Abstract
In order to maintain chromosomal stability during cell division, eukaryotic cells have evolved a number of surveillance mechanisms termed checkpoints. These checkpoints monitor the completion of essential molecular and cellular processes of one stage before entering another. The spindle checkpoint watches the bi-orientation attachment of spindle microtubules to all condensed chromosomes before initiation of nuclear division during mitosis. Histones are subject to a number of post-translational modifications during the cell cycle, which may in turn modify or facilitate cell cycle progression. Recent studies suggest that mitotic proteins including Bub1 and Sgo1 that are involved in the spindle checkpoint also play a major role in the regulation of histone modifications and chromatin remodeling. This mini-review summarizes emerging information about the new role of spindle checkpoint proteins in chromatin remodeling.
Keywords: Mitotic Checkpoint, Chromatin Remodeling, Sgo1, mitosis, Bub1, Review
2. INTRODUCTION
Equal segregation of chromosomes during mitosis and meiosis is pivotal for the maintenance of genomic stability. Failure of accurate chromosome segregation inevitably leads to cell death or malignant transformation. To faithfully maintain the genetic content and ensure species survival, eukaryotic cells have evolved checkpoint systems to monitor nuclear division during mitosis and meiosis. Recent studies have shown that many of the components that are critical for spindle checkpoint control are also involved in regulating chromatin remodeling. In fact, these proteins seem to coordinate histone modifications and chromatin remodeling with cell cycle progression during mitosis and meiosis. This review attempts to summarize recent developments in the area of spindle checkpoint regulation and chromatin remodeling.
Chromatin is the network of fibers of DNA and proteins that make up the chromosome in eukaryotic cells. The protein components essentially consist of four well-defined histones (1). Chromatin is classified into euchromatin and heterochromatin primarily based on the tightness of the DNA wrapping around the core histone complex (2). In eukaryotic cells, chromatin structures are dynamic and need to be constantly altered to accommodate DNA replication, gene transcription and stress responses. Alterations in the interaction between DNA and histones, together with the recruitment of nuclear proteins, cause changes in the chromatin structure, a process which is commonly referred to as chromatin remodeling(3). The amino acid sequences for four core histones are remarkably conserved across a wide spectrum of evolution. The sequence conservation of histones suggests that these molecules among all eukaryotes have very similar three-dimentsional conformations which are thought to be optimized for a variety of molecular and cellular processes including DNA replication and chromatin dynamics during cell division(1). Histone tails are subject to multiple post-translational modifications such as phosphorylation, methylation, acetylation, and ubiquitination. It has been suggested that the combination of these distinct covalent modifications of histones constitutes the “the histone code” that regulates a variety of cellular processes, including mitosis and meiosis (4). Covalent histone modifications are essential for chromatin remodeling and they also impact mitosis through modulation of the activity and subcellular localization of proteins important to spindle checkpoint regulation (5–7). For example, tri- methylation of histone H3 lysine 9 (H3K9) is tightly related to heterochromatinization (2) and recruitment of checkpoint proteins to centromeres (5–7).
The spindle checkpoint monitors the bi-orientation attachment of microtubules to condensed chromosomes before initiation of anaphase entry. A number of conserved proteins have been identified and characterized that are required for the checkpoint function. These proteins include Bub1, Bub3; Mad1, Mad2 and Mad3 (8–12). In addition to the orthologs of Bub and Mad families that consist of core components of the spindle checkpoint in mammalian cells, several additional gene products including Shugoshin, Aurora B, Plk1 and PP2A also play a role in spindle checkpoint control (13–18).
3. SPINDLE CHECKPOINT COMPONENTS
3.1.Bub1
Bub1 was originally isolated in a screen for budding yeast mutants that were sensitive to a spindle destabilizing drug benomyl (8). It was later elucidated as a serine/threonine protein kinase which is involved in spindle checkpoints during the cell cycle (19–20). The kinase has an N-terminal tetratricopeptide repeat domain followed by a Gle2-binding-sequence motif which mediates the interaction with Bub3 (21). There are two KEN boxes that precede the C-terminal kinase domain (22), suggesting that it is a potential target of anaphase promotion complex/cyclosome (APC/C). Extensive analyses have shown that the conserved N-terminal region is essential for the recruitment of BUB1 to kinetochores whereas KEN boxes serve as docking motifs necessary for substrate recognition (22–23). Recent studies show that Bub1 also regulates loading of important mitotic proteins to kinetorchores (24–25). For example, Bub1 promotes the assembly of outer kinetochore components including CENP-F and BubR1 (a yeast Mad3 homolog), thus functioning as a master organizer of the inner centromeric region (ICR) (26).The kinase activity of Bub1 is required for recruitment of chromosomal passenger complex and Sgo1 to ICR (26). A recent study by Kawashima et al. has demonstrated that H2A-S121 in fission yeast is phosphorylated by Bub1 and that the phosphorylation is regulated in a cell cycle dependent manner. H2A-S121 is phosphorylated along entire chromosome arms in G2 phase whereas its phosphorylation is limited to centromeres during mitosis, suggesting that the phorsphorylation status may play a role in the maintenance of sister chromatid cohesion. This regulatory relationship between Bub1 and histones is conserved in human cells as well (25).
3.2.Shugoshin 1 (Sgo1)
Shugoshin, meaning “Spirit Guardian” in Japanese, was first identified and characterized as a protector of the centromeric cohesion in the fission yeast (27–28). Sgo2, a Sgo1 paralog, was also characterized in fission yeast (27–28). Given their functional conservation, Shugoshin orthologs have been identified and studied in higher eukaryotes including Xenopus, mouse and human (29–31). Intriguingly, Mei-S332, a Drosophila homolog of Sgo1, was studied rather extensively long before Shugoshin family members were identified. It was reported to maintain sister chromatid cohesion during meiosis (32–33). Despite their functional conservation, the Shugoshin family proteins identified from various eukaryotic species only share a limited similarity at the level of amino acid sequences. On the other hand, conserved moitifs including a coiled coil structure and a basic domain termed the SGO motif are identified which are confined to N-terminus and C-terminus, respectively (27).
3.2.1.Splice variants and subcellular localization
Splicing variants of Sgo1 were first reported in both human and mouse (30). It was thought that splice variants of Sgo1 might function in a tissue-specific manner (30). However, there are two major forms of Sgo1 mRNA that are predominantly present in various types of mammalian cells (34). These splice variants in human cells encode a long isoform with 527 amino acid (Sgo1) and a short form with 292 amino acid (sSgo1), respectively. The shorter isoform lacks 268 amino acids that are primarily encoded by exon 6 (34–35). The long form Sgo1, which is mostly studied in higher eukaryotic cells, localizes at the inner centromeric region (25, 27, 30, and 36). Temporally, Sgo1 starts to accumulate at centromeres during prophase, and its peak accumulation is detected at metaphase. Only residual Sgo1 is visible in early anaphase (37). Sgo1 also localizes to centrosomes during interphase and to spindle poles during mitosis (34). Subsequent studies reveal that sSgo1 is primarily localized to centrosome/spindle poles but not to kinetochores (34–35), suggesting that it plays a role in centrosome dynamics during the cell cycle.
3.2.2.Biochemical and molecular functions
3.2.2.1.Kinetochores
The cohesion complex functions as molecular “glue” that connects sister chromatids soon after DNA replication. During early mitosis, cohesion dissociates from chromosome arms by the so-called “prophase pathway” (37). However, centromeric cohesion is retained until the onset of anaphase (38). Researchers were puzzling about the molecular basis by which sister centromeric cohesion is protected. Since the identification of Sgo1 in various eukaryotic cells, it had been proposed that Sog1 may function as a protector for centromeric cohesion before anaphase entry. A series of studies from several laboratories around the world have confirmed the function of Sgo1 as a protector of centromeric cohesion during meiosis in yeast and during mitosis in high eukaryotes because suppression of Sgo1 function results in premature separation of sister chromatids in both meiosis and mitosis (30–31). Given severe phenotypes such as mitotic arrest and catastrophe induced as the result of Sgo1 depletion, it has been proposed that Sgo1 can serve as a molecular target for induction of apoptosis in cancer cells. Indeed, delivery of a competitive peptide interferes with cellular Sgo1 function, resulting in a change in cell cycle progression as well as reduced cell viability in HeLa and A549 cells (39).
3.2.2.2.Spindle poles
Depletion of Sgo1 through RNA interference (RNAi) results in the formation of extra centrosomal foci and premature separation of paired mother and daughter centriols (35). Extensive analyses reveal that sSgo1, but not Sgo1, functions in centrosome dynamics during the cell cycle (35). It has been proposed that sSgo1 may serve as a protector of a “cohesive factor” at the centrosomes. Rad21, a cohesin subunit cleavable by separase, has been also found to be transiently associated with the centrosomes during metaphase and anaphase in Drosophila (40); it is thus reasonable to speculate that sSgo1 at the spindle poles may work in a fashion similar to that of Sgo1 at centromeres by protecting cohesion(35).
3.2.3.Molecular regulation of Sgo1
3.2.3.1Phosphorylation
In Drosophila, Mei-S332 is regulated by phosphorylation by POLO and serine 234 and threonine 331 are the primary phosphorylation (41). In human cells, Sgo1 during mitosis undergoes a dramatic shift on denaturing gels and pretreatment with λ phophatase collapses it to the interphase form, suggesting that it is subjected to protein phosphorylation (35). The short form of Sgo1 (sSgo1) appears to be subject to phosphorylation as well because the localization of sSgo1 to spindle poles is regulated by Plk1 (35). Depletion of Plk1 or expression of Sgo1 mutants devoid of putative Plk1-phorphorylation sites abolished the spindle poles localization of ectopically expressed GFP-sSgo1 (35).
In vitro kinase assays carried by several independent research groups have identified some phosphorylation sites. Aurora B, an important mitotic kinase, phosphorylates Mei-S332 at serine 124,125 and 126 (Figure 1). Expression of Sgo1 mutant with these serine sites mutated to alanines leads to significant reduction in centromeric signals (13). Moreover, depletion of Aurora B through RNAi results in translocalization of Sgo1 from the centromeres to the chromosome arms in HeLa cells. The change of Sgo1 subcelluar localization leads to ectopic protection of cohesin on chromosome arms with concomitant loosening of centromeric cohesion and loss of the primary constriction (14). Sgo1 is also a substrate of Nek2A which phosphorylates human Sgo1 in vitro at serines 14 and 507 (Figure 1). Phosphorylation by Nek2A is not required for assembly of Sgo1 at the kinetochore, however, expression of non-phosphorylatable mutant human Sgo1 perturbs chromosome congression, leading to a significant increase in microtubule attachement errors including an increase of syntelic and monotelic attachments (42).
Figure 1.
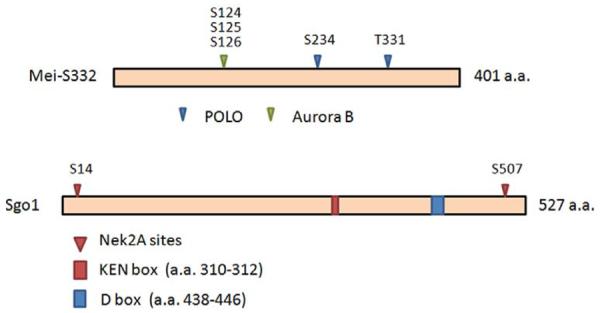
Known or proposed posttranslational modification sites of Sgo1. (A) Amino acid residues subject to posttranslational modifications in Drosophila Mei-S332. (B) Amino acid residues subject to posttranslational modifications in mammalian Sgo1. Phosphorylation sites confirmed by in vitro kinase assays are denoted by filled triangles. Amino acid residues whose phosphorylation have not been verified are denoted by open triangles. KEN box is highlighted in red and confirmed D box is highlighted in dark blue. Putative D boxes are highlighted in light blue. Refer to Table 1 for Additional details.
Consistent with the importance of Sgo1 phosphorylation in its function during mitosis, protein phosphatase 2A (PP2A) can significantly affect the activity and subcellular localization of Sgo1. PP2A physically interacts with Sgo1, and the interaction is required for normal function of Sgo1 during mitosis (17, 43–44). Depletion of PP2A delocalizes centromeric Sgo1 and induces chromosome mis-segregation (17,44). Interestingly, there's a functional interaction between PP2A and Plk1 in the regulation of Sgo1 because Plk1 depletion by RNAi restores centromeric localization of Sgo1 in cells depleted of PP2A and suppresses chromosome mis-segregation induced by the absence of PP2A (44). To date, it remains unknown whether Sgo1 is a direct substrate of PP2A.
3.2.3.2. Ubiquitination
Like many mitotic protein, the level of Sgo1 is also subject to regulation during the cell cycle. Sgo1 protein levels are low during G1 and S phases of the cell cycle; it peaks in mitosis and is degraded after exiting from mitosis (45). Sgo1 degradation depends on APC/C (45–46). Human Sgo1 contains one putative KEN (Lys-Glu-Asn) box (a.a. 310–316) and three putative destruction boxes (D box, a.a. 192–200, a.a. 438–446, and a.a. 457–465). Among the three putative destruction boxes, only the second one (a.a. 438–446) is conserved among human, mouse and Xenopus. Removal of either the KEN box or the D box is not sufficient to stabilize Sgo1. However, Sgo1 mutant with deletion of both KEN and D boxes is more stable in cells (45).Surprisingly, mitosis is not perturbed in the presence of non-degradable Sgo1, suggesting that Sgo1 degradation is required neither for sister-chromatid separation nor mitotic exit (45–46). Figure 1 summarizes various posttranslational modifications of Sgo1.
3.3.Sgo2
Sgo2, a paralogue of Sgo1, plays a distinct role in mitosis and meiosis. While Sgo1 is expressed only around meiosis I in fission yeast Sgo2 is expressed in both mitosis and meiosis. Unlike Sgo1, Sgo2 is dispensable for centromeric protection of cohesion in yeast (27); however, it interacts with Bir1/Survivin and promotes localization of Aurora kinase to the pericentromeric region, which is implicated to correct erroneous attachment of microtubule to kinetochores (27, 43). In contrast to the role in yeast, Sgo1 in higher eukaryotes is essential in the protection of centromeric cohesion in mitosis (30–31). On the other hand, mammalian Sgo2 is highly expressed in oocytes and plays a predominant role in protecting centromeric cohesion in meiosis I (31).
In mammalian cells, PP2A is required for localization of human Sgo1 at the centromeric region (17, 44). However, there is controversy regarding whether in yeast the association of PP2A-π with centromeres is regulated in a Sgo1-dependent manner (17). Kitajima et al. have shown that human Sgo2 (Sgo2 coprecipitates with PP2A and that centromeric localization of PP2A is abolished in Sgo2-dificient mitotic cells, which is accompanied by precocious separation of sister chromatids (43). Thus, Sgo2 may be responsible for recruitment of PP2A to mitotic chromosomes in human cells (43).
3.4.Other components
3.4.1. Aurora B
Aurora B is a member of the conserved protein kinases of the Aurora family (47). It is also characterized as a chromosome passenger protein which mediates spindle checkpoint functions during mitosis (48–49). Aurora B level fluctuates and its subcellular localization varies as well during the cell cycle. Aurora B peaks at the G2/M transition and its kinase activity is high throughout mitosis (47). Aurora B localizes to centromeres during mitosis and the centromeric localization during metaphase is dynamic with a constant exchange between centromeric Aurora B and the soluble pool in the nucleocytoplasm. During anaphase Aurora B concentrates at the mid-zone of mitotic spindles (48). The kinase activity of Aurora B is required for stable activation of the spindle checkpoint; more importantly, Aurora B is primarily responsible for phosphorylation of histone H3 serine 10 (H3S10) during mitosis (47). In fact, H3S10 phosphorylation is the major mitosis-specific phosphorylation of histone molecules and is thought to play a role in super-condensation and super-compaction of chromosomes during mitosis in higher eukaryotic cells (50).
3.4.2. Plk1
Polo like kinases are named after POLO, a gene encoding a protein serine/threonine kinase in Drosophila. (51) Plk1 is the best characterized member of the Polo kinase family. The protein level of Plk1 and its kinase activity are also regulated in a cell cycle dependent fashion, peaking during mitosis (52). Extensive studies in the past have identified many targets that are involved in cell cycle regulation. Depletion of Plk1 results in mitotic arrest that is partly due to alterations of important cell cycle molecules including APC/C (51). During prophase, Plk1 is also involved in controlling so-called the “prophase pathway” that regulates arm cohesion of sister chromatids through direct phosphorylation of Scc1, an integral component of the cohesin complex37. Cohesin not only plays a major role in the regulation of sister chromatid cohesion during the cell cycle but also functions as a transcriptional insulator for the genome (53).
3.4.3.PP2A
Protein phosphatase 2A (PP2A) exists primarily as a heterotrimeric complex which is composed of the scaffolding A subunit (PP2A-A), the variable regulatory subunit B (PP2A-B), and the catalytic subunit (PP2A-C) (54). As a serine/threonine phosphatase, PP2A regulates numerous molecular processes through dephosphorylating various substrates (55). Recent studies indicate that PP2A also localizes centromeres during mitosis and that its activity is essential for the maintenance of centromeric cohesion of sister chromatids before anaphase entry (17, 43–44). In mammalian cells, the localization of Sgo1 to centromeres is PP2A-dependent (43–44).
4.THE FUNCTIONAL INTERACTION OF SPINDLE CHECKPOINT AND CHROMATIN
Chromatin remodeling is a constant process that accompanies the cell cycle and it is especially dynamic during mitosis as duplicated chromosomes super-condense and super-compact in preparation for nuclear division. Many kinetochore proteins that are assembled at the centromeres during mitosis are part of heterochromatin, the function of which remains largely unknown. For example, inactivation of heterochromatin formation by genetic mutations abolishes centromere-directed location of cohesion (38). Moreover, heterochromatin protein (HP1), a fundamental units of heterochromatin; has several functions at centromeres including silencing gene expression, recombination, loading and retaining cohesin, promoting kinetochore assembly and preventing erroneous microtubule attachement to the kinetochores (5). Soon after Sgo1 is identified in fission yeast, the physical interaction between HP1 (Swi6 as the homologue in fission yeast) and Sgo1 is demonstrated (5, 56). Yamagishi et al. has shown that a mutated form of Sgo1 compromises the interaction between Swi6 and Sgo1 (V242E) which in turn impairs the centromeric localization and function of Sgo1. Sgo1-VE expression provokes nondisjunction in meiosis II, which is similar to the phenotype observed in Swi6 deficient cells (5). Furthermore, forced centromeric localization of Sgo1 is capable of restoring proper meiotic chromosome segregation in cells deficient in Swi6 (5). These observations strongly suggest that the heterochromatin plays a primary role of recruiting Sgo1 to the centromeres.
Heterochromatin components other than HP1 are also temporally involved in Sgo1 subcellular localization. In Suv39 null murine embryonic fibroblasts (MEFs), histone H3 lysine 9(H3K9) cannot undergo trimethylation, and consequently HP1 dislocates from heterochromatin (6). Interestingly, Sgo1 fails to localize to the centromeres during G2 phase of the cell cycle in these cells although it is targeted back to centromeres at the onset of mitosis in a Bub1 kinase activity-dependent manner (7).
Bub1 is known to play an important role in targeting Sgo1 to centromeric heterochromatin (24, 57). However, the mechanism by which Bub1 regulates Sgo1 subcellular localization is not entirely clear. It has been shown that Sgo1 can be largely displaced along the entire length of the chromosome in Bub1-repressed cells (57). Given that Bub1 is a protein kinase, it is likely that the phosphorylation status of Sgo1, which is directly or indirectly regulated by Bub1, would modulate the activity of Sgo1. To date, there is no evidence indicating that Bub1 directly phosphorylates Sgo1. On the other hand, a recent study shed light on the regulatory relationship between Bub1 and Sgo1. Bub1 is capable of phorphorylating histone H2A at Serine 121(S121) and the phosphorylated site is critical for centromeric localization of Sgo1 (25). In fission yeast, expression of Bub1 kinase dead (Bub1-KD) cells abolishes Shugoshin localization. This phenotype is mimicked by expression of an H2A mutant in which S121 has been replaced a non-phosphorylatable form (25). In fact, Sgo1 is associated with nucleosomes containing phosphorylated H2A-S121 (p-H2A-S121) through the conserved SGO motif; forced localization of Shugoshin at centromeric heterochromatin in Bub1-deficient cells restores Shugoshin localization and its biological function (25). Consistent with the role of Bub1 in targeting Sgo1 via chromatin, H2A-S121 is phosphorylated along entire chromosome length in G2 phase but its phosphorylation is limited to centromeres in M phase (25). Similar results have also been found in human cells (25). Therefore, Bub1 phosphorylates H2A on S121, which in turn recruits Shugoshin/Sgo1 to chromosomes. Additional factors like Suv39 may also affect Shugoshin localization during the cell cycle (6–7).
5. AN INTEGRATED MODEL
Recent studies revealed that spindle checkpoint proteins also played a significant role in regulating histone modifications, suggesting a crosstalk between two seemingly discrete molecular processes during mitosis. The following model is proposed to illustrate regulatory relationship between mitotic components and histone modifications (Figure 2). After DNA replication, the cohesin complex functions to “hold” together sister chromatids. Centromeric cohesin is retained or protected by the combined action of Sgo1 and PP2A before anaphase initiation. Suv39 tri-methylates H3K9 in heterchromatin, leading to the retention or recruitment of Sgo1; therefore results in the protection of centromeric cohesin. Moreover, Bub1 phosphorylates H2A T121 (H2A S120 in yeast), which in turn recruits Sgo1 to the centromeres, reinforcing the integrity of the cohesin complex. Intriguingly, Bub1 appears to also function to suppress arm-localization of Sgo1 albeit its mechanism remains unclear. Therefore, Bub1 and Suv39 recruit Sgo1 to the centromeres through phosphorylation of H2A T121 and tri-methylation of H3K9, respectively, which reinforces the spindle checkpoint control during mitosis.
Figure 2.
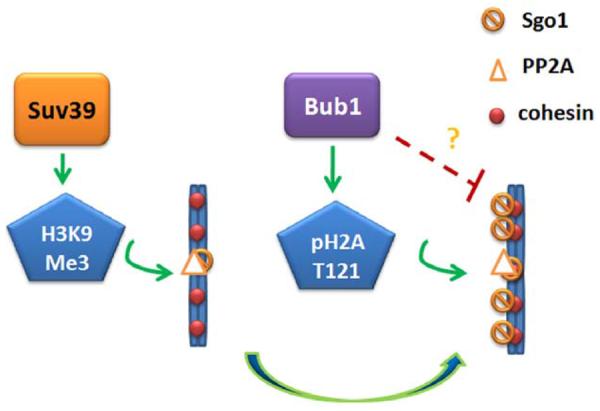
A model illustrating regulatory network of spindle checkpoint control and histone remodeling. Arrows indicate positive regulation and red blocks indicate negative regulation. The question marks denote the regulatory relationship remains unknown.
Table 1.
Known or proposed sites of Sgo1 subject to phosphorylation or ubiquitination
| Mmodification | Putative site(s) | Enzyme | In vitro assay | Mutant assay | Molecular function |
|---|---|---|---|---|---|
| phosphorylation | Mei-S332 Serine 124,125,126 |
Aurora B | yes | yes | Required for centromere localization in meiosis13 |
| phosphorylation | Mei-S332 Serine 234, Threonine 331 |
POLO | yes | yes | Delocalization from chromosome41 |
| phosphorylation | Human Sgo1 Serine 14, 507 |
NEK2A | yes | yes | Mutants lead to abnormal chromosome congression42 |
| phosphorylation | Human Sgo1 192–195,317–320,457–460 |
Aurora | no | no | Reviewed in Ref.30 |
| phosphorylation | Human short form Sgo1 serine 73, threonine 146 | Plk1 | no | yes | Localization to spindle poles35 |
| phosphorylation | Human Sgo1 365–370 | Plk1 | no | no | Reviewed in Ref.30 |
| phosphorylation | Human Sgo1 456–459, 476–479 |
PKA | no | no | Reviewed in Ref.30 |
| phosphorylation | Human Sgo1 346–349, 461–465 |
Cdk | no | no | Reviewed in Ref.30 |
| ubiquitination | KEN | APC/Cdh1 | yes | yes | Needed with D box 438–446 for ubiquitination45 |
| ubiquitination | D box 438–446 | APC/Cdh1 | yes | yes | Needed with KEN box for ubiquitination45 |
| ubiquitination | D box 192–200 | APC/Cdh1 | no | yes | Ectopic expression level is too low to determin45 |
| ubiquitination | D box 457–565 | APC/Cdh1 | no | yes | Mutant leads to defects of chromosome alignment and segregation46 |
ACKNOWLEDGEMENTS
We thank Dazhong Xu and other co-workers in the laboratory for various assistance and helpful discussion. We also thank Nedda Tichi for her administrative assistance. This work was supported in part by US Public Service Award to WD (CA090658 and CA113349). This work is also supported in part by HIEHS center fund and Superfund grants (ES000260 and ES010344).
7. REFERENCES
- 1.Lodish H, A.B., Kaiser CA, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira P. Transcriptional Control of Gene Expression. In: Ahr Katherin, Tontonoz Matthew, FROST Erica Pantages, Rice Elizabeth., editors. Molecular Cell Biology. 2006. [Google Scholar]
- 2.Elgin SC. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 3.Hogan C, Varga-Weisz P. The regulation of ATP-dependent nucleosome remodelling factors. Mutat Res. 2007;618:41–51. doi: 10.1016/j.mrfmmm.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Xu D, Bai J, Duan Q, Costa M, Dai W. Covalent modifications of histones during mitosis and meiosis. Cell Cycle. 2009;8:3688–3694. doi: 10.4161/cc.8.22.9908. [DOI] [PubMed] [Google Scholar]
- 5.Yamagishi Y, Sakuno T, Shimura M, Watanabe Y. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 2008;455:251–255. doi: 10.1038/nature07217. [DOI] [PubMed] [Google Scholar]
- 6.Koch B, Kueng S, Ruckenbauer C, Wendt KS, Peters JM. The Suv39h-HP1 histone methylation pathway is dispensable for enrichment and protection of cohesin at centromeres in mammalian cells. Chromosoma. 2008;117:199–210. doi: 10.1007/s00412-007-0139-z. [DOI] [PubMed] [Google Scholar]
- 7.Perera D, Taylor SS. Sgo1 establishes the centromeric cohesion protection mechanism in G2 before subsequent Bub1-dependent recruitment in mitosis. J Cell Sci. 2010;123:653–659. doi: 10.1242/jcs.059501. [DOI] [PubMed] [Google Scholar]
- 8.Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 9.Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 10.Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J Cell Biol. 1996;132:111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cahill DP, Lenqauer C, Yu J, Riqqins GJ, Wilson JK, Markowitz SD, Kinzler KW, Vogelstein B. Mutations of mitotic checkpoint genes in human cancers. Nature. 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 12.Olesen SH, Thykjaer T, Orntoft TF. Mitotic checkpoint genes hBUB1, hBUB1B, hBUB3 and TTK in human bladder cancer, screening for mutations and loss of heterozygosity. Carcinogenesis. 2001;22:813–815. doi: 10.1093/carcin/22.5.813. [DOI] [PubMed] [Google Scholar]
- 13.Resnick TD, Satinover DL, Maclsaac F, Stukenberg PT, Earnshaw WC, Orr-Weaver WC, Carmena M. INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev Cell. 2006;11:57–68. doi: 10.1016/j.devcel.2006.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai J, Sullivan BA, Higgins JM. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell. 2006;11:741–750. doi: 10.1016/j.devcel.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 15.Yao X, Anderson KL, Cleveland DW. The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol. 1997;139:435–447. doi: 10.1083/jcb.139.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mistry HB, MacCallum DE, Jackson RC, Chaplain MA, Davidson FA. A pharmacodynamic model of Aurora kinase inhibitors in the spindle assembly checkpoint. Front Biosci. 2010;15:249–258. doi: 10.2741/3619. [DOI] [PubMed] [Google Scholar]
- 17.Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, Helmhart W, Galova M, Petronczki M, Gregan J, Cetin B, Mudrak I, Ogris E, Mechtler K, Pelletier L, Buchholz F, Shirahiqe K, Nasmyth K. Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 2006;441:53–61. doi: 10.1038/nature04664. [DOI] [PubMed] [Google Scholar]
- 18.Takaki T, Trenz K, Costanzo V, Petronczki M. Polo-like kinase 1 reaches beyond mitosis--cytokinesis, DNA damage response, and development. Curr Opin Cell Biol. 2008;20:650–660. doi: 10.1016/j.ceb.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 19.Roberts BT, Farr KA, Hoyt MA. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taylor SS, McKeon F. Kinetochore localization of murine Bub1 is required for normal mitotic timing and checkpoint response to spindle damage. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 21.Larsen NA, Al-Bassam J, Wei RR, Harrison SC. Structural analysis of Bub3 interactions in the mitotic spindle checkpoint. Proc Natl Acad Sci U S A. 2007;104:1201–1206. doi: 10.1073/pnas.0610358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang J, Yang M, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, Yu H. Structure and substrate recruitment of the human spindle checkpoint kinase Bub1. Mol Cell. 2008;32:394–405. doi: 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolanos-Garcia VM, Kiyomitsu T, D'Arcy S, Chirgadze DY, Grossmann JG, Matak-Vinkovic D, Venkitaraman AR, Yanaqida M, Robinson CV, Blundell TL. The crystal structure of the N-terminal region of BUB1 provides insight into the mechanism of BUB1 recruitment to kinetochores. Structure. 2009;17:105–116. doi: 10.1016/j.str.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang Z, Sun Y, Harley SE, Zou H, Yu H. Human Bub1 protects centromeric sister-chromatid cohesion through Shugoshin during mitosis. Proc Natl Acad Sci U S A. 2004;101:18012–18017. doi: 10.1073/pnas.0408600102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawashima SA, Yamagishi Y, Honda T, Ishiguro K, Watanabe Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science. 2010;327:172–177. doi: 10.1126/science.1180189. [DOI] [PubMed] [Google Scholar]
- 26.Boyarchuk Y, Salic A, Dasso M, Arnaoutov A. Bub1 is essential for assembly of the functional inner centromere. J Cell Biol. 2007;176:919–928. doi: 10.1083/jcb.200609044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitajima TS, Kawashima SA, Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- 28.Rabitsch KP, Gregan J, Schleiffer A, Javerzat JP, Eisenharber F, Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 29.Rivera T, Losada A. Shugoshin regulates cohesion by driving relocalization of PP2A in Xenopus extracts. Chromosoma. 2009;118:223–233. doi: 10.1007/s00412-008-0190-4. [DOI] [PubMed] [Google Scholar]
- 30.McGuinness BE, Hirota T, Kudo NR, Peters JM, Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:e86. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee J, Kitajima TS, Tanno Y, Yoshida K, Morita T, Miyano T, Miyake M, Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- 32.Davis BK. Genetic analysis of a meiotic mutant resulting in precocious sister-centromere separation in Drosophila melanogaster. Mol Gen Genet. 1971;113:251–272. doi: 10.1007/BF00339546. [DOI] [PubMed] [Google Scholar]
- 33.Goldstein LS. Mechanisms of chromosome orientation revealed by two meiotic mutants in Drosophila melanogaster. Chromosoma. 1980;78:79–111. doi: 10.1007/BF00291909. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Yang Y, Dai W. Differential subcellular localizations of two human Sgo1 isoforms: implications in regulation of sister chromatid cohesion and microtubule dynamics. Cell Cycle. 2006;5:635–640. [PubMed] [Google Scholar]
- 35.Wang X, Yang Y, Duan Q, Jiang N, Huang Y, Darzynkiewicz Z, Dai W. sSgo1, a major splice variant of Sgo1, functions in centriole cohesion where it is regulated by Plk1. Dev Cell. 2008;14:331–341. doi: 10.1016/j.devcel.2007.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JY, Dej KJ, Lopez JM, Orr-Weaver TL. Control of centromere localization of the MEI-S332 cohesion protection protein. Curr Biol. 2004;14:1277–1283. doi: 10.1016/j.cub.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Dai W. Shugoshin, a guardian for sister chromatid segregation. Exp Cell Res. 2005;310:1–9. doi: 10.1016/j.yexcr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y, Wang X, Dai W. Human Sgo1 is an excellent target for induction of apoptosis of transformed cells. Cell Cycle. 2006;5:896–901. doi: 10.4161/cc.5.8.2691. [DOI] [PubMed] [Google Scholar]
- 40.Warren WD, Steffensen S, Lin E, Coelho P, Loupart M, Cobbe N, Lee JY, McKay MJ, Orr-Weaver T, Heck MM, Sunkel CE. The Drosophila RAD21 cohesin persists at the centromere region in mitosis. Curr Biol. 2000;10:1463–1466. doi: 10.1016/s0960-9822(00)00806-x. [DOI] [PubMed] [Google Scholar]
- 41.Clarke AS, Tang TT, Ooi DL, Orr-Weaver TL. POLO kinase regulates the Drosophila centromere cohesion protein MEI-S332. Dev Cell. 2005;8:53–64. doi: 10.1016/j.devcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Fu G, Ding X, Yuan K, Aikhionbare F, Yao J, Cai X, Jiang K, Yao X. Phosphorylation of human Sgo1 by NEK2A is essential for chromosome congression in mitosis. Cell Res. 2007;17:608–618. doi: 10.1038/cr.2007.55. [DOI] [PubMed] [Google Scholar]
- 43.Kitajima TS, Sakuno T, Ishiquro K, Iemura S, Natsume T, Kwashima SA, Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- 44.Tang Z, Shu H, Qi W, Mahmood NA, Mumby MC, Yu H. PP2A is required for centromeric localization of Sgo1 and proper chromosome segregation. Dev Cell. 2006;10:575–585. doi: 10.1016/j.devcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Karamysheva Z, Diaz-Martinez LA, Crow SE, Li B, Yu H. Multiple anaphase-promoting complex/cyclosome degrons mediate the degradation of human Sgo1. J Biol Chem. 2009;284:1772–1780. doi: 10.1074/jbc.M807083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fu G, Hua S, Ward T, Ding X, Yang Y, Guo Z, Yao X. D-box is required for the degradation of human Shugoshin and chromosome alignment. Biochem Biophys Res Commun. 2007;357:672–678. doi: 10.1016/j.bbrc.2007.03.204. [DOI] [PubMed] [Google Scholar]
- 47.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 48.Adams RR, Carmena M, Earnshaw WC. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 2001;11:49–54. doi: 10.1016/s0962-8924(00)01880-8. [DOI] [PubMed] [Google Scholar]
- 49.Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat Rev Mol Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- 50.Johansen KM, Johansen J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 2006;14:393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- 51.Dai W, Wang Q, Traganos F. Polo-like kinases and centrosome regulation. Oncogene. 2002;21:6195–6200. doi: 10.1038/sj.onc.1205710. [DOI] [PubMed] [Google Scholar]
- 52.Anger M, Kues WA, Klima J, Mielenz M, Kubelka M, Motlik J, Esner M, Dvorak P, Carnwath JW, Niemann H. Cell cycle dependent expression of Plk1 in synchronized porcine fetal fibroblasts. Mol Reprod Dev. 2003;65:245–253. doi: 10.1002/mrd.10289. [DOI] [PubMed] [Google Scholar]
- 53.Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- 54.Ingebritsen TS, Cohen P. The protein phosphatases involved in cellular regulation. 1. Classification and substrate specificities. Eur J Biochem. 1983;132:255–261. doi: 10.1111/j.1432-1033.1983.tb07357.x. [DOI] [PubMed] [Google Scholar]
- 55.Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 56.Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y. Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol. 2005;15:353–359. doi: 10.1016/j.cub.2004.12.044. [DOI] [PubMed] [Google Scholar]


