Abstract
B-Precursor acute lymphoblastic leukemia (B-ALL) is the most common childhood cancer. Although 80% of B-ALL patients are able to be cured, significant challenges persist. Significant disparities in clinical outcomes and mortality rates exist between racial/ethnic populations. The objective of this study was to determine whether gene expression levels significantly differ between ethnic populations. We compared gene expression levels between four ethnic populations (Whites, Blacks, Hispanics, and Asians) in the United States. Additionally, we performed network and pathway analysis to identify gene networks and pathways. Gene expression data involved 198 samples distributed as follows: 126 Whites, 51 Hispanics, 13 Blacks, and 8 Asians. We identified 300 highly significantly (P < 0.001) differentially expressed genes between the four ethnic populations. Among the identified genes included the genes PHF6, BRD3, CRLF2, and RNF135 which have been implicated in pediatric B-ALL. We identified key pathways implicated in B-ALL including the PDGF, PI3/AKT, ERBB2-ERBB3, and IL-15 signaling pathways.
Keywords: leukemia gene expression variation pediatric B-ALL
Introduction
B-precursor acute lymphoblastic leukemia (B-ALL) is the most common childhood cancer and the leading cause of cancer-related death in children and young adults.1,2 B-ALL is curable with chemotherapy in approximately 80% of patients,3 however not all children and adolescents have, unfortunately, benefitted equally from this progress. Significant health disparities exist between populations in incidence, treatment outcomes, and mortality rates.4–7 Hispanics and Whites (Caucasians) tend to have the highest incidences of B-ALL,4–7 however Hispanics and Blacks (African Americans) tend to have worse outcomes.7–10 The molecular basis of health disparities in B-ALL between populations is unknown. In addition, the causes of treatment failure in the remaining 20% of patients are largely unknown. Therefore, there is an urgent need to identify biomarkers and to eliminate health disparities. Although drug treatment and prevention are clearly the most pressing public health priorities, resolving the genetics of B-ALL and eliminating health disparities are important long-term goals. Across a broad front, technical and analytic advances have now created a reasonably clear strategy that can be used to move towards that goal.
The majority of B-ALL cases harbor recurring structural chromosomal rearrangements which are important initiating events in leukemogenesis.11 These genetic mechanisms, however, are insufficient to explain the molecular basis of health disparities and the mechanisms underlying resistance to drug treatment. Advances in microarray technologies in recent years have made possible both the classification of pediatric B-ALL and the identification of molecular signatures in B-ALL.12–16 The desire to dissect the genomic architecture of pediatric cancer such as B-ALL and to identify clinically actionable biomarkers has motivated the launch of the National Cancer Institute funded Therapeutically Applicable Research to Generate Effective Treatments (TARGET)17 initiative and the privately funded Pediatric Cancer Genome Project.18 With the launch of these two initiatives, opportunities for understanding pediatric cancer are now unprecedented, as advances in genomics are harnessed to obtain robust foundational knowledge about the genomic landscape of the disease in different populations and about the molecular basis of health disparities.
Key to the success of genomic research in improving pediatric oncology care is translation of genomic discoveries into clinical practice. In a clinical setting, responses to medical therapies, such as drugs, are often compared among populations that are divided according to traditional medicine. With continued advances in genomic research, information technology, molecular diagnostic, and other biotechnologies, it is anticipated that pediatric oncology treatment will shift towards a more personalized paradigm of medicine. Realizing this vision requires a clear understanding of the patterns of gene expression variation within and between ethnic populations diagnosed with B-ALL. Such information would be critical for stratifying patients, development of targeted therapies, and realization of personalized medicine.
The primary objective of this study was to determine whether gene expression levels differ significantly between racial/ethnic populations, and to characterize patterns of gene expression variation within and between pediatric patient populations. A secondary but equally important objective was to identify gene regulatory networks and biological pathways that are dysregulated and shared among the pediatric patient populations diagnosed with B-ALL in the U.S.
Material and Methods
Gene expression data
Gene expression data generated under the auspices of the National Cancer Institute’s TARGET project (http://www.target.cancer.gov)17 was used in our analysis. The samples used for generating gene expression data have been described previously in detail.16 Here, we provide a brief but detailed description of the characteristics and distribution of the samples used in this study.
The original gene expression data included 207 uniformly treated pediatric B-ALL patients.16 The samples used to generate gene expression data were cryopreserved pretreatment B-ALL samples obtained from the Children’s Oncology Group (COG) P9906 clinical trial for patients with newly diagnosed high risk (HR) B-ALL between March 2000 and April 2003. Patients between the ages of 1–21 were eligible if they met specific combinations of higher white blood cell count and generally older age, though any child with a white blood cell count >100,000/μL was eligible and thus a more selected group of children who would have been predicted to have higher risk B-ALL were included. Ineligible HR patients included those who expressed the BCR/ABL fusion gene or who were known to be hypodiploid (DNA index <0.95) or who were induction failures. All the data was processed using the Affymetrix platform using the Human GeneChip U133Plus 2.0, applying standard Affymetrix protocols. Expression data (average scaled difference values) were processed and normalized using the Affymetrix Microarray Analysis Software (MAS 5.0). The data was filtered out to remove spiked control genes. In addition, subjects without specified ethnicity were removed from the data. The final data matrix consisted of expression profiles of approximately 54,000 probes measured on 198 patient samples. The population distribution of gene expression data was as follows: Whites N = 126, Hispanic N = 51, Blacks N = 13, and Asians N = 8. Information on race/ethnicity was obtained by self-reporting, and therefore does not necessarily represent the genotype, a weakness which we readily acknowledge. However, in this study we used gene expression levels as intermediate phenotypes, meaning that the genes themselves are the variables and the expression levels are the measurements. Although this is an unbalanced design, the samples sizes were adequate to detect differences in expression profiles at P < 0.05 with a power of greater than 95%.19 The data was transformed to log2 prior to analysis.
Data analysis
We used a combination of methods for data analysis. As a first step, we partitioned data into four subsets representing the four racial/ethnic populations under study (Whites, Blacks, Hispanics, and Asians). We performed supervised analysis using a t-test comparing gene expression levels between ethnic populations (ie, Whites vs. Hispanics, Whites vs. Blacks, Whites vs. Asians, Hispanics vs. blacks, Hispanics vs. Asians, and Asians vs. Blacks) on the partitioned data. The goal of this analysis was to determine whether gene expression levels differ significantly (P < 0.05) between ethnic populations, and to identify significantly differentially expressed genes distinguishing the ethnic populations under study. In addition, because of the significant admixing of the White and Hispanic subpopulations, we combined gene expression data on the two subpopulations and treated them as one population (White-Hispanics) and then performed analysis using a t-test comparing gene expression levels between Blacks and White-Hispanics, and between Asians and White-Hispanics. Permutation test was used to calculate the empirical P-values. Empirical P-values and those computed using t-test were found to be identical. The false discovery rate (FDR) was used to correct for multiple hypothesis testing.20 Genes were ranked by P-values and the significantly differentially expressed genes were selected. For each comparison of gene expression levels performed between two populations, we used a threshold of P < 0.001 and an FDR of <1% to select the significantly differentially expressed genes. This was done to ensure uniformity and reliability as well as to ensure that the results are comparable. Because of small sample sizes for some ethnic populations, the data set was not divided into test and validation sets. Instead, out of sample validation, a leave-one-out procedure21 was used to assess the predictive power of the identified sets of genes in each comparison. To assess variability in gene expression levels in all the four populations, we used analysis of variance (ANOVA)22 focusing on the differently expressed genes.
To investigate gene expression variability within and between the pediatric patient populations, we used the coefficient of variation (CV). We first sought to examine whether the genes have a similar level of within population variation in different populations. For each gene, we quantified the within-population expression variability by calculating its CV, which is the ratio of the standard deviation of its expression (across individuals within a population) to the mean value.23,24 Specifically, the CV for the ith gene measured across patients within the kth population was calculated as CVik = σik/μik, where σik and μik are the standard deviation and mean expression value, respectively.23,24 Although other metrics can be used to quantify the expression variability, the coefficient of variation is known to be one of the most robust and unbiased metrics23 and has been used for assessing variation in natural human populations.25–27 Our group has successfully used this metric for estimation of sample sizes and statistical power in microarray experiments.19 A larger CVik indicates higher expression variability for a particular gene across individuals within a population, while a significant reduction in CVik indicates that the gene may be dosage sensitive and thus under severe selection pressure to minimize expression variability.
Comparison of gene expression levels between ethnic populations may be criticized because the allocation of individual patients in racially predefined groups imposes a pre-existing structure and may influence the outcome of the genetic study. Furthermore, populations are defined in many (often arbitrary) ways in this case by self-identification. Therefore, we performed unsupervised analysis using hierarchical clustering to characterize the patterns of gene expression profiles. The goal of this analysis was to identify genes and patients with similar expression profiles. We computed the genetic similarity between all possible pairs of genes and between all possible pairs of individual patients using the Pearson correlation coefficients as the distance measure. The genes and samples were then grouped by a hierarchical clustering algorithm using the complete linkage method, as implemented in the GenePattern System,28 to identify clusters of co-expressed genes and clusters of patients who are most similar to one another. Prior to clustering, the data was normalized, standardized, and centered using the standard procedure.29
We used the gene ontology (GO) information30 to identify functionally related genes among the genes differentially expressed between ethnic populations. The GO Consortium has developed three separate categories (molecular function, biological process, and cellular component) to describe the attributes of gene products. Molecular function defines what a gene product does at the biochemical level without specifying where or when the event actually occurs or its broader context, biological process describes the contribution of the gene product to the biological objective, while the cellular component refers to where in the cell a gene product functions. Because our goal in this study was to gain biological insights about the broader context in which the genes distinguishing the ethnic populations and contributing to both between and within ethnic population variation operate, we considered all the three GO categories.
We performed network and pathway analysis and visualization using the Ingenuity Pathway Analysis (IPA) System ( http://www.ingenuity.com).31 The goal was to identify gene regulatory networks and biological pathways that are shared among ethnic populations. The Human Genome Organization (HUGO) Gene Nomenclature Committee (HGNC) gene identifiers were mapped to networks available in the IPA database and ranked by score. The score indicates the likelihood of the genes in a network being found together by random chance. Using a 99% confidence interval, scores of ≥3 are considered significant. Validation of predicted pathways and identification of other downstream target genes was achieved through the literature and database mining module implemented in the IPA System. The feature allows identification of other genes that are functionally related or interact with input genes.
Results
Differences in gene expression levels between ethnic populations
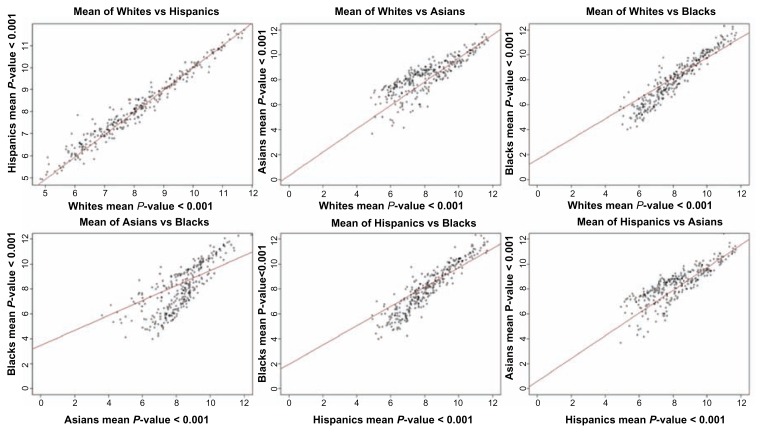
One of the objectives of this study was to determine whether gene expression levels significantly differ between ethnic populations in pediatric B-ALL patients. Our working hypothesis was that gene expression levels differ significantly between populations. We tested this hypothesis by comparing gene expression levels in each population against another. After correcting for multiple hypothesis testing, we identified 300 highly significantly (P < 10−3) differentially expressed genes, and therefore confirming our hypothesis. The estimates of P-values including the FDR for all the 300 genes are presented in Table A, provided as supplementary data. Also presented in Table A are the estimates of P-values and FDR based on ANOVA. The results showing mean expression levels for the 300 genes are presented in Figure 1. Overall, the differences in gene expression levels were generally moderate, ranging from P ~ 10−2 to P ~ 10−5 depending on the pair of population compared (Fig. 1). Predictive modeling using out of sample validation revealed that individual patients could not fall neatly into one of the ethnic populations. Further examination of estimates of P-values and mean expression values for differentially expressed genes revealed that most patient individuals could not be classified with 100% certainty into one of the ethnic populations (Fig. 1). There were significant overlaps in differential gene expression levels between racial/ethnic populations. The overlap in gene expression levels suggests that gene expression in different ethnic populations may be subjected to similar regulatory mechanisms. This pattern of shared variation has important implications for our understanding of population differences and similarities, and it also impacts the critical biomedical issue of patient treatment and classification by race or ethnicity. The results suggest that using race or ethnicity alone as a surrogate for patient classification may not be entirely accurate.
Figure 1.
Pair-wise population scatter plot diagram showing the mean expression values on a log2 scale across the 300 genes significantly (P < 0.001) differentially expressed genes between populations.
Out of the 300 most significantly differentially expressed genes, the largest differences in gene expression levels were observed between Blacks and Asians (184 genes), and between the Asians and Hispanics (140 genes) (Table A). Differences in gene expression levels between Whites and Asians (130 genes) and Blacks and Hispanics (120 genes) were also large (Table A). Smaller differences in gene expression levels were found between Whites and Blacks (106 genes) while the smallest differences were between Whites and Hispanics (60 genes) (Table A). Among the highly significantly differentially expressed genes identified included the genes BRD3, PHF6, CRLF2, and RNF135 which have been directly implicated in pediatric B-ALL.1,32–34 Analysis of variance performed on the 300 genes produced 196 significantly differentially expressed genes (Table A). The observed small numbers in differentially expressed genes between Blacks and Whites, between Hispanics and Whites, and the overlap in differential gene expression levels could be explained in part by the admixing of the populations under study.35,36
Because of the high admixing of the Hispanic population with populations of European ancestry, and the small number of differentially expressed genes between the two racial/ethnic subpopulations, we combined gene expression data on the two subpopulations. When we compared the combined gene expression data on Whites-Hispanics to gene expression levels in Asians, we identified 133 significantly differentially expressed genes. Repeating the same analysis comparing gene expression on Whites-Hispanics to Blacks produced 111 genes (Table A).
These results demonstrate substantial variation in gene expression levels both within and between ethnic populations in B-ALL patients and show that population structure exists in levels of gene expression. Although there have been no systematic studies of gene expression variation within and between ethnic populations in pediatric B-ALL, the differences in gene expression levels between populations found in this study are consistent with literature reports on gene expression variation in human populations.22–27
Gene expression variation within and between ethnic populations
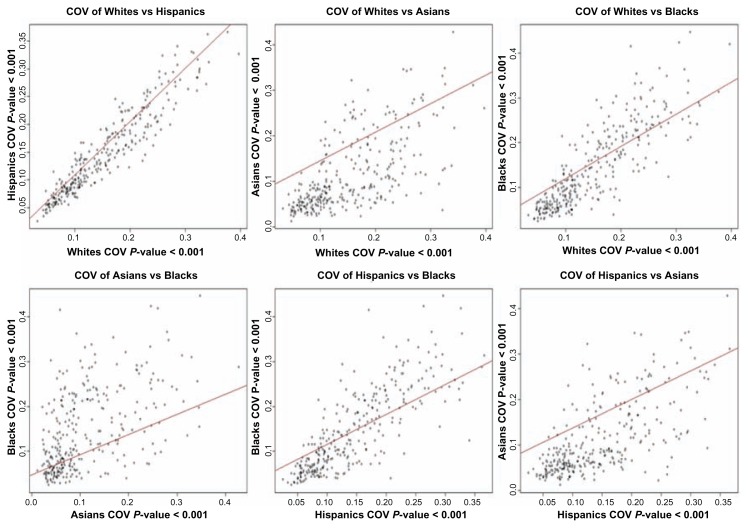
The second objective of this study was to investigate and characterize the patterns of gene expression variation within and between populations. The rationale is that knowledge of variation in gene expression levels within and between patient populations may be required for stratifying patients and setting ethnicity-specific reference intervals in a clinical setting. We investigated within population variation by computing the CV for individual genes, as explained in the methods section. The results showing the correlation between CV between pairs of populations for the 300 genes are presented in Figure 2. Gene expression variation between populations was assessed by reciprocal regression on the values of CV from each population using a regression model, which is analogous to computing a correlation between CVs from two populations. CVs for individual genes derived from gene expression data within each ethnic population are presented in Table B, provided as supplementary data.
Figure 2.
Pair-wise population scatter plot diagrams showing the coefficient of variation for expression values on a log2 scale across the 300 genes significantly (P < 0.001) differentially expressed genes between populations.
Between White and Hispanic populations, most of the genes exhibit similar levels of within population variability. Pairwise comparison of the coefficients of gene expression variation between all the four populations studied confirmed this trend (Fig. 2). Gene expression variability between Whites and Blacks, and between Hispanics and Blacks, also tended to be similar (Fig. 2). The strong correlations in within population expression variation between these populations suggests that either expression variability of most genes is subject to similar levels of constraints in these populations or could partially be explained by the admixing of the populations. The largest differences in within population variation were between Asians and Blacks, Asians and Whites, and between Hispanics and Asians (Fig. 2). In general, the within population variation was larger than the between population variation. The results found in this study are consistent with literature reports on human populations.23–27 The similarity in patterns of gene expression profiles between Whites and Blacks is consistent with an earlier report which found that between Whites and Yuruba populations, most of the human genes exhibited a similar level of within population variability.25–27 These data suggest that studies seeking to stratify B-ALL patients must consider the within population variation in gene expression and the effects of population structure. In general, the results are in agreement with the neutral theory of evolution, which states that the variation between populations is a positive function of within population variation,37 suggesting that much of the significant variation between ethnic populations may represent random genetic drift.22
The clinical significance of these results is that in searching for clinically actionable biomarkers among a group of differentially expressed genes, it is important to consider the biological variation due to inter-individual variation that are due to a clinically relevant phenotype or response to a particular treatment, rather than race and or ethnicity, which are social constructs. Because gene expression and allelic variation tend to be shared widely among ethnic populations, race is likely to be an inaccurate predictor of response to treatment. It would be far preferable to test directly the responsible alleles in affected individuals.
Functional relationships of identified genes
Identifying differentially expressed genes and assessing variation within and between ethnic populations may provide a limited view of the quantitative details of gene expression variation. The third objective of this study was, therefore, to characterize the global patterns of gene expression profiles in the populations under study to identify genes and patients with similar patterns of expression profiles. To begin to address this problem, we performed pattern recognition analysis using hierarchical clustering in order to group the genes and patients according to similarity in patterns of gene expression profiles. Clustering of all the significantly differentially expressed genes using samples from all the ethnic populations failed to accurately classify genes and patients. This was caused by considerable overlap in patterns of gene expression profiles in populations under study (results not presented).
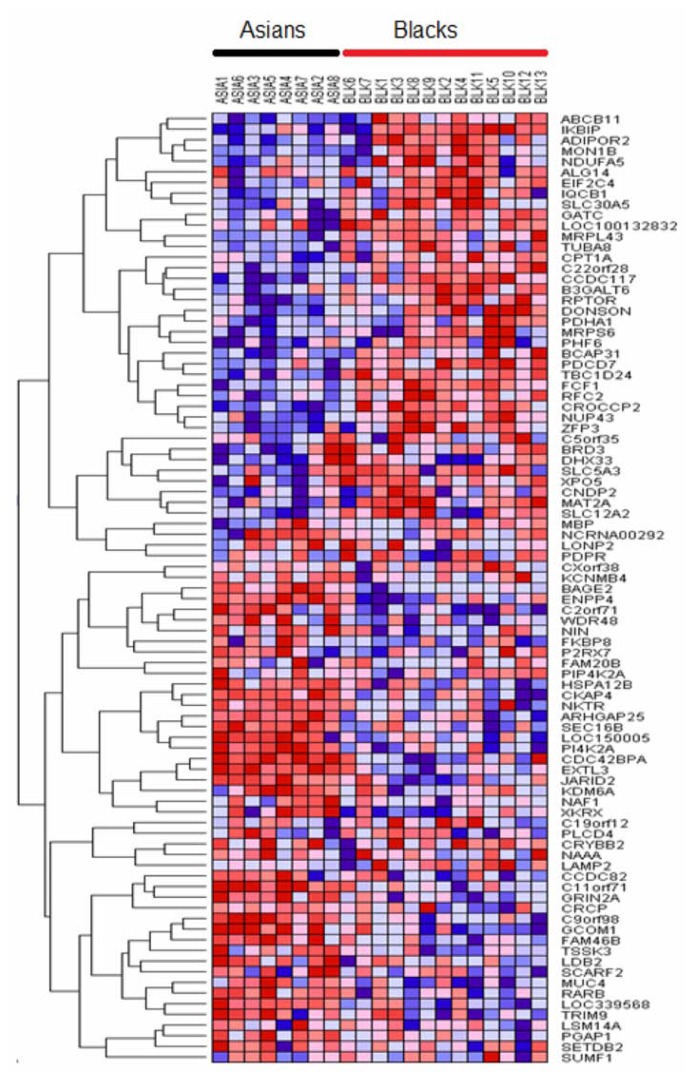
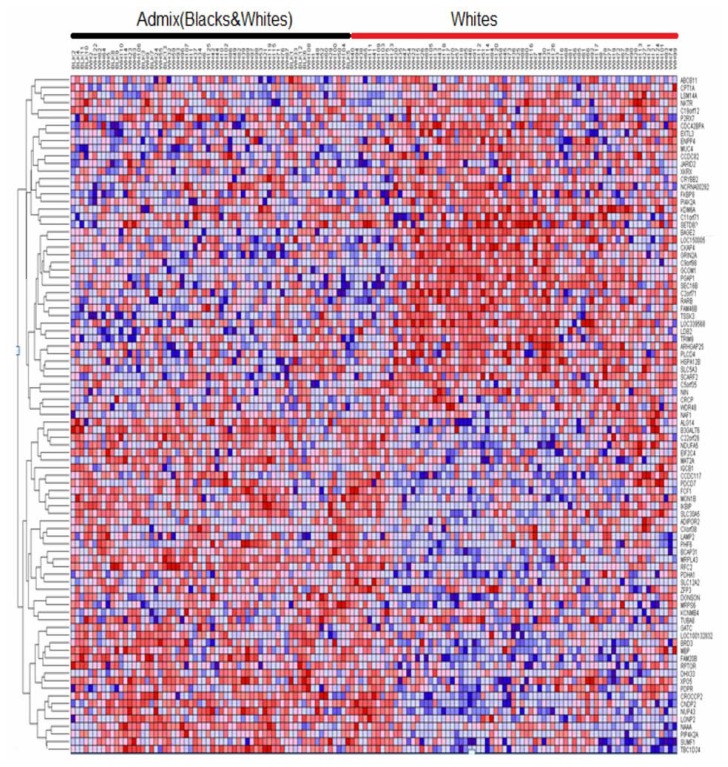
We therefore performed sequential clustering, clustering two populations at a time using the most highly significantly differentially expressed sets of genes. Interestingly, this analysis identified significantly differentially expressed up and down regulated genes with similar patterns of expression profiles. The results showing patterns of gene expression profiles between the ethnic populations under study are presented in Figure 3 through Figure 6. Figure 3 shows the patterns of gene expression profiles for Asian and Black populations. The results showing patterns of gene expression for Asians and Whites-Hispanics are presented in Figure 4. Figure 5 presents the results for Blacks and Whites-Hispanics. The results showing patterns of gene expression profiles between Blacks and Whites are presented in Figure 6. In all four figures, genes are represented in rows and B-ALL patients in columns. We also identified clusters of genes with similar patterns of gene expression profiles which distinguished one racial/ethnic population from another. However, there was considerable variation and overlap in patterns of gene expression profiles. Most of the variability and differences in patterns of gene expression profiles were among individual patients (Figs. 3–6). This demonstrates that genetic variation tends to be shared widely among populations, and that molecular perturbation in different ethnic populations is likely subjected to the same regulatory mechanisms.
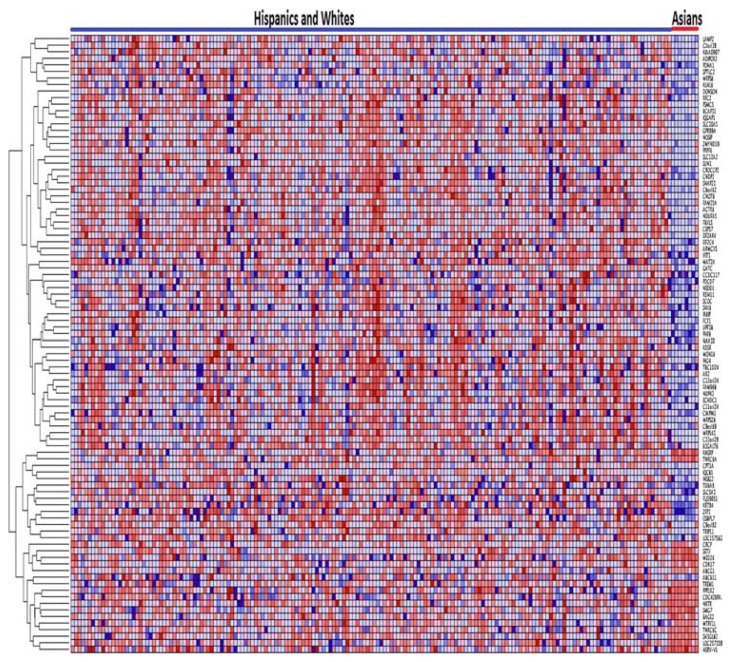
Figure 3.
Variation in patterns of gene expression profiles between Asians and Blacks in pediatric patient populations. The results were obtained by unsupervised analysis using hierarchical clustering. Genes are represented in rows and patients in columns. Red indicates up regulation and blue indicates down regulation.
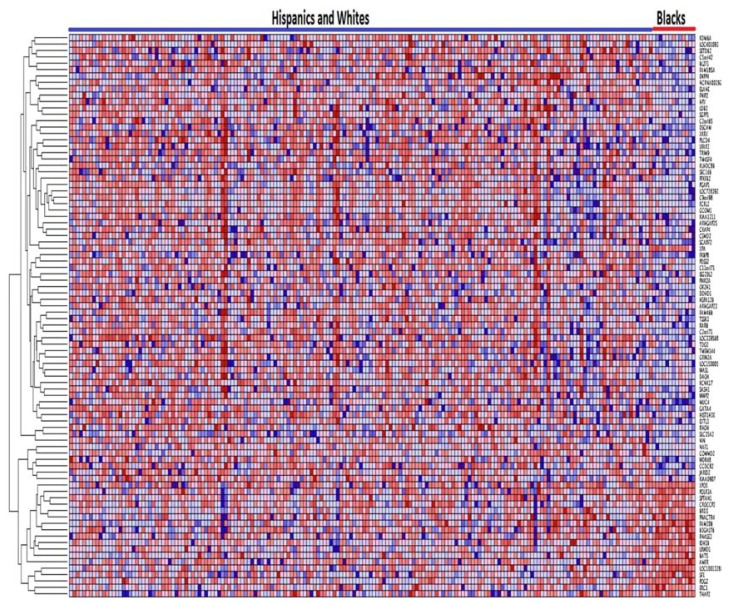
Figure 6.
Variation in patterns of gene expression profiles between Blacks and Whites in pediatric patient populations. The results were obtained by unsupervised analysis using clustering. Genes are represented in rows, and patients in columns. Significant admixing in patterns of gene expression observed. Red indicates up regulation and blue indicates down regulation.
Figure 4.
Variation in patterns of gene expression profiles between Asian and White-Hispanic in pediatric patient populations. The results were obtained by unsupervised analysis using clustering. Genes are represented in rows and patients in columns. Significant overlap in patterns of expression within the Hispanic population was observed. Red indicates up regulation and blue indicates down regulation.
Figure 5.
Variation in patterns of gene expression profiles between Blacks and White-Hispanics in pediatric patient populations. The results were obtained by unsupervised analysis using clustering. Genes are represented in rows and patients in columns. By a large measure, Whites clustered separately. Red indicates up regulation and blue indicates down regulation.
The most clearly distinguishable patterns of gene expression profiles were found between Asians and Blacks (Fig. 3), between Asians and White-Hispanics (Fig. 4), and between Blacks and Whites-Hispanic (Fig. 5). However, as expected, there were significant overlaps between the Hispanics and Blacks, Whites and Blacks, and between Whites and Hispanics (results not presented). This was not surprising given the high level of admixture in these populations. For these reasons we did not present the results of clustering based on Whites and Asians and Hispanics and Asians separately, but rather classified Whites and Hispanics as one race/ethnicity against the Asians (Fig. 4). The significant overlap in patterns of gene expression between and among these racial/ethnic populations demonstrates that race or ethnic populations are not discrete types. The substantial overlap in patterns of gene expression suggests that B-ALL patients may not be accurately stratified or classified strictly on the basis of race or ethnicity alone, and that other criteria such as outcomes may be more useful.
In order to put the observed overlap in patterns of gene expression profiles in context, we examined the literature on human populations. The significant overlap in patterns of gene expression profiles within and between the ethnic populations can be explained in part by the admixing of the populations.35,36 Asians have been less influenced by admixture and hence they tend to cluster separately from other ethnic populations.38 There has been significant gene flow between Whites and Blacks,38 and several studies have estimated the proportion of White admixture in BLakcs to be approximately 17%, ranging regionally from 12% to 23%.39 This is consistent with the results in this study (Fig. 6).
Indeed, with such proportions, admixed patients will tend to exhibit patterns of gene expression profiles that are similar to both ethnic populations (Fig. 6). The most complex patterns of gene expression profiles were observed between Hispanics and Blacks and between Hispanics and Whites. Although we initially considered Hispanics a separate population, the U.S. Census does not. Hispanics are typically a mix of Native American, White, and African/African American with the relative proportion varying regionally.38 Southwest Hispanics, who are primarily Mexican-American, appear to be largely White and Native-American; recent admixture estimates are 39% Native American, 58% White and 3% Black.40 By contrast, East Coast Hispanics are largely Caribbean in origin, and have a greater proportion of African admixture.41 Therefore, depending on geography, self-identified Hispanics could aggregate genetically with Blacks or with Whites (Fig. 5), with Native Americans, or form their own cluster. This is consistent with the results in this study. In this study we did not follow regional classification of the Hispanic population. However, the results in Figures 4 and 5 clearly show this admixing pattern of the Hispanic population with the Whites. Interestingly, although our investigations used gene expression profiles which are intermediate phenotypes, the results are consistent with findings in an earlier report involving nearly 4000 single-nucleotide polymorphisms (SNPs) mapped to 313 genes.38,42 These authors found distinct clusters for Whites, Blacks, and Asian; the Hispanic Americans did not form a separate cluster but were either grouped with Whites or not easily classified. This again is consistent with the results found in this study as well as the rationale of our decision to combine gene expression data on Whites and Hispanics in subsequent analysis.
To ascertain that the identified genes are involved in similar biological processes, molecular functions, and cellular components, we performed GO analysis as described in the methods section. All the 300 genes exhibiting significant differences in expression levels between ethnic populations were subjected to GO analysis. GO analysis revealed that the genes are functionally related and involved in multiple overlapping, but similar, biological processes and cellular components therefore confirming our hypothesis. A summary of functional information based on GO analysis is provided in Table C as supplementary material.
Gene network and pathway analysis
To gain insights into the broader context in which the significantly differentially expressed genes operate, we performed network modeling in order to test whether the genes are overrepresented in the networks and biological pathways. Our working hypothesis was that genes that are significantly differentially expressed between ethnic populations, and exhibiting significant variation in patterns of expression within and between populations, interact with one another, thereby affecting entire network states and biological pathways, and in turn affect the severity of the disease in different ethnic populations. To test this hypothesis we mapped the 89 most highly significantly differentially genes (P < 10−4) identified after each comparison between populations onto the networks and pathways as implemented in the IPA. The clinical significance of the identified networks and pathways was evaluated using information on published reports on pediatric B-ALL as explained in subsequent paragraphs in this section.
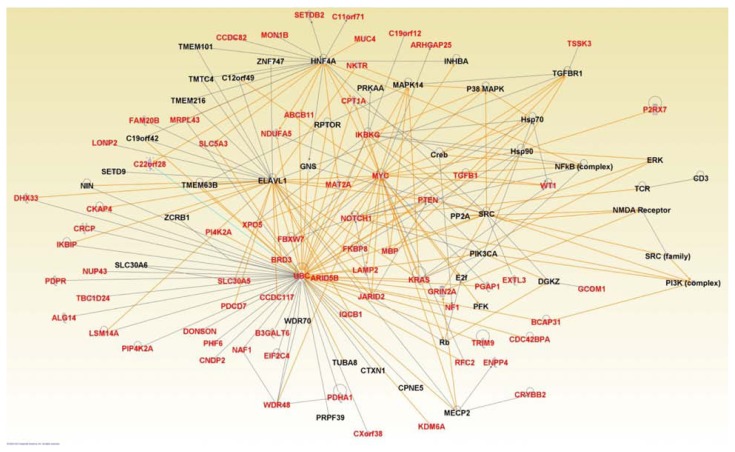
Network analysis revealed 5 multi-gene networks with IPA scores ranging from 17 to 61. In order to streamline the results and make them amenable to proper interpretation, we consolidated the results of the five networks into one large network by using the merge and design modules as implemented in IPA. The consolidated network enriched for the most highly significantly differentially expressed genes (in red font) between racial/ethnic populations is presented in Figure 7. We identified many genes with overlapping functions that were interacting in the network (Fig. 7). It is worth noting that not all the genes differentially expressed between populations are represented in the network because we pruned the network to remove those with spurious interactions as well as those not found to be interacting or functionally related with other genes. This action was taken to ensure the reliability of the identified network. In addition to differentially expressed genes enriching the network, we identified a set of novel genes (Fig. 7, in black font), which could not be identified through differential expression analysis. The results confirmed our hypothesis that differentially expressed genes between populations are functionally related and interact with each and their downstream targets in gene regulatory networks. The network contained genes involved in many biological processes including cancer, hematological and immunological diseases, molecular transport, neurological, carbohydrate metabolism, lipid metabolism, cell morphology, cell cycle, cellular movement, and tissue development.
Figure 7.
Graphical representation of a consolidated gene regulatory network enriched for genes significantly differentially expressed between ethnic populations (red) and novel functionally related genes (black). Genes in red font represent the significantly differentially expressed genes identified in this study. Genes in black font represent novel genes which are functionally related with differentially expressed genes. Solid lines indicate direct interactions and functional relationships.
Among the genes identified in the consolidated network (Fig. 7), many genes were implicated in pediatric ALL including UBC, NOTCH1, FBXW7, PIK3CA, PTEN, KRAS, and WT1. Although the UBC did not reach the threshold of differential expression between racial/ethnic populations in this study, the human CDC 34 UBC protein has been found to be expressed at 3–4 fold higher level in pediatric T-cells than in pre-B-cell acute lymphoblastic leukemia in two independent patient groups,43 suggesting that the human UBC could serve as a potential therapeutic target. NOTCH1 signaling has been shown to be responsible for the anti-leukemic effect of histone deacetylases inhibitors (HDACis) in B-ALL cells demonstrating its potential as a therapeutic target.44 The FBXW7 gene is a tumor suppressor which encodes a subunit of an ubiquitin protein ligase that targets numerous oncoproteins for proteasomal degradation.45 Regulation of FBXW7 in B-ALL is mediated by the miR-27a, which is involved in regulating cell cycle progression.45PTEN is a tumor suppressor gene which has been shown to reverse MDM2-mediated chemotherapy resistance by interacting with p53 in ALL cells.46PTEN has also been shown to suppress B-ALL development through downstream regulation of AKT1.47WT1 is expressed in majority of ALL.48 For example, in a study involving 14 B-ALL patients, WT1 was detected in 12 (86%) patients.48 In the same study involving 31 T-ALL patients, WT1 was detected in 21 (74%) of the patients.48 Interestingly, in patients diagnosed with ALL, WT1 has been used as a target for the detection of minimal residue diseases (MRD).49 Other genes implicated in leukemia found to be interacting in the network included BRD3, TGFB1, MYC, and RAS.2 In addition, the network contained the NF1 and AKT genes which have also been implicated in pediatric ALL.50 For example, activation of AKT is associated with poor prognosis and chemotherapeutic resistance in pediatric B-ALL.50
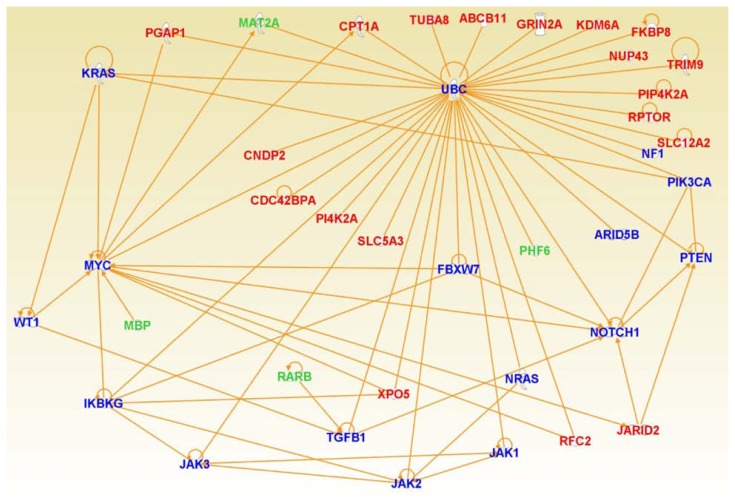
In silico confirmation using differentially expressed genes and genes implicated in ALL, revealed that the two sets of genes interact with each other in gene regulatory networks (Fig. 8). Interestingly, the differentially expressed genes were found to interact with the ARID5B gene (Fig. 8) which contains genetic polymorphism associated with B-ALL.51 The genetic polymorphisms in the ARID5B gene have been known to contribute to racial and ethnic disparities in incidence and treatment outcome in B-ALL.52 The ARID5B genetic variants have also been linked to inter-patient variability in the anti-leukemic drug (methotrexate) metabolism.52 This confirms our hypothesis that genes which are significantly differentially expressed between ethnic populations interact with one another and affect entire network states and biological pathways, and in turn affect the severity of the disease or response to treatment in different ethnic populations. Of particular interest is that these networks and pathways are shared between populations, suggesting that gene regulation is subject to the same constraints in different populations.
Figure 8.
Graphical representation of a consensus gene regulatory network enriched for genes significantly differentially expressed between ethnic populations (red) and genes from the literature that have been implicated B-ALL (blue). Genes in green font represent the significantly differentially expressed genes identified in this study that have been directly implicated in B-ALL. Solid lines indicate direct interactions and functional relationships.
To discern the biological meanings of the differentially expressed genes mapped to the networks, we evaluated them using GO information on molecular and cellular function as implemented in the IPA. We identified many genes involved in multiple overlapping functions and multiple biological processes. Out of the 89 most significantly differentially expressed genes evaluated, 24 genes were significantly associated with cellular development (P = 1.22E-07–2.52E-03), 23 genes were significantly associated with cellular growth proliferation (P = 2.22E-07–2.52E-03), 28 genes were significantly associated with cell death and survival (P = 1.39E-06–2.46E-03), 18 genes were significantly associated with cell cycle (P = 1.41E-06–2.41E-03), and 13 genes were significantly associated with gene expression (P = 1.52E-06–1.76E-03).
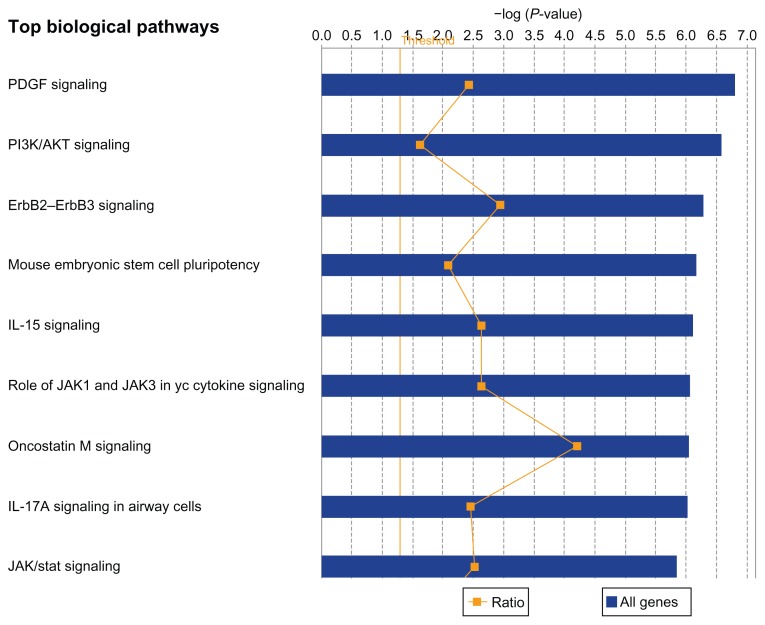
To further gain insights on the broader context in which differentially expressed genes and genes contributing to within and between population variation operate, pathway analysis were performed. The pathways were identified by mapping the genes onto Ingenuity pathways as implemented in the IPA System. The results of pathway analysis are presented in Figure 9. Interestingly, pathway predictions revealed significant overrepresentation of the genes in many canonical pathways. The top five pathways included the PDGF (P = 1.58E-07), PI3/AKT (P = 2.7E-07), ERBB2–ERBB3 (P = 5.28E-07), and IL-15 (P = 7.95E-07) signaling pathways and the mouse embryonic stem cell pluripotent (P = 6.72E-07) (Fig. 9). Other significant pathways included the role of JAK1 and JAK3 in γc cytokines signaling, Oncostatin M signaling, IL-17A signaling, and JAK/STAT signaling (Fig. 9). To assess the clinical significance of the pathways as potential therapeutic targets in B-ALL, we mined the literature associating these pathways to pediatric B-ALL. Literature findings revealed that all the top five pathways play important roles in leukemogenesis.53–58 The platelet-derived growth factor (PDGF) regulates clonal proliferation of malignant pre-B cell lines.53 The phosphatidylinositiol 3-kinase (PI3K) AKT, the mammalian target of rapamycin (mTOR) signaling pathway (PI3K/AKT/mTOR) is abnormally activated in childhood acute lymphoblastic leukemia.54 Most commonly, this abnormal activation occurs as a consequence of constitutive activation of AKT.54 Activation of AKT is associated with poor prognosis and chemotherapeutic resistance in pediatric B-ALL,55 which provides a compelling rationale for therapeutically targeting this pathway, particularly in the 20% of the patient population who are resistant to treatment. The ERBB2, also known as Her2/neu, has been implicated in ALL and is considered a potential therapeutic target.56 Genetic variants mapped to IL-15 have been associated with minimal residual disease (MRD).57 MRD has been integrated into risk stratification58,59 and MRD assays provide a direct assessment of early treatment response and are associated with final treatment outcome.60–63 Thus the IL-15 pathway could serve as a potential therapeutic target for early interventions. In summary, the results confirmed our hypothesis that gene expression differences between ethnic populations are regulated by a wide variety of pathways, and that these pathway are shared among ethnic populations.
Figure 9.
Graphical representation of the top most highly significant canonical pathways enriched for differentially expressed genes between ethnic populations. The ratio represents the number of genes that map to the canonical pathway relative to (blue) the number of genes in the canonical pathway. The threshold (thin straight vertical orange line) is the P-value on a log scale indicating the probability with which a set of genes were correctly assigned to a particular canonical pathway after correcting for multiple hypothesis testing.
Discussion
This study was conducted to determine whether gene expression levels differ between ethnic populations in order to characterize variation in patterns of gene expression profiles, and to identify gene regulatory networks and biological pathways that are dysregulated in different populations (Whites, Hispanics, Blacks, and Asians) diagnosed with B-ALL. To our knowledge this is the first study to compare the gene expression levels and delineate the patterns of gene expression variation within and between ethnic populations in pediatric B-ALL. The analysis revealed moderate but appreciable differences in gene expression levels between populations.
There was, however, considerable overlap in both differential expression levels and patterns of gene expression profiles among the four populations under study. Analysis of patterns of gene expression profiles revealed clusters of co-expressed genes. These clusters were correlated with race and ethnicity, but the correlations were imperfect as variation in patterns of gene expression tended to be distributed in a continuous, overlapping fashion among populations. These results indicate that in some cases, race or ethnicity may provide some insights about the disease state. Nevertheless, the study demonstrates that use of race or ethnicity alone as a surrogate for assessing the molecular basis of health disparities or classifying patients may be inaccurate because of overlap and admixing in the patterns of gene expression.
This study focused on using gene expression data to determine the difference in molecular perturbation between populations. However, it is worth noting that gene expression reflects the disease state and may be influenced by both genetic and non-genetic factors such as environment and socio-economic status. These factors should be taken into account when making decisions at the point of care or attempting to use race and/or ethnicity as a surrogate for stratification of patients or assessing outcomes. There is biological plausibility that the observed inconsistencies in patterns of gene expression profiles are partially attributable to the genetic heterogeneity inherent in B-ALL.11 Due to the small sample sizes for some ethnic populations, notably Asians and Blacks, we did not stratify B-ALL according to the subtypes to mitigate heterogeneity. It is also conceivable that sampling errors may partially have contributed to the observed outcome.
Studies of gene expression on pediatric B-ALL have been reported.12–16 The main difference between this study and reported studies, is that this study focused on characterizing variation in patterns of gene expression within and between populations. The clinical and translation significance of this study lies in the fact that responses to medical therapies, such as drug treatment, are often compared among populations that are divided according to traditional racial and ethnic groups. Therefore, delineating the patterns of gene expression profiles within and between these traditional racial and ethnic groups would provide insights about the use of race and/or ethnicity as surrogates for patient classification, assessment of treatment outcomes, and as a yardstick for assessing the molecular basis of health disparities. The overlap and shared patterns in gene expression profiles observed in this study have important implications for our understanding of population differences and similarities, and bears on critical biomedical issues including use of race as a surrogate for assessing response to treatment, outcome reporting, and assessing health disparities. For example, as demonstrated in this study, if the patterns of gene expression are similar and shared between populations, use of an individual’s race or ethnicity affiliation alone would be potentially a faulty indicator or classifier of the presence or absence of a biomarker related to diagnosis or response to treatment.
Recently, genetic polymorphisms conferring race and ethnic population-specific risk in B-ALL incidence and outcomes were found in the ARID5B gene using genome-wide association studies (GWAS).51,52 In this study we did not investigate the distribution of genetic variants or alleles among populations. However, pathway analysis including the ARID5B gene in network analysis revealed the broader context in which the SNPs in this gene may operate. To the extent that gene expression is regulated by genetic variants and allelic variation tends to be shared widely among populations, these results provide functional bridges between GWAS findings and the disease state. Importantly, the results in this study are consistent with previous studies in human populations that have analyzed both genetic variants and gene expression.64,65 Ideally, it would be far preferable to test directly the responsible alleles in affected individuals.42 Assessment of allelic variation was beyond the scope of this study, but is the hope for future studies as most of the genes and rare variants which contribute to B-ALL remain to be identified. In addition, non-genetic factors such as environmental factors and socio-economic status nearly always have an important and sometimes predominant role in susceptibility and outcomes.6,7 With the launch of the TARGET and the Pediatric Cancer Genome Projects,17,18 progress is being made in identifying and cataloguing genetic variants and mutations that underlie B-ALL. Rapid development in new technologies, such as clinical sequencing technologies, will provide efficient and far-ranging genetic assays for use at the point of care.
The results of this study show small but appreciable differences in gene expression between the populations under study. However, there are significant limitations to the study which are readily acknowledged, including lack of data on cancer-free controls and clinical information along with small sample sizes for the Asian and Black populations. The use of cancer free-controls would have enabled us to identify population-specific genes. However, addressing that issue would have required having cancer-free controls for each of the four populations under study, which was not feasible. The use of clinical information would have allowed us to assess gene expression using other variables in addition to race/ethnicity, an important strategy given the limitations of race/ethnicity as discussed in this report in the preceding sections. However, clinical information was not available on the data set used in this study. The use of small sample sizes for some populations, particularly the Asians and Blacks, could lead to sampling errors and some of the observed spuriousness in the results. This is an issue beyond the scope of this report, however. Lastly, the use of self-identified race/ethnicity could be prone to errors that could influence the results. Given these acknowledged limitations, we view the results reported in this study as exploratory and should be interpreted conservatively.
In conclusion, the analysis shows that gene expression profiling for the discovery of genetic markers holds great promise for understanding the molecular basis of health disparities in pediatric B-ALL. The analysis reveals that molecular perturbation in B-ALL patients tends to be shared widely, vary significantly, and substantially overlap within and between populations. The data further demonstrates that populations tend to have shared gene regulatory networks and biological pathways. We recommend that because genetic assessment alone will never be a panacea, there is a need to consider both genetic and non-genetic factors in clinical care.
Supplementary Tables
(Supplementary) Estimates of P-values and FDR obtained from comparing gene expression between and among populations for the 300 most highly significantly differentially expressed genes.
(Supplementary) Estimates of coefficients of variation (CV) for individual populations for the 300 most highly significantly differentially expressed genes between the four populations. Note: CV are expressed as ratios.
(Supplementary) Results of GO analysis for all the 300 most highly significantly differentially expressed genes between ethnic populations.
Acknowledgements
The authors wish to thank the University of Mississippi Cancer Institute and the Children’s Cancer Center for providing funding support for the project.
Footnotes
Author Contributions
Conceived and designed the experiments: CH, GM, LM. Analyzed the data: CH, TK, DR. Wrote the first draft of the manuscript: CH. Contributed to the writing of the manuscript: LY-G, TK, VV, GM, LM, DR. Agree with manuscript results and conclusions: CH, TK, LY-G, VV, GM, LM, DR. All authors were involved and jointly developed the structure and arguments for the paper. Made critical revisions and approved final version CH, TK, LM, GM. All authors reviewed and approved of the final manuscript.
Competing Interests
Authors disclose no potential conflicts of interest.
Disclosures and Ethics
As a requirement of publication the authors have provided signed confirmation of their compliance with ethical and legal obligations including but not limited to compliance with ICMJE authorship and competing interests guidelines, that the article is neither under consideration for publication nor published elsewhere, of their compliance with legal and ethical guidelines concerning human and animal research participants (if applicable), and that permission has been obtained for reproduction of any copyrighted material. This article was subject to blind, independent, expert peer review. The reviewers reported no competing interests.
References
- 1.Pui CH, Robinson LL, Look AT. Acute lymphoblastic leukemia. Lancet. 2008;371(9617):1030–43. doi: 10.1016/S0140-6736(08)60457-2. [DOI] [PubMed] [Google Scholar]
- 2.Mullighan CG. The molecular genetic makeup of acute lymphoblastic leukemia. Hematology. 2012:389–95. doi: 10.1182/asheducation-2012.1.389. [DOI] [PubMed] [Google Scholar]
- 3.Holleman A, Cheok MH, den Boer M, et al. Gene-expression patterns in drug-resistant acute lymphoblastic leukemia cell and response to treatment. New Eng J Med. 2004;351(6):533–42. doi: 10.1056/NEJMoa033513. [DOI] [PubMed] [Google Scholar]
- 4.Hunger SP, Lu X, Devidas M, et al. Improved survival for children and adolescents with acute lymphoblastic leukemia between 1990 and 2005: a report from the children’s oncology group. J Clin Oncol. 2012;30(14):1663–9. doi: 10.1200/JCO.2011.37.8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ries LA, Kosary CL, Hankey BF, et al. SEER Cancer Statistics Review, 1973–1996. Bathesda, MD: National Cancer Institute; 1999. Also available online. [Google Scholar]
- 6.Smith MA, Ries LA, Gurney JG, et al. Leukemia. In: Ries LA, Smith MA, Gurney, et al., editors. Cancer Incidences and Survival Among Children and Adolescents: United States SEER Program 1975–1995. Bathesda, MD: National Cancer Institute; 1999. pp. 17–34. SEER Program, NIH Pub. No. 99-4649. [Google Scholar]
- 7.Dores GM, Devesa SS, Curtis RE, et al. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood. 2012;119(1):34–43. doi: 10.1182/blood-2011-04-347872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pui C-H, Sandlund JT, Pei D, et al. Results of therapy for acute lymphoblastic leukemia in black and white children. JAMA. 2003;290(15):2001–7. doi: 10.1001/jama.290.15.2001. [DOI] [PubMed] [Google Scholar]
- 9.Kadam-Lottick NS, Ness KK, Bhatia S, Gurney JG. Survival variability by race and ethnic in childhood acute lymphoblastic leukemia. JAMA. 2003;290(15):2008–14. doi: 10.1001/jama.290.15.2008. [DOI] [PubMed] [Google Scholar]
- 10.Schultz KR, Pullen DJ, Sather HN, et al. Risk-and response-based classification of childhood leukemia B-precursor acute lymphoblastic leukemia: a combined analysis of prognostic markers from the Pediatric Oncology Group (POG) and Children’s Cancer Group (CCG) Blood. 2007;109(3):926–35. doi: 10.1182/blood-2006-01-024729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mullighan CG. Molecular genetics of B-precursor acute lymphoblastic leukemia. J Clin Invest. 2012;122(10):3407–15. doi: 10.1172/JCI61203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeoh E-J, Ross ME, Shurtleff SA, et al. Classification of, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1(2):133–43. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- 13.Ross ME, Zhou X, Song G, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102(8):2951–9. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 14.Yang JJ, Cheng C, Devidas M, et al. Genome-wide association study identifies germline polymorphisms associated with relapse of childhood acute lymphoblastic leukemia. Blood. 2012;120(20):4197–204. doi: 10.1182/blood-2012-07-440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harvey RC, Mullighan CG, Wang X, et al. Identification of novel cluster groups in pediatric high-risk B-precursor acute lymphoblastic leukemia gene expression profiling: Correlations with genome-wide copy number alterations, clinical characteristics, and outcome. Blood. 2010;116(23):4874–84. doi: 10.1182/blood-2009-08-239681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang H, Chen I-M, Wilson CS, et al. Gene expression classifiers for relapse-free survival and minimum residual disease improve risk classification and outcome prediction in pediatric B-precursor acute lymphoblastic leukemia. Blood. 2010;115(7):1394–405. doi: 10.1182/blood-2009-05-218560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loh ML, Zhang J, Harvey RC, et al. Tyrosine kinome sequencing of pediatric acute lymphoblastic leukemia: a report from the Children’s Oncology Group TARGET Project. Blood. 2013;121(3):485–8. doi: 10.1182/blood-2012-04-422691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Downing JR, Wilson RK, Zhang J, et al. The Pediatric Cancer Genome Project. Nat Genet. 2012;44(6):619–22. doi: 10.1038/ng.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu CC, Rao DC, Stormo G, Hicks C, Province MA. Role of gene expression microarray analysis in finding complex disease genes. Genet Epidemiol. 2002;23(1):37–56. doi: 10.1002/gepi.220. [DOI] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochberg Y. Controlling the false discovery rat: a practical and powerful approach to multiple testing. J Royal Stat Soc Series B. 1995;57(1):289–300. [Google Scholar]
- 21.Radmacher MD, McShane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9(3):505–11. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 22.Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32(2):261–6. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 23.Kaern M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6(6):451–64. doi: 10.1038/nrg1615. [DOI] [PubMed] [Google Scholar]
- 24.Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309(5743):2010–3. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Liu Y, Kim T, Min R, Zhang Z. Gene expression variability within and between human populations and implications toward disease susceptibility. PLoS Comput Biol. 2010;6(8):e1000910. doi: 10.1371/journal.pcbi.1000910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storey JD, Madeoy J, Strout JL, Wurfel M, Ronald J, Akey JM. Gene expression variation within and among human populations. Am J Hum Genet. 2007;80(3):502–9. doi: 10.1086/512017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fan HPY, Liao CD, Fu BY, Lam LC, Tang NL. Interindividual and interethnic variation in genomewide gene expression: Insights into the biological variation of gene expression and clinical implications. Clin Chem. 2009;55(4):774–85. doi: 10.1373/clinchem.2008.119107. [DOI] [PubMed] [Google Scholar]
- 28.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 29.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Nat Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11(8):1425–33. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ingenuity (IPA) System. Ingenuity Inc; [Accessibility verified Jun 8, 2013]. Available at: http://www.ingenuity.com/ [Google Scholar]
- 32.Mullighan CG, Goorha S, Radke I, et al. Genome-wide analysis of genetic alterations in acute lymphblastic leukemia. Nature. 2007;446(7137):758–64. doi: 10.1038/nature05690. [DOI] [PubMed] [Google Scholar]
- 33.Zhang J, Ding L, Holmfeldt L, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukemia. Nature. 2012;481(7380):157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahoney DH, Jr, Fernbach DJ, Glaze DG, Cohen SR. Elevated myelin basic protein levels in the cerebrospinal fluid of children with acute lymphoblastic leukemia. J Clin Onco. 1984;2(1):58–61. doi: 10.1200/JCO.1984.2.1.58. [DOI] [PubMed] [Google Scholar]
- 35.Alves I, Hanulova AS, Foll M, Excoffier L. Genomic data reveal a complex making of humans. PLoS Genetics. 2012;8(7):e1002837. doi: 10.1371/journal.pgen.1002837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shriner D, Adeyemo A, Ramos E, Chen G, Rotimi CN. Mapping of disease-associated variants in admixed populations. Genome Biol. 2011;12(5):223. doi: 10.1186/gb-2011-12-5-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nei M. Molecular Evolutionally Genetics. Columbia Univ Press; New York: 1987. [Google Scholar]
- 38.Risch N, Burchard E, Ziv E, Tang H. Categorization of humans in biomedical research: genes, race and disease. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-comment2007. comment2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parra EJ, Marcini A, Akey J, et al. Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63(6):1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tseng M, Williams RC, Maurer KR, Schanfield MS, Knowler WC, Everhart JE. Genetic admixture and gallbladder disease in Mexican Americans. Am J Phys Anthropol. 1998;106(3):361–71. doi: 10.1002/(SICI)1096-8644(199807)106:3<361::AID-AJPA8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 41.Hanis CL, Hewett-Emmett D, Bertin TK, Schull WJ. Origins of U.S. Hispanics. Implications for diabetes. Diabetes Care. 1991;14(7):618–27. doi: 10.2337/diacare.14.7.618. [DOI] [PubMed] [Google Scholar]
- 42.Stephens JC, Schneider JA, Tanguay DA, et al. Haplotype variation and linkage disequilibrium in 313 human genes. Science. 2001;293(5529):489–93. doi: 10.1126/science.1059431. [DOI] [PubMed] [Google Scholar]
- 43.Eliseeva E, Pati D, Diccinanni MB, et al. Expression and localization of the CDC34 ubiquitin-conjugating enzyme in pediatric acute lymphoblastic leukemia. Cell Growth Differ. 2001;12(8):427–33. [PubMed] [Google Scholar]
- 44.Shao N, Ma D, Wang J, Lu T, Guo Y, Ji C. Notch1 signaling is irresponsible for the anti-leukemic effect of HDAG is in B-ALL Nalm-6 cells. Ann Hematol. 2013;92(1):33–9. doi: 10.1007/s00277-012-1561-8. [DOI] [PubMed] [Google Scholar]
- 45.Lerner M, Lundgren J, Akhoondi S, et al. MiRNA-27a controls FBW7/hCDC4-depenedent cyclin E degradation and cell cycle progression. Cell Cycle. 2011;10(13):2172–83. doi: 10.4161/cc.10.13.16248. [DOI] [PubMed] [Google Scholar]
- 46.Zhou M, Gu L, Findley HW, Jiang R, Woods WG. PTEN reverses MDM2-mediated chemotherapy resistance by interacting with P53 in acute lymphoblastic leukemia cells. Cancer Res. 2003;6(19):6357–62. [PubMed] [Google Scholar]
- 47.Peng C, Chen Y, Yang Z, et al. PTEN is a tumor suppressor in CML stem cells and BCR-ABL-induced leukemias in mice. Blood. 2010;115(3):626–35. doi: 10.1182/blood-2009-06-228130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Menssen HD, Renkl HJ, Rodeck U, et al. Presence of Wilms’ tumor gene (WT1) transcripts and the WT1 nuclear protein in the majority of human acute leukemias. Leukemia. 1995;9(6):1060–7. [PubMed] [Google Scholar]
- 49.Kerst G, Bergold N, Gieseke F, et al. WT1 protein expression in childhood leukemia. Am J Hematol. 2008;83(5):382–6. doi: 10.1002/ajh.21123. [DOI] [PubMed] [Google Scholar]
- 50.Morishita N, Tsukahara H, Chayama K, et al. Activation of AKT is associated with poor prognosis and chemotherapeutic resistance in pediatric B-precursor acute lymphoblastic leukemia. Pediatr Blood Cancer. 2012;59(1):83–9. doi: 10.1002/pbc.24034. [DOI] [PubMed] [Google Scholar]
- 51.Healy J, Richer C, Bourgey M, Kritikou EA, Sinnett D. Replication analysis confirms the association of ARID5B with childhood B-cell acute lymphoblastic leukemia. Haematologica. 2010;95(9):1608–11. doi: 10.3324/haematol.2010.022459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu H, Cheng C, Devidas M, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol. 2012;30(7):751–7. doi: 10.1200/JCO.2011.38.0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ho CL, Hsu LF, Phyliky RL, Li CY. Autocrine expression of platelet-derived growth factor B in B cell chronic lymphoblastic leukemia. Acta Haematol. 2005;114(3):133–40. doi: 10.1159/000087886. [DOI] [PubMed] [Google Scholar]
- 54.Barrett D, Brown VI, Grupp SA, Teachey DT. Targeting the PI3K/AKT/mTOR signaling axis in children with hematologic malignancies. Paediatr Drugs. 2012;14(5):299–316. doi: 10.2165/11594740-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Morishita N, Tsukahara H, Chayama K, et al. Activation of the AKT is associated with poor prognosis and chemotherapeutic resistance in pediatric B-precursor acute lymphoblastic leukemia. Pediatr Blood Cancer. 2012;59(1):83–9. doi: 10.1002/pbc.24034. [DOI] [PubMed] [Google Scholar]
- 56.Muller MR, Grunebach F, Kayser K, et al. Expression of her2/neu on acute lymphoblastic leukemia: implications for the development of immunotherapeutic approaches. Clin Cancer Res. 2003;9(9):3448–53. [PubMed] [Google Scholar]
- 57.Yang JJ, Cheng C, Yang W, et al. Genome-wide interrogation of germline genetic variation associated with treatment response in childhood acute lymphoblastic leukemia. JAMA. 2009;301(4):393–403. doi: 10.1001/jama.2009.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Dongen JJ, Seriu T, Panzer-Grumayer ER, et al. prognostic value of minimal residual disease in acute lymphoblastic leukemia in childhood. Lancet. 1998;352(9142):1731–8. doi: 10.1016/S0140-6736(98)04058-6. [DOI] [PubMed] [Google Scholar]
- 59.Flohr T, Schrauder A, Cazzaniga C, et al. Minimal residual disease-directed risk stratification using real time quantitative PCR analysis of immunoglobulin and T-cell receptor gene rearrangements in the international multicenter trial AIEOP-BFM ALL 2000 for childhood acute lymphoblastic leukemia. Leukemia. 2008;22(4):771–82. doi: 10.1038/leu.2008.5. [DOI] [PubMed] [Google Scholar]
- 60.Cave H, van der Werff ten Bosch J, Suciu S, et al. European Organization for Research and Treatment of Cancer-Childhood Leukemia Cooperative Group. Clinical significance of minimal residual disease in childhood acute lymphoblastic leukemia. N Engl J Med. 1998;339:591–8. doi: 10.1056/NEJM199808273390904. [DOI] [PubMed] [Google Scholar]
- 61.Borowitz MJ, Pullen DJ, Shuster JJ, et al. Children’s Oncology Group Study. Minimal residual disease detection in childhood precursor-B-cell acute lymphoblastic leukemia: relation to other risk factors. A Children’s Oncology Group study. Leukemia. 2003;17(8):1566–72. doi: 10.1038/sj.leu.2403001. [DOI] [PubMed] [Google Scholar]
- 62.Coustan-Smith E, Sancho J, Hancock ML, et al. Clinical importance of minimal residual disease in childhood acute lymphoblastic leukemia. Blood. 2000;96(8):2691–6. [PubMed] [Google Scholar]
- 63.Zhou J, Goldwasser MA, Li A, et al. Dana-Farber Cancer Institute ALL Consortium. Quantitative analysis of minimal residual disease predicts relapse in children with B-lineage acute lymphoblastic leukemia in DFCI ALL Consortium Protocol 95-01. Blood. 2007;110(5):1607–11. doi: 10.1182/blood-2006-09-045369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39(2):226–31. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39(10):1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(Supplementary) Estimates of P-values and FDR obtained from comparing gene expression between and among populations for the 300 most highly significantly differentially expressed genes.
(Supplementary) Estimates of coefficients of variation (CV) for individual populations for the 300 most highly significantly differentially expressed genes between the four populations. Note: CV are expressed as ratios.
(Supplementary) Results of GO analysis for all the 300 most highly significantly differentially expressed genes between ethnic populations.