Abstract
ADP-ribosylation factor (Arf) and related small GTPases play crucial roles in membrane traffic within the exo- and endocytic pathways. Arf proteins in their GTP-bound state are associated with curved membrane buds and tubules, frequently together with effector coat proteins to which they bind. Here we report that Arf1 is found on membrane tubules originating from the Golgi complex where it colocalizes with COPI and GGA1 vesicle coat proteins. Arf1 also induces tubulation of liposomes in vitro. Mutations within the amino-terminal amphipathic helix (NTH) of Arf1 affect the number of Arf1-positive tubules in vivo and its property to tubulate liposomes. Moreover, hydrophilic substitutions within the hydrophobic part of its NTH impair Arf1-catalyzed budding of COPI vesicles in vitro. Our data indicate that GTP-controlled local induction of high curvature membranes is an important property of Arf1 that might be shared by a subgroup of Arf/Arl family GTPases.
Membrane traffic in eukaryotic cells involves the formation of curved membrane buds and tubules that carry cargo from a donor to an acceptor compartment (1–3). Frequently areas of positive membrane curvature are stabilized by cytoplasmic coat proteins including COPII, COPI, and clathrin polymers (2, 4, 5). Recruitment of coat proteins is regulated by small Ras-related GTPases that cycle between inactive GDP- and active GTP-bound conformations. ADP-ribosylation factors (Arfs) and Arf-related proteins (Arls) constitute one major branch of Ras-related trafficking small GTPases (6–8). Arf1, the best characterized member of the Arf subfamily, facilitates recruitment of COPI to Golgi membranes and of clathrin to late Golgi and endosomal compartments through its direct association with heterotetrameric (AP1, AP3, AP4) and monomeric (GGA1–3) adaptors (7, 9) and by activating lipid-modifying enzymes including phospholipase D and phosphatidylinositol-specific (PI)4 kinases (10). Thus, Arf1 might act as a master regulator of coated vesicle formation. Membrane targeting of Arf1 involves guanine-nucleotide exchange factor-mediated GTP loading on Arf1 (11) that is coupled to a conformational change resulting in membrane association of its N-myristoylated amino-terminal helix (NTH) (12) and the co-recruitment of Arf-GTP-binding effector proteins (compare also Fig. 1A). Additional interactions with acidic phospholipids (13) and membrane proteins including SNAREs may contribute to the spatiotemporal regulation of Arf1 recruitment to Golgi and endosomal membranes. Other Arf and Arl family members such as Arf6 may function similarly at other stations of membrane traffic (7, 8).
FIGURE 1.
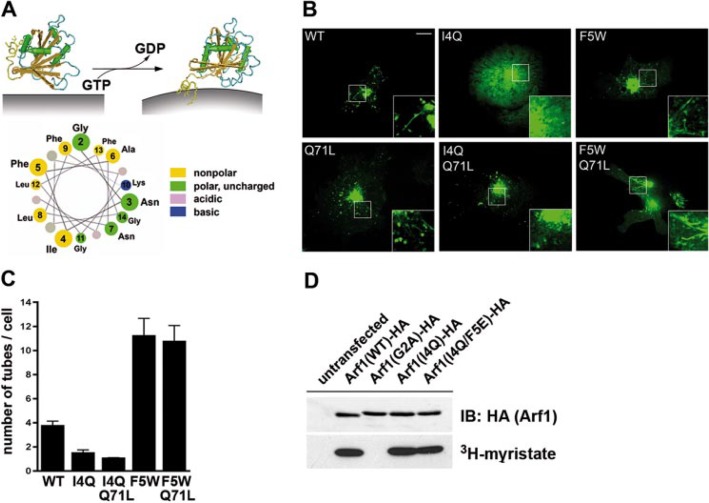
Arf1 decorates membrane tubules in living cells dependent on the integrity of its N-terminal amphipathic helix. A, structure-based model of Arf1-GTP-induced membrane curvature acquisition (top) (Protein Data Bank accession numbers 1RRF (Arf1-GDP) and 1J2J (Arf1-GTP)). Bottom, helical wheel representation of its NTH. B, Arf1-eGFP-decorated membrane tubules at the Golgi and spinning disc confocal images of live HeLa cells transiently expressing C-terminally eGFP-tagged wild-type (WT) Arf1, Arf1 (F5W), Arf1 (I4Q), GTP-locked Q71L, or GTP-locked I4Q/Q71L and F5W/Q71L double mutants. Inset, 2-fold magnified image of boxed area. Representative images from at least three independent experiments are shown. Scale bar, 10 μm. C, quantification of Arf1-eGFP-induced membrane tubulation in living cells (as in A). Given are the mean number of tubules (±S.E.) per cell (n = 3 for each Arf1 variant) averaged from 10 different time points. D, immu-noprecipitates (anti-HA) from Cos7 cells grown in the presence of [3H]myristate and transiently expressing C-terminally HA-tagged Arf1-wild-type, myristoylation-defective G2A, or tubulation-defective I4Q and I4Q/F5E mutants. Untransfected Cos7 cells were used as a control. Immunoprecipitated samples were analyzed by immunoblotting (anti-HA, top) or autoradiography (bottom).
Arf-triggered coat recruitment correlates with the induction of positive membrane curvature (2, 4) leading to the generation of free coated vesicles or tubules (14, 15). Based on the homology between Arf family proteins and the distantly related small GTPase Sar1 (16), one might speculate that nucleotide exchange and subsequent hydrolysis on Arf could serve as a driving force to complete the cycle of membrane deformation and budding. In the case of Sar1p it has been shown that exposure of its amphipathic NTH induced by GTP binding initiates membrane curvature during budding of COPII-coated vesicles (17). Additional support of this idea comes from the observation that many membrane-deforming proteins such as endophilin (18, 19), epsin (20), and amphiphysin (21, 22) contain amphipathic helices, which may insert into the lipid bilayer.
Here we report that Arf1 is found on phosphatidylinositol 4-phosphate (PI4P)-positive membrane tubules in vivo where it partially colocalizes with COPI and GGA1 vesicle coat proteins. We also show that Arf1 can deform lipid bilayers into tubules in vitro. The property of Arf1-GTP to associate with membrane tubules appears to be encoded within its NTH. Mutations within the NTH that affect association with membrane tubules in living cells lead to closely correlated changes in the ability of purified Arf1 to deform lipid membranes in vitro. Based on these data and on the observation that the NTHs of other Arf/ Arl family members also associate with tubular membranes in living cells we suggest that GTP-controlled local induction of high curvature membranes is an important property of Arf1 and related small GTPases.
EXPERIMENTAL PROCEDURES
Live Cell Imaging and Immunofluorescence Analysis—Confocal images of transfected HeLa or Cos7 cells (15–24 h post-transfection) were acquired using a Zeiss Axiovert 200M-based PerkinElmer Life Sciences UltraView ERS dual spinning disc system. Data were processed using Volocity software (Improvision). For indirect immunofluorescence microscopy cells were fixed in 2% paraformaldehyde, further processed, and analyzed using a Zeiss Axiovert 200M under control of the Stallion System (Intelligent Imaging). For quantification of Arf1-eGFP-decorated tubules living cells expressing modest levels of the transfected proteins were monitored for at least 5 min, and images were taken every 3 s. Ten different time points were chosen, distributed over the entire time of monitoring. For these the numbers of tubules displayed were determined, and an average number of tubules per cell was calculated. Data from at least three different cells were collected and averaged.
Electron Microscopy—Liposomes (26% palmitoyloleylphosphatidylserine, 26% phosphatidylethanolamine (brain-purified), 4.5% PI (liver purified), 26% palmitoyloleylphosphatidylcholine, 4.5% phosphoinositol 4,5-bisphosphate (PI(4,5)P2, 13% cholesterol) were prepared by dehydration/rehydration in GTPase buffer (20 mm HEPES, pH 7.4, 100 mm NaCl, 1 mm MgCl2) at 1 mg/ml. To fully resuspend the spontaneously grown liposomes, the tube was shortly vortexed at full power before removing the suspension. Samples for electron microscopy analysis were prepared essentially as described previously (18, 22).
Differential Interference Contrast Videomicroscopy of Membrane Sheets—Differential interference contrast video microscopy of membrane sheets was done essentially as described in Refs. 23 and 24.
COPI-Vesicle Budding Assay—200 μg of rat liver Golgi membranes (pretreated with 500 mm KCl), 10 μg of myristoylated Arf1 protein, and 25 μg of coatomer were incubated for 10 min at 37 °C in a total volume of 250 μl in assay buffer (25 mm HEPES, pH 7.4, 2.5 mm magnesium acetate, 50 mm KCl, 1.2 mm GTP, 300 mm sucrose, and 0.25 mm dithiothreitol), and 1% of the input was taken for Western blot analysis. The sample was subjected to 250 mm KCl to dissociate tethered COPI vesicles from the donor Golgi membranes, which were pelleted by a 10-min centrifugation at 14,000 rpm at 4 °C. The supernatant, containing COPI vesicles, was layered on top of two sucrose cushions in a Beckman SW55-minitube over 37.5% (50 μl) and 45% (5 μl) sucrose. After centrifugation for 50 min at 100,000 × g, the vesicles were taken from the 45-37-5% sucrose inter-phase, and 50% of the collected material was analyzed by immunoblotting.
Multiple Sequence Alignments—Multiple protein sequence alignments were performed using the ClustalW program.
Supplemental methods are available online for plasmids, transfection, antibodies, and reagents, protein expression, purification, nucleotide exchange, and determination of protein N-myristoylation, liposome and effector binding experiments, yeast transformation, and analysis of complementation.
RESULTS
Arf1 Association with Membrane Tubules in Living Cells Is Modulated by Its NTH—GTP binding to Sar1p has been shown to result in exposure of its NTH to initiate membrane curvature acquisition during budding of COPII-coated vesicles at ER exit sites (17). We thus hypothesized that in analogy to Sar1p the conformational transition triggered by GTP loading on Arf1 via insertion of its amphipathic NTH (12) would result in its association with tubular membranes, perhaps by direct induction of positive membrane curvature (Fig. 1A). To determine if Arf1 is indeed associated with tubular membrane structures in living cells we used spinning disc confocal microscopy. Arf1-eGFP localized to the perinuclear area and to puncta and tubules throughout the cytoplasm (Fig. 1B). Arf1-eGFP-decorated tubular structures were seen to emanate from the Golgi and frequently underwent fission (supplemental Fig. 1A and supplemental video 1). Arf1-eGFP-positive tubules were also seen in cells expressing GTP-locked Arf1 (Q71L) (Fig. 1B), whereas Arf1-GDP (T31N) remained largely cytosolic (supplemental Fig. 1C). To determine the contribution of the NTH to its association with membrane tubules we created Arf1 mutants in which hydrophobic amino acids Ile-4 and Phe-5 (lying on one side of the predicted amphipathic helix, compare with Fig. 4A) had been replaced by hydrophilic Arf1 (I4Q/F5E) or even more hydrophobic Arf1 (F5W) residues. Analogous mutants have been introduced previously into other amphipathic helix-containing proteins to analyze their role in membrane bending (18–22). Arf1-eGFP (I4Q) showed a reduced association with the Golgi area and with membrane tubules (Fig. 1, B and C; supplemental video 2), suggesting that the hydrophobic face of the NTH, perhaps in cooperation with the N-myristoyl group, stabilizes Arf1 at membranes. The reduced membrane association of Arf1 (I4Q) could be overcome by locking the protein in the GTP-bound state Arf1 (I4Q/Q71L). However, even in this case mutant Arf1-eGFP (I4Q/Q71L) was rarely observed on membrane tubules (Fig. 1, B and C). By contrast, Arf1-eGFP (F5W) and Arf1-eGFP (F5W/Q71L) (Fig. 1, B and C; supplemental Fig. 1B) decorated numerous membrane tubules originating from Golgi donor compartments (supplemental video 3) at a frequency about 3 times higher than that seen in cells expressing Arf1-eGFP (WT) (Fig. 1C). All Arf1 mutants underwent N-myristoylation similar to the WT protein (Fig. 1D). Neither Arf1 (I4Q) nor Arf1 (F5W) interfered with the overall Golgi structure, as monitored by double-labeling experiments with the Golgi matrix protein GM130 or p230, a peripheral protein associating with the trans-Golgi network (supplemental Fig. 2). These data suggest that Arf1 (I4Q) does not act as a dominant-negative mutant. This is underscored by our observation that yeast cells expressing yArf1 (F4Q) in addition to its endogenous counterpart do not exhibit defects in cell growth or viability (not shown; see also supplemental Fig. 3).
FIGURE 4.
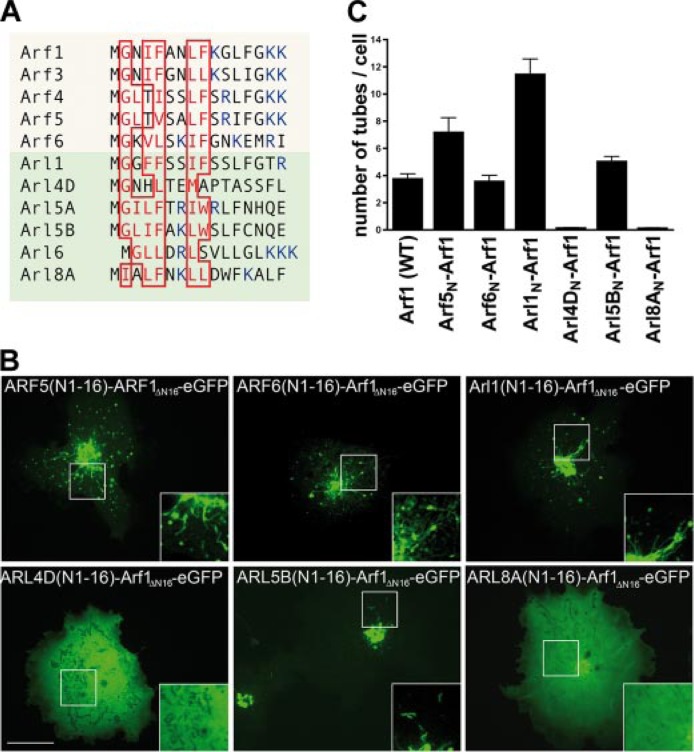
Membrane tubulation activity encoded within the amphipathic helix of Arf-family proteins. A, multiple sequence alignment of the N-terminal helices of Arf/Arl family members. Hydrophobic residues within the predicted amphipathic helices are boxed in red, positively charged residues are depicted in blue. B, live cell imaging of chimeric proteins comprising the first 16 residues of the NTHs of Arf5 and -6 or Arl1, -4D, -5B, and -8A fused to an Arf1-eGFP truncation mutant lacking its own NTH. Arf5N-, Arf6N-, Arl1N- and Arl5BN-Arf1, but not Arl4DN- or Arl8AN-Arf1, decorate numerous membrane tubules at the Golgi seen by spinning disc confocal live cell microscopy. Inset, 2-fold magnified image of boxed area. Representative images from at least three independent experiments are shown. Scale bar, 10 μm. C, quantification of Arf/ArlN-Arf1-induced membrane tubulation in living cells (as in B). Given are the mean numbers of tubules (±S.E.) per cell (n = 3 for each Arf/Arl variant) averaged from 10 different time points.
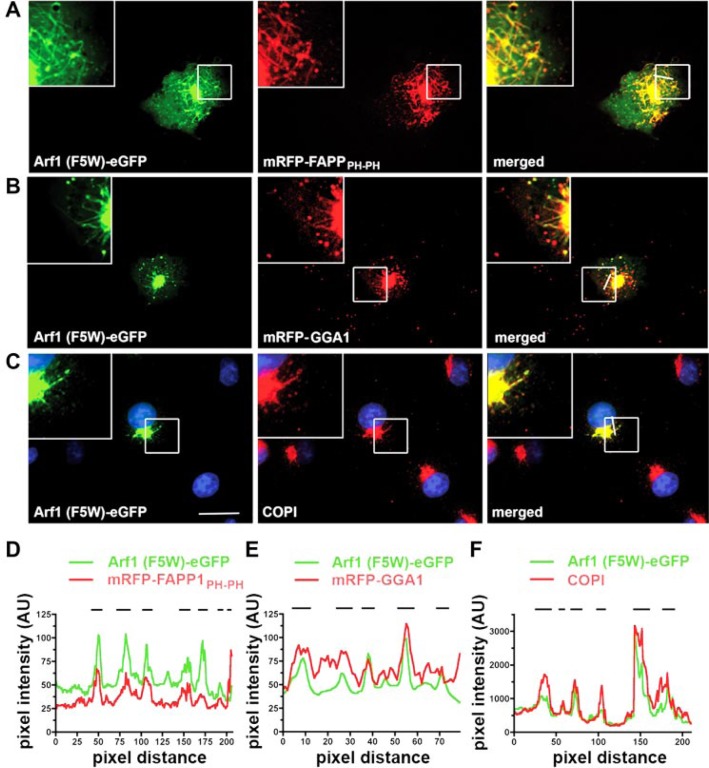
Arf1-eGFP-decorated Membrane Buds and Tubules Contain PI(4)P and Coat Proteins—An important function of Arf1 in vivo is to recruit coat proteins to budding membranes at the Golgi and the trans-Golgi network (TGN). To test whether coat proteins were associated with Arf1-decorated buds and tubules in living cells we analyzed the distribution of some of these proteins in cells expressing Arf1 (F5W)-eGFP. As stated above Arf1 (F5W)-eGFP decorated numerous tubular structures emanating from the Golgi area where it extensively colocalized with an mRFP fusion protein comprising a tandemly repeated PI(4)P-binding PH domain of FAPP1 (Fig. 2, A and D). Thus, membrane tubules decorated by Arf1 likely contain PI(4)P, although the Arf1 binding properties of FAPP-PH may also contribute to its localization. Consistent with its known functions in vivo strong colocalization was also seen between Arf1 (F5W)-eGFP and mRFP-GGA1 (Fig. 2, B and E) as well as COPI vesicle coat proteins (Fig. 2, C and F) on membrane buds and tubules. Similar results were seen for Arf1-eGFP (WT) (data not shown). These findings are also consistent with the recently reported presence of GGAs on membrane tubules in living cells (25). By contrast, p230, a TGN marker, and the cis-Golgi matrix protein GM130 were excluded from Arf1 (F5W)-induced membrane tubules (data not shown). In conclusion, our data suggest that Arf1 associates with and perhaps induces the formation of PI(4)P-positive membrane tubules containing GGA1 and COPI coat proteins.
FIGURE 2.
Arf1-effector proteins assemble on Arf1-induced membrane tubules. A and B, Arf1 (F5W)-induced membrane tubules are decorated by coexpressed mRFP-FAPP1PH-PH (A) or mRFP-GGA1 (B) as visualized by spinning disc confocal microscopy. C, endogenous COPI is found on Arf1 (F5W)-induced membrane tubules. Cells expressing Arf1 (F5W)-eGFP were fixed, and COPI was detected by indirect immunofluorescence using Alexa568-coupled secondary antibodies. Insets, 2.5-fold magnified image of boxed areas. Scale bar, 10 μm. D–F, fluorescence intensity profiles of the images depicted in A–C, respectively. The positions of membrane tubules are indicated by black lines.
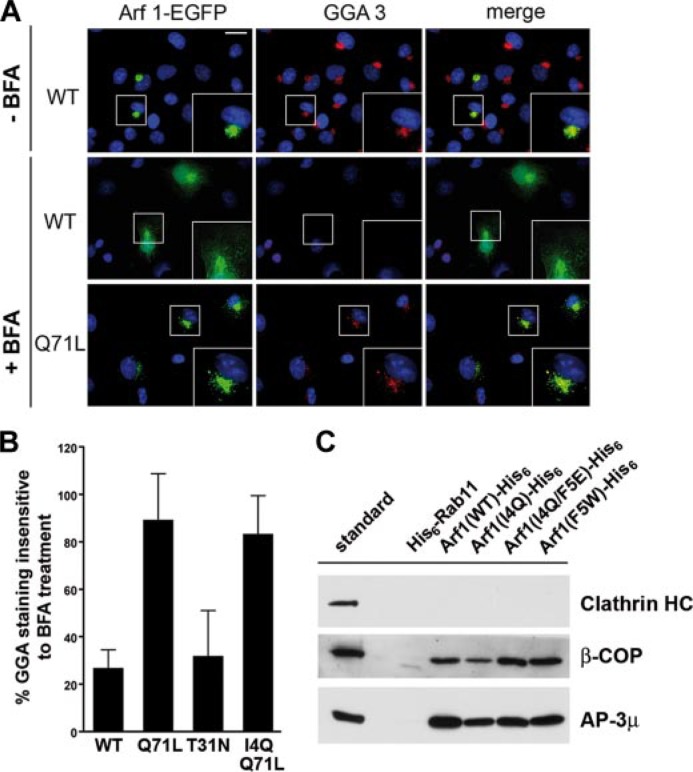
Given that Arf1-eGFP (F5W) (and to some extent also the WT) was associated with GGAs on numerous membrane tubules, we wanted to test whether the reduced number of Arf1 (I4Q)- or Arf1 (I4Q/ Q71L)-eGFP-decorated membrane tubules might be an indirect effect of an impaired association of mutant Arf1 with coat proteins, which may induce and/or stabilize tubules. To test this possibility, cells were treated with the fungal metabolite brefeldin A (BFA), an inhibitor of Arf-specific guanine nucleotide exchange factors, which prevents GTP loading of Arf1. Such treatment results in a rapid redistribution of endogenous GGAs to the cytosol (7, 8), a defect that can be rescued by overexpressed Arf1 (Q71L) locked in its GTP-bound state (Fig. 3A). As seen in Fig. 3B tubulation-defective Arf1-eGFP (I4Q/Q71L) was capable of stably recruiting GGA3 to the TGN of BFA-treated cells, similar to the activity of its WT counterpart in the absence of BFA. Consistent with these findings in living cells Arf1 (I4Q), Arf1 (I4Q/F5E), and Arf1 (F5W) interacted with COPI and AP-3 adaptor proteins in vitro with an efficiency indistinguishable from that of wild-type Arf1 (Fig. 3C). By contrast, purified Rab11 did not associate with any of these proteins. Thus, mutations within the NTH of Arf1 do not impair its ability to interact with coat proteins in vitro or in living cells. These results also rule out that mutations within the Arf1 NTH cause any gross folding defects or altered GTP binding. We note that BFA, which inhibits nucleotide exchange on Arf1, was also shown to induce membrane tubulation (26). However, BFA-induced tubules are driven by motors that pull the membrane along microtubular tracks (27), whereas GTP-Arf1 appears to act directly by producing membrane deformation.
FIGURE 3.
Effector binding by Arf1 mutants. A, recruitment of GGA3 by Arf1 (Q71L) in brefeldin A-treated living cells. Epifluorescent images of fixed Cos7 cells expressing Arf1-eGFP (WT) or the GTP-locked Q71L mutant counter-stained for endogenous GGA3 (red). Blue, 4′,6-diamidino-2-phenylindole (DAPI)-stained nuclei. Treatment with BFA results in dispersal of GGA3 from the Golgi area and a partial redistribution of Arf1-eGFP to the nucleus and cytoplasm. Arf1-Q71L renders GGA3 partially BFA-resistant. Scale bar, 20 μm. B, GGA3 retention at the TGN. Depicted is the relative amount of TGN-associated GGA3 remaining after brefeldin A treatment in cells expressing the indicated Arf1 variant. n = 25 cells were analyzed for each condition. Data are given as mean ± S.E. C, affinity purification from a rat brain extract using His6-Rab11 or Arf1-His6 bound to nickel-nitrilotriacetic acid-agarose in the presence of 200 μm GTPγS. The retained material was subjected to SDS-PAGE and subsequent immunoblotting for clathrin heavy chain (HC), β-COP, and AP-3μ. Similar results were observed for AP-1γ (data not shown).
Association of the NTHs of Select Arf/Arl Family Members with Membrane Tubules—The experiments described above imply that Arf1 associates with high curvature membranes in vivo and raise the possibility that this association may result from a membrane bending activity of its amphipathic NTH (28) (see below also). We hypothesized therefore that the amphipathic NTHs of Arf and Arl proteins in general (16) might be important determinants for their association with tubular membranes in living cells. Multiple sequence alignments (Fig. 4A) revealed that the NTHs of many Arf/Arl family members contain hydrophobic residues that are predicted to lie on one side of an amphipathic helix. Based on these sequence comparisons we predicted that the NTHs of some members of the Arf/Arl family including Arf6 and Arl1 would be able to associate with high curvature membranes, whereas others, such as Arl4D or Arl8a would not. Arl8a lacks the critical Gly residue in position 2 and therefore is predicted not to undergo N-myristoylation. Within the NTH of Arl4D a hydrophilic amino acid (His) is present at position 4 corresponding to Ile-4 within Arf1, a substitution expected to compromise its association with tubules and membranes per se (compare B and C of Fig. 1). To test our hypothesis that the NTHs of Arf/Arl family proteins are indeed important determinants for their association with membrane tubules, we created chimeric proteins comprising the first 16 residues of the NTHs (N) of Arf5 and -6, or Arl1, -4D, -5B, and -8A fused to an Arf1-eGFP truncation mutant lacking its own NTH. These proteins were analyzed for their ability to associate with membrane tubules in fibroblasts. As predicted, expression of Arl1N-Arf1 led to the growth of numerous membrane tubules from the Golgi complex region which were even more numerous than those seen for Arf1 (WT) (Fig. 4, B and C). This might be due to the substitution of Ile-4 within the Arf1-NTH by a much bulkier Phe residue in the NTH of Arl1. Likewise, we saw extensive tubulation in cells expressing fusion proteins comprising the NTHs of Arf5, Arf6, and Arl5B (Fig. 4, B and C). By contrast, Arl4DN- (containing a His in position 4 instead of Ile in Arf1) or Arl8AN-Arf1 (the latter presumably lacking the N-myristate, see above) did not lead to membrane tubulation and instead remained largely cytosolic (Fig. 4, B and C). Thus, the ability to associate with curved membranes might be a property common to a subset of Arf/Arl family proteins. Based on the observed close correlation between the presence of an intact amphipathic NTH and the association of Arf1 with tubular structures in living cells we hypothesized that Arf1 may in fact induce membrane tubulation itself.
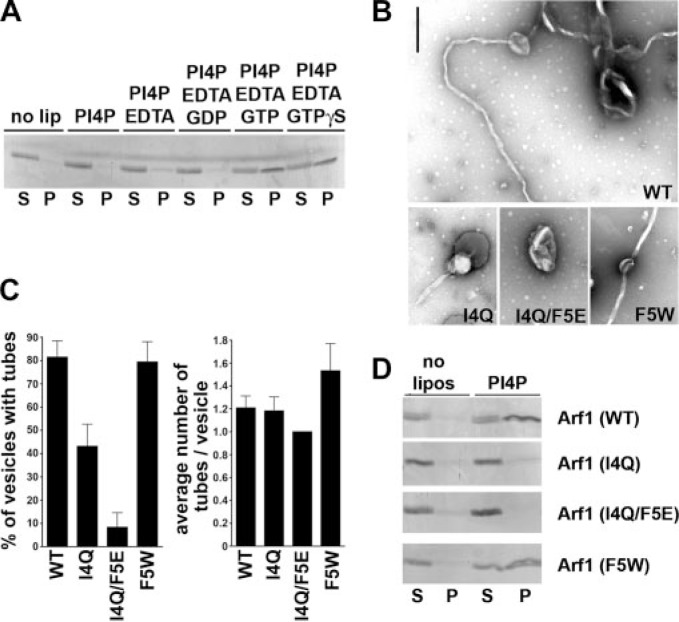
Arf1-GTP Is Sufficient to Induce Tubule Formation on Liposomes—Because of the results shown above (Fig. 4) we decided to directly investigate the membrane tubulating properties of Arf1 in vitro. To this aim we purified untagged N-myristoylated human Arf1 (myrArf1) from bacteria co-expressing human N-myristoyltransferase from a bicistronic plasmid. Resulting Arf1 preparations were more than 90% pure, and about 80% of this protein had also undergone N-myristoylation (not shown (30)). As a premise to test our hypothesis we investigated the membrane binding properties of myrArf1. Liposomes of different compositions were incubated with purified myrArf1, and protein binding was monitored by a cosedimentation assay. MyrArf1 specifically associated with liposomes containing PI(4)P or PI(4,5)P2(13) and to a lesser extent with other negatively charged phospholipids (data not shown). Membrane association was seen only for GTP-loaded myrArf1 consistent with in vivo data (Fig. 5A), whereas myrArf1-GDP remained largely soluble. To determine if myrArf1 could induce positive membrane curvature, we incubated phosphoinositide-containing liposomes with purified GTP- or GDP-loaded wild-type (WT) or mutant Arf1 proteins and analyzed the samples by negative stain electron microscopy. We observed extensive tubulation of liposomal membranes by Arf1-GTP (WT) (Fig. 5, B and C) but not Arf1-GDP (data not shown). The average external diameter of the tubules was 45 ± 5 nm, slightly larger than those induced by Sar1p (17). Substitution of residue Ile-4 by glutamine reduced the number of liposomes with tubular extensions (Fig. 5, B and C). Exchange of both Ile-4 and Phe-5 by hydrophilic residues led to a complete loss in the ability of Arf1 to cause membrane bending (Fig. 5, B and C). This correlated with a strongly reduced association with liposomal membranes of both Arf1 (I4Q) and Arf1 (I4Q/F5E) (Fig. 5D), in agreement with our observations in living cells. Again, these data support the view that the hydrophobic face of the NTH stabilizes the membrane-bound state, similar to what has been reported for other membrane-deforming proteins including endophilin (18) and epsin (20). By contrast, Arf1 (F5W) displayed a slightly increased number of tubules per vesicle (Fig. 5C).
FIGURE 5.
Arf1 induces tubulation of liposomes in vitro. A, liposome cosedimentation assay. N-Myristoylated Arf1 (5 μg) was incubated with liposomes (50 μg) of defined composition containing 10% (w/w) phosphatidyl-serine, phosphatidic acid, and PI(4)P. P, liposomal pellet (100%); S, supernatant (15%). Control (no lip), no liposomes present. 1 mm EDTA and 100 μm GTPγS were added to enable nucleotide exchange. A representative Coomassie Blue-stained gel from one of three independent experiments is shown. Similar patterns were observed when phosphatidylinositol 4,5-bisphosphate was used instead of PI4P (data not shown). B, electron micrographs of liposomes incubated with 15 μm myristoylated GTP-loaded Arf1 (WT) or the indicated mutants. Arf1 (I4Q/F5E) lacks the ability to deform membranes. A mutant with increased hydrophobicity (F5W) displays slightly enhanced tubulating ability. For the non-tubulating Arf1 mutant (I4Q/F5E) higher concentrations of up to 50 μm were also tested with very similar results. Scale bar, 500 nm. C, quantification of Arf1-induced tubule formation. Multiple electron micrograph images (n = 10) were analyzed with regard to the number of vesicles and the number of tubules emanating thereof. Given is the mean fraction of vesicles containing tubules (in %, ±S.E.; left panel) and the average number of tubules per vesicle displaying tubulation (±S.E.; right panel). All samples were processed and analyzed in parallel. Results were confirmed qualitatively in additional independent experiments. D, liposome cosedimentation assay. Myristoylated Arf1 (7.5 μg of protein) was incubated with liposomes (50 μg) of defined composition containing 10% (w/w) PI(4)P in the presence of 1 mm EDTA and 100 μm GTPγS. For controls sample Arf1 was incubated under the same conditions in the absence of liposomes. Arf1 (WT) and mutants within its amphipathic NTH (decrease in hydrophobicity by Arf1 (I4Q) or Arf1 (I4Q/F5E), increase in hydrophobicity by Arf1 (F5W)) were analyzed. A representative Coomassie Blue-stained gel from one of three independent experiments is shown. P, liposomal pellet (100%); S, supernatant (15%).
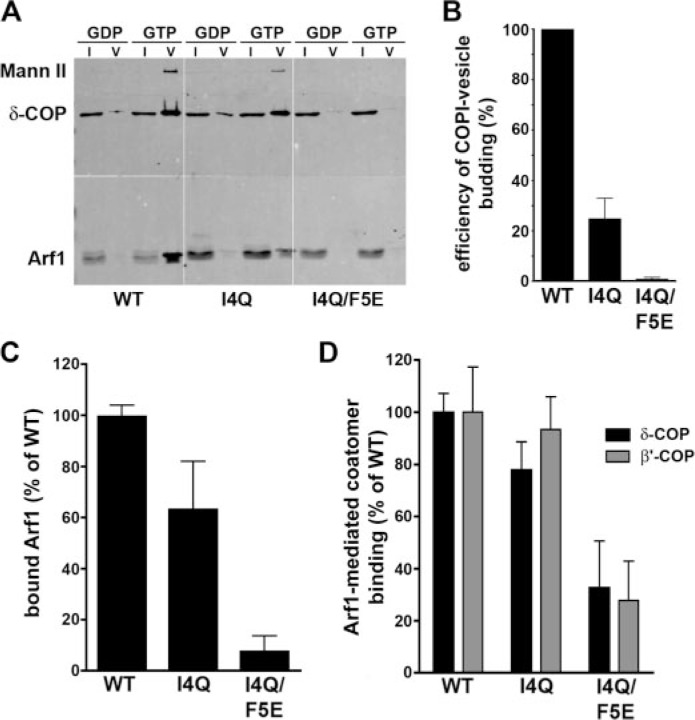
Given that Arf1 is required for formation of COPI-coated vesicles in vivo and in vitro we wanted to determine the effect of the mutations in an in vitro budding assay. Isolated Golgi membranes were incubated with purified coatomer and Arf1 (WT), Arf1 (I4Q), or Arf1 (I4Q/F5E) in the presence of GTP, and vesicle formation was assayed by immunoblotting of density gradient-purified COPI vesicle fractions (Fig. 6A). Quantitative analysis revealed that myrArf1 (I4Q) and even more pronounced myrArf1 (I4Q/F5E) displayed a strongly reduced ability to support COPI vesicle formation (Fig. 6B), despite their association with COPI coatomer (Fig. 3C). In the presence of coatomer and GTPγS, myrArf1 (I4Q) associated with Golgi membranes, albeit with slightly reduced efficiency when compared with myrArf1 (WT) (Fig. 6C), resulting in stable recruitment of COPI coat proteins to the membrane (Fig. 6D). MyrArf1 (I4Q/F5E) failed to bind to Golgi membranes under these conditions and displayed a strongly reduced ability to facilitate coatomer recruitment. Thus, hydrophobic residues within the NTH of Arf1 are required for stable membrane association, tubulation, and COPI coat-mediated vesicle budding.
FIGURE 6.
Hydrophobic residues within the NTH of Arf1 are required for COPI coat-mediated vesicle budding. A, GTP-dependent budding of COPI-vesicles from Golgi-derived membranes. Budded vesicles were isolated and analyzed by SDS-PAGE and subsequent immunoblotting for a cargo protein (Mann II, mannosidase II), COPI, and Arf1. I, input; V, budded COPI vesicle fraction. B, the GTP-dependent budding efficiency in the presence of different Arf1 variants was determined by quantifying the amount of mannosidase II detected in the budded vesicle fraction. Data were normalized with regard to the budding efficiency observed in the presence of Arf1 (WT) (in %; mean ± S.E.; n = 3). C and D, analysis of Arf1 and COPI-coatomer association with Golgi membranes. Salt-washed rat liver Golgi membranes (10 μg) were incubated with purified COPI coatomer (2.5 μg) for 15 min at 37 °C in the presence of Arf1 (WT or mutants) and 100 μm GTPγS (in a total volume of 100 μl). Golgi membranes were reisolated by pelleting through a sucrose (15% w/v) cushion, and membrane association of Arf1 (C) or COPI-coatomer (D) was assayed by immunoblotting. Data were quantified by densitometric scanning (in %; mean ± S.E.; n = 3).
To further support this finding, we made use of yeast genetics. Yeast mutants lacking yARF1 and yARF2 are non-viable but can be rescued by expressing a plasmid-borne copy of yARF1 from its own promoter (31). Yeast Δarf1/Δarf2 double knockout cells kept alive by yARF1 encoded on a URA3-containing plasmid were transformed with LEU2-based expression plasmids for yARF1 (WT) or yARF1 (F4Q), a mutation corresponding to I4Q in mammalian Arf1, and spotted onto media containing 5-fluoroorotic acid to counterselect against the URA3-plasmid. As expected, yARF1 (WT) or a hexahistidine tagged version of it was able to complement loss of the URA3-yARF1 plasmid, but the F4Q mutant was not (supplemental Fig. 3A), although yARF1-His6 (WT) and yARF1-His6 (F4Q) were expressed to similar levels in the absence of 5-fluoroorotic acid and underwent proper N-myristoylation (supplemental Fig. 3B). Thus, hydrophobic residues within the amphipathic NTH of yARF1 are required for viability of yeast cells in vivo, further suggesting that properties which critically require these residues such as membrane binding and deformation are physiologically important.
DISCUSSION
In summary, our data indicate that Arf1-GTP induces positive curvature on membranes, and this property appears to be related to its role in COPI vesicle budding. This conclusion is based on several lines of evidence. First, Arf1-eGFP is associated with membrane tubules that also contain coat proteins such as COPI, GGA1, and the PI(4)P- and Arf-binding adaptor FAPP1. The frequency at which such Arf1-decorated tubules are observed is dependent on hydrophobic residues within the amphipathic NTH of Arf1, suggesting that tubules might at least in part be generated by Arf1 itself. Second, we find that purified untagged myristoylated Arf1 can induce membrane tubules on liposomes, a property affected by mutations in its NTH. Third, an Arf1 mutant, in which a hydrophobic residue found to be critical for the induction of high curvature on membranes has been exchanged for a hydrophilic amino acid (I4Q), is compromised in its ability to facilitate budding of COPI-coated vesicles from native Golgi membranes in vitro, although it is perfectly well able to bind to and recruit coatomer to the membrane.
We therefore propose that Arf1 and, based on our experiments involving chimeric proteins (compare Fig. 4), also other members of the Arf/Arl subfamily of GTPases such as Arf5 and -6 (29) and Arl1 and -5B act as GTP-regulated membrane benders during tubulovesicular membrane traffic. Pre-existing curvature could also facilitate membrane insertion in a positive feedback mechanism that may facilitate membrane tubulation. Based on our findings as well as on previous work (17) we suggest that GTP-controlled local induction of high curvature membranes is a critical function shared by a subset of small GTPases in membrane transport. The abilities of Arfs to drive membrane deformation and to recruit proteins (i.e. GGAs) or protein complexes that aid membrane bending and/or stabilize and sense curved membranes are likely to be functionally linked. In this context it seems worthwhile to note that Arf-GAP1 has been postulated to sense membrane curvature and this in turn may stimulate its enzymatic activity (4) thereby providing negative feedback regulation. Thus, Arf-induced membrane bending could reflect a common principle underlying several of the cell physiological activities of Arf family proteins including the formation and budding of clathrin- and COPI-coated vesicles (2, 4, 9), the extension and fission of TGN-derived tubular carriers (25), and internalization of macropinocytic vacuoles. In all of these cases the mechanisms of membrane deformation or budding have remained largely elusive. Coupling amphipathic helix insertion into membranes to the Arf/Arl GTPase activity may thus be a means to impose directionality and provide spatiotemporal control to membrane deformation.
Acknowledgments
We thank Dr. A. Nakano for providing the Δarf1/Δarf2 yeast strain.
Footnotes
This work was supported in part by German Research Funding Agency DFG Grants HA2686/4-1, SFB 740/TP C6 (to V. H.), and SFB 638/TP A10 (to F. T. W.) and European Molecular Biology Organization (EMBO) YIP Programme (to V. H.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: PI, phosphatidylinositol; NTH, amino-terminal amphipathic helix; PI4P, phosphatidylinositol 4-phosphate; PI(4,5)P2, phosphoinositol 4,5-bisphosphate; eGFP, enhanced green fluorescent protein; WT, wild type; TGN, trans-Golgi network; BFA, brefeldin A; GTPγS, guanosine 5′-O-(thiotriphosphate); HA, hemagglutinin; COPI, coat protein complex I; GGA, Golgi-localized γ-ear containing adaptor; PH, pleckstrin homology.
The on-line version of this article (available at http://www.jbc.org) contains supplemental methods, supplemental Figs. 1–3, and supplemental videos 1–3.
REFERENCES
- 1.Rothman JE. Nat Med. 2002;8:1059–1062. doi: 10.1038/nm770. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino JS, Glick BS. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 3.Cai H, Reinisch K, Ferro-Novick S. Dev. Cell. 2007;12:671–682. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Antonny B. Curr Opin Cell Biol. 2006;18:386–394. doi: 10.1016/j.ceb.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 6.Burd CG, Strochlic TI, Gangi Setty SR. Trends Cell Biol. 2004;14:687–694. doi: 10.1016/j.tcb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 7.D’Souza-Schorey C, Chavrier P. Nat Rev Mol Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 8.Gillingham A, Munro S. Annu Rev Cell Dev Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 9.Robinson MS. Trends Cell Biol. 2004;14:167–174. doi: 10.1016/j.tcb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, Iurisci C, Luini A, Corda D, De Matteis MA. Nat Cell Biol. 1999;1:280–287. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- 11.Bos JL, Rehmann H, Wittinghofer A. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Antonny B, Beraud-Dufour S, Chardin P, Chabre M. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- 13.Randazzo PA. J Biol Chem. 1997;272:7688–7692. [PubMed] [Google Scholar]
- 14.McMahon HT, Gallop JL. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- 15.Farsad K, De Camilli P. Curr Opin Cell Biol. 2003;15:372–381. doi: 10.1016/s0955-0674(03)00073-5. [DOI] [PubMed] [Google Scholar]
- 16.Kahn RA, Cherfils J, Elias M, Lovering RC, Munro S, Schurmann A. J Cell Biol. 2006;172:645–650. doi: 10.1083/jcb.200512057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 18.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT. EMBO J. 2006;25:2898–2910. doi: 10.1038/sj.emboj.7601174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- 21.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 22.Takei K, Slepnev VI, Haucke V, De Camilli P. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- 23.Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dev. Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Roux A, Uyhazi K, Frost A, De Camilli P. Nature. 2006;441:528–531. doi: 10.1038/nature04718. [DOI] [PubMed] [Google Scholar]
- 25.Polishchuk RS, San Pietro E, Di Pentima A, Tete S, Bonifacino JS. Traffic. 2006;7:1092–1103. doi: 10.1111/j.1600-0854.2006.00453.x. [DOI] [PubMed] [Google Scholar]
- 26.Sciaky N, Presley J, Smith C, Zaal KJ, Cole N, Moreira JE, Terasaki M, Siggia E, Lippincott-Schwartz J. J Cell Biol. 1997;139:1137–1155. doi: 10.1083/jcb.139.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lippincott-Schwartz J, Donaldson JG, Schweizer A, Berger EG, Hauri HP, Yuan LC, Klausner RD. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 28.Losonczi JA, Prestegard JH. Biochemistry. 1998;37:706–716. doi: 10.1021/bi9717791. [DOI] [PubMed] [Google Scholar]
- 29.Caplan S, Naslavsky N, Hartnell LM, Lodge R, Polishchuk RS, Donaldson JG, Bonifacino JS. EMBO J. 2002;21:2557–2567. doi: 10.1093/emboj/21.11.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Z, Anderl F, Frohlich K, Zhao L, Hanke S, Brugger B, Wieland F, Bethune J. Traffic. 2007;8:582–593. doi: 10.1111/j.1600-0854.2007.00554.x. [DOI] [PubMed] [Google Scholar]
- 31.Yahara N, Ueda T, Sato K, Nakano A. Mol. Biol. Cell. 2001;12:221–238. doi: 10.1091/mbc.12.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]