353-362.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szybalski W. Initiation and patterns of transcription during phage development. Proc Can Cancer Conf. 1969;8:183–215. [PubMed] [Google Scholar]
- WIBERG J. S., BUCHANAN J. M. STUDIES ON LABILE DEOXYCYTIDYLATE HYDROXYMETHYLASES FROM ESCHERICHIA COLI B INFECTED WITH TEMPERATURE-SENSITIVE MUTANTS OF BACTERIOPHAGE T4. Proc Natl Acad Sci U S A. 1964 Mar;51:421–428. doi: 10.1073/pnas.51.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WIBERG J. S., DIRKSEN M. L., EPSTEIN R. H., LURIA S. E., BUCHANAN J. M. Early enzyme synthesis and its control in E. coli infected with some amber mutants of bacteriophage T4. Proc Natl Acad Sci U S A. 1962 Feb;48:293–302. doi: 10.1073/pnas.48.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. J., Bose S. K. Bacteriophage-induced inhibition of host functions. I. Degradation of Escherichia coli deoxyribonucleic acid after T4 infection. J Virol. 1968 Apr;2(4):327–334. doi: 10.1128/jvi.2.4.327-334.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg J. S. Amber mutants of bacteriophage T4 defective in deoxycytidine diphosphatase and deoxycytidine triphosphatase. On the role of 5-hydroxymethylcytosine in bacteriophage deoxyribonucleic acid. J Biol Chem. 1967 Dec 25;242(24):5824–5829. [PubMed] [Google Scholar]
- Wiberg J. S. Mutants of bacteriophage T4 unable to cause breakdown of host DNA. Proc Natl Acad Sci U S A. 1966 Mar;55(3):614–621. doi: 10.1073/pnas.55.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W. B., Edgar R. S., King J., Lielausis I., Henninger M. Bacteriophage assembly. Fed Proc. 1968 Sep-Oct;27(5):1160–1166. [PubMed] [Google Scholar]
- Wood W. B., Henninger M. Attachment of tail fibers in bacteriophage T4 assembly: some properties of the reaction in vitro and its genetic control. J Mol Biol. 1969 Feb 14;39(3):603–618. doi: 10.1016/0022-2836(69)90148-x. [DOI] [PubMed] [Google Scholar]



