Abstract
Many forms of cellular stress cause an elevation of endogenous ceramide levels leading to growth arrest or apoptosis. Ceramidases (CDase) play a critical role in regulating apoptosis by hydrolyzing ceramide into sphingosine, a precursor for promitogenic sphingosine-1-phosphate. Growth factor induction of neutral CDase (nCDase) has been shown to have a cytoprotective effect against cytokine-induced increases in ceramide levels. To further define the physiological regulation of nCDase, we identified a 200 bp promoter region and demonstrated that serum activated this proximal promoter, which correlated with a serum-induced increase in human nCDase mRNA expression. Computational analysis revealed a putative cis-element for AP-1, a transcription factor activated by serum. Electrophoretic mobility shift assays demonstrated that the identified transcriptional response element binds to AP-1 transcription factors. RNA interference-mediated knockdown of the AP-1 subunit, c-Jun, inhibited the activity of the human nCDase proximal promoter, whereas, c-Jun overexpression increased promoter activity, which directly correlated with human nCDase mRNA transcription, decreased ceramide mass, and protection against caspase 3/7-dependent apoptosis. Taken together, our findings suggest that c-Jun/AP-1 signaling may, in part, regulate serum-induced human nCDase gene transcription.
Keywords: ceramidase, ceramide, sphingosine, promoter, transcriptional regulation, AP-1
1. Introduction
Mounting evidence has demonstrated that sphingolipid metabolites, including ceramide (Cer), sphingosine (Sph), and sphingosine-1-phosphate (So1P), are critical modulators of cellular function. Cer and Sph are second messengers shown to induce cell growth arrest or apoptosis [1, 2]. In contrast, So1P typically promotes cell growth and differentiation [3, 4] and can suppress the apoptotic effects of Cer [5]. It is postulated that overall balance between sphingolipid metabolites, in part, determines cellular responses.
Sph, the precursor for So1P, is formed through the catalytic activity of ceramidases (CDases) and reverted back to its unphosphorylated state via So1P phosphatases. CDases cleave the N-acyl linkage of Cer, yielding Sph and free fatty acids as products [6]. CDases are classified into three subtypes according to their pH optima: acidic, neutral, and alkaline. Evidence suggests that neutral CDases (nCDases) are critical physiological enzymes in regulating the balance of sphingolipid metabolites. Human, rat, and mouse nCDases have all been defined as integral membrane proteins mainly localizing to the plasma membrane [7, 8]. Furthermore, rat kidney nCDase was shown to be enriched in lipid rafts, which are structured membrane microdomains [9]. Not only has nCDase been shown to localize to these lipid rafts, but exogenously delivered Cer [10] as well as endogenous ceramide, sphingosine and the enzyme sphingosine kinase 1 [11]. We and others have previously shown that Cer recruits and activates a downstream target, PKCζ, within these lipid rafts [12, 13]. The abilities of nCDase to modulate lipid mediated signaling in response to various cytokine and growth factor stimuli are supported by the co-localization of enzyme, substrate, and targets within lipid rafts.
The physiological consequences of altering Cer mass are becoming more apparent. Many forms of cellular stress, including, but not limited to, heat shock, ionizing radiation, ultraviolet light, Fas ligand, growth factor removal, and oxidative stress cause an elevation of endogenous Cer levels [3, 14]. Similarly, many anticancer drugs mediate apoptosis through elevating levels of Cer [15, 16]. Evidence has accumulated demonstrating the importance of nCDase in regulating Cer concentration in response to cytokine- and growth factor- mediated signaling. We have previously shown that platelet derived growth factor increased nCDase activity in rat mesangial cells [17]. In addition, nCDase activity was regulated by the cytokine interleukin-1β (IL-1β) in a bimodal manner in rat hepatocytes [18]. Furthermore, after an initial release of Cer, long-term exposure of mesangial cells to IL-1β results in cytoprotective up-regulation of nCDase mRNA expression and protein synthesis [19]. Degradation of nCDase induced by nitric oxide (NO) results in apoptosis from increased Cer levels [20]. Moreover, nCDase was shown to protect against TNFα-induced Cer accumulation and apoptosis in primary hepatocytes and against TNFα-induced hepatotoxicity in a rat in vivo model [21]. Despite the increasing evidence of the critical role of nCDase in cellular function, little is known concerning the transcriptional regulation of this enzyme. Putative transcriptional response elements (TRE) have been identified, but not confirmed, in the mouse nCDase promoter [22]. In concurrently submitted work, we identified the proximal promoter region of the human nCDase gene and also, identified several important TRE within this proximal promoter region. In the present study, we investigated the physiological transcriptional regulation of human nCDase promoter in human embryonic kidney (HEK 293) and coronary artery vascular smooth muscle cell models, focusing on the serum-responsive AP-1 TRE. Identifying the physiological mechanisms underlying transcriptional regulation of nCDase may identify new targets in diseases of dysfunctional ceramide metabolism, including vascular diseases, diabetes and cancer.
2. Materials and Methods
2.1 Cell Culture
Human embryonic kidney 293 (HEK 293) cells were obtained from American Type Culture Collection (Rockville, MD). Passages 5-20 were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS). Human coronary artery smooth muscle cells (HCASMC) were obtained from Cascade Biologics, Inc. (Portland, Oregon). Passages 3-10 were maintained in Medium-231(Cascade Biologics, Inc.) supplemented with Smooth Muscle Cell Growth Supplement (Cascade Biologics, Inc.).
2.2 Sequence Analysis
Putative transcription factor binding sites were predicted with Transcriptional Element Search Software (TESS) (http://cbil.upenn.edu/tess/).
2.3 Construction of Reporter Genes
Specific sequences of the 5′-flanking regions of the human nCDase gene were amplified by PCR. The nucleotide range of each amplified region and the respective 5′-forward primers used to amplify them are as follows: -3000/-1 (CPF3-KpnI: 5′-GAGCGGTACCTATCAATACTCTTTAATCTCATAC-3′), -500/-1 (CPF8-KpnI: 5′-AGTCGGTACCAGTCCATGACCACAGAGTACTGTC-3′), -200/-1 (CPF-200-KpnI: 5′-AGTCGGTACCAATTGATTGGTCTTGTCTGCCATG-3′), -50/-1 (CPF-50-KpnI: 5′-AGTAGGTACCTTCTTCCTCTTCAGTATTTCTTCT-3′). The 3′ reverse primer used to amplify all regions was CPR1-XhoI (5′-TGACCTCGAGTTCTTCTCAGGTACAGCAGAGATG). All aforementioned and subsequent primers were synthesized by IDT (Coralville, IA). The underlined bases are restriction digest sites for KpnI for the 5′ primers and XhoI for the 3′ primer. PCR were performed using the pGEM-T vector containing the 5′-flanking region as a template. PfuUltra High Fidelity DNA polymerase (Stratagene, La Jolla, CA) was used according to manufacturer’s instructions. The cycling parameters were 95°C for 30 sec, 60°C for 30 sec, and 72°C for 1 min/kb, for 30 cycles. The DNA fragments generated were cloned into a TOPO vector using Zero Blunt TOPO PCR Cloning Kit (Invitrogen), propagated in Ecoli, followed by plasmid isolation with Qiaprep Spin Mini Kit (Qiagen, Valencia, CA). The plasmids containing the 5′-flanking regions were digested with KpnI and XhoI, and the specific regions were purified by gel electrophoresis and cloned into the firefly luciferase expression vector pGL3-Basic vector (Promega) using the same restriction sites.
2.4 Preparation of Nuclear Extract
Nuclear extracts were prepared using the Pierce Nuclear and Cytoplasmic Extraction Reagent Kit (NE-PER) (Pierce/Thermo Fisher Scientific, Rockford, IL) according to manufacturer’s instructions with slight modifications. Briefly, HEK 293 cells were grown to 90% confluency in a T175 flask, detached with trypsin, and isolated by centrifugation to yield a packed cell volume of approximately 300 μL. After the addition of 3 ml of CERI (cytoplasm extraction reagent I) buffer, the pellet was vortexed and incubated on ice for 10 min. CERII (165 uL) was then added followed by vortexing and an additional incubation on ice. The nuclear fraction was then separated by centrifugation and the supernatant (cytoplasmic extract) was pipetted off. The pellet containing the nuclei was then resuspended in 375 μL NER (Nuclear extraction reagent) buffer and incubated on ice for 40 min with periodic vortexing. The suspension was centrifuged a final time and the supernatant (nuclear extract) was removed and dialyzed in 4% glycerol, 10 mM Tris (pH 7.5) buffer for two hours to remove excess salt. The nuclear extract was stored at −80°C, and the protein concentration was measured using the Bio-Rad Protein Assay Kit.
2.5 Electrophoretic Mobility Shift Assays (EMSA)
All DNA oligonuceotides were synthesized by IDT (Table 1). The Gel Shift Assay System (Promega) was used according to manufacturer’s instructions. Synthetic complementary nucleotides were annealed, end-labeled with [γ-32P]ATP (7000Ci/mmol, American Radiolabeled Chemicals, St. Louis, MO) and T4 polynucleotide kinase, and purified with Sephadex G-25 Quick Spin Columns (Roche Diagnostics, Indianapolis, IN).
Table 1.
Sequence (5′-3′) of double-stranded oligonucleotides used in EMSA analyses
| Ap1/ccaat | sense | CTGTAATTGATTGGTCTTGTCT |
| antisense | AGACAAGACCAATCAATTACAG | |
| AP1 (AP-1 Consensus) | sense | CGCTTGATGAGTCAGCCGGAA |
| antisense | TTCCGGCTGACTCATCAAGCG |
Bold letters in oligonucleotide sequences indicate putative TF binding sites.
EMSA assays were performed with 35-40 fmoles of double-stranded end-labeled probes and 12-15 ug of HEK 293 nuclear protein for 20 minutes at room temp. For competition assays, nuclear protein was preincubated with a 50-fold excess of unlabeled double-stranded EMSA probes for 10 min before the addition of labeled probe. Sequences of labeled probes are given in Table 1. Gel-loading buffer without dye was added before the protein/DNA complexes were resolved on Novex 6% DNA retardation gels (Invitrogen) using 1/2x TBE buffer (Sigma Aldrich, St. Louis, MO) at 200 V for 20 min. After electrophoresis, gels were dried and autoradiographed.
2.6 Transient Transfection and Luciferase Assays
Human embryonic kidney 293 (HEK 293) cells (40-60% confluent) were transfected with each reporter construct (250 ng) and the Renilla luciferase expression vector phRL-null (100 ng, Promega) using Lipofectamine 2000 (3:1, μLLipofectamine: μgDNA) (Invitrogen) in 250 μL Opti-MEM (Invitrogen) in 12 well plates. HEK 293 cells were grown in 1 ml DMEM supplemented with 10% FBS, and the media was not changed after the addition of the transfection reagents. Twenty-four hours after transfection, cells were lysed by the addition of 250 μL of Passive Lysis Buffer (Promega). For serum induced promoter activity, 30-40% confluent HEK 293 cells were transfected overnight and serum deprived with basal media the next morning for 24 hours. FBS was then added to a final concentration of 10% v/v, followed by lysing at the time points given in the results section. The luciferase activity in the cell lysates was determined using the Dual Luciferase Reporter System (Promega). Firefly luciferase activities of the human nCDase promoter and luciferase gene chimeras were normalized to that of Renilla luciferase (or to total protein where noted) and expressed relative to the activity of the pGL3-Basic plasmid. For overexpression of c-Jun and c-Fos, 500 ng of each vector was co-transfected with the reporter vectors, while maintaining a 3:1 ratio (μL: μg) of Lipofectamine 2000:DNA. For c-Jun knockdown, 10 pmol of c-Jun siRNA (JUN Stealth™ RNAi DuoPak, Invitrogen) was delivered with the reporter DNA in 3 μL of Lipofectamine 2000 per well of a 12-well plate. Knockdown was confirmed by Western blot and quantified via densitometry using ImageJ.
2.7 Quantitative Real-Time RT-PCR
HCASMC and HEK 293 cells were seeded onto 60 mm tissue culture dishes and six well plates, respectively, and grown to 70-85% (HCASMC) or 60-70% (HEK 293) confluency. Cells were then serum starved in their respective basal media for 24 hours. After 24 hours, cells were treated with either vehicle or 10% FBS for the time points given prior to RNA extraction. RNA was extracted using the Qiagen RNeasy Mini Kit (Qiagen, Inc, Valencia, CA) according to manufacturer’s protocol. RNA quantity and quality were assessed using the Agilent 2100 Bioanalyzer with the RNA 6000 Nano Assay (Agilent, Palo Alto, CA). First strand complementary DNA (cDNA) was transcribed from 1 μg RNA using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA) according to manufacturer’s protocol. The relative expression of human nCDase was quantified by quantitative real-time polymerase chain reaction (QRT-PCR) assay using the ABI PRISM 7900HT Sequence Detection System (Applied Biosystems), maintained at Penn State College of Medicine Functional Genomics Core Facilities. TaqMan gene expression assays containing both primers and probes for each gene were purchased from Applied Biosystems.
Quantitative PCR was carried out on a real-time detection instrument (ABI 7900HT Sequence Detection System) in 384-well optical plates using Taqman Universal PCR Master Mix and Assay on Demand primers and probes (Applied Biosystems) similar to previously described methods [23, 24]. Reactions were carried out in 10 μL volume, containing 2X TaqMan Universal PCR Master Mix with UNG, 25 ng cDNA from reverse transcribed RNA, 450 nM unlabeled Taqman gene specific primers, and 125 nM FAM dye-labeled Taqman MGB probe. The reaction was held at 50ºC for 2 min and then held at 95ºC for 10 min before undergoing 40 cycles of a denaturation step at 95ºC for 15 sec and an annealing/extension step at 60ºC for 1 min. Relative quantities were calculated using ABI SDS 2.2.2 RQ software (Applied Biosystems, Inc.) and the 2ΔΔCt analysis method [25] with GAPDH as the endogenous control. Final results are given as relative expression normalized to vehicle/serum starved samples.
2.8 Lipid Quantification by Mass Spectroscopy
Sphingolipids from HEK 293 cells were analyzed by electrospray ionization-tandem mass spectrometry (ESI-MS/MS) based on the method described by Merrill et al. [26] and modified by us [27]. Samples were chromatographically separated on an Agilent 1100 HPLC system and the eluate was analyzed on an ABI 4000 QTrap (Applied Biosystems, Foster City, CA) mass spectrometer equipped with a turbo ion spray source. The peak areas for the different sphingolipid subspecies were compared with that of the internal standards obtained from Avanti Polar Lipids (Alabaster, AL).
2.9 Caspase Assay
HEK 293 cells were seeded to a density of 3.0 x 103 cells/well in 96-well plates and grown for 48 h in culture media containing 10% FBS. Cells were then transfected with vectors expressing c-Jun, c-Fos, or non-expressing for 8 h, followed by 12 h of serum starvation. Cells were then further serum starved for an additional 24 h or treated with 10% FBS for 24 h. Caspase-3/7 enzymatic activity levels were measured using the Apo-ONE homogenous caspase-3/7 assay and performed following the instructions of the manufacturer. The kit provides a caspase-3/7 substrate and rhodamine 110, bis-(N-CBZ-L-aspartyl-L-glutamyl-L-valyl-L-aspartic acid amide), which is cleaved by enzymatically active caspase-3/7 resulting in a fluorogenic cleavage product.
2.10 Statistical Analysis
The results are expressed as mean ± standard error of the mean of at least three independent experiments. Probability (p) values ≤ 0.05 (Student’s t-test or one way ANOVA) were considered to indicate statistically significant differences.
3. Results
3.1 Serum induces human nCDase mRNA expression through proximal regulatory sequences
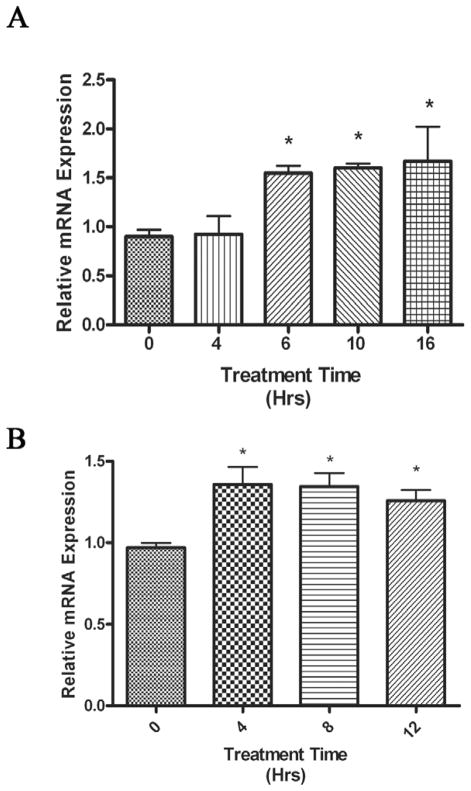
As Cer metabolism can produce the promitogenic phosphorylated metabolite SoP through the initial production of sphingosine, we investigated if serum treatment induced nCDase expression. We initially chose human coronary artery smooth muscle cells (HCASMC) as a model system, since we have previously reported that balloon angioplasty induced smooth muscle cell migration and proliferation, which correlated with increased promitogenic signaling cascades and metabolism of Cer [28, 29]. Treatment with fetal bovine serum (FBS) induced approximately a 60% increase in nCDase mRNA expression in HCASMC after 6 hours of treatment (Fig. 1A). This increase in nCDase expression remained significantly elevated for 10 and 16 hours after treatment. In the next series of experiments, we investigated serum-regulated human nCDase in HEK 293 cells, a more “transfection-friendly” cell line. FBS elevated nCDase mRNA expression approximately 35% in HEK 293 cells (Fig. 1B). Significant increases in nCDase expression occurred four hours after treatment and persisted through 12 hours. Taken together, these data suggested that serum components transcriptionally upregulate the expression of nCDase.
Figure 1. Serum induces human nCDase mRNA expression in HCASMC and HEK 293 cells.
Log phase growing cells were serum deprived in basal media for 24 h and then treated with serum (10% final concentration) for the indicated number of hours. Following serum treatment, total RNA was extracted and used as the initial template in quantitative real-time RT-PCR analysis. Relative quantities were calculated using the 2ΔΔCt analysis method [25] with GAPDH as the endogenous control. Final results are shown as relative expression normalized to serum starved samples treated for 0 hours with serum. Results represent the mean ± S.E.M of four individual experiments, with treatments done in triplicate for each experiment. Significant changes in relative expression are marked with an asterisk. A) Relative nCDase expression in HCASMC in response to serum was measured over a 16 hr time period. B) Relative nCDase expression in HEK 293 cells in response to serum was measured over a 12 hr time period.
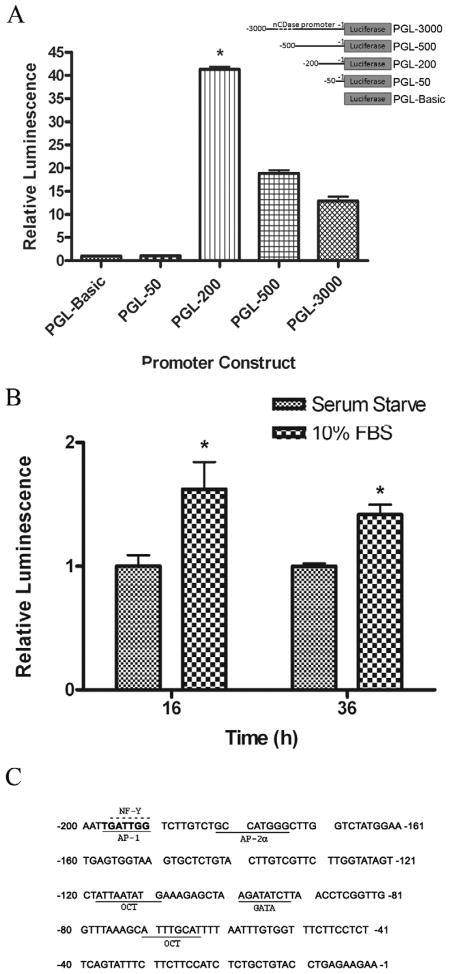
In order to better understand the molecular mechanisms by which serum induces transcription of human nCDase, we examined the promoter region of the human nCDase gene. In our concurrently manuscipt, we reported that a 1-200 bp promoter sequence is the proximal promoter of human nCDase and identified distinct cis-elements by EMSA and mutational studies. In the present manuscript we have verified that the major positive regulatory elements of the human nCDase gene reside within this short 200 bp region of the 5′-untranslated sequence (Figure 2A). We then investigated if this proximal promoter region could be transcriptionally activated by FBS. For this, HEK 293 cells were transiently transfected with PGL-200 and then serum deprived for 24 h. The transfected cells were then treated with 10% FBS and luciferase activity was measured at various time points. After 16 hours, FBS increased luciferase activity over 60% compared to serum starved cells (Fig. 2B). Furthermore, FBS induced luciferase activity was significantly elevated 36 h post treatment.
Figure 2. Serum activates the proximal promoter of the human nCDase gene.
A) Deletion analysis of the 5′-untranslated region of the human nDase gene verified the major positive regulatory elements of the human nCDase gene reside within a 200 bp minimal essential promoter region. Luciferase expression vectors containing different regions of the nCDase promoter were cotransfected into HEK 293 cells with a plasmid (phRL-null) containing the renilla luciferase gene. Following 24 h incubation with DMEM containing FBS, firefly luciferase (fLuc) and renilla luciferase (rLuc) activities were measured. The fLuc/rLuc ratios were determined, and means for a minimum of three transfections were calculated. Values are expressed as mean ± S.E.M-fold increase relative to the luminescence ratio observed in pGL3-Basic transfected cells (relative value = 1). B) Luciferase activity of the proximal promoter is increased through serum treatment. HEK 293 cells cotransfected overnight with PGL-200, the 200 bp promoter/reporter construct, were serum starved for 24 hours and then treated with serum or basal media for the indicated time points. Values are expressed as mean ± S.E.M-fold increase in luminescence relative to serum starved PGL-200 transfected cells (relative value = 1). C) The minimal essential promoter region of the human nCDase gene contains a major potential binding site for the serum regulated transcription factor, AP-1. Nucleotide sequence of the region from −200 to −1 was analyzed for consensus transcription factor-binding elements using TESS analysis. Putative cis-elements are underlined or over scored (over-lined) in the case of overlapping sites.
3.2 The human nCDase proximal promoter contains a functional AP-1 binding site
The above data indicate that one or more of the positive regulatory elements within the 200 bp region are activated by FBS. In order to determine the transcription factors that may play a role in serum-induced activation of the proximal promoter, the human nCDase gene 5′-flanking region was analyzed using Transcription Element Search Software (TESS). This software is designed to search nucleotide sequences for potential transcription factor binding sites using site or consensus strings and positional weight matrices from TRANSFAC 6.0. TESS analysis indicated that the region from −200 to −1 contained, in addition to other cis-elements described in our concurrent manuscript, a major potential serum/growth factor regulated transcription factor binding site, AP-1, which overlapped a CCAAT box (Figure 2C).
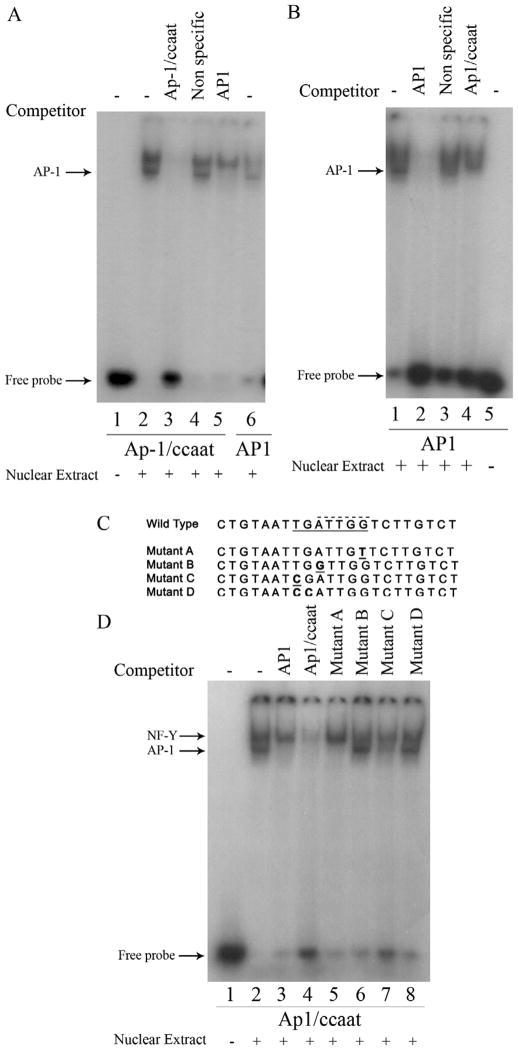
We focused our interest on the overlapping putative AP-1 and CCAAT box cis-elements for two reasons. First, serum and growth factors are known to activate the transcription factor AP-1. Second, CCAAT boxes have been shown to have important roles in regulating TATA-less genes [30, 31]. In our concurrent manuscript, we demonstrated that the transcription factor NF-Y, which has a well established binding affinity to CCAAT boxes [30], is able to bind the overlapping AP-1/CCAAT box site in the human nCDase proximal promoter. To determine if the putative AP-1/CCAAT site of the human nCDase gene can functionally bind AP-1 transcription factors, electrophoretic mobility shift assays (EMSAs) were performed using nuclear extracts prepared from HEK 293 cells and a 32P-radiolabeled 22-nucleotide probe (bp −204 to −183) containing the putative AP-1/CCAAT site centrally positioned, designated “Ap1/ccaat” (see Table 1 for sequence). In the aforementioned manuscript and in this body of work, EMSA results revealed the formation of two distinct nuclear protein/DNA complexes, visualized as two shifted bands (Fig. 3A, lane 2) compared to the control labeled probe without nuclear extract (Fig. 3A, lane 1). Preincubating the nuclear extract with 50-molar excess of unlabeled Ap-1/ccaat oligonucleotide completely inhibited the binding of nuclear proteins to the labeled probe (Fig. 3A, lane 3), indicating the specific interaction of potential transcription factors with the putative AP-1/CCAAT binding site. Pre-incubation with 50-molar excess of an unlabeled, nonspecific oligonucleotide (bp −108 to −87 of the proximal promoter) did not affect the shifted bands (Fig. 3A, lane 4), ruling out non-specific interactions. These results indicated the putative AP-1/CCAAT binding site binds specifically to nuclear proteins.
Figure 3. Competition EMSAs revealed the proximal promoter of the human nCDase gene contains overlapping Ap-1 and NF-Y binding sites with different nucleotide specificity for binding.
Nuclear extracts from HEK 293 cells were subjected to EMSA with radiolabeled Ap1/Ccaat probe (Table 1) containing nucleotides −204 to −183 or with radiolabeled AP-1 probe (Table 1) containing the AP-1 consensus sequence. Lanes containing each labeled probe are underlined with the corresponding labeled probe marked below the underlined lanes. Oligonucleotides used as competitors are marked above the corresponding lanes. Unlabeled Ap1/Ccaat oligonucleotide, non-specific oligonucleotide, and AP-1 consensus oligonucleotide were used as competitors in a 50-fold excess in the corresponding lanes. A) EMSA demonstrate that unlabeled Ap-1 consensus oligonucleotide is a competitor against binding of nuclear extract proteins to labeled probe containing the putative Ap-1 binding site (probe Ap1/Ccaat). B) Conversely, EMSA also demonstrated that unlabeled Ap1/Ccaat oligonucleotide competitively inhibits binding of labeled AP-1 consensus probe to nuclear extract proteins. C) Alignment of wild-type sequence (WT) of region −204/−183 found in the human nCDase promoter and oligonucleotides used as competitors (labeled Mutant A-D) is shown. The introduced mutations in oligonucleotides Mutant A-D are underlined. These unlabeled oligonucleotides were used as additional competitors in D, where the WT oligonucleotide was used as the labeled probe. The putativeAP-1 binding site is underlined with a solid line in the WT sequence, whereas the previously identified NF-Y site is over scored with a dashed line. D) Nuclear extracts were subjected to EMSA with radiolabeled WT (Ap1/Ccaat) probe as described above. In addition, mutated oligonucleotides were used as competitors in a 50-fold excess in the corresponding lanes.
Data from our concurrent manuscript demonstrated that the upper complex is the result of binding of the transcription factor NF-Y to the putative Ap-1/ccaat site, however the protein(s) responsible for the formation of the lower complex was still unknown. Being that a putative AP-1 site overlapped the CCAAT box, we hypothesized that the unknown protein in the faster migrating protein/DNA complex is AP-1. In order to determine AP-1 as the transcription factor that directly binds to AP-1/CCAAT DNA, competition EMSAs were performed using an unlabeled 21 bp oligonucleotide containing the AP-1 consensus binding sequence as a competitor (Table 1, AP1). As shown in Fig. 3A, an excess of unlabeled AP1 oligonucleotide completely inhibited the lower band while not affecting the upper band (Fig. 3A, lane 5). This suggested that the lower band was the result of the binding of AP-1 to the Ap1/ccaat probe. In order to further confirm that the lower band is indeed the AP-1 transcription factor binding to the Ap-1/ccaat probe, additional EMSAs were performed using 32P-labeled AP1 consensus oligonucleotide as a probe. Labeled AP1 probe formed a complex that migrated with the lower band of labeled Ap1/ccaat (Fig. 3A, lane 6 vs. lane 2). More importantly, a 50-molar excess of unlabeled Ap-1/ccaat oligonucleotide inhibited formation of the labeled AP-1 co-migrating band (Fig. 3B, lane 4). Taken together, these results indicate that the putative AP-1/CCAAT sequence in the human nCDase is able to bind with AP-1 transcription factors. However, with AP-1 and NF-Y both binding closely overlapping cis-elements, the question arises on whether or not the binding of AP-1 is dependent on NF-Y
3.3 The AP-1 binding site overlaps, but is not dependent on, a functional NF-Y binding CCAAT box
We next utilized mutant oligonucleotide competitors to decipher the binding of distinct trans-activating factors to the functional overlapping AP-1/CCAAT box. These studies were designed to determine which nucleotides are necessary for AP-1 and/or NF-Y binding. We performed competition EMSAs with excess unlabeled mutant oligonucleotides and labeled Ap1/ccaat probe. One or two base-pair mutations (mutants A-D) were introduced into the Ap-1/ccaat oligonucleotide (Fig. 3C) to determine the necessity of those nucleotides for binding AP-1 or NF-Y. Repetitious of Fig 3B, unlabeled AP1 consensus probe completely competed out the binding of the lower, AP-1/labeled probe complex band (Fig. 3D, lane 3 vs. lane 2). The unlabeled wild type probe (lane 4) effectively competed for binding to both complexes, while the unlabeled AP1 consensus probe competed for only the lower AP-1 complex band (lane 3). The putative AP-1 binding sequence is –197 T G A T T G G –191, while the putative CCAAT-binding factor sequence is –196 G A T T G G –191. Mutation of nucleotide –191 from a G to a T (Mutant A) competed with the labeled wild type probe for binding with AP-1, but not NF-Y (Fig. 3D, lane 5). Simultaneous mutation of nucleotides –190 and –189 from T and C to G and A, respectively, also prevented NF-Y binding but did not inhibit AP-1 binding (data not shown), thus demonstrating that specific nucleotides directly adjacent to the CCAAT box are necessary for binding of NF-Y [30, 31]. In contrast, mutating position –195 from an A to G (mutant B) dramatically decreased the oligonucleotide’s affinity for AP-1, but not NF-Y. A mutation at nucleotide –197 from T to C (mutant C) slightly decreased the affinity of the oligonucleotide for both AP-1 and NF-Y (Fig. 3D, lane 7). Mutant D contained two mutations (nucleotides –197 T to C and –196 G to C), prevented binding of the oligonucleotide to both AP-1 and NF-Y (Fig. 3D, lane 8). These results further support the binding of both AP-1 and NF-Y to an overlapping AP-1/CCAAT binding site. Furthermore, these results indicated that, despite an almost complete overlap of binding site nucleotides, the nucleotides necessary for binding of each transcription factor are partially independent.
3.4 c-Jun, but not c-Fos, regulates the AP-1 transcriptional element on human nCDase proximal promoter
To address the importance of the AP-1 element to the overall activity of the proximal promoter, we introduced a 9 bp deletion into the PGL-200 reporter vector, eliminating the entire AP-1 element. As reported in our concurrent manuscript, this deletion significantly reduced luciferase activity by 14% compared with the wild-type proximal reporter. Therefore, the proximal promoter of the human nCDase 5′-flanking region retains substantial promoter activity without this AP-1 site; however, it is unable to reach the maximal promoter activity of the reporter that contains this site. These data indicate that the AP-1 site is necessary, but not sufficient, for maximal promoter activity.
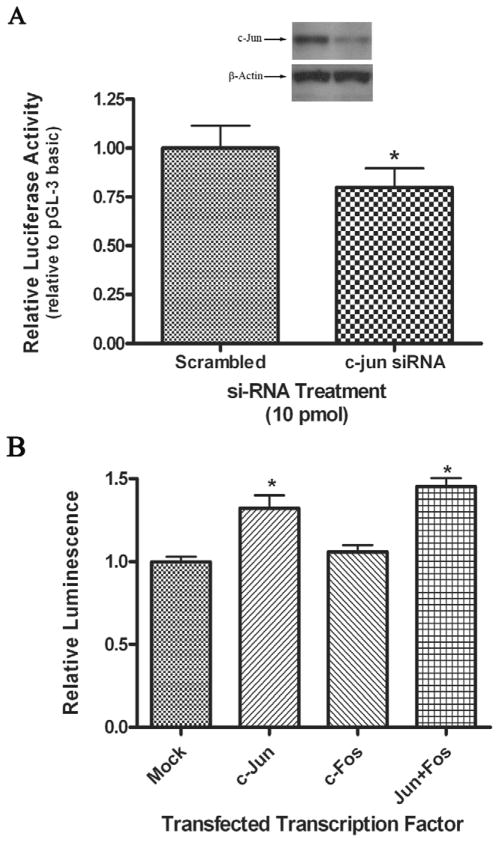
As we identified a functional AP-1 TRE in the proximal promoter of human nCDase, we next investigated the role of serum-responsive AP-1 transcription factors in the regulation of the proximal promoter. We initially chose to investigate c-Jun as it is the most common subunit of AP-1 transcription factors. We co-transfected dual siRNAs directed against c-Jun with PGL-200 into proliferating HEK 293 cells and assayed for luciferase activity. We show that c-Jun siRNA successfully knocked down c-Jun protein levels 40-60% as determined by densitometry analyses (Fig. 4A insert). Corresponding to this decrease in c-Jun protein levels, treatment with c-Jun siRNA reduced the luciferase activity by 20% compared to cells co-transfected with scrambled siRNA and PGL-200 (Fig. 4A). The data correlated well to the level of reduction in promoter activity observed when the AP-1 binding site was deleted from the promoter in our concurrent work. This result demonstrated that a functional AP-1 protein, consisting of at least one c-Jun subunit, is important to achieve maximal human nCDase promoter activity.
Figure 4. AP-1 is important for maximal human nCDase proximal promoter activity and sufficient to induce its activity.
A) HEK 293 cells were co-transfected with siRNA against c-Jun or scrambled siRNA and the PGL-200 luciferase reporter in DMEM containing FBS. fLuc activity was measured 24 h post-transfection and normalized to rLuc activity. The c-Jun siRNA-induced decrease in luciferase activity is expressed relative to the activity from scrambled siRNA-treated cells, where an asterisk marks a significantly different decrease in activity compared to scrambled siRNA treated cells. Results represent the mean ± S.E.M of at least three individual experiments. (A insert) Cell lysates were analyzed by western blots to determine c-Jun knockdown efficiency. Representative autoradiograph from Western analyses demonstrates the knockdown of c-Jun. Knockdown levels were quantified using Image J for densitometry analyses.) B) Overexpression of c-Jun indicated that AP-1 is sufficient to increase the activity of the human nCDase proximal promoter. HEK 293 cells were co-transfected with the PGL-200 luciferase reporter vector and either c-Jun expression vector, c-Fos expression vector, both c-Jun and c-Fos vectors, or empty vector. fLuc activity was measured 24 h post-transfection and normalized to rLuc activity. Reporter activity is expressed relative to the luciferase activity from cells co-transfected with the empty vector (Mock). Results represent the mean ± S.E.M of at least three individual experiments. Significant changes in relative expression are marked with an asterisk.
The role of AP-1 to induce the proximal promoter from the human nCDase gene was further tested using c-Jun and c-Fos expression vectors, co-transfected with PGL-200, into proliferating HEK 293 cells. The transcription factor AP-1 consists of Jun-family/Jun-family dimers or Jun-family/Fos-family dimers. Overexpression of c-Jun induced a 34% increase in luciferase activity relative to the activity of cells transfected with PGL-200 alone (Fig. 4B). Overexpression of c-Fos had no significant effect on luciferase activity. Co-transfection of both c-Jun and c-Fos with PGL-200 induced an approximate 45% increase in luciferase activity compared to PGL-200 alone (Fig. 4B), which was not significantly different from the activation with c-Jun alone. These data suggested that AP-1 transcription factors induces the human nCDase promoter. Specifically, AP-1 with c-Jun subunit(s) was sufficient to mediate this induction, whereas a c-Fos subunit appeared to have little functional importance.
3.5 c-Jun, but not c-Fos, induces nCDase mRNA transcription
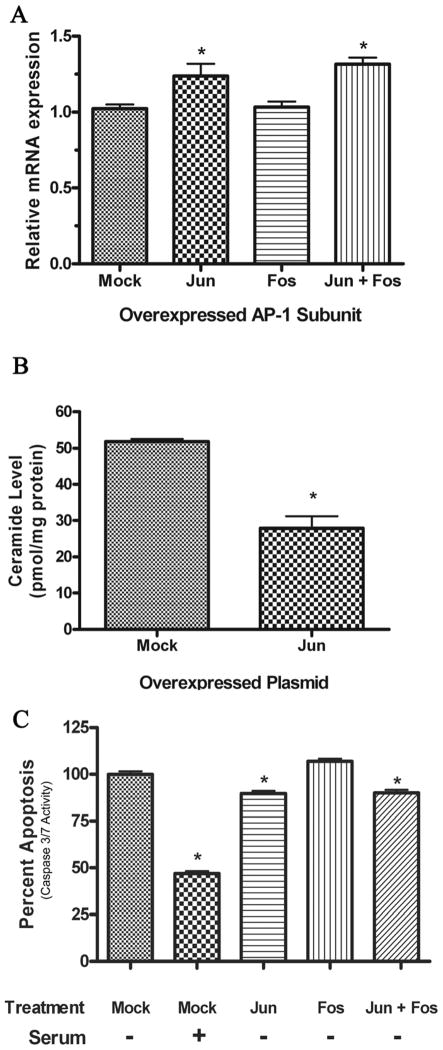
We next asked if AP-1/c-Jun regulation of promoter activity correlates with expression, activity, and biology of nCDase. Initially, we determined if c-Jun-induced proximal promoter activity correlated with transcription of human nCDase mRNA. Log-phase HEK293 cells were transfected with c-Jun, c-Fos, or both c-Jun and c-Fos for 24 hours after which nCDase mRNA levels were quantified by RT-PCR. Overexpression of c-Jun induced a 22% increase in human nCDase mRNA expression, whereas overexpression of c-Fos had no effect on nCDase transcription (Fig. 5A). Furthermore, co-expression of c-Jun and c-Fos resulted in a 30% increase in nCDase expression, though not statistically different from cells overexpressing c-Jun alone. These results suggest that c-Jun-induced activation of the nCDase proximal promoter can translate into increased nCDase mRNA transcription, indicating that AP-1, containing c-Jun subunits, contributes to nCDase transcription.
Figure 5. Physiological significance of c-Jun overexpression and activation of human nCDase proximal promoter.
A) Effects of AP-1 subunit overexpression on human nCDase transcription. Log phase growing HEK 293 cells were transfected with the AP-1 subunits c-Jun and c-Fos as described in the methods. Twenty-four hours after transfection, total RNA was extracted and used as the initial template in quantitative real-time RT-PCR analysis. Relative quantities were calculated using the 2ΔΔCt analysis method [25] and normalized to GAPDH. Final results are expressed as mean ± S.E.M levels relative to nCDase mRNA in mock transfected cells for 4 individual experiments, each replicated in triplicate. Single asterisk represents significance compared to mock transfected cells. B) Effects of c-Jun overexpression on Cer metabolism. Log phase growing HEK 293 cells were transfected with c-Jun for 24 h. Following transfection, lipids were extracted and Cer mass was determined by electrospray ionization tandem mass spectrometry. Final results are expressed as mean ± S.E.M Cer levels from 3 individual experiments replicated in triplicate. Single asterisk represents significance compared to mock transfected cells. C) Effects of AP-1 subunit overexpression on apoptosis. Log phase growing HEK 293 cells were transfected with the AP-1 subunits c-Jun and c-Fos and then serum starved to induce apoptosis. In addition, serum-starved mock transfected cells were treated with FBS to inhibit apoptosis. Apoptosis was quantified by measuring caspase 3/7 activity and normalized to serum starved, mock transfected cells (set at 100% apoptosis). Final results from 3 individual experiments with each treatment replicated in triplicate are expressed as mean ± S.E.M percent apoptosis with a single asterisk representing significance compared to mock transfected cells.
3.6 c-Jun overexpression reduces Cer mass
To determine if a c-Jun-induced increase in nCDase transcription correlates with enzyme activity and a change in Cer mass, mass spectrometry analyses were performed as described in the materials and methods. HEK 293 cells overexpressing c-Jun had an average Cer mass of 28 pmol/mg protein (Fig. 5B), which was significantly decreased from Cer levels in mock transfected cells (52 pmol/mg). Lipidomic analysis revealed decreases in all major species of ceramide including C16:0, C18:0 and C24:0. Our results, showing a decrease in Cer mass after c-Jun overexpression, correlate with an increase in nCDase transcription but do not rule out c-Jun overexpression altering other Cer metabolizing enzymes.
3.7 c-Jun, but not c-Fos, protects against apoptosis
Finally, to confirm if regulation of nCDase transcription and activity by AP-1 leads to a measureable biological correlate, we assessed the ability of c-Jun and c-Fos to protect against stress-induced caspase 3/7-dependent apoptosis. HEK293 cells overexpressing c-Jun, c-Fos, and both c-Jun and c-Fos were serum starved to induce apoptosis. As seen in Fig. 5C, replenishing FBS to serum-starved cultures attenuates the relative amount of apoptosis by 53%. HEK 293 cells overexpressing c-Jun alone responded to serum starvation with a significant 10% decrease in caspase 3/7-dependent apoptosis compared to mock transfected cells. Overexpressing c-Fos did not decrease caspase 3/7-dependent apoptosis. HEK 293 cells overexpressing both c-Jun and c-Fos responded similarly to HEK293 cells overexpressing c-Jun alone. Taken together, the ability of c-Jun, but not c-Fos, to inhibit serum-induced caspase 3/7-dependent apoptosis in HEK 293 cells (Fig. 5C) correlates with c-Jun-induced nCDase proximal promoter activity (Fig. 4C) and resulting nCDase mRNA transcription (Fig. 5A) and bioactivity (Fig. 5B).
4. Discussion
The present study characterizes the physiological regulation of human nCDase. EMSA analyses revealed a putative cis-element for AP-1 within the proximal promoter region of nCDase gene that is regulated by serum in two cell models. We chose to focus on the transcriptional factors that regulate this overlapping AP-1/CCAAT element because the activation of AP-1 by growth factors and serum and the role of AP-1 in proliferation and transformation is well documented [32]. AP-1 trans-activators are homo- or heterodimer transcription factors comprising members of the Jun family (c-Jun, JunB, and JunD) and the Fos family (c-Fos, FosB, Fra-1, and Fra-2), with c-Jun being the most abundant AP-1 subunit [32]. Our studies demonstrated that overexpressing c-Jun increased reporter activity, revealing that AP-1 is sufficient to induce the human nCDase proximal promoter. Consistent with this finding, c-Jun overexpression was also sufficient to induce human nCDase mRNA synthesis, which correlated with decreased Cer mass and protection from stress-induced, caspase 3/7-dependent apoptosis. Interestingly, the degree of decrease in Cer mass was significantly greater than the reduction of human nCDase transcription, raising the possibility that AP-1 may regulate other Cer metabolizing enzymes. In contrast, we observed that overexpression of c-Fos did not affect promoter reporter activity or nCDase mRNA transcription, nor did it protect against apoptosis. These results may not be surprising, considering that c-Fos does not form homodimers and, therefore, cannot bind DNA without a Jun-family subunit, since dimer formation is a prerequisite to DNA binding. In agreement with this, co-transfection of AP-1 dependent reporters with a c-Fos expression vector resulted in no significant transcription of the reporters in cells lacking c-Jun [33]. Taken together, these studies suggest that growth factor-induced c-Jun/Ap-1 signaling reduces ceramide mass and protect against apoptosis via transcriptional regulation of human nCDase..
In our study, we also focused on the interplay between overlapping TREs in the human nCDase proximal promoter. As an example, we found that the AP-1 binding site overlaps, but is not dependent on, a functional NF-Y binding CCAAT box. To interpret this observation, it is of interest that the putative AP-1 binding site in the human nCDase gene promoter differs from the consensus AP-1 binding sequence, TGA(C/G)TCA. The nCDase AP-1 TRE sequence does, however, match exactly to one of the known AP-1 binding site variants, TGATTGG [34]. The first three nucleotides form the first half of the AP-1 consensus sequence, whereas the latter two thirds of the site actually form a CCAAT box site in an inverted orientation, e.g. 5′-ATTGG-3,′ on the coding strand. In fact, ATTGG is the predominant orientation when it functions as an NF-Y binding element in TATA-less promoters [30]. Interestingly, the CCAAT box is invariantly flanked by at least one functionally important cis-element [31], which supports the concurrent binding of NF-Y along with AP-1 to the identified AP-1/CCAAT site in the human nCDase proximal promoter. Recently, a different overlapping NF-Y and AP-1 binding site, TATTGGTCAT was discovered in the follicle-stimulating hormone beta (FSHβ) subunit promoter [35]. In this site, the NF-Y binding reverse CCAAT box, ATTGG, preceded, yet partially coincided with, the latter half of the AP-1 consensus sequence, GTCA. Interestingly, in the FSHβ promoter, the binding of AP-1 to the novel AP-1 site was contingent upon an intact CCAAT box [35], whereas a mutation of the CCAAT box in the human nCDase promoter had no effect on AP-1 binding, yet prevented the binding of NF-Y (Fig. 3C). Similarly, other mutations of the nCDase AP-1 cis-element could disrupt AP-1 binding, without affecting NF-Y binding (Fig. 3C).
The mutual interactions between multiple trans-activating factors (including AP-1) and NF-Y have been shown to be essential for optimal regulation of transcription [31]. NF-Y has the capacity to enhance transactivation through direct protein-protein interactions and/or facilitate the positioning of transcription factors through DNA conformation changes. Although spatial constraints must exist in the binding of two transcription factors to directly adjacent binding sites, our current work and the work by Coss et al [35], clearly show that such constraints do not prohibit concurrent binding of AP-1 and NF-Y. Structural and binding characteristics of AP-1 and NF-Y allow hypothetical circumvention of the spatial constraints. The subunits of an AP-1 dimer form a flexible fork, which could yield to other transcription factors bound in close proximity [35, 36]. Subunits of AP-1 bind their corresponding cis-element at the major groove, with only four amino acids in contact with the DNA, while vast portions of the rest of the subunits lie perpendicular to the DNA [36], thus possibly leaving enough space along the DNA for binding of other transactivators [35]. Coincidently, NF-Y has been shown to bind to the minor groove of DNA [37], opposite that of AP-1, and hence further minimizing the spatial constraints of AP-1 and NF-Y binding adjacent to each other [35].
Despite having the strongest promoter activity of the nCDase 5′-flanking region and containing a binding site for the potentially serum-activated AP-1, the proximal promoter was only mildly induced by serum, suggesting strict regulation of the nCDase gene. Direct interaction of NF-Y and AP-1 at the AP-1/CCAAT element could help regulate the extent to which serum-activated AP-1 induced transcription. It is also possible that AP-1 and/or NF-Y could interact with other proximate transcription factors. Several other cis-elements in close proximity to the AP-1/NF-Y site were shown to bind their respective transcription factors, such as Oct and GATA factors, which could presumably interact synergistically or opposingly with NF-Y or AP-1. As examples, OCT-1 represses the c-Jun-dependent activation of rat 3α-dydroxysteroid oxidoreductase gene promoter [38] or rat CYP4A2 gene promoter [39]; yet Oct-1 and c-Jun act synergistically with each other in the regulation of human thromboxane A2 receptor [40], H3.3B histone [41], IL-2, and IL-5 [42] gene promoters.
In our concurrent work, we found that the 5′-flanking region of the human nCDase gene lacks a TATA box, an important sequence in the initiation of transcription, found in many, if not the majority, of eukaryotic genes. However, we observed a CCAAT box, which has been shown to associate with proteins within the transcription initiation complex [43], within the human nCDase promoter. Interestingly, the lack of a TATA box [44] and the presence of an in inverted CCAAT box [30] within a promoter are typical features of tightly regulated genes. Thus, it is not surprising that we observe tight transcriptional control of mRNA expression with serum in both HCASMC and HEK 293 cells. Furthermore, signal transduction through sphingolipid second messengers is very sensitive to sphingolipid balance, so minute changes in sphingolipid species can have dramatic cellular effects, which behoove the tight regulation of sphingolipid metabolizing enzymes. Therefore, tight transcriptional control of nCDase mRNA maintains homeostatic control over highly bioactive sphingolipid metabolites.
In this work, we identified a TRE for a serum inducible transcription factor, AP-1, in the proximal promoter of the human nCDase gene. We demonstrated that AP-1 binds to this cis-element, which overlaps a NF-Y binding element. Correlating with the fact that the identified trans-activating factor AP-1 is serum activated, serum regulated both proximal promoter activity and nCDase mRNA expression. In particular, we demonstrated that c-Jun/AP-1 signaling might, in part, regulate human nCDase gene transcription, which correlated with nCDase mRNA expression, decreased Cer mass, and protection from apoptosis. Our studies indicate a mechanism by which growth factor stimulated c-Jun/Ap-1 signaling may generate promitogenic sphingosine metabolites at the expense of growth arresting ceramide. These studies have direct clinical implications in diseases such as cancer and atherosclerosis, where ceramide metabolites contribute to pro-survival and pro-mitogenic phenotypes.
Research Highlights.
Serum increases nCDase mRNA expression
Serum-induced nCDase mRNA is mediated through an AP-1 binding site in the promoter
Knockdown of the AP-1 subunit, c-jun, inhibits nCDase transcription
Overexpression of c-jun increases nCDase expression and protects from apoptosis.
Acknowledgments
NIH Grants HL66371 and HL76789 to MK supported this work. We would like to acknowledge the Molecular Genetics Core Facility at the Penn State College of Medicine for their assistance in DNA sequencing analyses. Core Facility services and instruments used in this project were funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement Funds. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions. We would also like to thank Dr. Tom Curran, Professor, Dept of Pathology & Laboratory Medicine, University of Pennsylvania, Philadelphia, PA for graciously giving us the c-Jun and c-Fos expression vectors and Nicole Divittore for assistance with manuscript preparation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kolesnick R. J Clin Invest. 2002;110:3–8. doi: 10.1172/JCI16127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cuvillier O. Biochim Biophys Acta. 2002;1585:153–62. doi: 10.1016/s1388-1981(02)00336-0. [DOI] [PubMed] [Google Scholar]
- 3.Spiegel S, Foster D, Kolesnick R. Curr Opin Cell Biol. 1996;8:159–67. doi: 10.1016/s0955-0674(96)80061-5. [DOI] [PubMed] [Google Scholar]
- 4.Spiegel S, Kolesnick R. Leukemia. 2002;16:1596–602. doi: 10.1038/sj.leu.2402611. [DOI] [PubMed] [Google Scholar]
- 5.Pyne S. Subcell Biochem. 2002;36:245–68. [PubMed] [Google Scholar]
- 6.Gatt S. J Biol Chem. 1963;238:3131–3. [PubMed] [Google Scholar]
- 7.Hwang YH, Tani M, Nakagawa T, Okino N, Ito M. Biochem Biophys Res Commun. 2005;331:37–42. doi: 10.1016/j.bbrc.2005.03.134. [DOI] [PubMed] [Google Scholar]
- 8.Tani M, Iida H, Ito M. J Biol Chem. 2003;278:10523–30. doi: 10.1074/jbc.M207932200. [DOI] [PubMed] [Google Scholar]
- 9.Mitsutake S, Tani M, Okino N, Mori K, Ichinose S, Omori A, Iida H, Nakamura T, Ito M. J Biol Chem. 2001;276:26249–59. doi: 10.1074/jbc.M102233200. [DOI] [PubMed] [Google Scholar]
- 10.Stover T, Kester M. J Pharmacol Exp Ther. 2003;307:468–75. doi: 10.1124/jpet.103.054056. [DOI] [PubMed] [Google Scholar]
- 11.Hengst JA, Guilford JM, Fox TE, Wang X, Conroy EJ, Yun JK. Arch Biochem Biophys. 2009;492:62–73. doi: 10.1016/j.abb.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox TE, Houck KL, O’Neill SM, Nagarajan M, Stover TC, Pomianowski PT, Unal O, Yun JK, Naides SJ, Kester M. J Biol Chem. 2007;282:12450–12457. doi: 10.1074/jbc.M700082200. [DOI] [PubMed] [Google Scholar]
- 13.Hajduch E, Turban S, Le Liepvre X, Le Lay S, Lipina C, Dimopoulos N, Dugail I, Hundal HS. Biochem J. 2008;410:369–379. doi: 10.1042/BJ20070936. [DOI] [PubMed] [Google Scholar]
- 14.Hannun YA. Science. 1996;274:1855–9. doi: 10.1126/science.274.5294.1855. [DOI] [PubMed] [Google Scholar]
- 15.Kobayashi E, Motoki K, Uchida T, Fukushima H, Koezuka Y. Oncol Res. 1995;7:529–34. [PubMed] [Google Scholar]
- 16.Lucci A, Han TY, Liu YY, Giuliano AE, Cabot MC. Int J Oncol. 1999;15:541–6. doi: 10.3892/ijo.15.3.541. [DOI] [PubMed] [Google Scholar]
- 17.Coroneos E, Martinez M, McKenna S, Kester M. J Biol Chem. 1995;270:23305. doi: 10.1074/jbc.270.40.23305. [DOI] [PubMed] [Google Scholar]
- 18.Nikolova-Karakashian M, Morgan ET, Alexander C, Liotta DC, HMA J Biol Chem. 1997;272:18718. doi: 10.1074/jbc.272.30.18718. [DOI] [PubMed] [Google Scholar]
- 19.Franzen R, Pautz A, Brautigam L, Geisslinger G, Pfeilschifter J, Huwiler A. J Biol Chem. 2001;276:35382. doi: 10.1074/jbc.M102153200. [DOI] [PubMed] [Google Scholar]
- 20.Franzen R, Fabbro D, Aschrafi A, Pfeilschifter J, Huwiler A. J Biol Chem. 2002;277:46184–90. doi: 10.1074/jbc.M204034200. [DOI] [PubMed] [Google Scholar]
- 21.Osawa Y, Uchinami H, Bielawski J, Schwabe RF, Hannun YA, Brenner DA. J Biol Chem. 2005;280:27879–87. doi: 10.1074/jbc.M503002200. [DOI] [PubMed] [Google Scholar]
- 22.Tani M, Okino N, Mori K, Tanigawa T, Izu H, Ito M. J Biol Chem. 2000;275:11229–34. doi: 10.1074/jbc.275.15.11229. [DOI] [PubMed] [Google Scholar]
- 23.Bowyer JF, Frame LT, Clausing P, Nagamoto-Combs K, Osterhout CA, Sterling CR, Tank AW. J Pharmacol Exp Ther. 1998;286:1074. [PubMed] [Google Scholar]
- 24.Maley D, Mei J, Lu H, Johnson DL, Ilyin SE. Comb Chem High Throughput Screen. 2004;7:727. doi: 10.2174/1386207043328300. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Merrill AH, Jr, Sullards MC, Allegood JC, Kelly S, Wang E. Methods. 2005;36:207–224. doi: 10.1016/j.ymeth.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Wijesinghe DS, Allegood JC, Gentile LB, Fox TE, Kester M, Chalfant CE. J Lipid Res. 2010;51:641–651. doi: 10.1194/jlr.D000430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Charles R, Sandirasegarane L, Yun J, Bourbon N, Wilson R, Rothstein RP, Levison SW, Kester M. Circ Res. 2000;87:282–8. doi: 10.1161/01.res.87.4.282. [DOI] [PubMed] [Google Scholar]
- 29.O’Neill SM, Olympia DK, Fox TE, Brown JT, Stover TC, Houck KL, Wilson R, Waybill P, Kozak M, Levison SW, Weber N, Karavodin LM, Kester M. Vasc Dis Prev. 2008;5:200. doi: 10.2174/156727008785133809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mantovani R. Nucleic Acids Res. 1998;26:1135. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mantovani R. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 32.Angel P, Karin M. Biochim Biophys Acta. 1991;1072:129. doi: 10.1016/0304-419x(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 33.Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. Cell. 1988;54:541. doi: 10.1016/0092-8674(88)90076-1. [DOI] [PubMed] [Google Scholar]
- 34.Mermod N, Williams TJ, Tjian R. Nature. 1988;332:557. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- 35.Coss D, Jacobs SB, Bender CE, Mellon PL. J Biol Chem. 2004;279:152–162. doi: 10.1074/jbc.M304697200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glover JN, Harrison SC. Nature. 1995;373:257. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- 37.Ronchi A, Bellorini M, Mongelli N, Mantovani R. Nucleic Acids Res. 1995;23:4565. doi: 10.1093/nar/23.22.4565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin HK, Penning TM. Cancer Res. 1995;55:4105. [PubMed] [Google Scholar]
- 39.Fiala-Beer E, Lee AC, Murray M. Int J Biochem Cell Biol. 2007;39:1235. doi: 10.1016/j.biocel.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 40.Coyle AT, Kinsella BT. Febs J. 2005;272:1036. doi: 10.1111/j.1742-4658.2004.04538.x. [DOI] [PubMed] [Google Scholar]
- 41.Witt O, Albig W, Doenecke D. FEBS Lett. 1997;408:255. doi: 10.1016/s0014-5793(97)00436-5. [DOI] [PubMed] [Google Scholar]
- 42.Thomas MA, Mordvinov VA, Sanderson CJ. Eur J Biochem. 1999;265:300. doi: 10.1046/j.1432-1327.1999.00732.x. [DOI] [PubMed] [Google Scholar]
- 43.Bellorini M, Lee DK, Dantonel JC, Zemzoumi K, Roeder RG, Tora L, Mantovani R. Nucleic Acids Res. 1997;25:2174. doi: 10.1093/nar/25.11.2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.La Thangue NB, Rigby PJW. In: Frontiers in Molecular Biology: Transcription and Splicing. Hames D, Glover DM, editors. IRL Press; Washington, DC: 1988. pp. 1–42. [Google Scholar]