Abstract
This chapter describes a method to generate plasma membrane sheets that are large enough to visualize the membrane architecture and perform quantitative analyses of protein distributions. This procedure places the sheets on electron microscopy grids, parallel to the imaging plane of the microscope, where they can be characterized by transmission electron microscopy. The basic principle of the technique is that cells are broken open (“ripped”) through mechanical forces applied by the separation of two opposing surfaces sandwiching the cell, with one of the surfaces coated onto an EM grid. The exposed inner membrane surfaces can then be visualized with electron dense stains and specific proteins can be detected with gold conjugated probes.
Keywords: Plasma membrane, Transmission electron microscopy, Protein distribution
1. Introduction
The plasma membrane is the outer border of a cell and physically separates its interior from the surrounding environment. However, the plasma membrane is not an inert shell. Rather, it is utilized in many cellular processes, and therefore its composition and structure are of great interest to many researchers. One of the early attempts to describe the plasma membrane was the “fluid mosaic” model by Singer and Nicholson (1). In this model, the plasma membrane is a homogeneous two-dimensional lipid bilayer in which proteins can diffuse freely. Over the last decades, this view has been revised, as it became clear that diffusion in the plasma membrane is slower than expected, molecules are not evenly distributed over the cell surface, and molecules are confined to membrane domains and/or surface areas with dimensions of less than one micrometer. These findings led to more complex models, including the “lipid raft” (2), “picket fence” (3), and “protein island” models (4). However, the actual organization of the plasma membrane and its associated molecules remains controversial.
Many biological processes in immune cells utilize and reorganize the plasma membrane. Most immune cells are activated via cell surface receptors, which in some cases is accompanied by the reorganizations of the plasma membrane, most dramatically seen in the formation of the immunological synapse (5). The systematic functions of immune cells often involve the plasma membrane, e.g., endocytosis, phagocytosis, and secretion. Here, we describe a method based on Sanan et al. (6) that has been successfully used in the analyses of the two-dimensional architecture of the plasma membrane in T cells (4, 7), B cells (8), mast cells (9, 10), and other cell types (6, 11, 12). It is based on the generation of plasma membrane sheets attached to EM grids (Fig. 1), by breaking (“ripping”) cells open through forces applied by separating two opposing surfaces sandwiching the cells of interest (Fig. 2). This procedure has been used with a variety of modifications, each optimized for the study of a particular question. In this chapter, the method is divided into four steps: (1) Surface choices and preparations, (2) Cell binding to primary surface, (3) Generation of plasma membrane sheets, and (4) Labeling of plasma membrane sheets. For each step, up to three possible protocols are shown, which can be “mixed and matched,” and further adapted, to suit any specific study aim.
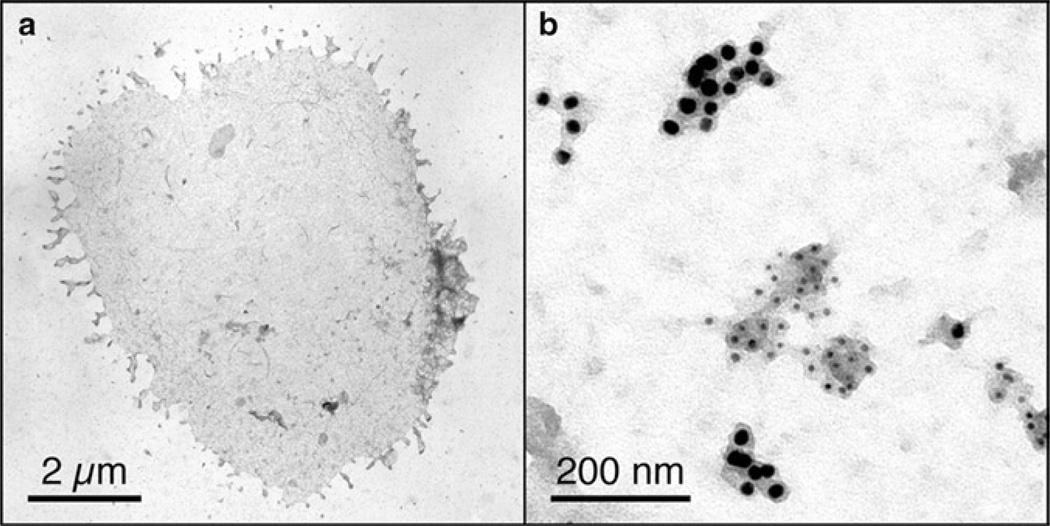
Fig. 1.
(a) Whole plasma membrane sheet from an activated T cell bound to an EM grid coated with stimulatory ligands (peptide-MHC II and co-stimulatory B7.1 molecules). (b) Magnified area of a plasma membrane sheet from a quiescent T cell bound to an EM grid coated with poly-l-lysine. Ectopically expressed myristoylated “non-raft” and myristoylated + palmitoylated “raft” markers are labeled with 5 and 10 nm gold particles, respectively.
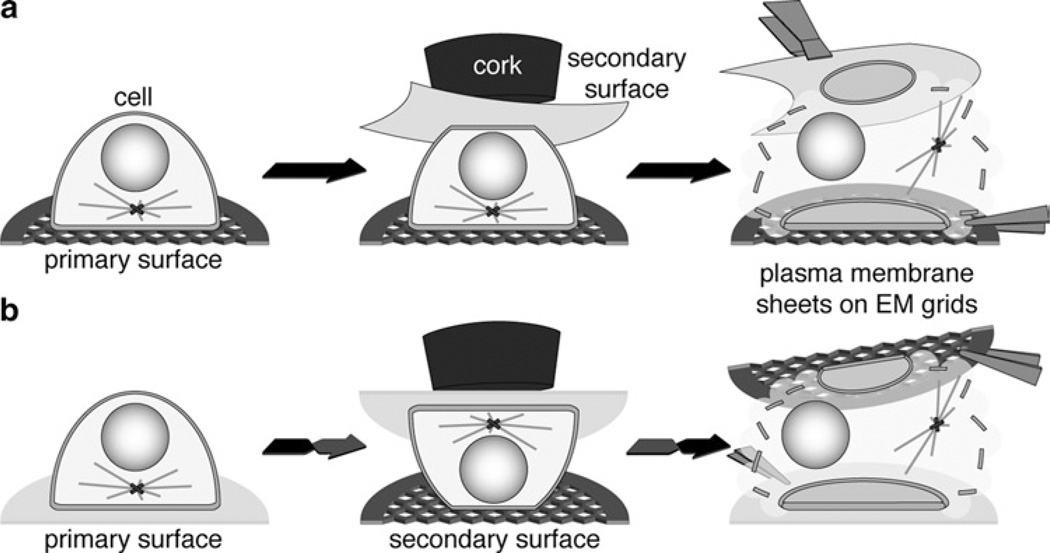
Fig. 2.
Simplified three-step illustrations of the “ripping” procedures to obtain plasma membrane sheets. (For simplification, working surface, filter papers and acetate disks are not included in the illustration.) (a) Example for the generation of plasma membrane sheets from the adherent cell side attached to a coated EM grid (primary surface). A PVDF membrane is used as secondary surface. (b) Example for the generation of plasma membrane sheets from the solvent exposed cell side attached to a PLL-coated EM grid (secondary surface). A PLL-coated cover glass is used as primary surface.
2. Materials
2.1. Equipment
Carbon vacuum evaporator.
Electric glow discharger (e.g., Bench Top Turbo III from Denton Vacuum).
Film casting device.
Forceps (nonmagnetic) type 7 and N5.
Glass beakers (50 ml).
Glass plate (at least 10 × 10 cm).
Hydrophobic slide marker.
Incubator or slide warmer.
Rubber cork #3 or #5.
Vacuum tip connected to a liquid trap.
2.2. Consumables
Acetate disks (0.22 µm pore size; 25 mm diamerter).
Acetic acid.
Acetone ACS and EM grade.
Ashless filter paper.
Biotin-N-hydroxysuccinimide ester (NHS-biotin).
Bovine serum albumin (BSA).
Coverslips (25 mm diameter).
Deionized water.
EM grids (hexagonal).
Fetal calf serum (FCS).
Formvar (1%) in ethylene dichloride.
Glass slides (75 × 25 mm).
Glutaraldehyde (4%).
Glycine.
Hank’s balanced saline solution (HBSS).
Methanol.
Osmium tetroxide.
ParafilmM®™.
Paraformaldehyde (16%).
Petri dishes.
Phosphate-buffered saline (PBS).
Poly-l-lysine HBr (PLL) (150–300 kDa).
Polyvinylidene fluoride membranes (PVDF).
Sodium cacodylate.
Streptavidin.
Syringes + 0.2 µm filters.
Tannic acid.
Uranyl acetate.
2.3. Study-Specific Reagents
Biotinylated ligands and/or antibodies to adhere cells.
Tissue culture media.
Stimulatory antibodies/reagents.
Primary antibodies/probes (possibly gold conjugated) to detect molecules of interest.
Differently sized gold-conjugated secondary antibodies/streptavidin.
2.4. Buffers
HEPES buffer (+Ca/Mg): 20 mM HEPES pH 7.4, 150 mM sodium chloride (+2.5 mM magnesium chloride, 0.5 mM calcium chloride).
Cytosol buffer: 20 mM HEPES pH 7.4, 140 mM potassium chloride, 10 mM sodium chloride, 2.5 mM magnesium chloride, 0.5 mM calcium chloride, 50 µM β-mercaptoethanol.
3. Methods
3.1. Surface Choices and Preparations
The generation of plasma membrane sheets requires the simultaneous interaction of cells with two opposing surfaces (Fig. 2). Therefore, for each study suitable surfaces have to be found or developed. The “primary” surface is initially used to capture and adhere cells as efficiently as possible. In the case of adherent cells, this can be achieved through simply cultivating the cells on surfaces. Alternatively, immobilized ligands to surface receptors, or affinity reagents, like antibodies or streptavidin, can capture and adhere cells. The latter is generally faster and occurs with higher affinities. These primary surfaces can also be used to manipulate (activate, polarize, etc.) the cells of interest. The “secondary” surface is added so that the forces necessary to “rip” the cells can be applied, and in most cases, binds the cells nonspecifically, quickly, and only for a short period of time. In the methods described here, the secondary surfaces interact with the cells for 10–20 s, but up to 15 min have been reported (12). Depending on which surface is used to coat the EM grid, plasma membrane sheets originating from the adherent (Fig. 2a) or solvent exposed side (Fig. 2b) can be analyzed while attached to a surface. This section describes the preparation of glass surfaces (Subheading 3.1.1) or EM grids (Subheading 3.1.2), which can then be coated with two different methods (PLL, Subheading 3.1.3 and immobilized proteins, Subheading 3.1.4). Alternatively, polyvinylidene fluoride (PVDF) membranes (Subheading 3.1.5) can be used instead of the above.
3.1.1. Pretreatment of Glass Coverslips for Efficient Coating
Round glass coverslips are cleaned by sequentially dipping and gently agitating them in deionized water, 20% acetic acid, deionized water, acetone, and three times deionized water.
Place coverslips on top of ashless filter paper inside an auto-clavable container, cover with aluminum foil, and dry autoclave for 20 min.
Just prior to coating with primary or secondary surfaces, electric glow discharge the top of coverslips for 90 s under air of 50–100 mTorr to negatively charge the surface.
3.1.2. Preparation of EM Grids for Efficient Coating
Wash (and if available sonicate) EM grids sequentially in deionized water, 20% acetic acid, three times deionized water, and two times EM grade acetone. Air-dry EM grids and protect from dust (see Notes 1 and 2).
Score formvar film (generated on a glass slide with a film casting device) with a razor blade, and slowly dip the slide perpendicularly into deionized water to float the film ontothe water surface. Place multiple EM grids with their rough side down on top of the film.
Carefully place a clean piece of parafilm on top, sandwiching the EM grids between formvar and parafilm. This is best done by “rolling” the parafilm slowly onto the EM grids, starting on one side of the formvar film and without water running on the top of the parafilm.
Place the sandwich with the formvar side up on ashless filter paper, gently remove any trapped air with an ashless filter wedge, and air-dry.
Slightly carbon coat EM grids in a vacuum evaporator, and just before coating with the primary or secondary surfaces, glow discharge for 30 s under air of 50–100 mTorr to negatively charge the surface.
3.1.3. Poly-l-Lysine as Primary and/or Secondary Surfaces
PLL surfaces bind cells nonspecifically due to the ion bonds between the positively charged lysines and negative charges on the cell surface.
Incubate the glow discharged glass coverslips or EM grids with 0.2 mg/ml PLL in deionized water for 20–30 min at room temperature (RT). EM grids are incubated either by placing them on a glass slide with the parafilm side attached to the glass and placing liquid on top, or by removing them from their parafilm support and inverting them onto droplets (see Note 3). Up to four EM grids can be conveniently incubated on a single droplet.
Rinse coverslips or EM grids with deionized water, remove excess water by touching the edge of the EM grid with an ashless filter paper and air-dry them for at least 1 h. EM grids that have been removed from their parafilm support can be washed by inverting and floating them on water in a small Petri dish.
3.1.4. Immobilized Ligands and/or Antibodies as Primary Surfaces
These surfaces bind specific receptors or other molecules on the cell surface and can be used to activate the signaling pathway downstream of a specific receptor. In the case of T cells, specific major histocompatibility complex II molecules together with the co-stimulatory receptor B7.1 have been used successfully (4, 7). In this case, T cell signaling was initiated by the surfaces, and the T cells polarized toward the adhesion site. In different studies, other endogenous ligands or antibodies against surface molecules can be used in similar fashion.
Recombinantly expressed ligands or antibodies are purified, and chemically or enzymatically biotinylated. Using site-specific biotinylation on a single amino acid is preferred, as it will orientate the protein on the surface and prohibit functional loss due to high biotin incorporation.
PLL is modified at 10 mg/ml with 50-fold excess of NHS-biotin in 50 mM sodium phosphate pH 7.8 for 2 h at RT. Nonsoluble NHS-biotin is removed by centrifugation. The biotinylated PLL is then filtered through a 0.2 µm cut-off syringe filter and dialyzed against deionized water. (Biotinylated PLL can be stored at −20°C).
Glow discharged coverslips or EM grids are incubated with 0.1 mg/ml biotinylated PLL for 20–30 min at RT.
Rinse coverslips or EM grids with deionized water, remove excess water by touching the edge of the surface with an ashless filter paper, and air-dry them for at least 1 h.
Block surfaces with HEPES buffer + 10% FCS or alternatively + 1% BSA for 1 h at RT in a humidity chamber, and wash three times with HEPES buffer.
Incubate surface with 20–50 µg/ml streptavidin in HEPES buffer + 0.2% BSA for 1 h at RT in a humidity chamber and wash three times with HEPES buffer.
Incubate with a combined concentration of biotinylated ligands and/or antibodies of approximately 400 pmol/ml in HEPES buffer + 0.2% BSA for 1 h at RT in a humidity chamber and wash three times with HEPES buffer. This step should be optimized (time and protein concentrations) for different ligands and antibodies.
After steps 4, 5, 6, or 7, the surfaces can be stored for up to several days under HEPES buffer, with or without BSA, in a humiditybox at 4°C. However, some recombinant ligands might not be stable under these conditions for an extended period of time.
3.1.5. Polyvinylidene Fluoride Membranes as Secondary Surface
PVDF membranes have high protein binding capacity and bind well to cells. Due to their flexibility, they are an easy material for the generation of membrane sheets. PVDF membranes do not allow TEM analysis of the plasma membrane attached to them, and are only suitable as secondary surface.
PVDF membranes are activated by soaking them in methanol for ~2 min.
Wash and equilibrate membranes in HEPES buffer + Ca/Mg for at least 5 min.
3.2. Cell Binding to Primary Surfaces
Plasma membrane sheets are bound to EM grids via the extracellular leaflet and the cytosolic leaflet is exposed to solvent. Thus, they can only be sufficiently labeled with reagents that recognize the cytosolic portion of molecules. Proteins or other markers, fused to intracellular tags (HA-tag, Myc-tag, GFP, etc.), can be ectopically expressed in cells and detected in plasma membrane sheets with the appropriate antibodies. In studies that require thedetection of extracellular molecules, gold-conjugated probes have to be used prior to the ripping procedure. Live cells can be labeled when bound to the primary surface if a study focuses on the solvent exposed cell side, or alternatively in suspension for either side. However, in this latter case, the label may inhibit binding to the primary surface. Multivalent probes can multimerize their targets and/or induce endocytosis in live cells, which will affect the results of the TEM analyses. In studies, where cells are activated by receptor cross-linking this can be a desired effect. Multimerization and activation can be prevented by fixation of the cells prior to labeling. However, the degree of fixation has to be optimized to avoid multimerization, but still allow binding to the surfaces and ripping of the cell. Here, three possible conditions that have been successfully used to bind cells to primary surfaces are described.
3.2.1. Short-Term Binding of Suspension Cells to Primary Poly-l-Lysine Surface
Grow up enough cells to cover 60–80% of the primary surfaces. If EM grids are the primary surface, bind them with the attached parafilm proximal to the glass slide and draw a border around them with a hydrophobic slide marker. For the appropriate number of cells, take the additional area surrounding the EM grid into account.
Wash the cells two times either in tissue culture media without serum or in HBSS. Resuspend the cells in enough media or buffer to cover the primary surface with an appropriate amount of liquid.
Apply the cells to the primary surface and place in a tissue culture incubator with humidity control, or on a slide warmer surrounded by moist tissues (to avoid drying of the samples) at 37°C for 30–60 min. The duration has to be optimized for different cell types.
Rinse and remove excess cells by sequentially dipping and then gently agitating the surface two times in HEPES buffer + Ca/Mg and continue with the ripping procedure.
3.2.2. Cell Activation and Binding to Immobilized Ligand Surfaces
This procedure is similar to the binding of cells to PLL surfaces (Subheading 3.2.1). However, some adjustments have to be considered.
The cell number has to be increased so that 60–80% of the primary surfaces are covered within the time used to activate cells with immobilized ligands. For example, to obtain the same number of T cell membrane sheets on surfaces coated with immobilized ligands in 5 min as on PLL coated surfaces in 30–60 min the T cell concentration has to be tripled.
Any necessary supplement for the activation and adhesion of the cells on immobilized ligand surfaces (Calcium, Magnesium, FCS, etc.) has to be added to the media or HBSS.
Before the ripping procedure, protein in the media or buffer has to be removed to increase the binding efficiency to the secondary surface.
3.2.3. Cell Growth on Primary Surface and Labeling of Extracellular Receptors
Cells are grown on PLL surface (coverslips or EM grids) or directly on glass coverslips under the same conditions as used in normal cell culture condition. Coverslips can be submerged under media in tissue culture plates with wells ~1.5 times wider than the coverslip itself. Cells are grown on EM grids in a similar fashion as described for the binding to PLL surfaces (Subheading 3.2.1). Alternatively, EM grids can be bound to the bottom of a tissue culture plates or wells via the attached parafilm.
- Continue with either a or b.
Rinse and remove excess cells by sequentially dipping and gently agitating the surface two times in HEPES buffer + Ca/Mg and continue with the ripping procedure.
3.3. Generation of Plasma Membrane Sheets
Depending on what surface is used to coat the EM grid, the plasma membrane attached to either the primary surface (adherent cell side; Fig. 2a) or secondary surface (solvent exposed cell side; Fig. 2b) can be analyzed. In general, the primary surfaces yield larger, more intact and higher numbers of plasma membrane sheets. Therefore, if the study permits it, the EM grid is ideally coated with the primary surface. The ripping procedure can take place on ice or at RT. If cold conditions are required, place all beakers with buffers in and the ripping surface on compressed ice in a large insulated tray. Under any conditions, excessive amounts of drying that would destroy the membrane sheets or condensation that affects buffer concentrations should be avoided (see Note 4). Here, two procedures are described that were successfully used to generate plasma membrane sheets from the adherent (Subheading 3.3.1) and the solvent exposed side (Subheading 3.3.2) of T cells.
3.3.1. Ripping Procedure for the Analysis of Adherent Cell Side (Fig. 2a; Steps 5–14 Are Also Used in Subheading 3.3.2)
Place an ashless filter paper on a clean glass surface and equilibrate it thoroughly with HEPES buffer + Ca/Mg. Place an acetate disk on top of filter paper and remove excess buffer with a vacuum tip.
Equilibrate EM grids (two can be easily ripped simultaneously) coated with primary surface and bound cells (Subheading 3.2) by dipping and gently agitating it in HEPES buffer + Ca/Mg (see Note 5).
Place EM grids with the cell side up on the acetate disk (Fig. 2a, left). Remove any excess liquid carefully with a vacuum tip without drying the surface.
Place a PVDF membrane or other secondary surface (Subheading 3.1) equilibrated with HEPES buffer + Ca/Mg and excess liquid removed (by touching the edge of the surface with an ashless filter paper) on top of EM grids (PLL surface can be placed dry on top of cells). Make sure no air is trapped between EM grids and secondary surfaces.
Place another acetate disk, previously equilibrated in HEPES buffer + Ca/Mg, and a rubber cork on top of the sandwich (Fig. 2a, middle or Fig. 2b middle for Subheading 3.3.2).
Apply vertical pressure to the sandwich for 5–20 s by firmly bearing down with the cork and at the same time remove excess buffer with a vacuum tip. This step requires practice and differs between cell and surface types. If not enough force is applied, the secondary surface does not bind the cells and no ripping occurs; and if too much force is applied, the membrane sheets become damaged.
Remove the rubber cork while keeping the rest of the sandwich in place with a pair of forceps.
Immerse the whole sandwich thoroughly in cytosol buffer. This will avoid drying of the specimen while the sandwich is disassembled.
Remove the top acetate disk with a pair of forceps while keeping the rest of the sandwich in place with another pair of forceps.
Lift the secondary (or primary for Subheading 3.3.2) surface with a pair of forceps while pushing down on the bottom acetated disk with another pair of forceps. The EM grids should stick to the surface. If EM grids remain on bottom acetate disk, immerse them quickly in cytosol buffer and continue with step 12. This can happen if too few cells are used or the secondary surface is not binding well enough to the cells.
Remove the EM grids vertically from the surface with a fine tipped pair of forceps. If the EM grid accidentally slides along the secondary (or primary for Subheading 3.3.2) surface, the cells might rip in a way that results in damaged plasma membrane sheets (Fig. 2a, right or Fig. 2b right for Subheading 3.3.2; see Note 6).
Quickly invert and float EM grids on cytosol buffer in a small Petri dish for 30 s to 1 min. This will remove cell debris from the ripping procedure.
Float EM grids on cytosol buffer + 4% paraformaldehyde + 0.25% glutaraldehyde for 20 min at RT. Depending on the labeling method, the concentrations of the fixatives and time of fixation have to be optimized. Some antibodies do not recognize their epitopes after stringent fixation (see Note 7).
Float EM grids for 10 min in PBS + 25 mM glycine in order to quench excess fixation reagents and continue with labeling of plasma membrane sheets.
3.3.2. Ripping Procedure for the Analysis of Solvent-Exposed Cell Side (Fig. 2b)
Place an ashless filter paper on a clean glass surface and equilibrate it thoroughly with HEPES buffer + Ca/Mg. Place an acetate disk on top of filter paper and remove excess buffer with a vacuum tip.
Equilibrate EM grid coated with the secondary surface (Subheading 3.1) with HEPES buffer + Ca/Mg and place it on top of the acetate disk with the coated side facing up (two can easily be ripped simultaneously, and PLL-coated EM grids can be placed dry). Remove excess buffer carefully with a vacuum tip.
Equilibrate primary surface with bound cells (Subheading 3.2) by dipping and gently agitating it in HEPES buffer + Ca/Mg (Fig. 2b, left; see Note 5). Remove excess liquid by touching the edge of the surface with an ashless filter paper.
Place secondary surface with the bound cells facing down on top of the EM grid and continue with steps 5–14 as described in Subheading 3.3.1.
3.4. Labeling of Plasma Membrane Sheets
Multiple membrane-associated molecules of interest can be labeled with different sized gold-conjugated detection reagents (Fig. 1b). However, more than two different sizes can make the identification of the gold species difficult during analyses. Probes specific to certain molecules can be directly conjugated to colloidal gold, thus no secondary label is required.
Rinse EM grids with the fixed plasma membrane sheets attached twice by floating on PBS.
Invert the EM grids onto droplets (50–100 µl) of the primary staining reagents in PBS + 0.2% BSA and incubate between 1h and overnight at RT in a humidity chamber. Antibodies have been successfully used in concentrations of 10–100 µg/ml; however, this has to be optimized for every probe. If multiple probes are used, they should be applied as a mixture to minimize effects due to steric inhibition of binding (see Note 7).
Rinse EM grids three times for 5 min at RT by floating them on PBS.
Invert EM grids onto droplets (50–100 µl) of mixtures of gold-conjugated secondary probes in PBS + 0.2% BSA for 1–3 h at RT in a humidity chamber. Gold reagents are mostly stored at concentrations of an OD520nm of ~1.0 and are used in 5- to 20-fold dilutions.
Rinse EM grids three times for 5 min at RT by floating them on PBS.
Postfix EM grids on a droplet of 2% glutaraldehyde in PBS for 10 min at RT in a humidity chamber.
Rinse EM grids sequentially in PBS + 25 mM glycine, PBS, and 100 mM cacodylate for 5 min at RT.
Incubate grids on droplets of 100 mM cacodylate + 1% osmium tetroxide for 10 min at RT in a humidity chamber under a chemical hood. Rinse one time on 100 mM cacodylate and twice on deionized water for 5 min each time at RT.
Incubate grids on droplets of 1% tannic acid (freshly filtered) for 10 min at RT in a humidity chamber and rinse three times on deionized water for 5 min at RT.
Incubated grids on droplets of 1% uranyl acetate (freshly filtered) for 10 min at RT in a humidity chamber and rinse for 30 s to 1 min on deionized water.
Remove excess liquid by touching the edge of the EM grid with an ashless filter paper, air-dry for ~15 min on ashless filter paper with the coated side facing up, and store at RT for analysis by TEM.
The specimens are imaged using a transmission electron microscope (TEM), and the gold distribution in the resulting images can be analyzed for clustering, sizes of clusters, co-localization, and other parameters by a multitude of statistical methods (e.g., (13, 14)).
Acknowledgments
The authors thank Dr. Bridget S. Wilson for advice on TEM and plasma membrane sheet preparation, and Dr. Fleur E. Tynan for comments on the manuscript.
Footnotes
EM grids are best handled with nonmagnetic, fine tipped and curved type 7 forceps, and coverslips are most conveniently manipulated with reverse action type N5 forceps.
The choice of EM grids is crucial for the preparation of plasma membrane sheets. The EM grid should distribute the force during the attachment of the secondary surface equally onto the carbon-coated formvar sheet. Hexagonal nickel EM grids have wide metal bands forming the mesh, which is ideal for this procedure. EM grids made from wire cut the formvar/carbon sheet and are not suited for this procedure.
Due to surface tension of liquid trapped between the two tines, EM grids easily become attached to the surface of forceps during their release. Therefore, when removing liquid by touching the edge of the EM grid with an ashless filter paper, touch the gap between the tines simultaneously, which will remove any liquid between them. When floating EM grids on liquid, make sure that the EM grid is in contact with liquid surface during the opening of the tines, which will ensure that it floats onto the liquid.
The ripping conditions can be optimized by TEM analyses of plasma membrane sheets labeled with the electron dense stains only or by the analyses of cells labeled with a fluorescent membrane marker (e.g., DiO, DiI, or DiD) on an inverted fluorescence microscope. For the latter, submerge the EM grid with the plasma membrane sheet side down in a microscopy chamber.
The efficiency of the ripping procedure can be increased for some cells, by incubating them in hypotonic buffer (25–75 mM salt) for a short period of time prior and during the ripping procedure. However, this potentially induces changes in the cell morphology and may activate stress-related responses.
If PVDF membranes are used as secondary surfaces, it is easier to pick the EM grids with forceps, when the membrane is bend between two fingers with the EM grid on the outside of the arch. If glass coverslips are used, place them on top of the EM grids with the edge of the EM grid slightly extended past the surface of the coverslip. This makes the removal of the EM grids from the coverslips easier.
Fixation and binding condition for the labeling can be optimized by preliminary experiments using fluorescence-activated cell sorting (FACS). This enables many conditions to be examined quickly.
References
- 1.Singer SJ, Nicolson GL. The fluid mosaic model of the structure of cell membranes. Science. 1972;175(23):720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.van Meer G, Simons K. Viruses budding from either the apical or the basolateral plasma membrane domain of MDCK cells have unique phospholipid compositions. Embo J. 1982;1(7):847–852. doi: 10.1002/j.1460-2075.1982.tb01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sako Y, Kusumi A. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J Cell Biol. 1994;125(6):1251–1264. doi: 10.1083/jcb.125.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proc Natl Acad Sci USA. 2006;103(50):18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 1998;395(6697):82–86. doi: 10.1038/25764. [DOI] [PubMed] [Google Scholar]
- 6.Sanan DA, Anderson RG. Simultaneous visualization of LDL receptor distribution and clathrin lattices on membranes torn from the upper surface of cultured cells. J Histochem Cytochem. 1991;39(8):1017–1024. doi: 10.1177/39.8.1906908. [DOI] [PubMed] [Google Scholar]
- 7.Lillemeier BF, Mortelmaier MA, Forstner MB, Huppa JB, Groves JT, Davis MM. TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat Immunol. 2010;11(1):90–96. doi: 10.1038/ni.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim JH, Cramer L, Mueller H, Wilson B, Vilen BJ. Independent Trafficking of Ig-{alpha}/Ig-{beta} and {micro}-Heavy Chain Is Facilitated by Dissociation of the B Cell Antigen Receptor Complex. J Immunol. 2005;175(1):147–154. doi: 10.4049/jimmunol.175.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson BS, Pfeiffer JR, Oliver JM. Observing FcepsilonRI signaling from the inside of the mast cell membrane. J Cell Biol. 2000;149(5):1131–1142. doi: 10.1083/jcb.149.5.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson BS, Pfeiffer JR, Surviladze Z, Gaudet EA, Oliver JM. High resolution mapping of mast cell membranes reveals primary and secondary domains of Fc(epsilon)RI and LAT. J Cell Biol. 2001;154(3):645–658. doi: 10.1083/jcb.200104049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prior IA, Muncke C, Parton RG, Hancock JF. Direct visualization of Ras proteins in spatially distinct cell surface microdomains. J Cell Biol. 2003;160(2):165–170. doi: 10.1083/jcb.200209091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morone N, Fujiwara T, Murase K, et al. Three-dimensional reconstruction of the membrane skeleton at the plasma membrane interface by electron tomography. J Cell Biol. 2006;174(6):851–862. doi: 10.1083/jcb.200606007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ripley BD. Modeling spatial patterns. J. R. Stat. Soc. 1977;B39:172–212. [Google Scholar]
- 14.Zhang J, Leiderman K, Pfeiffer JR, Wilson BS, Oliver JM, Steinberg SL. Characterizing the topography of membrane receptors and signaling molecules from spatial patterns obtained using nanometer-scale electron-dense probes and electron microscopy. Micron. 2006;37(1):14–34. doi: 10.1016/j.micron.2005.03.014. [DOI] [PubMed] [Google Scholar]