Abstract
Normal development of the immune system requires regulated processing of NF-κB2 p100 to p52, which activates NF-κB2 signaling. Constitutive production of p52 has been suggested as a major mechanism underlying lymphomagenesis induced by NF-κB2 mutations, which occur recurrently in a variety of human lymphoid malignancies. To test the hypothesis, we generated transgenic mice with targeted expression of p52 in lymphocytes. In contrast to their counterparts expressing the tumor-derived NF-κB2 mutant p80HT, which develop predominantly B cell tumors, p52 transgenic mice are not prone to lymphomagenesis. However, they are predisposed to inflammatory autoimmune disease characterized by multiorgan infiltration of activated lymphocytes, high levels of autoantibodies in the serum, and immune complex glomerulonephritis. p52, but not p80HT, represses Bim expression, leading to defects in apoptotic processes critical for elimination of autoreactive lymphocytes and control of immune response. These findings reveal distinct signaling pathways for actions of NF-κB2 mutants and p52 and suggest a causal role for sustained NF-κB2 activation in the pathogenesis of autoimmunity.
NF-κB2 is a member of the NF-κB family of transcription factors that also includes NF-κB1 (p105/p50), RelA (p65), RelB, and c-Rel (1). The full-length NF-κB2 protein p100 contains an N-terminal Rel homology domain, responsible for dimerization, nuclear translocation, and DNA binding. Its C-terminal region contains an ankyrin repeat domain with IκB activity. Under basal conditions, NF-κB2 p100 forms inhibitory complexes with Rel proteins (2–4). Phosphorylation of the C terminus of p100 by IκB kinase α and NF-κB inducing kinase leads to proteolytic processing of p100 to p52 (5, 6). The resulting p52-Rel protein heterodimers then translocate into the nucleus and activate transcription of their target genes. This alternative NF-κB signaling pathway is activated by engagement of the receptors for B cell-activating factor, lymphotoxin-β, and CD40 ligand (3, 7–9). Previous studies with NF-κB2−/− mice demonstrate a crucial role of NF-κB2 in B cell development and secondary lymphoid organogenesis. These mice present a marked decrease in the peripheral B cell population and an absence of discrete perifollicular marginal and mantle zones and of germinal centers in the spleen (10, 11). More recently, it has been shown that NF-κB2 signaling is essential for the development of medullary thymic epithelial cells that function as antigen-presenting cells in negative selection of autoreactive T cell clones (12).
The NF-κB2 gene is recurrently mutated in a variety of human lymphoid malignancies, including T-cell lymphoma, chronic lymphocytic leukemia, multiple myeloma, and B cell lymphoma (13). A cardinal feature of these genetic alterations is the generation of C-terminally truncated NF-κB2 mutants that lack various portions of the ankyrin repeat domain (14–20). To determine whether NF-κB2 mutation can directly initiate lymphomagenesis, we generated transgenic mice with targeted expression in lymphocytes of p80HT, a lymphoma-associated NF-κB2 mutant (17, 18). The transgenic mice display defects in lymphocyte apoptosis, leading to a marked expansion of the peripheral B cell population and the development of predominantly B cell tumors. p80HT directly activates the expression of TRAF1 that encodes an anti-apoptotic protein also implicated in lymphoid malignancies (21). Importantly, TRAF1 knockdown abrogates the anti-apoptotic activity of p80HT, and TRAF1 deficiency re-establishes B cell homeostasis in p80HT mice. These findings demonstrate that NF-κB2 mutation can directly induce lymphomagenesis, with TRAF1 as a key component of this oncogenic pathway (22).
Tumor-derived NF-κB2 mutants, including p80HT, are constitutively localized in the nucleus (16, 18, 19, 23). Moreover, it has been shown that p80HT can directly bind to a κB probe in its unprocessed form and has a higher transcriptional activity compared with p52 (23–25). These observations suggest that NF-κB2 mutants may have gained a transactivation function responsible for their oncogenic activity. However, this view has been challenged by a number of observations. It is well known that NF-κB2 mutants manifest constitutive processing (13), most likely as a result of their loss of the C-terminal processing inhibitory domain (5). Indeed, lymphocytes from p80HT transgenic mice (22) and p80HT-expressing HuT78 T-lymphoma cells express high levels of p52 (supplemental Fig. S1). These findings raise the question of whether p80HT induces lymphomagenesis through constitutive production of p52 and, thus, sustained activation of NF-κB2. Consistent with the enhanced processing model, elevated levels of p52 have been observed in several types of human cancer (20, 26–29). More recently, an analysis of NF-κB2 mutant processing has led to the suggestion that it is the processed p52 that is responsible for the oncogenic activity of NF-κB2 mutants (30). To examine these alternative models for the action of NF-κB2 mutants, we generated transgenic mice with constitutive production of p52 in lymphocytes. These mice do not develop lymphomas but are predisposed to inflammatory autoimmune disease. At the molecular level, elevated levels of p52 repress the expression of Bim, a pro-apoptotic protein essential for eliminating autoreactive lymphocytes through activation-induced cell death (31–33). Our study provides genetic evidence for distinct NF-κB2 signaling pathways in the pathogenesis of lymphoma and autoimmune disease.
EXPERIMENTAL PROCEDURES
Mice—The coding sequence for human NF-κB2 p52 was amplified by PCR using NF-κB2 cDNA as the template. The primers 5NFKBsal (5′-GCGGTCGACATGGAGAGTTGCT-ACAACCCAG-3′) and 3NFKBp52 (5′-GCGGGATCCTCAT-CGCTGCAGCATCTCCGGGGC-3′) were designed to introduce a SalI site at the 5′ end, a termination codon after the amino acid 442, and a BamHI site at the 3′ end. The amplified p52 sequence was verified by sequencing and cloned into the SalI-BamHI sites of pHSE3′ (34), a vector containing an H-2Kb promoter and an immunoglobulin μ chain enhancer, which direct transgene expression specifically in lymphocytes. The construct was linearized by PvuI and microinjected into (C57BL/6J × SJL/J) F2 fertilized eggs (University of Michigan Transgenic Animal Core). Transgenic founders were identified by PCR amplification of a 1.3-kb fragment from mouse tail DNA using the primers detailed above. Two transgenic lines were established from the founder mice 434 and 452 by mating transgenic males to C57BL/6J ×SJL/J F1 females (Jackson Laboratory). Rag1−/− (recombination activation gene 1-deficient) mice were purchased from the Jackson Laboratory. All of the animal studies were preapproved by the Institutional Animal Care and Use Committee of University of Toledo Health Science Campus.
Immunoblotting—Immunoblotting was performed according to standard procedures. The following antibodies were used: mouse anti-NF-κB2 (05–361; Upstate Biotechnology Inc.; 1:500), rabbit anti-TRAF1 (H-132; Santa Cruz; 1:200), rabbit anti-Bim (AAP-330; Stressgen; 1: 500), and mouse anti-α-tubulin (B-5-1-2; Sigma; 1:2000). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit were used as secondary antibodies (ICN).
Electrophoretic Mobility Shift Assay—Nuclear extracts were prepared from mouse splenocytes using a NE-PER nuclear extraction kit (Pierce) and analyzed for κB binding activity as described previously (22, 35). For supershifting, 3 μg of extracts were incubated with 2 μg of either preimmune rabbit IgG or antibodies against NF-κB2 (06–413; Upstate), NF-κB1 (06–886; Upstate), RelA (SC-109×; Santa Cruz), RelB (SC-226×; Santa Cruz), or c-Rel (SC-71×; Santa Cruz) for 30 min at 4°C before the addition of the 32P-labeled κB probe 5′-CAGGGCTGGG-GATTCCCCATCTCCACAGTTTCACTTC-3′ (36).
Histology, Immunochemistry, and Immunofluorescence—Mouse tissues were fixed in 10% neutral buffered formalin, embedded in paraffin, sectioned at 5 μm, and stained with hematoxylin and eosin. For immunohistochemistry, the sections were deparaffinized, rehydrated, and boiled in 10 mm citrate buffer (pH 6.0) or 1 mm EDTA (pH 8.0) for 10 min for retrieval of B220 or CD3 antigen. Following quenching of endogenous peroxidase activity with H2O2 and blocking with normal serum, the sections were incubated for 60 min with rat anti-B220 (RA3-6B2, 5 μg/ml; BD Pharmingen), rat anti-CD3 (CD3-12, 10 μg/ml; Serotec), or an Isotype control antibody (10 μg/ml; BD Pharmingen). After washing, a biotinylated rabbit anti-rat secondary antibody (Vector Laboratories) was applied for 30 min. The sections were then incubated for 30 min with ABC reagent (Vector Laboratories), and immunostaining was visualized with 3,3′-diaminobenzidine (Sigma). The sections were counterstained with Hematoxylin. For immunofluorescent staining of glomerular immune complexes, cryostat sections of mouse kidneys were fixed in cold acetone for 15 min, rehydrated in phosphate-buffered saline, and blocked with 10% goat serum, 3% bovine serum albumin in phosphate-buffered saline for 2 h at room temperature. The sections were then incubated with Alexa Fluor 568 goat anti-mouse IgG (1:500; Molecular Probes) for 1 h at room temperature.
Proteinuria and Autoantibody Detection—Urinary protein levels of 8-month-old p52 and wild-type mice were assessed using Urinalysis reagent strips (Labstix; Bayer Corporation) and graded semi-quantitatively (0, none; 1, 30–100 mg/dl; 2, 100–300 mg/dl; 3, 300–2000 mg/dl; and 4, >2,000 mg/dl). For analysis of autoantibodies to double-stranded DNA (dsDNA),4 serum samples were collected from 8–12-month-old p52 and wild-type mice and examined using an enzyme-linked immunosorbent assay kit (Alpha Diagnostic). The values are expressed as μg/ml.
Splenocyte Transfer—Splenocytes from 7-month-old p52 or wild-type mice were suspended in phosphate-buffered saline, and 4 × 107 cells were injected into tail vein of sublethally irradiated (350 rads) Rag1−/− mice (C57BL/6J, Jackson Laboratory). The mice were sacrificed 3 months after the transfer and analyzed for lung infiltration, glomerular immune complexes, and serum autoantibodies against dsDNA as described above.
Flow Cytometry—Single-cell suspensions were prepared from mouse lymphoid organs according to standard procedures. Lung-infiltrating cells were isolated as described (12). The cells were stained with fluorescein isothiocyanate-conjugated rat anti-mouse B220 (RA3-6B2), CD4 (GK1.5), CD44 (IM7), hamster anti-mouse CD69 (H1.2F3), allophycocyanin-conjugated hamster anti-mouse CD3e (145-2C11), R-phycoerythrin-conjugated rat anti-mouse CD4 (RM4-5), CD8a (53-6.7), and IgM (R6-60.2) (all from BD Pharmingen). The cells were then sorted on Epics Elite (Beckman-Coulter), and the data were analyzed with WinMDI 2.8 software.
In Vitro Lymphocyte Proliferation and Survival Assays—Splenic B and T cells were purified from 6–8-week-old mice using mouse B- and T-immunocolumns (Cedarlane), respectively. The purified cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 250 μm l-asparagine, and 50 μm 2-mercaptoethanol at 105/well (96-well plate). B cells were stimulated with F(ab’)2 goat anti-mouse IgM (20 μg/ml; Jackson ImmunoResearch) or lipopolysaccharide (LPS; 20 μg/ml, Sigma); T cells were stimulated with plate-bound anti-CD3e (145-2C11) plus anti-CD28 (37.51) antibodies (10 μg/ml for each antibody; BD Pharmingen) or phorbol 12-myristate 13-acetate (PMA; 2 ng/ml; Sigma) plus ionomycin (250 ng/ml; Calbiochem). After 48 h, the cells were either harvested for cell cycle analysis or pulsed for 12 h with [3H]thymidine (1 μCi/well; Amersham Biosciences). Incorporation of [3H]thymidine was measured using a Scintillation counter. For survival assays, B cells were either untreated or treated with LPS (20 μg/ml) or doxorubicin (0.5 μg/ml; Ben Venue Laboratories); T cells were either untreated or treated with PMA (2 ng/ml) or doxorubicin (0.5 μg/ml). Viable cells were determined daily by trypan blue exclusion assays.
Real Time PCR—Total RNA was extracted from cultured cells or mouse splenocytes using the TRI reagent (Molecular Research). The primers and probe were designed to detect all the three isoforms of Bim transcripts as follows: 5′-CGGATCGGAGACGAG-TTCA (forward primer), 5′-TTCAGCCTCGCGGTAATCA (reverse primer), and 5′-CGAAACTTACACAAGGAGGGT-GTTTGCAA (probe) labeled with the fluorescent reporter dye FAM at the 5′end and the fluorescent quencher dye TAMRA at the 3′ end. PCRs in triplicate were performed using the Taq-Man Universal PCR Master Mix (Applied Biosystems) and run on an Applied Biosystems 7500 real time PCR system. The glyceraldehyde-3-phosphate dehydrogenase gene was assayed in parallel as control.
Luciferase Reporter Assay—293T cells in 6-well plates were transfected with 0.6 μg of luciferase reporter constructs with or without the 0.8-kb mouse Bim promoter sequence (37), 0.3 μg of pSV-β-galactosidase plasmid, and 0.3 μg of pcDNA3 or pcDNA3-p52 using a Lipofectamine Plus kit (Invitrogen). The cells were lysed 40 h after transfection. Luciferase and β-galactosidase activities were assayed using a kit (Promega). Luciferase activity was normalized to β-galactosidase activity to account for the difference in the transfection efficiency.
Retroviral Infection and Anti-IgM-induced Apoptosis of WEHI-231 Cells—Murine WEHI-231 B lymphoma cells (ATCC CRL-1702) were cultured in high glucose Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 50 μm 2-mercaptoethanol. The cells were infected with retroviruses produced from the construct MSCV-IRES-GFP or MSCV-p52-IRES-GFP and sorted by flow cytometry based on the expression of GFP. For down-regulation of Bim, synthesized 64-bp oligonucleotides encoding mouse Bim siRNA (5′-AGCAACCTTCTGATGTAAG, position 8–27 bp relative to the start codon, GenBank™ NM_207680) were cloned into pSuper-retro-puro (Oligo-Engine) for producing retroviruses as described (22). The infected cells were selected with puromycin (1 μg/ml, 3 days). For B cell antigen receptor ligation-induced apoptosis, WEHI-231 cells were treated with F(ab’)2 goat anti-mouse IgM (1 μg/ml) for 48 h and analyzed for apoptosis by annexin-V staining and trypan blue exclusion assays.
RESULTS
Generation of p52 Transgenic Mice—To determine whether constitutive production of p52 can recapitulate the lymphoma-inducing activity of p80HT, we generated transgenic mice with targeted expression in lymphocytes of a human NF-κB2 transgene coding for the N-terminal 442 amino acids. The exact processing site for the generation of p52 is unknown, but NF-κB2 molecules of similar size have been used extensively for characterization of p52 activity (23–25, 38) and for generation of p52 “knock-in” mice (39). To be comparable, the same expression vector, pHSE3′, was used to generate p80HT and p52 mice, which also have the same genetic background (C57BL/6J × SJL/J). Three founder mice were found to carry various copy numbers of the p52 transgene (data not shown), and two independent p52 transgenic mouse lines were successfully established. Mice from both transgenic lines were examined that displayed very similar phenotypes, as presented below.
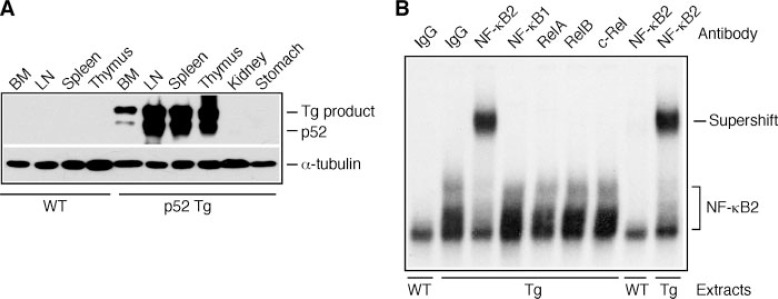
Major organs were obtained from 4–6-week-old p52 mice and their wild-type littermates for immunoblot analysis of tissue expression of p52. As expected, only lymphoid organs of p52 mice expressed high levels of the transgene product and fully processed p52 (Fig. 1A). The p52 expression levels were comparable between p52 mice and the previously reported 808 line of p80HT transgenic mice (22) (data not shown).
FIGURE 1.
Characterization of p52 transgenic mice. A, immunoblot analysis of tissue-specific expression of the transgene (Tg) product and processed p52, using an antibody against the N-terminal region of human NF-κB2. The levels of α-tubulin are shown as loading control. BM, bone marrow; LN, lymph nodes. B, electrophoretic mobility shifting assay for κB binding activity in nuclear extracts of splenocytes from p52 transgenic (Tg) and wild-type (WT) mice. The NF-κB2-κB complex is indicated, which contains no significant levels of Rel proteins, based on antibody-mediated supershift analysis. Preimmune rabbit IgG was used as control.
We next examined κB binding activity in nuclear extracts from unstimulated p52 and wild-type splenocytes. No significant levels of κB binding activity were detected in extracts from wild-type cells, whereas p52 extracts contained high levels of constitutive κB binding activity. The κB-binding complexes in p52 extracts could be supershifted by an antibody against human NF-κB2 (Fig. 1B). In contrast to p80HT, which binds to the same κB probe as heterodimers containing RelA or c-Rel (22), the p52-κB complex contained no significant levels of Rel proteins, because preincubation of p52 extracts with antibodies against RelA, RelB, or c-Rel failed to super-shift or disrupt the κB complex (Fig. 1B). These findings suggest that p52 exists primarily as homodimers in lymphocytes from p52 mice.
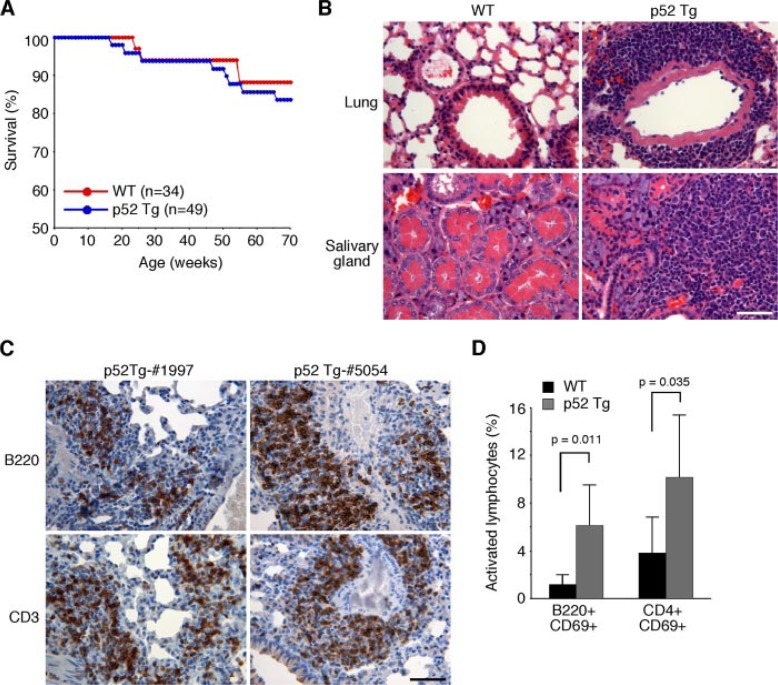
Development of Inflammatory Autoimmune Disease in p52 Transgenic Mice—By 70 weeks of age, p52 transgenic mice showed a mortality rate slightly higher than their wild-type littermates (16.3% versus 11.2%; Fig. 2A). Histological examination revealed that only one of the eight deceased p52 mice had apparent thymoma (data not shown). Thus, constitutive production of p52 did not significantly predispose mice to lymphomagenesis. This is in striking contrast to p80HT mice, which have a significantly higher mortality rate (up to 79% during the same period) and are highly prone to lymphoma development (22). Taken together, these data suggest that constitutive processing to p52 is unlikely a major mechanism underlying the lymphoma-inducing activity of p80HT.
FIGURE 2.
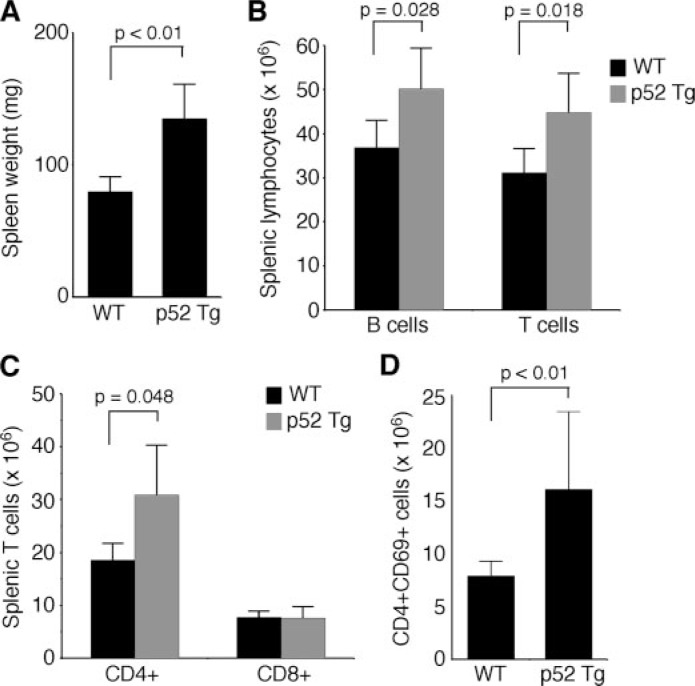
Multiorgan inflammation in p52 transgenic mice. A, survival curves of p52 transgenic mice and their wild-type (WT) littermates. The numbers of mice for each genotype are indicated. B, hematoxylin and eosin staining of formalin-fixed sections of the lung and salivary gland from one representative p52 transgenic mouse (1-year-old), with an age-matched wild-type mouse as control. Scale bar, 100 μm. C, immunohistochemical staining of formalin-fixed lung sections from two 1-year-old p52 transgenic (Tg) mice (mice 1997 and 5054). Most infiltrating cells stained positively for lymphocyte markers (CD3, a T-cell marker; B220, a B cell marker). Scale bar, 100 μm. D, flow cytometry analyzing of lung infiltrating lymphocytes from 1-year-old p52 transgenic mice showing an increase in the frequency of activated (CD69+) lymphocytes in comparison to age-matched wild-type mice. The data represent the means ±S.D. from six mice for each genotype. Student’s t test was used for statistical analyses, with p values indicated.
Although p52 mice did not develop lymphomas, a detailed histological examination revealed that ∼76% of p52 mice (n=33) at 6–12 months of age had inflammation in the lungs, salivary glands, and, to a lesser degree, kidneys. The inflammation was characterized by marked infiltration of lymphocytes and macrophages (Fig. 2B). In contrast, only 2 of 12 (17%) age-matched wild-type littermates showed modest lymphocyte infiltration in the lungs. Immunohistochemical staining of p52 mouse lung sections showed that infiltrating lymphocytes consisted of both B and T cells (Fig. 2C). We also isolated infiltrating cells from the lungs of p52 mice (n=6) and performed flow cytometry analysis for the frequency of activated lymphocytes (Fig. 2D). Staining for the activation marker CD69 revealed an average of 5-fold increase in the percentage of activated B cells in the lungs of p52 mice. The percentage of CD69+CD4+ T cells was also increased by an average of 2.7-fold. These data suggest that lymphocytic infiltration in the lungs of p52 mice was a result of an ongoing immune response.
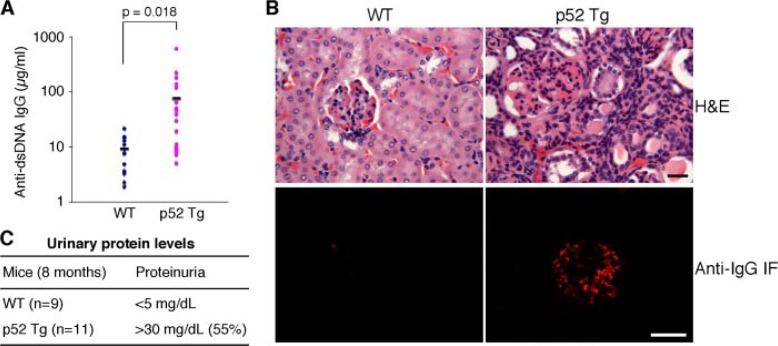
Given the apparent inflammatory phenotype of p52 mice, we further examined these mice for signs of autoimmune disease. An analysis of serum samples from 8–12-month-old p52 mice (n = 24) and their age-matched wild-type littermates (n = 14) revealed that p52 mice had an average of 8.2-fold increase in the levels of circulating autoantibodies against dsDNA (Fig. 3A). Approximately half of p52 mice also showed pathological evidence of glomerulopathy, including increased cellularity in glomeruli and diffuse interstitial lymphocyte infiltration (Fig. 3B). Immunofluorescent staining of cryostat-sectioned renal tissues using anti-IgG revealed deposits of immune complexes with a granular pattern in glomeruli (Fig. 3B). Moreover, 55% of 8-month-old p52 mice (n = 11) showed elevated protein levels in the urine (Fig. 3C), indicative of glomerular dysfunction. Together, these data indicate that p52 transgenic mice are highly susceptible to the development of immune complex glomerulonephritis, an autoimmune renal disease.
FIGURE 3.
Autoimmune glomerulonephritis in p52 transgenic mice. A, increased production of anti-dsDNA autoantibody in 8–12-month-old p52 transgenic (Tg) mice (n = 24) and wild-type (WT) littermates (n = 14). Student’s t test was used for statistical analyses, with p values indicated. B, development of autoimmune renal disease in p52 transgenic mice. Upper panels, hematoxylin and eosin (H&E) staining of formalin-fixed renal sections from a 1-year-old p52 transgenic mouse and an age-matched wild-type littermate, showing increased cellularity in glomeruli and diffuse interstitial lymphocytic infiltration in the renal cortex of the p52 mouse. Lower panels, immunofluorescent (IF) staining of cryostat renal sections with anti-mouse IgG, showing the presence of IgG-containing immune complexes in glomeruli of the same p52 mouse. Shown is representative staining of tissue sections from six mice for each genotype. Scale bars, 50 μm. C, elevated urinary protein levels in p52 transgenic mice, indicating renal dysfunction.
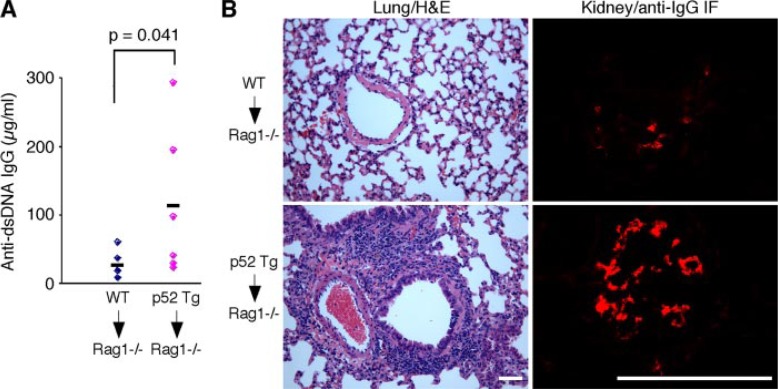
Autoimmune Inflammation in p52 Transgenic Mice Is Lymphocyte Autonomous—To determine whether the autoimmune inflammation in p52 mice is caused by a defect within the lymphoid compartment, we transferred 4 × 107 splenocytes from 7-month-old p52 transgenic mice (n= 6) and wild-type littermates (n = 6) into sublethally irradiated Rag1−/− mice. Three months after transplantation, three of the six Rag1−/− recipients of p52 splenocytes showed higher levels of circulating autoantibodies against dsDNA, with an average of 4.4-fold increase relative to the Rag1−/− recipients of wild-type splenocytes (Fig. 4A). In addition, all of the Rag1−/− mice that received p52 cells developed lung inflammation characterized by lymphocytic infiltration and had deposits of immune complexes with a granular pattern in glomeruli (Fig. 4B). In contrast, none of the Rag1−/− recipients of wild-type splenocytes showed any signs of autoimmune inflammation (Fig. 4). These data indicate that splenocytes from p52 mice were sufficient to induce autoimmunity.
FIGURE 4.
Inflammatory autoimmune disease in p52 transgenic mice is lymphocyte autonomous. Splenocytes from 7-month-old p52 transgenic and wild-type (WT) mice (n = 6 for each genotype) were individually transferred into sublethally irradiated Rag1−/− mice. Three months after transfer, the Rag1−/− recipients were examined for anti-dsDNA autoantibody in the serum (A), infiltration in the lung (B, left panels), and IgG containing immune complexes in the kidney (B, right panels). Only the Rag1−/− recipients of p52 splenocytes showed evidence of autoimmune inflammation. Student’s t test was used for statistical analysis (A), with p values indicated. H&E, hematoxylin and eosin; IF, immunofluorescent. Scale bars, 50 μm.
Expansion of Lymphocyte Populations in p52 Transgenic Mice—To gain insights into the mechanism whereby p52 induces inflammatory autoimmune disease, we examined lymphoid organs and lymphocyte populations of p52 mice and their wild-type littermates at ages of 6–24 weeks. Thymi from p52 and wild-type mice were similar in size, in the numbers of total thymocytes, and in the ratios of major thymocyte subsets (data not shown), indicating that constitutive production of p52 had not significant effect on thymocyte development. However, compared with wild-type mice, p52 mice showed a significant increase in sizes of spleens (∼69%; Fig. 5A) and lymph nodes (∼71%; data not shown). Consistent with the increased sizes of lymphoid organs, flow cytometry analysis revealed that p52 mice had a significant increase in numbers of total splenic B (36%) and T cells (44%) (Fig. 5B), demonstrating that constitutive production of p52 promoted expansion of splenic lymphocyte populations. We further examined the major subtypes of splenic T cells in p52 mice. There was no difference in the number of CD8+ cytotoxic T cells between p52 and wild-type mice; however, p52 mice showed a significant increase (66%) in the number of CD4+ helper T cells (Fig. 5C), primarily as a result of a 2-fold increase in the number of CD4+ CD69+ activated helper T cells (Fig. 5D). The elevated levels of activated helper T cells suggest a sustained immune response in p52 mice, which may drive the inflammatory autoimmune disease by excess production of inflammatory cytokines (40).
FIGURE 5.
Expansion of peripheral lymphocyte populations in p52 transgenic mice. A, splenomegaly in p52 transgenic (Tg) mice at ages of 6–24 weeks, with the spleens from age-matched wild-type (WT) mice as control. B–D, expansion of splenic lymphocyte populations in p52 mice. Shown are numbers of total splenic T and B cells (B), CD4+ and CD8+ T cells (C), and CD4+ CD69+ activated helper T cells (D) in 6–24-week-old p52 transgenic mice and their age-matched wild-type littermates. The data in A–D represent the means ±S.D. of spleens or cells from six mice of each genotype. Student’s t test was used for statistical analyses, with p values indicated.
Lymphocytes from p52 Mice Show Normal Proliferative Responses but Are Resistant to Certain Apoptotic Stimuli—To understand the cellular processes responsible for the expansion of peripheral lymphocyte populations in p52 mice, we examined the growth properties of purified splenic B and T cells from 6-week-old p52 mice. Freshly isolated B and T cells showed no significant proliferation, as determined by cell cycle analysis (Fig. 6, A and B). To assess whether p52 expression enhances their proliferative responses to mitogens, purified B cells were treated for 2 days with either LPS or the anti-μ chain antibody F(ab’)2; purified T cells were treated for 2 days with antibodies against CD3 and CD28 or PMA plus ionomycin. Flow cytometry analysis revealed that the percentages of cells in all cell cycle phases were similar between p52 and wild-type lymphocytes (Fig. 6, A and B). Also, we observed no significant difference in [3H]thymidine incorporation levels between p52 and wild-type lymphocytes following mitogen stimulation (data not shown). Thus, p52 expression did not cause splenic lymphocytes to proliferate autonomously or enhance their proliferative response to mitogens.
FIGURE 6.
Lymphocytes from p52 transgenic mice are resistant to certain apoptotic stimuli. A, cell cycle analysis of splenic B cells that were either untreated or treated for 48 h with LPS (20 μg/ml) or with F(ab’)2 goat anti-mouse IgM (20 μg/ml). Percentages of cells in each phase of the cell cycle are shown. B, cell cycle analysis of splenic T cells that were either untreated or treated for 48 h with plate-bound anti-CD3e plus anti-CD28 (10 μg/ml of each antibody) or with PMA (2 ng/ml) plus ionomycin (250 ng/ml). Percentages of cells in each phase of the cell cycle are shown. C–H, in vitro survival assays of splenic lymphocytes. B cells were either untreated (C) or treated with 20 μg/ml of LPS (D) or with 0.5 μg/ml of doxorubicin (E); T cells were either untreated (F) or treated with 2 ng/ml of PMA (G) or with 0.5 μg/ml of doxorubicin (H). Viability was determined by trypan blue dye exclusion assay. The data in A–H represent the means ± S.D. of cells from at least three mice of each genotype. WT, wild type; Tg, transgenic.
Apoptosis also plays a critical role in maintaining lymphocyte homeostasis (41). We examined the survival of purified splenic B and T cells from p52 and wild-type mice under a variety of conditions. When cultured in the absence of cytokines (no treatment) or treated with LPS, p52 B cells survived much better than wild-type B cells (Fig. 6, C and D). Similarly, compared with wild-type T cells, p52 T cells showed a significant increase in survival in the absence of cytokines (no treatment) or following PMA treatment (Fig. 6, F and G). However, p52 splenic B and T cells were essentially as sensitive as the wild-type cells to doxorubicin, a DNA-damaging drug (Fig. 6, E and H), indicating that p52 expression protects lymphocytes from some, but not all, apoptotic stimuli. Together, these data indicate that constitutive generation of p52 specifically promotes survival of lymphocytes in the absence of cytokines or following mitogenic stimulation, which likely contributes to the expansion of peripheral lymphocyte populations in p52 mice.
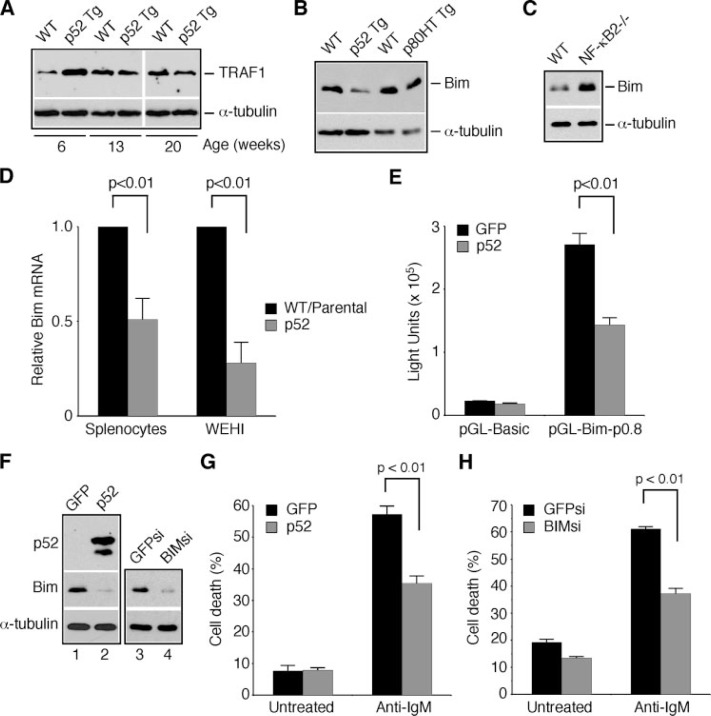
p52 Represses Bim Expression and Inhibits Activation-induced Apoptosis—We next investigated the molecular mechanism by which p52 protects lymphocytes from apoptosis. We have shown previously that up-regulation of the anti-apoptotic gene TRAF1 is a major mechanism underlying the anti-apoptotic activity of p80HT and the expansion of lymphocyte populations in p80HT mice (22). Splenic lymphocytes from 6-week-old p52 mice also showed an increase in TRAF1 levels; however, the up-regulation was transient, because lymphocytes from older p52 mice expressed similar levels of TRAF1 as wild-type cells (Fig. 7A). This transient up-regulation of TRAF1 in p52 lymphocytes is in striking contrast to what was observed in p80HT mice: both lymphocytes from young p80HT mice and lymphoma cells from old p80HT mice displayed a marked increase in TRAF1 expression (22). Of note, our previous study showed that overexpression of p52 actually resulted in a modest inhibition of TRAF1 promoter-directed luciferase expression (22). Together, these observations suggest that at the molecular level, p52 functions distinctly from p80HT, and TRAF1 is not likely a key target gene of p52. We also examined the expression of several anti-apoptotic genes implicated as NF-κB targets, including Bcl-Xl, cIAP2, TRAF2, and XIAP (42–45), and found no difference in their expression levels between p52 and wild-type splenocytes (data not shown).
FIGURE 7.
p52 represses Bim expression and inhibits BCR ligation-induced apoptosis. A–C, immunoblot analysis of the expression of TRAF1 (A) and Bim (B and C) in splenocytes from wild-type (WT), p52 transgenic (Tg), and NF-κB2−/− mice at 6–24 weeks of ages. The levels of α-tubulin are shown as loading control. The data are representatives of four independent experiments with samples from four mice for each genotype. D, real time PCR analysis of Bim mRNA levels in splenocytes from WT and p52 transgenic mice and in WEHI-231 B cells with or without p52 overexpression. The Bim mRNA levels in wild-type splenocytes and parental WEHI-231 B cells are defined as 1.0. The data represent the means ± S.D. from three mice for each genotype or from three independent analyses of WEHI-231 B cells. E, reporter assays for luciferase expression under the control of mouse Bim promoter in 293T cells. Luciferase values were normalized to β-galactosidase activity to account for differences in the transfection efficiency. The data represent the means ±S.D. from three independent assays. F, immunoblot analysis of p52 and Bim expression in WEHI-231 B cells infected with retroviruses expressing GFP, p52, GFP siRNA (GFPsi), or Bim siRNA (BIMsi). The levels of α-tubulin are shown as loading control. G and H, analysis of anti-IgM-induced apoptosis in WEHI-231 B cells overexpressing GFP or p52 (G) or GFP siRNA or BIM siRNA (H) by annexin-V staining and trypan blue dye exclusion assays. The data represent the means ± S.D. from three independent experiments. Student’s t test was used for statistical analyses (D, E, G, and H), with p values indicated.
It is well documented that p52 homodimers can repress gene expression (18, 38, 46). Because p52 apparently exists as homodimers in lymphocytes from p52 mice (Fig. 1B), we investigated the possibility that p52 may repress the expression of pro-apoptotic genes critical for maintaining lymphocyte homeostasis. One potential target is Bim, a pro-apoptotic protein critical for lymphocyte apoptosis (47). Lymphocytes from Bim−/− mice are resistant to apoptosis triggered by cytokine deprivation or mitogen stimulation but sensitive to DNA damage-induced apoptosis, and Bim−/− mice show expansion of peripheral lymphocyte populations and develop autoimmune renal disease (31). These are the phenotypes shared by p52 transgenic mice, as presented above. Immunoblot analysis revealed that Bim expression was markedly down-regulated in splenic lymphocytes from p52 mice compared with the lymphocytes from wild-type littermates (Fig. 7B). No repression of Bim expression was observed in splenic lymphocytes from age-matched p80HT mice (Fig. 7B), providing further evidence that p52 functions distinctly from p80HT. Importantly, splenic lymphocytes from NF-κB2−/− mice showed significant up-regulation of Bim (Fig. 7C), indicating a physiological role of p52 in repressing Bim expression.
To confirm the immunoblotting data, we performed real time PCR analysis of Bim mRNA levels in splenocytes from p52 and wild-type mice, which revealed an ∼50% reduction in Bim mRNA levels in the p52 cells compared with the wild-type cells (Fig. 7D, Splenocytes). Similarly, overexpression of p52 in WEHI-231 B lymphoma cells resulted in a 72% reduction in Bim mRNA levels relative to control WEHI-231 cells (Fig. 7D, WEHI). These data suggest that p52-mediated Bim repression occurs at the transcription level.
We next performed luciferase assays using a reporter construct in which luciferase expression is under the control of a 800-bp sequence immediately upstream of mouse Bim exon 1, which contains all major elements for the control of Bim expression (37). The Bim promoter reporter construct (pGL-Bim-p0.8) exhibited robust activity in 293T cells, leading to an 86-fold increase in luciferase activity relative to the control basic reporter construct (pGL-basic). However, co-transfection of 293T cells with pcDNA3-p52 and pGL-Bim-p0.8 resulted in an ∼2-fold reduction in the luciferase activity (Fig. 7E). Similar results were also obtained with human fibrosarcoma HT1080 cells overexpressing p52 (data not shown). These results suggest that p52 acts on the Bim promoter to repress Bim expression.
The observation that p52 represses Bim transcription in WEHI-231 B cells provided a system for examining the effect of p52-mediated Bim down-regulation on apoptosis induced by B cell receptor engagement, a model for activation-induced apoptosis in B cells (48, 49). Consistent with the results of real time PCR analysis (Fig. 7D), Bim protein levels were also significantly decreased in WEHI-231 B cells overexpressing p52 relative to the control cells expressing GFP (Fig. 7F, lanes 1 and 2). In addition, we generated WEHI-231 B cells with stable expression of Bim siRNA, which resulted in a marked down-regulation of Bim expression compared with the cells expressing siRNA against GFP (Fig. 7F, lanes 3 and 4). Down-regulation of Bim, either by p52 or by siRNA, significantly protected WEHI-231 B cells from apoptosis induced by B cell receptor engagement (Fig. 7, G and H). Together, our results suggest that repression of Bim expression is an important mechanism for p52 to protect lymphocytes from activation-induced apoptosis.
DISCUSSION
C-terminal truncations and rearrangements of the NF-κB2 gene occur recurrently in a variety of human lymphoid malignancies (13). How these structural alterations affect NF-κB2 signaling and contribute to tumorigenesis is not well understood. One of the key questions is whether the resulting NF-κB2 mutants function directly as oncogenic proteins or merely serve as altered precursors for constitutive production of p52, the active form of NF-κB2. In this report, we present genetic evidence that constitutive production of p52 is not a major mechanism for the tumorigenic activity of NF-κB2 mutants. Mice with targeted expression of p52 in their lymphocytes, unlike their counterparts expressing the tumor-derived NF-κB2 mutant p80HT (22), are not prone to lymphoma development. Instead, p52 transgenic mice are highly susceptible to the development of inflammatory autoimmune disease. These findings reveal distinct NF-κB2 signaling pathways in the pathogenesis of lymphoid malignancies and autoimmune diseases.
Previous studies using reporter assays have shown that p80HT has a higher transactivation activity than p52 (23–25). Also, it does not have the transcriptional repressor function normally associated with p52 homodimers (18, 23). Interestingly, our electrophoretic mobility shifting assay results indicate that p80HT forms heterodimers with either RelA or c-Rel in lymphocytes (22), whereas p52 exists predominantly as homodimers. The distinct compositions of p80HT and p52 dimers probably underlie the differences in their transcriptional activities and target genes, which may contribute to the distinct phenotypes of p80HT and p52 transgenic mice. Pretumor lymphocytes and lymphoma cells from p80HT mice showed sustained up-regulation of TRAF1, a direct target gene of p80HT essential for its anti-apoptotic and tumorigenic activity (22). In contrast, lymphocytes from p52 transgenic mice showed only transient up-regulation of TRAF1. The molecular basis for the transient TRAF1 up-regulation in p52 lymphocytes is not clear at present. In cell-based assays, p52 overexpression actually inhibited TRAF1 promoter-directed luciferase expression (22). Regardless of the mechanism involved, the inability of p52 to sustain TRAF1 up-regulation in lymphocytes is probably one of the major reasons for the essentially tumor-free phenotype of p52 mice.
Nevertheless, constitutive production of p52 is pathogenic. With age, p52 mice develop inflammatory autoimmune disease characterized by multiorgan infiltration of activated lymphocytes, high levels of autoantibodies in the serum, and spontaneous development of autoimmune glomerulonephritis. At cellular levels, p52 mice display a significant increase in the numbers of peripheral B and T cells. For splenic T cell subsets, it is particularly interesting to note the accumulation of activated helper T cells, which may drive the disease by excess production of inflammatory cytokines (40). The increase in lymphocyte populations in p52 mice most likely results from enhanced survival. Constitutive production of p52 has no significant effect on the proliferation of lymphocytes but protects them from apoptosis induced by cytokine deprivation or following mitogenic stimulation. We further demonstrate that p52 can protect WEHI-231 B cells from B cell receptor ligation-induced apoptosis, a model for activation-induced apoptosis responsible for deletion of autoreactive B cells (48, 49). We suggest that the defects in lymphocyte apoptosis result in a breakdown in eliminating autoreactive lymphocytes and/or in keeping immune responses in check, which eventually leads to autoimmune disease.
The transcriptional repressor activity of p52 is well documented (18, 38, 46). However, the physiological significance of this repressor activity has been unclear, because few target genes of p52 repression have been identified. We show that Bim is a target of p52 repression in both in vivo and in vitro systems. More importantly, we show that Bim expression is up-regulated in lymphocytes from NF-κB2−/− mice, suggesting that repression of Bim expression is a physiological function of NF-κB2 signaling. Bim is a member of the Bcl-2 homology 3-only subgroup of the Bcl-2 family with pro-apoptotic activity and is critically important for apoptosis of lymphocytes (47). Bim-deficient mice display defects in activation-induced apoptosis (31–33, 50). These mice also show expansion of peripheral lymphocyte populations and develop autoimmune renal disease (31). These are phenotypes shared by p52 transgenic mice, suggesting that Bim repression is an important mechanism underlying the pathogenesis of autoimmune disease in p52 mice. We want to point out that p52-mediated Bim repression is not complete, which may explain why the autoimmune phenotype of p52 transgenic mice is less severe than that of Bim−/− mice, which often develop fatal autoimmune disease (31). As reported here, p52 transgenic mice have a life span similar to their wild-type littermates.
In summary, our study with p52 transgenic mice suggests a gain of oncogenic activity for NF-κB2 mutants in lymphomagenesis and a causal role for sustained NF-κB2 activation in the pathogenesis of inflammation and autoimmunity. This mouse model, in combination with patient samples, should enable further analysis of the role of NF-κB2 signaling pathway in human inflammatory autoimmune disease.
Acknowledgments
We thank Philippe Bouillet and Jerry Adams for the Bim promoter-luciferase construct; Goleeta Alam for the Bim siRNA construct; Tom Sawyer and Karen Domenico for flow cytometry; and William Gunning, Judy Meredith, and Connie Nowak for histology analysis.
Footnotes
The abbreviations used are: dsDNA, double-stranded DNA; PMA, phorbol 12-myristate 13-acetate; siRNA, small interfering RNA; LPS, lipopolysaccharide; GFP, green fluorescent protein.
This work was supported by American Cancer Society Grant RSG-03-173-01-CCG and National Cancer Institute Grant R01 CA106550 (to H.-F. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
REFERENCES
- 1.Gilmore TD. Oncogene. 2006;25:6680–6684. doi: 10.1038/sj.onc.1209954. [DOI] [PubMed] [Google Scholar]
- 2.Solan NJ, Miyoshi H, Carmona EM, Bren GD, Paya CV. J Biol Chem. 2002;277:1405–1418. doi: 10.1074/jbc.M109619200. [DOI] [PubMed] [Google Scholar]
- 3.Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, Klaus GG, Johnston LH, Ley SC. EMBO J. 2002;21:5375–5385. doi: 10.1093/emboj/cdf542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basak S, Kim H, Kearns JD, Tergaonkar V, O’Dea E, Werner SL, Benedict CA, Ware CF, Ghosh G, Verma IM, Hoffmann A. Cell. 2007;128:369–381. doi: 10.1016/j.cell.2006.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao G, Harhaj EW, Sun SC. Mol Cell. 2001;7:401–419. doi: 10.1016/s1097-2765(01)00187-3. [DOI] [PubMed] [Google Scholar]
- 6.Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, Karin M. Science. 2001;293:1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 7.Dejardin E, Droin NM, Delhase M, Haas E, Cao Y, Makris C, Li ZW, Karin M, Ware CF, Green DR. Immunity. 2002;17:525–535. doi: 10.1016/s1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 8.Claudio E, Brown K, Park S, Wang H, Siebenlist U. Nat Immunol. 2002;3:958–965. doi: 10.1038/ni842. [DOI] [PubMed] [Google Scholar]
- 9.Kayagaki N, Yan M, Seshasayee D, Wang H, Lee W, French DM, Grewal IS, Cochran AG, Gordon NC, Yin J, Starovasnik MA, Dixit VM. Immunity. 2002;17:515–524. doi: 10.1016/s1074-7613(02)00425-9. [DOI] [PubMed] [Google Scholar]
- 10.Caamano JH, Rizzo CA, Durham SK, Barton DS, Raventos-Suarez C, Snapper CM, Bravo R. J Exp Med. 1998;187:185–196. doi: 10.1084/jem.187.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franzoso G, Carlson L, Poljak L, Shores EW, Epstein S, Leonardi A, Grinberg A, Tran T, Scharton-Kersten T, Anver M, Love P, Brown K, Siebenlist U. J Exp Med. 1998;187:147–159. doi: 10.1084/jem.187.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Wang Z, Ding J, Peterson P, Gunning WT, Ding HF. J Biol Chem. 2006;281:38617–38624. doi: 10.1074/jbc.M606705200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courtois G, Gilmore TD. Oncogene. 2006;25:6831–6843. doi: 10.1038/sj.onc.1209939. [DOI] [PubMed] [Google Scholar]
- 14.Neri A, Chang C-C, Lombardi L, Salina M, Corradini P, Maiolo AT, Chaganti RSK, Dalla-Favera R. Cell. 1991;67:1075–1087. doi: 10.1016/0092-8674(91)90285-7. [DOI] [PubMed] [Google Scholar]
- 15.Fracchiolla NS, Lombardi L, Salina M, Migliazza A, Baldini L, Berti E, Cro L, Polli E, Maiolo AT, Neri A. Oncogene. 1993;8:2839–2845. [PubMed] [Google Scholar]
- 16.Migliazza A, Lombardi L, Rocchi M, Trecca D, Chang C-C, Antonacci R, Stefano N, Ciana P, Maiolo AT, Neri A. Blood. 1994;84:3850–3860. [PubMed] [Google Scholar]
- 17.Thakur S, Lin HC, Tseng WT, Kumar S, Bravo R, Foss F, Gelinas C, Rabson AB. Oncogene. 1994;9:2335–2344. [PubMed] [Google Scholar]
- 18.Zhang J, Chang CC, Lombardi L, Dalla-Favera R. Oncogene. 1994;9:1931–1937. [PubMed] [Google Scholar]
- 19.Derudder E, Laferte A, Ferreira V, Mishal Z, Baud V, Tarantino N, Korner M. Biochem Biophys Res Commun. 2003;308:744–749. doi: 10.1016/s0006-291x(03)01474-8. [DOI] [PubMed] [Google Scholar]
- 20.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, Dave S, Hurt EM, Tan B, Zhao H, Stephens O, Santra M, Williams DR, Dang L, Barlogie B, Shaughnessy JD, Jr, Kuehl WM, Staudt LM. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zapata JM, Lefebvre S, Reed JC. Adv Exp Med Biol. 2007;597:188–201. doi: 10.1007/978-0-387-70630-6_15. [DOI] [PubMed] [Google Scholar]
- 22.Zhang B, Wang Z, Li T, Tsitsikov EN, Ding HF. Blood. 2007;110:743–751. doi: 10.1182/blood-2006-11-058446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang CC, Zhang J, Lombardi L, Neri A, Dalla-Favera R. Mol Cell Biol. 1995;15:5180–5187. doi: 10.1128/mcb.15.9.5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Epinat JC, Kazandjian D, Harkness DD, Petros S, Dave J, White DW, Gilmore TD. Oncogene. 2000;19:599–607. doi: 10.1038/sj.onc.1203376. [DOI] [PubMed] [Google Scholar]
- 25.Kim KE, Gu C, Thakur S, Vieira E, Lin JC, Rabson AB. Oncogene. 2000;19:1334–1345. doi: 10.1038/sj.onc.1203432. [DOI] [PubMed] [Google Scholar]
- 26.Bours V, Dejardin E, Goujon-Letawe F, Merville MP, Castronovo V. Biochem Pharmacol. 1994;47:145–149. doi: 10.1016/0006-2952(94)90448-0. [DOI] [PubMed] [Google Scholar]
- 27.Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS. Oncogene. 2000;19:1123–1131. doi: 10.1038/sj.onc.1203412. [DOI] [PubMed] [Google Scholar]
- 28.Dejardin E, Bonizzi G, Bellahcene A, Castronovo V, Merville MP, Bours V. Oncogene. 1995;11:1835–1841. [PubMed] [Google Scholar]
- 29.Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, Braggio E, Henry T, Zhu YX, Fogle H, Price-Troska T, Ahmann G, Mancini C, Brents LA, Kumar S, Greipp P, Dispenzieri A, Bryant B, Mulligan G, Bruhn L, Barrett M, Valdez R, Trent J, Stewart AK, Carpten J, Bergsagel PL. Cancer Cell. 2007;12:131–144. doi: 10.1016/j.ccr.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qing G, Qu Z, Xiao G. Proc Natl Acad Sci U S A. 2007;104:5324–5329. doi: 10.1073/pnas.0609914104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 32.Bouillet P, Purton JF, Godfrey DI, Zhang LC, Coultas L, Puthalakath H, Pellegrini M, Cory S, Adams JM, Strasser A. Nature. 2002;415:922–926. doi: 10.1038/415922a. [DOI] [PubMed] [Google Scholar]
- 33.Hildeman DA, Zhu Y, Mitchell TC, Bouillet P, Strasser A, Kappler J, Marrack P. Immunity. 2002;16:759–767. doi: 10.1016/s1074-7613(02)00322-9. [DOI] [PubMed] [Google Scholar]
- 34.Pircher H, Mak TW, Lang R, Ballhausen W, Ruedi E, Hengartner H, Zinkernagel RM, Burki K. EMBO J. 1989;8:719–727. doi: 10.1002/j.1460-2075.1989.tb03431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Cui H, Schroering A, Ding JL, Lane WS, McGill G, Fisher DE, Ding HF. Nat Cell Biol. 2002;4:888–893. doi: 10.1038/ncb872. [DOI] [PubMed] [Google Scholar]
- 36.Finco TS, Beg AA, Baldwin AS., Jr Proc Natl Acad Sci U S A. 1994;91:11884–11888. doi: 10.1073/pnas.91.25.11884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bouillet P, Zhang LC, Huang DC, Webb GC, Bottema CD, Shore P, Eyre HJ, Sutherland GR, Adams JM. Mamm Genome. 2001;12:163–168. doi: 10.1007/s003350010242. [DOI] [PubMed] [Google Scholar]
- 38.Bours V, Franzoso G, Azarenko V, Park S, Kanno T, Brown K, Siebenlist U. Cell. 1993;72:729–739. doi: 10.1016/0092-8674(93)90401-b. [DOI] [PubMed] [Google Scholar]
- 39.Ishikawa H, Carrasco D, Claudio E, Ryseck RP, Bravo R. J Exp Med. 1997;186:999–1014. doi: 10.1084/jem.186.7.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reiner SL. Cell. 2007;129:33–36. doi: 10.1016/j.cell.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 41.Rathmell JC, Thompson CB. Cell. 2002;109(suppl):S97–S107. doi: 10.1016/s0092-8674(02)00704-3. [DOI] [PubMed] [Google Scholar]
- 42.Chu Z-L, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW. Proc Natl Acad Sci U S A. 1997;94:10057–10062. doi: 10.1073/pnas.94.19.10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C-Y, Mayo MV, Korneluk RG, Goeddel DV, Baldwin AS. Science. 1998;281:1680–1683. doi: 10.1126/science.281.5383.1680. [DOI] [PubMed] [Google Scholar]
- 44.Tang G, Minemoto Y, Dibling B, Purcell NH, Li Z, Karin M, Lin A. Nature. 2001;414:313–317. doi: 10.1038/35104568. [DOI] [PubMed] [Google Scholar]
- 45.Chen C, Edelstein LC, Gelinas C. Mol Cell Biol. 2000;20:2687–2695. doi: 10.1128/mcb.20.8.2687-2695.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang CC, Zhang J, Lombardi L, Neri A, Dalla-Favera R. Oncogene. 1994;9:923–933. [PubMed] [Google Scholar]
- 47.Adams JM, Cory S. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benhamou LE, Cazenave PA, Sarthou P. Eur J Immunol. 1990;20:1405–1407. doi: 10.1002/eji.1830200630. [DOI] [PubMed] [Google Scholar]
- 49.Hasbold J, Klaus GG. Eur J Immunol. 1990;20:1685–1690. doi: 10.1002/eji.1830200810. [DOI] [PubMed] [Google Scholar]
- 50.Enders A, Bouillet P, Puthalakath H, Xu Y, Tarlinton DM, Strasser A. J Exp Med. 2003;198:1119–1126. doi: 10.1084/jem.20030411. [DOI] [PMC free article] [PubMed] [Google Scholar]