FLX4 physically interacts with FLX to form a transcription activation module in the FRIGIDA complex, which is required for the activation of FLC expression in winter-annual Arabidopsis.
Abstract
Many naturally occurring Arabidopsis (Arabidopsis thaliana) are very late flowering, unless flowering is promoted by a prolonged period of cold (e.g. winter) known as vernalization. In these winter-annual strains, flowering prior to winter is blocked by the synergistic interaction of FRIGIDA (FRI) and FLOWERING LOCUS C (FLC). FLC acts as a strong floral inhibitor, and FRI is required for high levels of FLC expression. Vernalization, in turn, leads to an epigenetic down-regulation of FLC expression. Most rapid-cycling Arabidopsis carry loss-of-function mutations in FRI, leading to low levels of FLC and rapid flowering in the absence of vernalization. Recent work has shown that FRI acts as a scaffolding protein for the assembly of a FRI complex (FRI-C) that includes both general transcription and chromatin-modifying factors, as well as FRI-specific components such as FRI-LIKE1, FRI ESSENTIAL1 (FES1), SUPPRESSOR OF FRI4 (SUF4), and FLC EXPRESSOR (FLX). Here, we show that FLX-LIKE4 (FLX4) is a novel component of the FRI-C and is essential for the activation of FLC by FRI. Both FLX and FLX4 contain leucine zipper domains that facilitate interaction with FRI. In addition, FLX and FLX4 interact with each other and show synergistic transcription activation activity. Interestingly, we show that FLX, FLX4, FES1, and SUF4 are required for basal levels of FLC expression in the absence of FRI. Thus, components of the FRI-C play a role in the regulation of FLC expression in both FRI-containing winter annuals, as well as fri-null rapid-cycling strains.
Arabidopsis (Arabidopsis thaliana) is an important model for the study of flowering-time regulation. Most naturally occurring Arabidopsis accessions can be divided into two categories based on the need for a prolonged period of cold exposure, known as vernalization, to promote early flowering. Rapid-cycling types, including most laboratory strains, flower rapidly without vernalization, whereas flowering is strongly delayed in the absence of vernalization in winter-annual accessions. Winter annuals are thus well suited to temperate climates, where flowering is inhibited until the vernalizing cold temperatures of winter have passed. Early studies revealed that two genes, FRIGIDA (FRI) and FLOWERING LOCUS C (FLC), confer the winter-annual habit in Arabidopsis (Napp-Zinn, 1987; Burn et al., 1993; Clarke and Dean, 1994; Koornneef et al., 1994; Lee et al., 1994). FRI is a coiled-coil domain-containing protein (Johanson et al., 2000), and FLC is a MADS-box transcription factor that acts as a potent floral repressor (Michaels and Amasino, 1999; Sheldon et al., 1999). In winter annuals, FRI activates FLC expression (Michaels and Amasino, 2001); in turn, FLC represses the expression of downstream floral promoters, such as SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1, FLOWERING LOCUS T (FT), and TWIN SISTER OF FT (Borner et al., 2000; Lee et al., 2000; Samach et al., 2000; Hepworth et al., 2002; Michaels et al., 2005). In contrast to winter annuals, most rapid-cycling accessions contain naturally occurring loss-of-function alleles of fri. Therefore, FLC expression is low, allowing the plants to flower rapidly. Mutant screens conducted in rapid-cycling strains have identified a group of genes, known collectively as the autonomous floral promotion pathway, that are required for the repression of FLC. Similar to FRI-containing winter annuals, recessive autonomous pathway mutants have elevated levels of FLC expression and are late flowering.
The late-flowering phenotype of naturally occurring winter annuals or autonomous pathway mutants is suppressed by vernalization, which results in a mitotically stable epigenetic repression of FLC expression (Michaels and Amasino, 1999; Sheldon et al., 1999). Vernalization results in the deposition of repressive histone modifications, such as histone 3 Lys 27 trimethylation (H3K27me3), at FLC chromatin (Bastow et al., 2004; Sung and Amasino, 2004; Finnegan and Dennis, 2007). Homologs of the Drosophila melanogaster Polycomb Repressive Complex2 (PRC2) and PRC1 have been implicated in the vernalization-triggered chromatin remodeling of the FLC locus. The deposition of H3K27me3 requires a PRC2-like complex containing the Suppressor of Zeste12 homolog VERNALIZATION2, the histone methyltransferases CURLY LEAF and SWINGER, the Extra Sex Combs homolog FERTILIZATION INDEPENDENT ENDOSPERM, and the p55 homolog MULTICOPY SUPPRESSOR OF IRA1 (Wood et al., 2006; De Lucia et al., 2008). In addition to the core PRC2 complex, three Plant Homeodomain finger proteins, VERNALIZATION INSENSITIVE3 (VIN3), VERNALIZATION5 (VRN5)/VIN3-Like1, and (VRN5/VIN3)-Like1/VIN3-LIKE2, as well as the Arabidopsis homolog of HETEROCHROMATIN PROTEIN1 (HP1), LIKE HP1, are also required for establishing and maintaining repressive FLC chromatin (Sung and Amasino, 2004; Mylne et al., 2006; Sung et al., 2006a, 2006b; Greb et al., 2007; Turck et al., 2007; De Lucia et al., 2008). Recently, it has been shown that long noncoding RNAs are also involved in vernalization response (Swiezewski et al., 2009; Heo and Sung, 2011). One long noncoding RNA, COLD ASSISTED INTRONIC NONCODING RNA, is transcribed from the first intron of FLC upon cold treatment and targets PRC2 activity to the FLC chromatin, which is vital for the maintenance of FLC repression by vernalization (Heo and Sung, 2011).
Genetic screens have identified a large number of genes required for the up-regulation of FLC by FRI. These can be divided into two groups based on the presence or absence of phenotypes in addition to flowering time. Mutations in the first-group genes lead to pleiotropic phenotypes, suggesting that their functions are not limited to FLC regulation. Many of these genes encode proteins that have, or are predicted to have, chromatin-associated functions (Kim and Sung, 2012). These include components of several complexes that incorporate activating histone modifications at target loci. A histone H2B monoubiquitination complex (Cao et al., 2008; Xu et al., 2008; Gu et al., 2009), an RNA polymerase II-associated factor 1 complex (Zhang and van Nocker, 2002; He et al., 2004; Oh et al., 2004; Park et al., 2010; Yu and Michaels, 2010), and a Complex Proteins Associated with Set1-like complex promote activating H3K4 and H3K36 methylation at FLC (Jiang et al., 2009, 2011). In addition to histone modification, the incorporation of histone variants, such as the substitution of H2A by H2A.Z, catalyzed by the SWI2/SNF2-RELATED1/SNF2-RELATED CBP ACTIVATOR PROTEIN complex, is also important for the regulation of FLC (Deal et al., 2007; March-Díaz et al., 2008; March-Díaz and Reyes, 2009).
The second group of genes implicated in the activation of FLC by FRI is often considered to act in a FRI-specific pathway, based on the fact that the phenotypes of these mutants are largely limited to flowering. These include the FRI homolog FRI-LIKE1 (FRL1), the zinc finger-containing proteins FRI ESSENTIAL1 (FES1) and SUPPRESSOR OF FRI4 (SUF4), and FLC EXPRESSOR (FLX), which contains a leucine zipper (Schmitz et al., 2005; Kim et al., 2006; Kim and Michaels, 2006; Andersson et al., 2008; Crevillén and Dean, 2011; Kim and Sung, 2012). It has been hypothesized that these proteins form a FRI transcription activator complex (FRI-C), with FRI acting as a scaffold to recruit the DNA-binding protein SUF4, the transactivating proteins FLX and FES1, and chromatin-modifying complexes to FLC chromatin (Choi et al., 2011).
Here, we report the identification of a new gene in the FRI-specific pathway. FLX-LIKE4 (FLX4) shows limited sequence similarity to FLX and, like FLX, is required for the up-regulation of FLC by FRI. We show that FLX4 physically interacts with FRI and FLX through distinct domains and that FLX and FLX4 show a synergistic enhancement of transcriptional activation. We also show that most genes of the FRI-C act to promote FLC expression, even in the absence of FRI. This suggests that genes such as FLX4, FLX, SUF4, and FES1 play a role in establishing basal levels of FLC expression in both winter-annual and rapid-cycling strains of Arabidopsis.
RESULTS AND DISCUSSION
FLX4 Is Required for FRI-Mediated Late Flowering
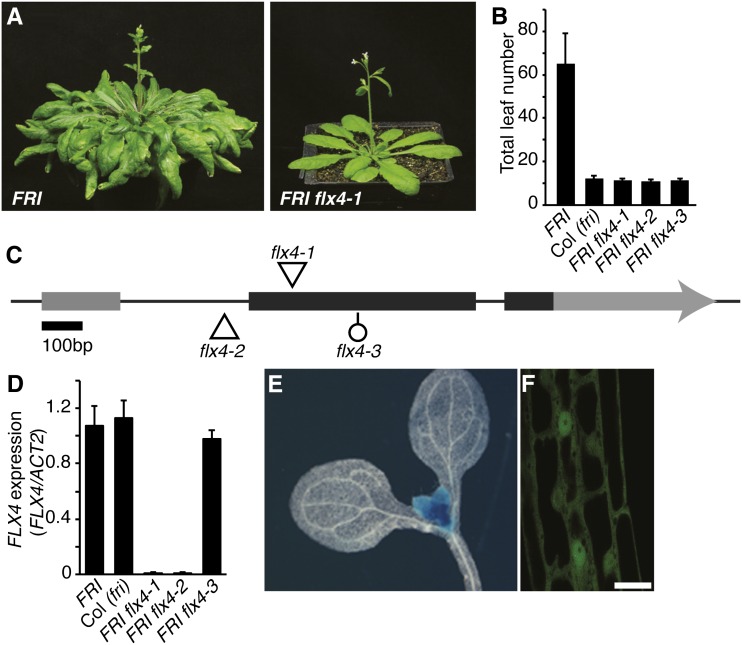
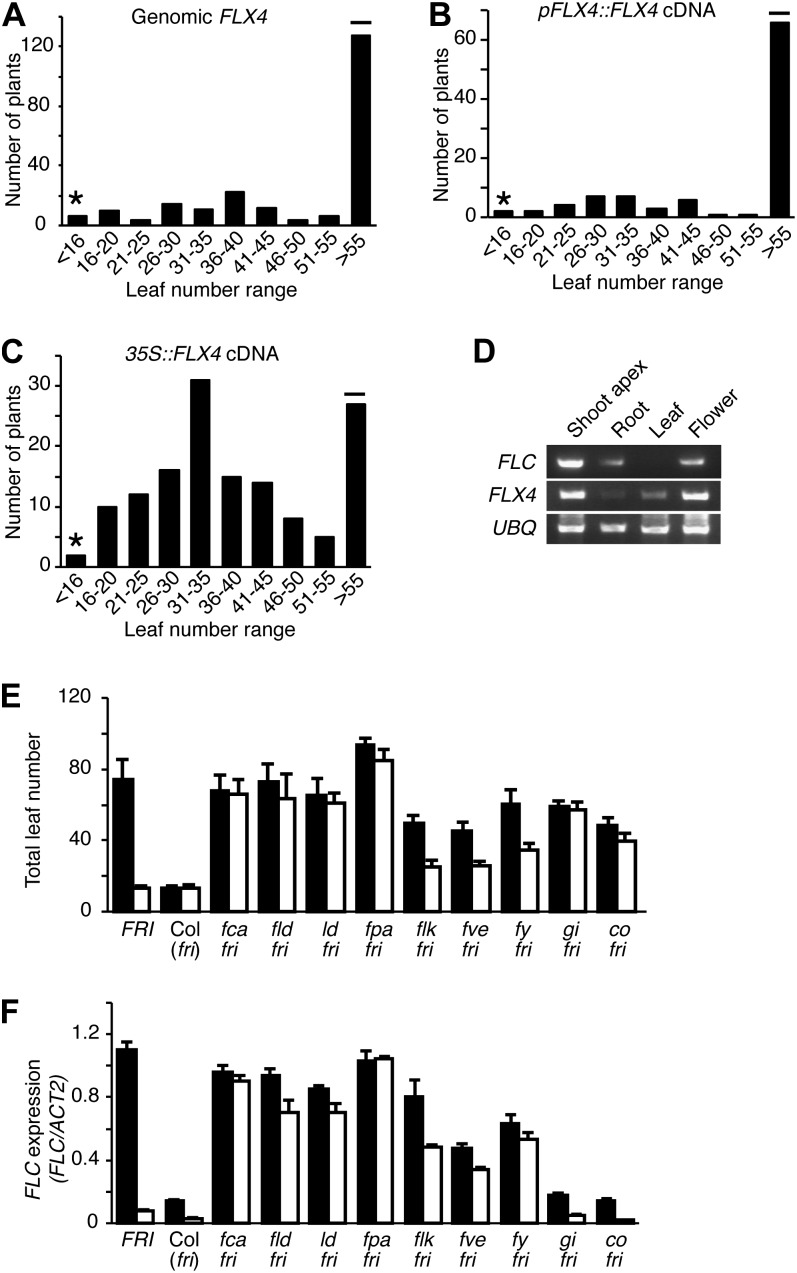
To identify additional genes required for FRI-mediated late flowering, we conducted a transfer DNA (T-DNA)-insertional mutant screen in a late-flowering background containing an active allele of FRI in the ecotype Columbia (Col-0) background (FRI-Col). We identified three allelic early-flowering mutants in the T2 (Fig. 1, A and B). Recovery of T-DNA flanking sequences revealed that two of the mutants (flx4-1 and flx4-2) have T-DNA insertions in At5g61920, FLX4 (Fig. 1C). Sequencing of FLX4 from flx4-3 showed that it contains a small deletion in the open reading frame (discussed below). FLX4 has previously been identified as an FLX-related gene (Choi et al., 2011), but its role in the regulation of flowering time has not yet been characterized. As expected from the T-DNA insertions, flx4-1 and flx4-2 showed a strong reduction in At5g61920 transcript levels, whereas flx4-3 levels were similar to the wild type (Fig. 1D). To ensure that the lesions in FLX4 are responsible for the early-flowering phenotype, flx4 mutants were transformed with either a genomic FLX4 clone, a clone containing the native promoter driving expression of the complementary DNA (cDNA), or the cDNA driven by the constitutive 35S promoter. All constructs were able to fully restore the late-flowering phenotype of FRI flx4 (Fig. 2, A–C). FLX4 belongs to a group of four proteins, including AT3G14750, AT1G55170, and AT1G67170, that share limited sequence similarity to FLX (Supplemental Fig. S1A; Choi et al., 2011). FLX4 contains a predicted coiled-coil domain as well as six conserved Leu residues in the amino terminal half of the protein that may function as a leucine zipper (Supplemental Fig. S1A). Similar to FLC, FLX4 is most highly expressed in the shoot apex and shows nuclear localization (Figs. 1, E and F, and 2D). Together, these data indicate that the detected mutations in FLX4 are responsible for the early-flowering phenotype of flx4 mutants.
Figure 1.
Mutations in flx4 suppress the late-flowering phenotype of FRI. A and B, The early-flowering phenotype of flx4 mutants. C, Drawing of the FLX4 locus. Thick lines represent exons; protein-coding regions are shown in black. The positions of mutations are indicated by triangles (T-DNA) and a circle (deletion). D, qRT-PCR analysis of FLX4 mRNA levels. E and F, Spatial expression (E, GUS) and subcellular localization (F, GFP) of FLX4. Error bars = 1 sd (B and D); scale bar = 25 μm.
Figure 2.
Molecular complementation and interaction of FLX4 with the autonomous floral promotion pathway. A to C, FLX4 constructs rescue the early-flowering phenotype of FRI flx4 mutants. Total leaf number of FRI flx4 T1 plants transformed with a genomic FLX4 construct (A), a construct containing the FLX4 cDNA driven by the FLX4 promoter (B), and 35S::FLX4 (C). The asterisks and horizontal bars indicate the flowering time of the FRI flx4 parent and FRI-Col, respectively. D, Reverse transcription-PCR analysis of FLX4 and FLC mRNA expression in various tissues. E, Flowering time of the indicated genotypes without (black bars) or with flx4 mutations (white bars). F, FLC expression as determined by qRT-PCR for the indicated genotypes without (black bars) or with flx4 mutations (white bars). Error bars = 1 sd (E and F).
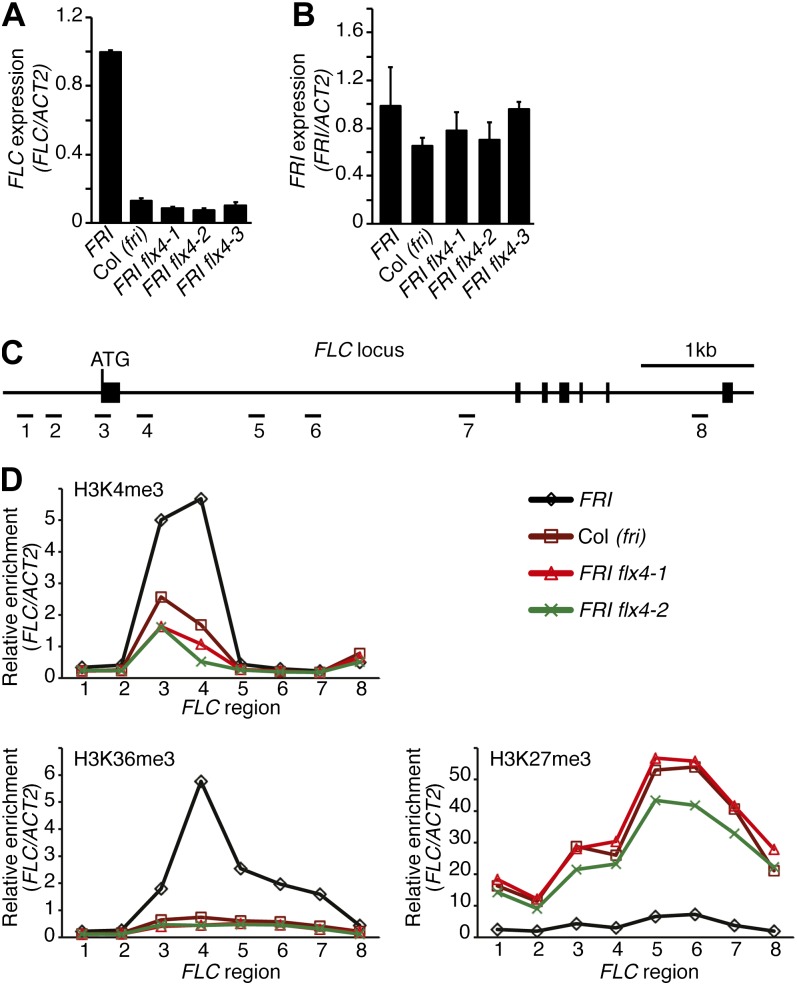
FRI acts to delay flowering through the up-regulation of the floral repressor FLC. To determine if FLX4 is required for the expression of FRI and/or FLC, we checked their expression levels by quantitative reverse transcription (qRT)-PCR (Fig. 3, A and B). FLC levels were strongly reduced in flx4 mutants, but FRI levels were not significantly different from the FRI-Col parent. To further investigate the effect of FLX4 on gene expression, we performed a microarray experiment comparing the wild type (FRI FLX4) to an flx4 mutant (FRI flx4-1). We found that FLX4 is required for the proper expression of a relatively small number of genes. Forty-six genes showed a greater than 2-fold change in expression with a P value < 0.05 (Supplemental Table S1). Of these, the largest change in expression was observed for FLC (approximately 70-fold reduction). We also used microarrays to examine the expression of FLX4-regulated genes in response to mutations in fri. If FLX4 acts in conjunction with FRI to regulate FLC, one would predict that there would be significant overlap between FLX4-regulated genes and those regulated by FRI. This is the case. Of the 46 genes regulated by FLX4, 71% (33 genes) also showed a statistically significant (P < 0.05) change in expression in fri mutants (Supplemental Table S1), indicating that FRI and FLX4 have closely related functions in gene regulation.
Figure 3.
FLX4 promotes FLC expression and activating histone modifications at the FLC locus. A and B, qRT-PCR analysis of FLC (A) and FRI (B) mRNA levels. Error bars = 1 sd. C, Drawing of the FLC locus. Thick lines represent exons, and numbered lines indicate fragments amplified in ChIP-quantitative PCR analysis. D, Analysis of histone modifications by ChIP-qPCR. sds are shown in Supplemental Figure S2.
The above results show that both FRI and FLX4 are required for the up-regulation of FLC; loss-of-function mutations in either gene result in a similar reduction in FLC expression and an early-flowering phenotype. In the presence of FRI, FLC chromatin is enriched in H3K4me3 and H3K36me3 and is highly expressed. In Col-0 (fri), by contrast, H3K4me3 and H3K36me3 are strongly reduced, and FLC expression is repressed by PRC2-mediated H3K27me3 (Fig. 3, C and D; Supplemental Fig. S2; Kim and Sung, 2012). In FRI flx4 plants, we also observe a reduction in H3K4me3/H3K36me3 and a strong increase in H3K27me3 (Fig. 3D; Supplemental Fig. S2). Although these experiments do not implicate FLX4 directly in chromatin remodeling, the observation that loss-of-function mutations in fri or flx4 result in similar changes in chromatin structure is consistent with the model that these two genes act in the same complex (i.e. FRI-C).
Recessive loss-of-function mutations in genes of the autonomous floral promotion pathway lead to high levels of FLC expression and delayed flowering in rapid-cycling backgrounds that lack active alleles of FRI. To determine if FLX4 is also required for the up-regulation of FLC in autonomous pathway mutants, we created double mutants between flx4 and various autonomous pathway mutants. The photoperiod pathway mutants gigantea and constans mutants were included as controls, as their late-flowering phenotypes are not dependent on FLC. Autonomous pathway mutants can be grouped into two categories based on flowering time. fca, flowering locus d (fld), fpa, and luminidependens (ld) show a stronger late-flowering phenotype than flowering locus k (flk), fve, and fy (Fig. 2, E and F; Veley and Michaels, 2008). Interestingly, the flx4-1 mutation had little effect on flowering time for fca, fld, fpa, or ld but partially suppressed the late-flowering phenotype of flk, fve, and fy. Thus, it appears that FLX4 plays a limited role in the activation of FLC expression in autonomous pathway mutant backgrounds. flx4 is similar to other FRI-suppressor mutations, such as frl1, fes1, and suf4, in that mutations strongly suppress the late-flowering phenotype of FRI but have weaker effects on autonomous pathway mutants. The level of suppression observed in autonomous pathway mutants varies. For example, frl1 and fes1 mutations show little or no effect on the autonomous pathway mutants tested (Michaels et al., 2004; Schmitz et al., 2005), whereas suf4 mutants show a stronger suppression of the late-flowering phenotype (Kim et al., 2006; Kim and Michaels, 2006).
FLX4 Physically Interacts with Both FRI and FLX
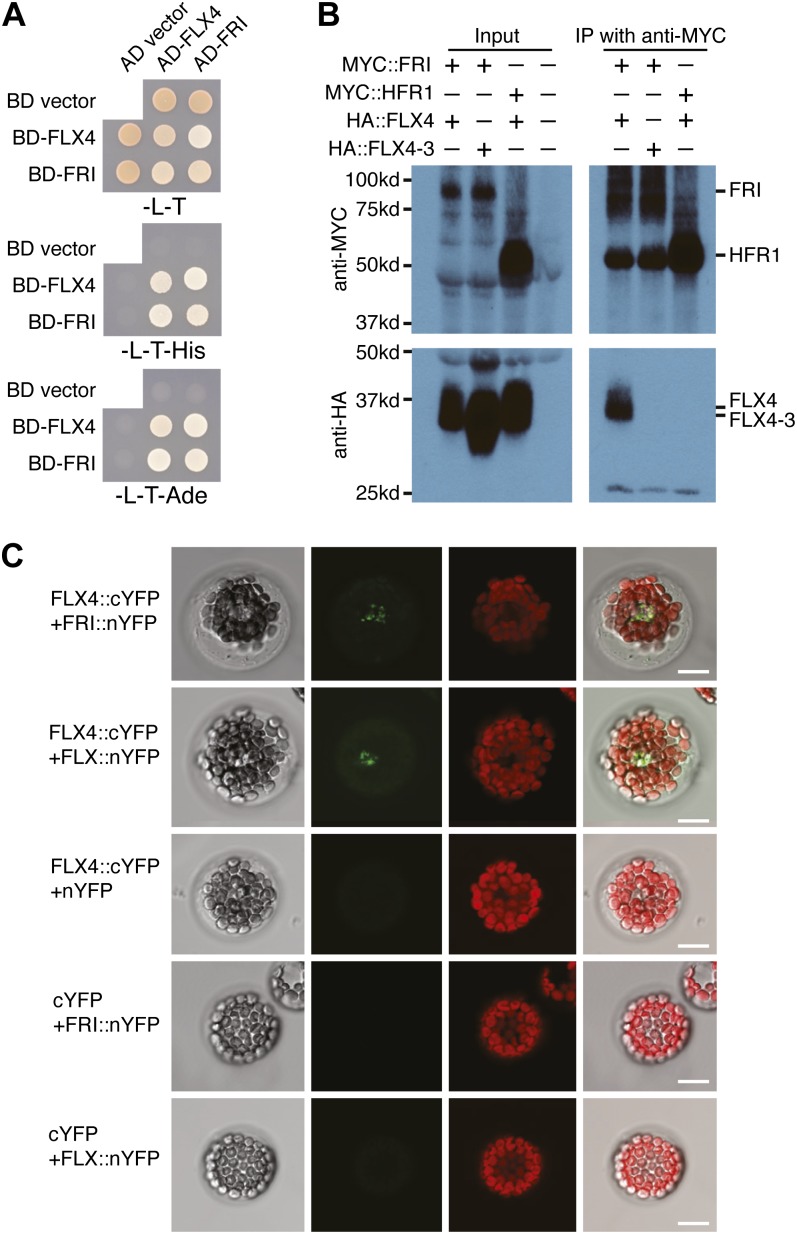
It has been proposed that FRI may act as a molecular scaffold to recruit FLC activators, including FRL1, FES1, SUF4, and FLX, to the FLC locus (Choi et al., 2011). In addition to FLX, it has been shown that the FLX-like genes AT3G14750, AT1G55170, AT1G67170, and FLX4 can interact with FRI in yeast (Saccharomyces cerevisiae) two-hybrid (Y2H) assays (Choi et al., 2011). Consistent with this result, we also found that FRI and FLX4 interact in Y2H assays (Fig. 4A). We also determined whether this interaction occurs in plants. First, we transiently coexpressed epitope-tagged FLX4 (HEMAGGLUTININ [HA]::FLX4) with either FRI (MYC::FRI) or LONG HYPOCOTYL IN FAR-RED (HFR1; MYC::HFR1, negative control) in tobacco (Nicotiana tabacum) leaves. HA::FLX4 coimmunoprecipitated using an anti-MYC antibody when coexpressed with MYC::FRI but not when coexpressed with the MYC::HFR1 negative control (Fig. 4B). Second, we used bimolecular fluorescence complementation to determine if FRI and FLX4 interact in Arabidopsis protoplasts. When FLX4 and FRI were fused to the C-terminal YELLOW FLUORESCENT PROTEIN (cYFP) and N-terminal (nYFP) portions of YFP, fluorescence was observed, indicating an interaction (Fig. 4C). Fluorescence was not observed when FLX4::cYFP or FRI::nYFP were cotransformed with the corresponding control plasmid. Thus, FRI and FLX4 can interact in plant cells.
Figure 4.
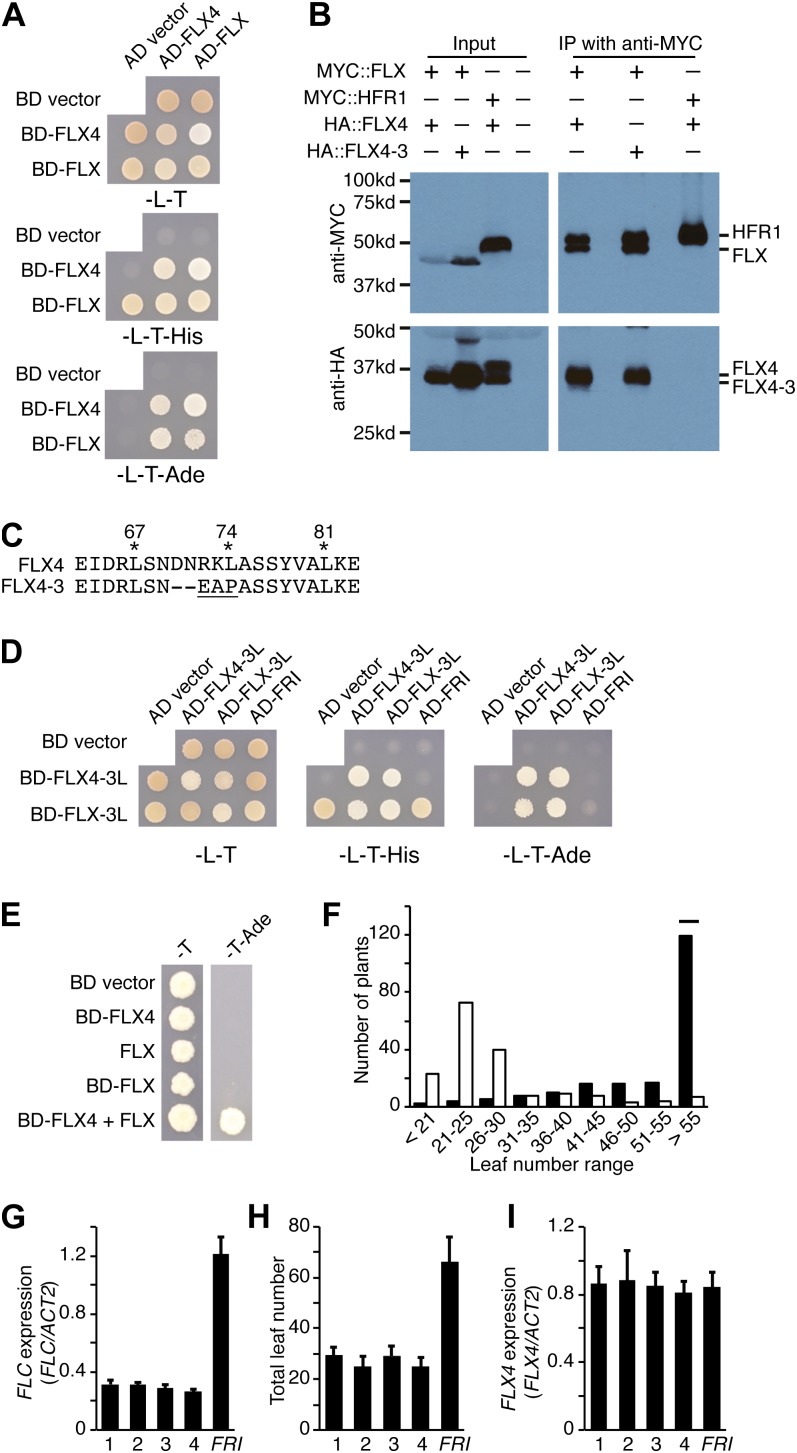
FLX4 interacts with FRI in yeast and in plants. A, FLX4 interacts with both FRI and itself in Y2H assays. The bait and prey plasmids confer growth in the absence of Leu (–L) and Trp (–T). An interaction is indicated by growth in the absence of His (–His) and/or adenine (–Ade). B, FLX4 protein coimmunoprecipitates with FRI when transiently expressed in tobacco. Binding is eliminated by the flx4-3 mutation. C, FLX4 interacts with FRI and FLX in bimolecular fluorescence complementation assays in Arabidopsis protoplasts. Bars = 10 μm.
In our Y2H assays, we also noticed FLX4 can interact with itself (Fig. 4A). This led us to determine if FLX4 can interact with other FLX-related proteins. In yeast, we found that FLX4 can interact with FLX but not with AT3G14750, AT1G55170, or AT1G67170 (Fig. 5A; Supplemental Fig. S3). Consistent with these interactions, both FLX and FLX4 are required for FRI activity (Andersson et al., 2008; Fig. 1), whereas mutations in AT3G14750, AT1G55170, or AT1G67170 do not affect flowering time (Choi et al., 2011). Similar to the experiments described above for the FLX4-FRI interaction, we found that FLX4 and FLX were able to physically interact in both coimmunoprecipitation assays using transient expression in tobacco leaves and in bimolecular fluorescence complementation assays in Arabidopsis protoplasts (Figs. 4C and 5B). Thus, FLX4 can physically interact with both FRI and FLX4 in plant cells. We did not detect interactions, however, between FLX4 and other proteins required for FRI activity, such as FRL1, FES1, or SUF4 (Supplemental Fig. S4).
Figure 5.
FLX4 and FLX physically interact and synergistically promote transcriptional activation. A, FLX4 interacts with FLX in Y2H assays. B, FLX4 protein coimmunoprecipitates with FLX when transiently expressed in tobacco. Binding is unaffected by the flx4-3 mutation. C, FLX4 protein sequence in the region affected by the flx4-3 mutation. Conserved Leu residues in the putative leucine zipper are marked by asterisks. Substituted amino acids are underlined. D, Y2H analysis of mutations in three conserved Leu residues in FLX4 (FLX4-3L) and FLX (FLX-3L). FLX4-3L and FLX-3L still interact with each other, but the interaction of FLX4-3 with FRI is eliminated and the interaction between FLX-3L and FRI is weakened (lack of growth on –l-T-Ade). E, FLX4 and FLX synergistically promote transcription activation. F, Addition of a repressor domain converts FLX4 from an FLC promoter to an inhibitor. Total leaf number of T1 plants transformed with 35S::FLX4 (black bars) or 35S::FLX4::RD (white bars). The flowering time of the untransformed FRI-Col parent is indicated by a horizontal bar. G and H, Reduced FLC expression as determined by qRT-PCR (G) and early-flowering phenotype (H) of T2 plants transformed with 35S::FLX4::RD. I, Endogenous FLX4 levels are unaffected by expression of the 35S::FLX4::RD transgene. Error bars = 1 sd. [See online article for color version of this figure.]
A Putative FLX4 Leucine Zipper Is Required for Interaction with FRI But Is Dispensable for Interaction with FLX
FLX4 and its homologs contain conserved Leu residues that may constitute a leucine zipper (Fig. 5C; Supplemental Fig. S1). These domains often function in mediating protein-protein interactions and typically feature Leu residues separated by six amino acids (e.g. L-X6-L-X6-L, where X = any amino acid). The flx4-3 lesion affects a 12-bp region, where 6 bp have been deleted and others have been substituted, resulting in the deletion of two amino acids and the substitution of three others, including a Leu that is absolutely conserved among FLX-related proteins (Supplemental Fig. S1). FLX4 mRNA levels in the flx4-3 mutant are similar to the wild type (Fig. 1D), suggesting that the defect in this allele may be due to compromised protein function rather than transcription. To determine if the flx4-3 lesion might disrupt the ability of FLX4 to interact with other proteins that are essential for activation of FLC, we tested the ability of the FLX4-3 protein to interact with FRI and FLX in plants. Interestingly, the flx4-3 lesion eliminated the interaction with FRI but had no effect on the interaction with FLX (Figs. 4B and 5B).
To further investigate the role of the putative leucine zipper in the function of FLX4, we used site-directed mutagenesis to change three conserved Leu residues to Ser in FLX4 (L67S, L74S, and L81S, referred to as FLX4-3L; Fig. 5C). Consistent with the result with FLX4-3, FLX4-3L also failed to interact with FRI in yeast (Fig. 5D). Interestingly, the FLX4-3L mutant still interacted with itself. Thus, the self-interaction of FLX4 can occur even when these conserved Leu residues are absent in both the bait and prey constructs. This led us to investigate whether the putative leucine zipper is also important for the interactions of FLX. We used site-directed mutagenesis to mutate the corresponding Leu residues to Ser in FLX (L53S, L60S, and L67S, referred to as FLX-3L; Supplemental Fig. S1). Similar to FLX4-3L, FLX-3L is still able to self-interact (Fig. 5D). In addition, the interaction between FLX and FLX4 was unaffected by the Leu mutations. The interaction between FLX and FRI, however, was weakened in FLX-3L (Fig. 5D). Thus, the putative leucine zippers of FLX and FLX4 are dispensable for self-interactions or interactions with each other. By contrast, these conserved Leu residues are essential for the interaction between FLX4 and FRI and facultative for the interaction between FLX and FRI. In these experiments, we focused on three Leu residues that are absolutely conserved among FLX family members and have canonical L-X6-L-X6-L spacing. It should be noted, however, that several additional Leu residues are conserved between FLX4 and FLX and may contribute to the weak interaction between FLX-3L and FRI (Supplemental Fig. S1).
The FLX4/FLX Module May Provide Transcription Activation Activity to the FRI Complex
Previous work has shown that FLX can activate transcription in yeast (Choi et al., 2011), suggesting that FLX may provide transcription activation activity to the FRI-C. To further investigate the transcription activation potential of FLX4 and FLX, we expressed each protein fused to the DNA binding domain (BD) of GALACTOSE4 (GAL4) in yeast. In the case of FLX4, we saw no activation of the His or Ade reporter genes, as evidenced by the lack of growth on –His and –Ade media (BD-FLX4 + activation domain [AD] vector; Fig. 4A). For FLX, we observed growth on –His but not –Ade media, indicating that BD-FLX was able to activate expression of the His reporter gene but not the Ade reporter (BD-FLX + AD vector; Fig. 5A). This is consistent with our general experience that greater transcriptional activation activity is required to activate the Ade reporter than the His reporter. Thus, when tested individually, FLX shows moderate transcriptional activation, whereas no activation was detected with FLX4.
Given the moderate transcription activation activity of FLX alone, we wondered how the physical interaction with FLX4 might affect the activity of FLX. The transcription activation activity of FLX or FLX4 is insufficient to activate expression of the Ade reporter gene (Fig. 5E). To test if FLX and FLX4 might have a synergistic effect on transcription activation, we coexpressed BD-FLX4 with an untagged FLX. This line, in which neither protein is fused to the GAL4 activation domain, showed robust growth on –Ade media (Fig. 5E). Thus, FLX and FLX4 do show a synergistic effect on transcription activation. This suggests that FLX and FLX4 may form a module that provides transcription activation activity to the FRI complex.
The observation that FLX alone can activate transcription in yeast suggests that the FRI-C might retain partial transcription activation activity in the absence of FLX4. Our data, however, shows that loss of FLX4 results in a complete loss of FRI activity (Figs. 1, A and B, and 3, A and B). Thus, the effect of flx4 mutations on flowering time cannot be explained solely in terms of FLX4’s synergistic effect with FLX on transcription activation. It is possible that FLX4 may have addition roles that are required for the structural stability or biochemical activity of the FRI-C.
To further test the model that the FLX and FLX4 play a role in the transcriptional activation of FLC by the FRI complex, we attempted to convert FLX4 into a transcriptional repressor using Chimeric Repressor Gene Silencing Technology. In this approach, a transcriptional activator is fused to a 12-amino acid (LDLDLELRLGFA) ERF-associated amphiphilic repression (EAR) motif repression domain (SRDX). Chimeric Repressor Gene Silencing Technology has successfully been applied in a number of cases to create transcription factor-SRDX fusions that act to repress the expression of genes that are normally activated by the native transcription factor (Hiratsu et al., 2003; Fujita et al., 2005; Takase et al., 2007; Koo et al., 2010). Constructs containing FLX4 alone or fused to the repressor domain (FLX4::RD) were transformed into FRI-Col plants. In this background, the FRI complex activates FLC expression, leading to a late-flowering phenotype. The majority of T1 transformants containing FLX4 alone showed a late-flowering phenotype similar to the FRI-Col parent (Fig. 5F). By contrast, most FLX4::RD transformants flowered much earlier than FRI. We examined FLC expression in four FLX4::RD lines and found that rapid flowering was correlated with reduced FLC expression (Fig. 5, G and H). Thus, the addition of the SRDX repressor domain to FLX4 is sufficient to repress FLC expression. To ensure that the rapid-flowering phenotype of FLX4::RD plants was not due to suppression of endogenous FLX4 expression, we tested expression of the native copy of FLX4 and found it to be similar in FRI-Col and early-flowering T2 plants (Fig. 5I).
FRI Complex Components Promote FLC Expression in the Absence of FRI
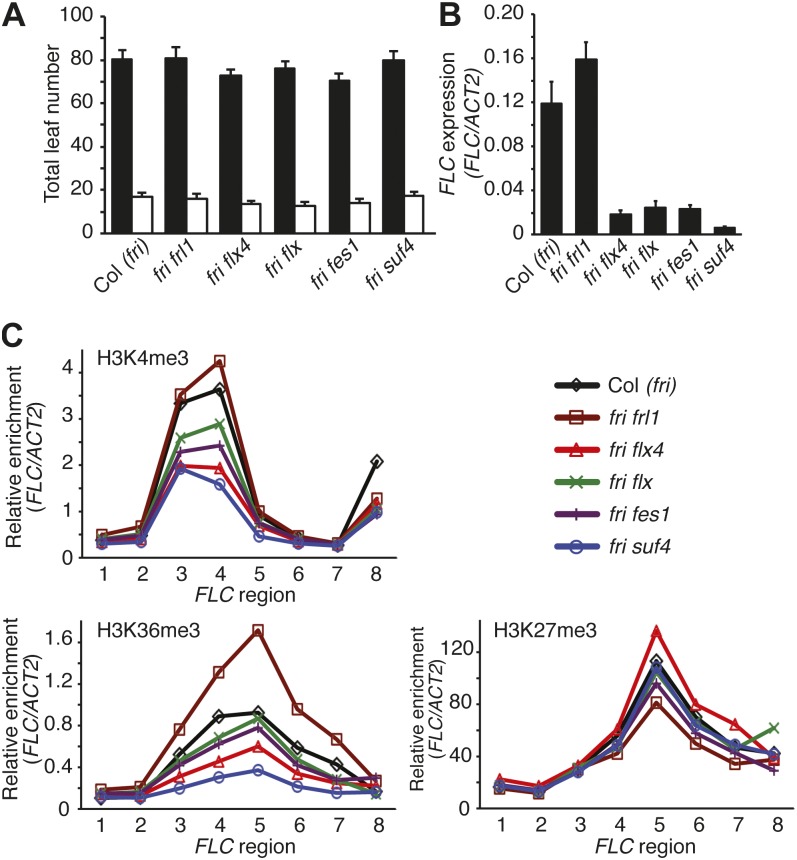
Loss-of-function mutations in FRL1, FES1, SUF4, FLX, and now FLX4 strongly suppress FLC expression in FRI-containing backgrounds but lack additional pleiotropic phenotypes. For this reason, these genes are though to act in a FRI-specific pathway. To determine if these genes play any role in the regulation of flowering time and/or FLC expression in the absence of FRI, we examined frl1, fes1, suf4, flx, and flx4 in Col-0, which contains a naturally occurring null allele of fri. With regard to flowering time, there was no significant difference between the wild type and any of the mutants in either long or short days (Fig. 6A). Interestingly, however, FLC expression was significantly reduced in fes1, suf4, flx, and flx4 (P < 0.01; Fig. 6B). For frl1, we observed a modest, but reproducible (P < 0.05), increase in FLC expression (Fig. 6B). Thus, it appears that most genes in the FRI-specific pathway (FES1, SUF4, FLX, and FLX4) play a role in the activation of FLC, even in the absence of FRI, whereas FRL1 may play a minor role in FLC repression.
Figure 6.
FRI-independent promotion of FLC expression by FLX4, FLX, FES1, and SUF4. A, Flowering time of FRI pathway mutants in Col-0 (fri) under short (black bars) or long days (white bars). B, FLC expression as determined by qRT-PCR. Error bars = 1 sd (A and B). C, Analysis of histone modifications by ChIP-qPCR. sds are shown in Supplemental Figure S5.
In late-flowering winter-annual Arabidopsis, FLC expression is up-regulated by the FRI complex, which causes increased H3K4me3/H3K36me3 and decreased H3K27me3 at the FLC locus. In the absence of FRI (e.g. wild-type Col-0), H3K4me3/H3K36me3 is reduced and H3K27me3 is increased, leading to lower levels of FLC expression (Fig. 3C; Kim and Sung, 2012). The further reduction in FLC expression observed in fes1, suf4, flx, and flx4 mutants in the Col-0 background suggests that these genes may act to promote H3K4me3/H3K36me3, repress H3K27me3, or both, even in the absence of FRI. In general, we found that fes1, suf4, flx, and flx4 had a larger effect on H3K4me3 and H3K36me3 than on H3K27me3 (Fig. 6C; Supplemental Fig. S5). suf4, for example, showed the lowest levels of FLC expression (Fig. 6B) and showed a clear decrease in H3K4me3 and H3K36me3 at FLC, particularly in the first exon and the beginning of the first intron. H3K27me3, by contrast, showed no significant difference between suf4 and Col-0 (Fig. 6C). These results suggest that the reduction in FLC expression in fes1, suf4, flx, and flx4 mutants in the Col-0 background is primarily due to a reduction in activating histone modifications rather than an increase in repressive H3K27me3. Interestingly, frl1 mutants showed a strong increase in H3K36me3 that may account for the slight increase in FLC expression (Fig. 6C).
CONCLUSION
Some two decades after the genetic mapping of the FRI locus in Arabidopsis, we are developing a detailed molecular understanding of the role of this important determinant of the flowering habit. The FRI protein acts as a scaffold for the assembly of DNA-binding proteins, chromatin remodelers, and transcriptional activators that are essential for the activation of FLC expression in winter-annual Arabidopsis. Our work has shown that FLX4, an FLX homolog, is a novel component of the FRI-C. In addition to physically interacting with FRI, FLX4 also interacts with FLX. Both FLX4 and FLX share a conserved leucine zipper domain, which facilitates their interaction with FRI, but is dispensable for their interaction with each other. Previous work has shown that FLX may provide transcription activation to the FRI complex. Interestingly, we find that, together, FLX and FLX4 have greater transcription activation potential than either protein alone, suggesting that FLX4 and FLX may form an activation module in the FRI-C. Although both FLX4 and FLX contribute to transcription activation, they likely have distinct activities. Both flx4 and flx mutants show an early-flowering phenotype similar to fri mutants; thus, neither protein alone is insufficient to produce a fully functional FRI-C. It is possible that, in planta, neither FLX4 nor FLX have sufficient activation potential alone to up-regulate FLC expression. Alternatively, the physical interaction between FLX4 and FLX may be important for their association with FRI and/or assembly of the FRI-C.
Another interesting finding is that many components of the FRI-C promote FLC expression in the absence of FRI. The FRI-C likely evolved in winter-annual Arabidopsis to up-regulate FLC and inhibit flowering prior to winter. Rapid-cycling strains, such as Col-0, contain naturally occurring null alleles of fri, which strongly reduces FLC expression and allows for early flowering. One unanswered question regarding rapid-cycling Arabidopsis, however, is whether the loss of FRI completely eliminates function of the FRI-C or whether the remaining components retain any residual activity in the absence of FRI. Interestingly, flx4, flx, fes1, and suf4 mutants in Col-0 all showed significant decreases in FLC expression, suggesting that the FRI-C does retain some ability to up-regulate FLC without FRI. Thus, these remaining FRI-C components may play a role in setting basal levels of FLC expression in rapid-cycling Arabidopsis. The decreased FLC expression in flx4, flx, fes1, and suf4 mutants is associated with lower levels of activating H3K4me3 and H3K36me3, whereas repressive H3K27me3 was relatively unchanged. This suggests that FLX4, FLX, FES1, and SUF4 promote basal levels of FLC expression by promoting activating histone modifications rather than by reducing repressive ones. The mechanism by which these proteins promote H3K4me3 and H3K36me3 at FLC, however, is unclear. Given that FLX4, FLX, FES1, and SUF4 are not predicted to possess histone-modifying activities, the mechanism is likely to be indirect, possibly by recruiting histone modifiers.
The evolution of rapid-cycling Arabidopsis from winter annuals suggests that the FRI-independent roles of FRI-C components may be important. Despite the fact that loss-of-function mutations in FRI, FLX4, FLX, FES1, FRL1, or SUF4 produce similar rapid-flowering phenotypes, the analysis of a large number of naturally occurring accessions has shown that the vast majority of rapid-cycling accessions evolved from winter annuals through loss-of-function mutations in FRI (Johanson et al., 2000). This suggests that there may be selective pressure to retain the activity of the other components of the FRI-C, possibly to maintain low levels of basal FLC expression. It is interesting to note that although flx4, flx, fes1, and suf4 mutants have lower levels of FLC expression in Col-0, flowering time is not dramatically affected under our laboratory conditions. It is possible that the fine-tuning of FLC expression in the absence of FRI may be important for reasons other than, or in addition to, flowering-time regulation. FLC has also been implicated in circadian rhythms and germination (Salathia et al., 2006; Chiang et al., 2009); thus, it is possible that basal levels of FLC expression, maintained by FLX4, FLX, FES1, FRL1, and SUF4, are important for the proper regulation of these processes.
MATERIALS AND METHODS
Plant Material and Growth Conditions
FRI-Col (Lee et al., 1994), frl1-1 (Michaels et al., 2004), fes1-3 (Schmitz et al., 2005), flx-2 (Choi et al., 2011), suf4-2 (Kim and Michaels, 2006), and autonomous pathway mutants (Veley and Michaels, 2008) have been described previously. Plants were grown in controlled growth rooms under cool-white fluorescent light (approximately 120 µmol m–2 sec–1) either under long days (16-h light/8-h dark) or short days (8-h light/16-h dark). T-DNA mutagenized populations have been described (Michaels and Amasino, 1999).
Gene Expression and Chromatin Immunoprecipitation Analysis
Total RNA was isolated using the Spectrum Plant Total RNA Kit (Sigma). cDNA was synthesized from 5 µg total RNA using an oligo(dT) primer as described previously (Michaels et al., 2004). qRT-PCR was performed using the Platinum SYBR Green qPCR SuperMix-UDG Kit (Invitrogen) as described by the manufacturer. Primers for FLC and ACTIN2 (ACT2) have been reported previously (Yu and Michaels, 2010). Microarray experiments, including growth conditions, were conducted as previously described (Veley and Michaels, 2008). Briefly, plants were grown until the 10-leaf stage, at which point the aboveground portions were harvested and RNA was extracted. Because of differences in flowering time, plants were grown under short days to ensure that all plants remained in the vegetative stage of development at the time of harvest. For each genotype, RNA from four biological replicates was used to prepare labeled complementary RNA, which was hybridized to Affymetrix GeneChip Arabidopsis (Arabidopsis thaliana) ATH1 genome arrays. Chromatin immunoprecipitation experiments were performed as reported previously (Yu and Michaels, 2010). Antibodies used are anti-H3K4me3 (Millipore, 17-614), H3K36me3 (Abcam, ab9050), and anti-H3K27me3 (Millipore, 07-449).
Bimolecular Fluorescence Complementation
Proteins of interest were fused to the C-terminal or N-terminal portions of YFP as described previously (Walter et al., 2004). Pairs of constructs, or a construct and corresponding control plasmid, were cotransformed into Arabidopsis mesophyll protoplasts as described (Yoo et al., 2007). Transfected protoplasts were incubated overnight at room temperature and imaged using a Leica SP5 confocal microscope.
Y2H and Transcription Activation Assays
For most Y2H assays, the bait constructs were fused to the Gal4 DNA-binding domain in pBridge (Figs. 4A and 5A; Supplemental Fig. S4) or pGBKT7 (Fig. 5D), and the Gal4 activation domain was fused to prey construct using pGADT7 (Figs. 4A and 5, A and D; Supplemental Fig. S4; Clontech). Experiments testing interactions between FLX family proteins (Supplemental Fig. S3) were performed using pDEST22 and pDEST32 (Invitrogen). For activation assays, FLX4 and FLX were expressed from pBridge with or without fusion to the DNA-binding domain, respectively.
Co-immunoprecipitation
FLX4 and FLX4-3 were fused with a 3×HA tag, and FRI and FLX were fused with a 5×Myc tag and cloned into pTA7002 (Aoyama and Chua, 1997). Constructs were transformed into Agrobacterium tumefaciens strain GV3101, and paired constructs were cotransformed into tobacco (Nicotiana tabacum) leaves. Forty-eight hours after infiltration, protein expression was induced by spraying with 50 µm dexamethasone and proteins were extracted 8 h after induction. Protein extraction, immunoprecipitation, and immunoblotting were performed as described (Shao et al., 2003; DeYoung et al., 2012)
Constructs
For complementation, an FLX4 genomic fragment containing 1,921 bp upstream of the start codon and 679 bp downstream of the stop codon was cloned into the binary vector pPZP211 (Hajdukiewicz et al., 1994). For cDNA expression using the native promoter, the 1,921-bp upstream fragment was used to drive expression of the FLX4 cDNA. The 35S::FLX4 construct was made by cloning the genomic coding region of FLX4 into pPZPY112 (Yamamoto et al., 1998). 35S::FLX4::RD was created by ligating the FLX4 cDNA into pPZPY112RD, a vector created by inserting the RD motif into pPZPY112.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers FLX, At2g30120; FLX4, At5g61920; FCA, At4g16280; FLD, At3g10390; FLK, At3g04610; FPA, At2g43410; FVE, At2g19520; FY, At5g13480; LUMINIDEPENDENS, At4g02560; FRI, At4g00650; GIGANTEA, AT1G22770; CONSTANS, At5g15840; FLC, At5g10140; UBIQUITIN, At4g05320; ACT2, At3g18780; HFR1, At1g02340; FRL1, At5g16320; FES1, At2g33835; and SUF4, At1g30970.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. FLX4 and related proteins contain putative leucine zipper domains.
Supplemental Figure S2. FLX4 promotes FLC expression and activating histone modifications at the FLC locus.
Supplemental Figure S3. FLX4 interacts with FLX but not other FLX-related proteins in Y2H analysis.
Supplemental Figure S4. FLX4 does not interact with FRL1, FES1, or SUF4 in Y2H analysis.
Supplemental Figure S5. FLX4, FLX, FES1, and SUF4 promote activating histone modifications at the FLC locus.
Supplemental Table S1. Microarray analysis of flx4 mutants.
Acknowledgments
We thank Stephanie Deyoung, Yangna Gu, Laurel Bender, Michael Galley, Ying Zhao, and Sibo Tao for their technical assistance and Xuhong Yu and Wei Feng for many useful discussions.
Glossary
- T-DNA
transfer DNA
- Col-0
ecotype Columbia
- cDNA
complementary DNA
- qRT
quantitative reverse transcription
- Y2H
yeast two-hybrid
- ChIP
Chromatin immunoprecipitation
References
- Andersson CR, Helliwell CA, Bagnall DJ, Hughes TP, Finnegan EJ, Peacock WJ, Dennis ES. (2008) The FLX gene of Arabidopsis is required for FRI-dependent activation of FLC expression. Plant Cell Physiol 49: 191–200 [DOI] [PubMed] [Google Scholar]
- Aoyama T, Chua NH. (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C. (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Borner R, Kampmann G, Chandler J, Gleissner R, Wisman E, Apel K, Melzer S. (2000) A MADS domain gene involved in the transition to flowering in Arabidopsis. Plant J 24: 591–599 [DOI] [PubMed] [Google Scholar]
- Burn JE, Smyth DR, Peacock WJ, Dennis ES. (1993) Genes conferring late flowering in Arabidopsis thaliana. Genetica 90: 147–155 [Google Scholar]
- Cao Y, Dai Y, Cui S, Ma L. (2008) Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20: 2586–2602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang GC, Barua D, Kramer EM, Amasino RM, Donohue K (2009) Major flowering time gene, FLOWERING LOCUS C, regulates seed germination in Arabidopsis thaliana Proc Natl Acad Sci USA 106: 11661–11666 [DOI] [PMC free article] [PubMed]
- Choi K, Kim J, Hwang HJ, Kim S, Park C, Kim SY, Lee I. (2011) The FRIGIDA complex activates transcription of FLC, a strong flowering repressor in Arabidopsis, by recruiting chromatin modification factors. Plant Cell 23: 289–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke JH, Dean C. (1994) Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol Gen Genet 242: 81–89 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Dean C. (2011) Regulation of the floral repressor gene FLC: the complexity of transcription in a chromatin context. Curr Opin Plant Biol 14: 38–44 [DOI] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C. (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105: 16831–16836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB. (2007) Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung BJ, Qi D, Kim SH, Burke TP, Innes RW. (2012) Activation of a plant nucleotide binding-leucine rich repeat disease resistance protein by a modified self protein. Cell Microbiol 14: 1071–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan EJ, Dennis ES. (2007) Vernalization-induced trimethylation of histone H3 lysine 27 at FLC is not maintained in mitotically quiescent cells. Curr Biol 17: 1978–1983 [DOI] [PubMed] [Google Scholar]
- Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K. (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17: 3470–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, Mylne JS, Crevillen P, Geraldo N, An H, Gendall AR, Dean C. (2007) The PHD finger protein VRN5 functions in the epigenetic silencing of Arabidopsis FLC. Curr Biol 17: 73–78 [DOI] [PubMed] [Google Scholar]
- Gu X, Jiang D, Wang Y, Bachmair A, He Y. (2009) Repression of the floral transition via histone H2B monoubiquitination. Plant J 57: 522–533 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- He Y, Doyle MR, Amasino RM. (2004) PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18: 2774–2784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S. (2011) Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331: 76–79 [DOI] [PubMed] [Google Scholar]
- Hepworth SR, Valverde F, Ravenscroft D, Mouradov A, Coupland G. (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21: 4327–4337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. (2003) Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J 34: 733–739 [DOI] [PubMed] [Google Scholar]
- Jiang D, Gu X, He Y. (2009) Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21: 1733–1746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Kong NC, Gu X, Li Z, He Y. (2011) Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet 7: e1001330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kim DH, Sung S. (2012) Environmentally coordinated epigenetic silencing of FLC by protein and long noncoding RNA components. Curr Opin Plant Biol 15: 51–56 [DOI] [PubMed] [Google Scholar]
- Kim S, Choi K, Park C, Hwang HJ, Lee I. (2006) SUPPRESSOR OF FRIGIDA4, encoding a C2H2-Type zinc finger protein, represses flowering by transcriptional activation of Arabidopsis FLOWERING LOCUS C. Plant Cell 18: 2985–2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Michaels SD. (2006) SUPPRESSOR OF FRI 4 encodes a nuclear-localized protein that is required for delayed flowering in winter-annual Arabidopsis. Development 133: 4699–4707 [DOI] [PubMed] [Google Scholar]
- Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, Park KM, Seo JS, Grbic V, Balasubramanian S, Schmid M, et al. (2010) Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box Gene AGAMOUS-LIKE6. Plant J 62: 807–816 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Blankestijn-de Vries H, Hanhart C, Soppe W, Peeters T. (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J 6: 911–919 [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. (2000) The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes Dev 14: 2366–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Michaels SD, Masshardt AS, Amasino RM. (1994) The late-flowering phenotype of FRIGIDA and mutations in LUMINIDEPENDENS is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J 6: 903–909 [Google Scholar]
- March-Díaz R, García-Domínguez M, Lozano-Juste J, León J, Florencio FJ, Reyes JC. (2008) Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J 53: 475–487 [DOI] [PubMed] [Google Scholar]
- March-Díaz R, Reyes JC. (2009) The beauty of being a variant: H2A.Z and the SWR1 complex in plants. Mol Plant 2: 565–577 [DOI] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. (2001) Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM. (2004) FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA 101: 3281–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Himelblau E, Kim SY, Schomburg FM, Amasino RM. (2005) Integration of flowering signals in winter-annual Arabidopsis. Plant Physiol 137: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, Jacobsen SE, Fransz P, Dean C. (2006) LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. Proc Natl Acad Sci USA 103: 5012–5017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napp-Zinn K (1987) Vernalization: environmental and genetic regulation. In JG Atherton, ed, Manipulation of Flowering. Butterworths, London, pp 123–132 [Google Scholar]
- Oh S, Zhang H, Ludwig P, van Nocker S. (2004) A mechanism related to the yeast transcriptional regulator Paf1c is required for expression of the Arabidopsis FLC/MAF MADS box gene family. Plant Cell 16: 2940–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Oh S, Ek-Ramos J, van Nocker S. (2010) PLANT HOMOLOGOUS TO PARAFIBROMIN is a component of the PAF1 complex and assists in regulating expression of genes within H3K27ME3-enriched chromatin. Plant Physiol 153: 821–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathia N, Davis SJ, Lynn JR, Michaels SD, Amasino RM, Millar AJ. (2006) FLOWERING LOCUS C-dependent and -independent regulation of the circadian clock by the autonomous and vernalization pathways. BMC Plant Biol 6: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Hong L, Michaels S, Amasino RM. (2005) FRIGIDA-ESSENTIAL 1 interacts genetically with FRIGIDA and FRIGIDA-LIKE 1 to promote the winter-annual habit of Arabidopsis thaliana. Development 132: 5471–5478 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW. (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES. (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. (2004) Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164 [DOI] [PubMed] [Google Scholar]
- Sung S, He Y, Eshoo TW, Tamada Y, Johnson L, Nakahigashi K, Goto K, Jacobsen SE, Amasino RM. (2006a) Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38: 706–710 [DOI] [PubMed] [Google Scholar]
- Sung S, Schmitz RJ, Amasino RM. (2006b) A PHD finger protein involved in both the vernalization and photoperiod pathways in Arabidopsis. Genes Dev 20: 3244–3248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Takase T, Yasuhara M, Geekiyanage S, Ogura Y, Kiyosue T. (2007) Overexpression of the chimeric gene of the floral regulator CONSTANS and the EAR motif repressor causes late flowering in Arabidopsis. Plant Cell Rep 26: 815–821 [DOI] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette ML, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V. (2007) Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veley KM, Michaels SD. (2008) Functional redundancy and new roles for genes of the autonomous floral-promotion pathway. Plant Physiol 147: 682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA. (2006) The Arabidopsis thaliana vernalization response requires a polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE 3. Proc Natl Acad Sci USA 103: 14631–14636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Menard R, Berr A, Fuchs J, Cognat V, Meyer D, Shen WH (2008) The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana Plant J 57: 279–288 [DOI] [PubMed]
- Yamamoto YY, Matsui M, Ang LH, Deng XW. (1998) Role of a COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10: 1083–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
- Yu X, Michaels SD. (2010) The Arabidopsis Paf1c complex component CDC73 participates in the modification of FLOWERING LOCUS C chromatin. Plant Physiol 153: 1074–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, van Nocker S. (2002) The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J 31: 663–673 [DOI] [PubMed] [Google Scholar]