Abstract
Genetic and biochemical analyses of RNA interference (RNAi) and microRNA (miRNA) pathways have revealed proteins such as Argonaute/PIWI and Dicer that process and present small RNAs to their targets. Well validated small RNA pathway cofactors, such as the Argonaute/PIWI proteins show distinctive patterns of conservation or divergence in particular animal, plant, fungal, and protist species. We compared 86 divergent eukaryotic genome sequences to discern sets of proteins that show similar phylogenetic profiles with known small RNA cofactors. A large set of additional candidate small RNA cofactors have emerged from functional genomic screens for defects in miRNA- or siRNA-mediated repression in C. elegans and D. melanogaster1,2 and from proteomic analyses of proteins co-purifying with validated small RNA pathway proteins3,4. The phylogenetic profiles of many of these candidate small RNA pathway proteins are similar to those of known small RNA cofactor proteins. We used a Bayesian approach to integrate the phylogenetic profile analysis with predictions from diverse transcriptional coregulation and proteome interaction datasets to assign a probability for each protein for a role in a small RNA pathway. Testing high-confidence candidates from this analysis for defects in RNAi silencing, we found that about half of the predicted small RNA cofactors are required for RNAi silencing. Many of the newly identified small RNA pathway proteins are orthologues of proteins implicated in RNA splicing. In support of a deep connection between the mechanism of RNA splicing and small RNA-mediated gene silencing, the presence of the Argonaute proteins and other small RNA components in the many species analysed strongly correlates with the number of introns in that species.
Proteins with similar patterns of conservation or divergence across phylogeny are more likely to act in the same pathways5. To identify proteins that share an evolutionary history with validated small RNA pathway proteins, we determined the phylogenetic profiles of approximately 20,000 proteins encoded by C. elegans genes in 85 genomes, representing diverse taxa of the eukaryotic tree of life: 33 animals, 6 land plants, 1 alga, 31 Ascomycota fungi, 3 Basidiomycota fungi, and 12 protists. Of the ∼20,000 C. elegans proteins, 10,054 show homologues in non-nematode eukaryotic genomes (Supplementary Table 1). Following correlation and clustering, this analysis sorts genes into clades of conservation and relative divergence or, loss in the various organisms as suites of genes are maintained from common ancestors or diverge in select lineages6. Protein divergence or loss in particular taxonomic clades is not random; entire suites of proteins can diverge or be lost as particular taxa specialize to no longer require ancestral functions. The correlated loss of proteins has been used to assign roles for nuclear-encoded mitochondrial proteins7 and eukaryotic cilia-associated proteins8. We developed a non-binary method of phylogenetic profiling to cluster all protein sequences encoded by C. elegans genes. Blast scores were normalized to the length of the query sequence and for relative phylogenetic distance between C. elegans and the queried organism9.
The matrix of 864,644 conservation scores for the 10,054 C. elegans proteins in the 86 genomes was queried either with a single protein to generate a ranking of other C. elegans proteins with the most similar pattern of conservation values or using a more global hierarchical clustering method (Figure 1A). Members of the same protein families exhibit similar patterns of phylogenetic conservation and therefore tend to group together in the hierarchical clustering. However, many phylogenetic clusters include members with no sequence similarity; only their conservation or divergence in genomes is correlated. The ability of this non-binary method of phylogenetic profiling to cluster proteins based on function is exemplified by the clustering of proteins known to act as members of complexes: for example the protein components of the cilated sensory ending in organisms with or without cilia clusters these components whereas the extraordinary high and universal conservation of ribosomal and translation factor proteins clusters many of these translation components (Supplementary Figure 1A,B).
Figure 1. Phylogenetic profiling analysis shows correlated conservation patterns ofC. elegans proteins.
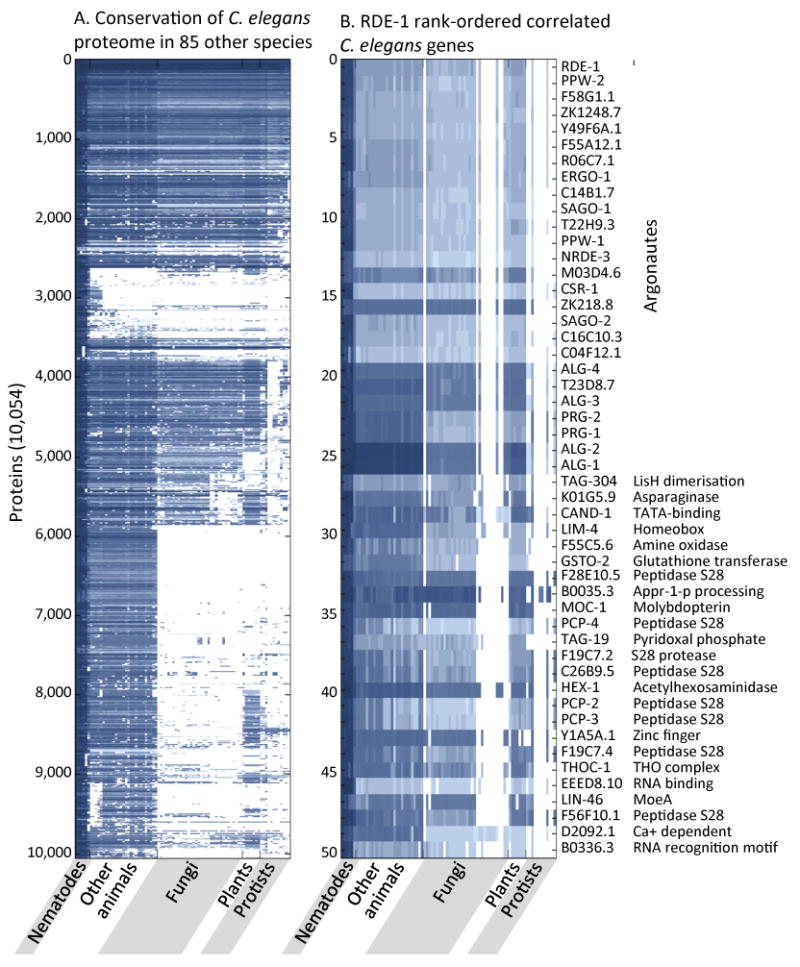
A. Phylogenetic profiles of 10,054 conserved C. elegans proteins across 85 other eukaryotic genomes. For each C. elegans query protein, the normalized ratio of the blastp score for the top scoring protein sequence similarity is listed in the column corresponding to each genome. Values range from 0 (white, no similarity) to 1 (blue, 100% similarity). B. Phylogenetic profiles of validated RNAi factor RDE-1 and the 49 most correlated proteins in rank order.
With a simple query of one of the central proteins in RNAi, the RDE-1 Argonaute protein, we generated a rank-ordered list of proteins with phylogenetic profiles most similar to that of RDE-1 (Figure 1B). The 26 other C. elegans Argonautes represented the top correlated proteins, a trivial consequence of protein sequence similarity within the Argonaute family. The signature phylogenetic profile of the Argonaute proteins is that they are absent in 9 of 31 Ascomycota species, 1 of 3 Basidiomycota species, and 6 of 14 protist species, but have not been lost in any of the 33 animal or 6 land plant species compared. The retention of Argonaute proteins correlates with the ability to inactivate genes by RNA interference10 and the loss of RNAi in about half of the sequenced Ascomycota fungi is correlated with the killer RNA virus 11. Additional proteins that cluster with the Argonautes but show no sequence similarity include an asparaginase/threonine aspartase/taspase encoded by K01G5.9, the CAND-1 elongation factor, and another elongation factor, the THO complex protein THOC-1. THO complex members have emerged from genetic screens for defective transgene and RNAi silencing in Arabidopsis thaliana12.
Another validated RNAi protein is MUT-2, a polyA polymerase implicated in a step downstream of the production of primary siRNAs by Dicer13. Of the 50 proteins with phylogenetic profiles most closely correlated with MUT-2 (Supplementary Figure 1C), 10 are Argonautes, which bear no sequence similarity to MUT-2, demonstrating the efficacy of this approach to detect validated small RNA pathway proteins. Also scoring with a similar phylogenetic profile are the splicing components MAG-1, RSP-8, RNP-4, RSP-5, and DDB-1 and translation factors EIF-3.D and EIF-3.E, many of which score in the validation tests below. Similarly, of the proteins most correlated with the C. elegans orthologue of Dicer (DCR-1), a cofactor for processing of siRNAs and miRNAs, 3 Argonaute proteins emerge among the top 50 positions (Supplementary Figure 1D, Supplementary Table 2).
The RNA-dependent RNA polymerases (RdRPs)14, siRNA-amplifying cofactors, are present in in only 5 of 27 animals, all nematodes and, surprisingly, the tick, all of the land plants, not in green algea, 2 of 4 Basidiomycota fungi, 18 of 27 Ascomycota fungi, and 4 of 14 protists. A query of the RdRP RRF-3 (Supplementary Figure 1E) revealed the cofactor-independent phosphoglycerate mutase, F57B10.3, as a dramatically correlated non-homologous protein (R = 0.93). Inactivation of this phosphoglycerate mutase gene caused defects in the endogenous siRNA response as well as transgene silencing, validating its role in RNA silencing (Supplementary Table 2). It is possible that either the biochemical substrate or product of this glycolysis pathway protein or the actual enzymatic activity as a phosphatase couples it to small RNA pathways.
To identify candidate small RNA pathway proteins more comprehensively, we globally ranked proteins based on phylogenetic profile correlation with multiple validated siRNA and miRNA cofactors. After assigning all conserved C. elegans proteins into hierarchical clusters, we defined for each protein a score reflecting its phylogenetic clustering with the validated set of small RNA proteins (Supplementary Figure 2). The phylogenetic profiles of 101 proteins cluster most closely with validated siRNA and miRNA pathway proteins (Figure 2), 61 of which have not yet been implicated in small RNA pathways.
Figure 2. Phylogenetic clusters of candidate small RNA pathway proteins.
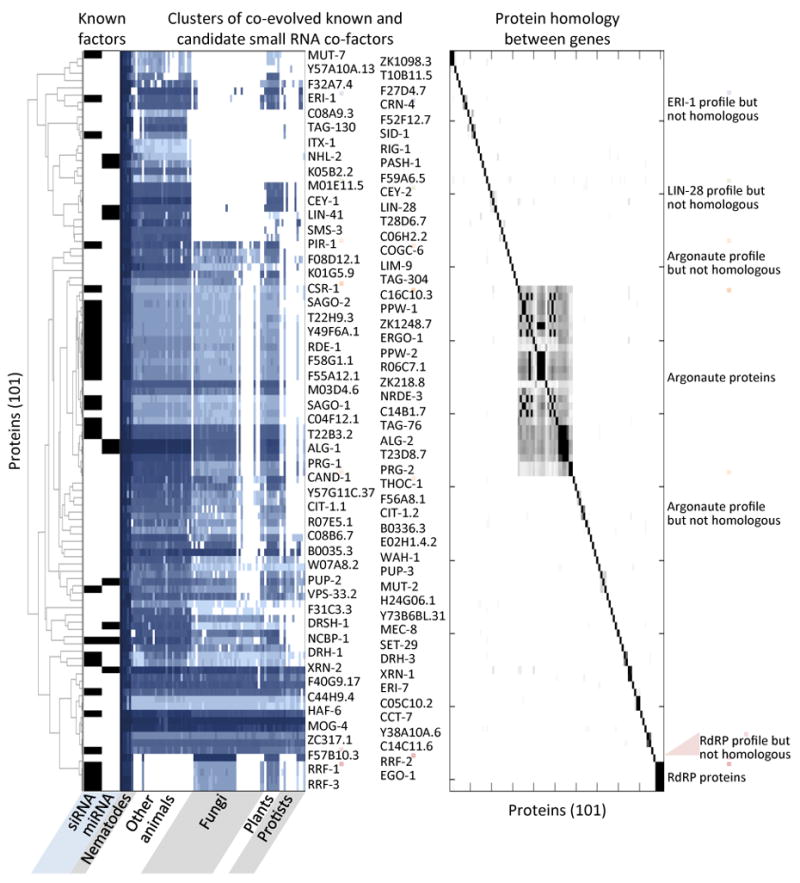
Validated miRNA and siRNA pathway proteins map non-randomly on the phylogenetic profile; proteins that map to the same clusters are likely to function in small RNA pathways. Left panel: Clusters enriched for validated miRNA and siRNA pathway proteins, black boxes. Darker blue represents higher protein sequence similarity. Right panel: Pairwise local protein sequence alignment of all pairs of proteins in the cluster; black represents significant similarity and white no similarity.
The validated siRNA and miRNA protein cofactors identified to date likely constitute a small fraction of the total number of proteins that mediate small RNA function. Full genome RNAi screens for defects in siRNA or miRNA pathway function have identified hundreds of additional candidate small RNA pathway proteins. We integrated ten genome-scale studies into the phylogenetic cluster analysis: five C. elegans gene inactivation screens for defects in RNAi or miRNA function1,15,16, C. elegans orthologues of Drosophila genes identified in two full-genome RNAi screens for impaired siRNA or miRNA response2, and three proteomic studies of complexes containing the known RNAi proteins DCR-14, ERI-117, and AIN-218. Candidate genes identified in these studies show little overlap (Supplementary Table 3; Supplementary Figure 3A,B). However, the candidates from the different studies have similar phylogenetic profiles to each other and to validated small RNA cofactors (Figure 3, Supplementary Figure 3C,D; Supplementary Table 4).
Figure 3. Select phylogenetic clusters enriched with hits from proteomic and functional genomic small RNA screens.
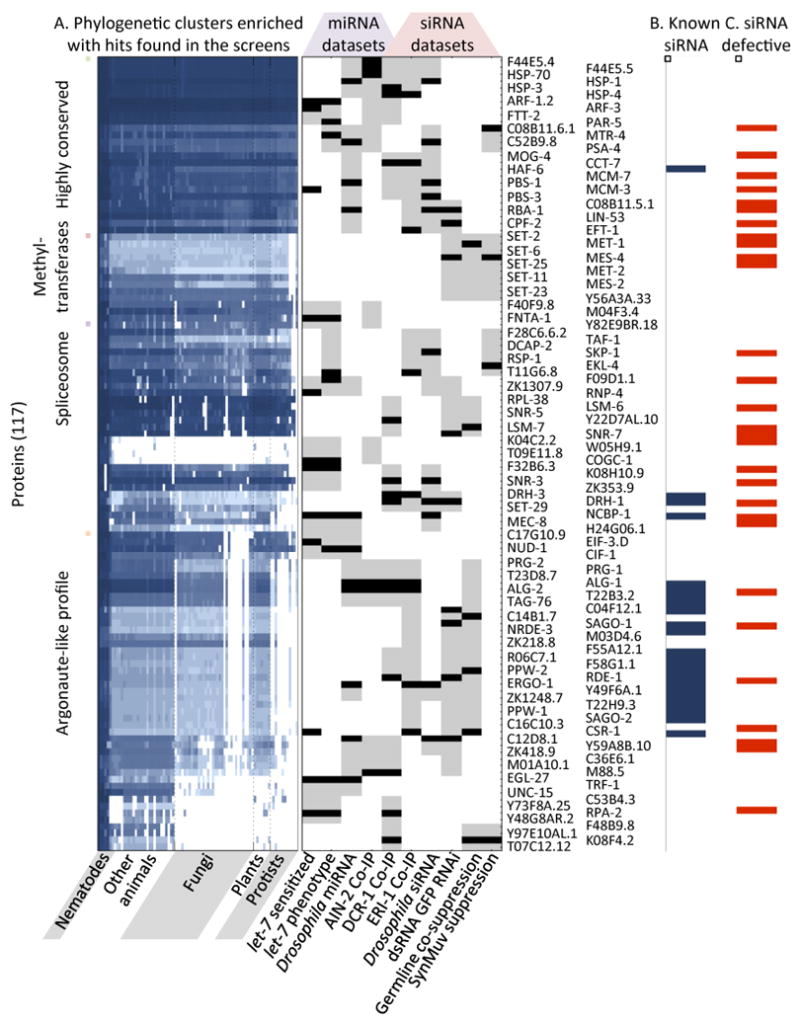
A. The phylogenetic profile matrix was clustered and a Max Ratio score (MRS) was calculated for every protein in each screen; 117 proteins scored significantly in miRNA (56 genes) or siRNA (75 genes) functional genomic screens, or both (14 genes). Middle panel, black tick, hit in screens; gray tick, significant MRS. B. Blue boxes, the 23 known small RNA pathway genes identified. C. From the 117 genes predicted by the phylogenetic profile, 28 genes (blue bars) show defects in siRNA silencing (p-value < 3x10-15).
We used a Naïve Bayesian Classifier to assign predictive values to six genome-scale studies of RNAi cofactors and five of miRNA cofactors (see Supplementary Methods)19,20. To the phylogenetic profiles, we added a score for each C. elegans gene that is co-expressed on microarrays21 or whose encoded gene product interacts with validated small RNA pathway proteins22. The top 105 genes identified by this analysis were enriched with 41 well-validated siRNA pathway genes (Supplementary Figure 7, Supplementary Table 2). The other genes on this list are excellent candidates to mediate siRNA or related small RNA functions. More than 20 of these genes encode RNA recognition motifs including RNP (p-value 2x10-06) and helicase (p-value 1.4x10-05), a ∼20-fold enrichment relative to the entire dataset. Nine proteins from this list constitute components of the spliceosome (Supplementary Figure 3).
We tested a set of the top predictions from phylogenetic profiling (Figures 1-3) and Bayesian analysis using two different tests for defects in RNAi. Transgene silencing in the somatic cells of the enhanced RNAi mutant eri-1(mg366) is mediated by an RNAi mechanism1. We tested a set of 87 predicted small RNA pathway genes in this strain, and 43 scored as significantly RNAi defective (Supplementary Table 2, Figure 4A). We also tested candidate genes using a GFP-based sensor for the abundant C. elegans endogenous siRNA 22G siR-123 to monitor whether any of the gene inactivations affect the production or response to this endogenous siRNA. Thirty-three out of 87 genes tested scored in this assay (Supplementary Table 2, Figure 4B). Eight of the nine predicted splicing components scored strongly in these validation screens.
Figure 4. Inactivation of genes implicated in RNAi pathways reanimates transgenes that are silenced by RNAi.
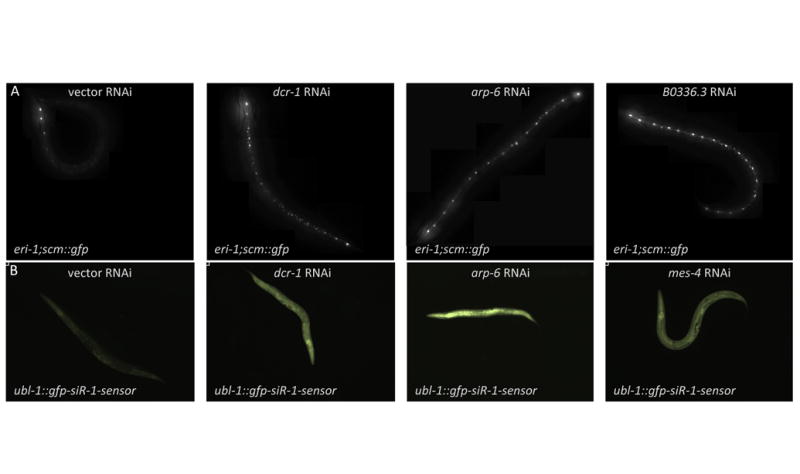
A. Expression of scm::gfp in the seam cells of an eri-1(mg366) mutant, where it is normally silenced by RNAi. Animals shown were treated with control, dcr-1, arp-6, or B0336.3 RNAi. B. GFP expression from the ubl-1::gfp-siR-1 sensor transgene, which is normally silenced by the siR-1 endogenous siRNA. Animals shown were treated with control, dcr-1, arp-6, or mes-4 RNAi.
The enrichment for RNA splicing components (Supplementary Figure 4) points to a close mechanistic connection between splicing and small RNA regulation. Among the Ascomycota and protist species that have lost the Argonaute proteins, most exhibit an extreme loss of introns, from 104-105 introns in species with Argonautes to 102 introns or less introns in most species without Argonautes (Supplementary Figure 5). We screened for defects in RNAi a cherry-picked gene inactivation sublibrary of C. elegans orthologues of known splicing factors that have emerged from biochemical and genetic screens for splicing components from other systems. From a set of 46 C. elegans genes annotated in KEGG to encode the orthologues of known splicing proteins that could be tested for roles in RNAi in our assays, 16 and 22 of these splicing factor genes scored strongly in the eri-1 transgene desilencing assay and the endogenous 22G siR-1 sensor assay. Many of the splicing components that scored strongly in these screens show a phylogenetic profile similar to the Argonaute proteins (Supplementary Figure 6, Supplementary Table 6). However, a subset of splicing factors that are well conserved across phylogeny also scored strongly in these assays.
We used the eri-1 transgene desilencing system to conduct a full genome screen for gene inactivations that disable transgene silencing and identified 855 genes required for transgene silencing, with more than 200 scoring above 3 on a scale of 0 to 4 for desilencing (Supplementary Table 7). Among gene inactivations that caused the greatest desilencing, 11% correspond to the highest ranked predictions from the siRNA Naïve Bayesian analysis, a 30-fold enrichment (p-value = 4.7x10-13 using hypergeometric test) for positives. Of the 84 splicing factors that have been assigned to specific splicing steps, 49 scored in the full genome screen as required for transgene silencing, and 32 showed phylogenetic profiles clustering with known small RNA factors. The splicing factors that couple to small RNA pathways were not isolated to any particular step of RNA splicing. Splicing factor mutations in S. pombe disrupt the RNAi based centromeric silencing24. Both splicing proteins and siRNA/miRNA pathway proteins co-localize to cytoplasmic processing bodies (P-bodies) and nuclear Cajal bodies25, further supporting the possibility of functional crosstalk between splicing and RNAi.
Early genome sequence comparisons of S. pombe, S. cerevisiae, and a small set of eukaryotes suggested that loss of introns and splicing components is highly correlated with loss of Argonaute proteins26. One interpretation was that the loss of RNAi in S. cerevisiae allowed viral invasion and a subsequent loss of introns via reverse transcription of genes by the invading viral replication enzymes. However, such a scenario would not predict that inactivation of splicing components in a species bearing the RNAi apparatus would cause an RNAi deficient phenotype. One model is that splicing could regulate RNAi indirectly by modulating spliced isoforms of key RNAi factors. However, the observations that only a subset of splicing cofactors are required for RNAi and the co-immunoprecipitation of splicing factors and DCR-1, ERI-1 and AIN-2 disfavors this indirect model. Rather, a mechanistic coupling between RNAi and RNA splicing explains these new data better. RNAi factors also affect splicing: Dicer is required for efficient spliceosomal RNA maturation in C. albicans27. If RNAi engages introns intimately by, for example, engaging nascent transcripts through the Argonaute NRDE-3 before splicing28, then the selective advantage of introns may fade once the RNAi pathway is lost.
Our data suggest that a large subset of the proteins that mediate steps in the maturation of mRNAs bearing introns are also required for RNAi, and those genomes that have lost most of their introns no longer require the RNAi pathway. Superimposed on the mRNA splicing pathway is an RNA surveillance system that eliminates aberrantly processed or mutant pre-mRNAs and mRNAs. It is possible that RNAi constitutes another level of mRNA surveillance that acts in parallel to and using many of the same components as the splicing quality control surveillance pathways.
Methods summary
Informatics
The Normalized Phylogenetic Profile data matrix (NPP) was clustered via MATLAB statistical toolbox using the average linkage method and Pearson correlation coefficient as a similarity measure. Clustering was performed on the rows of the matrix. To identify C. elegans proteins with phylogenetic profiles similar to published small RNA co-factors (Supplementary Table 9), the fraction of the validated genes in each phylogenetic cluster was calculated and optimized to define a Max Ratio Score (MRS), (Supplementary Figure 2).
Supplementary Material
Acknowledgments
We thank Thomas Duchaine for access to his ERI-1 proteomic data before it was published and to Sylvia Fischer, Chi Zhang, and Tai Montgomery for helpful discussions. The work was supported by NIH GM088565 and the Pew Charitable Trusts (J.K.K.) and NIH GM44619 and GM098647 (G.R.).
Footnotes
Author contributions
Y.T., J.K.K., and G.R. designed experiments and Y.T. developed analytical tools, analyzed data and Y.T., A.C.B., G.D.H., M.A.N., S.M.G., H.G., R.K., J.K.K., designed and performed experiments. O.Z. gave statistical support and conceptual advice. Y.T. K.Y, B.C. M.B. wrote code. Y.T., A.C.B., J.K.K., G.R. wrote the paper. G.R. J.K.K., supervised the project.
References
- 1.Kim JK, et al. Functional genomic analysis of RNA interference in C. elegans. Science. 2005;308:1164–1167. doi: 10.1126/science.1109267. 1109267. 10.1126/science.1109267. [DOI] [PubMed] [Google Scholar]
- 2.Zhou R, et al. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. S1097-2765(08)00734-X. 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meister G, et al. Identification of novel argonaute-associated proteins. Current biology : CB. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 4.Duchaine TF, et al. Functional proteomics reveals the biochemical niche of C. elegans DCR-1 in multiple small-RNA-mediated pathways. Cell. 2006;124:343–354. doi: 10.1016/j.cell.2005.11.036. S0092-8674(05)01394-2. 10.1016/j.cell.2005.11.036 . [DOI] [PubMed] [Google Scholar]
- 5.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci U S A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabaldon T. Evolution of proteins and proteomes: a phylogenetics approach. Evol Bioinform Online. 2005;1:51–61. [PMC free article] [PubMed] [Google Scholar]
- 7.Pagliarini DJ, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. S0092-8674(08)00768-X. 10.1016/j.cell.2008.06.016 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avidor-Reiss T, et al. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. S009286740400412X. [pii] [DOI] [PubMed] [Google Scholar]
- 9.Enault F, Suhre K, Abergel C, Poirot O, Claverie JM. Annotation of bacterial genomes using improved phylogenomic profiles. Bioinformatics. 2003;19(Suppl 1):i105–107. doi: 10.1093/bioinformatics/btg1013. [DOI] [PubMed] [Google Scholar]
- 10.Drinnenberg IA, et al. RNAi in budding yeast. Science. 2009;326:544–550. doi: 10.1126/science.1176945. 1176945. 10.1126/science.1176945 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drinnenberg IA, Fink GR, Bartel DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yelina NE, et al. Putative Arabidopsis THO/TREX mRNA export complex is involved in transgene and endogenous siRNA biosynthesis. Proc Natl Acad Sci U S A. 2010;107:13948–13953. doi: 10.1073/pnas.0911341107. 0911341107. 10.1073/pnas.0911341107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ketting RF, Plasterk RH. A genetic link between co-suppression and RNA interference in C. elegans. Nature. 2000;404:296–298. doi: 10.1038/35005113. [DOI] [PubMed] [Google Scholar]
- 14.Simmer F, et al. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 15.Cui M, Kim EB, Han M. Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet. 2006;2:e74. doi: 10.1371/journal.pgen.0020074. 10.1371/journal.pgen.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parry DH, Xu J, Ruvkun G. A whole-genome RNAi Screen for C. elegans miRNA pathway genes. Curr Biol. 2007;17:2013–2022. doi: 10.1016/j.cub.2007.10.058. S0960-9822(07)02155-0. 10.1016/j.cub.2007.10.058 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thivierge C, et al. Tudor domain ERI-5 tethers an RNA-dependent RNA polymerase to DCR-1 to potentiate endo-RNAi. Nature structural & molecular biology. 2012;19:90–97. doi: 10.1038/nsmb.2186. 10.1038/nsmb.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, et al. Systematic identification of C. elegans miRISC proteins, miRNAs, and mRNA targets by their interactions with GW182 proteins AIN-1 and AIN-2. Mol Cell. 2007;28:598–613. doi: 10.1016/j.molcel.2007.09.014. S1097-2765(07)00626-0. 10.1016/j.molcel.2007.09.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Calvo S, et al. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet. 2006;38:576–582. doi: 10.1038/ng1776. ng1776. 10.1038/ng1776 . [DOI] [PubMed] [Google Scholar]
- 20.Jansen R, et al. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. 10.1126/science.1087361. 302/5644/449. [DOI] [PubMed] [Google Scholar]
- 21.Hibbs MA, et al. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. btm403. 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- 22.Simonis N, et al. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery TA, et al. PIWI associated siRNAs and piRNAs specifically require the Caenorhabditis elegans HEN1 ortholog henn-1. PLoS genetics. 2012;8:e1002616. doi: 10.1371/journal.pgen.1002616. 10.1371/journal.pgen.1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bayne EH, et al. Splicing factors facilitate RNAi-directed silencing in fission yeast. Science. 2008;322:602–606. doi: 10.1126/science.1164029. 10.1126/science.1164029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pontes O, Pikaard CS. siRNA and miRNA processing: new functions for Cajal bodies. Current opinion in genetics & development. 2008;18:197–203. doi: 10.1016/j.gde.2008.01.008. 10.1016/j.gde.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aravind L, Watanabe H, Lipman DJ, Koonin EV. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:11319–11324. doi: 10.1073/pnas.200346997. 10.1073/pnas.200346997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bernstein DA, et al. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:523–528. doi: 10.1073/pnas.1118859109. 10.1073/pnas.1118859109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guang S, et al. An Argonaute transports siRNAs from the cytoplasm to the nucleus. Science. 2008;321:537–541. doi: 10.1126/science.1157647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


