Gene expression profiling in two seed compartments uncovers two transcriptional phases during seed germination that are separated by testa rupture.
Abstract
Seed germination is a critical stage in the plant life cycle and the first step toward successful plant establishment. Therefore, understanding germination is of important ecological and agronomical relevance. Previous research revealed that different seed compartments (testa, endosperm, and embryo) control germination, but little is known about the underlying spatial and temporal transcriptome changes that lead to seed germination. We analyzed genome-wide expression in germinating Arabidopsis (Arabidopsis thaliana) seeds with both temporal and spatial detail and provide Web-accessible visualizations of the data reported (vseed.nottingham.ac.uk). We show the potential of this high-resolution data set for the construction of meaningful coexpression networks, which provide insight into the genetic control of germination. The data set reveals two transcriptional phases during germination that are separated by testa rupture. The first phase is marked by large transcriptome changes as the seed switches from a dry, quiescent state to a hydrated and active state. At the end of this first transcriptional phase, the number of differentially expressed genes between consecutive time points drops. This increases again at testa rupture, the start of the second transcriptional phase. Transcriptome data indicate a role for mechano-induced signaling at this stage and subsequently highlight the fates of the endosperm and radicle: senescence and growth, respectively. Finally, using a phylotranscriptomic approach, we show that expression levels of evolutionarily young genes drop during the first transcriptional phase and increase during the second phase. Evolutionarily old genes show an opposite pattern, suggesting a more conserved transcriptome prior to the completion of germination.
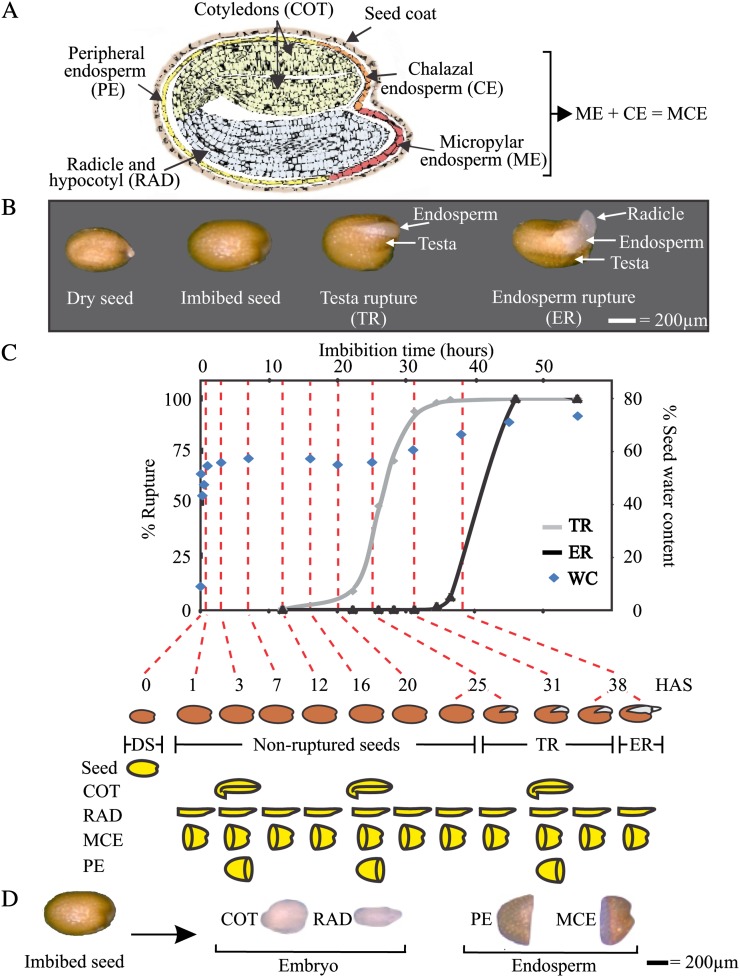
Seeds are important in the plant life cycle, since they represent the link between two successive generations. They are stress-resistant structures that help to bridge unfavorable periods and allow dispersal. Seed formation starts with a double fertilization event, and in Arabidopsis (Arabidopsis thaliana), it takes approximately 20 d to form a mature dry seed (Debeaujon et al., 2007; Ohto et al., 2007). At maturity, three major seed compartments can be distinguished (Holdsworth et al., 2008a; Belmonte et al., 2013): the testa (seed coat), a dead tissue that forms a protective outer layer; the endosperm, a single cell layer of tissue positioned directly underneath the testa; and the embryo (enclosed by the testa and endosperm), which emerges to become the future plant (Rajjou et al., 2012; Fig. 1A). A dry seed is a unique structure in the sense that it allows severe dehydration (desiccation tolerance) and enters a phase of quiescence, bringing processes occurring in “living” organisms to a halt without affecting viability (Farrant and Moore, 2011; Rajjou et al., 2012). Upon imbibition of water, the dry mature seed swells and metabolic activity resumes, marking the start of seed germination and the end of the quiescent state. Arabidopsis germination consists of two visible sequential events (Holdsworth et al., 2008a; Weitbrecht et al., 2011). First, the testa splits (testa rupture [TR]) due to underlying expansion of the endosperm and embryo. Thereafter, the radicle (RAD; embryonic root) protrudes through the endosperm (endosperm rupture [ER]), completing germination sensu stricto (Fig. 1B).
Figure 1.
Seed compartments and seed germination kinetics of Arabidopsis seeds. A, A section through an Arabidopsis seed depicting the different seed compartments. B, Different stages during seed germination including TR (which exposes the underlying endosperm layer) and ER (also known as RAD protrusion or germination sensu stricto). C, Arabidopsis seed germination analyzed by measuring TR (gray line), ER (black line), and seed water content (WC; blue diamonds). Below the graph, the time points and physiological stages (dry, NR, TR, and ER) are indicated for each sample. The 29 samples that were analyzed are schematically shown below the germination graph by the yellow pictograms. D, The four seed sections that were used for transcriptome analysis.
There are two nonexclusive mechanisms proposed to explain seed germination (Nonogaki, 2006; Nonogaki et al., 2007). The first involves the increase in embryo growth potential leading to elongation of the proximal embryonic axis (hypocotyl and RAD) that overcomes the restraint of the covering tissues. The second involves the weakening of these covering layers (including the micropylar endosperm, positioned over the RAD tip; Fig. 1A) to ease the protrusion of the RAD (for review, see Finch-Savage and Leubner-Metzger, 2006). The endosperm has been shown to affect germination even in species with a thin endosperm layer, such as Arabidopsis (Müller et al., 2006; Bethke et al., 2007; Lee et al., 2010). Genome-wide expression studies have been previously applied to gain insight into several aspects of seed biology (Holdsworth et al., 2008a, 2008b; Le et al., 2010), including temporal changes during Arabidopsis germination (Nakabayashi et al., 2005; Preston et al., 2009; Narsai et al., 2011) and in spatial differences between embryo and endosperm (Penfield et al., 2006; Endo et al., 2012). Nevertheless, a detailed knowledge of the temporal changes in gene expression in the different compartments of the Arabidopsis seed is thus far missing, but it is essential to understanding the control of the timing of germination as well as the underlying molecular processes contributed by these different seed compartments. Therefore, we have analyzed the Arabidopsis transcriptome by sampling 11 points along the germination time course, including those that allow an analysis of gene expression changes at the key events of germination (TR and ER), with a focus on the micropylar endosperm and the RAD.
RESULTS AND DISCUSSION
Arabidopsis Seed Imbibition, Germination Kinetics, and Transcriptome Analyses
We characterized Arabidopsis seed germination by scoring TR and ER over time. TR started around 20 h after sowing (HAS), and at 31 HAS almost all seeds were fully ruptured. From 31 HAS onward, ER was observed, which was completed in the entire seed population by 45 HAS (Fig. 1C). Microarray experiments were performed using dry seeds and seeds at nine time points along the germination time course until the completion of germination (Fig. 1C). The time points 25 and 38 HAS showed a mixture of nonruptured (NR) and TR seeds and TR and ER seeds, respectively; at these time points, both classes were separated and collected as distinct samples, which enabled us to map the transcriptome changes induced by TR and ER. To capture spatial dynamics, imbibed seeds were dissected into four parts. The key compartments for germination, the RAD (including a large part of the hypocotyl to ensure that it encompasses the region that elongates [Sliwinska et al., 2009]) and the micropylar end of the endosperm (which is a combination of micropylar and chalazal endosperm [MCE]), were sampled at all time points. At three time points (3, 16, and 31 HAS), the cotyledons (COT) and the remainder of the endosperm (peripheral endosperm [PE]) were collected (Fig. 1, A, C, and D; Supplemental Fig. S1). The 29 samples, with four replicates for each sample, were analyzed using Affymetrix ATH1 gene chips. Plotting probe set values in a histogram showed clearly distinguishable peaks for noise and signal and revealed that an appropriate cutoff for considering a gene as potentially expressed was 5 on a log2 scale (Supplemental Fig. S1). The percentage of genes detected in the different seed compartments was within the same range described for other Arabidopsis seed transcriptome analyses (Nakabayashi et al., 2005; Penfield et al., 2006; Belmonte et al., 2013). In total, 14,317 genes (67.2% of the 21,313 genes on the chip) were found to be expressed at least once in the 29 samples, of which 11,298 (78.8%) were shared between all compartments (Supplemental Fig. S2A).
At the start of the time course, a lower number of genes were found to be expressed, and this number increased during the time course in all tissues, most notably during the first 12 to 16 HAS (Supplemental Fig. S2B). We identified gene sets that were tissue specifically expressed by considering genes as specifically expressed in one tissue when expressed above 6 (on a log2 scale) in that tissue and expressed below 5 in all the other tissues (which, therefore, is in the noise region). This resulted in 415 genes specific to the endosperm and 546 genes specific to the embryo in our data set (Supplemental Fig. S2; Supplemental Data Set S1), which overlaps with previously published data sets (Penfield et al., 2006; Le et al., 2010; Supplemental Fig. S3). In total, 12,856 genes are expressed above 6 in either tissue, with 10,801 expressed above 6 in both tissues. Thus, according to this definition, 84.01% of the genes are shared between both tissues, while 3.22% are specific to the embryo and 4.24% are specific to the endosperm. The remaining genes (8.53%) are expressed over 6 in one tissue but between 5 and 6 in another tissue and so are not classed as being highly specific to any one tissue. Interrogation using overrepresentation analysis (ORA) revealed that the endosperm gene set was overrepresented for genes related to response to abscisic acid, defense response, cell wall macromolecule metabolism/catabolism, and cell death as well as genes associated with the regulation of transcription (Supplemental Fig. S2D), in agreement with recent findings (Endo et al., 2012). In the embryo, the largest class was related to plant development. Other Gene Ontology (GO) classes that were overrepresented included cell division, hormone metabolic process, protein amino acid phosphorylation, signaling, and regulation of transcription (Supplemental Fig. S2E). Thus, different GO classes were found to be overrepresented in each tissue, with regulation of transcription/gene expression appearing in both. Both tissue-specific gene sets are enriched for transcription factors (Supplemental Fig. S2F). In the endosperm, transcription factors of NAC, WRKY, and C3H classes, and in the embryo, transcription factors of bHLH, G2-like, and HB classes, are particularly enriched (Supplemental Fig. S2F). Compartment-specific gene sets containing 106, 47, 21, and two genes were identified for the RAD, COT, MCE, and PE (Supplemental Fig. S2; Supplemental Data Set S1), respectively, and quantitative reverse transcription-PCR confirmed the compartment-specific expression of 20 genes (Supplemental Fig. S4).
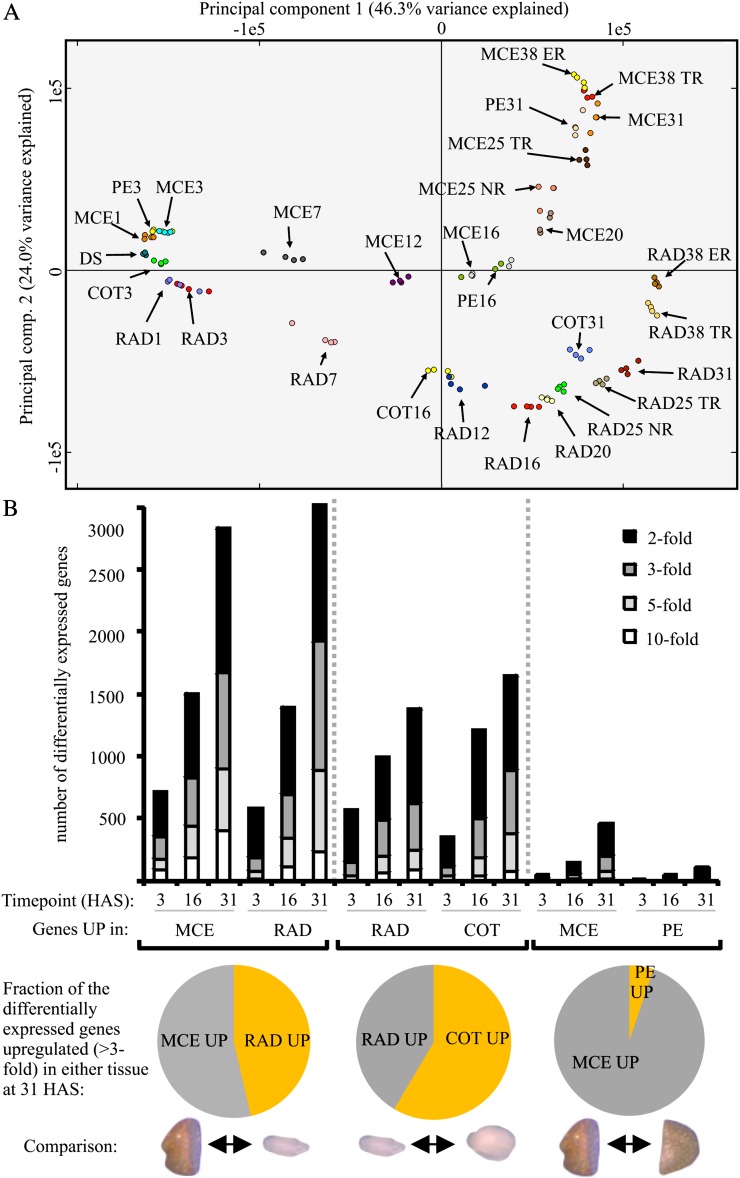
In order to globally compare gene expression between the samples, all 116 arrays were plotted using principal component analysis (PCA; Fig. 2A). In general, the largest transcriptome differences were observed between the endosperm and embryo (MCE versus RAD) followed by the comparison between both embryo parts (RAD versus COT). The smallest differences were found between both endosperm (MCE versus PE) parts (Fig. 2). The quality controls (Supplemental Fig. S1), the high correlation between the replicates (Supplemental Table S1), and the confirmation by quantitative reverse transcription-PCR of compartment-specific expression (Supplemental Fig. S4) indicate that this is a robust data set revealing transcriptome changes during seed rehydration and the developmental switch from a quiescent dry seed to germination in both temporal and spatial detail.
Figure 2.
Transcriptional differences between seed compartments. A, PCA of the 116 samples. The four replicates of all 29 samples are indicated by color. B, Tissue differences are represented by the number of differentially expressed genes at three time points during imbibition (3, 16, and 31 HAS, the time points in which all four tissues were sampled). Comparisons were made between endosperm and embryo (MCE versus RAD), between embryo tissues (RAD versus COT), and between both endosperm samples (MCE versus PE). The bars show the number of differentially expressed genes at a 2-, 3-, 5-, and 10-fold cutoff. The pie diagrams below the graph indicate the fraction of the total number of differentially expressed genes (at a 3-fold cutoff level) in either of the two tissues that were compared at 31 HAS.
Generation of Coexpression Networks and Data Visualization Tools
We generated coexpression networks (Bassel et al., 2011) for the endosperm (EndoNet) and the RAD samples (RadNet). We identified compact clusters of genes in the networks (Supplemental Data Set S1) that were further scrutinized with the network topological analyzer, TopoGSA (http://www.topogsa.net/; Glaab et al., 2010; Supplemental Fig. S5). Interactive visualizations of both networks are available online at http://vseed.nottingham.ac.uk. Compared with our previous visualization tool (Bassel et al., 2011), these visualizations offer improved performance and more advanced gene selection options, such as the highlighting of individual genes or entire clusters, searching for genes by name or descriptive keywords, and visualization of gene expression using our new Electronic Fluorescent Pictograph browser (Winter et al., 2007).
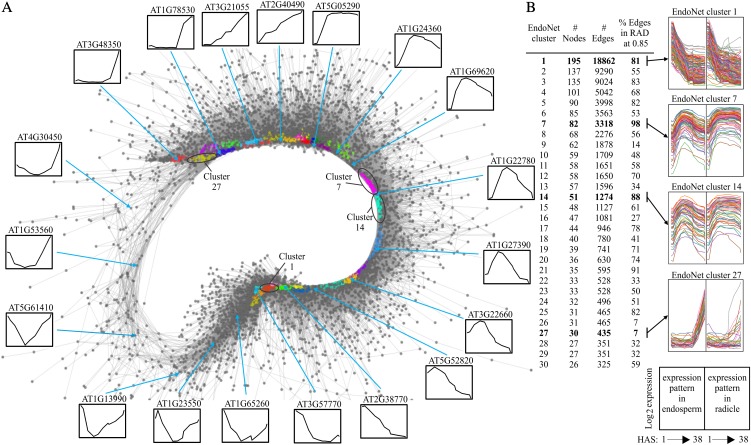
EndoNet shows a ring-like display, a result of the scarcity of genes with constant expression (Fig. 3A). This indicates that the regulation of gene expression is very dynamic in the endosperm during germination. The largest 30 EndoNet clusters are spread around the network and thus represent the major gene expression profiles. ORA revealed cluster-specific overrepresentation of specific biological processes (Supplemental Fig. S6). These clusters consist of 26 to 195 genes and contain at least 99.7% of all possible edges within them (Fig. 3), indicating that genes within such clusters have very similar expression patterns. Genes of some clusters (e.g. EndoNet cluster 1) are also coexpressed in RadNet (81% of the edges in cluster 1 are also found in RadNet at a 0.85 correlation) and show similar expression patterns in both compartments, while other genes (such as EndoNet cluster 27) show an endosperm-specific expression pattern and have few edges in common with RadNet (Fig. 3B). On the other hand, almost all connections in EndoNet clusters 7 and 14 (98% and 88%, respectively) are also present in RadNet (Fig. 3B). Despite strong coexpression between both networks, the expression profiles in these clusters are different between the two compartments, being induced in both but subsequently repressed in the endosperm.
Figure 3.
The endosperm coexpression network, EndoNet. A, Sample layout of EndoNet. The nodes (genes) are indicated by gray circles, and edges (gray lines) are drawn between two nodes if their correlation of expression is above 0.932. The 30 largest clusters are indicated by different colors. To visualize the gene expression profiles captured in the network, the expression profiles of exemplar genes are shown around the network. B, Details of the largest 30 clusters are shown, including the number of nodes, edges, and the percentage of edges that are shared with RadNet (at a cutoff of 0.85). The expression profiles of genes in the EndoNet clusters 1, 7, 12, and 27 are shown (the positions of these clusters in EndoNet are shown A). The right side of the graph depicts the expression profiles of the same set of genes in the RAD samples.
Arabidopsis Seed Germination Is Composed of Two Transcriptional Phases
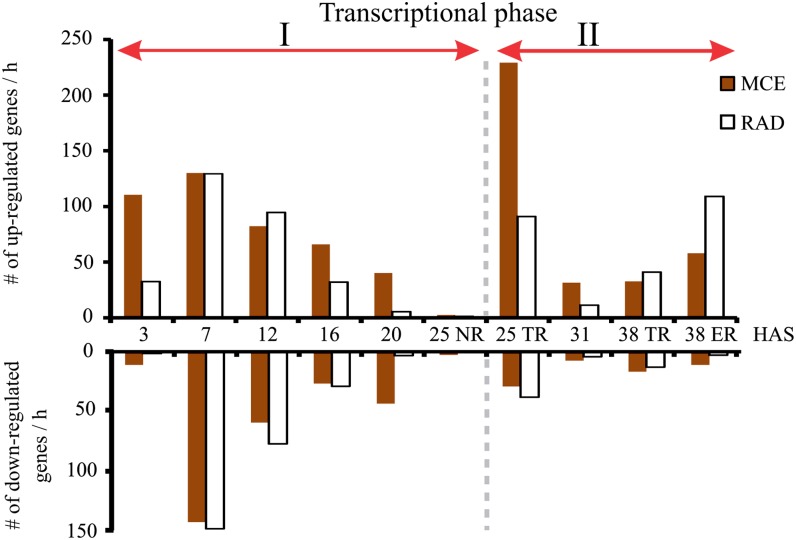
Analyzing the transcriptional dynamics between consecutive time points of the germination time course revealed two transcriptional phases (Fig. 4). The first phase runs from 1 to 25 HAS NR and is characterized by large transcriptional changes in both up- and down-regulated genes. At the end of this first phase, the number of differentially expressed genes was reduced (Fig. 4). The second phase, which runs from TR to the completion of germination, was marked by resumption of differential gene expression, most notably at TR. During the second phase, the majority of the differentially expressed genes are induced rather than repressed, in contrast to the first phase.
Figure 4.
Arabidopsis seed germination is characterized by two transcriptional phases. The number of differentially expressed genes (both up- and down-regulated) between consecutive time points (3 was compared with 1, 7 with 3, 12 with 7, etc.) in the MCE (white bars) and RAD (brown bars) with a reasonable fold change (taking a 3-fold difference as the cutoff) are presented. The two transcriptional phases, phase I from 1 to 25 HAS NR and phase II from 25 HAS NR to 38 HAS ER, are indicated by the red arrows.
The First Transcriptional Phase Is Characterized by an Inversion of the Seed Maturation Transcriptional Program
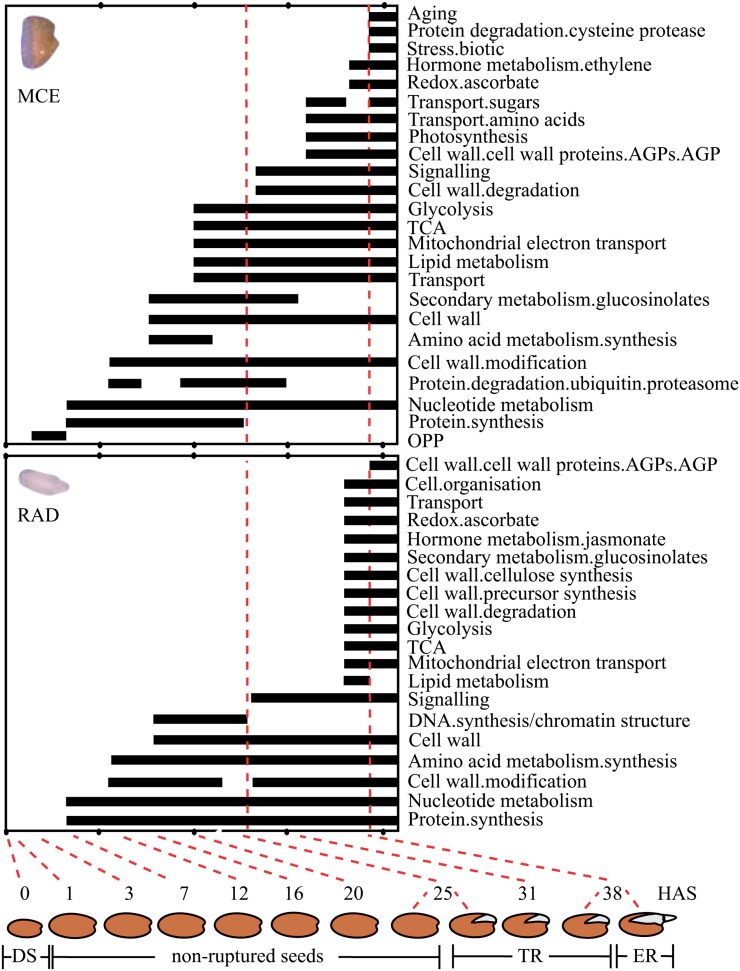
Between 1 and 3 HAS, differential gene expression was observed, particularly in the MCE (Fig. 4). In comparison, the response of the RAD was delayed, which could be due to its slower imbibition kinetics compared with the more outward-positioned MCE (Fig. 4). Large transcriptional changes occurred in the first 16 HAS. ORA of this phase suggests a large overlap in the functional classes that are activated in the MCE and RAD (i.e. genes related to cell wall function, nucleotide metabolism, amino acid metabolism, and protein translation; Fig. 5). A major difference is the activation of classes related to transport and energy metabolism (lipid metabolism, glycolysis, TCA, and mitochondrial electron transport) that are specifically activated in the MCE from 20 HAS, in agreement with findings that storage lipids are more rapidly mobilized in the endosperm compared with the embryo (Penfield et al., 2005).
Figure 5.
Temporal differences between endosperm and embryo using ORA. The overrepresented gene categories of the up-regulated genes of the germination time course (all time points were compared with 1 HAS) were identified in the MCE (top graph) and the RAD (bottom graph) using PageMan (Usadel et al., 2006). Selected categories are summarized in the graphs, and black bars show the time points during germination at which the indicated gene categories are overrepresented. OPP, Oxidative pentose phophate pathway.
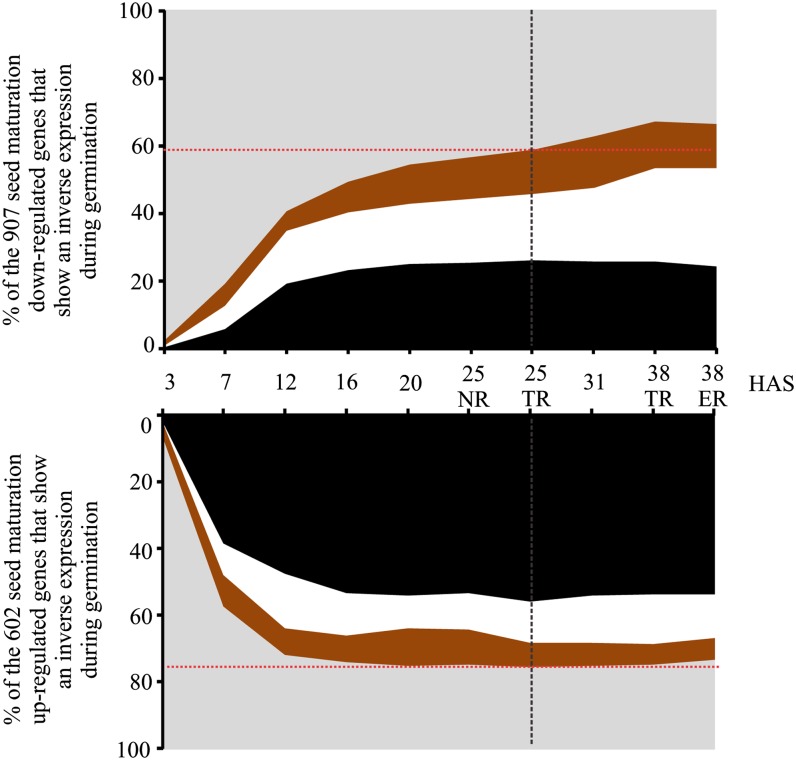
We compared gene expression during seed germination with gene expression during seed development and identified two gene sets containing 602 and 907 genes (Supplemental Data Set S1) that were strongly up- and down-regulated, respectively, between the embryo COT phase (early seed maturation) and the postmature green stage (late maturation) from a publicly available data set (Le et al., 2010). The expression of the two gene sets was analyzed during germination, and the majority of the genes of both sets showed inverse expression patterns during seed germination (Fig. 6). The largest overlap (75%) was found between genes that were up-regulated during seed maturation and those down-regulated during germination. Additionally, 67% of the genes from the set that were down-regulated during seed maturation showed an inverse expression pattern (were induced) during germination. The reinduction of these seed maturation down-regulated genes during germination was slower than the removal of the seed maturation-induced genes. Nevertheless, the majority of the seed maturation-repressed genes were reactivated in the first transcriptional phase rather than the second transcriptional phase.
Figure 6.
Inverse expression of seed maturation genes during germination in temporal and spatial detail. The top panel shows the percentage of up-regulated genes during germination among a set of 907 genes that are down-regulated during seed maturation. The bottom panel shows the percentage of down-regulated genes during germination among a set of 602 genes that are up-regulated during seed maturation. Genes expressed specifically in the MCE (in brown), in the RAD (in white), and in both (in black) are indicated.
TR Is Marked by High Transcriptional Activity That Overlaps in Part with a Response to Touch-Induced Signaling
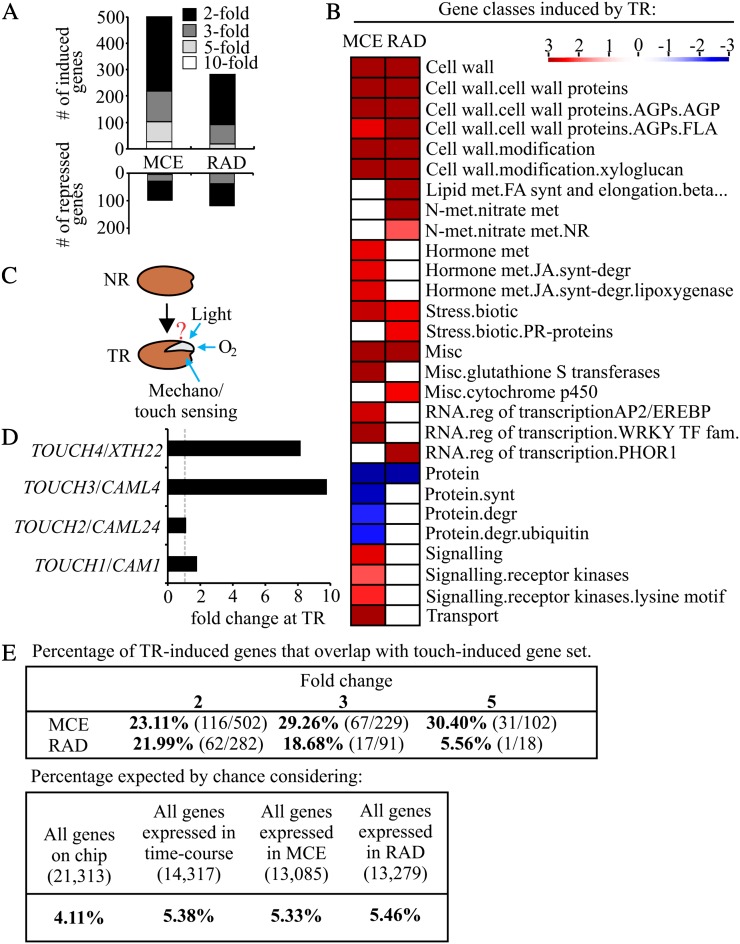
TR is characterized by a large number of differentially expressed genes when compared with NR seeds at 25 HAS, mostly genes that are up-regulated in the MCE (Fig. 7A). At TR, 104 genes were over 5-fold up-regulated in the MCE (Supplemental Data Set S1), 30 of which are related to cell wall function. Other classes induced by TR in the MCE include genes related to biotic stress, hormone metabolism, regulation of transcription, signaling (receptor kinases), and transport (Fig. 7B). Possible reasons for these large transcriptional changes between NR and TR seeds include an enhanced access to oxygen, light signaling, and/or a touch (mechano)-sensing response (Fig. 7C). ORA did not reveal a clear indication of the involvement of either oxygen or light. However, the gene set included TOUCH3 and TOUCH4 (both more than 8-fold induced), which are known to respond rapidly to touch (Braam, 2005; Fig. 7D). To investigate whether the transcriptional up-regulation at TR resembles touch sensing, we compared our MCE TR up-regulated data set with genes up-regulated upon touch in aerial parts of plants (Lee et al., 2005). We reanalyzed a published touch data set (Lee et al., 2005; Supplemental Materials and Methods S1) and found a 30% overlap with our TR-induced set in the MCE and the touch up-regulated genes, with a lower overlap between the touch data set and the TR-induced genes in the RAD (Fig. 7E). The overlap between the gene sets induced by TR in the MCE and touch was more striking when the gene classes were considered. Touch-induced signaling resulted in a relatively higher abundance of genes related to the GO classes cell wall associated, calcium binding, disease resistance, kinase, and transcription factor (Lee et al., 2005), which match well with the classes identified at TR (Fig. 7B). We also observed that gene expression associated with jasmonate biosynthesis was activated upon TR in the MCE; this plant hormone was recently shown to be a key regulator of plant morphogenesis and enhanced pest resistance upon touch (Chehab et al., 2012). It has been hypothesized that gene expression in the endosperm during germination might be affected by touch/mechano sensing (Martínez-Andújar et al., 2012), and this transcriptome study provides a strong suggestion that touch signaling is indeed, at least in part, responsible for the induction of gene expression in the endosperm.
Figure 7.
Genes induced with respect to TR show a large overlap with touch-induced signaling. A, Number of differentially expressed genes at 25 HAS TR (compared with 25 HAS NR) in the MCE and RAD at different fold change cutoffs. B, Gene classes overrepresented in the TR-induced gene sets in the MCE and RAD. C, Schematic presentation of effectors that could be responsible for the large gene expression changes observed at TR. D, Expression behavior of four TOUCH genes at TR in the MCE. E, Table shows the percentage of the TR up-regulated genes in the MCE and the RAD (at 2-, 3-, and 5-fold cutoff) that overlap with the 934 touch up-regulated genes. The percentage expected by chance is indicated using the number of genes present on the chip, genes expressed in the germination time course, genes expressed in the MCE, and genes expressed in the RAD. degr, Degradation; FA, fatty acid; fam, family; FLA, fasciclin-like arabinogalactan; JA, jasmonic acid; met, metabolism; misc, miscellaneous; NR, non-ruptured; PR, pathogenesis-related; reg, regulation; synt, synthesis; TF, transcription factor.
The Second Transcriptional Phase Highlights Distinct Fates for the Embryo and the Endosperm
The second transcriptional phase starts at TR and includes gene expression changes related to the completion of germination. Using ORA, we analyzed the temporal changes in the MCE and the RAD (Fig. 5) as well as gene sets that are more highly expressed within the MCE or RAD along the time course (Supplemental Fig. S7). This revealed that, in the MCE genes related to secondary metabolism, amino acid metabolism and protein synthesis are overrepresented transiently (Fig. 5). Genes more highly expressed in the MCE than the RAD are enriched for protein degradation, transport, and stress-related genes (although the latter are overrepresented in the MCE over the whole time course; Supplemental Fig. S7). The RAD, particularly at the later stage, is enriched for cellular metabolism related to DNA, RNA, and proteins compared with the MCE (Supplemental Fig. S7). ORA suggests that energy metabolism (lipid metabolism, glycolysis, TCA, and mitochondrial electron transport) is activated by 38 HAS. At this stage, genes for cell wall biosynthesis, transport, and secondary metabolism are activated, notably just prior to ER (Fig. 5). In addition, genes related to the cell cycle and lipid and amino acid metabolism are overrepresented within genes more highly expressed in the RAD than the MCE (Supplemental Fig. S7), which are all classes supporting tissue growth. The GO gene class “aging” becomes overrepresented in the latter part of the germination time course in the MCE (Fig. 5; Supplemental Fig. S7). This is in agreement with the down-regulation of key cellular metabolic pathways and the induction of gene classes related to remobilization, reminiscent of the transcriptional changes described for senescence (Lim et al., 2007; Breeze et al., 2011).
The Transition from a Dry Quiescent to a Hydrated and Germinating Seed Coincides with Increased Transcriptional Differences between Seed Compartments
From the PCA of all 116 arrays (Fig. 2A), we conclude that the transcriptome differences between seed compartments are small during early germination and increase with time. This is in agreement with the observation that the number of endosperm- and embryo-specific genes expressed increased along the time course from approximately 40 to 400 (Supplemental Fig. S8A). This may be explained by the fact that the majority of genes induced in seed maturation and that are subsequently removed during germination are shared by the MCE and RAD (72%) and that seed maturation-repressed genes (reactivated during germination) are, in contrast, mostly specific to either the RAD or the MCE (Fig. 6). Presumably, the repression of genes related to development and differentiation is a more general response for an organism passing through a desiccated state, as is shown for the expression of genes involved in stomatal development (in the COT samples) and root development (in the RAD samples). Many of these genes are induced (sometimes transiently) during germination, with low or no expression initially (Supplemental Fig. S8B).
Differential Gene Expression in the Endosperm Is Concentrated at the Micropylar End
The observation that the transcriptional differences increase with time between seed compartments is also shown, besides the PCA, in the number of differentially expressed genes between the seed compartments. The number of differentially expressed genes was least between both endosperm compartments. At 31 HAS, about 200 genes were differentially expressed (more than 3-fold difference), with the majority of these (95%) being up-regulated in the MCE (Fig. 2B) compared with the PE. Such a skewed division was not observed for other comparisons (Fig. 2B). The micropylar endosperm is hypothesized to possess an inhibitory role in germination, and endosperm changes, in particular of cell wall properties, are suggested to be important for germination control (Nonogaki et al., 2007). Recently, using in situ cell wall epitope detection, Arabidopsis endosperm cell walls were shown to have a different structure compared with the embryo cell wall, and the endosperm walls were shown to contain cellulose, unesterified homogalacturonan, arabinan, and xyloglucan polymers (Lee et al., 2012). However, no spatial or temporal heterogeneity in cell wall polymers was observed prior to germination (Lee et al., 2012). This could indicate that cell wall changes leading to germination are modifications that are not detectable by in situ analysis and/or that occur very locally. We compared both endosperm samples and found many differentially expressed genes between the MCE and PE (Supplemental Data Set S1). The largest differences were found close to the point of germination (31 HAS) in the MCE, and this set was investigated for candidates that are potentially involved in ER.
Several transcription factors were found to be highly expressed in the MCE compared with the PE that may function in gene regulation in this particular compartment. Genes related to cell wall function, including peroxidases, a pectin lyase-like superfamily protein, chitinase family protein, and ARABINOGALACTAN PROTEIN31 were identified in this set, and these could be potential candidates for affecting cell wall properties to enable seed germination. It is notable that one of the most highly differentially expressed (more than 20-fold) genes in the MCE is INFLORESCENCE DEFICIENT IN ABSCISSION-LIKE1 (IDL1). This encodes a putative ligand that promotes cell separation and floral organ abscission via the interaction with receptor-like kinases (Stenvik et al., 2008). Recently, it has been reported that the INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) peptide and its receptors HAESA (HAE) and HAESA-LIKE2 (HSL2) are also important for cell separation during lateral root emergence (Kumpf et al., 2013), suggesting that Arabidopsis seed germination may occur via a cell separation event that is potentially regulated by the IDA/IDL-HAE/HSL signaling module. This detailed data set allowed the identification of transcription factors, cell wall-related genes, and genes related to cell separation, although further research is needed to investigate their potential role in seed germination.
Seed Germination Is Characterized by Coordinated Expression of Evolutionarily Old and Young Genes
Recently, it has been shown that, like animal embryogenesis, plant embryogenesis involves a passage through a conserved and evolutionarily old transcriptional stage (Quint et al., 2012). This so-called phylotypic stage is mainly caused by the repression of evolutionarily young genes and is proposed to help the spatiotemporal organization and differentiation of multicellular life (Quint et al., 2012). Since we observed a largely inverse expression pattern during germination of gene sets that are up- and down-regulated during seed development (Fig. 6), we asked whether (1) seed germination is also characterized by the coordinated expression of evolutionarily old and young genes and (2), if so, whether these patterns are linked to the two transcriptomic phases we observed. To answer these questions, we first applied the phylostratigraphic approach (Domazet-Loso et al., 2007; Domazet-Loso and Tautz, 2010; Quint et al., 2012), in which we ordered the Arabidopsis genome into 12 evolutionary age classes (phylostrata; designated PS1–PS12). Each Arabidopsis gene is BLASTed against all genomes underlying the 12 phylostrata and is sorted in its phylostratum, defined as the most distant phylogenetic node containing at least one species with a detectable homolog (Quint et al., 2012). This resulted in the phylostratigraphic map in which PS1 (cellular organisms) contains the evolutionarily oldest genes and PS12 (Arabidopsis) contains the youngest genes that are specific to Arabidopsis, with no homologs detected in any of the other species (Supplemental Fig. S9A).
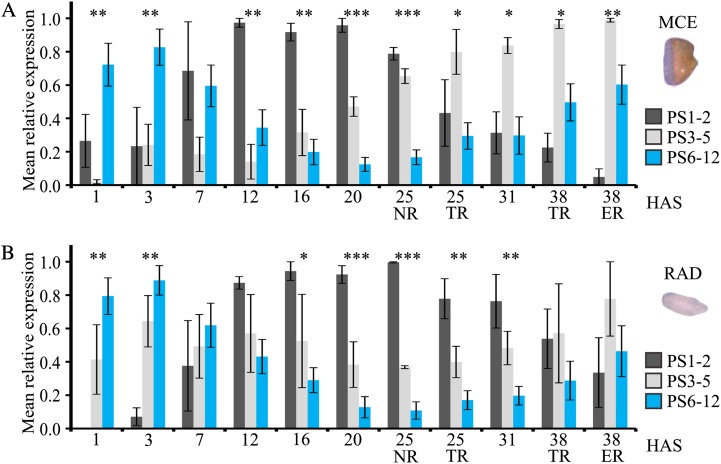
Next, we interrogated the gene expression data of the MCE and plotted the relative expression values of (1) genes that arose before plant evolution (PS1 and PS2 combined), (2) genes that arose during early plant evolution (algae and non-seed-bearing plants; PS3–PS5), and (3) the evolutionarily youngest genes (which evolved in seed-bearing plants; PS6–PS12). The analysis shows that in the MCE, the relative expression of evolutionarily young genes is high shortly after imbibition but drops during the first transcriptional phase, followed by an increase in the second transcriptional phase (Fig. 8A; Supplemental Fig. S9). Interestingly, the oldest genes (PS1 and PS2) showed an inverse behavior, starting low at the beginning of germination, peaking at the end of the first transcriptional phase, followed by a decrease in the second transcriptional phase. Genes of PS3 to PS5 show a different pattern, starting low and increasing during the course of germination. Comparable results were obtained for the RAD during germination (Fig. 8B; Supplemental Fig. S9). The patterns in both seed parts, particularly the inverse patterns of the evolutionarily old and young genes, suggest that seeds not only pass through an evolutionarily conserved stage during seed development but also during the successive germination phase. Coordinated expression of evolutionarily old and young genes (and, in this way, passage through a conserved transcriptional state) may help to channel large physiological transitions.
Figure 8.
Relative expression of evolutionarily old and young genes across the Arabidopsis germination time course. Plotted are the relative expression levels ± se of genes of PS1 and PS2, PS3 to PS5, and PS6 to PS12 across the Arabidopsis germination time course in the MCE (A) and RAD (B) compartments. The significance between the relative expression levels between the groups is indicated at each time point by asterisks: *P < 0.05, **P < 0.01, ***P < 0.001. For the phylostratigraphic map and the mean relative expression of individual phylostrata, see Supplemental Figure S9.
CONCLUSION
This study revealed two separate transcriptional phases for seed germination that are separated by TR and provides a strong indication that mechano-induced signaling affects gene expression at TR in the MCE. It also shows that time is an important determinant for spatial expression differences. Surprisingly, we found similar patterns of expression of evolutionarily old and young genes in seed development and seed germination, suggesting that plants passing through a transcriptional old and conserved stage may not be limited to embryogenesis. In addition to the novel biological insight, we are convinced that these detailed transcriptome data, including the tools developed for data visualization and mining, provide a powerful resource to gain further understanding of the roles of different seed compartments in germination, novel regulators, and gene networks underlying seed germination.
MATERIALS AND METHODS
Plant Material, Sampling, and Microarray Analysis
For this experiment, the Arabidopsis (Arabidopsis thaliana) accession Columbia-0 (N60000) was used. Seeds were sown on 0.7% water agarose (Eurogentec) and incubated in a germination cabinet at 22°C with continuous light. Germination curves (for both testa and endosperm rupture) were assessed by scoring germination in time. After the indicated HAS, seeds were harvested and dissected using forceps and a scalpel knife. For the isolation of RNA, a commercial kit (Absolutely RNA Nanoprep Kit; Agilent Technologies) was used. In total, 100 ng of RNA was used to synthesize biotin-labeled copy RNA (using the Affymetrix 3′ IVT-Express Labeling Kit), which was hybridized on the Affymetrix GeneChips Arabidopsis ATH1 Genome Array. The raw .cel files were background corrected and normalized using the Robust Microarray Averaging procedure (Irizarry et al., 2003). A detailed version of the methods used is available as Supplemental Materials and Methods S1.
The microarray data used in this article have been deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus with accession number GEO 41212.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. ATH1 Genechip quality assessment and reproducibility.
Supplemental Figure S2. General expression numbers.
Supplemental Figure S3. Comparisons with two other seed microarray datasets.
Supplemental Figure S4. RT-qPCR confirms tissue-specific expression found in the microarray dataset.
Supplemental Figure S5. Topological features of the EndoNet and RadNet.
Supplemental Figure S6. Overrepresentation analysis of the 30 largest clusters from the EndoNet co-expression network.
Supplemental Figure S7. ORA using Pageman of genes that are either higher expressed in the MCE or the RAD.
Supplemental Figure S8. Seed tissues differentiate during germination.
Supplemental Figure S9. Expression of evolutionary old and young genes during Arabidopsis seed germination.
Supplemental Figure S10. The node degree distribution for the correlation networks.
Supplemental Table S1. Correlations between the sample replicates.
Supplemental Table S2. Primer information of the genes tested by RT-qPCR.
Supplemental Materials and Methods S1. Supplemental Materials and Methods.
Supplemental Data Set S1. Collection of gene lists.
Acknowledgments
We thank Kieran Lee (University of Leeds) for providing the image used in Figure 1A, Janet Braam (Rice University) and Dennis Lee for providing the touch transcriptome data set, Nicolas Provart (University of Toronto) and Hardeep Nahal (University of Toronto) for help with developing the Electronic Fluorescent Pictograph browser, and Henk Hilhorst (Wageningen University) and Maarten Koornneef (Max Planck Institute for Plant Breeding Research) for critically reading earlier versions of the manuscript.
Glossary
- TR
testa rupture
- ER
endosperm rupture
- HAS
hours after sowing
- NR
nonruptured
- RAD
radicle
- MCE
micropylar and chalazal endosperm
- COT
cotyledons
- PE
peripheral endosperm
- ORA
overrepresentation analysis
- GO
Gene Ontology
- PCA
principal component analysis
References
- Bassel GW, Lan H, Glaab E, Gibbs DJ, Gerjets T, Krasnogor N, Bonner AJ, Holdsworth MJ, Provart NJ. (2011) Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc Natl Acad Sci USA 108: 9709–9714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. (2013) Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci USA 110: E435–E444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke PC, Libourel IG, Aoyama N, Chung YY, Still DW, Jones RL. (2007) The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol 143: 1173–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J. (2005) In touch: plant responses to mechanical stimuli. New Phytol 165: 373–389 [DOI] [PubMed] [Google Scholar]
- Breeze E, Harrison E, McHattie S, Hughes L, Hickman R, Hill C, Kiddle S, Kim YS, Penfold CA, Jenkins D, et al. (2011) High-resolution temporal profiling of transcripts during Arabidopsis leaf senescence reveals a distinct chronology of processes and regulation. Plant Cell 23: 873–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Yao C, Henderson Z, Kim S, Braam J. (2012) Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Curr Biol 22: 701–706 [DOI] [PubMed] [Google Scholar]
- Debeaujon I, Lepiniec L, Pourcel L, Routaboul J-M (2007) Seed coat development and dormancy. In Annual Plant Reviews, Vol 27: Seed Development, Dormancy and Germination. Blackwell Publishing, Oxford, pp 25–49 [Google Scholar]
- Domazet-Loso T, Brajković J, Tautz D. (2007) A phylostratigraphy approach to uncover the genomic history of major adaptations in metazoan lineages. Trends Genet 23: 533–539 [DOI] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. (2010) A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468: 815–818 [DOI] [PubMed] [Google Scholar]
- Endo A, Tatematsu K, Hanada K, Duermeyer L, Okamoto M, Yonekura-Sakakibara K, Saito K, Toyoda T, Kawakami N, Kamiya Y, et al. (2012) Tissue-specific transcriptome analysis reveals cell wall metabolism, flavonol biosynthesis, and defense responses are activated in the endosperm of germinating Arabidopsis thaliana seeds. Plant Cell Physiol 53: 16–27 [DOI] [PubMed] [Google Scholar]
- Farrant JM, Moore JP. (2011) Programming desiccation-tolerance: from plants to seeds to resurrection plants. Curr Opin Plant Biol 14: 340–345 [DOI] [PubMed] [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. (2006) Seed dormancy and the control of germination. New Phytol 171: 501–523 [DOI] [PubMed] [Google Scholar]
- Glaab E, Baudot A, Krasnogor N, Valencia A. (2010) TopoGSA: network topological gene set analysis. Bioinformatics 26: 1271–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ. (2008a) Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol 179: 33–54 [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Finch-Savage WE, Grappin P, Job D. (2008b) Post-genomics dissection of seed dormancy and germination. Trends Plant Sci 13: 7–13 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Kumpf RP, Shi CL, Larrieu A, Stø IM, Butenko MA, Péret B, Riiser ES, Bennett MJ, Aalen RB. (2013) Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc Natl Acad Sci USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le BH, Cheng C, Bui AQ, Wagmaister JA, Henry KF, Pelletier J, Kwong L, Belmonte M, Kirkbride R, Horvath S, et al. (2010) Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. Proc Natl Acad Sci USA 107: 8063–8070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Polisensky DH, Braam J. (2005) Genome-wide identification of touch- and darkness-regulated Arabidopsis genes: a focus on calmodulin-like and XTH genes. New Phytol 165: 429–444 [DOI] [PubMed] [Google Scholar]
- Lee KJD, Dekkers BJW, Steinbrecher T, Walsh CT, Bacic A, Bentsink L, Leubner-Metzger G, Knox JP. (2012) Distinct cell wall architectures in seed endosperms in representatives of the Brassicaceae and Solanaceae. Plant Physiol 160: 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turecková V, Strnad M, Lopez-Molina L. (2010) A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc Natl Acad Sci USA 107: 19108–19113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim PO, Kim HJ, Nam HG. (2007) Leaf senescence. Annu Rev Plant Biol 58: 115–136 [DOI] [PubMed] [Google Scholar]
- Martínez-Andújar C, Pluskota WE, Bassel GW, Asahina M, Pupel P, Nguyen TT, Takeda-Kamiya N, Toubiana D, Bai B, Górecki RJ, et al. (2012) Mechanisms of hormonal regulation of endosperm cap-specific gene expression in tomato seeds. Plant J 71: 575–586 [DOI] [PubMed] [Google Scholar]
- Müller K, Tintelnot S, Leubner-Metzger G. (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo-induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47: 864–877 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K, Okamoto M, Koshiba T, Kamiya Y, Nambara E. (2005) Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J 41: 697–709 [DOI] [PubMed] [Google Scholar]
- Narsai R, Law SR, Carrie C, Xu L, Whelan J. (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol 157: 1342–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki H. (2006) Seed germination: the biochemical and molecular mechanisms. Breed Sci 56: 93–105 [Google Scholar]
- Nonogaki H, Chen F, Bradford KJ (2007) Mechanisms and genes involved in germination sensu stricto. In Annual Plant Reviews, Vol 27: Seed Development, Dormancy and Germination. Blackwell Publishing, Oxford, pp 264–304 [Google Scholar]
- Ohto M-a, Stone SL, Harada JJ (2007) Genetic control of seed development and seed mass. In Annual Plant Reviews, Vol 27: Seed Development, Dormancy and Germination. Blackwell Publishing, Oxford, pp 1–24 [Google Scholar]
- Penfield S, Graham S, Graham IA. (2005) Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem Soc Trans 33: 380–383 [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18: 1887–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J, Tatematsu K, Kanno Y, Hobo T, Kimura M, Jikumaru Y, Yano R, Kamiya Y, Nambara E. (2009) Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: a comparative study on dormant and non-dormant accessions. Plant Cell Physiol 50: 1786–1800 [DOI] [PubMed] [Google Scholar]
- Quint M, Drost HG, Gabel A, Ullrich KK, Bönn M, Grosse I. (2012) A transcriptomic hourglass in plant embryogenesis. Nature 490: 98–101 [DOI] [PubMed] [Google Scholar]
- Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D. (2012) Seed germination and vigor. Annu Rev Plant Biol 63: 507–533 [DOI] [PubMed] [Google Scholar]
- Sliwinska E, Bassel GW, Bewley JD. (2009) Germination of Arabidopsis thaliana seeds is not completed as a result of elongation of the radicle but of the adjacent transition zone and lower hypocotyl. J Exp Bot 60: 3587–3594 [DOI] [PubMed] [Google Scholar]
- Stenvik GE, Tandstad NM, Guo Y, Shi CL, Kristiansen W, Holmgren A, Clark SE, Aalen RB, Butenko MA. (2008) The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitbrecht K, Müller K, Leubner-Metzger G. (2011) First off the mark: early seed germination. J Exp Bot 62: 3289–3309 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]