Differential nuclear trapping of a transcription factor between root epidermal cells at distinct positions causes differential accumulation of the protein and affects cell fate decision.
Abstract
Cell fate determination and differentiation are central processes in the development of multicellular organisms, and the Arabidopsis (Arabidopsis thaliana) root epidermis provides a model system to study the molecular basis of these processes. A lateral inhibition mechanism mediated by an R3 single-repeat MYB protein, CAPRICE (CPC), has been proposed to explain the specification of the two types of root epidermal cells (hair cells and nonhair cells). However, it is not clear how CPC acts preferentially in the H-position cells, rather than the N-position cells, where its gene is expressed. To explore this issue, we examined the effect of misexpressed CPC on cell fate specification and CPC localization in the root epidermis. We show that CPC is able to move readily within the root epidermis when its expression level is high and that CPC can induce the hair cell fate in a cell-autonomous manner. We provide evidence that CPC is capable of moving from the stele tissue in the center of the root to the outermost epidermal layer, where it can induce the hair cell fate. In addition, we show that CPC protein accumulates primarily in the nuclei of H-position cells in the early meristematic region, and this localization requires the H-cell-expressed ENHANCER OF GLABRA3 (EGL3) basic helix-loop-helix transcription factor. These results suggest that cell-cell movement of CPC occurs readily within the meristematic region of the root and that EGL3 preferentially traps the CPC protein in the H-position cells of the epidermis.
The formation of a complex multicellular organism from a single cell requires exquisite spatial and temporal regulation of gene expression within distinct cell lineages. Positional information represents a major factor establishing differential gene expression during multicellular development, and in turn, some of the regulated genes themselves encode additional positional signals that further refine the gene expression patterns via cell-cell communication events (Rudel and Sommer, 2003; Moreno-Risueno et al., 2012). Given the importance of positional information, there is great interest in defining the molecular nature and action of the molecules that mediate cell-cell communication during development.
The Arabidopsis (Arabidopsis thaliana) root epidermis provides an attractive model system for studying how cells communicate with one another to establish distinct gene expression patterns during development (Schiefelbein et al., 2009; Benítez et al., 2011; Grebe, 2012). The epidermis is the outermost tissue in the root, and it is composed of two cell types: root hair-bearing cells (hair cells) and nonhair cells. Both cell types originate from the epidermis/lateral root cap initials in the root meristem, and they differentiate according to their relative position to the underlying cortical cells. The epidermal cells located on the periclinal cell wall of the underlying cortical cells and contacting a single cortical cell (the N position) differentiate into a nonhair cell, while the epidermal cells located over the anticlinal cell wall of the underlying cortical cells and contacting two cortical cells (the H position) differentiate into a hair cell (Dolan et al., 1993; Galway et al., 1994).
Molecular genetic studies have revealed many regulators of root epidermal cell fate specification, including many transcription factors (WEREWOLF [WER, R2R3-MYB protein; Lee and Schiefelbein, 1999], MYB23 [R2R3-MYB protein; Kang et al., 2009], GLABRA3/ENHANCER OF GLABRA3 [GL3/EGL3, basic helix-loop-helix (bHLH) protein; Bernhardt et al., 2003, 2005], GLABRA2 [GL2, homeodomain (HD)-ZIP protein; Masucci et al., 1996], CAPRICE [CPC, R3-MYB protein; Wada et al., 1997], TRIPTYCHON [TRY, R3-MYB protein; Schellmann et al., 2002], and ENHANCER OF TRY AND CPC1 [ETC1, R3-MYB protein; Simon et al., 2007]), a WD-repeat protein, TRANSPARENT TESTA GLABRA1 (TTG1; Masucci and Schiefelbein, 1996), a Leu-rich repeat receptor-like kinase, SCRAMBLED (SCM; Kwak et al., 2005; Kwak and Schiefelbein, 2008), and a transmembrane protein, QUIRKY (QKY; Fulton et al., 2009). In the root epidermis of the scm mutant and the qky mutant, the position-dependent cell specification is disrupted, and either cell type may be formed at a given position. Among the other regulators, WER, MYB23, GL3, EGL3, GL2, and TTG1 are involved in nonhair cell fate specification, so that mutations at these genes lead to hair cell fate specification in the root epidermis regardless of the relative position. CPC and two CPC-related genes, TRY and ETC1, specify the hair cell fate in a partially redundant manner. In the root epidermis of the cpc mutant, many of the H-position cells differentiate into a nonhair cell rather than a hair cell, and try or etc1 mutation enhances the phenotype of the cpc single mutant.
Studies on the molecular genetic interactions between these genes have led to a proposed model for cell fate specification based on a lateral inhibition mechanism (Lee and Schiefelbein, 2002; Kwak and Schiefelbein, 2007, 2008). According to this model, a yet unidentified positional signal regulates WER expression (a positive regulator of the nonhair cell fate) through SCM in a position-dependent manner. As a result, WER is preferentially expressed in the N-position cells, where it induces (together with TTG1 and GL3/EGL3) the expression of CPC and GL2. The CPC protein moves from the N-position cell to the neighboring H-position cell, where it inhibits WER action (and thus GL2 expression) to prevent this H-position cell from adopting the nonhair cell fate. Thus, it appears that the relative level of CPC protein to the level of WER protein in a specific cell is critical to determine its fate (Song et al., 2011).
Although CPC plays a central role in the lateral inhibition and cell fate decision, there are several unresolved issues surrounding CPC’s action. In particular, it is not clear how CPC preferentially affects H-position cells rather than the N-position cells where it is produced. There are several possible explanations. First, CPC could move to the H-position cells via a targeted, unidirectional mechanism, which prevents its accumulation in N cells. Second, CPC may require cell-cell movement to be active. Third, a companion protein(s) that CPC requires for its function may be present only in the H-position cells. Fourth, a protein(s) that inhibits CPC action may be present in the N-position cells.
In this study, we explored the role of CPC and its intercellular movement in relation to root epidermal cell specification. We discovered that when misexpressed in the H-position cells, CPC was able to induce the hair cell fate in a cell-autonomous manner and it was able to move to the N-position cell and induce the hair cell fate when expressed at a high level. We further show that, when misexpressed in different tissues in the root, CPC was able to induce the hair cell fate in the root epidermis, indicating long-distance movement of CPC. In addition, we found that CPC protein accumulates preferentially in the nucleus of the H-position epidermal cells in an EGL3-dependent manner. These results indicate that CPC can move more readily through the Arabidopsis root than previously believed, and they suggest that the regulated accumulation of CPC protein is important in hair cell fate specification.
RESULTS
CPC Expressed in H-Position Cells Acts within Those Cells and Moves to the N-Position Cells in a Concentration-Dependent Manner
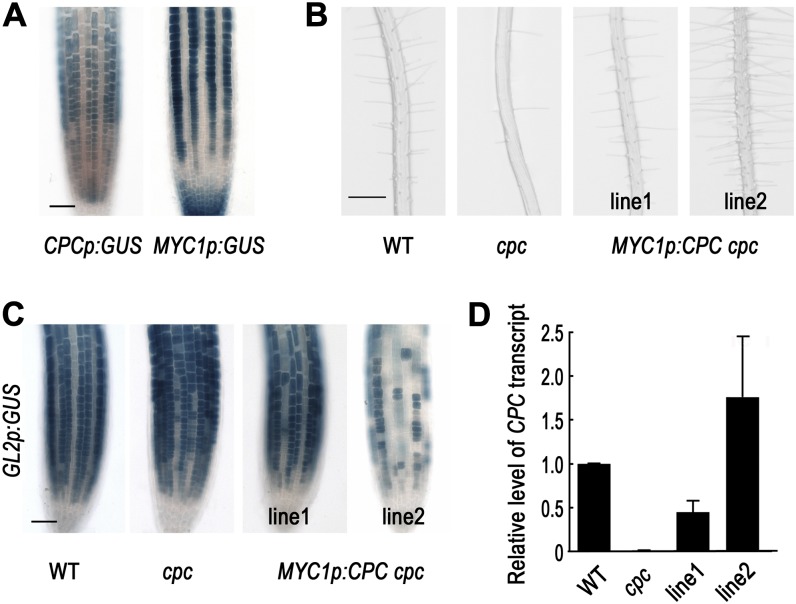
The MYC1 promoter is active in the H-position epidermal cells, the lateral root cap cells, and the central root cap cells based on the expression pattern of the MYC1p:GUS reporter gene (Fig. 1A; Bruex et al., 2012), while the CPC promoter is active in the N-position epidermal cells (Lee and Schiefelbein, 2002; Wada et al., 2002). In the developing root epidermis, those two promoters are active in complementary locations. To examine the effect of CPC expressed in the opposite cell position, we expressed the CPC coding region under the control of the MYC1 promoter in the cpc mutant (MYC1p:CPC cpc). Among the 10 transgenic lines analyzed in detail, six of them (including the representative line MYC1p:CPC -1 cpc) showed full complementation of the cpc mutant root hair phenotype, while two lines showed partial rescue and two lines (including the representative line MYC1p:CPC-2 cpc) showed a hairy phenotype, indicating CPC activity in the N-cell position (Fig. 1B; Table I). To determine whether these phenotypes were accompanied by corresponding changes in GL2 expression, we examined GL2p:GUS reporter gene expression in these lines (Fig. 1C). In the fully complemented lines (e.g. MYC1p:CPC-1 cpc), the expression pattern of GL2p:GUS in the root epidermis was completely restored, with appropriate N-position-specific expression as in the wild-type root. On the contrary, GL2p:GUS in the lines showing a hairy phenotype (e.g. MYC1p:CPC-2 cpc) was expressed only in a small number of the epidermal cells, which is consistent with the root hair phenotype in those lines. The CPC transcript level in the MYC1p:CPC-1 cpc plant root was about 45% of the level in the wild-type plant root, while in the MYC1p:CPC-2 cpc plant root, the CPC transcript level was 3.9-fold higher than the level in the MYC1p:CPC-1 cpc plant root (Fig. 1D). This suggests that the phenotypic differences between these lines were caused by differences in the expression level of the CPC construct.
Figure 1.
CPC expressed in the H-position cells functions in a cell autonomous manner. A, The promoter activity of CPC (CPCp:GUS) and MYC1 (MYC1p:GUS) in the root epidermis of 4-d-old seedlings shows a complementary pattern. Root tip of the promoter-GUS reporter lines stained for GUS activity using the X-gluc substrate. Bar = 50 μm. B, Effects of CPC expressed in the H-position cells on the root hair phenotype in 4-d-old seedlings. Line numbers represent independent transgenic lines. Bar = 200 μm. C, Expression of the GL2 promoter-GUS reporter gene (GL2p:GUS) in the root tip of the MYC1p:CPC cpc plants. Four-day-old seedlings were stained for GUS activity. Bar = 50 μm. D, Quantitative real-time RT-PCR analysis to measure the steady-state level of CPC transcript in each transgenic line. [See online article for color version of this figure.]
Table I. Specification of cell types in the root epidermis misexpressing CPC by various enhancers and promoters.
Values indicate the mean ± sd of three independent experiments. In each experiment, at least 10 seedlings were included per a plant line.
| Genotype | H Position |
N Position |
||
|---|---|---|---|---|
| Hair Cell | Nonhair Cell | Hair Cell | Nonhair Cell | |
| % | % | % | % | |
| Wild type | 98.0 ± 0.3 | 2.0 ± 0.3 | 1.3 ± 1.1 | 98.7 ± 1.1 |
| cpc | 27.0 ± 0.7 | 73.0 ± 0.7 | 0.0 ± 0.0 | 100.0 ± 0.0 |
| MYC1p:CPC -1 cpc | 93.2 ± 1.1 | 6.8 ± 1.1 | 0.0 ± 0.0 | 100 ± 0.0 |
| MYC1p:CPC -2 cpc | 98.5 ± 1.0 | 1.5 ± 1.0 | 79.3 ± 1.0 | 20.7 ± 1.0 |
| MYC1p:CPC-GFP -1 cpc | 61.2 ± 1.9 | 38.8 ± 1.9 | 0.0 ± 0.0 | 100.0 ± 0.0 |
| MYC1p:CPC-GFP -2 cpc | 98.3 ± 1.2 | 1.7 ± 1.2 | 4.0 ± 0.9 | 96.0 ± 0.9 |
| MYC1p:CPC-GFP -3 cpc | 99.2 ± 0.5 | 0.8 ± 0.5 | 61.3 ± 0.8 | 38.7 ± 0.8 |
| 35Sp:CPC | 99.2 ± 0.3 | 0.8 ± 0.3 | 91.2 ± 2.7 | 8.8 ± 2.7 |
| UASp:CPC | 95.6 ± 2.2 | 4.4 ± 2.2 | 6.7 ± 3.4 | 93.3 ± 3.4 |
| J1092>>CPC | 97.6 ± 0.4 | 2.4 ± 0.4 | 2.4 ± 1.0 | 97.6 ± 1.0 |
| J2301>>CPC | 100.0 ± 0.0 | 0.0 ± 0.0 | 99.7 ± 0.5 | 0.3 ± 0.5 |
| J0571>>CPC | 99.7 ± 0.5 | 0.3 ± 0.5 | 98.6 ± 1.7 | 1.4 ± 1.7 |
| J1721>>CPC | 99.8 ± 0.4 | 0.2 ± 0.4 | 42.0 ± 4.9 | 58.0 ± 4.9 |
| Q0990>>CPC | 99.4 ± 0.6 | 0.6 ± 0.6 | 41.4 ± 8.4 | 58.6 ± 8.4 |
| J0571>>CPC cpc | 100.0 ± 0.0 | 0.0 ± 0.0 | 97.6 ± 1.5 | 2.4 ± 1.5 |
| J1721>>CPC cpc | 62.5 ± 4.2 | 37.5 ± 4.2 | 1.1 ± 1.0 | 98.9 ± 1.0 |
| Q0990>>CPC cpc | 58.9 ± 7.3 | 41.1 ± 7.3 | 0.0 ± 0.0 | 100.0 ± 0.0 |
| J0571>>CPC-GFP cpc | 100.0 ± 0.0 | 0.0 ± 0.0 | 36.4 ± 5.9 | 63.6 ± 5.9 |
| Q0990>>CPC-GFP cpc | 46.5 ± 2.1 | 53.5 ± 2.1 | 0.0 ± 0.0 | 100.0 ± 0.0 |
| cpc try | 0.0 ± 0.0 | 100.0 ± 0.0 | 0.0 ± 0.0 | 100.0 ± 0.0 |
| J0571>>CPC cpc try | 99.6 ± 0.3 | 0.4 ± 0.3 | 97.0 ± 2.9 | 3.0 ± 2.9 |
| Q0990>>CPC cpc try | 25.0 ± 2.2 | 75.0 ± 2.2 | 0.3 ± 0.5 | 99.7 ± 0.5 |
| CPCp:CPC-GFP cpc | 85.6 ± 3.9 | 14.4 ± 3.9 | 14.6 ± 5.8 | 85.4 ± 5.8 |
| SHRp:CPC-GFP cpc | 78.3 ± 1.2 | 21.7 ± 1.2 | 0.5 ± 0.5 | 99.5 ± 0.5 |
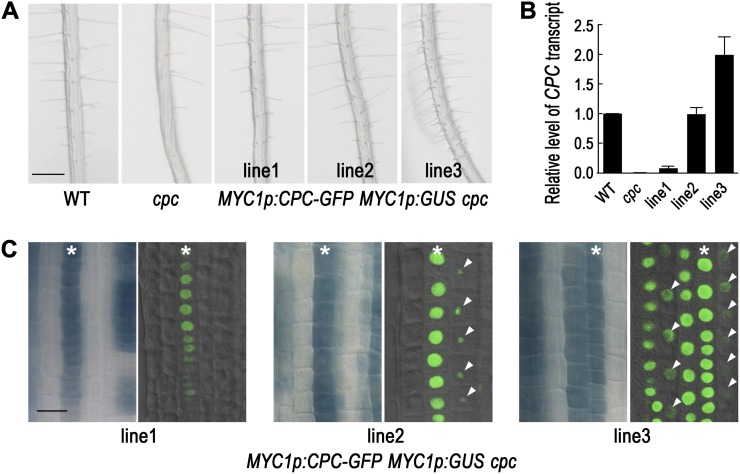
To extend these findings, we analyzed CPC expression and CPC protein accumulation by generating MYC1p:CPC-GFP MYC1p:GUS cpc transgenic plants. First, we analyzed the effect of the transgene on the root epidermis phenotype (Fig. 2A; Table I). As observed in the MYC1p:CPC cpc transgenic plants, some of the MYC1p:CPC-GFP MYC1p:GUS cpc transgenic plants (five lines out of 13 total lines analyzed, including the representative line MYC1p:CPC-GFP-1 MYC1p:GUS cpc) showed partial rescue of the cpc mutant root hair phenotype, some of the transgenic plants (five lines out of 13 lines, including MYC1p:CPC-GFP-2 MYC1p:GUS cpc) showed full complementation, and the other three lines (including MYC1p:CPC-GFP-3 MYC1p:GUS cpc) showed an overcomplemented (hairy) phenotype. Furthermore, like the MYC1p:CPC cpc lines, the phenotypic differences observed in the independent MYC1p:CPC-GFP MYC1p:GUS cpc transgenic lines were correlated with the CPC transcript level in the root tip (Fig. 2B). Next, we examined the MYC1 promoter activity and the CPC-GFP protein accumulation in the same plants (Fig. 2C). In the MYC1p:CPC-GFP-1 MYC1p:GUS cpc plant, we detected MYC1 promoter activity in a subset of the H-position epidermal cells, and the CPC-GFP protein was observed in the same cells. In the MYC1p:CPC-GFP-2 MYC1p:GUS cpc plant, the MYC1 promoter activity was detected in most of the H-position cells and not in the N-position epidermal cells. Interestingly, the CPC-GFP protein was detected not only in the CPC-GFP expressing cells in the H-position, but also in the neighboring N-position cells, albeit at a reduced level (Fig. 2C). This result indicates that the CPC-GFP protein moves from the H cells to the neighboring N-position cells. Finally, in the MYC1p:CPC-GFP-3 MYC1p:GUS cpc plant, the MYC1 promoter activity was detected in the H-position cells and in some of the neighboring N-position epidermal cells. The CPC-GFP protein accumulated in each of the MYC1p:GUS-expressing cells (no matter their position) with a high level, and it also was detected at a lower level in some of the cells not expressing the MYC1p:GUS.
Figure 2.
CPC expressed in the H-position cells moves to the neighboring N-position cells. A, Effects of CPC-GFP expressed in the H-position cells (MYC1p:CPC-GFP cpc) on the root hair phenotype in 4-d-old seedlings. Line numbers represent independent transgenic lines. Bar = 200 μm. B, Quantitative real-time RT-PCR analysis to measure the steady-state level of CPC transcript in each transgenic line. C, Expression of the MYC1 promoter-GUS reporter gene (MYC1p:GUS) in the root tip of the MYC1p:CPC-GFP MYC1p:GUS cpc plants shows where CPC gene is expressed. Confocal image for the CPC-GFP shows where the CPC-GFP protein accumulates. Asterisks indicate the H position, and arrowheads indicate CPC-GFP fluorescence in cells lacking MYC1 promoter activity. Bar = 20 μm. Note that only a portion of the GUS-expressing cell files show a fluorescence signal in these images due to the limited focal plane. [See online article for color version of this figure.]
Taken together, these results suggest that CPC is able to induce the hair cell fate in a cell-autonomous manner, because the same cells that express the MYC1p:GUS marker also accumulate the CPC-GFP protein and their proportions are consistent with the fraction of root hair cells. Furthermore, these results indicate that CPC is capable of moving from the H-position epidermal cell to the neighboring N-position cell in a concentration-dependent manner, and this H-cell-derived CPC is able to induce the hair cell fate in the N cells.
The CPC Protein Preferentially Accumulates in the Nuclei of H-Position Root Epidermal Cells
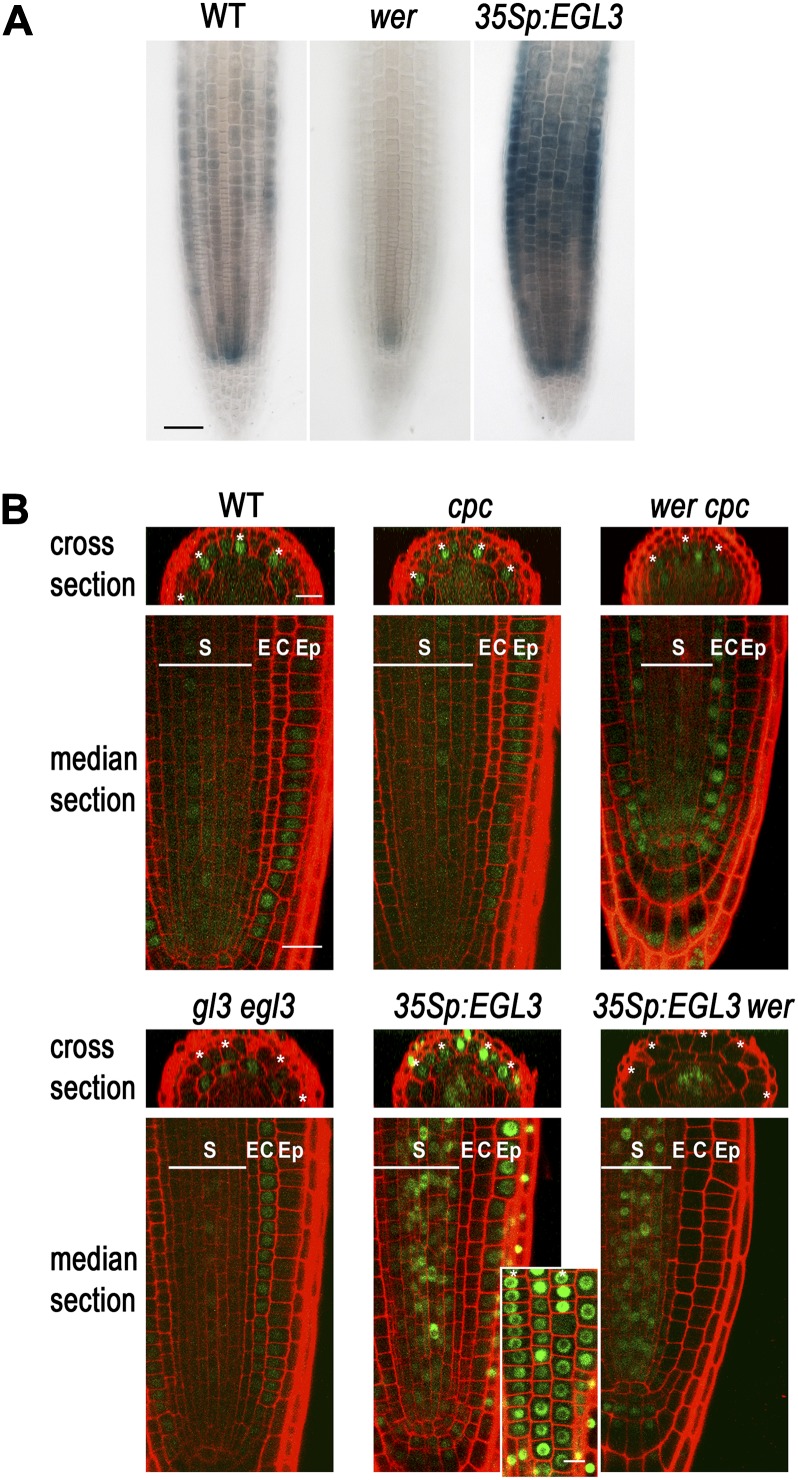
CPC is normally expressed in the differentiating stele cells as well as the N-position epidermal cells in the root meristematic region, and WER positively regulates CPC expression in the root epidermis (Lee and Schiefelbein, 2002; Wada et al., 2002; Ryu et al., 2005). However, WER is not expressed in the stele cells (Lee and Schiefelbein, 1999), and accordingly, the wer mutation does not cause great reduction of CPC promoter activity in the stele cells, unlike in the epidermal cells (Fig. 3A; Lee and Schiefelbein, 2002). We have previously reported that about 70% of the root epidermal cells differentiate into a hair cell in the wer cpc double mutant, while more than 90% of the root epidermal cells differentiate into a hair cell in the wer mutant (Lee and Schiefelbein, 1999). To explain this CPC-dependent hair cell specification in the wer mutant background, we considered the possibility that CPC expressed in the stele might move out to the epidermis. To test this, we introduced a CPCp:CPC-GFP reporter gene into the cpc mutant and the wer cpc double mutant (Fig. 3). This translational fusion was able to rescue the cpc mutant phenotype to the wild-type level (Supplemental Fig. S1A), indicating that the CPC-GFP protein functions like the native CPC and the flanking DNA fragments are sufficient for the proper control of its expression. The CPC-GFP protein in the cpc mutant root accumulated in the nucleus of the developing root epidermal cells (Fig. 3B). Notably, the H-position epidermal cells exhibited a higher level of CPC-GFP protein than the N-position epidermal cells in the early meristematic region. In the wer cpc double mutant root, the CPC-GFP protein was found in the stele cells, as expected. In addition, the CPC-GFP protein was observed in the early epidermis of this double mutant root tip at a low level, preferentially in the H-position cells, even though there was no detectable CPC promoter activity in the epidermis (Fig. 3A). These results suggest that the CPC-GFP expressed in the stele cells moves out to the epidermis and affects the cell fate specification in this tissue.
Figure 3.
GL3/EGL3 affects the CPC movement in the root. A, The wild type, the wer mutant, and the 35Sp:EGL3 transgenic plant roots bearing the CPCp:GUS transgene were stained for GUS activity. Bar = 50 μm. B, GFP expression in the developing root epidermis of various plants harboring CPCp:CPC-GFP translational reporter gene. The H-position epidermal cells are marked with an asterisk. The inset shows the surface view of CPC-GFP protein accumulation in the 35Sp:EGL3 plant root. S, Stele; E, endodermis; C, cortex; Ep, epidermis. Bar = 20 μm.
CPC Expressed in Subepidermal Tissues Accumulates Primarily in the H-position Epidermal Cells to Affect Cell Fate Specification in the Root Epidermis
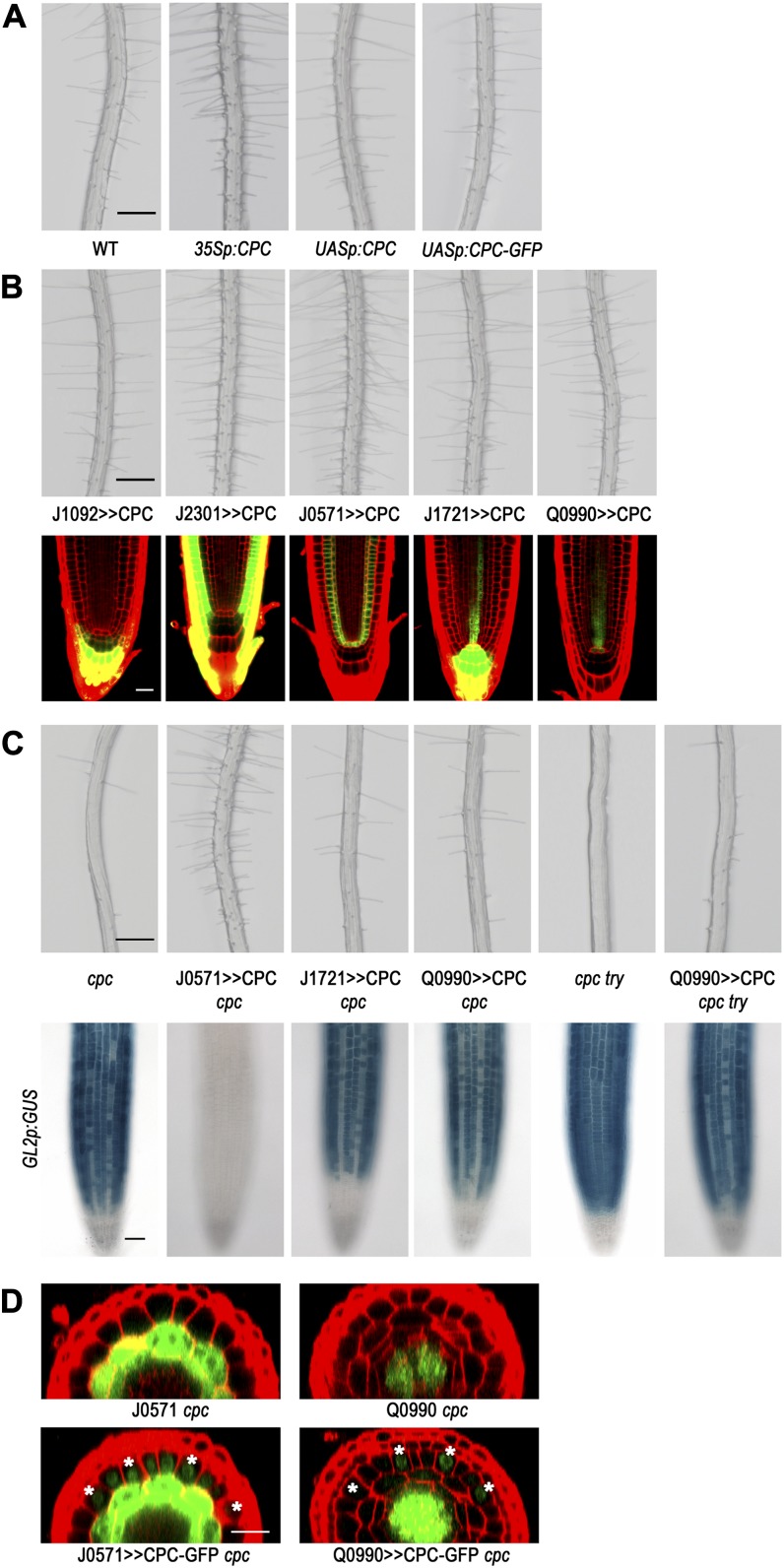
To further examine the capacity of CPC for long-range movement to the root epidermis, we drove CPC expression in distinct root tissues using the GAL4-Upstream Activation Sequence (UAS) ectopic expression system (Brand and Perrimon, 1993; Haseloff, 1999). We used a plant harboring the UASp:CPC construct, which had been reported previously (Song et al., 2011) and does not exhibit any defect in cell fate specification in the root epidermis (Fig. 4A). As a positive control, we expressed CPC using an enhancer trap line, J2301, which shows strong GFP expression throughout the root epidermis in the early meristematic region. We found that most of the root epidermal cells in this transgenic plant (J2301>>CPC) adopted the hair cell fate, similar to plants harboring 35Sp:CPC (Fig. 4, A and B; Wada et al., 1997). Next, to express CPC in the central root cap located at the distal tip, we crossed the UASp:CPC plant with the enhancer trap line J1092. The resulting plants (J1092>>CPC) did not show any detectable defect in cell fate specification in the root epidermis (Fig. 4B; Table I). Next, we used the J0571 enhancer trap line to express CPC in the cortex and endodermis, and these plants (J0571>>CPC) exhibited a change in cell fate specification, with most of the N-position cells adopting the hair cell fate (Fig. 4B; Table I). We also expressed CPC in further inner tissues of the root by introducing the UASp:CPC into the J1721 enhancer trap line (J1721>>CPC) to express CPC in the root protoxylem and central root cap and into the Q0990 enhancer trap line (Q0990>>CPC) to express CPC only in the stele cells in the meristematic region. In these plants, some of the N-position cells adopted the hair cell fate instead of the nonhair cell fate, which caused a hairy phenotype (Fig. 4B; Table I).
Figure 4.
CPC expressed in different tissue layers using the GAL4-UAS system affects cell fate specification in the root epidermis. A, Four-day-old seedlings illustrating root hair production. Bar = 200 μm. B, Root hair phenotype for the plants expressing CPC in a specific tissue in the wild-type root (top; bar = 200 μm). The plant harboring the UASp:CPC was crossed with the each enhancer trap line. Confocal microscope images indirectly show the CPC-expressing tissues (bottom; bar = 20 μm). C, Root hair phenotype (top) and GL2p:GUS expression pattern (bottom) in the plants expressing CPC in a specific tissue in the cpc mutant background. Bars = 200 μm (top) and 50 μm (bottom). D, CPC moves from the stele cells to the epidermis in the root. The strong GFP signal throughout the cell represents the signal from the endogenous GFP of the enhancer trap lines. The GFP fluorescence signal in the nucleus of the cells outside the ground tissue (in the case of the J0571 cpc) and the cells outside the stele cells (in the case of the Q0990 cpc) were detected only in the plants harboring UASp:CPC-GFP, implying that this signal represents the CPC-GFP protein. The H-position epidermal cells are marked with an asterisk. Bar = 20 μm.
Although the above results suggest that CPC produced in diverse locations, including the center of the root, is capable of moving to the epidermis and affecting cell fate decisions, the presence of endogenous CPC in the transgenic lines prevented us from determining whether epidermal cells at both positions were equally affected by the ectopically expressed CPC. To address this, we analyzed the effect of ectopically expressed CPC in the cpc mutant background using the same enhancer trap lines used in the wild-type background (J0571, J1721, and Q0990; Fig. 4C; Table I). In the cpc mutant background, J0571>>CPC caused almost every epidermal cell to adopt the hair cell fate, regardless of its relative position. By contrast, CPC expressed in the protoxylem and the central root cap or in the stele cells in the cpc mutant (J1721>>CPC cpc and Q0990>>CPC cpc) only partially rescued the cpc mutant phenotype in the root epidermis. Interestingly, the CPC in these plants preferentially affected the fate of the H-position cells, causing more cells at the H position to adopt the hair cell fate. We considered the possibility that the functionally related TRY gene (Schellmann et al., 2002) may be responsible for this preferential effect, so we further examined the effect of Q0990>>CPC in the cpc try double mutant to eliminate the effect of the CPC-like activity provided by TRY. We discovered that even in this double mutant background, ectopic expression of CPC by the Q0990>>CPC only altered the fate of H-position cells and not N-position cells (Table I).
Next, we wished to examine whether CPC produced in the ground tissue or the stele accumulates in the epidermis by analyzing GFP localization in J0571>>CPC-GFP cpc plants and in Q0990>>CPC-GFP cpc plants, respectively (Fig. 4D). We first fused a CPC-GFP chimeric gene to the 3′ end of the 5× UAS promoter (UASp:CPC-GFP) and introduced it into the wild-type background to confirm that this construct alone did not alter cell patterning in the root epidermis (Fig. 4A). We then generated the J0571>>CPC-GFP cpc and Q0990>>CPC-GFP cpc lines by genetic crosses. Comparing the GFP signal in the J0571 enhancer trap control and the J0571>>CPC-GFP cpc plants, we detected epidermal cell accumulation of GFP only in the J0571>>CPC-GFP cpc roots, with the GFP signal visible in the nucleus of the epidermal cells at both positions in addition to the ground tissue (Fig. 4D). In these J0571>>CPC-GFP cpc plants, all of the H-position cells and 36.4% of the N-position cells differentiated into a hair cell, showing a hairy phenotype (Table I). Comparing the GFP signal in the Q0990 enhancer trap control line to the Q0990>>CPC-GFP cpc plants, we observed epidermal cell accumulation of GFP only in the Q0990>>CPC-GFP cpc roots, with preferential GFP signal in the nucleus of the H-position epidermal cells (Fig. 4D). This corresponds with the H-cell-specific effect of the Q0990>>CPC-GFP on cell fate in the cpc background; only H-position epidermal cells were converted to the hair cell type (Table I). Together, these results suggest that the ability of ectopically expressed CPC to alter cell fate specification in the epidermis of the J0571>>CPC cpc plant and the Q0990>>CPC cpc plant is due to CPC movement to the epidermis.
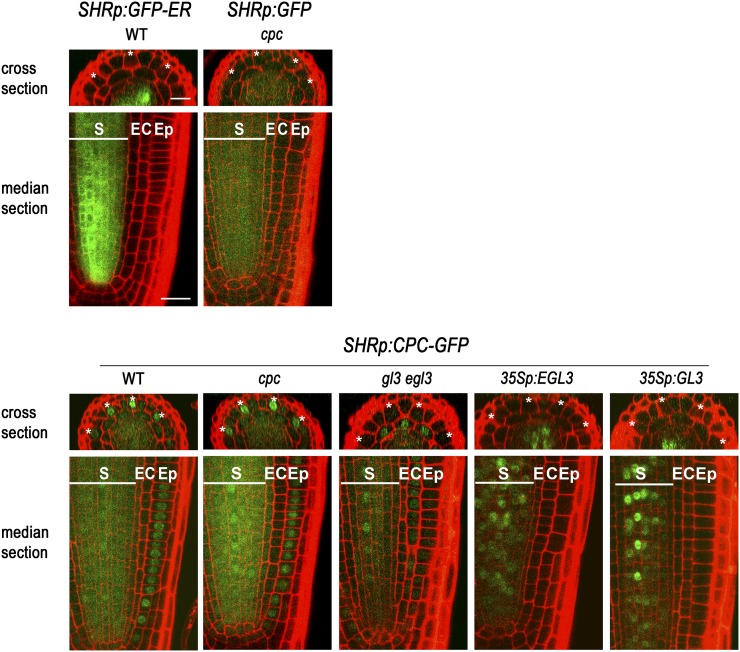
The above results seem to be inconsistent with a previous report that CPC expressed in the stele cells using the promoter of SHORT ROOT (SHR) gene was not able to move to the epidermis and affect cell fate specification (Kurata et al., 2005a). Thus, we reexamined this issue by assessing the effect of CPC expressed in the stele using the SHR promoter. We fused our CPC-GFP chimeric gene to the 3′ end of the SHR promoter and introduced it into the cpc mutant (Fig. 5). This line exhibited increased hair cell specification in the H-position cells relative to the cpc mutant (Table I), and it showed preferential CPC-GFP accumulation in the nuclei of H-position cells (Fig. 5; Table I). By contrast, the soluble GFP expressed under the control of the SHR promoter (SHRp:GFP) accumulated highly in the stele cells where it is expressed (Fig. 5). We did observe some GFP accumulation in the epidermis of these plants; however, the level was very low, and there was no preferential accumulation of GFP between the epidermal cell types. Taken together, these results suggest that CPC is able to move from inner cell layers to the epidermis, primarily accumulating in the H-position cell, and influence epidermal cell fate.
Figure 5.
Movement of CPC expressed under the control of SHR promoter to the epidermis was affected by EGL3. GFP expression was analyzed in the developing root epidermis of various plants harboring the SHRp:CPC-GFP translational fusion construct. Note that CPC-GFP accumulation in the epidermis decreased in the gl3 egl3 mutant, and 35Sp:EGL3 and 35Sp:GL3 made CPC-GFP accumulate in the nucleus of the stele cells where it is expressed. The H-position epidermal cells are marked with an asterisk. S, Stele; E, endodermis; C, cortex; Ep, epidermis. Bar = 20 μm.
The CPC Protein Is Trapped in the Specific Cells by GL3/EGL3
We were intrigued by our observation of preferential accumulation of CPC in the H-position cells. Previously, CPC has been reported to interact with the bHLH proteins GL3 and EGL3 in yeast (Saccharomyces cerevisiae) and plant cells (Bernhardt et al., 2003; Wester et al., 2009; Song et al., 2011). Therefore, we tested whether GL3/EGL3 might play a role in CPC localization by examining CPC-GFP accumulation in the gl3 egl3 double mutant (CPCp:CPC-GFP gl3 egl3; Fig. 3B). In these plants, CPC-GFP accumulated in the epidermal cells at a very low level as expected, because CPC promoter activity is greatly reduced in the gl3 egl3 double mutant (Bernhardt et al., 2003), while the accumulation pattern of CPC-GFP in the wild type was quite similar to that in the cpc mutant. Furthermore, unlike in the wer cpc mutant root epidermis, no preferential accumulation of CPC-GFP in the H-position cells was observed in the gl3 egl3 background.
Next, we examined CPC-GFP accumulation in plants expressing EGL3 in all root tissues (CPCp:CPC-GFP 35Sp:EGL3). In these plants, most of the root epidermal cells accumulated CPC-GFP regardless of their relative position, but the accumulation was not uniform across the root epidermis (Fig. 3B). Some of the epidermal cells showed much higher accumulation than others, and these were found at both positions. The CPC-GFP protein was also observed in the nuclei of the lateral root cap cells, which was not seen without 35Sp:EGL3. We examined the CPC promoter activity in the 35Sp:EGL3 plants and found that the CPCp:GUS expression pattern was similar to the CPC-GFP accumulation pattern in the epidermis and the lateral root cap (Fig. 3A). Another difference in the CPC-GFP accumulation between the wild-type root and the 35Sp:EGL3 plant root was observed in the stele cells, where CPC-GFP was localized to the nucleus in the 35Sp:EGL3 plant root rather than in the cytoplasm (Fig. 3B). In the wer mutant root expressing EGL3 ubiquitously (CPCp:CPC-GFP 35Sp:EGL3 wer), CPC-GFP was also exclusively localized to the nucleus in the stele cells (Fig. 3B), but no CPC-GFP was observed in the epidermis, as was observed in the wer cpc mutant root (CPCp:CPC-GFP wer cpc; Fig. 3B). However, changes in the CPC promoter activity by GL3/EGL3 (Bernhardt et al., 2003) and WER (Lee and Schiefelbein, 2002) and changes in the endogenous GL3/EGL3 expression by CPC (Bernhardt et al., 2005) make it difficult to interpret these results. Therefore, to further clarify the function of GL3/EGL3 in the localization of CPC, we expressed CPC-GFP under the control of the unrelated SHR promoter (SHRp:CPC-GFP). We examined SHRp:CPC-GFP accumulation in plants expressing excess GL3/EGL3 or lacking GL3/EGL3. In SHRp:CPC-GFP plants containing either 35Sp:GL3 or 35Sp:EGL3 to overexpress the GL3/EGL3 (SHRp:CPC-GFP 35Sp:GL3 and SHRp:CPC-GFP 35Sp:EGL3 lines), the only detectable CPC-GFP accumulation was in the nuclei of the stele cells (the site of SHR promoter activity), implying that the CPC-GFP is not able to move from the cells in which it is produced (Fig. 5). In gl3 egl3 double mutant plants containing the SHRp:CPC-GFP construct, the CPC-GFP protein was observed in the cytoplasm and the nuclei of stele cells and in the nucleus of the root epidermis at a low level (Fig. 5). However, the CPC-GFP did not accumulate in a cell position-dependent manner in the root epidermis, indicating that GL3/EGL3 is required for this pattern (Fig. 5). These observations are reminiscent of the proposed TTG1 nuclear trapping by GL3 in Arabidopsis trichomes (Balkunde et al., 2011). Similarly, it may be that GL3/EGL3 are important for nuclear trapping of CPC, leading to preferential accumulation of CPC in the H-position cells and restricting further protein movement.
The EGL3 Protein Preferentially Accumulates in the H-Position Cells
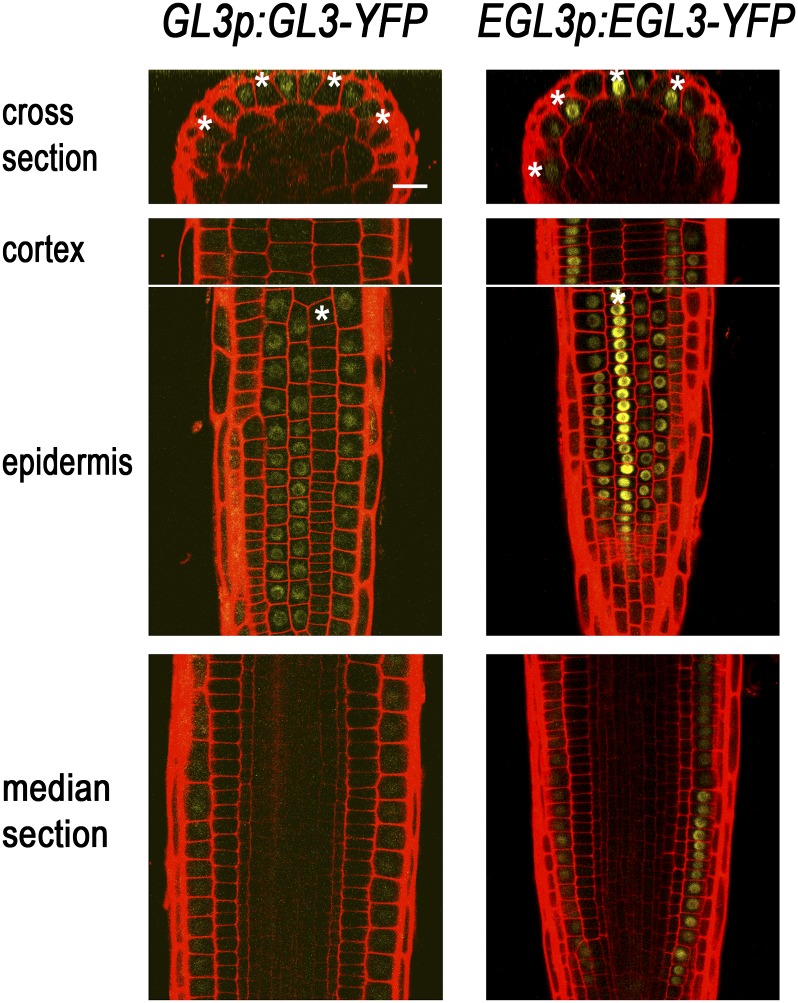
Given the potential importance of GL3/EGL3 in mediating the preferential accumulation of CPC, we considered the location of GL3/EGL3 expression and GL3/EGL3 protein accumulation. In prior studies, promoter-reporter fusions and in situ RNA hybridization experiments indicated that GL3 and EGL3 are transcribed in the H-position cells of the root epidermis, yet a GL3-yellow fluorescent protein (YFP) chimeric protein preferentially accumulates in the N-position epidermal cells, implying movement of GL3 from the H- to the N-position cells (Bernhardt et al., 2005). Given the functional redundancy between GL3 and EGL3, it has been assumed that EGL3 also moves to the N-position cells. To test this, we analyzed a EGL3p:EGL3-YFP translational fusion reporter line (Fig. 6). The EGL3p:EGL3-YFP construct was able to function effectively in the root epidermis based on the root hair phenotype of the gl3 egl3 EGL3p:EGL3-YFP plant (Supplemental Fig. S1B). EGL3-YFP fluorescence was observed preferentially in the nuclei of the developing H-position epidermal cells (Fig. 6), in contrast to the GL3-YFP accumulation in the N-position cells (Bernhardt et al., 2005).
Figure 6.
Subcellular localization of the EGL3 protein. GFP expression in the developing root epidermis of plants harboring the GL3p:GL3-YFP or the EGL3p:EGL3-YFP translational fusion construct. EGL3-YFP preferentially accumulated in the nucleus of the developing H-position cells, while GL3-YFP accumulated preferentially in the N-position cells. The H-position epidermal cells are marked with an asterisk. Bar = 20 μm.
DISCUSSION
The CPC-mediated lateral inhibition pathway in the root epidermis provides a paradigm for understanding cell-cell communication during cell fate determination in plants. To further define CPC’s action in this system, we have conducted a series of misexpression and localization experiments, and these have led to new insights into CPC protein movement in root tissues, cell type-specific accumulation of CPC, and the regulation of CPC nuclear accumulation.
Movement of CPC Protein within and between Tissues in the Developing Root
Many proteins and RNAs appear capable of moving between plant cells, presumably through plasmodesmata, and this molecular mobility is believed to enable cells to communicate with each other and coordinate their development (Jackson et al., 1994; Lucas et al., 1995; Sessions et al., 2000; Nakajima et al., 2001; Kim et al., 2003; Bernhardt et al., 2005; Schlereth et al., 2010; Zhou et al., 2013). The CPC protein was proposed to move from N-position cells to H-position cells in the developing root epidermis based on CPC gene expression and CPC RNA accumulation in N-position cells but CPC-GFP protein fusion accumulation in both H- and N-position cells and cpc mutant defects in H-position cells (Wada et al., 1997; Lee and Schiefelbein, 2002; Wada et al., 2002). In a subsequent study, a CPC-GFP chimeric gene was expressed under the control of the EGL3 promoter (active in the H-position epidermal cells; Bernhardt et al., 2005) and CPC-GFP accumulation was detected in all epidermal cells (Kurata et al., 2005a). Also, the CPC-GFP chimeric gene was expressed under the control of the SHR promoter (active in the stele; Helariutta et al., 2000), and CPC-GFP was detected only in the stele cells and not in the epidermis (Kurata et al., 2005a). These findings suggested that CPC is able to move between cells in the epidermis but not between cells in different tissues, seemingly consistent with a report that protein movement is more restricted laterally within layers in shoot apex than it is between different layers (Wu et al., 2003).
Here, we used a different H-cell-specific promoter (MYC1), and we examined both MYC1 promoter activity and CPC-GFP fluorescence signal in the same MYC1p:CPC-GFP roots to obtain compelling evidence that CPC protein is able to move in an opposite direction (from the H-position cells to the N-position cells), depending on its expression level. This extends findings described in previous reports that CPC-Dendra2 (a photoconvertible fluorescent protein) was able to move isodiametrically within the root epidermis (Wu et al., 2011) and that fluorescence signal was detected in all epidermal cells in EGL3p:CPC-GFP plants (Kurata et al., 2005a). Furthermore, we showed that CPC/CPC-GFP expressed in the root ground tissue or in the stele led to CPC-GFP accumulation in the root epidermis and alterations in epidermal cell fate specification, consistent with a report that fluorescence signal was detected in the epidermis and the stele when CPC-mCherry was expressed in root ground tissue (Rim et al., 2011). From this, we conclude that the CPC protein is able to move out from inner tissue layers of the root to the epidermis to affect cell fate specification.
The difference between our findings and those of Kurata et al. (2005a) regarding inner- to outer-layer CPC movement may be due to the size difference in the CPC-GFP recombinant proteins employed. In our experiments, the 1× GFP was used for the CPC-GFP translational fusion (total size = approximately 38 kD), whereas 2× GFP was used in the previous report (total size = approximately 65 kD). If this is the cause, then it further implies that the size exclusion limit (SEL) for plasmodesmatal movement of the CPC-GFP between these tissue layers is between 38 and 65 kD, assuming plasmodesmatal-mediated movement. Interestingly, this SEL is lower than the size of the SHR-GFP chimeric protein (approximately 86 kD), which is able to move between tissue layers (Nakajima et al., 2001).
The SEL for plasmodesmatal movement can differ based on the movement type, nontargeted or targeted. Targeted movement likely results from direct interactions with plasmodesmata and results in increased SEL (Kurata et al., 2005b). It had been suggested that the CPC-GFP protein moves in the root epidermis in a targeted manner because its movement depends on specific sequences in the CPC protein (Kurata et al., 2005a). Furthermore, multimeric GFP fused to CPC was able to move from the N-position epidermal cells to the neighboring H-position cells, while free 2× GFP was not able to move in the root epidermis (Kurata et al., 2005a). However, it now appears that the CPC-GFP protein may move in a nontargeted manner between root tissues, given that monomeric GFP fused to CPC and free monomeric GFP in the stele was able to move to the root epidermis (our experiments) and 2× GFP fused to CPC was not able to move out from the stele to the root epidermis (Kurata et al., 2005a).
Preferential Accumulation of CPC Protein in the H-Position Cells
Prior studies concluded that CPC-GFP protein accumulates in the nucleus of every root epidermal cell regardless of its position (Kurata et al., 2005a; Lee et al., 2006). However, our results showing the CPC-GFP protein accumulation and the effect of the CPC/CPC-GFP protein on the cell fate decision in various lines suggest that CPC primarily accumulates in the nucleus of the H-position cells, but if the amount of CPC protein is excessive, CPC will also accumulate in the nucleus of the N-position cells. This preferential accumulation of CPC protein in the H-position epidermal cells is consistent with results from the immunostaining of haemagglutinin-tagged CPC in the root (Kurata et al., 2005a).
In wild-type roots, CPC is expressed not only in the N-position epidermal cells, but also in the root stele cells (Lee and Schiefelbein, 2002; Wada et al., 2002; Ryu et al., 2005). It will be interesting to determine the proportion of the CPC protein in the H-position cell that is contributed by the neighboring N-position cells and by the stele cells and to what extent, if any, the CPC from the stele cells contributes to hair cell fate specification. Further experiments, for example, mosaic analyses using the Cre-Lox system, will be required to address these issues.
Nuclear Trapping of CPC by EGL3
Recent studies indicate that the nuclear localization of a protein seems to control its intercellular movement. For example, the homeodomain of KNOTTED1 (KN1) is necessary and sufficient for its movement, and removal of the nuclear localization signal in this homeodomain abolishes KN1 movement (Kim et al., 2005). TTG1 moves into trichome cells from the neighboring nontrichome cells, and nuclear localization of TTG1 protein in the trichome cells by GL3 controls its movement (Bouyer et al., 2008; Balkunde et al., 2011). Also, nuclear localization of SHR protein in the endodermal cells by SCARECROW (SCR) is important for proper SHR movement (Gallagher et al., 2004; Cui et al., 2007; Gallagher and Benfey, 2009).
Our results suggest that EGL3 is responsible for the preferential accumulation of the CPC-GFP protein in the nucleus of the H-position epidermal cells, possibly via direct protein-protein interaction (Bernhardt et al., 2003; Wester et al., 2009; Song et al., 2011), as a kind of nuclear trapping mechanism. This regulation of CPC movement by EGL3-mediated nuclear trapping is analogous to the regulation of SHR movement by SCR. SHR moves out from the root stele cells into the neighboring endodermis, where it accumulates in the nucleus in a SCR-dependent manner, which prevents further SHR movement (Cui et al., 2007). Consistent with this, misexpression of SCR in the stele cells blocked SHR movement out of the stele cells (Koizumi et al., 2012).
In addition to the nuclear trapping of SHR by SCR, it has been proposed that a balance between nuclear import and nuclear export in the stele cells is important, and nuclear accumulation of SHR in the stele cells may confer a competence for movement upon the SHR protein (Gallagher and Benfey, 2009). It seems that nuclear trapping in the target cells could be an important factor for CPC accumulation, because CPC-GFP protein expressed in the stele cells did not accumulate efficiently in the epidermis without the GL3/EGL3 activity and CPC-GFP protein expressed under the control of the CPC promoter is localized mainly to the nucleus in the epidermis and is not detected in the cytoplasm.
Although GL3 appears to also trap CPC in a nucleus, CPC accumulated less efficiently in the N-position cell where GL3 accumulates than in the H-position cell, perhaps due to the difference in the level of the two bHLH proteins in the N-position and H-position cells (EGL3 appears to be much more abundant in the H-position cells than GL3 in the N-position cells, based on the translational reporters) or to a possible difference in the affinity of CPC for the EGL3 versus GL3. The proposed EGL3-dependent nuclear trapping of CPC seems to be at odds with the lack of an egl3 single mutant phenotype in the root epidermis (Bernhardt et al., 2003). This may be explained by the action of (a small amount of) GL3 in the H-position cells of the egl3 mutant, which may not have been detected in our GL3-YFP fusion line, but may be sufficient to generate different CPC:WER ratios at the two cell positions.
An Updated Model for Cell Fate Specification in the Root Epidermis
Cell fate in the developing root epidermis is flexible and is dependent on numerous regulatory proteins, including CPC (Wada et al., 1997). Recently, we suggested that a critical factor in determining the cell fate is the ratio of the CPC level to the WER level within a given cell’s nucleus (Song et al., 2011). In this study, we have obtained additional insight into the distribution and function of CPC in this system. We showed that the translocation of CPC was not necessary for its function in cell specification, unlike KN1 and LEAFY that require their movement between cells for their proper function (Kim et al., 2003; Wu et al., 2003). We also showed that CPC accumulates preferentially in the nucleus of the H-position epidermal cells, even though it is capable of moving from H-position cells to N-position cells in the epidermis and vice versa. Therefore, we suggest that preferential localization of CPC protein to the nucleus of H-position cells represents a new and important factor for proper cell fate specification in the root epidermis.
Together with results from previous reports (Wada et al., 1997, 2002; Lee and Schiefelbein, 1999, 2002; Kurata et al., 2005a; Ryu et al., 2005; Kwak and Schiefelbein, 2007), we propose an updated model. In response to an unidentified positional signal, SCM inhibits WER expression in the H-position cells, causing differential accumulation of WER between the two cell positions and increased GL2 and CPC expression in the N-position epidermal cells. The CPC protein moves through plasmodesmata in a targeted manner and is trapped in the nucleus of the H-position cells by binding to EGL3, which causes a relative difference in the CPC protein level (and thus the WER:CPC ratio) between the cells located in the two positions and helps define the cell fates. This model provides an explanation for the puzzling lack of CPC action in the cells in which its gene is expressed (the N-cell position); this is proposed to be due to H-cell nuclear trapping, which prevents CPC from accumulating in the N-cell position.
Functional Divergence of the GL3 and EGL3 bHLH Genes
The GL3 and EGL3 are closely related bHLH genes in Arabidopsis that function redundantly in trichome and root hair specification (Bernhardt et al., 2003; Zhang et al., 2003; Morohashi et al., 2007; Zhao et al., 2008). Here, we suggest that these do not share identical roles in the root epidermis. EGL3 preferentially accumulated in the nucleus of the H-position epidermal cells, while the GL3 protein is in the nucleus of the N-position epidermal cells, and EGL3 plays a role in the preferential localization of CPC to the nucleus of the H-position cell. This situation is reminiscent of the relationship between the Myb proteins WER and GL1, which despite their biochemical equivalence, play different roles during plant development due to the different expression patterns caused by cis-regulatory sequences (Lee and Schiefelbein, 2001). Interestingly, the differential localization of GL3 and EGL3 is likely due to differences in their amino acid sequence, given the similar patterns of promoter activity and transcript accumulation in the H-position epidermal cells (Bernhardt et al., 2005), suggesting another possible mechanism of functional divergence of duplicated genes.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The cpc mutant, the wer mutant, the gl3 egl3 double mutant, and the cpc try double mutant have been previously described (Wada et al., 1997; Lee and Schiefelbein, 1999; Schellmann et al., 2002; Bernhardt et al., 2003; Zhang et al., 2003). The transgenic plants harboring 35Sp:CPC (Wada et al., 1997), GL2p:GUS (Masucci et al., 1996), GL3p:GL3-YFP (Bernhardt et al., 2005), and EGL3p:EGL3-YFP (Zhao et al., 2008) were also described.
The enhancer trap lines J1092, J2301, J0571, J1721, and Q0990 (http://data.plantsci.cam.ac.uk/Haseloff/assembly/page167/index.html) were obtained from Arabidopsis Biological Resource Center.
For plant growth, seeds were surface sterilized, germinated, and grown vertically on agarose-solidified medium containing mineral nutrients at 22°C under continuous light condition (Schiefelbein and Somerville, 1990).
Analysis of Epidermal Cell Patterning
For each line, we analyzed at least 10 4-d-old seedlings and performed three independent experimental repeats. We observed seedling roots using differential interference contrast microscopy and scored the 30 H-position cells and the 30 N-position cells for each seedling.
Gene Expression Analyses
GUS activity was histochemically defined by staining 4-d-old seedlings as described (Lee and Schiefelbein, 2002). Four-day-old seedlings were stained for 2 h in the case of the GL2p:GUS reporter gene and for 6 h in the case of the MYC1p:GUS reporter gene and the CPCp:GUS reporter gene. In Figure 3, the seedlings were stained for only 3 h to observe the GUS activity in the stele cells without the strong staining in the epidermis.
To measure the steady-state level of the CPC transcript, total RNA was extracted from the root tips of 4-d-old seedlings and used for the quantitative real-time reverse transcription (RT)-PCR analysis as described previously (Kang et al., 2009). EONGATION FACTOR1 (EF1) was used as an internal reference to normalize the relative level of each transcript. Each experiment was repeated three times, and each time, the experiment included triplicate samples. The nucleotide sequences of the primers were 5′-AGCAAAGCCAAGGCTTCTTGTTCC-3′ (forward primer) and 5′-CGACGCCGTGTTTCATAAGCCAAT-3′ (reverse primer) for the CPC gene and 5′-TTGACCAGATCAACGAGCCCAAGA-3′ (forward primer) and 5′-ACTCGTGGTGCATCTCAACAGACT-3′ (reverse primer) for the EF1 gene.
Confocal Microscopy
To detect GFP expression, seedlings were counterstained with 5 μg mL–1 propidium iodide solution for 5 min and examined using a Zeiss LSM510 Meta confocal microscope with excitation at 488 nm and detection with a 500- to 530-nm band-path filter for GFP and with excitation at 543 nm and detection with a 560-nm long-path filter for propidium iodide (Lee and Schiefelbein, 1999).
Gene Constructs and Plant Transformation
To examine the promoter activity of MYC1, the GUS gene was inserted between the 2.7-kb 5′ flanking region of MYC1 and the nopaline synthase (NOS) terminator (pMYC1:GUS).
To express CPC under the control of the MYC1 promoter, the coding region of CPC was PCR amplified and inserted into the same expression cassette (pMYC1:CPC). To express the CPC-GFP chimeric protein instead of CPC, the DNA fragments of the CPC coding region were PCR amplified and fused to the 5′ end of soluble GFP in frame. This chimeric CPC-GFP was inserted into the cassette (pMYC1:CPC-GFP).
To generate UASp:CPC-GFP for a targeted expression of CPC-GFP, the chimeric gene was fused to the 3′ end of the 5× UAS promoter as the CPC coding region was inserted previously (Song et al., 2008).
For the expression of CPC-GFP and GFP under the control of the SHR promoter, the 2.5-kb SHR promoter was PCR amplified (Nakajima et al., 2001) and fused to the 5′ end of CPC-GFP, GFP (soluble GFP), and GFP5-ER (endoplasmic reticulum-targeted GFP), respectively (SHRp:CPC-GFP, SHRp:GFP, and SHRp:GFP-ER). The above-mentioned constructs were introduced into the pCB302 binary vector (Xiang et al., 1999) containing the NOS terminator for plant transformation.
CPC-GFP was also inserted between the 2.0-kb CPC promoter and CPC terminator, and then this construct was introduced into the pBIN19 binary vector (CPCp:CPC-GFP).
All the DNA fragments PCR amplified using iProof High-Fidelity DNA polymerase (Bio-Rad) were sequenced and confirmed to be error free.
Plant transformation was achieved by the floral dip method as previously described (Clough and Bent, 1998).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under accession numbers CPC (At2g46410), EGL3 (At1g63650), GL2 (At1g79840), GL3 (At5g41315), WER (At5g14750), MYC1 (At4g00480), and SHR (At4g37650).
Supplemental Data
The following material is available in the online version of this article.
Supplemental Figure S1. Complementation of the root hair defect of the mutants using the translational reporter genes.
Acknowledgments
We thank Kyoung Hee Nam for helpful advice and suggestions.
Glossary
- SEL
size exclusion limit
- RT
reverse transcription
References
- Balkunde R, Bouyer D, Hülskamp M. (2011) Nuclear trapping by GL3 controls intercellular transport and redistribution of TTG1 protein in Arabidopsis. Development 138: 5039–5048 [DOI] [PubMed] [Google Scholar]
- Benítez M, Monk NAM, Alvarez-Buylla ER. (2011) Epidermal patterning in Arabidopsis: models make a difference. J Exp Zoolog B Mol Dev Evol 316: 241–253 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Lee MM, Gonzalez A, Zhang F, Lloyd A, Schiefelbein J. (2003) The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 130: 6431–6439 [DOI] [PubMed] [Google Scholar]
- Bernhardt C, Zhao M, Gonzalez A, Lloyd A, Schiefelbein J. (2005) The bHLH genes GL3 and EGL3 participate in an intercellular regulatory circuit that controls cell patterning in the Arabidopsis root epidermis. Development 132: 291–298 [DOI] [PubMed] [Google Scholar]
- Bouyer D, Geier F, Kragler F, Schnittger A, Pesch M, Wester K, Balkunde R, Timmer J, Fleck C, Hülskamp M. (2008) Two-dimensional patterning by a trapping/depletion mechanism: the role of TTG1 and GL3 in Arabidopsis trichome formation. PLoS Biol 6: e141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Bruex A, Kainkaryam RM, Wieckowski Y, Kang YH, Bernhardt C, Xia Y, Zheng X, Wang JY, Lee MM, Benfey P, et al. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui H, Levesque MP, Vernoux T, Jung JW, Paquette AJ, Gallagher KL, Wang JY, Blilou I, Scheres B, Benfey PN. (2007) An evolutionarily conserved mechanism delimiting SHR movement defines a single layer of endodermis in plants. Science 316: 421–425 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B. (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Fulton L, Batoux M, Vaddepalli P, Yadav RK, Busch W, Andersen SU, Jeong S, Lohmann JU, Schneitz K. (2009) DETORQUEO, QUIRKY, and ZERZAUST represent novel components involved in organ development mediated by the receptor-like kinase STRUBBELIG in Arabidopsis thaliana. PLoS Genet 5: e1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Benfey PN. (2009) Both the conserved GRAS domain and nuclear localization are required for SHORT-ROOT movement. Plant J 57: 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher KL, Paquette AJ, Nakajima K, Benfey PN. (2004) Mechanisms regulating SHORT-ROOT intercellular movement. Curr Biol 14: 1847–1851 [DOI] [PubMed] [Google Scholar]
- Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein JW. (1994) The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol 166: 740–754 [DOI] [PubMed] [Google Scholar]
- Grebe M. (2012) The patterning of epidermal hairs in Arabidopsis—updated. Curr Opin Plant Biol 15: 31–37 [DOI] [PubMed] [Google Scholar]
- Haseloff J. (1999) GFP variants for multispectral imaging of living cells. Methods Cell Biol 58: 139–151 [DOI] [PubMed] [Google Scholar]
- Helariutta Y, Fukaki H, Wysocka-Diller J, Nakajima K, Jung J, Sena G, Hauser MT, Benfey PN. (2000) The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567 [DOI] [PubMed] [Google Scholar]
- Jackson D, Veit B, Hake S. (1994) Expression of maize Knotted1 related homeobox genes in the shoot apical meristem predicts patterns of morphogenesis in the vegetative shoot. Development 120: 405–413 [Google Scholar]
- Kang YH, Kirik V, Hulskamp M, Nam KH, Hagely K, Lee MM, Schiefelbein J. (2009) The MYB23 gene provides a positive feedback loop for cell fate specification in the Arabidopsis root epidermis. Plant Cell 21: 1080–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Rim Y, Wang J, Jackson D. (2005) A novel cell-to-cell trafficking assay indicates that the KNOX homeodomain is necessary and sufficient for intercellular protein and mRNA trafficking. Genes Dev 19: 788–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Yuan Z, Jackson D. (2003) Developmental regulation and significance of KNOX protein trafficking in Arabidopsis. Development 130: 4351–4362 [DOI] [PubMed] [Google Scholar]
- Koizumi K, Hayashi T, Wu S, Gallagher KL. (2012) The SHORT-ROOT protein acts as a mobile, dose-dependent signal in patterning the ground tissue. Proc Natl Acad Sci USA 109: 13010–13015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurata T, Ishida T, Kawabata-Awai C, Noguchi M, Hattori S, Sano R, Nagasaka R, Tominaga R, Koshino-Kimura Y, Kato T, et al. (2005a) Cell-to-cell movement of the CAPRICE protein in Arabidopsis root epidermal cell differentiation. Development 132: 5387–5398 [DOI] [PubMed] [Google Scholar]
- Kurata T, Okada K, Wada T. (2005b) Intercellular movement of transcription factors. Curr Opin Plant Biol 8: 600–605 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Schiefelbein J. (2007) The role of the SCRAMBLED receptor-like kinase in patterning the Arabidopsis root epidermis. Dev Biol 302: 118–131 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Schiefelbein J. (2008) A feedback mechanism controlling SCRAMBLED receptor accumulation and cell-type pattern in Arabidopsis. Curr Biol 18: 1949–1954 [DOI] [PubMed] [Google Scholar]
- Kwak SH, Shen R, Schiefelbein J. (2005) Positional signaling mediated by a receptor-like kinase in Arabidopsis. Science 307: 1111–1113 [DOI] [PubMed] [Google Scholar]
- Lee JY, Colinas J, Wang JY, Mace D, Ohler U, Benfey PN. (2006) Transcriptional and posttranscriptional regulation of transcription factor expression in Arabidopsis roots. Proc Natl Acad Sci USA 103: 6055–6060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (1999) WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99: 473–483 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (2001) Developmentally distinct MYB genes encode functionally equivalent proteins in Arabidopsis. Development 128: 1539–1546 [DOI] [PubMed] [Google Scholar]
- Lee MM, Schiefelbein J. (2002) Cell pattern in the Arabidopsis root epidermis determined by lateral inhibition with feedback. Plant Cell 14: 611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Bouché-Pillon S, Jackson DP, Nguyen L, Baker L, Ding B, Hake S. (1995) Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW. (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- Masucci JD, Schiefelbein JW. (1996) Hormones act downstream of TTG and GL2 to promote root hair outgrowth during epidermis development in the Arabidopsis root. Plant Cell 8: 1505–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Benfey PN. (2012) Transcriptional switches direct plant organ formation and patterning. Curr Top Dev Biol 98: 229–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morohashi K, Zhao M, Yang M, Read B, Lloyd A, Lamb R, Grotewold E. (2007) Participation of the Arabidopsis bHLH factor GL3 in trichome initiation regulatory events. Plant Physiol 145: 736–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima K, Sena G, Nawy T, Benfey PN. (2001) Intercellular movement of the putative transcription factor SHR in root patterning. Nature 413: 307–311 [DOI] [PubMed] [Google Scholar]
- Rim Y, Huang L, Chu H, Han X, Cho WK, Jeon CO, Kim HJ, Hong JC, Lucas WJ, Kim JY. (2011) Analysis of Arabidopsis transcription factor families revealed extensive capacity for cell-to-cell movement as well as discrete trafficking patterns. Mol Cells 32: 519–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudel D, Sommer RJ. (2003) The evolution of developmental mechanisms. Dev Biol 264: 15–37 [DOI] [PubMed] [Google Scholar]
- Ryu KH, Kang YH, Park YH, Hwang I, Schiefelbein J, Lee MM. (2005) The WEREWOLF MYB protein directly regulates CAPRICE transcription during cell fate specification in the Arabidopsis root epidermis. Development 132: 4765–4775 [DOI] [PubMed] [Google Scholar]
- Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, Thumfahrt J, Jürgens G, Hülskamp M. (2002) TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J 21: 5036–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein J, Kwak SH, Wieckowski Y, Barron C, Bruex A. (2009) The gene regulatory network for root epidermal cell-type pattern formation in Arabidopsis. J Exp Bot 60: 1515–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein JW, Somerville C. (1990) Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2: 235–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlereth A, Möller B, Liu W, Kientz M, Flipse J, Rademacher EH, Schmid M, Jürgens G, Weijers D. (2010) MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 464: 913–916 [DOI] [PubMed] [Google Scholar]
- Sessions A, Yanofsky MF, Weigel D. (2000) Cell-cell signaling and movement by the floral transcription factors LEAFY and APETALA1. Science 289: 779–782 [DOI] [PubMed] [Google Scholar]
- Simon M, Lee MM, Lin Y, Gish L, Schiefelbein J. (2007) Distinct and overlapping roles of single-repeat MYB genes in root epidermal patterning. Dev Biol 311: 566–578 [DOI] [PubMed] [Google Scholar]
- Song S-K, Hofhuis H, Lee MM, Clark SE. (2008) Key divisions in the early Arabidopsis embryo require POL and PLL1 phosphatases to establish the root stem cell organizer and vascular axis. Dev Cell 15: 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song SK, Ryu KH, Kang YH, Song JH, Cho YH, Yoo SD, Schiefelbein J, Lee MM. (2011) Cell fate in the Arabidopsis root epidermis is determined by competition between WEREWOLF and CAPRICE. Plant Physiol 157: 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Kurata T, Tominaga R, Koshino-Kimura Y, Tachibana T, Goto K, Marks MD, Shimura Y, Okada K. (2002) Role of a positive regulator of root hair development, CAPRICE, in Arabidopsis root epidermal cell differentiation. Development 129: 5409–5419 [DOI] [PubMed] [Google Scholar]
- Wada T, Tachibana T, Shimura Y, Okada K. (1997) Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113–1116 [DOI] [PubMed] [Google Scholar]
- Wester K, Digiuni S, Geier F, Timmer J, Fleck C, Hülskamp M. (2009) Functional diversity of R3 single-repeat genes in trichome development. Development 136: 1487–1496 [DOI] [PubMed] [Google Scholar]
- Wu S, Koizumi K, Macrae-Crerar A, Gallagher KL. (2011) Assessing the utility of photoswitchable fluorescent proteins for tracking intercellular protein movement in the Arabidopsis root. PLoS ONE 6: e27536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Dinneny JR, Crawford KM, Rhee Y, Citovsky V, Zambryski PC, Weigel D. (2003) Modes of intercellular transcription factor movement in the Arabidopsis apex. Development 130: 3735–3745 [DOI] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ. (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40: 711–717 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhao M, Morohashi K, Hatlestad G, Grotewold E, Lloyd A. (2008) The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135: 1991–1999 [DOI] [PubMed] [Google Scholar]
- Zhou J, Wang X, Lee JY, Lee JY. (2013) Cell-to-cell movement of two interacting AT-hook factors in Arabidopsis root vascular tissue patterning. Plant Cell 25: 187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]