A class of plasma membrane-localized and highly Pro-rich glycoproteins are essential for abscisic acid- and stress-mediated repression of rice root elongation.
Abstract
In the root of rice (Oryza sativa), abscisic acid (ABA) treatment, salinity, or water deficit stress induces the expression of a family of four genes, REPETITIVE PROLINE-RICH PROTEIN (RePRP). These genes encode two subclasses of novel proline-rich glycoproteins with highly repetitive PX1PX2 motifs, RePRP1 and RePRP2. RePRP orthologs exist only in monocotyledonous plants, and their functions are virtually unknown. Rice RePRPs are heavily glycosylated with arabinose and glucose on multiple hydroxyproline residues. They are significantly different from arabinogalactan proteins that have glycan chains composed of arabinose and galactose. Transient and stable expressions of RePRP-green fluorescent protein reveal that a fraction of this protein is localized to the plasma membrane. In rice roots, ABA treatment increases RePRP expression preferentially in the elongation zone. Overexpression of RePRP in transgenic rice reduces root cell elongation in the absence of ABA, similar to the effect of ABA on wild-type roots. Conversely, simultaneous knockdown of the expression of RePRP1 and RePRP2 reduces the root sensitivity to ABA, indicating that RePRP proteins play an essential role in ABA/stress regulation of root growth and development. Moreover, rice RePRPs specifically interact with a polysaccharide, arabinogalactan, in a dosage-dependent manner. It is suggested that RePRP1 and RePRP2 are functionally redundant suppressors of root cell expansion and probably act through interactions with cell wall components near the plasma membrane.
Extreme climate changes such as drought, high temperature, and flooding have caused significant crop losses in recent years. In addition, crop productivity will need to be increased to meet the demands of the growing human population in the next half century (Tilman et al., 2002). Therefore, understanding how plants survive and minimizing the impact of abiotic stresses on yield are receiving considerable attention.
The phytohormone abscisic acid (ABA), a well-recognized stress hormone, is up-regulated to turn on stress response genes in cells to help plants cope with unsuitable environments (Christmann et al., 2006; Yamaguchi-Shinozaki and Shinozaki, 2006; Qin et al., 2011). For example, osmotic stress stimulates the biosynthesis and accumulation of ABA in guard cells (Yamaguchi-Shinozaki and Shinozaki, 2006). The complex of ABA and receptor interacts with protein phosphatase 2C proteins and switches off the inhibition of SUCROSE-NONFERMENTING1-RELATED PROTEIN KINASE2 (SnRK2; Umezawa et al., 2009). The active SnRK2 then phosphorylates certain transcription factors to promote the transcription of downstream ABA-responsive genes (Cutler et al., 2010; Kline et al., 2010; Raghavendra et al., 2010). In guard cells, SnRK2E (also known as OPEN STOMATA1 [OST1]) activated by ABA signaling phosphorylates POTASSIUM CHANNEL IN ARABIDOPSIS THALIANA1 (KAT1) to allow the entry of K+ (Sato et al., 2009). In addition, SLOW ANION CHANNEL-ASSOCIATED1 (SLAC1), which is also phosphorylated by the activated SnRK2E/OST1, interacts with calcium-dependent protein kinases 21 and 23 to stimulate the efflux of Cl− and malate2− from guard cells (Negi et al., 2008; Geiger et al., 2009, 2010). The combined action of these proteins results in turgor pressure and ionic changes and ultimately induces the closure of stomata to slow down transpiration.
Most ABA response genes have common cis-acting regulatory elements called ABA-responsive elements in their promoter regions that are directly controlled by bZIP transcription factors referred to as ABA-RESPONSIVE ELEMENT-BINDING PROTEINS/FACTORS (AREB/ABFs; Guiltinan et al., 1990; Shen et al., 1993; Foster et al., 1994; Uno et al., 2000). Overexpression of ABF3 and ABF4 in Arabidopsis (Arabidopsis thaliana) resulted in improved drought tolerance (Kang et al., 2002). ABI5, another important AREB/ABF, induces the LATE EMBRYOGENESIS ABUNDANCE (LEA) gene family during seed maturation and in vegetative tissues under drought conditions (Yamaguchi-Shinozaki and Shinozaki, 2006). LEA proteins are thought to protect cells by acting as hydration buffers, ion sequestration agents, or molecular chaperones (Dure, 1993; Close, 1996; Ingram and Bartels, 1996; Wise, 2003; Olvera-Carrillo et al., 2011). Other important transcription factors, such as APETALA2/ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR and the NAC family, have been shown to activate stress-related genes involved in the cell cycle and metabolic and physiological balance to maintain homeostasis under stress (Negrao et al., 2011; Mizoi et al., 2012; Nakashima et al., 2012). Overexpression of OsNAC5 and OsNAC6 in rice (Oryza sativa) increases plant tolerance to drought and salinity conditions (Nakashima et al., 2007; Takasaki et al., 2010). Although ectopic expression of stress-responsive genes in plants can improve stress tolerance, growth retardation and reduced productivity are often observed. In order to enhance stress tolerance without compromising crop yield, it is important to understand how ABA/stress-responsive genes coordinately regulate physical processes under unfavorable conditions.
Plant roots play an important role in water and nutrient transport from the soil to support plant growth. Root growth is significantly affected by environmental stimuli. Salinity and water stress induce ABA accumulation in roots (Zhang and Davies, 1987; Jia et al., 2002). The involvement of ABA in root development under water deficit conditions has also been examined (Sharp and LeNoble, 2002; Sharp et al., 2004). Inhibition of endogenous ABA synthesis in wild-type plants by fluridone or in the ABA-deficient mutants vp5 and vp14 markedly suppressed root elongation under water deficit conditions compared with wild-type plants (Saab et al., 1990). Root elongation was restored back to normal by the addition of exogenous ABA. In addition, the root length increase of the maize (Zea mays) vp mutant was significantly lower than that in the wild type under water deficit conditions, indicating that ABA is required for maintaining rather than completely repressing root development under water stress (Sharp, 2002; Sharp and LeNoble, 2002). The effects of ABA application on rice root have been observed to include the swelling of young root tips, abundant formation of root hairs, and initiation of lateral roots close to tips (Chen et al., 2006). These morphological changes require the involvement of calmodulin and de novo protein synthesis, but the mechanism is still unclear.
Plant hydroxyproline-rich glycoproteins (HRGPs) are the major structural proteins in cell walls (Showalter, 1993; Nothnagel, 1997; Cassab, 1998). Common features in HRGPs include enrichment in certain amino acids, repetitive sequence domains, and highly diverse carbohydrate contents. HRGPs can be classified into three groups based on domain features, including Pro-rich proteins with (Pro)3XYLys repeats, extensin-type proteins with Ser(Pro)3-5 repeats, and arabinogalactan proteins (AGPs) with central domains rich in (S/A/T)Pro repeats (Showalter, 1993; Nothnagel, 1997). Moreover, according to the “Hyp contiguity hypothesis,” most contiguous Pro repeats are attached to nonbranched Ara oligosaccharides on Hyp. In contrast, noncontiguous Pro repeats in AGPs carry large branched arabinogalactan (AG) polysaccharides (Kieliszewski, 2001; Kieliszewski and Shpak, 2001; Xu et al., 2008a). Over the years, the completion of genome sequence in many plants has revealed many nonclassical HRGPs with complicated domain structures. “Chimeric HRGPs” contain one known HRGP domain and other unrelated motifs in proteins, while “hybrid HRGPs” contain two different HRGP domains (Schultz et al., 2002). Of the HRGPs, AGPs have been extensively studied and play many roles in plant growth and development (Seifert and Roberts, 2007). The features of classical AGPs are (1) a mass of type II AGs (5–25 kD) attached to Hyp within (S/A/T)Pro repeats; (2) the presence of glycosylphosphoinositol (GPI) in the hydrophobic C-terminal region; and (3) the ability to bind β-glycosyl Yariv reagent (Showalter, 1993; Ellis et al., 2010). In functional studies using specific chemicals and in molecular genetic studies, it has been demonstrated that both the membrane-bound and secreted AGPs participate in cell division, cell expansion, programmed cell death, floral abscission, pollen tube guidance, pollen incompatibility, and plant-microbe interactions (Seifert and Roberts, 2007).
Rice is one of the most important cereal crops in the world and feeds close to 3.5 billion people (International Rice Research Institute [http://irri.org/], November 2012). Rice production yield is directly proportional to the development of the panicle and spikelet during reproductive stages. However, plants in the reproductive phase are sensitive to environmental stress, such as drought and high salinity (Negrao et al., 2011). In order to identify genes specifically regulated by ABA in rice roots, we performed microarray-based transcriptomic analysis and focused on the study of a family of ABA-induced root-specific genes encoding highly Pro-rich proteins with unknown function, REPETITIVE PROLINE-RICH PROTEIN (RePRP). These proteins are specifically up-regulated by stresses and ABA in roots and demonstrated that rice RePRPs are necessary and sufficient for ABA regulation of root growth. Their distinct Pro-repeat pattern, failure to interact with β-Glc Yariv reagent, and the presence of diverse sugar components in glycan on Hyp residues distinguish rice RePRP proteins as a novel family of the HRGPs. Our discovery that rice RePRPs are able to interact with a polysaccharide, AG, provides new clues on the mechanism by which RePRPs control cell elongation.
RESULTS
ABA and Stress Up-Regulate a Novel Pro-Rich Protein Family in Rice Roots
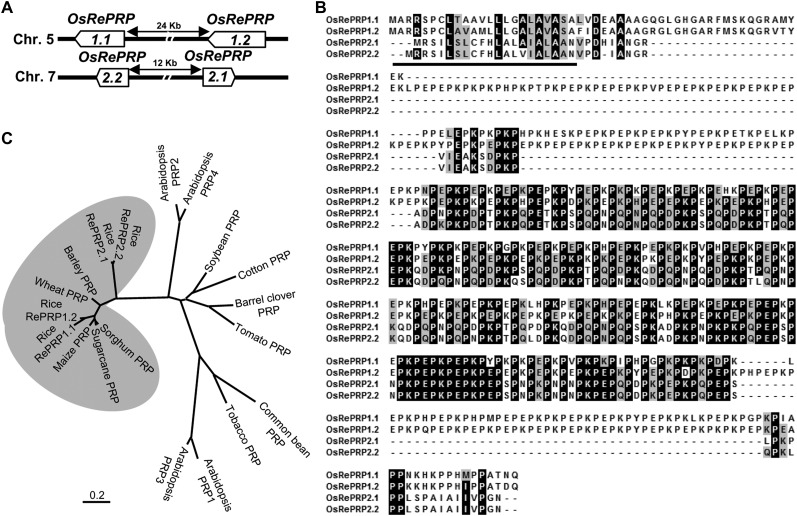
To isolate tissue-specific and ABA-inducible genes in rice, we carried out a gene expression profile analysis by the microarray technology using RNAs isolated from ABA-treated roots and shoots. AK105945 was notably up-regulated in root tissues but not in shoots 0.5 d after ABA treatment. AK105945 encodes a Pro-rich protein, a homolog of previously identified OsPRP1 in rice (Akiyama and Pillai, 2003). Based on The Institute for Genomic Research rice database (http://rice.plantbiology.msu.edu/) and the Rice Genome Annotation Database (http://ricegaas.dna.affrc.go.jp/rgadb/), four homologous intronless genes were found on two separate chromosomes; two genes are located closely together on chromosomes 5, and another two adjacent genes are located on chromosome 7 (Fig. 1A). All four of these genes contained highly repetitive PX1PX2 motifs; therefore, we designated this gene family as RePRP. RePRP genes on the same chromosome share an even higher similarity than genes on different chromosomes. The two RePRP genes on chromosome 5 share 84% sequence identity and are named RePRP1.1 (formerly called OsPRP1) and RePRP1.2, while another two genes located on chromosome 7 share 94% identity and are named RePRP2.1 (AK105945) and RePRP2.2.
Figure 1.
Rice RePRPs belong to the RePRP gene family found specifically in monocot species. A, Map of gene structure, direction of transcription, and chromosomal location of the rice RePRP gene family. B, Alignment of deduced protein sequences of four RePRPs. Identical residues are highlighted in black boxes. Similar residues are indicated in gray boxes. The predicted signal peptide is underlined. C, Dendrogram showing the relationships between various RePRPs in monocot species and other PRPs in dicots. The phylogenetic tree was generated by MEGA5 analysis of full-length amino acid sequences using the neighbor-joining method. The bar beneath the dendrogram represents evolutionary distance in units of the number of amino acid substitutions per site.
By comparing the deduced amino acid sequences, it was observed that each RePRP protein contained a signal peptide at the N terminus followed by a Pro-rich domain occupying approximately 70% of the protein. The Pro-rich domain contains high PX1PX2 repeats and constitutes the hydrophilic regions. The major difference among RePRPs is that the X residues of the PX1PX2 motif are often Lys or Glu in the RePRP1 subgroup, whereas most X residues are Lys, Glu, Asn, or Asp in the RePRP2 subgroup (Fig. 1B). Using the repetitive PX1PX2 motif to search for orthologs in other species, potential candidates were found only in monocots. The phylogenetic tree shows that rice RePRP1.1 and RePRP1.2 are evolutionarily close to maize, wheat (Triticum aestivum), sugarcane (Saccharum officinarum), sorghum (Sorghum bicolor), and barley (Hordeum vulgare) PRPs, which all contain the conserved repetitive proline-rich motif (Fig. 1C; Supplemental Fig. S1). Due to a high proportion of Gln, Asp, and Asn in the Pro-rich domain of rice RePRP2.1 and RePRP2.2, these proteins lie outside the main RePRP clade (Fig. 1C). Intriguingly, proteins that contained repetitive Pro-rich structure have not been found in the Hyp-rich protein family in Arabidopsis (Schultz et al., 2002; Showalter et al., 2010) in a search based on criteria not including unique PX1PX2 motifs. Similarly, using the same search criteria, RePRPs were not identified in the rice genome (Ma and Zhao, 2010; Showalter et al., 2010). Thus, the presence of a large number of PX1PX2 motifs is a unique feature of RePRPs, which is not shared with other subclasses in the Hyp-rich protein family.
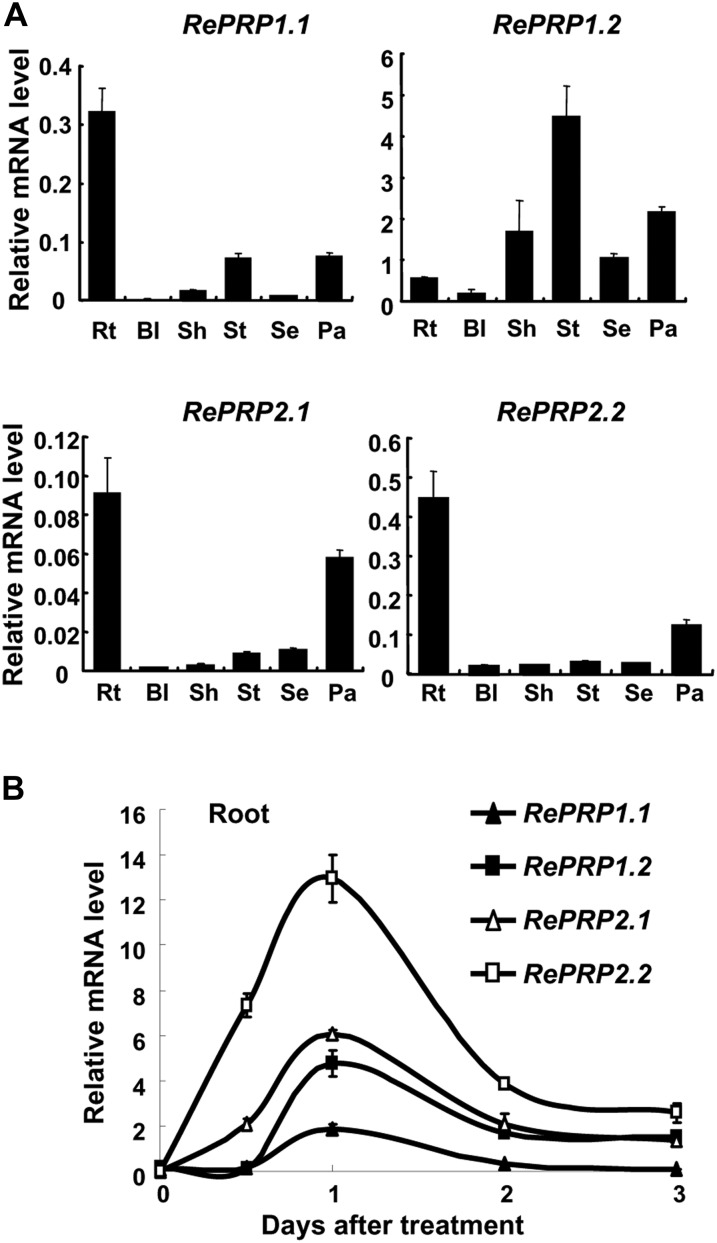
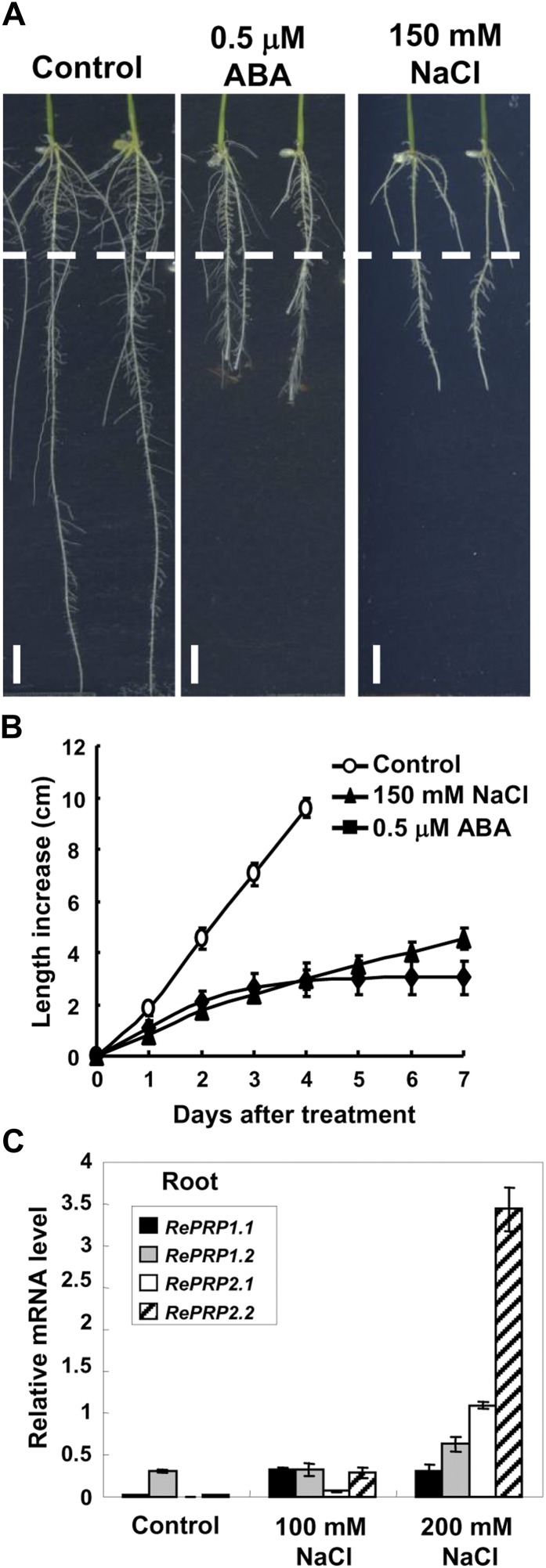
Rice RePRPs were differentially expressed in various tissues throughout vegetative and reproductive developmental stages, with RePRP1.1, RePRP2.1, and RePRP2.2 transcripts preferentially expressed in roots and panicles. In contrast, RePRP1.2 transcripts were highly expressed in stem, sheath, and panicles but not in roots (Fig. 2A). All four genes were highly induced by ABA in roots, and their expression reached highest levels 1 d after ABA treatment, especially for ReRPR2.1 and RePRP2.2 genes (Fig. 2B). In addition to ABA, salinity and drought also induced the expression of RePRPs in roots. While rice seedlings grew on the medium with high concentrations of NaCl or polyethylene glycol, RePRP transcripts were notably elevated (Fig. 3C; Supplemental Fig. S2C). Salinity and dehydration treatment significantly reduced root growth, which was similar to the effects of ABA on roots, even for a concentration as low as 0.5 μm ABA (Fig. 3B; Supplemental Fig. S2B). These studies suggest that the rice RePRP family may participate in the regulation of root growth under ABA and abiotic stresses.
Figure 2.
ABA induces the expression of four RePRP genes in rice roots. A, Relative levels of RePRP mRNA (means ± sd) in different tissues of wild-type rice were determined by quantitative RT-PCR and normalized with the ACTIN mRNA. Rt, Root; Bl, blade; Sh, sheath; St, stem; Se, seed; Pa, young panicle. B, Roots were collected from 30 individuals of 2-week-old wild-type rice plants treated with 20 μm ABA for 0.5 to 3 d, and relative levels of RePRP mRNA (means ± sd) were determined by quantitative RT-PCR using gene-specific primers.
Figure 3.
Salinity reduces rice root growth but enhances RePRP expression. A, Root images of wild-type seedlings grown on Murashige and Skoog medium containing 0.5 μm ABA or 150 mm NaCl for 7 d. The dashed line indicates the root tip position on day 0 of treatment. B, Root length increase (means ± sd) was measured from 30 individual plants under each condition. Experiments were repeated at least twice with similar results. C, Relative levels of RePRP mRNA (means ± sd) of wild-type rice grown under the normal condition or in medium containing 100 or 200 mm NaCl determined by quantitative RT-PCR and normalized with the internal control ACTIN mRNA.
RePRPs Are Glycoproteins with O-Glycan on Hyp Residues But Are Not Typical AGPs
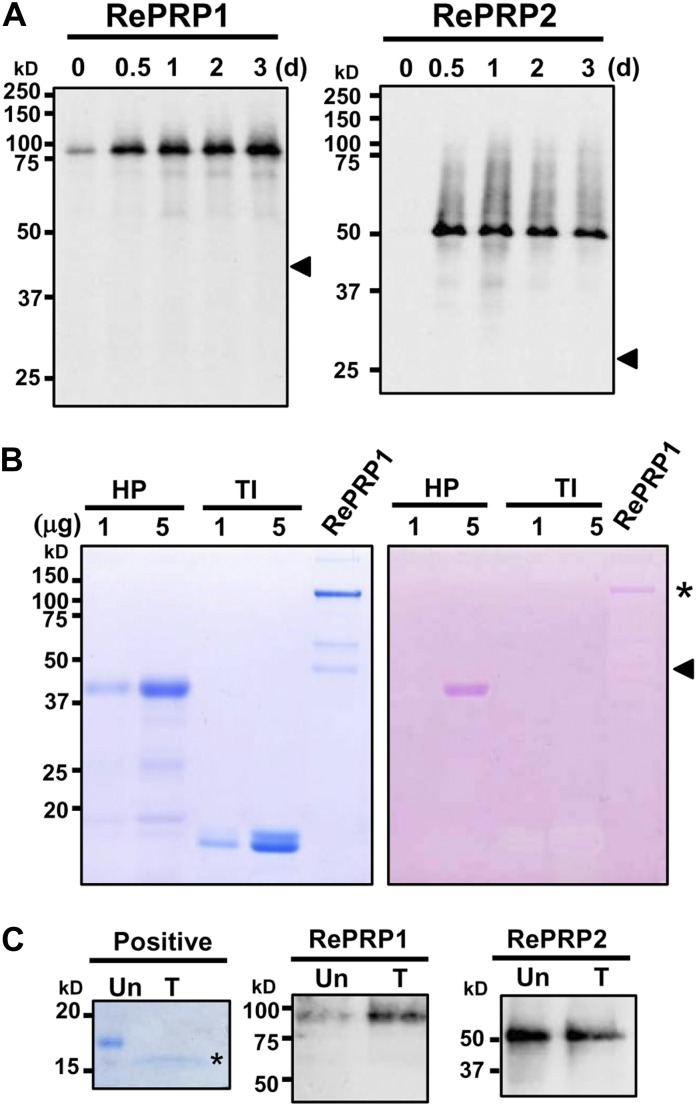
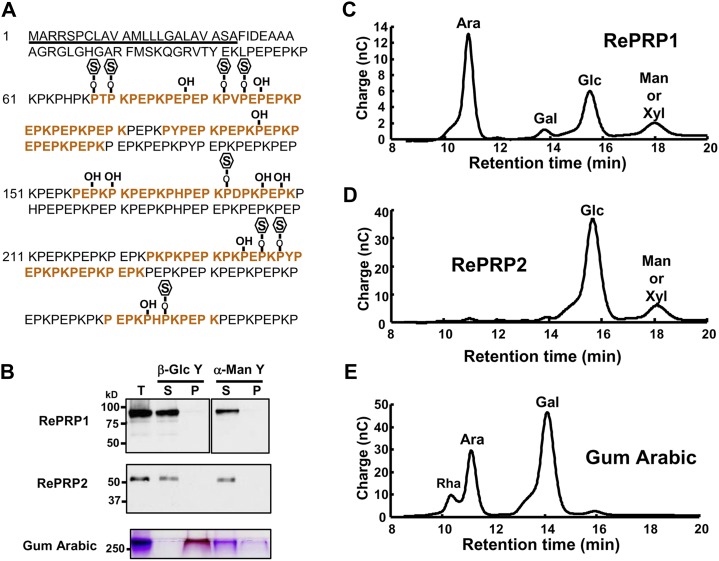
To facilitate the analysis of RePRP proteins, two specific polyclonal antibodies were raised against recombinant RePRP1 or RePRP2 proteins. The RePRP1 antibodies simultaneously recognized RePRP1.1 and RePRP1.2, while RePRP2 antibodies could recognize RePRP2.1 and RePRP2.2 at the same time. Without ABA treatment, little RePRP1 and RePRP2 were detected. However, the expression of these RePRPs was increased greatly after treatment (Fig. 4A). Interestingly, high-molecular-mass protein bands were detected by western blots. Figure 4A indicated that rice RePRP1.1 and RePRP1.2 are expected to have molecular masses of 40 and 46.5 kD, respectively, and RePRP2 proteins are expected to have a molecular mass of 26.7 kD (Fig. 4A, arrowheads). However, a greater than 75-kD protein band was detected by RePRP1 antibodies, and an approximately 50-kD protein band was detected by RePRP2 antibodies. In order to verify the protein identities, high-molecular-mass protein bands were purified and analyzed from overexpressing transgenic rice. Two protein bands above 100 and 50 kD from purified RePRP1 proteins (Fig. 4B, left; bands from purified RePRP1) and the 50-kD protein band from purified RePRP2 proteins were isolated and subjected to liquid chromatography (LC)-tandem mass spectrometry (MS/MS) analysis. The hit peptides with top scores matched RePRP proteins in the rice database (Supplemental Tables S1 and S2), indicating that these proteins were indeed rice RePRPs. Interestingly, quite a few Pro residues of RePRPs were hydroxylated (Supplemental Tables S1 and S2), which is an important feature for glycosylation in plant HRGPs.
Figure 4.
Glycosylated RePRPs accumulate in ABA-treated roots. A, RePRPs were detected in 2-week-old wild-type roots treated with 20 μm ABA for 0.5 to 3 d by western-blot analysis using gene-specific antibodies. Thirty and 15 μg of total proteins were loaded for RePRP1 and RePRP2 protein detection, respectively. RePRP proteins were detected by RePRP1- or RePRP2-specific antibodies, and protein bands were developed on x-ray film for 20 and 10 s for RePRP1 and RePRP2 proteins, respectively. The apparent sizes of major bands were higher than the expected protein size (arrowheads). B, RePRP1s were purified from transgenic rice overexpressing RePRP1 and analyzed on an SDS-polyacrylamide gel. The gel was stained with Coomassie Brilliant Blue (left) or periodic acid-Schiff reagent (right). The pink bands (asterisk) indicate the presence of glycoproteins. The protein band located at the predicted size (arrowhead) could not be detected by periodic acid-Schiff stain. Horseradish peroxidase (HP) and trypsin inhibitor (TI) were used as positive and negative controls, respectively. C, RePRPs treated with (T) or without (Un) PNGase F were detected by western-blot analysis. The N-glycosylated protein was used as a positive control and analyzed on an SDS-polyacrylamide gel stained with Coomassie Brilliant Blue stain. After PNGase F treatment, the protein size of an N-glycosylated protein was reduced by 2 to 15 kD (asterisk), whereas that of RePRPs was not changed.
To examine the possibility that glycosylation causes the increase of molecular mass in rice RePRPs, purified RePRP1 proteins were analyzed on an SDS denaturing gel and stained with a periodic-Schiff reagent. The major band above 100 kD of purified RePRP1 showed magenta color as a typical glycoprotein (Fig. 4B, asterisk), whereas the expected 46-kD RePRP1 was not recognized by Schiff reagent (Fig. 4B, arrowhead). These results suggest that rice RePRPs are likely heavily glycosylated, thereby appearing as a higher molecular mass band than the expected size on the gel. To clarify the types of glycosylation, the endoglycosidic enzyme peptide N-glycosidase F (PNGase F) was employed to verify the N-linked glycosylation of rice RePRPs. Figure 4C shows that PNGase F treatment did not reduce the size of RePRP. To evaluate O-linked glycosylation, methylamine was used for the removal of O-linked glycans from proteins and glycosylation sites were determined by LC-MS/MS. Since RePRP1 could be purified in sufficient quantity, further analysis was carried out with this protein. Analysis with purified RePRP1 showed that some Pro residues were hydroxylated and some were linked with methylamine (Fig. 5A). For RePRP1, among the 62 Pro residues detected by mass spectrometry (MS), 27% of them were hydroxylated (17 Hyp) and 13% of them were glycosylated (eight methylamine-labeled Hyp; Fig. 5A). Therefore, the increased molecular mass of rice RePRP could be caused by the addition of multiple glycan chains on hydroxylated Pro residues.
Figure 5.
Rice RePRPs are glycosylated on Hyp residues. A, Modification of RePRP1 in rice. The predicted signal peptide is underlined. Red letters indicate peptides detected in LC-MS/MS analysis. Hyp residues are marked as P-OH. Glycosylated Hyp residues labeled by methylamine are shown as P-O-S. B, Yariv reagent interaction assay. Gum arabic used as a positive control could be precipitated by the β-Glc Yariv reagent and detected in the pellet fraction (P) on the SDS-polyacrylamide gel by periodic acid-Schiff stain. RePRPs were detected only in the supernatant fraction (S) after incubation with the β-Glc Yariv reagent by western-blot analysis. Total proteins (T) before incubation were loaded in the first lane. α-Man Yariv, which does not interact with the gum arabic, was used as a negative control. C to E, Carbohydrate components in RePRP glycoproteins and gum arabic were detected by HPAEC-pulsed-amperometric detection analysis. Carbohydrates used were Rha, Ara, Gal, Glc, Man, and Xyl. nC, Nanocoulombs.
In Pro-rich glycoproteins, the significant change in Mr is found in the AGP subfamily that carries multiple large AG glycans linked to Hyp and specifically interacts with the β-Glc Yariv reagent (Yariv et al., 1967; Seifert and Roberts, 2007; Kitazawa et al., 2013). Due to the presence of similar discontinuous Pro repeats in RePRPs, the β-Glc Yariv reagent was used to verify whether rice RePRPs are new members of the AGP subfamily. The AGP standard gum arabic could be precipitated by β-Glc Yariv (Fig. 5B, bottom), whereas both OsRePRPs were only detected in the supernatant fraction after incubation with β-Glc Yariv (Fig. 5B). Therefore, OsRePRPs do not share the same properties as the AGP family. Furthermore, analysis of sugar composition in OsRePRPs is another way to classify glycoproteins. To further examine the sugar component in the RePRP glycoproteins, the glycan chains were hydrolyzed into monosaccharides and analyzed by high-performance anion-exchange chromatography (HPAEC). The results indicated that RePRP1 contained abundant Ara (56.1%) and Glc (28.4%; Fig. 5C), whereas Glc (82%) was the major component in RePRP2 (Fig. 5D). One minor peak present in both RePRP proteins could be Man or Xyl (Fig. 5, C and D). As expected, gum arabic contained abundant Ara and Gal, which is different from the carbohydrate composition of RePRPs (Fig. 5E; Supplemental Table S3). The differential sugar composition could be responsible for the failure of RePRPs to interact with the β-Glc Yariv reagent, which specifically binds β-1, 3-galactan chains (Kitazawa et al., 2013). Thus, RePRPs are a novel family of HRGPs but not typical AGPs, according to the distinct Pro repeat patterns, failure to interact with β-Glc Yariv, and diverse sugar components in glycan linked to Hyp residues.
RePRPs Are Localized to the Plasma Membrane
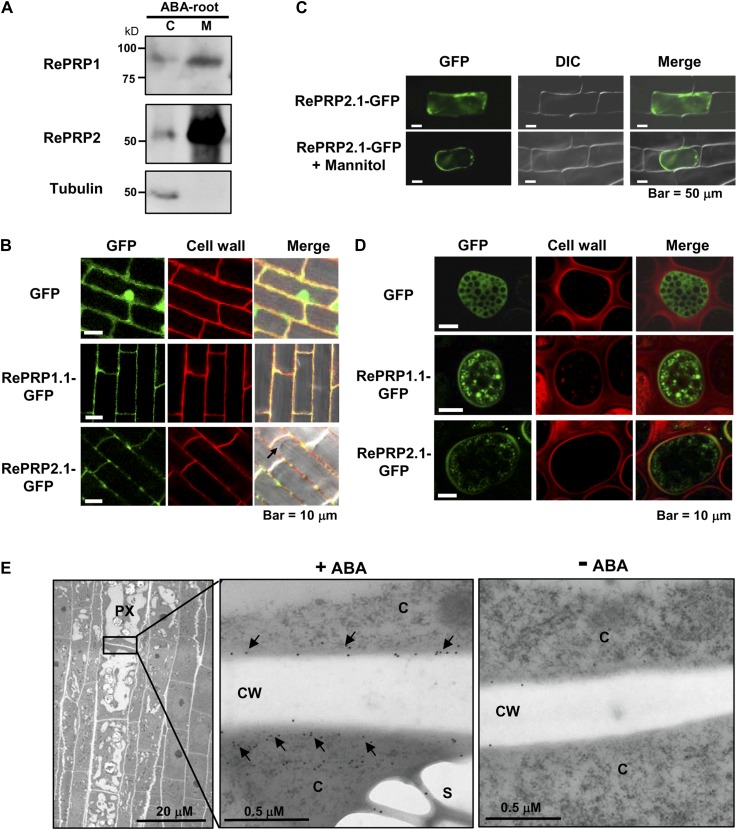
Some HRGPs contain signal peptide and GPI to assist either protein localization to the plasma membrane or secretion to the cell wall. To determine the subcellular localization of RePRPs, cytosolic and membrane-associated proteins were extracted from ABA-treated rice roots for western-blot analysis, and RePRPs were found to be enriched in the cell membrane fraction (Fig. 6A). To determine the subcellular localization of RePRPs, RePRP1.1 and RePRP2.1 were fused to GFP and expressed in transgenic rice. In transgenic rice roots, RePRP1.1-GFP and RePRP2.1-GFP were detected mainly at the cell boundary and colocalized with cell walls stained by propidium iodide (Fig. 6B). To distinguish whether RePRPs are localized on the plasma membrane or the cell wall, RePRP2.1-GFP was introduced into the onion (Allium cepa) epidermal cells, and cells were treated with 0.8 m mannitol. RePRP2. 1-GFP appeared to associate with the plasma membrane and dissociate from the cell wall in the plasmolyzed onion cells (Fig. 6C), suggesting that RePRP-GFP was targeted to the plasma membrane. A third system, barley aleurone cells, is also suitable for the study of the cellular localization of RePRPs, due to the presence of an extensive endomembrane system and the lack of a large central vacuole. While GFP alone was uniformly expressed in the cytosol (Fig. 6D, top row), RePRP1.1-GFP and RePRP2.1-GFP were present in small membrane vesicles and on the plasma membrane (Fig. 6D, middle and bottom rows), suggesting that the secretory pathway is involved in the localization to the plasma membrane.
Figure 6.
RePRP proteins are localized to the plasma membrane. A, Cytosolic (C) and membrane (M) proteins were extracted from 20 μm ABA-treated rice roots. RePRP1 and RePRP2 were detected mainly in the membrane fraction by western-blot analysis. Tubulin was used as a control for the cytosolic fraction. B, GFP and RePRP-GFP were stably expressed in rice. GFP was detected in cytosol and nucleus, whereas RePRP1.1-GFP and RePRP2.1-GFP were detected in cell boundary but not in cytosol (arrow) in root cells. Cell walls were stained by propidium iodide. C, GFP and RePRP2.1-GFP were transiently expressed in onion epidermal cells by particle bombardment-mediated transfection. RePRP2.1-GFP remained inside cells after plasmolysis by mannitol treatment. D, GFP and RePRP-GFP were transiently expressed in barley aleurone layers via particle bombardment. Cell wall was stained by propidium iodide. E, Localization of RePRPs in ABA-treated roots was detected by immunogold labeling with anti-RePRP2 antibody and observed by TEM. TEM images show a longitudinal section of the elongation region of ABA-treated young roots (left). Magnification of the boundary region between protoxylem (PX) cells shows gold particles (arrows) in cytoplasm (C) and the plasma membrane but not in cell walls (CW; middle). Only a few gold particles were detected in roots without ABA treatment (right).
In order to further clarify the membrane localization of RePRP in rice roots, young root tips were dissected from ABA-treated rice seedlings and longitudinal sections were used for immunodetection with anti-RePRP2 antibodies and colloidal gold particle-conjugated secondary antibodies. The specimens were examined by transmission electron microscopy (TEM). The TEM images showed that only a few gold particles were detected in root samples without ABA treatment (Fig. 6E, right). However, abundant particles were observed in protoxylem cells in the elongation zone of ABA-treated roots (Fig. 6E, left). The gold particles were localized in the cytoplasm and cell boundary, where the plasma membrane is expected to be, but not in the cell wall (Fig. 6E, middle), suggesting that RePRP proteins were localized on the plasma membrane but not in the cell wall.
RePRPs Specifically Interact with AG
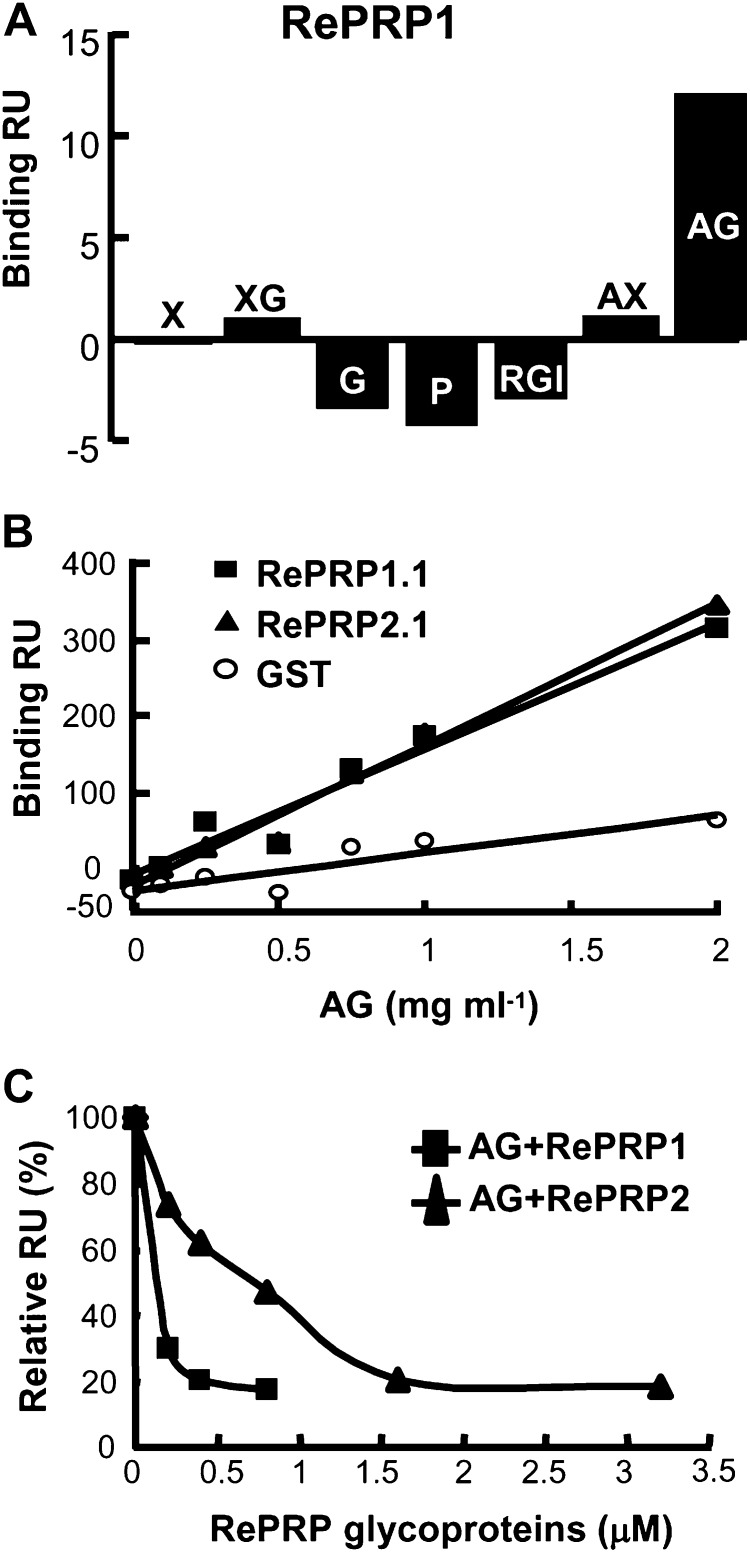
We have attempted to identify RePRP-interacting proteins by immunoprecipitation assays with anti-RePRP antibodies, but no interacting protein was detected. Therefore, other possible interacting compounds, such as cell wall polysaccharides, were tested using surface plasmon resonance (SPR) technology. Briefly, purified RePRPs were indirectly coated on the sensor chip through the association with anti-RePRP antibodies bound to the chip surface. Subsequently, seven selected cell wall polysaccharides, xylan, xyloglucan, glucan, pectin, rhamnogalacturonan I, arabinoxylan, and AG, were passed through the chip channels separately, and each binding response was measured in real time. The results revealed that only AG exhibited a notable binding response with the rice RePRP1 proteins (Fig. 7A).
Figure 7.
Rice RePRPs interact specifically with AG. A, Interactions between rice RePRP1 and cell wall polysaccharides were analyzed by SPR technology. Purified RePRP1 was captured by anti-RePRP antibodies immobilized on a CM5 chip. The interaction (Binding response unit [RU]) of RePRP1 with each cell wall polysaccharide (0.5 mg mL−1) shows typical results from two experimental repeats. Polysaccharides are as follows: xylan (X), xyloglucan (XG), glucan (G), pectin (P), rhamnogalacturonan I (RGI), arabinoxylan (AX), and AG. B, Interactions of recombinant RePRPs and AG were detected by SPR technology. Recombinant RePRP proteins and GST were immobilized on separate channels of a CM5 chip. Recombinant RePRP1.1 and RePRP1.2 proteins showed higher binding response (Binding RU) for AG than for GST. C, Interactions of RePRP glycoproteins with AG were analyzed by their competition against recombinant RePRP1.1 protein. The interaction of AG with recombinant RePRP1.1 protein was competed with various concentrations of native RePRP1 or RePRP2. Free AG interacting with recombinant RePRP1 was measured and converted to percentage, with AG alone set as 100%. Shown are the typical results from two experimental repeats.
To investigate whether the interaction with AG occurred on the glycan or the protein backbone of RePRPs, recombinant RePRPs (i.e. without glycosylation) produced from Escherichia coli were coated on the sensor chip, and their interactions with different concentrations of AG were examined. As the concentration of AG increased (from 0.1 to 2 mg mL−1), the binding response was significantly elevated with recombinant RePRP1.1 and RePRP2.1 proteins but not with glutathione S-transferase (GST; Fig. 7B; Supplemental Fig. S3), indicating that AG is able to interact with the RePRP protein backbone specifically.
The role of the glycosylation of RePRPs in the interaction with AG was also examined. Since RePRP glycoproteins could not be coated on chips efficiently, recombinant RePRP1.1 produced from E. coli was immobilized on the chip and its interaction with AG was competed by various concentrations of RePRP glycoproteins extracted from rice. As the concentration of RePRP1 or RePRP2 glycoprotein increased, the binding responses of AG with recombinant RePRP1.1 were notably reduced (Fig. 7C). The concentration of the AG-glycoprotein complex was elevated with the concentration of RePRP1 and RePRP2 and reached the maximum equilibrium level at 0.8 and 3 μm, respectively (Supplemental Fig. S4), signifying that RePRPs were able to interact with the AG and that the binding affinity of RePRP2 was weaker than that of RePRP1. Taken together, in rice cells, the highly glycosylated RePRPs are translocated to the plasma membrane and likely interact with cell wall AG to execute their functions.
RePRPs Are Essential for Mediating ABA-Dependent Inhibition of Root Growth
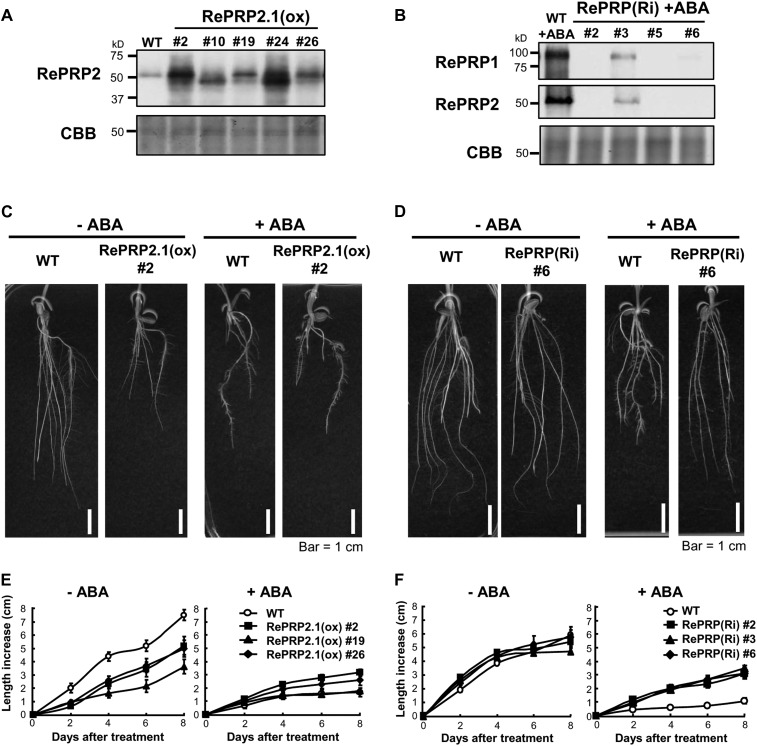
Root elongation was inhibited while numbers of lateral roots were increased in seedlings treated with 0.5 µm ABA or 150 mm NaCl or polyethylene glycol (Fig. 3A; Supplemental Fig. S2). The significant up-regulation by ABA and abiotic stresses in rice roots implies that RePRPs might play important roles in the root response to abiotic stresses. To ascertain the function of RePRPs in rice root growth under abiotic stress, RePRPs were ectopically expressed as well as knockdown expressed in transgenic rice. Protein levels of RePRPs in roots of transgenic rice were confirmed by western-blot analysis (Fig. 8, A and B).
Figure 8.
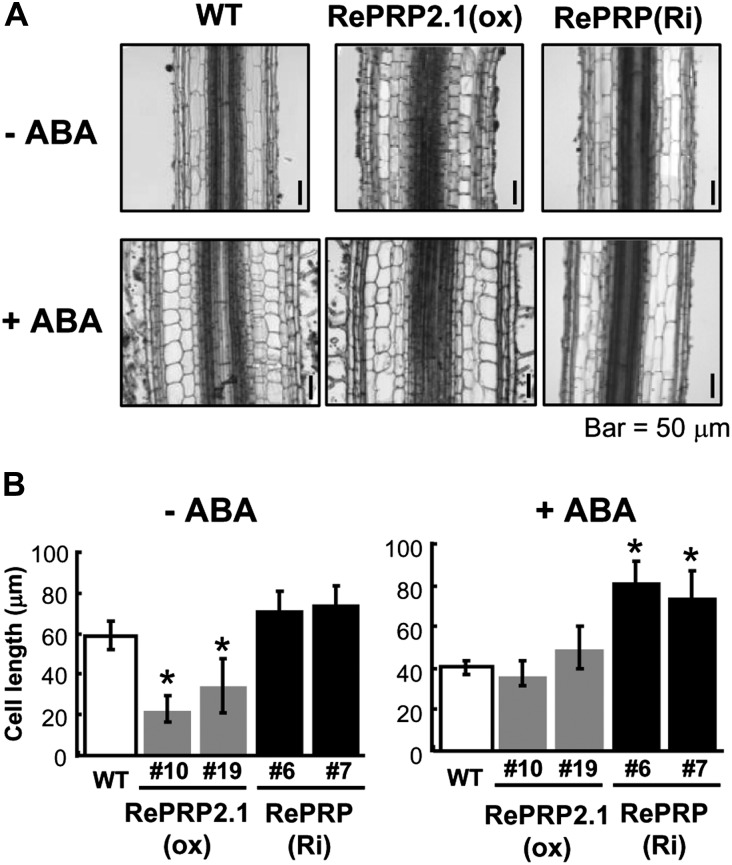
Effects of ABA on root growth of RePRP2.1(ox) and RePRP knockdown lines. A and B, Protein levels of RePRPs were detected in roots of wild-type (WT) and RePRP2.1(ox) plants, or ABA-treated roots of RePRP(Ri) plants, by western-blot analysis. Fifteen micrograms of protein was analyzed on an SDS-polyacrylamide gel stained by Coomassie Brilliant Blue (CBB). C and D, Roots of wild-type, RePRP2.1(ox), and RePRP(Ri) plants grown in medium without ABA (−ABA) or with 1 μm ABA (+ABA) for 8 d. E and F, Root length increase of wild-type, RePRP2.1(ox), and RePRP(Ri) plants in response to ABA. Each point indicates the average value (mean ± se) of eight individuals for each line. Shown are the typical results from triplicate experimental repeats.
Ectopic expression of RePRP2.1 led to shorter and thicker root phenotypes that were similar to ABA treatment on wild-type roots (Fig. 8C). The reduced root growth rate in RePRP2.1-overexpressing [RePRP2.1(ox)] lines was also similar to that in the ABA-treated wild type (Fig. 8E). These results indicate that overexpression of RePRP2 alone is sufficient to mimic ABA action in the suppression of rice root growth. However, ABA was still able to effectively inhibit root growth in RePRP2 RNA interference (RNAi) transgenic plants (Supplemental Materials and Methods S1; Supplemental Fig. S5), suggesting that functional redundancy of RePRP1 and RePRP2 genes is likely involved in the same ABA-response process. Indeed, simultaneously reducing the expression of both RePRP1 and RePRP2 genes by RNAi [RePRP(Ri)] resulted in ABA insensitivity of root growth. Radicle and crown roots of RePRP(Ri) lines were significantly longer than those of the wild type in the presence of ABA (Fig. 8D). Although these results seem to suggest that RePRP1 and RePRP2 are functionally redundant in the ABA suppression of rice root growth, it is surprising that the transfer DNA activation-tagged mutant for RePRP1 overexpression (Supplemental Materials and Methods S1), reprp1, displayed normal root growth and ABA response as in wild-type rice (i.e. without the short-root phenotype in RePRP2.1(ox) plants; Supplemental Fig. S6). This is probably because OsRePRP2 contributes more than OsRePRP1 to the ABA repression of root growth.
RePRPs Suppress Root Cell Expansion
The shorter and thicker roots observed in ABA-treated wild-type and RePRP2.1(ox) lines could result from either changes of the cell file arrangement or cell numbers. To further understand the morphological changes at the cellular level, the tissue organization in the mature region of rice roots was examined. As compared with the wild type under the normal condition, significant reduction in length and increase in width were observed in the epidermis, exodermis, sclerenchyma, and cortex of ABA-treated roots (Fig. 9A, left panels). Similar cellular changes, with even shorter and wider cells, were observed in roots of RePRP2.1(ox) lines (Fig. 9A, middle panels). The change of cell length was not detected in RePRP(Ri) lines regardless of the presence or absence of ABA (Fig. 9A, right panels). These studies suggest that the shorter and thicker roots are the consequence of reduced cell length and increased cell width. The reduction of cell length caused by ABA was determined by the measurement of cortex cell size (Fig. 9B). Interestingly, although cell length was not affected, cell width was still increased by ABA in roots of RePRP(Ri) lines. These studies demonstrated that RePRPs mediate the ABA-dependent inhibition of root cell elongation.
Figure 9.
RePRPs are involved in the regulation of cell expansion by ABA. A, Tissue sections of root mature region in wild-type (WT), RePRP2.1(ox), and RePRP(Ri) plants grown in medium without ABA (−ABA) or with 1 μm ABA (+ABA) for 8 d. B, Average cortex cell lengths (means ± se) of wild-type, RePRP2.1(ox), and RePRP(Ri) plants. The average cell length was calculated based on 100 cells per tissue section. Asterisks indicate significant differences compared with the wild type (P < 0.01).
ABA Specifically Induces RePRP Expression in the Root Elongation Zone
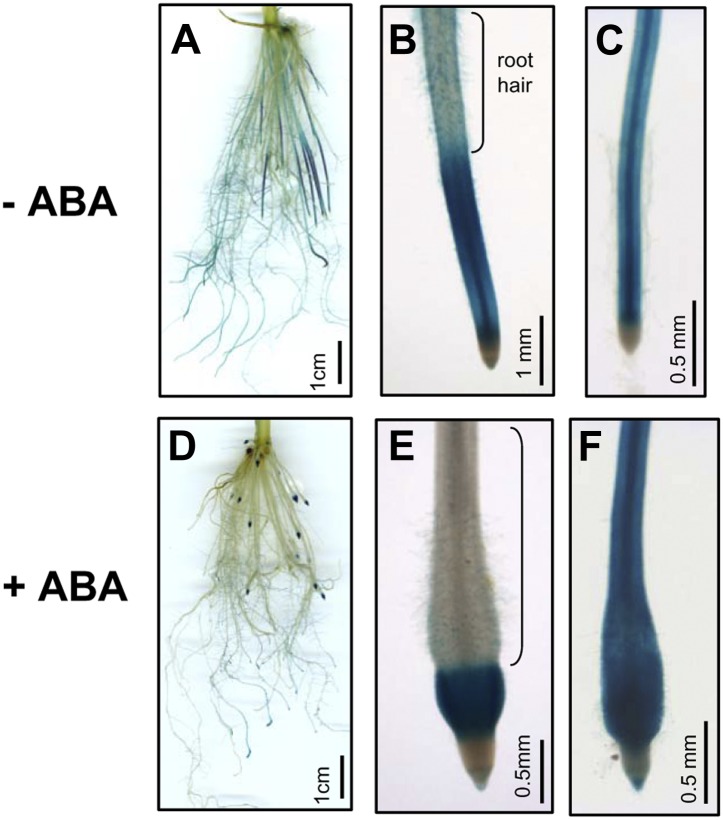
Due to rice RePRP members being highly expressed in roots (Fig. 2B), the tissue-specific expression of RePRPs was examined. The GUS reporter gene was driven by RePRP1.2 and RePRP2.1 promoters and expressed in transgenic rice. Both rice RePRP promoters directed similar GUS expression patterns in rice roots. Without ABA treatment, significant signals were detected in young roots that do not have lateral roots (Fig. 10A). In young roots, strong signal appeared above the division zone and gradually disappeared toward the root hair zone (Fig. 10B). However, in mature roots with a well-developed lateral root system, lower levels of GUS expression were detected, mainly in vascular tissues in root tips (Fig. 10C), and gradually decreased toward the basal region. In the basal regions of primary roots, GUS signals were only detected in lateral roots. One day after ABA treatment, the GUS expression patterns were unchanged in mature roots, which is consistent with the insensitivity of mature roots to ABA (Fig. 10F). However, GUS expression in young roots was shorter and restricted to the distal part of the swollen region in root tips (Fig. 10D), just between the division zone and the root hair zone (Fig. 10E). Specifically in young roots, the region with GUS expression between the division zone and the root hair zone is the elongation zone, where cells predominantly lengthen vertically (Fig. 10, B and E). Taken together, these results demonstrate that ABA induces RePRP expression in the elongation zone, where cell expansion is inhibited.
Figure 10.
RePRP genes are specifically expressed in root elongation zones. Rice RePRP1.2 promoter was used to drive GUS expression in transgenic rice. GUS was expressed in roots of transgenic plants grown in medium without ABA (−ABA; A–C) or with 10 μm ABA (+ABA; D–F). In young roots, GUS was most prominently observed in the elongation zone below the root hair zone (B and E). In mature roots, GUS was detected in primary roots behind the meristem (C and F).
DISCUSSION
Plant root systems are designed to take up water and soil nutrients so that the plant can develop, function, and reproduce. In addition to providing physical anchoring, plant roots are highly responsive to soil environmental changes such as drought, salinity, and nutrient deficiencies. Environmental stresses and the stress hormone ABA are known to alter the development and function of plant roots. As part of our effort to decipher the functions of genes involved in ABA/stress-mediated regulation of rice root development, we have identified rice RePRPs as ABA/stress-induced and root-specific genes encoding a highly Pro-rich protein with unusual PX1PX2 sequence motifs yet without known function. This work is focused on investigating the functions of RePRPs following a multidisciplinary approach combining protein biochemistry, cell biology, and reverse genetics.
RePRPs Are a Novel Class of Highly Pro-Rich Glycoproteins Existing Only in Monocotyledonous Plants
A highly Pro-rich glycoprotein in rice was first reported by Akiyama and Pillai (2003) and named OsPRP1. This protein was detected primarily in shoot tissues rather than in roots, and its expression was suppressed by submergence, ABA, or methyl jasmonate treatment. In this study, it was observed that OsPRP1 (RePRP1.1) and its homologs are encoded by two pairs of very similar genes: RePRP1.1 and RePRP1.2 appear to be direct repeats located on chromosome 5, and RePRP2.1 and RePRP2.2 are inverted repeats localized on chromosome 7. The protein products of these two subfamilies, RePRP1 and RePRP2, share 60% of similarities at the amino acid level; however, similarity levels between members in the same subfamily are as high as 94%, suggesting that subfamily members are probably products of recent gene duplication events.
Although quite a few Pro-rich proteins have been reported, the RePRP proteins described in this work are unique (Showalter, 1993; Cassab, 1998). First, the Pro content of rice RePRPs is around 40% for RePRP1 and 38% for RePRP2, significantly higher than that in the Pro-rich proteins so far reported. Second, RePRPs contain multiple copies of a unique repetitive motif, PX1PX2, where the X residues are highly hydrophilic amino acids Lys, Glu, Asn, or Gln. The unique repetitive PX1PX2 motifs set RePRP proteins apart from existing subclasses of PRPs, hence the suggestion of adopting the new name highlighting this feature. Third, rice RePRPs are glycoproteins but not typical AGPs because (1) they fail to bind to β-Yariv reagent that has been used to identify AGPs and (2) RePRPs are heavily glycosylated with Ara and Glc, instead of Ara and Gal that are expected from AGPs. Lastly, as shown in Figure 1C, rice RePRPs and their orthologs belong to a unique clade in the PRP subfamily present only in monocotyledonous plants. In maize, transcripts of a RePRP ortholog, ZmPRP, were found in xylem in the root maturation region. Because the expression pattern of ZmPRP was similar to that of genes involved in lignin biosynthesis, ZmPRP has been suggested to participate in secondary cell wall formation (Vignols et al., 1999). Another ortholog in wheat, WPRP1, shares high similarity with maize ZmPRP in protein sequence, and high levels of WPRP1 transcripts were detected in rapidly dividing tissues in shoots (Raines et al., 1991). In dicotyledonous plants, no HRGPs have repetitive PX1PX2 motifs.
RePRPs Are Localized to the Plasma Membrane
RePRPs have a standard signal peptide in the N-terminal region, which is sufficient for directing RePRPs to endoplasmic reticulum and related vesicles. Two lines of evidence suggest that at least a fraction of RePRPs are localized to the membranes, especially the plasma membrane. First, results of cell fractionation experiments indicate that RePRPs are preferentially enriched in microsomal membrane fractions (Fig. 6A). Second, RePRP-GFPs have been shown to be localized to the plasma membrane in three different cell types (i.e. stably transformed rice root cells, onion epidermal cells, and barley aleurone cells; Fig. 6, B–D). In all three cases, a fraction of RePRP-GFPs are localized to a putative plasma membrane region. This notion is confirmed in onion cells, as RePRP-GFPs stay with the plasma membrane when the cells are plasmolyzed (Fig. 6C). This observation also indicates that no detectable RePRPs are secreted into cell walls. Due to the presence of an extensive endomembrane system and the lack of a large central vacuole in barley aleurone cells, it is revealed that RePRPs are also associated with membrane vesicles (Fig. 6D). Although the nature of these membrane vesicles is not clear, it is conceivable that some of them may be involved in secretion. The nature of the rice RePRP association with membrane remains unclear. Due to the presence of a large number of Pro residues and charged polar amino acids in RePRPs, it is unlikely that membrane-spanning α-helices could be formed in RePRPs. One possible mechanism for RePRPs to be localized to the plasma membrane is by interacting with one of more integral membrane proteins even without having membrane-spanning α-helices themselves. Since RePRPs are capable of binding to the cell wall polysaccharide AG, which is similar to the AG glycan on AGPs, it is conceivable that RePRPs could become associated with the plasma membrane through its interactions with a membrane-bound AGP. Alternatively, RePRPs could be modified with a lipophilic moiety for anchoring in membranes. Membrane-bound AGPs are known to have a GPI anchor, which is added to the C terminus of a protein after its synthesis is completed (Borner et al., 2002, 2003). All known GPI-anchoring proteins from diverse organisms have an N-terminal signal sequence for targeting to the endoplasmic reticulum, a hydrophobic C-terminal sequence, and no internal transmembrane helices (Borner et al., 2002). Although RePRPs seem to fit with at least two of these criteria (i.e. N-terminal signal peptide and no internal transmembrane helices), analyses of RePRP sequences in the GPI prediction program by the big-PI Plant Predictor for GPI modification site prediction (http://mendel.imp.ac.at/gpi/plant_server.html) indicate that RePRPs are not likely to have a C-terminal sequence that is hydrophobic enough for the addition of a GPI anchor. Furthermore, RePRPs also do not contain sequences needed for attaching to other lipid anchors, such as farnesylation and prenylation. Nevertheless, future experimental determination of a potential lipophilic anchor is warranted for investigating the nature of the RePRP association with membrane.
RePRPs Are Essential for ABA/Stress-Regulated Root Development
RePRPs are up-regulated in roots by abiotic stress or ABA treatment that also inhibits root growth in rice. It has been demonstrated that under severe drought stress, the overall length of maize and soybean (Glycine max) roots is reduced (Sharp et al., 2004; Yamaguchi et al., 2010). It is important to note that shorter roots are the consequence of reduced growth rate in the elongation zone rather than in the cell division zone (Sharp et al., 2004; Yamaguchi et al., 2010). We have observed that ABA treatment also leads to shorter and more branched roots, which is consistent with previously published work (Chen et al., 2006). Our findings that ABA treatment restricts RePRP expression in the root elongation zone and not in the division zone suggest that RePRPs may be responsible for the inhibitory effect of ABA/stress on root elongation. Furthermore, transgenic rice lines overexpressing RePRP2.1 display a shorter root phenotype, which supports the notion that RePRPs could be involved in the inhibition of root growth. On the other hand, knockdown expression of all RePPRs by RNAi designed for both the RePRP1 and RePRP2 subfamilies in rice reduces the inhibitory effect of ABA on root growth. Taken together, these observations suggest that RePRPs are essential for root growth inhibition by ABA or stress.
RePRPs Are Involved in the Regulation of Cell Expansion
This work has also addressed the cause of altered root growth at the cellular level. When treated with ABA, the morphology of rice roots becomes shorter and wider in epidermis, exodermis, sclerenchyma, and cortex in the elongation zone, but few morphological changes were observed in other regions (Chen et al., 2006). Such phenotypes are also observed in the roots of RePRP2.1(ox) transgenic rice plants without ABA treatment. In ABA-treated roots of double RNAi lines targeting the RePRPs, the cell shape remains long as in the wild type. These observations suggest that RePRP proteins participate in the regulation of cell elongation.
Cell elongation is regulated by turgor pressure and cell wall expansion. However, the turgor pressure along the elongation zone was equally reduced in both well-watered and water deficit-stressed conditions, suggesting that cell wall extensibility is the major factor in the regulation of root cell growth (Spollen and Sharp, 1991; Zhu et al., 2007). In water-stressed roots, a reduced pH was detected in the apical region (Wu et al., 1996; Fan and Neumann, 2004; Fan et al., 2006; Zhu et al., 2007). The acidic pH could be responsible for the acid-induced extensibility that maintains the cell growth rate. In contrast, cell wall extensibility progressively decreased above the apical region associated with the lower growth rate. The extensibility of roots from RePRP2.1(ox) and RePRP-RNAi lines could be examined to gain further insights into the involvement of these proteins in regulating root growth.
The glycoprotein AGPs have been demonstrated to participate in the regulation of cell expansion. For example, root growth was inhibited and the epidermal cells above the meristem were abnormally expanded in Arabidopsis seedlings grown in medium treated with β-Yariv, which specifically interacts with AGPs and interferes with their functions (Willats and Knox, 1996; Ding and Zhu, 1997). Similarly, elongation of cultured carrot (Daucus carota) cells was also inhibited by β-Yariv supplementation (Willats and Knox, 1996; Ding and Zhu, 1997). In addition, AGPs also regulate cell expansion in the moss Physcomitrella patens (Lee et al., 2005), and moss AGP1 knockout mutants showed reduction of cell length in protonemal filaments. The protein product of SALT OVERLY SENSITIVE5 (SOS5), one of the AGP genes in Arabidopsis, is localized to the plasma membrane and is likely required for cell adhesion. Under high-salinity conditions, sos5 mutants showed root tip swelling with abnormally expanded epidermal cells and cortical cells (Shi et al., 2003). FEI is a Leu-rich repeat receptor kinase, deletion of which leads to defective synthesis of cell wall polymers and causes abnormal cell expansion in high-Suc conditions (Xu et al., 2008b). Interestingly, even more severe defects were observed in sos5/fei1/fei2 mutants than in individual mutants, suggesting that SOS5 and FEI could be in the same pathway controlling cell expansion. Similarly, overexpression of RePRP in rice roots causes shorter and wider cells in the epidermis, exodermis, sclerenchyma, and cortex, suggesting that RePRPs are one of the important regulators controlling cell expansion.
Significance of the Specific Binding of RePRPs to the Polysaccharide AG
Plant cell walls are composed of cellulose microfibrils cross linked with hemicelluloses, and cell expansion is tightly controlled by cell wall extensibility (Cosgrove, 2005). Cell wall loosening is the crucial factor in facilitating cell expansion during growth and development. In roots, the highest cell expansion rate occurs in the elongation zone. Expansins, xyloglucan endotransglycosylases, and endo-(1,4)-β-d-glucanases have been suggested as candidates for loosening cell walls through interfering cross linkages between celluloses and hemicelluloses (Geitmann and Ortega, 2009). In addition, methyl esterification of pectin plays an important role in the regulation of root cell expansion. A low degree of methyl esterification on the carboxyl groups of homogalacturonan permits pectin molecules to cross link each other easily through Ca2+ and makes the cell wall more rigid (Proseus and Boyer, 2007). In Arabidopsis, nondividing cells of the quiescent center contain most unesterified pectins, but dividing and elongating cells carry abundant methyl-esterified pectins in cell walls, suggesting that pectin modification is involved in root cell expansion (Dolan et al., 1997). It has been demonstrated in this work that rice RePRP proteins specifically interact with AG in a dosage-dependent manner. Exudate gums with abundant AG and AGPs are known to be synthesized and secreted when plant cells are wounded and stressed (Verbeken et al., 2003). It is likely that the plasma membrane-localized RePRPs interact with AG in cell walls, affect the wall properties, and thus lead to the suppression of cell elongation.
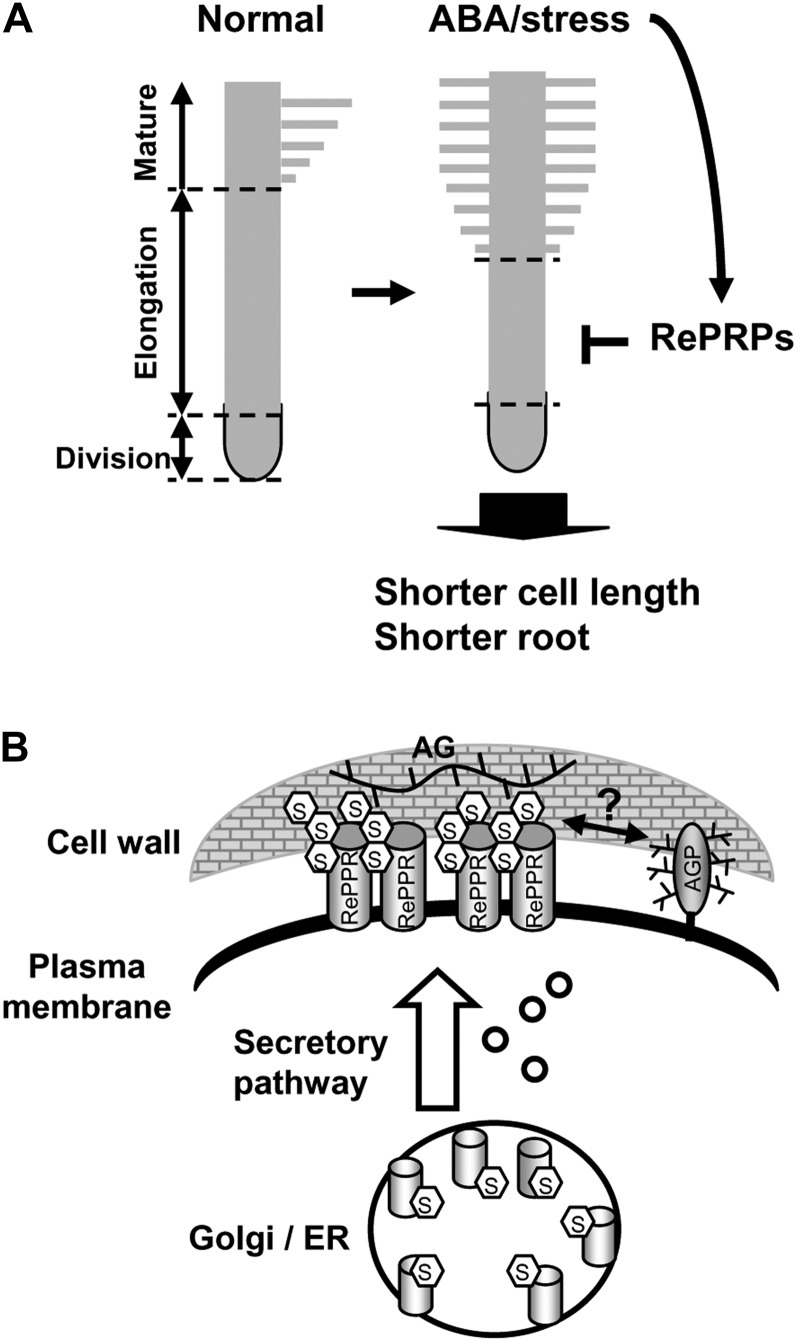
Based on observations made in this work, the potential role of RePRPs is proposed as shown in Figure 11. The synthesis of RePRPs is induced by ABA/stress in rice roots, especially in the elongation zone of young growing roots. RePRPs are modified with O-glycosylation on Hyp residues and are rich in Ara and Glc. Some RePRPs are localized to the plasma membrane, where they interact with the polysaccharide AG in the walls, which is produced when plant cells are wounded or under stress, leading to the repression of cell elongation. Alternatively, RePRPs could exert their regulatory role on the function of cell walls by binding to the AG moiety of plasma membrane-localized AGP, which has recently been shown to be linked to cell wall polysaccharides arabinoxylan and pectin (Tan et al., 2013). Proper O-glycosylation of RePRPs could be important for its function, similar to the role of O-glycosylation on HRGP regulation of root hair development (Velasquez et al., 2011).
Figure 11.
Role of rice RePRPs in ABA/stress repression of root elongation. A, ABA strongly induces RePRP expression specifically in the elongation region in young roots to inhibit length-wise cell expansion, causing roots to be shorter than those grown under normal conditions. B, RePRPs are glycosylated in the Pro-rich domain and localized on the plasma membrane. The glycosylated RePRPs are able to interact with cell wall polysaccharide AG or AGPs to potentially interfere with root cell expansion. ER, Endoplasmic reticulum.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Seeds of rice (Oryza sativa ‘Tainung 67’, a japonica type) were sterilized with 2% (w/v) sodium hypochlorite for 30 min and washed thoroughly with distilled water. To obtain uniform germination, rice seeds were soaked in distilled water at 37°C for 1 d in darkness and then germinated in petri dishes (20 cm) containing distilled water at 37°C. Transgenic rice seeds were germinated in water containing hygromycin B (30 μg mL−1; Invitrogen) at 37°C for 3 d to select for transgene-containing seedlings. Uniformly germinated seeds were then selected and cultivated in a beaker containing one-half-strength Kimura B solution (Hsu and Kao, 2003). The hydroponically cultivated seedlings were grown at 28°C and 90% relative humidity in a 14-h-light/10-h-dark condition.
Generation of Transgenic Rice Lines
For the generation of RePRP2.1(ox) transgenic plants, the rice RePRP2.1 coding region was cloned behind the maize (Zea mays) Ubi1 promoter, with its first intron in the pPZP binary vector (Hajdukiewicz et al., 1994). For the generation of RePRP-RNAi lines, the coding regions of rice RePRP1.1 and RePRP2.1 were isolated by genomic PCR and fused together in pCR8/GW/TOPO. The combined fragment was cloned into the pANDA binary vector (Miki and Shimamoto, 2004) by site-specific recombination (LR Clonase; Invitrogen). For the generation of RePRP-GFP transgenic plants, each RePRP1.1 and RePRP2.1 coding region fused with GFP was controlled by the maize Ubi1 promoter in the pANDA backbone. For the generation of RePRPprom:GUS lines, rice RePRP1.2 (2.7 kb) and RePRP2.1 (2.4 kb) promoters upstream of the transcription start site were isolated from the rice genome and cloned into pCAMBA1381Z vector to control the expression of the GUS reporter gene. Transgenic rice was generated via Agrobacterium tumefaciens-mediated transformation as described (Hong et al., 2004).
Reverse Transcription-PCR and Quantitative Reverse Transcription-PCR Analysis
Primer sets used for reverse transcription (RT)-PCR and quantitative real time-PCR are provided in Supplemental Table S4. Total RNA was isolated from different tissues using the TRIzol reagent (Invitrogen) according to the supplier’s recommendations. The first-strand complementary DNA was synthesized using the SuperScript III first-strand synthesis system (Invitrogen). Quantitative RT-PCR was carried out using the SYBR Advantage qPCR Premix (Clontech) with the 7500 Real-Time PCR System (Applied Biosystems). The thermal cycling conditions were 95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. Gene-specific primers were designed from the 3′ untranslated region of the rice RePRP genes. The rice ACTIN gene was used for normalization.
Western-Blot Analysis
Specific anti-RePRP antibodies were generated against full-length RePRP1.1 or RePRP2.1 recombinant proteins produced from Escherichia coli (LTK Biolaboratories). In order to improve the antibody specificity, antisera were further purified through RePRP recombinant conjugated resin. Antibody specificity tests by western blot were performed: anti-RePRP1 antibody was able to recognize RePRP1.1 and RePRP1.2 recombinant proteins simultaneously, and anti-RePRP2 antibody recognized both RePRP2.1 and RePRP2.2 recombinant proteins but slightly cross reacted with recombinant RePRP1.1 and RePRP1.2 proteins. For the detection of rice endogenous RePRP proteins, total proteins were extracted from leaves and roots of rice seedlings in extraction buffer (50 mm Tris-HCl, pH 8.5, 2% [w/v] SDS, 2% [v/v] β-mercaptoethanol, and protease inhibitor mix [one tablet of protease inhibitor cocktail in 50 mL; Roche]). Fifteen micrograms of total proteins was loaded on 10% or 12% (w/v) SDS-polyacrylamide gels and analyzed by western blotting. Rice endogenous RePRP1 and RePRP2 proteins were detected by specific anti-RePRP1 and anti-RePRP2 rabbit polyclonal antibodies, respectively, and horseradish peroxidase-conjugated anti-rabbit antibodies (Perkin-Elmer) were used as secondary antibodies.
β-Glc Yariv Binding Assay
The binding assay was carried out according to an AGP purification protocol with slight modification (Schultz et al., 2000). Shoot proteins were extracted from 2-week-old seedlings in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% [v/v] Triton X-100, 1 mm EDTA, and 1% [w/v] sodium deoxycholate), and the buffer was exchanged with 1% (w/v) NaCl by ultrafiltration (Vivaspin 4, molecular weight cutoff 10 kDa; Sartorius Stedim). Fifty micrograms of gum arabic (no. 100-7; Biosupplies) was used as a standard, and 100 µg of shoot total proteins was incubated with 50 µg of β-d-Glc Yariv (no. 100-2; Biosupplies) or α-d-Man Yariv (no. 100-5; Biosupplies) at 4°C overnight. Supernatant and pellet fractions were separated by centrifugation at 10,000g for 1 h. Proteins in supernatant or pellet fraction were dissolved in 0.1 n NaOH and analyzed by 12% (w/v) SDS-PAGE. Gum arabic proteins in SDS-PAGE gel were detected by periodic acid-Schiff stain according to the supplier’s recommendations (Pierce Glycoprotein Staining Kit; Thermo Scientific), and RePRP proteins were detected by western-blot analysis.
Protein Identification and Analysis of Methylamine-Treated Proteins by LC-MS/MS
In transgenic rice overexpressing RePRPs, rice RePRP1 and RePRP2 proteins were purified by immunoaffinity chromatography from shoots and roots, respectively. Anti-RePRP antibodies were covalently linked to protein G magnetic Sepharose, and the beads were used to purify RePRP proteins from tissue extracts according to the supplier’s recommendations (GE Healthcare). One to 10 μg of proteins was analyzed by SDS-PAGE, and protein bands were treated with methylamine prior to trypsin digestion for LC-MS/MS analysis as described previously (Hanisch et al., 2001). An LTQ mass spectrometer coupled with an online capillary LC system from Thermo Fisher Scientific was utilized for protein identification and analysis. The capillary LC system equipped with a C18 trap cartridge (Zobax 300SB-C18, 5 µm, 5 × 0.3 mm; Agilent Technologies) and a C18 reverse-phase column (BioBasic C18, 75 µm × 10 cm, PicoFrit column; New Objective) was used to deliver solvent and tryptic peptides with a linear gradient from 5% to 40% (v/v) acetonitrile in 0.1% (v/v) formic acid for 60 min at nanoflow (approximately 300 nL min−1) rate. The PicoFrit column was coupled to a nanoelectrospray ionization source, and the acquisition of the data was performed with a full MS scan followed by four MS/MS scans of the top four precursor ions from the MS scan. The acquired MS/MS data were analyzed using a SEQUEST search program (BioWorks Browser 3.3; Thermo Fisher Scientific) against a rice database downloaded from the National Center for Biotechnology Information. The matched peptides were accepted when they passed multiple filters: cross-correlation score ≥ 1.5 for singly charged ions (z = 1), 2.0 for doubly charged ions (z = 2), 2.5 for triply charged ions (z = 3), difference in cross-correlation score ≥ 0.1, and peptide probability ≤ 1 × 10−3. Matched proteins were accepted only when they had at least two distinct peptide hits.
Sugar Composition Analysis
Purified rice RePRP glycoproteins were treated with 4 n trifluoroacetic acid at 110°C for 4 h to hydrolyze glycan chains into monosaccharides as described (Packer et al., 1999). Vacuum-dried sugars were dissolved in distilled water for composition analysis by HPAEC (ICS-5000; Dionex), where 25 μL of sample was injected into the system with a CarboPac PA10. Sugar was analyzed with 10 mm NaOH for 20 min. The column was recharged with 200 mm NaOH for 10 min and balanced with 10 mm NaOH for 20 min.
Polysaccharide Interaction Analysis
Xylan, glucan, pectin, and AG were purchased from Sigma. Xyloglucan, rhamnogalacturonan I, and arabinoxylan were purchased from Megazyme. Interaction response was monitored by SPR with a Biacore-T100 system (GE Healthcare). All experiments were performed at room temperature using 1× HEPES buffered saline-sodium chloride (HBS-N) buffer (GE Healthcare) containing 1 mm CaCl2 and 1 mm MgCl2 as the running buffer for dissociation. To examine the interaction of rice RePRP glycoproteins with polysaccharides, anti-RePRP antibodies were immobilized on a CM5 sensor chip using standard amine-coupling chemistry. Purified rice RePRP proteins were first captured by antibodies, and each polysaccharide dissolved in the running buffer (5 mg mL−1 in 1× HBS-N, 1 mm CaCl2, and 1 mm MgCl2) was injected for 60 s (flow rate of 10 or 30 μL min−1). Running buffer was used for dissociation (30 μL min−1 for 120 s), and sodium acetate (10 mm, pH 4.6) was used for regeneration (30 μL min−1 for 60 s). To study the interaction between RePRP recombinant proteins and AG, RePRP1, RePRP2, and GST were immobilized on a CM5 sensor chip using the amine-coupling procedure. Serial concentrations of AG were injected through a sensor chip for 5 min to measure the binding response. Gly (10 mm, pH 2.5) was used for regeneration (30 μL min−1 for 30 s). To study the binding affinity of AG with rice RePRP proteins, AG (1 mg mL−1 in 1× HBS-N, 1 mm CaCl2, and 1 mm MgCl2) was incubated with various concentrations of purified RePRP1 or RePRP2 for 16 h at 4°C. The mixture was injected through a recombinant RePRP1-coated CM5 sensor chip for 3 min.
Soluble Versus Membrane Protein Fractionation
ABA-treated roots (0.5 g) were ground to fine powder with liquid nitrogen and mixed well with 4 mL of extraction buffer (15 mm Tris-HCl, pH 7.8, 0.25 m Suc, 1 mm EDTA, 2 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, 0.6% [w/v] polyvinylpyrrolidone, and protease inhibitor mix [one tablet of protease inhibitor cocktail in 50 mL; Roche]). The homogenate was initially centrifuged at 10,000g for 15 min at 4°C, and then the supernatant was further centrifuged at 100,000g for 1 h at 4°C. The cytosolic proteins were in the supernatant fraction. The pellet containing proteins in the membrane fraction was dissolved in 4% (w/v) SDS completely. Each fraction was used to detect rice RePRP proteins by western-blot analysis.
Subcellular Localization Analysis
For the detection of RePRP-GFP in rice cells, root tissues of 5-d-old rice seedlings were used for GFP observation. Rice roots were counterstained with 5 mg L−1 propidium iodide solution for 10 min and examined with a Zeiss META 510 confocal microscope. The images were captured in the 505- to 530-nm range for GFP and the 535- to 617-nm range for propidium iodide after excitation at 488 nm with an argon laser beam.
For onion (Allium cepa) transformation, the inner epidermis of an onion scale was peeled and placed inside up on an agar plate containing Murashige and Skoog medium with 0.3% (w/v) phytagel, then the bombardment was performed according to the procedure provided with the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad). One-micrometer-diameter gold particles were coated with 5-μg plasmid constructs for three shots of bombardment according to the manufacturer’s protocol. Bombarded epidermal cells were incubated at 25°C in the dark for 16 h.
For barley (Hordeum vulgare) transformation, barley embryoless half-seeds were prepared and transformed transiently by particle bombardment as described (Shen et al., 1993). Briefly, imbibed, deembryonated half-seeds from Himalaya barley were incubated in buffer for 2 d at 27°C, and then the pericarp and testa were removed. Each RePRP1.1 and RePRP2.1 coding region fused with GFP was controlled by the maize Ubi1 promoter in a pANDA-mini background. Signal peptides of RePRPs, amino acid positions 1 to 23 of RePRP1 and 1 to 20 of RePRP2, were fused with GFP for the signal peptide at the N-terminal region of rice RePRP-GFP. Three-microgram plasmids for each construct were bombarded into barley embryoless half-seeds. After particle bombardment, the half-seeds were incubated for 24 to 48 h at 25°C in the dark. After the starchy endosperm was removed, the isolated aleurone layers were used for GFP observation.
TEM
Young root tips (2 mm) were dissected from 12-d-old rice seedlings with or without 20 μm ABA treatment for 1 d. These root tips were fixed in 4% (v/v) paraformaldehyde and 0.1% (v/v) glutaraldehyde in 0.1 m phosphate buffer, pH 7.0, at 4°C for 4 h. After three 20-min buffer rinses, the samples were dehydrated in an alcohol series. LR White resin was used for infiltration and embedding. Ultrathin longitudinal sections (90–120 nm) were cut using a Reichert Ultracut S or Lecia EM UC6 and collected using 100-mesh nickel grids. For immunogold labeling, the individual grids were floated on Tris-buffered saline (TBS) for 15 min and then TBS and 1% (w/v) bovine serum albumin for 15 min. The grids were incubated with anti-RePRP2 polyclonal antibodies (diluted five times in TBS and 1% [w/v] bovine serum albumin) for 1 h. After being washed four times with TBS, the grids were floated on an excess amount (1:20 dilution) of 12-nm colloidal donkey anti-rabbit IgG (Jackson ImmunoResearch) at room temperature for 1 h. Then the grids were washed sequentially with three droplets of TBS, followed by three washes with distilled, deionized water. After immunogold labeling, the sections were stained with 5% (w/v) uranyl acetate in water for 10 min and 0.4% (w/v) lead citrate for 6 min. Sections were observed using a Philips CM 100 Transmission Electron Microscope at 80 kV, and the images were captured with a Gatan Orius CCD camera.
Root Growth in Response to Hormone and Stress Treatments
For the measurement of RePRP transcripts in response to ABA, hydroponically cultivated 2-week-old rice seedlings were treated with 20 μm ABA for 0.5 to 3 d, and roots and shoots were collected separately for quantitative RT-PCR analysis.
For the assessment of root growth in response to ABA and salinity stress, 4-d-old seedlings grown on the surface of Murashige and Skoog agar plates were transferred to Murashige and Skoog agar supplemented with 0.5 μm ABA or 150 mm NaCl. Seedlings were grown on agar vertically in the closed chamber with saturated humidity in a 14-h-light/10-h-dark condition. Root length was recorded and calculated by the software ImageJ (http://rsb.info.nih.gov/ij/).
For the examination of root growth in response to ABA, 6-d-old seedlings were treated with 1 μm ABA in a hydroponic growth system for 8 d. Root images were taken and medium was replaced every 2 d. Three independent lines of transgenic plants and eight individuals of the wild type or each transgenic line were taken for each experiment. Root length was calculated from images by ImageJ.
Histochemical Analysis
For GUS staining, whole rice seedlings were incubated in GUS staining solution at 37°C for 30 to 60 min as described (Jefferson et al., 1987). Tissues were cleared with 70% (v/v) ethanol and examined with a Nikon SMZ 1500 stereoscopic microscope.
Sequence data from this article can be found in GenBank/EMBL libraries with the following accession numbers: rice OsRePRP1.1 (BAF16879), OsRePRP1.2 (BAF16881), OsRePRP2.1 (BAF21387), and OsRePRP2.2 (BAF21386), maize PRP (Q9ZNY1), wheat PRP (Q01979), sorghum PRP (ABR08569.1), sugarcane PRP (hybrid cv CP65-357; AF331851_1), barley PRP (BAJ85705.1), Arabidopsis AtPRP1 to AtPRP4 (AAF64548, AAF64549, AAF64550, and AAF64551), common bean (Phaseolus vulgaris) PRP (CAA42942.1), soybean PRP (AAA66287.1), Medicago truncatula PRP (AES71726.1), tomato (Solanum lycopersicum) PRP (CAA43666.1), tobacco (Nicotiana alata) PRP (CAA49895.1), and cotton (Gossypium hirsutum) PRP (AFH57274.1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Alignment of deduced protein sequences of rice RePRP, monocot orthologs, and AtPRP.
Supplemental Figure S2. Drought reduces rice root growth and enhances RePRP expression.
Supplemental Figure S3. RePRP recombinant proteins specifically interact with AG.
Supplemental Figure S4. Binding affinity of AG with RePRP proteins.
Supplemental Figure S5. Effects of ABA on root growth in RePRP2 knockdown lines.
Supplemental Figure S6. Effects of ABA on plant growth in reprp1 mutants.
Supplemental Table S1. RePRP1 protein identification by LC-MS/MS.
Supplemental Table S2. RePRP2 protein identification by LC-MS/MS.
Supplemental Table S3. Glycosyl composition of RePRP proteins.
Supplemental Table S4. Primer sets for RT-PCR and quantitative RT-PCR.
Acknowledgments
We are grateful to the Proteomics Core Facility (Institute of Plant and Microbial Biology, Academia Sinica) for LC-MS/MS analyses and the Plant Cell Biology Core Laboratory (Institute of Plant and Microbial Biology, Academia Sinica) for TEM analyses. We also appreciate the able assistance with HPAEC-pulsed-amperometric detection analyses by Dr. Wen-Ben Yang (Genomics Research Center, Academia Sinica), Hsia-Jun Yen (Institute of Plant and Molecular Biology, Academia Sinica), and Yu-Ching Wu (Metabolomics Core Facility, Institute of Plant and Microbial Biology, Academia Sinica). We are also thankful for the valuable consultations about SPR analyses provided by Dr. Shy-Chuan Jao (Biophysics Core Facility, Academia Sinica) and Dr. Chuan-Fa Chang (Department of Medical Technology, National Chen Kung University). We thank our colleagues at Academia Sinica, Shu-Hsing Wu, Paul E. Verslues, Jychian Chen, and Yi-Fang Tsay, for helpful discussions during the course of this research.
Glossary
- ABA
abscisic acid
- HRGP
hydroxyproline-rich glycoprotein
- AGP
arabinogalactan protein
- AG
arabinogalactan
- GPI
glycosylphosphoinositol
- LC
liquid chromatography
- MS/MS
tandem mass spectrometry
- MS
mass spectrometry
- HPAEC
high-performance anion-exchange chromatography
- TEM
transmission electron microscopy
- SPR
surface plasmon resonance
- GST
glutathione S-transferase
- RNAi
RNA interference
- RePRP2.1(ox)
RePRP2.1-overexpressing
- RePRP(Ri)
RNA interference of RePRP1 and RePRP2
- RT
reverse transcription
- TBS
Tris-buffered saline
References
- Akiyama T, Pillai MA. (2003) Isolation and characterization of a gene for a repetitive proline rich protein (OsPRP) down-regulated during submergence in rice (Oryza sativa). Physiol Plant 118: 507–513 [Google Scholar]
- Borner GH, Sherrier DJ, Stevens TJ, Arkin IT, Dupree P. (2002) Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a genomic analysis. Plant Physiol 129: 486–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GHH, Lilley KS, Stevens TJ, Dupree P. (2003) Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: a proteomic and genomic analysis. Plant Physiol 132: 568–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassab GI. (1998) Plant cell wall proteins. Annu Rev Plant Physiol Plant Mol Biol 49: 281–309 [DOI] [PubMed] [Google Scholar]
- Chen CW, Yang YW, Lur HS, Tsai YG, Chang MC. (2006) A novel function of abscisic acid in the regulation of rice (Oryza sativa L.) root growth and development. Plant Cell Physiol 47: 1–13 [DOI] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E. (2006) Integration of abscisic acid signalling into plant responses. Plant Biol (Stuttg) 8: 314–325 [DOI] [PubMed] [Google Scholar]
- Close TJ. (1996) Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plant 97: 795–803 [Google Scholar]
- Cosgrove DJ. (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. (2010) Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679 [DOI] [PubMed] [Google Scholar]
- Ding L, Zhu J-K. (1997) A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 203: 289–294 [DOI] [PubMed] [Google Scholar]
- Dolan L, Linstead P, Roberts K. (1997) Developmental regulation of pectic polysaccharides in the root meristem of Arabidopsis. J Exp Bot 48: 713–720 [Google Scholar]
- Dure L III. (1993) A repeating 11-mer amino acid motif and plant desiccation. Plant J 3: 363–369 [DOI] [PubMed] [Google Scholar]
- Ellis M, Egelund J, Schultz CJ, Bacic A. (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol 153: 403–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Linker R, Gepstein S, Tanimoto E, Yamamoto R, Neumann PM. (2006) Progressive inhibition by water deficit of cell wall extensibility and growth along the elongation zone of maize roots is related to increased lignin metabolism and progressive stelar accumulation of wall phenolics. Plant Physiol 140: 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Neumann PM. (2004) The spatially variable inhibition by water deficit of maize root growth correlates with altered profiles of proton flux and cell wall pH. Plant Physiol 135: 2291–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH. (1994) Plant bZIP proteins gather at ACGT elements. FASEB J 8: 192–200 [DOI] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Marten I, Ache P, Matschi S, Liese A, Wellmann C, Al-Rasheid KA, Grill E, Romeis T, Hedrich R. (2010) Guard cell anion channel SLAC1 is regulated by CDPK protein kinases with distinct Ca2+ affinities. Proc Natl Acad Sci USA 107: 8023–8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger D, Scherzer S, Mumm P, Stange A, Marten I, Bauer H, Ache P, Matschi S, Liese A, Al-Rasheid KA, et al. (2009) Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc Natl Acad Sci USA 106: 21425–21430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geitmann A, Ortega JKE. (2009) Mechanics and modeling of plant cell growth. Trends Plant Sci 14: 467–478 [DOI] [PubMed] [Google Scholar]
- Guiltinan MJ, Marcotte RW, Jr, Quatrano RS. (1990) A plant leucine zipper protein that recognizes an abscisic acid response element. Science 250: 267–271 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hanisch F-G, Jovanovic M, Peter-Katalinic J. (2001) Glycoprotein identification and localization of O-glycosylation sites by mass spectrometric analysis of deglycosylated/alkylaminylated peptide fragments. Anal Biochem 290: 47–59 [DOI] [PubMed] [Google Scholar]
- Hong CY, Cheng KJ, Tseng TH, Wang CS, Liu LF, Yu SM. (2004) Production of two highly active bacterial phytases with broad pH optima in germinated transgenic rice seeds. Transgenic Res 13: 29–39 [DOI] [PubMed] [Google Scholar]
- Hsu YT, Kao CH. (2003) Role of abscisic acid in cadmium tolerance of rice (Oryza sativa L.) seedlings. Plant Cell Environ 26: 867–874 [DOI] [PubMed] [Google Scholar]
- Ingram J, Bartels D. (1996) The molecular basis of dehydration tolerance in plants. Annu Rev Plant Physiol Plant Mol Biol 47: 377–403 [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Wang Y, Zhang S, Zhang J. (2002) Salt-stress-induced ABA accumulation is more sensitively triggered in roots than in shoots. J Exp Bot 53: 2201–2206 [DOI] [PubMed] [Google Scholar]
- Kang JY, Choi HI, Im MY, Kim SY. (2002) Arabidopsis basic leucine zipper proteins that mediate stress-responsive abscisic acid signaling. Plant Cell 14: 343–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieliszewski MJ. (2001) The latest hype on Hyp-O-glycosylation codes. Phytochemistry 57: 319–323 [DOI] [PubMed] [Google Scholar]
- Kieliszewski MJ, Shpak E. (2001) Synthetic genes for the elucidation of glycosylation codes for arabinogalactan-proteins and other hydroxyproline-rich glycoproteins. Cell Mol Life Sci 58: 1386–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa K, Tryfona T, Yoshimi Y, Hayashi Y, Kawauchi S, Antonov L, Tanaka H, Takahashi T, Kaneko S, Dupree P, et al. (2013) β-Galactosyl Yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiol 161: 1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline KG, Barrett-Wilt GA, Sussman MR. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc Natl Acad Sci USA 107: 15986–15991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KJD, Sakata Y, Mau S-L, Pettolino F, Bacic A, Quatrano RS, Knight CD, Knox JP. (2005) Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. Plant Cell 17: 3051–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhao J. (2010) Genome-wide identification, classification, and expression analysis of the arabinogalactan protein gene family in rice (Oryza sativa L.). J Exp Bot 61: 2647–2668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2012) AP2/ERF family transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 86–96 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Takasaki H, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. (2012) NAC transcription factors in plant abiotic stress responses. Biochim Biophys Acta 1819: 97–103 [DOI] [PubMed] [Google Scholar]
- Nakashima K, Tran LS, Van Nguyen D, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Yamaguchi-Shinozaki K. (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51: 617–630 [DOI] [PubMed] [Google Scholar]
- Negi J, Matsuda O, Nagasawa T, Oba Y, Takahashi H, Kawai-Yamada M, Uchimiya H, Hashimoto M, Iba K. (2008) CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 452: 483–486 [DOI] [PubMed] [Google Scholar]
- Negrao S, Courtois B, Ahmadi N, Abreu I, Saibo N, Oliveira MM. (2011) Recent updates on salinity stress in rice: from physiological to molecular responses. Crit Rev Plant Sci 30: 329–377 [Google Scholar]
- Nothnagel EA. (1997) Proteoglycans and related components in plant cells. Int Rev Cytol 174: 195–291 [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Luis Reyes J, Covarrubias AA. (2011) Late embryogenesis abundant proteins: versatile players in the plant adaptation to water limiting environments. Plant Signal Behav 6: 586–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer N, Ball M, Devine P (1999) Glycoprotein detection of 2-D separated proteins. In A Link, ed, 2-D Proteome Analysis Protocols, Vol 112. Humana Press, Totawa, NJ, pp 341-352 [DOI] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS. (2007) Tension required for pectate chemistry to control growth in Chara corallina. J Exp Bot 58: 4283–4292 [DOI] [PubMed] [Google Scholar]
- Qin F, Shinozaki K, Yamaguchi-Shinozaki K. (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52: 1569–1582 [DOI] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. (2010) ABA perception and signalling. Trends Plant Sci 15: 395–401 [DOI] [PubMed] [Google Scholar]
- Raines CA, Lloyd JC, Chao SM, John UP, Murphy GJ. (1991) A novel proline-rich protein from wheat. Plant Mol Biol 16: 663–670 [DOI] [PubMed] [Google Scholar]
- Saab IN, Sharp RE, Pritchard J, Voetberg GS. (1990) Increased endogenous abscisic acid maintains primary root growth and inhibits shoot growth of maize seedlings at low water potentials. Plant Physiol 93: 1329–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A, Sato Y, Fukao Y, Fujiwara M, Umezawa T, Shinozaki K, Hibi T, Taniguchi M, Miyake H, Goto DB, et al. (2009) Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem J 424: 439–448 [DOI] [PubMed] [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A. (2000) The classical arabinogalactan protein gene family of arabidopsis. Plant Cell 12: 1751–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. (2002) Using genomic resources to guide research directions: the arabinogalactan protein gene family as a test case. Plant Physiol 129: 1448–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert GJ, Roberts K. (2007) The biology of arabinogalactan proteins. Annu Rev Plant Biol 58: 137–161 [DOI] [PubMed] [Google Scholar]
- Sharp RE. (2002) Interaction with ethylene: changing views on the role of abscisic acid in root and shoot growth responses to water stress. Plant Cell Environ 25: 211–222 [DOI] [PubMed] [Google Scholar]
- Sharp RE, LeNoble ME. (2002) ABA, ethylene and the control of shoot and root growth under water stress. J Exp Bot 53: 33–37 [PubMed] [Google Scholar]
- Sharp RE, Poroyko V, Hejlek LG, Spollen WG, Springer GK, Bohnert HJ, Nguyen HT. (2004) Root growth maintenance during water deficits: physiology to functional genomics. J Exp Bot 55: 2343–2351 [DOI] [PubMed] [Google Scholar]
- Shen Q, Uknes SJ, Ho TH. (1993) Hormone response complex in a novel abscisic acid and cycloheximide-inducible barley gene. J Biol Chem 268: 23652–23660 [PubMed] [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. (2003) The arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15: 19–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. (1993) Structure and function of plant cell wall proteins. Plant Cell 5: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM, Keppler B, Lichtenberg J, Gu D, Welch LR. (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol 153: 485–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spollen WG, Sharp RE. (1991) Spatial distribution of turgor and root growth at low water potentials. Plant Physiol 96: 438–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki H, Maruyama K, Kidokoro S, Ito Y, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K, Nakashima K. (2010) The abiotic stress-responsive NAC-type transcription factor OsNAC5 regulates stress-inducible genes and stress tolerance in rice. Mol Genet Genomics 284: 173–183 [DOI] [PubMed] [Google Scholar]
- Tan L, Eberhard S, Pattathil S, Warder C, Glushka J, Yuan C, Hao Z, Zhu X, Avci U, Miller JS, et al. (2013) An arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25: 270–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilman D, Cassman KG, Matson PA, Naylor R, Polasky S. (2002) Agricultural sustainability and intensive production practices. Nature 418: 671–677 [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. (2009) Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA 106: 17588–17593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K. (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci USA 97: 11632–11637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasquez SM, Ricardi MM, Dorosz JG, Fernandez PV, Nadra AD, Pol-Fachin L, Egelund J, Gille S, Harholt J, Ciancia M, et al. (2011) O-glycosylated cell wall proteins are essential in root hair growth. Science 332: 1401–1403 [DOI] [PubMed] [Google Scholar]
- Verbeken D, Dierckx S, Dewettinck K. (2003) Exudate gums: occurrence, production, and applications. Appl Microbiol Biotechnol 63: 10–21 [DOI] [PubMed] [Google Scholar]
- Vignols F, José-Estanyol M, Caparrós-Ruiz D, Rigau J, Puigdomènech P. (1999) Involvement of a maize proline-rich protein in secondary cell wall formation as deduced from its specific mRNA localization. Plant Mol Biol 39: 945–952 [DOI] [PubMed] [Google Scholar]
- Willats WG, Knox JP. (1996) A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of beta-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. Plant J 9: 919–925 [DOI] [PubMed] [Google Scholar]
- Wise MJ. (2003) LEAping to conclusions: a computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinformatics 4: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Sharp RE, Durachko DM, Cosgrove DJ. (1996) Growth maintenance of the maize primary root at low water potentials involves increases in cell-wall extension properties, expansin activity, and wall susceptibility to expansins. Plant Physiol 111: 765–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Tan L, Lamport DT, Showalter AM, Kieliszewski MJ. (2008a) The O-Hyp glycosylation code in tobacco and Arabidopsis and a proposed role of Hyp-glycans in secretion. Phytochemistry 69: 1631–1640 [DOI] [PubMed] [Google Scholar]
- Xu SL, Rahman A, Baskin TI, Kieber JJ. (2008b) Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki K, Shinozaki K. (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57: 781–803 [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Valliyodan B, Zhang J, Lenoble ME, Yu O, Rogers EE, Nguyen HT, Sharp RE. (2010) Regulation of growth response to water stress in the soybean primary root. I. Proteomic analysis reveals region-specific regulation of phenylpropanoid metabolism and control of free iron in the elongation zone. Plant Cell Environ 33: 223–243 [DOI] [PubMed] [Google Scholar]
- Yariv J, Lis H, Katchalski E. (1967) Precipitation of arabic acid and some seed polysaccharides by glycosylphenylazo dyes. Biochem J 105: 1C–2C [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Davies WJ. (1987) Increased synthesis of ABA in partially dehydrated root tips and ABA transport from roots to leaves. J Exp Bot 38: 2015–2023 [Google Scholar]
- Zhu J, Alvarez S, Marsh EL, Lenoble ME, Cho IJ, Sivaguru M, Chen S, Nguyen HT, Wu Y, Schachtman DP, et al. (2007) Cell wall proteome in the maize primary root elongation zone. II. Region-specific changes in water soluble and lightly ionically bound proteins under water deficit. Plant Physiol 145: 1533–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]