Abstract
The unfolded phrotein response is a mechanism to cope with endoplasmic reticulum stress. In Saccharomyces cerevisiae, Ire1 senses the stress and mediates a signaling cascade to upregulate responsive genes through an unusual HAC1 mRNA splicing. The splicing requires interconnected activity (kinase and endoribonuclease) of Ire1 to cleave HAC1 mRNA at the non-canonical splice sites before translation into Hac1 transcription factor. Analysis of the truncated kinase domain from Ire1 homologs revealed that this domain is highly conserved. Characterization by domain swapping indicated that a functional ATP/ADP binding domain is minimally required. However the overall domain compatibility is critical for eliciting its full endoribonuclease function.
Keywords: Unfolded protein response, Ire1, Domain swapping, HAC1 splicing, protein kinase, endoribonuclease
1. Introduction
The endoplasmic reticulum (ER) is the site for the folding and assembly of secretory and membranous proteins. The environment inside the lumen of the ER is optimized to facilitate folding of the nascent polypeptide into a correct tertiary structure prior to trafficking to the Golgi. Perturbation of the homeostatic environment inside the ER results in accumulation of unfolded proteins that disrupts ER function leading to cell death [1]. An exquisite quality control system ensures that only properly folded proteins exit the ER, and those that are irreparably misfolded are directed to degradation through ER-associated proteasomal degradation and/or autophagy. The cell has evolved the ability to adjust the protein-folding capacity to meet fluctuations in the protein-folding demand [2]. The unfolded protein response (UPR) is a set of adaptive signaling pathways that evolved to limit accumulation of misfolded proteins by 1) reducing the load on the ER by inhibiting mRNA translation initiation, 2) inducing the transcription of genes encoding protein folding, processing, and trafficking functions, and 3) increasing the degradation rate of irreversibly misfolded proteins. Activation of the UPR is initiated through three ER-localized transmembrane protein sensors Ire1, Perk and ATF6 [3–5]. These proximal effectors of the UPR monitor the protein folding status in the ER through detecting the free pool of the molecular chaperone BiP that is available to facilitate protein folding [6].
Ire1 is the most ancient UPR sensor and is conserved in all eukaryotes. The Ire1 signaling has been well characterized in Saccharomyces cerevisiae [6–8]. Its amino terminus resides in lumen ER lumen to sense the protein folding status. The carboxyl terminus resides in the cytosol to initiate a unique signaling through its serine/threonine kinase and endoribonuclease (RNase) activities to induce an unconventional splicing reaction that removes a 252 nucleotide intron from HAC1 mRNA, encoding a transcription factor that activates UPR-responsive genes [9,10]. The Ire1 kinase comprises 11 subdomains folded into two distinct lobes. A small lobe at the amino terminus is mainly involved in nucleotide binding domain whereas a large lobe at the carboxyl terminus participates in substrate binding [8,11]. Ire1 activation is mediated by oligomerization and trans-autophosphorylation leading to a conformational change that fully elicits the RNase activity [10,12,13].
Here, we describe an inducible Ire1 platform that is amendable for modulating the UPR response in S. cerevisiae. Through domain swapping we demonstrate that specific coordination within the kinase domain is critical role for activating the RNase function.
2. Materials and Methods
2.1. Strains and manipulations
S. cerevisiae AWY14 (W303-1A, UPRE-CYC1-LacZ, UPRE-CYC1-LEU 2), S. cerevisiae AWY 19 (same as AWY14 except Δire1∷kanamycinr) [14], Saccharomyces carlsbergensis (Sca), Saccharomycodes ludwigii (Sl), Pachysolen tannophilus (Pt), Torulasopora delbrueckii (Td), Wickerhamia fluorescens (Wf) were maintained in YEPD. Transformation of recombinant plasmids into the AWY19 strain was performed by the standard lithium acetate method. Strains carrying the plasmid were grown on uracil dropout medium. Eschericia coli DH5α was used for propagation and construction of all plasmids.
2.2. Construction of Ire1 expression plasmid
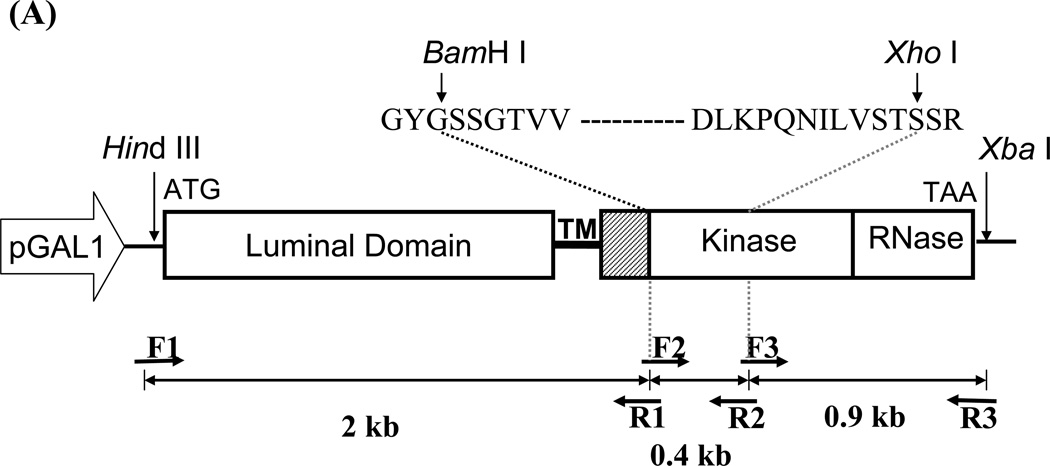
The entire Open reading frame of the IRE1 gene was PCR amplified from S. cerevisiae genomic DNA into 3 fragments by Vent DNA polymerase (New England Biolab) using primers shown in Figure 1 and Table 1. Silent mutations were introduced to the gene to create two unique restriction recognition sites, BamH I and Xho I, at positions 2047 and 2421 (relative to the translation initiation site). The fragments were sequentially assembled into pYES-2 plasmid (Invitrogen) downstream of the GAL1 promoter resulting in pYES-IRE1. The nucleotide sequence was verified by automated DNA sequencing.
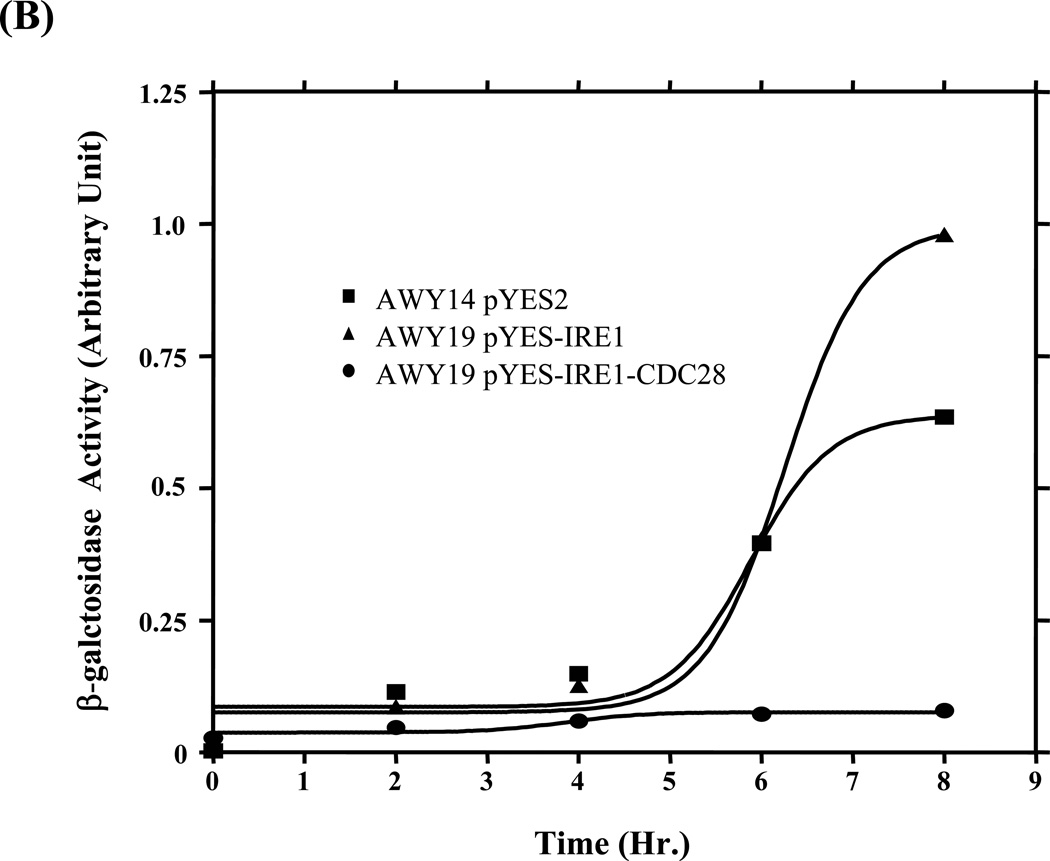
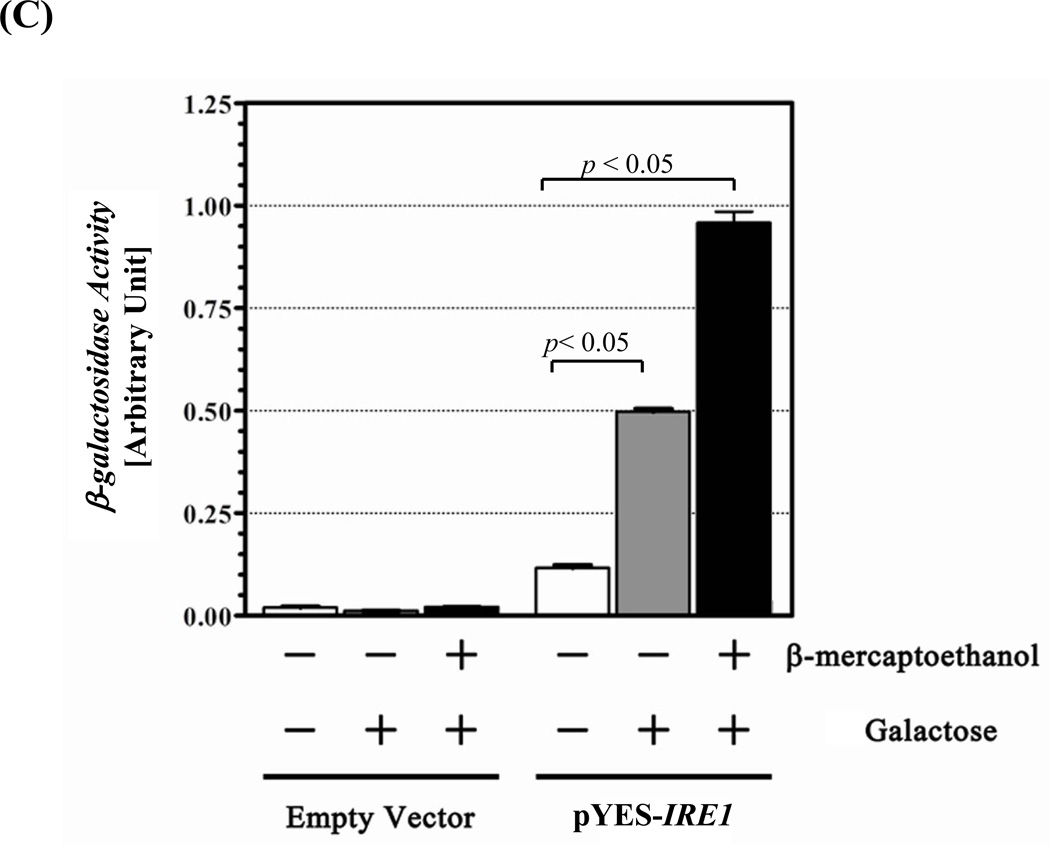
Figure 1. UPR restoration by inducible Ire1.
(A) Construction of the inducible Ire1 expression plasmid. The S. cerevisiae IRE1 gene was PCR amplified as 3 consecutive fragments then cloned under GAL 1 promoter in pYES-2 plasmid. BamH I and Xho I restriction sites (indicate by arrows) were introduced into the kinase subdomains I and VI, respectively. Positions of primers and the approximate size of the amplified fragments are indicated. GYGSSGTVV and DLKPQNILVSTSSR are conserved amino acid sequences in subdomain I and VI used to design degenerate primers. (B) Measure of UPR reporter gene activity. Wild type (AWY14) or ire1 null (AWY19) strains with indicated plasmids were cultured uracil dropout containing 2% raffinose, 1% galactose and 10 mM β-mercaptoethanol. β-galactosidase activity was measured at indicated time. (C) The UPR activity of the recombinant Ire1 in the presence of absence of ER stress was determined (Mean ± SEM) (n=3). ** represent significant difference of the reporter (P< 0.05, n = 3) by one-way ANOVA test.
Table 1.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| F1 | |
| R1 | |
| F2 | |
| R2 | |
| F3 | |
| R3 | |
| Degenerate F1 | |
| Degenerate R1 | 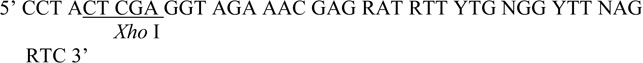 |
| Degenerate CDC28 F | |
| Degenerate CDC28 R | 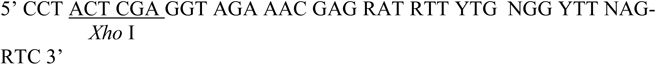 |
| HAC1F | 5’ GCC CAA GAG TAT GCC GAT TCC G 3’ |
| HAC1R | 5’ ACC CTC GAG CGA TTG TCT TCA TG 3’ |
| ACT1F | 5’ GCC GGA TCC GCC GGT GAC GAC GCT CC 3’ |
| ACT1R | 5’ TCG TCG AAT TCT TGT TTT GAG ATC C 3’ |
2.3. Cloning of IRE1 homologs
Partial fragments of the IRE1 homologs were PCR amplified from genomic DNA of various yeast species using Vent DNA polymerase (New England Biolabs) and Degenerate F1 and Degenerate R1 primers designed from the amino acid sequence of the S. cerevisiae Ire1 kinase in subdomain I and VI (Table 1). The homologous region from CDC28 from S. cerevisiae DNA was amplified by Degenerate CDC28 F and Degenerate CDC28 R primers.
To construct chimeric IRE1s, the 0.4 kb of BamH I and Xho I fragment encoding kinase subdomain I–VI in pYES-IRE1 was substituted with the homologous region from CDC28 or IRE1 homologs. DNA sequencing was performed to determine their sequences and ensure appropriate translational frame.
2.4. β -Galactosidase assay
Yeast cells were grown in uracil dropout medium using 2% raffinose as carbon source. Activation of Ire1 expression was performed by addition of galactose (1% final) for 4 hr. 10 mM β-mercaptoethanol (or 2 µg/ml tunicamycin) was added to induce ER stress for 4 hr (or 6hr) before the β-galactosidase assay [15]. Total protein in the cell extract was measured by Bradford (BioRad) for normalization.
2.5. Reverse Tanscription–PCR
Total RNA was extracted from yeast strains using the phenol-chloroform method [19]. First strand cDNA synthesis was performed using oligo-dT primer and Imprompt II reverse transcriptase (Promega). PCR amplifications were carried out using specific primers (Table 1). HAC1 mRNA splicing was measured using HAC1F and HAC1R primers flanking the 252 base intron region. The level of IRE1 transcript was determined by multiplex PCR using F3 and R3 primer. The amplicon of Actin with ACT1F and ACT1R primers served as an internal control.
3. Results
3.1. UPR activation can be conferred by inducible Ire1 expression
The IRE1 gene was modified to harbor a BamH I site in the consensus kinase subdomain I motifs (GYG↓SSGTVV) and a XhoI site in subdomainVI (DLKPQNILVSTS↓SR) under an inducible GAL 1 promoter (pYES-IRE1) (Figure 1A). This plasmid restored UPR signaling in an ire1 null (AWY19) strain only when cultured in medium with raffinose and galactose but not glucose (figure 1C). The recombinant gene displayed comparable activity to the endogenous gene in AWY14 within the first 6 hr of induction (Figure1B). The recombinant Ire1 exhibited greater activity (~ 40%) when the induction was prolonged to 8 hours. Notably modulation of the recombinant Ire1 activity is constitutive however the magnitude was further increased under the stress condition (Figure 1C). In contrast the chimeric Ire1 harboring the kinase subdomain from an unrelated CDC28 kinase (Ire1-CDC28), serving as a negative control, failed to restore the UPR. Together this evidence shows that the inducible Ire1 system is a functional platform.
3.2. The nucleotide binding domain is highly conserved among Ire1 homologs
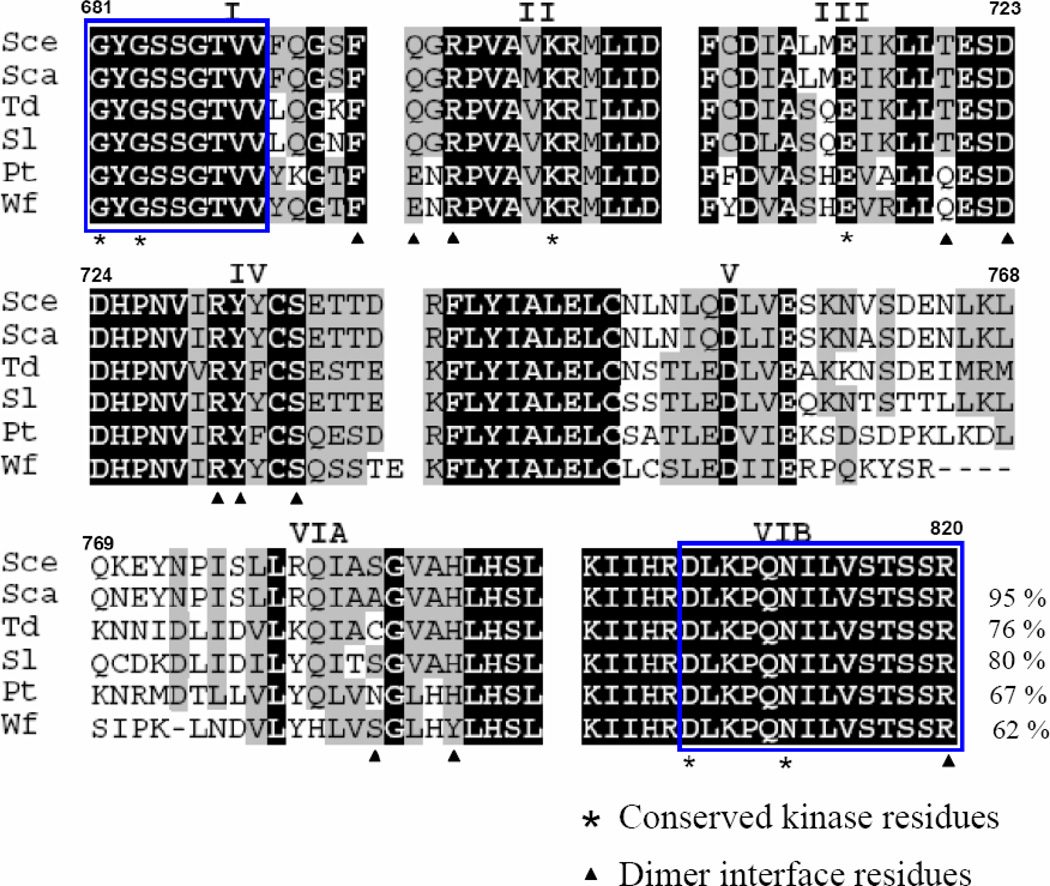
The cytoplasmic half of Ire1 consists of dual catalytic modes, a protein ser/thr kinase and an RNase [16–18]. Activation of the intrinsic kinase is prerequisite for activating its adjacent RNase, implicating a close communication between the two domains [16,17]. Using degenerate primers designed from sequences in subdomain I and VIB of Ire1, covering the entire small lobe and part of the big lobe designated as the ATP/ADP binding domain, we cloned the 0.4 kb homologous fragments from several yeast strains: S. carlsbergensis, S. ludwigii, P. tannophilus, T. delbrueckii and W. fluorescens. The deduced amino acids of these fragments showed highest homology (62–95%) to S. cerevisiae Ire1 (Figure 2). All (except W. fluorescens) sequences have subdomain organization and identical length as found in S. cerevisiae Ire1. The sequences in subdomains I, II, III and IV are highly conserved. In contrast, a high degree of variation was found clustered in subdomain V and VIA [8,11]. The homolog from S. carlsbergensis is almost identical to S. cerevisiae Ire1. Only 6 amino acid substitutions (V701M, L752I, V756I, V761A, K770N and S783A) of a total 130 residues were found. All substitutions (except V701A) are localized in subdomain V and VIA.
Fig 2. Amino acid alignment of Ire1 homologues.
Partial deduced amino acid sequences of Ire1 homologues from Saccharomyces carlsbergensis (accession number FN677757), Saccharomycodes ludwigii (accession number FN677758), Pachysolen tannophilus (accession number FN677759), Torulasopora delbrueckii Wf (accession number FN677760) and Wickerhamia fluorescens (accession number FN677761) were aligned to Saccharomyces cerevisiae Ire1. Roman numerals indicate kinase subdomains. Blocks represent the position of degenerate primers. Dark shading indicates identical residues. Light shading indicates conserved residues. Numbers indicate amino acid residues according to S. cerevisiae Ire1(Sce). Percent sequence identities to S. cerevisiae Ire1 are shown in the last line.
3.3. Swapping of nucleotide binding domain in chimeric Ire1 kinase alters UPR activity
The high conservation among the Ire1 homologs prompted us to investigate its significance for Ire1 function. These homologous 0.4 kb fragments were swapped into pYES-IRE1 for functional assessment. The chimeric Ire1 proteins derived from plasmid carrying the fragments from S. carlsbergensis, S. ludwigii, P. tannophilus, T. delbrueckii or W. fluorescens were designated as Ire1-Sca, Ire1-Sl, Ire1-Pt, Ire1-Td and Ire1-Wf, respectively.
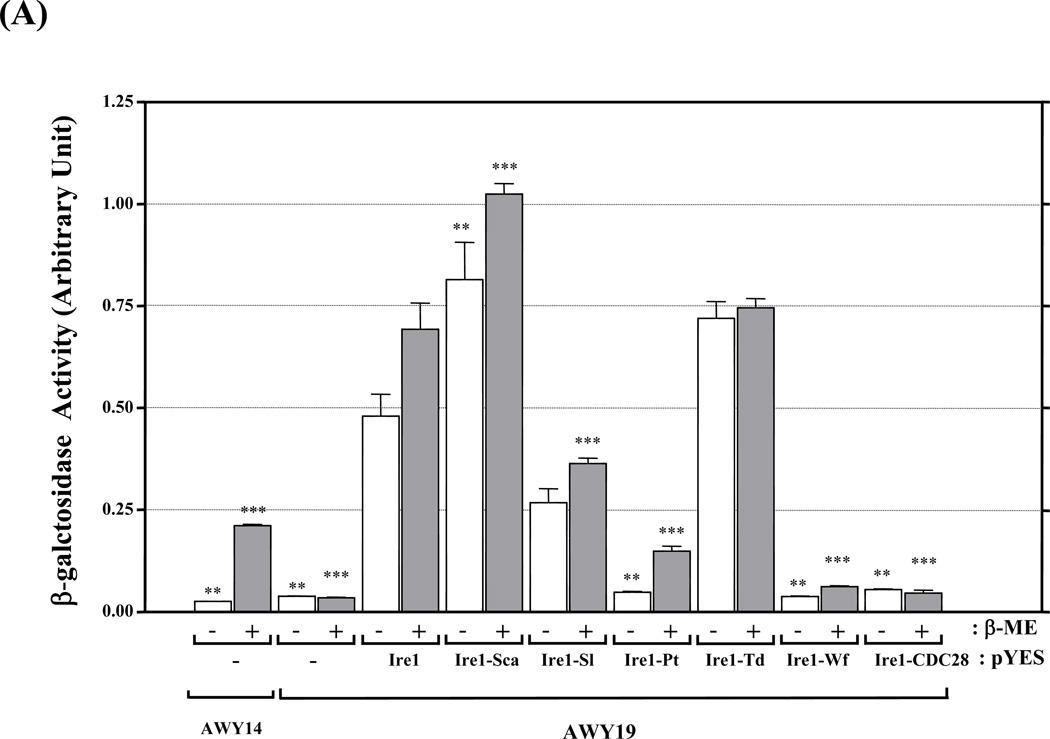
The UPR activity of the chimeric Ire1s could be divided into 3 groups (Figure 3A). First, the highly active chimeras (Ire1-Sca, Ire1-Sl or Ire1-Td as well as recombinant Ire1) exhibiting high and constitutive UPR activity regardless of ER stress.
Figure 3. UPR complementation by chimeric Ire1.
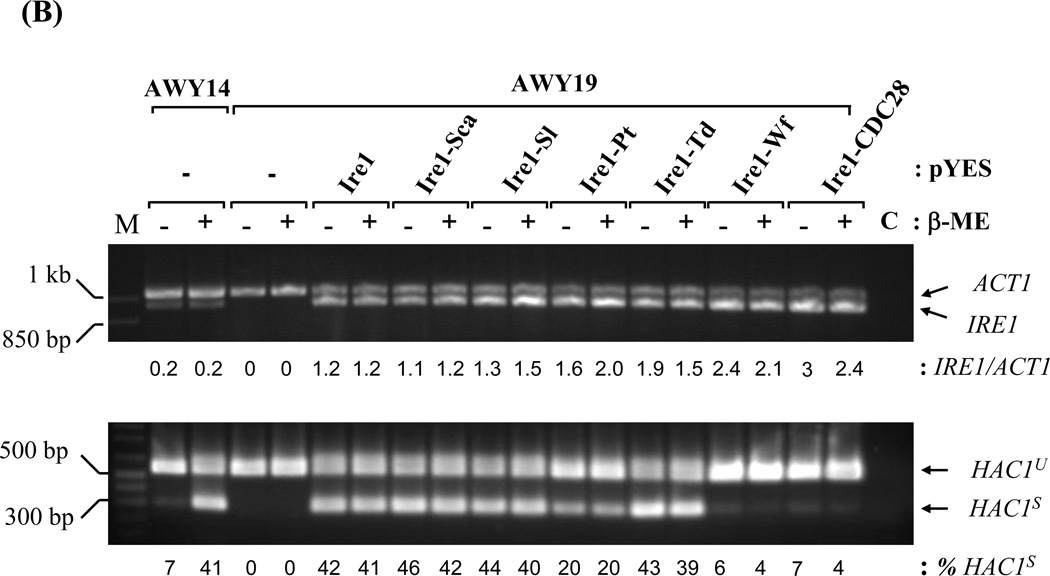
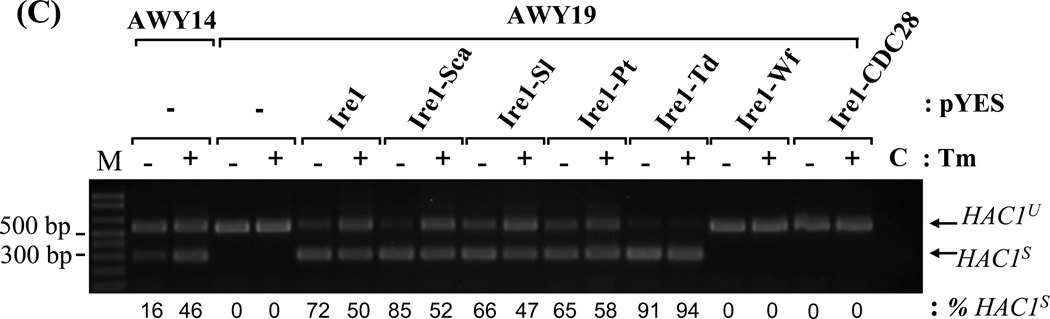
(A) UPR reporter gene assay. AWY14 or AWY19 strains harboring the indicated plasmids were cultured in medium containing galactose for 4 hr followed by addition of β-ME for 4 hr. The activities were reported as Mean ± SEM (n=3). ** and *** represent significant difference (P< 0.05) compared to recombinant Ire1 activity under the same conditions by one-way ANOVA. (B) Relative expression of chimeric IRE1 compared to ACT1 mRNA and efficiency of HAC1 mRNA splicing was analyzed by Scion Image program. HAC1 mRNA splicing upon 2µg/ml Tunicamycin (Tm) was shown in (C). C: RT-PCR without reverse transcriptase. HAC1u and HAC1s represent unspliced and spliced HAC1. M: 1 kb plus DNA ladder.
Intriguingly Ire1-Sca and Ire1-Td showed reproducibly stronger UPR activation than recombinant Ire1. Second, the moderately active chimera (Ire1-Pt) had dramatically reduced UPR activity but retained ER stress responsiveness. Finally the inactive group (Ire1-Wf and Ire1-CDC28) had almost undetectable activity.
3.4. Chimeric Ire1s with an intact nucleotide binding domain trigger HAC1 mRNA splicing
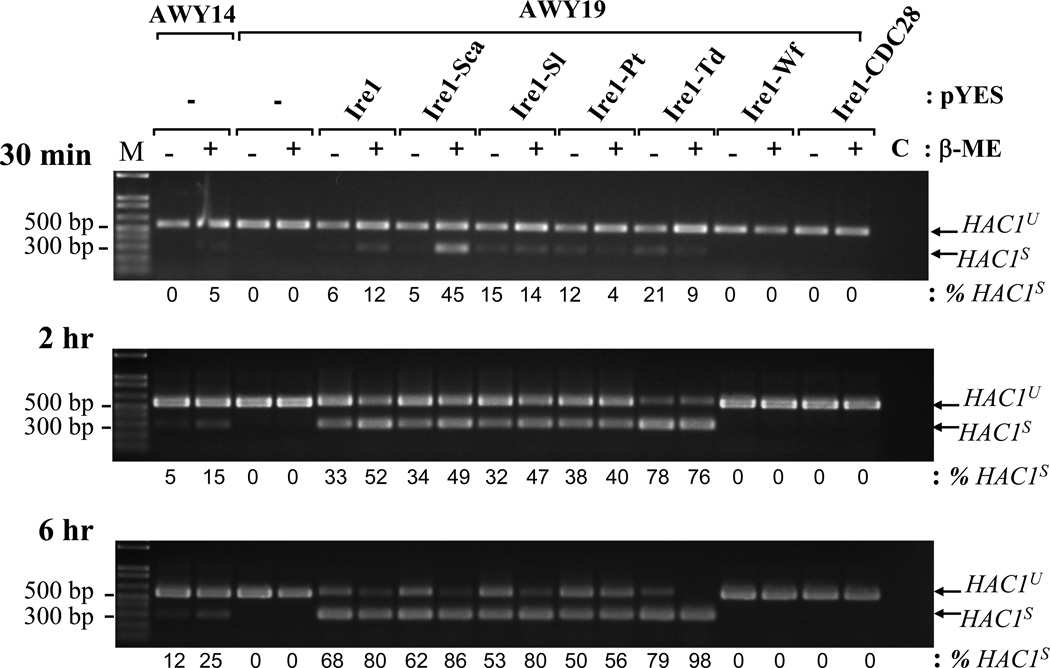
Lacking a sensitive Ire1 antibody for detection, RT-PCR was used to measure chimeric Ire1 expression at mRNA level. All chimeric IRE1 mRNAs were expressed at least 3 fold higher than the endogenous IRE1 in AWY14 (Figure 3B). Their expression was not altered by ER stress. The activities of the Ire1 chimeras were analyzed by their ability to restore HAC1 mRNA splicing in the AWY19. The highly- and moderately active Ire1 chimeras efficiently spliced HAC1 mRNA independent to β-mercaptoethanol stress (Figure 3B). Interestingly, these chimeras behave differently in modulating HAC1 splicing upon activating ER stress by blocking the N-linked glycosylation process by Tunicamycin (Figure 3C). While the HAC1 splicing meditated by the recombinant Ire1, and several chimeras (Ire1-Sca, Ire1-Sl and Ire1-Pt) were slightly compromised by this stressor, Ire1-Td mediated HAC1 splicing was not affected. Surprisingly, a very low reproducible level of HAC1s was detected in AWY19 expressing the Ire1-Wf chimera or the Ire1-CDC28 chimera comparable to the non-stressed AWY14. Unlike the endogenous Ire1, the Ire1-Wf and Ire1-CDC28 chimeras failed to respond to ER stress. Lastly, we tested if these chimeras modulate HAC1 mRNA splicing with different kinetic. Two Ire1 chimeras exhibited distinct kinetic from Ire1. Ire1-Sca swiftly activated the splicing at early time point (30 min) under the stress (Figure 4). Ire1-Td, on the other hand, exhibited more splicing efficiency at late time point (6 hr). Together these results indicate that all chimeric proteins were produced, however they exhibited distinct catalytic activities.
Figure 4. Kinetic of HAC1 mRNA splicing by chimeric Ire1.
AWY14 or AWY19 strains harboring the indicated plasmids were cultured in medium containing galactose for 4 hr. status of HAC1 mRNA splicing was monitored upon exposing to β-ME for indicated time. The efficiency of the splicing was analyzed by Scion Image program. M: 1 kb plus ladder.
4. Discussion
We established an inducible Ire1 expression platform to enable analysis of structure-function relationships. The transcriptional induction by the GAL 1 promoter generated high-level expression of the recombinant Ire1, thus causing constitutive UPR activation. This data correspond to previous findings of ER stress-independent activation of Ire1 upon over expression [12,16].
The architecture of the Ire1 catalytic domain comprises kinase and RNase domains in close proximity [11,16,17,19]. The kinase activity is required to elicit the RNase during ER stress [4,12,13]. Based on the S. cerevisiae Ire1 crystal structure, subdomains I to VIB span the entire small domain, ATP/ADP binding cleft and the N-terminal region of the large domain. It is most likely that all of the Ire1 homologues we studied possess a functional ATP/ADP binding site. The high conservation of amino acid sequences, subdomain organization and dimer/oligomer interface among these homologs implicates that they possess similar structures and mechanisms for modulation [17]. This also emphasizes, partly if not entirely, the role of kinase domain as a molecular switch for UPR signaling.
The dramatically difference among chimeric Ire1s activity suggests that amino acid variation in certain regions may be crucial for activity. It is worth noting that sequence variations are concentrated in subdomains V and VIA (residues 749–783) which are equivalent to part of ATP/ADP binding cleft and hinge region. Of these residues, only Asn751 was shown to interact with the ribose ring of ADP whereas the functional significance of the other residues remains obscure [16]. Comparison between chimeric Ire1-Sca and Ire1, 5 of 6 substitution residues were in subdomains V and VIA, resulting in a significant increase in the UPR activity. Similarly, comparison between Ire1-Sl and Ire1-Td (91% homology), 17 of 23 residues amino acid substitutions were concentrated in the same region resulting in ~ 50% difference in activity. Lastly, our finding that the chimeras Ire1-CDC28 and Ire1-Wf were capable of triggering a basal level of HAC1 mRNA splicing was unexpected. This suggests that a functional ATP/ADP binding domain per se is sufficient to elicit the RNase activity but their incompatibility within Ire1 backbone may prevent an overall conformational change required to fully activate Ire1.
In conclusion, this study provides the fist evidence showing that compatibility within Ire1 kinase subdomains is crucial for modulating its adjacent RNase activity. Unique amino acid sequences in subdomains V–VIA in the ADP/ATP binding domain may provide flexibility to fine tune the Ire1 kinase conformation that influences overall conformational plasticity of Ire1 during its transition into a fully active state.
Acknowledgements
We thank Dr. David Engelke, University of Michigan for providing yeast strains. Nucleotide sequences of these Ire1 homologs were submitted to EMBL. This work was partially supported by Thailand Research Fund to WT (BRG5080023). RJK is an Investigator of the Howard Hughes Medical Institute and supported by NIH grants DK42394, HL52173, and PO1 HL057346.
Abbreviations used
- UPR
unfolded protein response
- RNase
endoribonuclease
- RT-PCR
reverse transcribed-Polymerase chain reaction
- Sca
Saccharomyces carlsbergensis
- Sl
Saccharomycodes ludwigii
- Pt
Pachysolen tannophilus
- Td
Torulasopora delbrueckii
- Wf
Wickerhamia fluorescens
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- 2.Schroder M. Endoplasmic reticulum stress responses. Cell Mol Life Sci. 2008;65:862–894. doi: 10.1007/s00018-007-7383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harding HP, Zhang Y, Bertolotti A, Zeng H, Ron D. Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell. 2000;5:897–904. doi: 10.1016/s1097-2765(00)80330-5. [DOI] [PubMed] [Google Scholar]
- 4.Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K. ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol. 2000;20:6755–6767. doi: 10.1128/mcb.20.18.6755-6767.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bertolotti A, Zhang Y, Hendershot LM, Harding HP, Ron D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat Cell Biol. 2000;2:326–332. doi: 10.1038/35014014. [DOI] [PubMed] [Google Scholar]
- 7.Cox JS, Shamu CE, Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- 8.Mori K, Ma W, Gething MJ, Sambrook J. A transmembrane protein with a cdc2+/related kinase activity is required for signaling from the ER to the nucleus. Cell. 1993;74:743–756. doi: 10.1016/0092-8674(93)90521-q. [DOI] [PubMed] [Google Scholar]
- 9.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 10.Sidrauski C, Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 11.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 12.Shamu CE, Walter P. Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J. 1996;15:3028–3039. [PMC free article] [PubMed] [Google Scholar]
- 13.Welihinda AA, Kaufman RJ. The unfolded protein response pathway in Saccharomyces cerevisiae. Oligomerization and trans-phosphorylation of Ire1p (Ern1p) are required for kinase activation. J Biol Chem. 1996;271:18181–18187. doi: 10.1074/jbc.271.30.18181. [DOI] [PubMed] [Google Scholar]
- 14.Liu CY, Schroder M, Kaufman RJ. Ligand-independent dimerization activates the stress response kinases IRE1 and PERK in the lumen of the endoplasmic reticulum. J Biol Chem. 2000;275:24881–24885. doi: 10.1074/jbc.M004454200. [DOI] [PubMed] [Google Scholar]
- 15.Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- 16.Korennykh AV, Egea PF, Korostelev AA, Finer-Moore J, Zhang C, Shokat KM, Stroud RM, Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KP, Dey M, Neculai D, Cao C, Dever TE, Sicheri F. Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell. 2008;132:89–100. doi: 10.1016/j.cell.2007.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papa FR, Zhang C, Shokat K, Walter P. Bypassing a kinase activity with an ATP-competitive drug. Science. 2003;302:1533–1537. doi: 10.1126/science.1090031. [DOI] [PubMed] [Google Scholar]
- 19.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]