Class A1 and A2 heat shock factors possess overlapping and distinct functions in diverse abiotic stress responses and development.
Abstract
There are 21 heat shock factor (HSF) homologs in Arabidopsis (Arabidopsis thaliana), of which members of class A1 (HSFA1a/HSFA1b/HSFA1d/HSFA1e) play the major role in activating the transcription of heat-induced genes, including HSFA2. Once induced, HSFA2 becomes the dominant HSF and is able to form heterooligomeric complexes with HSFA1. However, whether HSFA2 could function independently as a transcription regulator in the absence of the HSFA1s was undetermined. To address this question, we introduced a Cauliflower mosaic virus 35S promoter:HSFA2 construct into hsfa1a/hsfa1b/hsfa1d/hsfa1e quadruple knockout (QK) and wild-type (Wt) backgrounds to yield transgenic lines A2QK and A2Wt, respectively. Constitutive expression of HSFA2 rescued the developmental defects of the QK mutant and promoted callus formation in A2QK, but not in A2Wt, after heat treatment. Transcriptome analysis showed that heat stress response genes are differentially regulated by the HSFA1s and HSFA2; the genes involved in metabolism and redox homeostasis are preferentially regulated by HSFA2, while HSFA1-preferring genes are enriched in transcription function. Ectopic expression of HSFA2 complemented the defects of QK in tolerance to different heat stress regimes, and to hydrogen peroxide, but not to salt and osmotic stresses. Furthermore, we showed that HSFA1a/HSFA1b/HSFA1d are involved in thermotolerance to mild heat stress at temperatures as low as 27°C. We also noticed subfunctionalization of the four Arabidopsis A1-type HSFs in diverse abiotic stress responses. Overall, this study reveals the overlapping and distinct functions of class A1 and A2 HSFs and may enable more precise use of HSFs in engineering stress tolerance in the future.
Plants have evolved complex response systems to cope with environmental stresses. Dynamic reprogramming of transcriptional activities constitutes one of the major events that enhance stress tolerance. Several transcription factor families are involved in complex and overlapping responses under different stress conditions (Singh et al., 2002). WRKY, MYB, APETALA2/ethylene response factor, bZIP, NAC, zinc-finger proteins, and heat shock factors (HSFs) are encoded by large gene families and have been intensively studied for their roles in stress responses (Singh et al., 2002; Saibo et al., 2009; Hirayama and Shinozaki, 2010; Santos et al., 2011; Scharf et al., 2012). Among these transcription factor families, HSFs are of particular interest because their functions in heat stress response and thermotolerance are highly conserved across all eukaryotes.
HSFs are characterized by a DNA-binding domain and hydrophobic heptad repeat regions (Wu, 1995; Morimoto, 1998; Åkerfelt et al., 2010). Current models suggest that in higher eukaryotes, typical HSFs assemble into an active, trimeric conformation via the hydrophobic heptad repeat regions in response to stress factors (Åkerfelt et al., 2010; Anckar and Sistonen, 2011). The trimerized transcription factors bind to the conserved heat shock cis-elements (GAAnnTTC) in the promoters of target genes via the DNA-binding domains and further recruit transcription machineries for gene expression. During nonstress or poststress periods, HSF activity is negatively regulated by attenuators such as HEAT SHOCK PROTEIN70 (HSP70), HSP90, and HSF binding protein (HSBP) or modulated by posttranslational modifications such as phosphorylation, acetylation, and sumoylation (Morimoto, 1998; Satyal et al., 1998; Pirkkala et al., 2001; Åkerfelt et al., 2010; Xu et al., 2012). In contrast to yeast (Saccharomyces cerevisiae) and animals, which contain only one to four HSFs (Åkerfelt et al., 2010), between 19 and 52 HSF homologs have been identified in the sequenced genomes of flowering plants (Scharf et al., 2012). The multiplicity of HSFs in plant species has been attributed to gene duplication and functional divergence during evolution (Scharf et al., 2012).
The Arabidopsis (Arabidopsis thaliana) genome encodes 21 HSF homologs that can be categorized into three major classes (A, B, and C) and 14 groups (A1–A9, B1–B4, and C1; Nover et al., 2001; von Koskull-Döring et al., 2007). There are four HSF genes in the A1 group, HSFA1a, HSFA1b, HSFA1d, and HSFA1e. HSFA1a and HSFA1d, and HSFA1b and HSFA1e both constitute pairs of duplicated genes diverged after a recent whole genome duplication event (Blanc et al., 2003). Studies on HSFA1a/HSFA1b/HSFA1d/HSFA1e quadruple knockout (KO) and four triple KO mutants showed that the four A1-type HSFs have overlapping functions in the development of seeds and cotyledons and HSFA1a, HSFA1b, or HSFA1d alone, but not HSFA1e, could trigger heat stress response and confer thermotolerance (Liu et al., 2011; Yoshida et al., 2011). The triple KO of HSFA1a/HSFA1b/HSFA1d and the quadruple KO (QK) mutants show dramatic defects in tolerance to different heat stress regimes, suggesting that HSFA1a/HSFA1b/HSFA1d are the master regulators of heat stress response in Arabidopsis. In addition, tolerance to salt, osmotic, and oxidative stresses were compromised in the QK mutant (Liu et al., 2011), suggesting that HSFA1s function under a broad range of adverse conditions that are not limited to heat stress. The roles of individual HSFA1s in response to the stress factors other than heat have not been determined.
In response to elevated temperature, HSFA1s trigger a transcriptional cascade by inducing the expression of diverse transcription regulators, including HSFs of other classes (class A2, A3, A7, B1, and B2), DREB2A, DREB2B, MBF1C, and bZIP28 (Liu et al., 2011; Yoshida et al., 2011; Liu and Charng, 2012). Several of these transcription regulators have been shown to be involved in heat stress response and thermotolerance (Sakuma et al., 2006; Charng et al., 2007; Gao et al., 2008; Larkindale and Vierling, 2008; Schramm et al., 2008; Suzuki et al., 2008; Yoshida et al., 2008; Ikeda et al., 2011). The heat inducibility of the HSFs is a feature unique to plants and is not found in yeast and animals (Nover et al., 2001; von Koskull-Döring et al., 2007), and of the HSFs, HSFA2 is the most highly induced upon heat stress (Busch et al., 2005). In Arabidopsis and tomato (Solanum lycopersicum), HSFA2 is structurally similar to the HSFA1s, in that they share conserved functional modules (Scharf et al., 2012). As a downstream gene of HSFA1, HSFA2 is involved in the late phase of heat stress response (Wunderlich et al., 2007), extending the effect of heat acclimation (Charng et al., 2007) and mediating the amplification of a subset of heat stress response genes (Schramm et al., 2006). In addition to heat stress, expression of HSFA2 is also positively regulated by HSFA1 under salt, osmotic, and oxidative stresses (Liu et al., 2011; Nishizawa-Yokoi et al., 2011), suggesting an epistatic relationship between these two types of HSFs under several stress conditions.
Transient expression of tomato HSFs in tobacco (Nicotiana tabacum) protoplasts indicated that the efficient nuclear import of HSFA2 requires interaction with HSFA1a (Scharf et al., 1998; Heerklotz et al., 2001). Interaction between tomato HSFA1a and HSFA2 leads to the formation of heterooligomeric complexes that can synergistically activate the expression of a small HSP (Chan-Schaminet et al., 2009). Interaction between Arabidopsis HSFA1a/HSFA1b and HSFA2 has also been demonstrated (Li et al., 2010). These observations raise the question of whether HSFA2 can function independently as a transcription regulator in the absence of the HSFA1s and, if so, to what extent. Moreover, the hetero- and homooligomers of HSFA1 and HSFA2 may preferentially activate different sets of genes. However, this notion is difficult to assess at the genomic level as the expression of HSFA2 strictly requires the presence of the HSFA1s (Mishra et al., 2002; Liu et al., 2011; Nishizawa-Yokoi et al., 2011; Yoshida et al., 2011).
To address the above questions, we produced transgenic plants constitutively expressing Arabidopsis HSFA2 in the background of the HSFA1 QK mutant and examined whether ectopic HSFA2 expression could rescue the defects of the mutant in development and stress tolerance under various conditions. Interestingly, complete or partial recovery from multiple defects was observed in the transgenic plants, suggesting that HSFA2 can at least partially replace the function of the HSFA1s. However, it could not rescue the defect of the QK mutant under salt or osmotic stresses. Genes preferentially activated by HSFA1 or HSFA2 upon heat stress were identified by microarray analysis of the transcriptomes of the wild type, hsfa2 (the transfer DNA KO mutant of HSFA2), the QK mutant, and the QK mutant transformed with recombinant 35S:HSFA2 (A2QK). We also showed that HSFA1a/HSFA1b/HSFA1d are involved in thermotolerance to mild heat stress at temperatures as low as 27°C. Furthermore, results from the response of the triple KO mutants of HSFA1s to salt, osmotic, and oxidative stresses suggest that Arabidopsis HSFA1s went through subfunctionalization after gene duplication.
RESULTS
Ectopic Expression of HSFA2 Complements the Defects of HSFA1 QK Mutant in Growth and Development
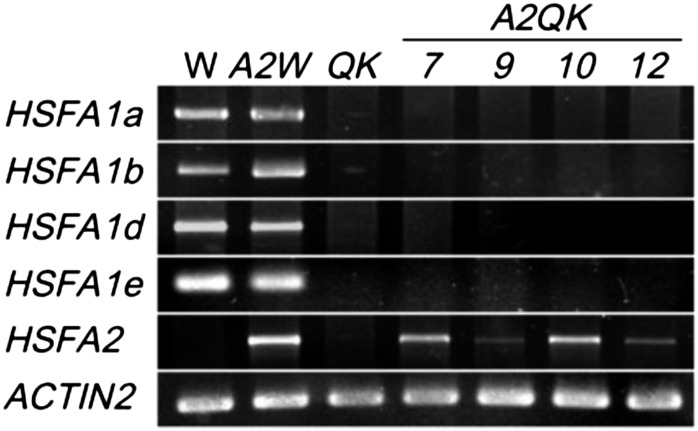
To examine whether and to what extent HSFA2 functions in the absence of the HSFA1s, we generated transgenic lines containing a recombinant HSFA2 complementary DNA fused to the Cauliflower mosaic virus 35S promoter in the HSFA1a/HSFA1b/HSFA1d/HSFA1e QK mutant background. Four transgenic lines, designated as A2QK-7, A2QK-9, A2QK-10, and A2QK-12, with a single insertion event were selected from 15 independent lines for further studies. As a comparison, the same transgene construct was introduced into the ecotype Columbia (Col-0) wild-type background to yield a transgenic line designated as A2Wt. Figure 1 shows the expression of HSFA2 and the HSFA1s in different transgenic lines as determined by reverse transcription (RT)-PCR to confirm their genotypes. Under normal conditions, transcripts of HSFA2 were detected in all the transgenic lines, but not in the wild type and QK mutant, indicating the constitutive expression of the transgene. A2QK-7/A2QK-10 and A2QK-9/A2QK-12 lines had high and low expression levels of HSFA2, respectively.
Figure 1.
Transcript levels of the four HSFA1s and HSFA2 in transgenic plants overexpressing HSFA2. RT-PCR analysis of the transcript levels of HSFA1a, HSFA1b, HSFA1d, HSFA1e, and HSFA2 under normal conditions in 7-d-old seedlings of wild-type (W), HSFA1s QK mutant, and transgenic plants transformed with 35S:HSFA2 in wild-type (A2W) or QK backgrounds (A2QK-7, A2QK-9, A2QK-10, and A2QK-12). ACTIN2 is shown as a loading control. For HSFA2 and ACTIN2, 29 cycles of PCR were performed. For HSFA1a/HSFA1b/HSFA1d/HSFA1e, 34 cycles of PCR were performed. The amplification products were resolved electrophoretically on 1% to 3% agarose gels.
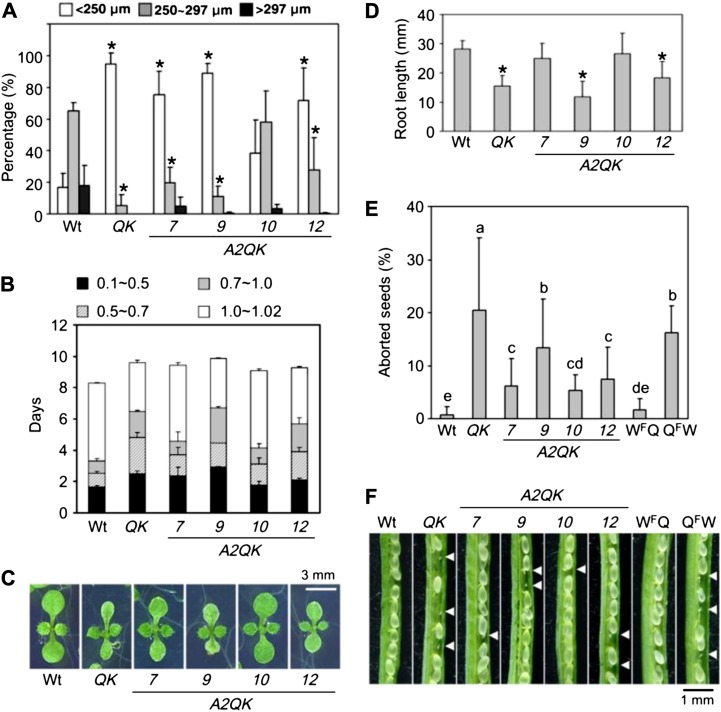
The growth and morphologies of the transgenic plants readily manifested the effect of constitutive expression of HSFA2 in the HSFA1 QK mutant. Previously, we showed that the seedlings of the QK mutant had abnormal cotyledons, grew significantly slower than the wild type, and produced smaller seeds (Liu et al., 2011). Expression of HSFA2 in the HSFA1 QK mutant restored at least partially the seed size, seedling growth rate, cotyledon morphology, and root length (Fig. 2, A–D). In this study, we additionally noticed that the QK mutant produced aborted seeds at a rate about 10-fold higher than that of the Col-0 wild type (Fig. 2, E and F). When crossing Col-0 and the QK mutant with different gametophyte combinations, the higher abortion rate was associated with the female gametophyte of the HSFA1 QK mutant. Ectopic HSFA2 expression in the QK background significantly reduced the seed abortion rate (Fig. 2, E and F). These results suggest that HSFA2 could at least partially replace the role of the HSFA1s in growth and development.
Figure 2.
Ectopic HSFA2 expression complements the defects of QK mutant in development of seed and seedling. A, Distribution of seed size of Col-0 wild-type (Wt), QK mutant, and A2QK transgenic plants (A2QK-7, A2QK-9, A2QK-10, and A2QK-12). Results are presented as mean values of five replicates ± sd (n ≥ 100 each). *P < 0.01 (versus the wild type, Student’s t test). B, Growth rate from the imbibed seed to the two rosette leaf stage grown in 0.5× MS medium plates containing 1% Suc. Growth stages are defined as follows: 0.1, seed imbibition; 0.5, radicle emergence; 0.7, hypocotyl and cotyledon emergence; 1.0, cotyledons fully opened; and 1.02, two rosette leaves greater than 1 mm. The growth stages are presented as mean values of four replicates ± sd (n = 45 each). C, Morphologies of the representative plants at growth stage 1.02. D, Root length of the 7-d-old seedlings. Results are presented as mean values of six replicates ± sd (n = 25). *P < 0.01 (versus the wild type, Student’s t test). E, The percentages of the aborted seeds in the immature siliques of wild-type, QK mutant, and A2QK transgenic plants and crosses with different gametophyte combinations of the wild type and QK mutant. WFQ is the hybrid with the female gametophyte of the wild type and the male gametophyte of the QK mutant. QFW is the hybrid with the female gametophyte of the QK mutant and the male gametophyte of the wild type. The percentages were averaged from more than 15 siliques of five independent plants. Groups with different alphabets indicate there is a statistically significant difference among them (lsd, α = 0.05). F, The development of seeds in immature siliques of wild-type, QK mutant, and A2QK transgenic plants and the hybrids of the wild type and QK mutant. The arrowheads indicate the aborted seeds.
Constitutive Expression of HSFA2 Induces Expression of HSPs in the Absence of HSFA1s
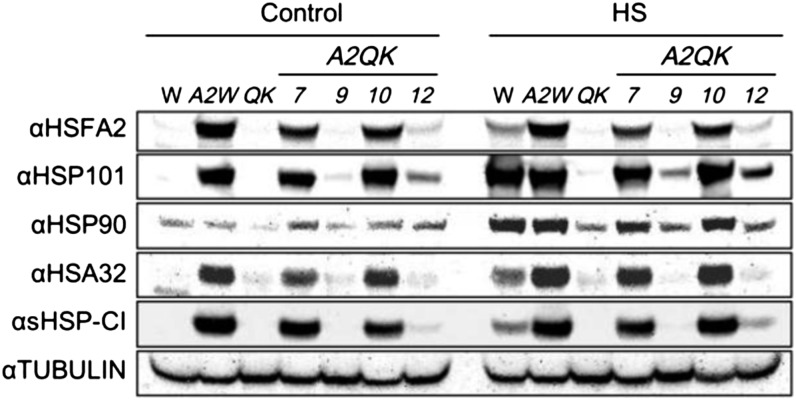
To investigate whether HSFA2 could activate the expression of HSPs in the absence of the HSFA1s, western-blot analysis was conducted to detect the levels of HSFA2, HSP101, HSP90, HEAT-STRESS-ASSOCIATED 32-kD PROTEIN (HSA32), and class I small HSPs (sHSP-CI) in A2QK lines. Under normal conditions, A2QK-7, A2QK-10, and A2Wt produced high levels of HSFA2 protein, while the A2QK-9 and A2QK-12 lines produced undetectable and low levels of HSFA2, respectively (Fig. 3). The wild type and QK mutant did not contain detectable HSFA2 under the same condition. The protein levels of HSFA2 in these plants were consistent with the transcript levels of HSFA2 as determined by RT-PCR (Fig. 1). The levels of HSFA2 correlated with the levels of HSP101, HSA32, and sHSP-CI, but not HSP90, in the transgenic lines under nonstressed conditions, suggesting that HSFA2 could activate the expression of some HSPs in the absence of the HSFA1s.
Figure 3.
Constitutive expression of HSFA2 induces expression of HSPs in the absence of HSFA1s. Western-blot analysis of HSFA2 and HSPs in wild-type (W), A2Wt (A2W), QK mutant, and A2QK transgenic plants. Seven-day-old plants grown on medium plates were treated at 22°C (Control) or 37°C (Heat-shocked) for 1 h and recovered at 22°C for 2 h before protein extraction. In each lane, approximately 100 μg of total protein was loaded. Tubulin serves as a loading control.
Strong Induction of HSFA2 in Response to Heat Stress Is Mainly Regulated by the HSFA1s Rather Than by Autoregulation
The strong induction of HSFA2 in response to heat stress depends on the function of the HSFA1s (Liu et al., 2011). However, it is not clear whether HSFA1s trigger an initial low-level expression of HSFA2, which then amplifies its own expression via autoregulation. We noticed that heat treatment at 37°C for 1 h did not further increase the levels of HSFA2 protein in the A2Wt and A2QK transgenic lines (Fig. 3), suggesting that the recombinant HSFA2 does not activate the endogenous HSFA2 gene in the transgenic lines. Because the antibody against HSFA2 could not distinguish the recombinant HSFA2 protein from the endogenous protein, RT-PCR analysis with discriminating primers was performed to address this possibility.
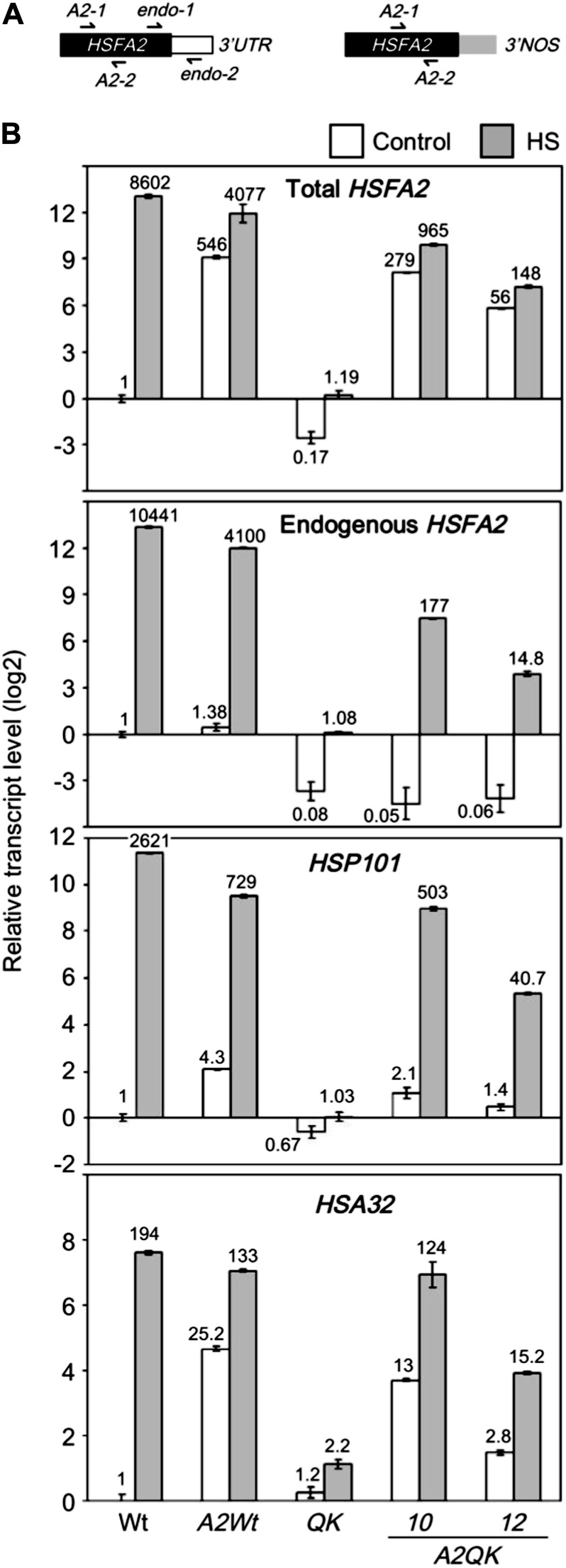
Two different primer pairs were designed for quantitative RT-PCR to reveal the transcript abundance of the endogenous HSFA2 gene and total HSFA2 transcripts derived from the combination of the endogenous and transgenic HSFA2 (Fig. 4A). Figure 4B shows the quantitative analysis of the expression levels of endogenous HSFA2 in the presence or absence of constitutively expressed HSFA2 transgene. Without heat treatment, the recombinant HSFA2, regardless of its abundance, was unable to up-regulate the endogenous HSFA2 in the presence or absence of the HSFA1s, as revealed by comparing the wild type to the A2Wt line and QK to the A2QK line. By contrast, the transcript levels of HSP101 and HSA32 were significantly higher in accordance with the higher level of total HSFA2 transcripts, which mainly came from the expression of the transgene (Fig. 4B). After heat treatment, the transcripts of endogenous HSFA2 were slightly increased in A2QK-10 and A2QK-12 but were much less abundant than in the wild type and A2Wt. No substantial difference was found between the total and endogenous transcripts of HSFA2 in the A2Wt line after heat treatment, which was about 2-fold lower than that in the wild type (Fig. 4B). These results suggest that the strong induction of HSFA2 in response to heat stress is mainly regulated by HSFA1s without substantial positive autoregulation by HSFA2.
Figure 4.
Effect of constitutive expression of HSFA2 on the transcript levels of HSP101, HSA32, and endogenous HSFA2. A, The schemes of the endogenous (left) and recombinant (right) HSFA2 mRNAs. Black, white, and gray blocks indicate the coding region of HSFA2, 3′ untranslated region of endogenous HSFA2, and 3′ untranslated region of the nopaline synthase (NOS) terminator of the transgene, respectively. The arrows indicate the locations of the primers used for quantitative RT-PCR. B, The relative transcript levels of HSFA2, HSP101, and HSA32 in wild-type (Wt), QK mutant, and 35S:HSFA2 transgenic plants (A2Wt, A2QK-10, and A2QK-12) with or without heat treatment. Total HSFA2 transcript was detected by the primer set A2-1 and A2-2, and endogenous HSFA2 transcript was detected by the primer set endo-1 and endo-2. White bars indicate the control samples without heat treatment, and gray bars indicate the samples treated at 37°C for 1 h. The numbers at the top or bottom of bars are the transcript level relative to the wild-type sample under control conditions.
Heat Stress Response Genes Are Differentially Regulated by the HSFA1s and HSFA2
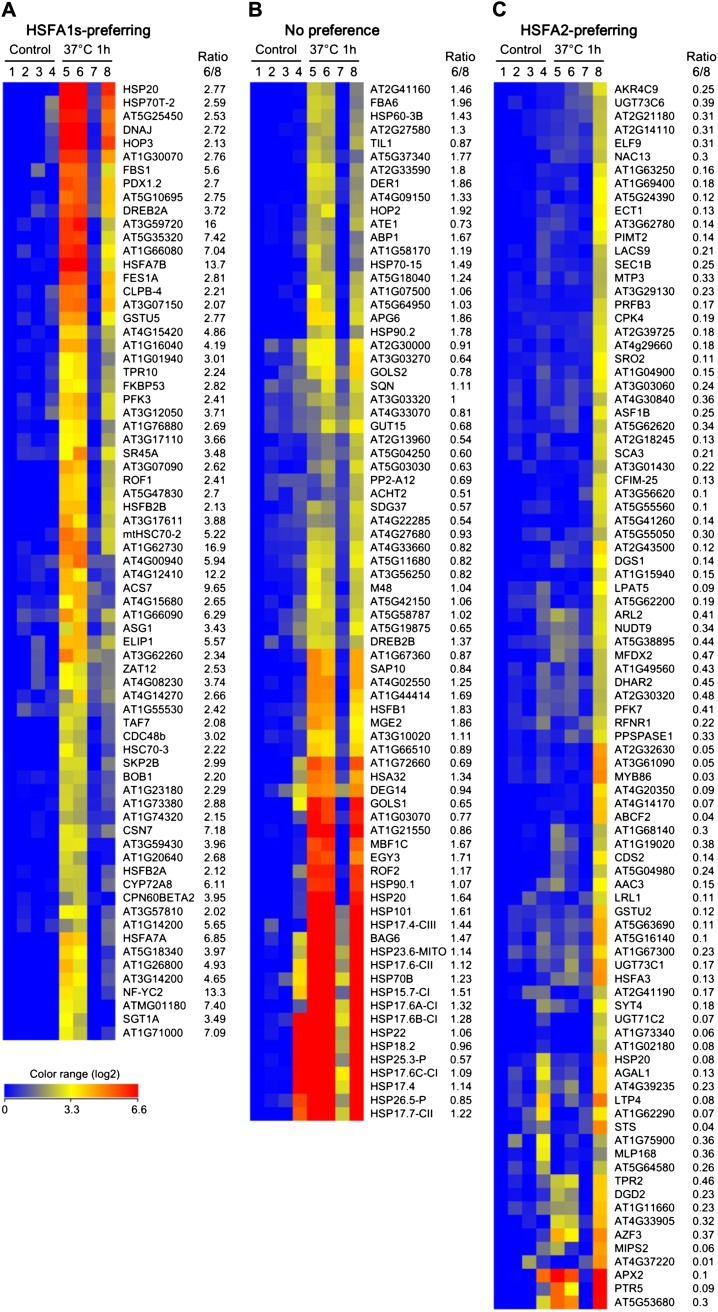
The results shown in Figure 4B indicate that the HSFA1s and HSFA2 differentially regulate heat stress response genes. To further explore this phenomenon at the level of the genome, we compared the transcriptomic profiles of hsfa2 (the KO mutant of HSFA2) and A2QK-10 (overexpression of HSFA2 in the absence of the HSFA1s) seedlings with or without heat shock treatment using the Affymetrix ATH1 microarray chips. The difference in heat stress response between hsfa2 and A2QK-10 was attributed to differential regulation by the HSFs and was identified as follows. First, the microarray data were combined with those of the Col-0 wild type and the QK mutant seedlings previously obtained under the same conditions (Liu et al., 2011). We found genes highly induced by heat by subjecting the signals of 22,810 probe sets on the microarray chip to two filters. Firstly, we eliminated 14,117 low-expressing probe sets whose normalized absolute expression values were lower than 100 in all samples. Secondly, 783 probe sets were selected that had at least a 5-fold change in any individual comparison with the Col-0 wild type under control conditions. Then, hierarchical clustering analysis was performed to group the coexpressed probe sets. We found 240 probe sets that were strongly heat induced in the wild type, hsfa2, or A2QK, but not in the QK mutant. The target preferences of the HSFA1s and HSFA2 were judged according to the ratio of expression level under heat stress conditions in the hsfa2 and A2QK-10 mutants (Supplemental Table S1). Genes with an expression level 2-fold higher in hsfa2 than in A2QK-10 were defined as HSFA1s preferring (Fig. 5A). HSFA2-preferring genes were defined as having a hsfa2/A2QK-10 expression ratio of less than 0.5-fold (Fig. 5C), whereas genes shown in Figure 5B with an expression ratio between 0.5- and 2-fold were defined as having no substantial preference for HSFA1s or HSFA2.
Figure 5.
Heat shock-induced genes differentially regulated by HSFA1s and HSFA2. Heat maps of selected microarray expression profiles in 7-d-old seedlings of wild-type, hsfa2, QK, and A2QK-10 transgenic plants under control and heat shock (37°C for 1 h) conditions. The 7-d-old seedlings of the wild type (lanes 1 and 5), hsfa2 (lanes 2 and 6), QK (lanes 3 and 7), and A2QK-10 (lanes 4 and 8) under control condition (lanes 1–4) or after heat shock treatment (lanes 5–8) were collected for the analysis of microarray expression profiles. A total of 239 selected genes (excluding HSFA2) depending on HSFA1s or HSFA2 for heat shock induction are clustered into three major groups. The genes preferentially regulated by HSFA1s (A) or HSFA2 (C) were identified by having a signal ratio of lanes 6/8 greater than 2 or less than 0.5, respectively. The genes with signal ratio between 0.5 and 2 were considered as having no preference for HSFA1s or HSFA2 (B). The color range represents the relative level (log2) compared with the wild-type sample under control conditions.
Based on known or predicted functions, we sorted the 240 selected genes into functional categories (Table I and S1). For the HSFA1s-preferring genes, the most enriched categories were “chaperone and cochaperone” and “transcription,” which contained 17 (23.6%) and 11 (15.3%) genes, respectively. For the genes with no preference for HSFA1s or HSFA2, the most enriched category was “chaperone and cochaperone” (27 genes; 35.1%). Among the genes with no preference, 14 genes were small HSPs. For the genes preferably regulated by HSFA2, 19 genes (20.9%) were found to be involved in various metabolic processes, eight genes (8.8%) were involved in redox homeostasis, and 13 genes (14.3%) had predicted enzyme functions. By contrast, only five genes (5.5%) were classified as chaperones and cochaperones.
Table I. Classifications of the heat-induced genes differentially regulated by HSFA1s and HSFA2.
Heat shock-induced genes in three major groups (HSFA1s preferring, no significant preference, and HSFA2 preferring) were categorized based on published information functions predicted in The Arabidopsis Information Resource (Lamesch et al., 2012).
| Functional Categories | HSFA1s Preferring |
No Significant Preference |
HSFA2 Preferring |
|||
|---|---|---|---|---|---|---|
| nos. | % | nos. | % | nos. | % | |
| Signaling | 2 | 2.8 | 3 | 3.9 | 4 | 4.4 |
| Transcription | 11 | 15.3 | 3 | 3.9 | 5 | 5.5 |
| Posttranscriptional regulation | 1 | 1.4 | 1 | 1.3 | 3 | 3.3 |
| Chaperone and cochaperone | 17 | 23.6 | 27 | 35.1 | 4 | 4.4 |
| Redox homeostasis | 4 | 5.6 | 2 | 2.6 | 8 | 8.8 |
| Metabolism | 3 | 4.2 | 4 | 5.2 | 19 | 20.9 |
| Enzyme | 5 | 6.9 | 8 | 10.4 | 13 | 14.3 |
| Development | 2 | 2.8 | ||||
| Heat tolerance | 3 | 3.9 | ||||
| Metal homeostasis | 1 | 1.1 | ||||
| Transporter | 4 | 4.4 | ||||
| Unknown | 27 | 38 | 26 | 33.8 | 30 | 33 |
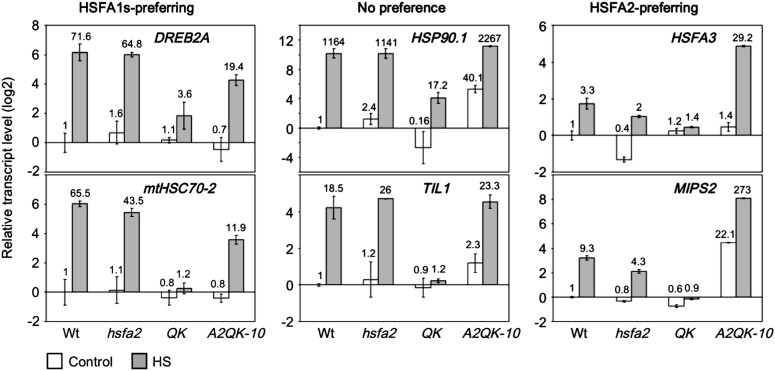
To confirm the results of the microarray, we performed quantitative RT-PCR analysis of the transcript levels of DREB2A and MITOCHONDRIAL HEAT SHOCK COGNATE 70-kD PROTEIN-2 (mtHSC70-2) (HSFA1s preferring), HSP90.1 and TEMPERATURE-INDUCED LIPOCALIN1 (TIL1; no preference), and HSFA3 and MYO-INOSITOL1-PHOSPHATE SYNTHASE2 (MIPS2; HSFA2 preferring). The expression ratios of heat-treated hsfa2 versus A2QK plants for DREB2A, mtHSC70-2, HSP90.1, TIL1, HSFA3, and MIPS2 were 3.34, 3.65, 0.5, 1.16, 0.07, and 0.016, respectively (Fig. 6). These data are consistent with those derived from the microarray.
Figure 6.
Confirmation of the target preference of HSFA1s and HSFA2 by quantitative RT-PCR. The relative transcript levels of selected genes with a preference for regulation by the HSFA1s or HSFA2 according to the microarray data were analyzed by quantitative RT-PCR and relative to the level of the wild type under control conditions. The samples and treatment conditions were the same as those in Figure 5. The relative transcript levels of each sample are indicated at the top of the bars. White bars indicate the control samples without heat treatment, and gray bars indicate the samples treated at 37°C for 1 h.
Ectopic HSFA2 Expression Confers Tolerance to Diverse Heat Stress Regimes and Promotes Heat-Induced Callus Formation in the Absence of HSFA1s
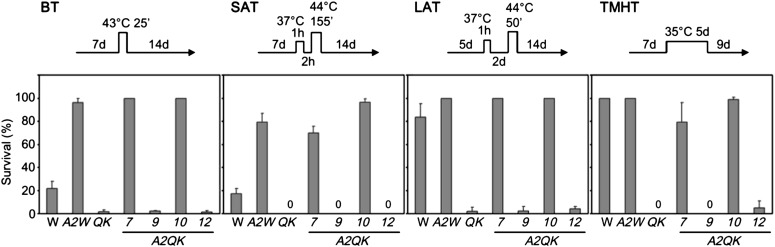
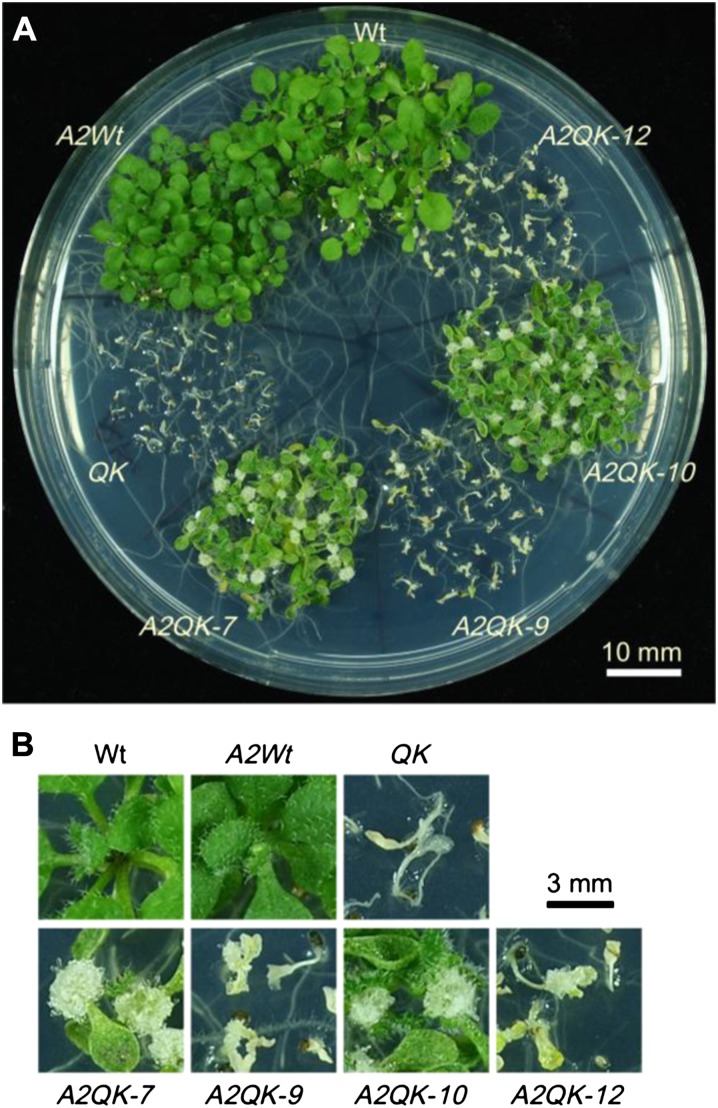
Given the differential regulation of heat stress response genes by the HSFA1s and HSFA2, it was of interest to know whether HSFA2 could replace the role of the HSFA1s in conferring tolerance to four different heat stress regimes that are commonly used to reveal thermotolerance diversity in Arabidopsis (Yeh et al., 2012). The results of thermotolerance assays showed that expression of HSFA2 in QK dramatically increased the basal thermotolerance (BT) of A2QK-7 and A2QK-10, which had a higher viability than the wild type (Fig. 7; Supplemental Fig. S1). Intriguingly, A2QK-7 and A2QK-10 also had a higher viability than the wild type under the assay conditions for short-term and long-term acquired thermotolerance and were comparable to that of the A2Wt (Fig. 7; Supplemental Fig. S1), suggesting that overexpression of HSFA2 is sufficient to confer a high level of acquired thermotolerance even in the absence of the master regulators. Expression of HSFA2 in the QK background also rescued the defect in thermotolerance to moderately high temperature (TMHT) but provided no obvious advantage as compared with the wild type (Fig. 7; Supplemental Fig. S1). By contrast, the low level of expression of HSFA2 in A2QK-9 and A2QK-12 could not rescue the thermotolerance defects of QK. Notably, the A2QK seedlings tended to form calluses at the apical meristems after the thermotolerance assays (Fig. 8). The number of calluses formed was positively correlated to the expression level of HSFA2. Calluses did not form in the A2Wt plants despite having amounts of HSFA2 similar to that of A2QK-7 and A2QK-10, suggesting that the heat-induced callus formation is mediated by overexpression of HSFA2 in the absence of the HSFA1s.
Figure 7.
Constitutive expression of HSFA2 conferred high level of thermotolerance in the absence of the HSFA1s under different heat stress regimes. The viabilities of the wild type, QK mutant, and 35S:HSFA2 transgenic lines (A2Wt, A2QK-7, A2QK-9, A2QK-10, and A2QK-12) after treatment with four different heat stress regimes (shown by the schemes) for BT, SAT, LAT, and TMHT. The viable plants were defined as those that generated new rosette leaves after heat stress. Images of SAT, BT, and TMHT are shown in Supplemental Figure S1, and an image of LAT is shown in Figure 8A. Results are presented as mean values of three replicates ± sd (n ≥ 40 each).
Figure 8.
Constitutive expression of HSFA2 promoted the formation of calluses after heat treatment in the absence of the HSFA1s. A, The phenotypes of wild-type, QK mutant, and 35S:HSFA2 transgenic plants (A2Wt, A2QK-7, A2QK-9, A2QK-10, and A2QK-12) after treatment with heat stress regime for LAT as indicated in Figure 7. B, Enlarged images of the representative plants in A.
QK and hsf1a-hsf1b-hsf1d Triple Mutants Are Hypersensitive to Prolonged Exposure to Temperatures as Low as 27°C
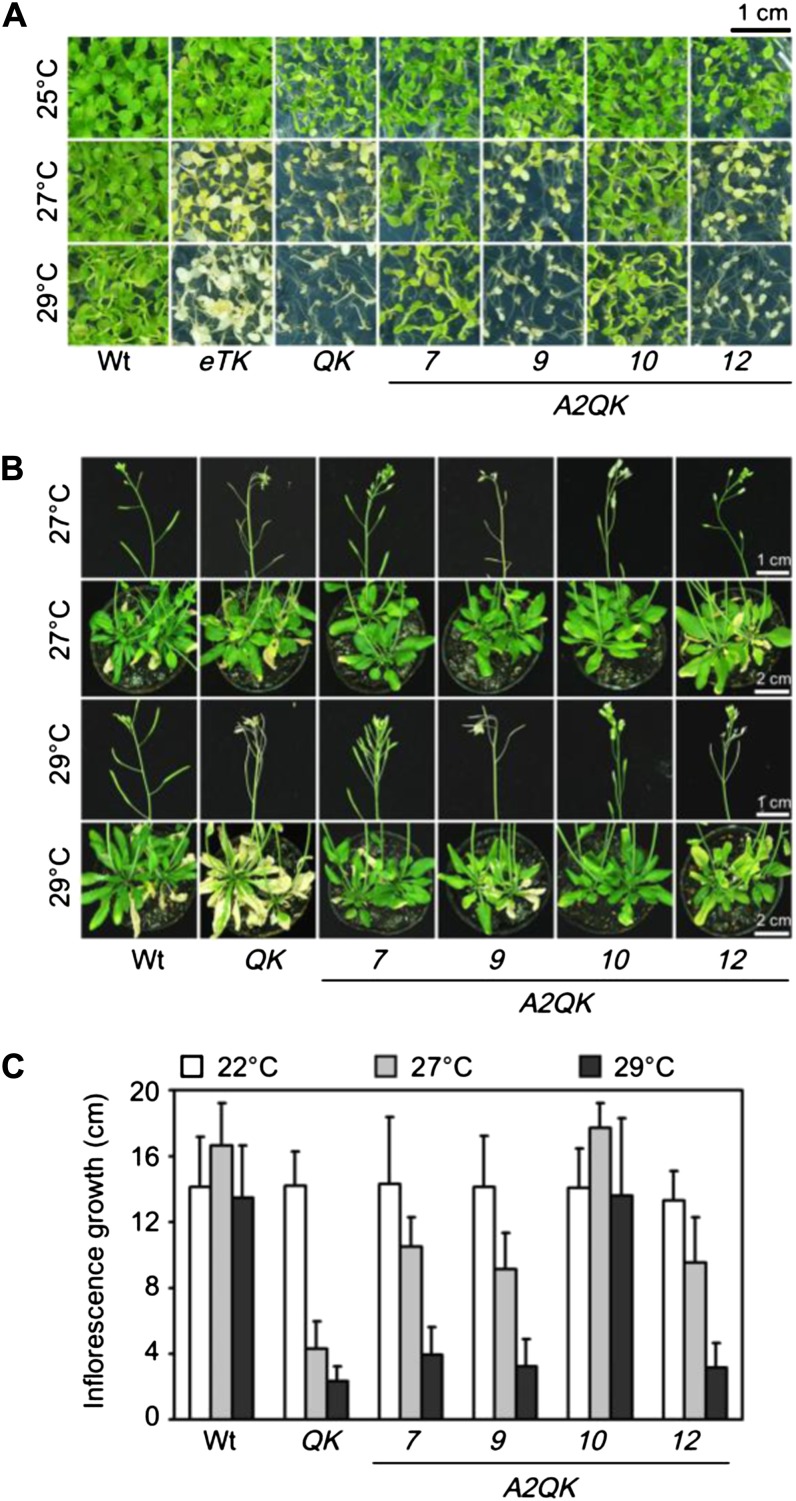
HSFA1a, HSFA1b, and HSFA1d redundantly confer tolerance to prolonged heat stress at 35°C in Arabidopsis (Liu et al., 2011). However, it is not known whether they are also important for tolerating long-term exposure to milder temperatures below 30°C, which do not kill Arabidopsis plants but result in adaptation by reprogramming the developmental processes (Franklin, 2009; Kumar and Wigge, 2010). To test this, phenotypes of the QK and the four triple KO mutants of the HSFA1s (labeled as aTK, bTK, dTK, and eTK, with the prefix letters representing the remaining functional HSFA1) were compared to that of the wild type after prolonged treatments at 25°C to 29°C. QK and eTK seedlings were not viable when grown continuously at temperatures above 27°C for 7 d, while the viability of the wild type, aTK, bTK, and dTK were not affected (Fig. 9A; Supplemental Fig. S2). Although HSFA2 is not required for viability in this temperature range (Supplemental Fig. S3), ectopic expression of HSFA2 in A2QK-7 and A2QK-10 complemented the function of the HSFA1s to confer tolerance to the mild heat stress (Fig. 9A). Similar results were obtained for the adult plants. In the wild type, the growth of rosette leaves, elongation of inflorescences, and development of siliques after switching growth temperature from 22°C to 27°C were similar to those of plants at 22°C (Fig. 9, B and C). The elongation of inflorescences was slightly enhanced at 27°C (Fig. 9C) but not at 29°C. The QK mutant showed obvious defects at 27°C, including stunted growth of inflorescences and aborted siliques, despite of normal growth of leaves (Fig. 9, B and C). These defects were more obvious at 29°C than at 27°C. The rosette leaves of the QK mutant were bleached at 29°C, while those of the wild type remained green. Ectopic expression of HFSA2 also complemented the thermotolerance defects of QK at these stages (Fig. 9, B and C). These results suggest that HSFA1a/HSFA1b/HSFA1d play pivotal roles in the thermotolerance of reproductive as well as vegetative tissues under chronic heat stress at temperatures as low as 27°C.
Figure 9.
HSFA1a/HSFA1b/HSFA1d are required for tolerance to chronic heat stress at temperatures above 27°C in seedlings and flowering plants. A, Phenotypes of 14-d-old seedlings of the wild type (Wt), eTK, QK, A2QK-7, A2QK-9, A2QK-10, and A2QK-12 after treatment at 25°C, 27°C, or 29°C for 7 d. B, Phenotypes of 40-d-old plants of indicated lines after treatment at 27°C or 29°C for 8 d. C, The elongated length of inflorescences of the plants in B after treatment at 22°C, 27°C, or 29°C for 8 d.
The HSFA1s and HSFA2 Have Differential Roles in Tolerance to Salt, Osmotic, and Oxidative Stresses
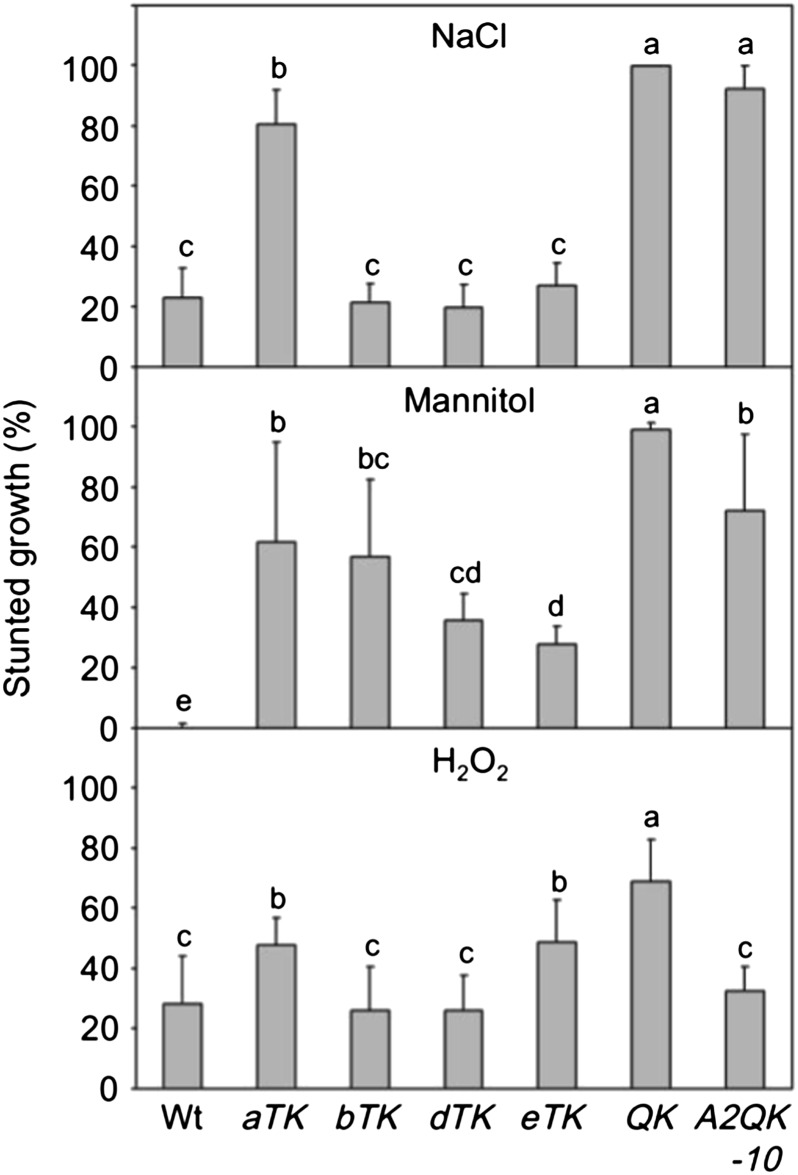
Previously, we demonstrated that the HSFA1s are involved in response and tolerance to salt, osmotic, and oxidative stresses during seedling establishment (Liu et al., 2011); however, the individual roles of the four HSFA1s were not clear. Overexpression of HSFA2 implicated the gene in salt and oxidative stress tolerance (Ogawa et al., 2007), but whether HSFA2 can function against these stress factors independently of the HSFA1s is not known. Here, we compared the responses of the wild type, QK mutant, HSFA1 triple KO mutant, and A2QK-10 seeds sown on the growth media containing 125 mm NaCl, 300 mm mannitol, or 5 mm H2O2. Consistent with a previous report (Liu et al., 2011), the QK mutant showed a significantly higher percentage of severely stunted growth than the Col-0 wild type after germination under stress conditions (Fig. 10). Unexpectedly, the four HSFA1 triple KO mutants showed differential responses to these stress factors. The aTK mutant showed severely stunted growth in response to high salt concentration, while the other three triple KO mutants behaved like the wild type (Fig. 10). The four triple KO mutants showed different degrees of defect under high concentrations of mannitol, with the eTK and dTK mutants least affected. In response to H2O2, the bTK and dTK mutants behaved like the wild type, while the growth of aTK and eTK were more significantly retarded, but to less severe extent than the QK mutant. Interestingly, overexpression of HSFA2 in the QK background significantly rescued the defect in tolerance to H2O2, but provided little or no protection to salt and osmotic stresses (Fig. 10). However, the phenotype of the hsfa2 mutant under these stress conditions was similar to that of the wild type (Supplemental Fig. S3), suggesting that HSFA2, at least for seedling establishment, does not play an important role in response to these stress conditions as compared with the functions of the HSFA1s.
Figure 10.
Contributions of individual HSFA1s and constitutive expression of HSFA2 in tolerance to salt, osmotic, and oxidative stresses. The growth of wild-type, HSFA1 triple KO mutant (aTK, bTK, dTK, and eTK), QK, and A2QK-10 transgenic plants sown on 0.5× MS medium with 125 mm NaCl, 300 mm mannitol, or 5.5 mm H2O2 was assessed after 12 d. Stunted growth was defined as plants without green cotyledons and true leaf emergence. The growth rates under stress conditions were normalized by the growth rate of each line sown on plates without the stress factors. Results are the mean values of six replicates ± sd (n = 45). Groups labeled with different letters of the alphabet indicate those with statistically significant differences (lsd, α = 0.05).
DISCUSSION
The presence of diverse HSF proteins in plant species is presumably associated with their success in adapting to changing environments that could have negative impact. Unraveling the biological function of each HSF may provide a better understanding of how plants cope with stresses and the evolution of plant HSFs. However, the roles of individual plant HSFs are masked by the redundant functions they share with other members of the protein family. Genetic disruption of one HSF gene may not necessarily produce an obvious phenotype, as exemplified in recent studies of Arabidopsis A1-type HSFs (Lohmann et al., 2004; Liu et al., 2011; Nishizawa-Yokoi et al., 2011; Yoshida et al., 2011). Stable or transient overexpression of a HSF transgene may also generate misleading results, as the overexpressed protein may derepress the endogenous master HSFs by competing with the attenuators, such as HSP70, HSP90, and HSBP (Voellmy, 2004; Fu et al., 2006; Yamada et al., 2007; Hsu et al., 2010; Hahn et al., 2011). For example, HSFA2 was shown to interact with HSP90 and HSBP (Meiri and Breiman, 2009; Hsu et al., 2010), and therefore overexpression of HSFA2 in the wild-type plant could derepress the HSFA1s, trigger a stress response, and enhance stress tolerance. This possibility makes interpreting overexpression results difficult. Here, to assess the independent function of HSFA2, we ectopically expressed it in an Arabidopsis mutant lacking all four HSFA1s. This strategy did not allow the exclusion of possible interference from the other remaining HSFs, but any such interference is likely to be much weaker in the absence of the HSFA1s, as the expression of several HSFs, such as HSFA3, HSFA7a/HSFA7b, HSFB1, and HSFB2a/HSFB2b, also requires the HSFA1s (Liu et al., 2011).
In this study, we demonstrated that HSFA2 could perform certain functions in the absence of the HSFA1s, such as triggering heat stress response and conferring thermotolerance. These results suggest that HSFA2 can act as a transcription activator independent of the HSFA1s, including in nuclear localization and transcription activation of heat stress genes. Our findings are in agreement with a previous study that demonstrated that transiently expressed Arabidopsis HSFA2 can shuttle between the cytoplasm and nucleus without HSFA1 (Kotak et al., 2004), a prerequisite for HSFA2 to act as transcription factor. Because the expression of HSFA2 in seedlings is strictly controlled by the HSFA1s upon stress, the independent role of HSFA2 can only be fulfilled after prolonged or reexposure to stress conditions or by ectopic overexpression, where HSFA2 preferentially targets a subset of genes with different physiological functions from those conferred by the HSFA1-preferring genes (Fig. 5).
In response to heat stress, HSFA2 seems to have higher activity than the HSFA1s in activating the genes involved in redox homeostasis (ASCORBATE PEROXIDASE2 [APX2] and ALDO-KETO REDUCTASE4 SUBFAMILY C9 [AKR4C9]) and the metabolism of carbohydrates (MIPS2) and lipids (LONG-CHAIN ACYL-COA SYNTHETASE9 [LACS9], CYTIDINEDIPHOSPHATE DIACYLGLYCEROL SYNTHASE2 [CDS2], DIGALACTOSYL DIACYLGLYCEROL DEFICIENT2 [DGD2], and DGD1 SUPPRESSOR1 [DGS1]; Fig. 5; Table I; Supplemental Table S1). According to the functions reported in previous studies (Kelly et al., 2003; Rossel et al., 2006; Simpson et al., 2009; Haselier et al., 2010; Moellering and Benning, 2010; Zhao et al., 2010; Valluru and Van den Ende, 2011), these genes may contribute to maintenance of cellular homeostasis during prolonged heat stress. In addition, HSFA2, when expressed at high levels, functioned in a similar manner to the HSFA1s regulating a large number of chaperones and cochaperones, such as HSP101, HSP90, HSP70B, HSP70-15, HSP60-3B, small HSPs, MITOCHONDRIAL GRPE2 (MGE2), and ROTAMASE FKBP2 (ROF2; Fig. 5; Table I; Supplemental Table S1), explaining its capacity to confer a wide spectrum of heat tolerance. By contrast, HSFA2, as compared with the HSFA1s, is a relatively weak activator of the heat-induced transcription regulators such as HSFA7a, HSFA7b, NUCLEAR FACTOR Y SUBUNIT C2 (NF-YC2), DREB2A, HSFB2b, and HSFA2 itself (Figs. 4 and 5; Table I; Supplemental Table S1), which may prevent these transcription factors from being amplified and overaccumulated. Overaccumulation of stress-related transcription regulators could be detrimental to growth and is not deemed preferable (Ogawa et al., 2007; Zhu et al., 2009). Recently, a heat-induced splice variant of HSFA2 was identified and suggested to be involved in the autoregulation of HSFA2 (Liu et al., 2013). However, the protein level of this HSFA2 variant was not determined, and whether it could exert an effect in heat stress response is unclear. Unlike the above-mentioned transcription factors, HSFA3 is preferentially regulated by HSFA2, which is in good agreement with its late induction profile under heat stress (Schramm et al., 2008). Interestingly, HSFA3 was shown to be partially regulated by DREB2A (Sakuma et al., 2006; Schramm et al., 2008). Hence, two pathways control the heat induction of HSFA3, one through HSFA1s-DREB2A and another through HSFA1s-HSFA2. It would be of interest to find out why HSFA3 is subjected to regulation via two independent pathways.
Several genes involved in diverse signaling pathways are also under the differential regulation of the HSFA1s and HSFA2. For example, CSN7 (COP9 signalosome, Dessau et al., 2008) and ACS7 (ethylene, Dong et al., 2011) are preferentially regulated by the HSFA1s; BAG6 (calcium, Kang et al., 2006) and ABP1 (auxin, Braun et al., 2008) are regulated by both HSFA1s and HSFA2; and UGT73C6 (brassinosteroids, Husar et al., 2011) and CPK4 (abscisic acid, Zhu et al., 2007) are preferentially regulated by HSFA2. These connections suggest that the HSFA1s and HSFA2 can differentially modulate these diverse signaling pathways despite the HSFs being considered the terminal components of signal transduction (Kotak et al., 2007). Due to the short duration of the single heat treatment performed in this study, we were not able to investigate the synergetic effect of heterooligomers formed by HSFA1s and HSFA2 as shown in tomato (Chan-Schaminet et al., 2009); this direction should be explored in the future.
Although we showed that HSFA2 can promote growth and development in the absence of the HSFA1s (Fig. 2), overexpression of HSFA2 in the wild-type background resulted in dwarfism (Ogawa et al., 2007). Consistently, we also observed that the A2Wt lines showed dwarf phenotype (Supplemental Fig. S4A) that is associated with reduced cell number, but not cell size, in expanded leaves (Supplemental Figure S4, B and C). These observations suggest that HSFA2 acts as a positive regulator in the absence of the HSFA1s but as a negative regulator if HSFA1s are present. In the absence of HSFA1s, it is possible that HSFA2 replaces the function of HSFA1s in growth and development due to the high degree of sequence homology shared between the two classes of HSF. However, it is not clear why a high level of HSFA2 suppresses the growth of the transgenic plants in the presence of the HSFA1s. Previous reports showed that disruption of HSFA2 does not affect growth and development of Arabidopsis plants under nonstress conditions (Nishizawa et al., 2006; Schramm et al., 2006; Charng et al., 2007). However, whether HSFA2 plays a role in regulating growth and development after being induced to a high level by heat stress has not been investigated.
The notion that HSFA2 affects development is supported by the association of HSFA2 overexpression and callus formation after heat stress (Fig. 8). A callus is a mass of dedifferentiated cells often induced in wounded tissues exposed to auxin-rich media (Skoog and Miller, 1957; Birnbaum and Sánchez Alvarado, 2008). It has been shown that heat treatment promotes callus formation in explants (Miyoshi, 1996; Parra-Vega et al., 2013) and that the expression of HSFA2 is up-regulated during the process of callus formation (Che et al., 2002). Interestingly, overexpressing HSFA2 in the wild type did not promote callus formation in heat-stressed seedlings (Fig. 8, compare the A2Wt and A2QK lines) but was shown to enhance formation of calluses in callus-inducing, auxin-rich medium (Ogawa et al., 2007). The phenotypic difference in the promotion of callus formation in HSFA2 overexpressing plants in the wild-type and QK backgrounds suggests that the HSFA1s affect the function of HSFA2. This is consistent with the observation mentioned above, i.e. that the presence or absence of the HSFA1s alters the function of HSFA2 as a regulator of growth and development. Taken together, these findings suggest that HSFA2 plays an important role in growth and development, but its intrinsic role and mechanism of regulation remain to be elucidated in the future.
In Drosophila spp., HSF is required for oogenesis and early larval development (Jedlicka et al., 1997). Absence of HSF1 in mice is not lethal but increases female infertility and prenatal lethality and retards growth (Xiao et al., 1999). Here, we showed that disruption of all four HSFA1s, in addition to impairing growth of young seedlings, increases the chances of seed abortion (Fig. 2E), which is associated with the female gametophyte of the QK mutant (Fig. 2F). These findings indicate a conserved role of HSF in the development of female organs and embryos in plants and animals. It would be of interest to know how HSFA1s control these processes. Of note, a similar seed abortion phenotype was shown in the KO mutant of HSBP, a negative regulator of the HSFA1s (Hsu and Jinn, 2010; Hsu et al., 2010). The seed abortion rate in the HSBP KO mutant is about 35%, higher than the 20% observed in the QK mutant (Fig. 2F). This phenotypic similarity suggests that HSFA1s and HSBP may function in concert in seed development despite their functions in heat stress response being antagonistic (Hsu and Jinn, 2010; Hsu et al., 2010). The small HSPs, HSP17.4 (AT3G46230) and HSP17.6A (AT1G59860), have been shown to be involved in early embryogenesis, and double KO of these genes leads to seed abortion (Dafny-Yelin et al., 2008). Because HSP17.4 and HSP17.6A are the targets of the HSFA1s in heat stress response (Fig. 5; Supplemental Table S1), it is likely that they are the target genes of HSFA1s during seed development as well. Overexpression of HSFA2 in the QK mutant was able to rescue the seed abortion phenotype (Fig. 2, E and F), probably due to its ability to interact with HSBP (Hsu and Jinn, 2010; Hsu et al., 2010), and was as proficient as the HSFA1s in inducing HSP17.4 and HSP17.6A (Fig. 5; Supplemental Table S1).
Overexpression of HSFA2 in Arabidopsis wild-type background was shown to enhance tolerance of seedlings germinating on medium with high salt or mannitol concentrations (Ogawa et al., 2007). However, our results showed that in the absence of HSFA1s, overexpression of HSFA2 does not seem to rescue the tolerance to salt stress and has little effect on osmotic stress tolerance (Fig. 10). This observation suggests that HSFA2 plays a very minor, if any, role in tolerance to these adverse conditions, at least at the stage of germinating young seedling. This finding correlates with the results showing that the hsfa2 null mutant is not more sensitive than the wild type to salt and osmotic stresses (Supplemental Fig. S3). Although hsfa2 mutant showed similar sensitivity to H2O2 as the wild type, overexpression of HSFA2 in QK mutant did restore tolerance to this oxidative agent (Supplemental Fig. S3), suggesting that a high level of HSFA2 could provide protection independent of the HSFA1s, probably by inducing genes involved in redox homeostasis as mentioned above. These results are in good agreement with evidence demonstrated previously that implicates in HSFA2 in oxidative stress response (Li et al., 2005).
In this study, we further investigated the capacity of individual HSFA1s to confer tolerance to salt, osmotic, and oxidative stresses by using the four triple KO mutants. To our surprise, among the four HSFA1s, HSFA1a, the most potent inducer of thermotolerance (Liu et al., 2011), is the least effective inducer of tolerance to salt and osmotic stresses (Fig. 10). By contrast, HSFA1e, which does not confer thermotolerance in the absence of HSFA1a, HSFA1b, and HSFA1d, strongly induces salt and osmotic stress tolerance. HSFA1d seems to be involved in the response to all three stress factors. These results suggest that during evolution, the four members of the HSFA1 group have evolved divergent functions in stress response by subfunctionalization. It is tempting to speculate that the existence of multiple, subfunctionalized HSFA1s are beneficial for plants to cope with combined stresses of varying degrees of severity. This hypothesis awaits testing with vigorous challenges of combined stresses on mutants containing different combinations of HSFA1s. Of note, several monocots, such as rice (Oryza sativa), sorghum (Sorghum bicolor), and Brachypodium distachyon, have a single HSFA1 in their genomes (Scharf et al., 2012). It will therefore be of interest to know whether or how the monocot HSFA1 can mediate multiple stress tolerances.
The finding that the HSFA1s have a role in tolerance to mild heat stress at ambient temperature around 27°C (Fig. 9) was quite unexpected, as this temperature is not linked to heat stress response, which is usually tested at temperatures 10°C to 15°C above that for optimum growth (Lindquist, 1986). It has been previously shown that Arabidopsis plants respond to high ambient temperature by promoting morphological and developmental alterations, such as petiole elongation, in which the basic helix-loop-helix transcription factor PHYTOCHROME-INTERACTING FACTOR4 (PIF4) plays a pivotal role (Koini et al., 2009; Franklin et al., 2011; Kumar et al., 2012; Sun et al., 2012). Because the eTK mutant still showed the phenotype of elongating petioles (Fig. 9) and the pif4 mutant seemed to be viable under this high ambient temperature (Koini et al., 2009), these results suggest that HSFA1a/HSFA1b/HSFA1d and PIF4 trigger two independent pathways for thermotolerance and morphological adjustment, respectively. Our data reveal a new type of thermotolerance, which constitutes thermotolerance diversity orchestrated by the master regulators of heat stress response (Yeh et al., 2012). It remains to be seen how HSFA1a/HSFA1b/HSFA1d regulate transcription reprogramming in this temperature range.
In conclusion, our work reveals common and divergent roles of Arabidopsis class A1 and A2 HSFs in development and response to different stresses. The approach shown here may be applicable to the elucidation of the functions of other Arabidopsis HSFs as well as the posttranslational modifications of HSFA2 recently identified (Cohen-Peer et al., 2010; Evrard et al., 2013).
MATERIALS AND METHODS
Plant Materials and Growth Condition
The mutant lines of QK, aTK, bTK, dTK, eTK, and hsfa2 used in this study were obtained as previously described (Charng et al., 2007; Liu et al., 2011). The normal growth conditions in Murashige and Skoog (MS) medium and soil were also as previously described (Liu et al., 2011).
Generating Transgenic Plants
The complementary DNA of HSFA2 was amplified by RT-PCR using mRNA isolated from heat-stressed samples (37°C, 1 h) of 7-d-old Col-0 wild type as a template. The PCR product was cloned into pCR8/GW/TOPO (Invitrogen) and then subcloned into Gateway overexpression vector pB2GW7 (Karimi et al., 2002) to yield the binary vector pYC125. The construct was then transferred into Agrobacterium tumefaciens GV3101 strain and transformed into QK mutant and the Col-0 wild type by the floral dip method to generate A2QK and A2Wt, respectively. The A2Wt homozygous lines with high levels of HSFA2 were sterile, and only few viable seeds could be obtained. Therefore, the heterozygous line of A2Wt was used for this study.
Evaluating Seed Size, Seed Abortion Rate, and Growth Rate of Young Seedlings
The method of seed size evaluation was as described in (Liu et al., 2011). The development of immature seeds in the ninth and tenth siliques from the top of inflorescences was observed under a stereomicroscope (Olympus), and the seed abortion rates of 15 siliques from five independent plants were scored. The growth rate from the imbibed seed to the two rosette leaves stage was measured as previously described (Liu et al., 2011). The root length of 7-d-old seedlings grown in 0.5× MS medium was calculated using ImageJ software (National Institutes of Health).
Thermotolerance Assays
The thermotolerance assays were performed as previously described with modifications (Charng et al., 2006; Liu et al., 2011). For the BT assay, 7-d-old seedlings were treated for 25 min at 43°C. For short-term acquired thermotolerance (SAT) assay, 7-d-old seedlings were acclimated for 1 h at 37°C, recovered for 2 h at 22°C, and then treated for 155 min at 44°C. For long-term acquired thermotolerance (LAT) assay, 5-d-old seedlings were acclimated for 1 h at 37°C, recovered for 2 d at 22°C, and then treated for 50 min at 44°C. After exposure to these heat stress regimes, the plants were recovered for 14 d at 22°C. For the TMHT assay, 7-d-old seedlings were treated for 5 d at 35°C under a light/dark cycle of 16 h/8 h (120 mmol m–2 s–1) and recovered for 9 d at 22°C. At the end of recovery, photos were taken and the survival rates were evaluated. To test the tolerance to chronic heat stress at 27°C or 29°C, 7-d-old plants grown in 0.5× MS medium plates were placed in a growth chamber set at the indicated temperature for 7 d under a light/dark cycle of 16 h/8 h (120 mmol m–2 s–1), then the phenotypes were recorded. Heat tolerance of 40-d-old plants grown in potted soil was assessed by incubating in a growth chamber set at the indicated temperature and treated for 8 d under a light/dark cycle of 16 h/8 h (120 mmol m–2 s–1). After treatment, the phenotypes were photographed and the elongating length of inflorescences was measured.
Oxidative, Osmotic, and Salt Stress Treatments
The seeds were sterilized and sown on 0.5× MS medium containing 1% (w/v) Suc with 125 mm NaCl and 300 mm mannitol or 0.1% (w/v) Suc with 5 mm H2O2 imbibed at 4°C for 3 d and then grown at 22°C for 18 d before photograph taking and evaluation of the growth phenotype.
Immunoblotting and Quantitative RT-PCR
The methods of protein extraction and immunoblotting were as described previously (Chi et al., 2009). Antibodies against HSFA2, HSP101, HSP90, HSA32, sHSP-CI, and tubulin were also previously described (Charng et al., 2006; Chi et al., 2009; Liu et al., 2011). Total RNA extraction and quantitative RT-PCR were performed as described in Liu et al. (2011). The primers used for RT-PCR are listed in Supplemental Table S2.
Microarray Analysis
Seven-day-old seedlings grown at 22°C on 0.5× MS plates containing 1% (w/v) Suc were first heat-shocked for 1 h at 37°C (Heat-shocked) or left at 22°C (control) and collected for RNA extraction. Total RNA used for microarray analysis was purified by the RNeasy procedure (Qiagen). Examination of the RNA quality and processing of ATH1 GeneChip arrays with 22,810 features (Affymatrix) were performed by Vita Genomics. Two independent biological replicates were processed for the analysis. Assessment of experimental quality and statistical analyses were performed as previously described (Liu et al., 2011). The microarray result, including hierarchical clustering, was analyzed using GeneSpring 11.5.1 (Agilent Technologies). The annotations of the Arabidopsis (Arabidopsis thaliana) genes were downloaded from The Arabidopsis Information Resource (http://www.arabidopsis.org). The microarray data can be accessed in the Gene Expression Omnibus at the National Center for Biotechnology Information (accession no. GSE44655).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbersHSFA1a (At4g17750), HSFA1b (At5g16820), HSFA1d (At1g32330), HSFA1e (At3g02990), and HSFA2 (At2g26150). The accession numbers of other genes mentioned in this article can be found in Supplemental Table S1.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Constitutive expression of HSFA2 conferred a high level of thermotolerance in the absence of HSFA1s.
Supplemental Figure S2. HSFA1a/b/d are required for tolerance to chronic heat stress at temperature above 27°C.
Supplemental Figure S3. HSFA2 is not essential for growth at 29°C, or tolerance to oxidative, salt, or osmotic stresses.
Supplemental Figure S4. Constitutive expression of HSFA2 in wild-type background caused dwarfism with small rosette leaves.
Supplemental Table S1. List of HSFA1s-preferring, no preference, and HSFA2-preferring genes shown in Figure 5.
Supplemental Table S2. Primers used in this study.
Acknowledgments
We thank Jen Sheen for critical reading of the manuscript, Yun-Ru Lai for technical support, Miranda Loney for English editing, and two anonymous reviewers for constructive comments.
Glossary
- KO
knockout
- Col-0
ecotype Columbia
- RT
reverse transcription
- MS
Murashige and Skoog
- BT
basal thermotolerance
- SAT
short-term acquired thermotolerance
- LAT
long-term acquired thermotolerance
- TMHT
thermotolerance to moderately high temperature
References
- Åkerfelt M, Morimoto RI, Sistonen L. (2010) Heat shock factors: integrators of cell stress, development and lifespan. Nat Rev Mol Cell Biol 11: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. (2011) Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem 80: 1089–1115 [DOI] [PubMed] [Google Scholar]
- Birnbaum KD, Sánchez Alvarado A. (2008) Slicing across kingdoms: regeneration in plants and animals. Cell 132: 697–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH. (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Rechenmann C, Fleming AJ. (2008) Conditional repression of AUXIN BINDING PROTEIN1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20: 2746–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W, Wunderlich M, Schöffl F. (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Chan-Schaminet KY, Baniwal SK, Bublak D, Nover L, Scharf K-D. (2009) Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J Biol Chem 284: 20848–20857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Chi WT, Wang CN, Chang SH, Wang TT. (2007) A heat-inducible transcription factor, HsfA2, is required for extension of acquired thermotolerance in Arabidopsis. Plant Physiol 143: 251–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charng YY, Liu HC, Liu NY, Hsu FC, Ko SS. (2006) Arabidopsis Hsa32, a novel heat shock protein, is essential for acquired thermotolerance during long recovery after acclimation. Plant Physiol 140: 1297–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH. (2002) Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2771–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi WT, Fung RWM, Liu HC, Hsu CC, Charng YY. (2009) Temperature-induced lipocalin is required for basal and acquired thermotolerance in Arabidopsis. Plant Cell Environ 32: 917–927 [DOI] [PubMed] [Google Scholar]
- Cohen-Peer R, Schuster S, Meiri D, Breiman A, Avni A. (2010) Sumoylation of Arabidopsis heat shock factor A2 (HsfA2) modifies its activity during acquired thermotolerance. Plant Mol Biol 74: 33–45 [DOI] [PubMed] [Google Scholar]
- Dafny-Yelin M, Tzfira T, Vainstein A, Adam Z. (2008) Non-redundant functions of sHSP-CIs in acquired thermotolerance and their role in early seed development in Arabidopsis. Plant Mol Biol 67: 363–373 [DOI] [PubMed] [Google Scholar]
- Dessau M, Halimi Y, Erez T, Chomsky-Hecht O, Chamovitz DA, Hirsch JA. (2008) The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell 20: 2815–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H, Zhen Z, Peng J, Chang L, Gong Q, Wang NN. (2011) Loss of ACS7 confers abiotic stress tolerance by modulating ABA sensitivity and accumulation in Arabidopsis. J Exp Bot 62: 4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrard A, Kumar M, Lecourieux D, Lucks J, von Koskull-Döring P, Hirt H. (2013) Regulation of the heat stress response in Arabidopsis by MPK6-targeted phosphorylation of the heat stress factor HsfA2. PeerJ 1: e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA. (2009) Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol 12: 63–68 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Rogowsky P, Nover L, Scanlon MJ. (2006) The maize heat shock factor-binding protein paralogs EMP2 and HSBP2 interact non-redundantly with specific heat shock factors. Planta 224: 42–52 [DOI] [PubMed] [Google Scholar]
- Gao H, Brandizzi F, Benning C, Larkin RM. (2008) A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA 105: 16398–16403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Bublak D, Schleiff E, Scharf K-D. (2011) Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 23: 741–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haselier A, Akbari H, Weth A, Baumgartner W, Frentzen M. (2010) Two closely related genes of Arabidopsis encode plastidial cytidinediphosphate diacylglycerol synthases essential for photoautotrophic growth. Plant Physiol 153: 1372–1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz D, Döring P, Bonzelius F, Winkelhaus S, Nover L. (2001) The balance of nuclear import and export determines the intracellular distribution and function of tomato heat stress transcription factor HsfA2. Mol Cell Biol 21: 1759–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. (2010) Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Hsu S-F, Jinn T-L. (2010) AtHSBP functions in seed development and the motif is required for subcellular localization and interaction with AtHSFs. Plant Signal Behav 5: 1042–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-F, Lai H-C, Jinn T-L. (2010) Cytosol-localized heat shock factor-binding protein, AtHSBP, functions as a negative regulator of heat shock response by translocation to the nucleus and is required for seed development in Arabidopsis. Plant Physiol 153: 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husar S, Berthiller F, Fujioka S, Rozhon W, Khan M, Kalaivanan F, Elias L, Higgins GS, Li Y, Schuhmacher R, et al. (2011) Overexpression of the UGT73C6 alters brassinosteroid glucoside formation in Arabidopsis thaliana. BMC Plant Biol 11: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Mitsuda N, Ohme-Takagi M. (2011) Arabidopsis HsfB1 and HsfB2b act as repressors of the expression of heat-inducible Hsfs but positively regulate the acquired thermotolerance. Plant Physiol 157: 1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jedlicka P, Mortin MA, Wu C. (1997) Multiple functions of Drosophila heat shock transcription factor in vivo. EMBO J 16: 2452–2462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang CH, Jung WY, Kang YH, Kim JY, Kim DG, Jeong JC, Baek DW, Jin JB, Lee JY, Kim MO, et al. (2006) AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ 13: 84–95 [DOI] [PubMed] [Google Scholar]
- Karimi M, Inzé D, Depicker A. (2002) GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kelly AA, Froehlich JE, Dörmann P. (2003) Disruption of the two digalactosyldiacylglycerol synthase genes DGD1 and DGD2 in Arabidopsis reveals the existence of an additional enzyme of galactolipid synthesis. Plant Cell 15: 2694–2706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kotak S, Larkindale J, Lee U, von Koskull-Döring P, Vierling E, Scharf K-D. (2007) Complexity of the heat stress response in plants. Curr Opin Plant Biol 10: 310–316 [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P. (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA. (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. (2010) H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. (2012) The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res 40: D1202–D1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkindale J, Vierling E. (2008) Core genome responses involved in acclimation to high temperature. Plant Physiol 146: 748–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Chen Q, Gao X, Qi B, Chen N, Xu S, Chen J, Wang X. (2005) AtHsfA2 modulates expression of stress responsive genes and enhances tolerance to heat and oxidative stress in Arabidopsis. Sci China C Life Sci 48: 540–550 [DOI] [PubMed] [Google Scholar]
- Li M, Berendzen KW, Schöffl F. (2010) Promoter specificity and interactions between early and late Arabidopsis heat shock factors. Plant Mol Biol 73: 559–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S. (1986) The heat-shock response. Annu Rev Biochem 55: 1151–1191 [DOI] [PubMed] [Google Scholar]
- Liu HC, Charng YY. (2012) Acquired thermotolerance independent of heat shock factor A1 (HsfA1), the master regulator of the heat stress response. Plant Signal Behav 7: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Liao HT, Charng YY. (2011) The role of class A1 heat shock factors (HSFA1s) in response to heat and other stresses in Arabidopsis. Plant Cell Environ 34: 738–751 [DOI] [PubMed] [Google Scholar]
- Liu J, Sun N, Liu M, Liu J, Du B, Wang X, Qi X. (2013) An autoregulatory loop controlling Arabidopsis HsfA2 expression: role of heat shock-induced alternative splicing. Plant Physiol 162: 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann C, Eggers-Schumacher G, Wunderlich M, Schöffl F. (2004) Two different heat shock transcription factors regulate immediate early expression of stress genes in Arabidopsis. Mol Genet Genomics 271: 11–21 [DOI] [PubMed] [Google Scholar]
- Meiri D, Breiman A. (2009) Arabidopsis ROF1 (FKBP62) modulates thermotolerance by interacting with HSP90.1 and affecting the accumulation of HsfA2-regulated sHSPs. Plant J 59: 387–399 [DOI] [PubMed] [Google Scholar]
- Mishra SK, Tripp J, Winkelhaus S, Tschiersch B, Theres K, Nover L, Scharf K-D. (2002) In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Genes Dev 16: 1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K. (1996) Callus induction and plantlet formation through culture of isolated microspores of eggplant (Solanum melongena L.). Plant Cell Rep 15: 391–395 [DOI] [PubMed] [Google Scholar]
- Moellering ER, Benning C. (2010) Phosphate regulation of lipid biosynthesis in Arabidopsis is independent of the mitochondrial outer membrane DGS1 complex. Plant Physiol 152: 1951–1959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto RI. (1998) Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev 12: 3788–3796 [DOI] [PubMed] [Google Scholar]
- Nishizawa A, Yabuta Y, Yoshida E, Maruta T, Yoshimura K, Shigeoka S. (2006) Arabidopsis heat shock transcription factor A2 as a key regulator in response to several types of environmental stress. Plant J 48: 535–547 [DOI] [PubMed] [Google Scholar]
- Nishizawa-Yokoi A, Nosaka R, Hayashi H, Tainaka H, Maruta T, Tamoi M, Ikeda M, Ohme-Takagi M, Yoshimura K, Yabuta Y, et al. (2011) HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol 52: 933–945 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf K-D. (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa D, Yamaguchi K, Nishiuchi T. (2007) High-level overexpression of the Arabidopsis HsfA2 gene confers not only increased thermotolerance but also salt/osmotic stress tolerance and enhanced callus growth. J Exp Bot 58: 3373–3383 [DOI] [PubMed] [Google Scholar]
- Parra-Vega V, Renau-Morata B, Sifres A, Seguí-Simarro J. (2013) Stress treatments and in vitro culture conditions influence microspore embryogenesis and growth of callus from anther walls of sweet pepper (Capsicum annuum L.). Plant Cell Tissue Organ Cult 112: 353–360 [Google Scholar]
- Pirkkala L, Nykänen P, Sistonen L. (2001) Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J 15: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, Pogson BJ. (2006) A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant Cell Environ 29: 269–281 [DOI] [PubMed] [Google Scholar]
- Saibo NJM, Lourenço T, Oliveira MM. (2009) Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot (Lond) 103: 609–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Qin F, Osakabe Y, Shinozaki K, Yamaguchi-Shinozaki K. (2006) Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc Natl Acad Sci USA 103: 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos AP, Serra T, Figueiredo DD, Barros P, Lourenço T, Chander S, Oliveira MM, Saibo NJM. (2011) Transcription regulation of abiotic stress responses in rice: a combined action of transcription factors and epigenetic mechanisms. OMICS 15: 839–857 [DOI] [PubMed] [Google Scholar]
- Satyal SH, Chen D, Fox SG, Kramer JM, Morimoto RI. (1998) Negative regulation of the heat shock transcriptional response by HSBP1. Genes Dev 12: 1962–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf K-D, Berberich T, Ebersberger I, Nover L. (2012) The plant heat stress transcription factor (Hsf) family: structure, function and evolution. Biochim Biophys Acta 1819: 104–119 [DOI] [PubMed] [Google Scholar]
- Scharf K-D, Heider H, Höhfeld I, Lyck R, Schmidt E, Nover L. (1998) The tomato Hsf system: HsfA2 needs interaction with HsfA1 for efficient nuclear import and may be localized in cytoplasmic heat stress granules. Mol Cell Biol 18: 2240–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm F, Ganguli A, Kiehlmann E, Englich G, Walch D, von Koskull-Döring P. (2006) The heat stress transcription factor HsfA2 serves as a regulatory amplifier of a subset of genes in the heat stress response in Arabidopsis. Plant Mol Biol 60: 759–772 [DOI] [PubMed] [Google Scholar]
- Schramm F, Larkindale J, Kiehlmann E, Ganguli A, Englich G, Vierling E, von Koskull-Döring P. (2008) A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J 53: 264–274 [DOI] [PubMed] [Google Scholar]
- Simpson PJ, Tantitadapitak C, Reed AM, Mather OC, Bunce CM, White SA, Ride JP. (2009) Characterization of two novel aldo-keto reductases from Arabidopsis: expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. J Mol Biol 392: 465–480 [DOI] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Oñate-Sánchez L. (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5: 430–436 [DOI] [PubMed] [Google Scholar]
- Skoog F, Miller CO. (1957) Chemical regulation of growth and organ formation in plant tissues cultured in vitro. Symp Soc Exp Biol 11: 118–130 [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. (2012) PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet 8: e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Bajad S, Shuman J, Shulaev V, Mittler R. (2008) The transcriptional co-activator MBF1c is a key regulator of thermotolerance in Arabidopsis thaliana. J Biol Chem 283: 9269–9275 [DOI] [PubMed] [Google Scholar]
- Valluru R, Van den Ende W. (2011) Myo-inositol and beyond—emerging networks under stress. Plant Sci 181: 387–400 [DOI] [PubMed] [Google Scholar]
- Voellmy R. (2004) On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones 9: 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Koskull-Döring P, Scharf K-D, Nover L. (2007) The diversity of plant heat stress transcription factors. Trends Plant Sci 12: 452–457 [DOI] [PubMed] [Google Scholar]
- Wu C. (1995) Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol 11: 441–469 [DOI] [PubMed] [Google Scholar]
- Wunderlich M, Doll J, Busch W, Kleindt C, Lohmann C, Schöffl F. (2007) Heat shock factors: regulators of early and late functions in plant stress response. Plant Stress 1: 16–22 [Google Scholar]
- Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. (1999) HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J 18: 5943–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-M, Huang D-Y, Chiu J-F, Lau ATY. (2012) Post-translational modification of human heat shock factors and their functions: a recent update by proteomic approach. J Proteome Res 11: 2625–2634 [DOI] [PubMed] [Google Scholar]
- Yamada K, Fukao Y, Hayashi M, Fukazawa M, Suzuki I, Nishimura M. (2007) Cytosolic HSP90 regulates the heat shock response that is responsible for heat acclimation in Arabidopsis thaliana. J Biol Chem 282: 37794–37804 [DOI] [PubMed] [Google Scholar]
- Yeh C-H, Kaplinsky NJ, Hu C, Charng YY. (2012) Some like it hot, some like it warm: phenotyping to explore thermotolerance diversity. Plant Sci 195: 10–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Ohama N, Nakajima J, Kidokoro S, Mizoi J, Nakashima K, Maruyama K, Kim J-M, Seki M, Todaka D, et al. (2011) Arabidopsis HsfA1 transcription factors function as the main positive regulators in heat shock-responsive gene expression. Mol Genet Genomics 286: 321–332 [DOI] [PubMed] [Google Scholar]
- Yoshida T, Sakuma Y, Todaka D, Maruyama K, Qin F, Mizoi J, Kidokoro S, Fujita Y, Shinozaki K, Yamaguchi-Shinozaki K. (2008) Functional analysis of an Arabidopsis heat-shock transcription factor HsfA3 in the transcriptional cascade downstream of the DREB2A stress-regulatory system. Biochem Biophys Res Commun 368: 515–521 [DOI] [PubMed] [Google Scholar]
- Zhao L, Katavic V, Li F, Haughn GW, Kunst L. (2010) Insertional mutant analysis reveals that long-chain acyl-CoA synthetase 1 (LACS1), but not LACS8, functionally overlaps with LACS9 in Arabidopsis seed oil biosynthesis. Plant J 64: 1048–1058 [DOI] [PubMed] [Google Scholar]
- Zhu S-Y, Yu X-C, Wang X-J, Zhao R, Li Y, Fan R-C, Shang Y, Du S-Y, Wang X-F, Wu F-Q, et al. (2007) Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. Plant Cell 19: 3019–3036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang Z, Jing Y, Wang L, Liu X, Liu Y, Deng X. (2009) Ectopic over-expression of BhHsf1, a heat shock factor from the resurrection plant Boea hygrometrica, leads to increased thermotolerance and retarded growth in transgenic Arabidopsis and tobacco. Plant Mol Biol 71: 451–467 [DOI] [PubMed] [Google Scholar]