Three bHLH-type transcription factors negatively regulate jasmonate responses.
Abstract
Jasmonates regulate transcriptional reprogramming during growth, development, and defense responses. Jasmonoyl-isoleucine, an amino acid conjugate of jasmonic acid (JA), is perceived by the protein complex composed of the F-box protein CORONATINE INSENSITIVE1 (COI1) and JASMONATE ZIM DOMAIN (JAZ) proteins, leading to the ubiquitin-dependent degradation of JAZ proteins. This activates basic helix-loop-helix-type MYC transcription factors to regulate JA-responsive genes. Here, we show that the expression of genes encoding other basic helix-loop-helix transcription factors, JASMONATE ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3, is positively regulated in a COI1- and MYC2-dependent manner in Arabidopsis (Arabidopsis thaliana). However, contrary to myc2, the jam1jam2jam3 triple mutant exhibited shorter roots when treated with methyl jasmonate (MJ), indicating enhanced responsiveness to JA. Our genome-wide expression analyses revealed that key jasmonate metabolic genes as well as a set of genes encoding transcription factors that regulate the JA-responsive metabolic genes are negatively regulated by JAMs after MJ treatment. Consistently, loss of JAM genes resulted in higher accumulation of anthocyanin in MJ-treated plants as well as higher accumulation of JA and 12-hydroxyjasmonic acid in wounded plants. These results show that JAMs negatively regulate the JA responses in a manner that is mostly antagonistic to MYC2.
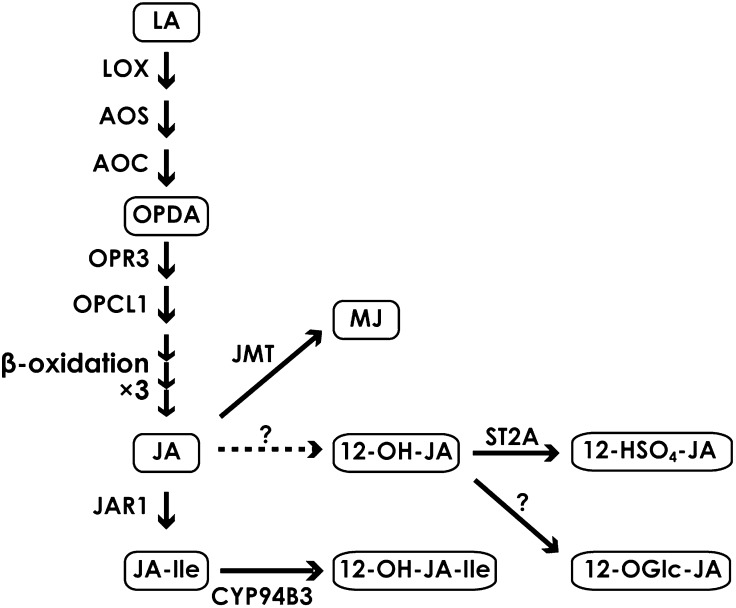
Jasmonic acid (JA) and its cyclic precursors and derivatives, so called jasmonates, are widely distributed in the plant kingdom. Jasmonates regulate a wide spectrum of plant processes, such as growth and development, as well as defense systems against biotic and abiotic stresses (Schilmiller and Howe, 2005; Browse and Howe, 2008; Zhang and Turner, 2008; Browse, 2009; Reinbothe et al., 2009). JA and jasmonoyl-l-isoleucine (JA-Ile), both derived from α-linolenic acid, accumulate in response to biotic and abiotic stress (Fig. 1; Wasternack, 2007; Gfeller et al., 2010). The accumulation of JA-Ile, which is the ligand for the CORONATINE INSENSITIVE1 (COI1)-JASMONATE ZIM DOMAIN (JAZ) receptor complex (Chini et al., 2007; Thines et al., 2007; Sheard et al., 2010), is followed by the generation of 12-hydroxyjasmonoyl-l-isoleucine (12-OH-JA-Ile), which has a lesser binding ability to the complex (Kitaoka et al., 2011; Koo et al., 2011; Heitz et al., 2012). 12-Hydroxyjasmonic acid (12-OH-JA) and 12-hydroxyjasmonic acid glucoside, on the other hand, have been described as inducers of tuber formation (Yoshihara et al., 1989). 12-OH-JA is also synthesized by the jasmonate pathway and has been detected in a wide range of plant species, especially after wounding (Glauser et al., 2008; Miersch et al., 2008). Biochemical pathways of JA-related compounds will be referred to as the jasmonate metabolic pathway in this paper (Fig. 1).
Figure 1.
Jasmonate metabolic pathway. Abbreviations not defined in the text are as follows: LOX, lipoxygenase; AOC, allene oxide cyclase; OPR3, OPDA reductase3; OPCL1, 3-oxo-2-[2′-pentenyl]-cyclopentane-1-octanoic acid (OPC-8:0) CoA ligase; CYP94B3, JA-Ile-12-hydroxylase; JMT, JA carboxyl methyltransferase; LA, linolenic acid; 12-HSO4-JA, 12-hydroxyjasmonic acid sulfate; 12-OGlc-JA, 12-OH-JA glucoside.
Increased JA and JA-Ile levels lead to transcriptional reprogramming. Upon the perception of JA-Ile by the COI-JAZ receptor complex, SCF-type E3 ubiquitin ligase (SCFCOI1) presumably ubiquitinates and degrades JAZ proteins via the 26S proteasome system (Chini et al., 2007; Thines et al., 2007; Staswick, 2008). This is followed by derepression of MYC2, a basic helix-loop-helix (bHLH) transcription factor that plays a central role in JA signaling, resulting in transcriptional activation of downstream target genes (Lorenzo et al., 2004, Chico et al., 2008; Katsir et al., 2008). However, the JA-insensitive phenotype in the myc2 mutant is weaker than that in coi1-1 (Xie et al., 1998; Lorenzo et al., 2004), suggesting the existence of other redundant factors. Indeed, MYC3 and MYC4 are the close homologs of MYC2, and these three transcription factors have a redundant function as the activators of JA-dependent defense responses (Cheng et al., 2011; Fernández-Calvo et al., 2011; Niu et al., 2011). MYC2, as well as MYC3 and MYC4, regulate two distinct JA-inducible gene expression patterns represented either by VSP2 or PDF1.2 (Lorenzo et al., 2004; Fernández-Calvo et al., 2011; Niu et al., 2011). For VSP2 expression, MYC2 is a positive regulator, but for PDF1.2, MYC2 plays a negative role (Lorenzo et al., 2004). MYC2 directly binds to the promoter region of ANAC019 that is a positive regulator of VSP2 expression (Bu et al., 2008; Zheng et al., 2012). In contrast, PDF1.2 expression is positively regulated by ERF1 and its homolog ORA59, both of which belong to the AP2/ERF domain family and are negatively regulated by MYC2 (Solano et al., 1998; Dombrecht et al., 2007; Pré et al., 2008; Zarei et al., 2011). Thus, the transcriptional cascade of MYC2 with its downstream transcription factors is important to regulate branched pathways downstream of JA-Ile perception.
In addition to VSP2 and PDF1.2 expression, JA (or often methyl jasmonate [MJ]) treatments simultaneously activate a number of biosynthetic pathways such as for Trp, indole glucosinolate, Ser, Cys, glutathione, ascorbate, anthocyanin, as well as jasmonates (Sasaki et al., 2001; Devoto et al., 2005; Sasaki-Sekimoto et al., 2005; Dombrecht et al., 2007; Pauwels et al., 2008; Shan et al., 2009). During this process, the expression patterns of the metabolic genes are tightly correlated with those of the corresponding regulatory transcription factors. For example, ATR1/MYB34 is a positive regulator of both Trp and indole glucosinolate biosynthetic genes, and the expression of those genes is induced by MJ treatments (Bender and Fink, 1998; Taki et al., 2005). MYB29 is a positive regulator of aliphatic glucosinolate biosynthetic genes, and their expression of genes is also induced by MJ treatments (Hirai et al., 2007). PAP1 encodes a MYB-type transcription factor that positively regulates anthocyanin biosynthesis (Borevitz et al., 2000), and its expression profile is closely correlated with that of anthocyanin biosynthetic genes such as DFR/TT3 (At5g42800), UGT79B1 (At5g54060), AAT (At3g29590), and GSTF12/TT19 (At5g17220; ATTED-II; Obayashi et al., 2011). Consistently, overexpression of PAP1 results in the simultaneous induction of anthocyanin biosynthetic genes (Tohge et al., 2005). These data also indicate that coexpression analysis is an efficient way to identify functionally correlated gene groups, including both genes encoding transcription factors and metabolic genes.
To identify novel regulators of the JA responses, we performed a gene network analysis using the expression profiles of JA-responsive genes. We found a bHLH-type transcription factor that shows high similarity to MYC2, and its expression pattern is tightly correlated with that of JA-responsive genes. We designated this bHLH-type transcription factor JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1). Arabidopsis (Arabidopsis thaliana) contains two close JAM1 homologs, which we designated as JAM2 and JAM3. The expression of JAM1, JAM2, and JAM3 genes is down-regulated in myc2 by MJ treatments. However, unlike myc2, the jam1jam2jam3 triple mutants show enhanced JA response phenotypes, such as shorter roots, higher accumulation of JA, 12-OH-JA, and anthocyanin, and higher expression of transcription factors that regulate JA-responsive metabolic pathways. Furthermore, most of phenotypes in the jam1jam2jam3 triple mutants are attenuated by the myc2 mutation. Thus, we propose that JAM1, JAM2, and JAM3 are novel factors that function mostly antagonistically to MYC2 in JA signaling, regulating various metabolic pathways in Arabidopsis.
RESULTS
Network Analysis of JA-Responsive Genes Identified JAM Genes
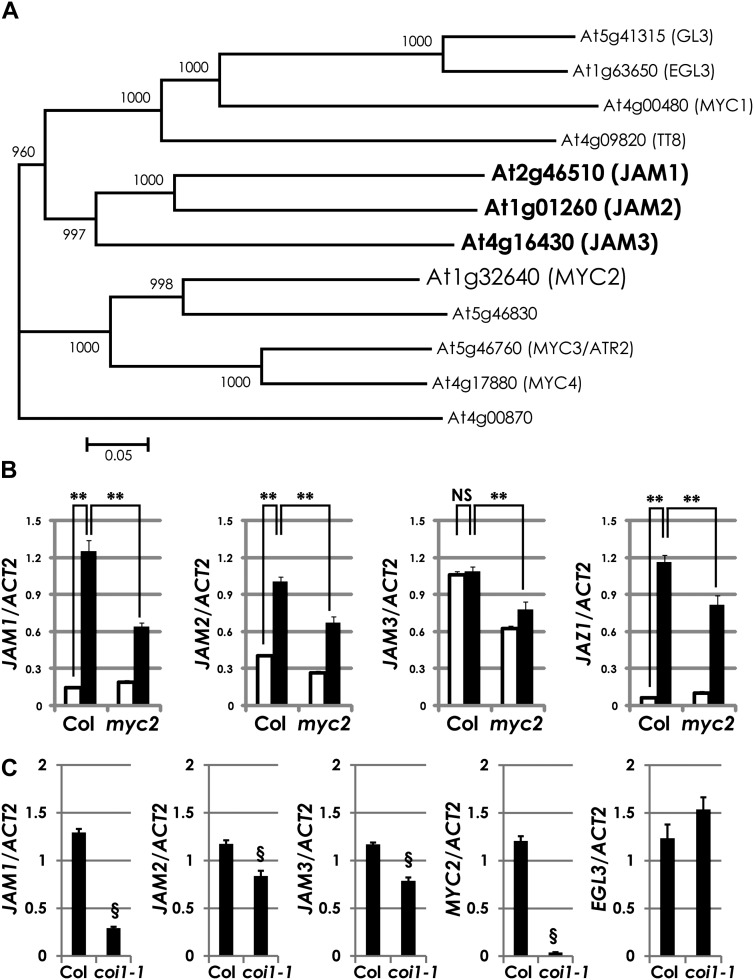
To identify new transcription factors that regulate JA signaling, we constructed coexpression gene networks with 2,220 JA-responsive genes (see “Materials and Methods”). For this analysis, we combined our previous array data (Taki et al., 2005) and the AtGenExpress data on stress (Kilian et al., 2007). Among the 2,220 genes, 233 genes were clustered into 11 groups based on their characteristic expression patterns (Supplemental Table S1). In this study, we focused on cluster 7, which includes 128 early and strong JA- and wound-responsive genes (Supplemental Fig. S1). MYC2 was included in cluster 7, consistent with its rapid induction after MJ treatments (Boter et al., 2004; Lorenzo et al., 2004). Hierarchical clustering of the 128 genes in cluster 7 revealed more detailed correlation of expression profiles. We found that JA biosynthetic genes OPR3 and OPCL1, which were previously shown to be expressed in a similar fashion to MYC2 (Sasaki et al., 2001; Koo et al., 2006), were in the same clade with MYC2, confirming that genes were properly clustered in this analysis. We also found that a gene encoding a bHLH transcription factor was in the nearest neighbor clade with JAZ genes (Supplemental Fig. S1). As the product of this gene shares 30% amino acid identity with MYC2, we designated this gene as JAM1 (At2g46510) and characterized it in detail.
Among the 133 bHLH transcription factors in Arabidopsis, both MYC2 and JAM1 are included in the group III bHLH proteins (Fig. 2A; Heim et al., 2003). MYC2 homologs MYC3 and MYC4 belong to the same clade as MYC2. On the other hand, GL3, EGL3, and TT8, which are involved in the regulation of flavonoid biosynthesis, are in another clade (Zhang et al., 2003). As JAM1 and its two closest homologs form a small clade, we designated the homologs JAM2 (At1g01260) and JAM3 (At4g16430). JAM2 and JAM3 are 47% and 34% identical to JAM1 at the amino acid level, respectively.
Figure 2.
COI1- and MYC2-dependent expression of JAM genes. A, A phylogenetic tree of JAMs and related bHLH proteins included in group III d to f of Arabidopsis. We performed ClustalW alignment (1,000 bootstrap runs) using whole proteins. B, Relative transcript levels of JAM1, JAM2, JAM3, and JAZ1 in 50 μm MJ-treated Col-0 and myc2 plants. Total RNA was isolated from 7-d-old plants, which were grown in liquid GM with 1% Suc. Plants were treated with 50 μm MJ for 1 h. The JAM1, JAM2, JAM3, and JAZ1 transcript levels were normalized to the expression of ACTIN2 measured in the same samples. Average data from three independent RNAs with error bars (se) are presented. **P < 0.01, NS = not significant (P > 0.05), by Tukey-Kramer multiple comparison test. White bars represent mock treatment, and black bars represent 50 μm MJ treatment. C, Relative transcript levels of JAM1, JAM2, JAM3, MYC2, and EGL3 in MJ-treated Col-0 and coi1-1 plants. Total RNA was isolated from 2-week-old plants, which were grown on GM containing 1% Suc and 50 μm MJ. The JAM1, JAM2, JAM3, MYC2, and EGL3 transcript levels were normalized to the expression of ACTIN2 measured in the same samples. Note that JAM expression data in coi1-1 without MJ were not provided due to no clear phenotype to distinguish coi1-1 homozygous plants in the sample population. Average data from three independent RNAs with error bars (se) are presented. §P < 0.01, by two-tailed Student’s t test.
As JAM1 was originally selected as a JA-responsive gene, we confirmed the gene expression profiles of JAM genes in wild-type (Columbia-0 [Col-0]) seedlings upon 50 μm MJ treatment. Consistent with our array data, the expression of JAM1 and JAM2 was clearly induced by the MJ treatment (Fig. 2B). In this experiment, JAZ1 expression levels were also increased by the MJ treatment, as reported previously (Pauwels et al., 2010). However, the expression levels of JAM3 in the MJ-treated plants were comparable to those of the mock treatments. Similar to JAZ1, the expression levels of JAM1, JAM2, and JAM3 were decreased in myc2. In addition, the expression levels of JAM1, JAM2, and JAM3 were also decreased in coi1-1 (Fig. 2C; Xie et al., 1998). As reported previously, EGL3 (At1g63650), which encodes a homolog of JAMs, shows coi1-independent gene expression (Qi et al., 2011). Therefore, we used EGL3 as a marker gene of constitutive expression in coi1 (Fig. 2C). These results indicate that the expression of JAM genes is positively regulated by COI1 and MYC2.
The jam1jam2jam3 Triple Mutant Showed Increased JA Responses
To elucidate the biological functions of JAM transcription factors, we obtained transferred DNA (T-DNA) insertion lines and confirmed the insertion sites by PCR and sequence analyses (Supplemental Fig. S2A). We did not observe the expression of the JAM1 coding region in jam1-1 and jam1-2 (Supplemental Fig. S2B). Similarly, we did not observe JAM3 expression in jam3-1 and jam3-2 (Supplemental Fig. S2C). Thus, we considered jam1-1, jam1-2, jam3-1, and jam3-2 as knockout mutants. We found a T-DNA insertion in the noncoding region of JAM2, and quantitative real-time (qRT)-PCR analysis revealed that JAM2 transcript levels were lower than those in the wild type upon MJ treatments (Supplemental Fig. S2D). Thus, we considered that this mutant is likely to be a knockdown mutant and designated it as jam2-1.
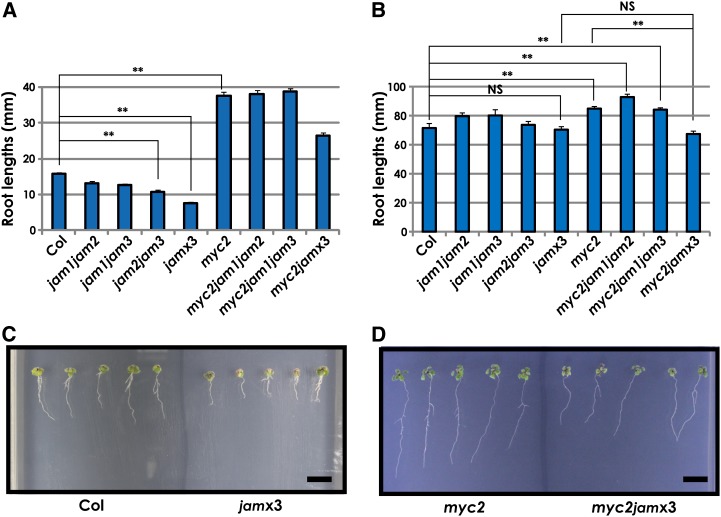
To investigate MJ sensitivity, we measured the root lengths of jam1-1, jam1-2, jam2-1, jam3-1, and jam3-2 plants grown on germination medium (GM) with or without 50 μm MJ for 2 weeks. The roots of jam3-2 grown on MJ-containing medium were slightly shorter than those of the wild type; however, the root lengths of other lines were almost comparable to the wild type (Supplemental Fig. S3, A–C). We also measured the root lengths of jam mutants in the absence of MJ and confirmed that their root lengths are almost comparable to the wild type (Supplemental Fig. S3D). As JAM1 and JAM2 exhibited similar MJ-inducible expression profiles, we hypothesized that JAM genes have redundant functions. Then, we crossed the jam single mutant lines jam1-2, jam2-1, and jam3-2 to produce jam double and triple mutants (Supplemental Fig. S2A). The jam1jam2jam3 triple mutants (jamx3 in this paper) exhibited shorter roots compared with the wild type when grown on 50 μm MJ-containing medium but not without MJ (Fig. 3, A–C). jam1jam2, jam1jam3, and jam2jam3 exhibited intermediate root lengths between wild-type and jamx3 plants (Fig. 3A). In this assay, the roots of myc2 were longer than those of the wild type, consistent with a previous report (Fig. 3A; Lorenzo et al., 2004). The myc2 phenotype was attenuated in the myc2jamx3 quadruple mutants, but no obvious effects were seen in the myc2jam1jam2 and myc2jam1jam3 triple mutants (Fig. 3, A and D), indicating that JAM1, JAM2, and JAM3 have redundant functions that are antagonistic to MYC2 in JA responses.
Figure 3.
Increased MJ responses in jamx3. A, Root lengths of Col-0, jam1jam2, jam1jam3, jam2jam3, jamx3, myc2, myc2jam1jam2, myc2jam1jam3, and myc2jamx3 seedlings grown on 1% Suc and 50 μm MJ-containing medium. Averaged primary root lengths are shown as means ± se for n = 107, 22, 13, 32, 35, 33, 23, 48, and 40 plants, respectively. (myc2, myc2-2; jam1, jam1-2; jam2, jam2-1; jam3, jam3-2.) **P < 0.01, by Tukey-Kramer multiple comparison test. B, Root lengths of Col-0, jam1jam2, jam1jam3, jam2jam3, jamx3, myc2, myc2jam1jam2, myc2jam1jam3, and myc2jamx3 seedlings grown on GM containing 1% Suc. Averaged primary root lengths are shown as means ± se for n = 8, 12, 4, 14, 12, 12, 16, 14, and 10 plants, respectively. **P < 0.01, NS = not significant (P > 0.05), by Tukey-Kramer multiple comparison test. C, Photographs of Col-0 and jamx3 mutant seedlings grown for 2 weeks on GM containing 1% Suc and 50 μm MJ. Bar = 10 mm. D, Photographs of myc2 and myc2jamx3 mutant seedlings grown for 2 weeks on GM containing 1% Suc and 50 μm MJ. Bar = 10 mm.
Identification of JAM Targets
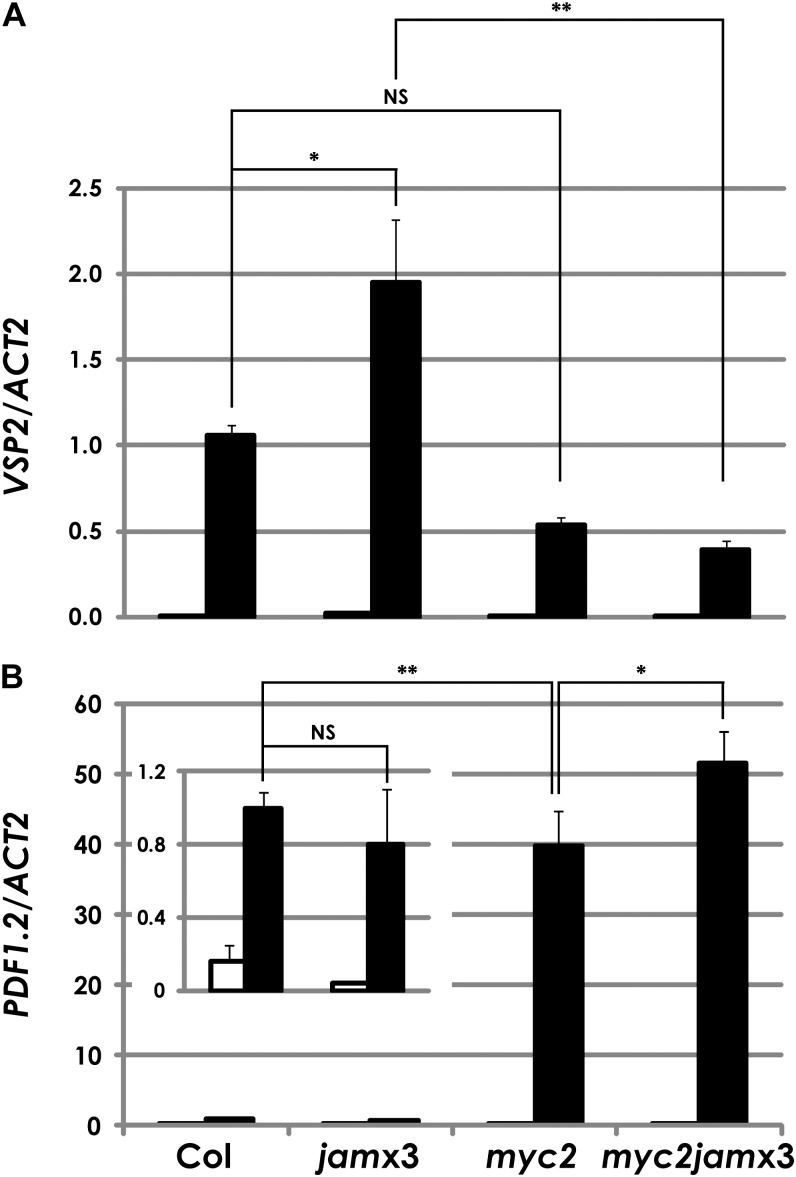
Next, we investigated the effects of the jam mutations on the expression patterns of JA-responsive marker genes. VSP2 expression was significantly increased in MJ-treated jamx3 plants. The reduction of the mean expression level of VSP2 in myc2 was not statistically significant in our experimental condition (GM with 1% Suc; see “Materials and Methods”), but the expression in jamx3 was attenuated by the myc2 mutation (Fig. 4A). On the other hand, PDF1.2 expression levels in MJ-treated jamx3 plants were almost comparable to the wild type (Fig. 4B). These data suggest that JAM genes are the negative regulators of VSP2 expression but not of PDF1.2.
Figure 4.
qRT-PCR analysis of VSP2 and PDF1.2 in Col-0, jamx3, myc2, and myc2jamx3 plants. VSP2 (A) and PDF1.2 (B) transcript levels were normalized to the expression of ACTIN2 measured in the same samples. For this analysis, total RNA was isolated from 7-d-old plants that were grown in liquid GM with 1% Suc and treated with mock (white bars) or 50 μm MJ (black bars) for 24 h. Results shown are means ± se for three biologically independent RNAs. **P < 0.01, *P < 0.05, NS = not significant (P > 0.05), by Tukey-Kramer multiple comparison test.
To identify other downstream target genes of JAM1, JAM2, and JAM3, we compared gene expression profiles between wild-type and jamx3 plants treated with 50 μm MJ or mock for 1 h using GeneChip. Supplemental Figure S4 represents scatterplots of “averaged intensities” (see “Materials and Methods”) from 50 μm MJ-treated wild-type and jamx3 plants. We hypothesized that the MJ-inducible expression of JAM downstream targets will be enhanced in jamx3, as is the case for VSP2 (Fig. 4A). We found that 82 probes showed enhanced MJ-inducible expression in jamx3 compared with the wild type (MJ:mock ratio > 2 in jamx3 and jamx3:wild type ratio > 1.75 in MJ-treated plants; pink dots in Supplemental Fig. S4; Supplemental Table S2A). We also found eight probes that showed enhanced MJ-responsive reduction in jamx3 compared with the wild type (MJ:mock ratio < 1:2 in jamx3 and jamx3:wild type ratio < 1:1.75 in MJ-treated plants; pale blue dots in Supplemental Fig. S4; Supplemental Table S2B). We also calculated P values by Student’s t test to estimate the statistical significance of differential gene expression (Supplemental Data Set S1). A previous report demonstrated that a straightforward approach of fold change ranking plus a nonstringent P cutoff is effective to identify reproducible gene lists (Shi et al., 2006). Thus, we mainly used the fold change information to screen the target genes of jamx3. The 97 genes corresponding to those 90 probes were regarded as the candidate downstream targets of JAMs and were characterized further in detail (Supplemental Table S2).
JAMs Negatively Regulate the Jasmonate Metabolic Pathway
Among the 98 candidate targets of JAMs, we found genes encoding allene oxide synthase (AOS), JA-Ile-12-hydroxylase (CYP94B3), JA carboxyl methyltransferase (JMT), and hydroxyjasmonate sulfotransferase (ST2A) in jamx3 (Fig. 1; Supplemental Table S2A; Seo et al., 2001; Park et al., 2002; Gidda et al., 2003; Koo et al., 2011). Consistent with our GeneChip results, qRT-PCR analyses confirmed that the expression of AOS, CYP94B3, JMT, and ST2A was clearly enhanced in jam1jam2 and jamx3 plants, but the lower expression of AOS, CYP94B3, and JMT was not statistically significant in MJ-treated myc2 compared with the wild type (Supplemental Fig. S5). In myc2jamx3 plants, the expression levels of AOS, CYP94B3, JMT, and ST2A were higher than those in the myc2 single mutant. These data indicate that JAMs negatively regulate the expression of AOS, CYP94B3, JMT, and ST2A upon MJ treatments (Supplemental Fig. S5; Supplemental Table S2A).
JAMs Modulate the Jasmonate Metabolic Pathway in Wounded Leaves
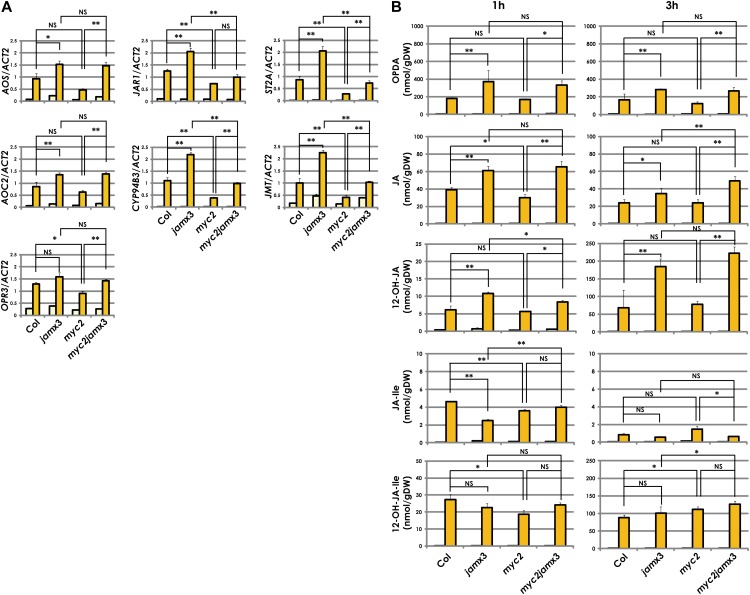
Because we observed that the jamx3 plants exhibited increased MJ responses (Fig. 3), we investigated the effects of jam mutations on jasmonate metabolism. As the jasmonate metabolic pathway is activated after wounding, we examined whether JAMs regulate the expression of genes involved in the jasmonate pathway in wounded leaves. Wild-type, jamx3, myc2, and myc2jamx3 plants were harvested 1 h after wounding, and the expression levels of AOS, AOC2, OPR3, JAR1, CYP94B3, JMT, and ST2A were quantified by qRT-PCR. These genes except for OPR3 exhibited enhanced expression in jamx3 (Fig. 5A). In contrast, OPR3, JAR1, CYP94B3, JMT, and ST2A showed reduced expression in myc2 compared with the wild type. Furthermore, the enhanced expression of JAR1, CYP94B3, JMT, and ST2A in jamx3 was attenuated by the myc2 mutation. Thus, opposite to MYC2, JAMs negatively regulated the expression of certain genes involved in the jasmonate metabolic pathway in wounded leaves.
Figure 5.
JAMs negatively regulate the jasmonate metabolic pathway in wounded leaves. A, Gene expression analyses of jasmonate metabolic genes after wounding. Col-0 and mutant plants were grown for 3 weeks on GM containing 1% Suc. Total RNA was isolated 1 h after wounding. Averaged data from three independent experiments with error bars (se) are presented. Yellow bars represent control (unwounded) samples, and orange bars represent wounded samples. B, Quantitative analyses of jasmonate contents in wounded plants. Col-0 and mutant plants were grown for 3 weeks on GM containing 1% Suc. Jasmonates were isolated from 1 and 3 h after wounding. Averaged data from three biologically independent experiments with error bars (se) are presented. Yellow bars represent control (unwounded) samples, and orange bars represent wounded samples. **P < 0.01, *P < 0.05, NS = not significant (P > 0.05), by Tukey-Kramer multiple comparison test.
To determine whether the transcriptional regulation of genes involved in the jasmonate metabolic pathway affects jasmonate contents in jam mutants, we simultaneously measured the contents of 12-oxo-phytodienoic acid (OPDA), JA, JA-Ile, 12-OH-JA-Ile, and 12-OH-JA in wounded and unwounded leaves. Wounding causes the accumulation of jasmonates in all genotypes (Fig. 5B). We observed that both OPDA and JA levels in jamx3 were significantly higher than in the wild type 1 and 3 h after wounding. The absence of MYC2 was less effective on the contents of OPDA and JA, while those contents in myc2jamx3 were higher than in myc2. Such accumulation profiles were consistent with the expression profiles of AOS and AOC2. 12-OH-JA contents in jamx3 and myc2jamx3 were also higher than those in the wild type and myc2, respectively, as was the case of OPDA and JA. Interestingly, JA-Ile levels in jamx3 were almost one-half of those in the wild type, although JAR1 expression levels in jamx3 were higher than in the wild type. Consistent with the lower JA-Ile levels in jamx3, the 12-OH-JA-Ile levels tended to be lower than in the wild type at 1 h after wounding, but this was not consistent with the expression levels of CYP94B3 in jamx3, for unknown reasons. Based on these results, we concluded that JAMs negatively regulate OPDA, JA, and 12-OH-JA accumulation in wounded leaves.
A previous report showed that 12-OH-JA did not cause JA responses as observed in MJ-treated tomato (Solanum lycopersicum; Miersch et al., 2008). To test whether 12-OH-JA is also inactive in Arabidopsis, we grew wild-type, jamx3, myc2, and myc2jamx3 plants on 12-OH-JA-containing medium and then measured their root lengths. The root lengths of 12-OH-JA-treated wild-type as well as jamx3, myc2, and myc2jamx3 plants were comparable to those of mock-treated plants (Supplemental Fig. S6). Thus, exogenous 12-OH-JA did not show JA activity on the root growth of not only the wild type but also jam mutants.
JAMs Negatively Regulate the Expression of Certain Transcription Factor-Encoding Genes
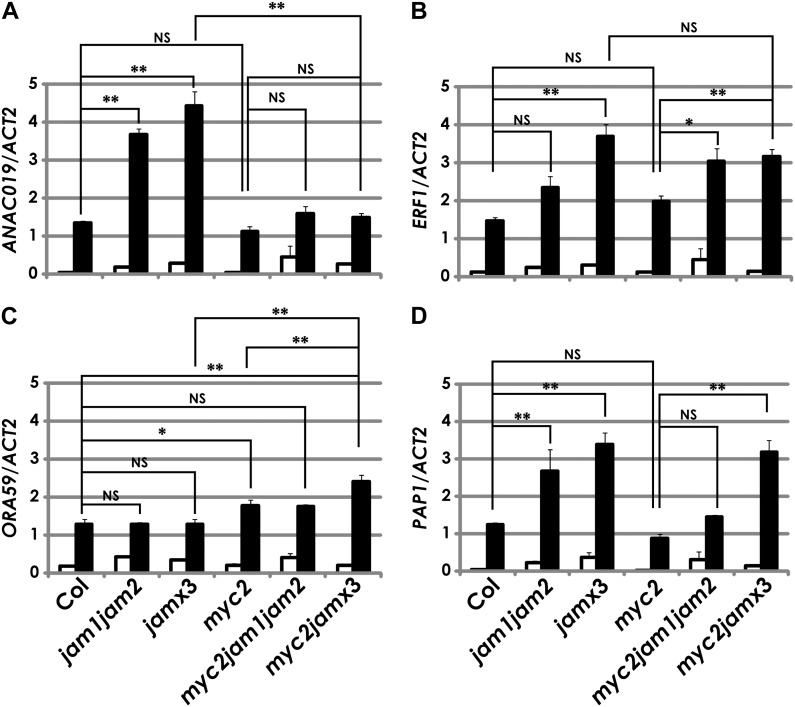
MYC2 regulates the expression of downstream metabolic genes through transcription factor-encoding genes such as ANAC019, ERF1, and ORA59. Out of the 98 genes regulated by JAMs, we found 14 genes that potentially encode transcription factors, including ANAC019 and ERF1 but not ORA59 (Supplemental Table S2). ANAC019 showed enhanced MJ-inducible expression in jam1jam2 and jamx3; however, the expression levels of ANAC019 in myc2jam1jam2 and myc2jamx3 were almost comparable to that in myc2 (Fig. 6A). Thus, JAMs negatively regulate ANAC019 expression, and its profile was consistent with VSP2 (Figs. 4A and 6A). ERF1 expression is induced by MJ treatments, as reported previously (Fig. 6; Lorenzo et al., 2003). The ERF1 expression levels in MJ-treated jam1jam2 and jamx3 were clearly higher than in the wild type. Furthermore, ERF1 was expressed higher in MJ-treated myc2jam1jam2 and myc2jamx3 than in myc2. Thus, JAMs also negatively regulate ERF1 expression in both the wild-type and myc2 backgrounds. However, the expression profiles of ERF1 and PDF1.2 were not consistent with those in jamx3, unlike the cases of ANAC019 and VSP2 (Figs. 4 and 6, A and B). As ORA59 is another positive regulator of PDF1.2 induction, we tested the expression profile of ORA59 (Pré et al., 2008). We found that the expression of ORA59 was independent from jamx3. Interestingly, ORA59 levels in myc2jamx3 were higher than that in myc2 and closely correlated to those of PDF1.2 (Figs. 4B and 6C). These data show that JAMs negatively regulate the expression of ANAC019 and ERF1. Therefore, ANAC019 and ERF1 can be the target points of JAMs to negatively regulate downstream metabolic genes.
Figure 6.
Expression analysis of ANAC019, ERF1, ORA59, and PAP1 in Col-0, jam1jam2, jamx3, myc2, myc2jam1jam2, and myc2jamx3 plants. For this analysis, total RNA was isolated from 7-d-old plants that were grown in liquid GM with 1% Suc and treated with mock (white bars) or 50 μm MJ (black bars) for 1 h. Results shown are means ± se for three biologically independent RNAs. **P < 0.01, *P < 0.05, NS = not significant (P > 0.05), by Tukey-Kramer multiple comparison test.
Correlated Expression between JAM-Regulated Transcription Factors and Metabolic Genes
Next, we focused on the other JAM-regulated transcription factors. Other than ANAC019 and ERF1, we found that JAMs negatively regulate the expression of genes encoding five MYB-type, two AP2/ERF-type, one WRKY-type, and one bHLH-type transcription factors (Supplemental Table S2A). As correlated expression of transcription factors and target metabolic genes was known during JA responses, we calculated the mutual rank of the Pearson’s correlation coefficient (MR) using ATTED-II (Obayashi and Kinoshita, 2009) and visualized the highly correlated genes with the JAM-regulated genes (MR < 50) by Cytoscape (Supplemental Fig. S7; see “Materials and Methods”; Smoot et al., 2011). We found that PAP1 expression was regulated by JAMs and strongly correlated with UGT75C1, a gene encoding UDP-glucosyltransferase 75C1, consistent with a previous report (Fig. 6D; Supplemental Fig. S7; Supplemental Table S2A; Tohge et al., 2005). Other than the correlated expression of PAP1 and anthocyanin biosynthetic genes, we found several tight correlations between transcription factor-encoding genes and metabolic genes. For example, the expression of MYB47, which encodes a MYB-type transcriptional factor, closely correlated with that of PAP1, AOS, and At1g52000 (encoding lectin family protein). Similarly, the expression RAP2.6, which encodes an AP2/ERF-type transcriptional factor (Zhu et al., 2010; Krishnaswamy et al., 2011), tightly correlated with the jasmonate metabolic genes CYP94B3 and ST2A as well as UGT76E12 and At2g38240 (putative oxygenase). In addition, At1g10585 encodes a putative bHLH domain-containing transcriptional factor and also shows a correlated expression with UGT76E12, At2g38240, ST2A, JRG21 (oxidoreductase activity), and TERPENE SYNTHASE04. Thus, UGT76E12, At2g38240, and ST2A are likely to be coregulated by At1g10585 and RAP2.6. In summary, PAP1, MYB47, RAP2.6, and At1g10585 can be the target points by JAMs to regulate JA-responsive metabolic genes.
JAMs Negatively Regulate the Anthocyanin Biosynthetic Pathway through the Transcriptional Cascade
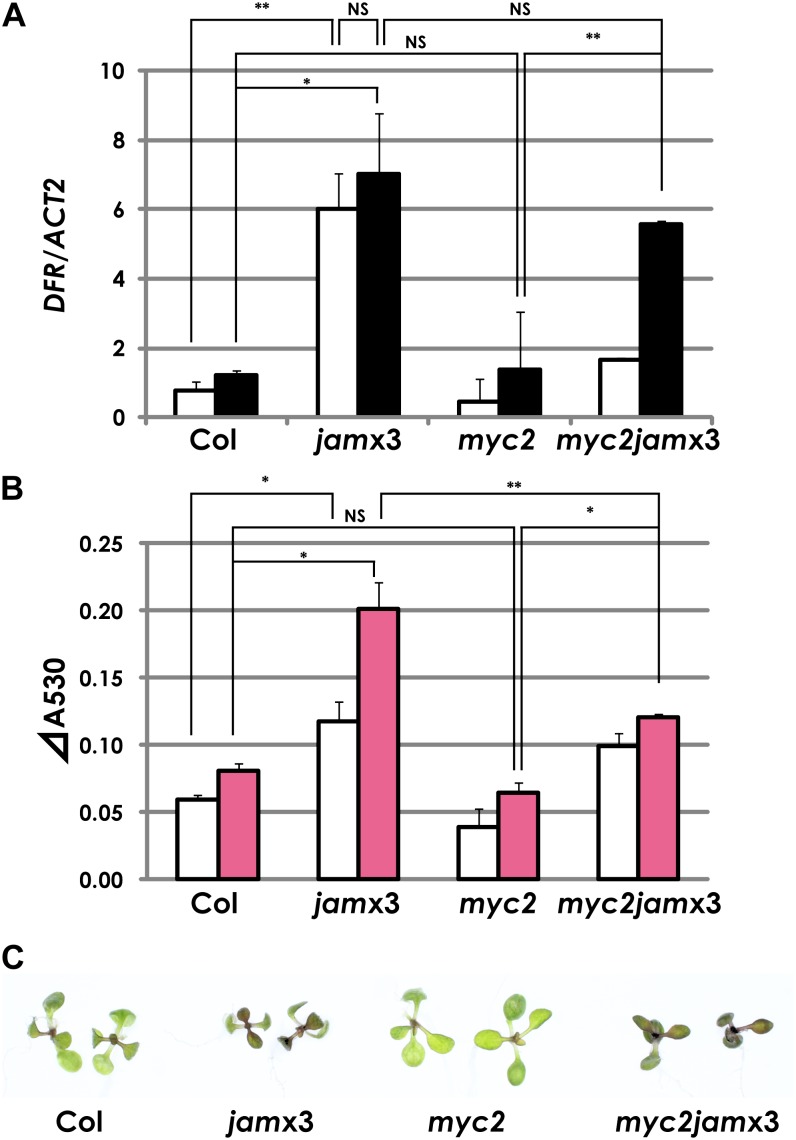
We tested whether the regulation of the transcriptional cascade by JAMs causes metabolic changes in plants. Since we observed enhanced PAP1, UGT75C1, and At3AT1/At3AT2 (At1g03940/At1g03495; anthocyanidin 3-O-glucoside acyl-CoA transferase) expression in MJ-treated jamx3 (Fig. 6D; Supplemental Table S2A), we also tested the expression profile of a known PAP1-regulated anthocyanin biosynthetic gene, DFR/TT3 (At5g42800), which encodes a dihydroflavonol reductase, a key step of anthocyanin biosynthesis (Shirley et al., 1995). The expression profile of DFR/TT3 is similar to that of PAP1 (Figs. 6D and 7A). Thus, JAMs negatively regulated a set of anthocyanin biosynthetic genes probably through the regulation of PAP1 expression. Consistently, anthocyanin levels in jamx3 plants were more than two times higher than in the wild type (Fig. 7, B and C). In this assay, the anthocyanin level in myc2 was almost comparable to that in the wild type in our experimental conditions. Furthermore, jamx3 mutations in the myc2 background increased anthocyanin contents, consistent with the expression profiles of anthocyanin regulatory and biosynthesis genes (Figs. 6D and 7, A and B). Interestingly, we noted that DFR expression levels, as well as the anthocyanin levels, in mock-treated jamx3 were higher than in the wild type (Fig. 7, A and B).
Figure 7.
JAMs negatively regulate anthocyanin biosynthesis. A, Expression profiles of the anthocyanin biosynthetic gene DFR in Col-0, jamx3, myc2, and myc2jamx3 plant seedlings. Col-0 and mutant plants were grown in liquid GM containing 1% Suc. Total RNA was isolated from plants 1 h after mock or 50 μm MJ treatment. Averaged data from three independent experiments with error bars (se) are presented. White bars represent mock treatment, and black bars represent 50 μm MJ treatment. B, Anthocyanin levels in Col-0, jamx3, myc2, and myc2jamx3 plant seedlings. Plants were grown in liquid GM containing 1% Suc for 7 d and then treated with mock or 50 μm MJ for 24 h. Results shown are means ± se of three independent experiments. White bars represent mock treatment, and pink bars represent 50 μm MJ treatment. **P < 0.01, *P < 0.05, NS = not significant (P > 0.05), by Tukey-Kramer multiple comparison test. C, Photographs of Col-0, jamx3, myc2, and myc2jamx3 plants that were grown in liquid GM with 1% Suc and treated with 50 μm MJ for 24 h.
As Suc treatments also cause anthocyanin accumulation (Teng et al., 2005), we tested the effect of Suc on jam mutants. In liquid medium without Suc, the wild type and jam mutants did not accumulate anthocyanin, as reported previously (Supplemental Fig. S8A; Loreti et al., 2008). Suc-treated jam1jam2 and jamx3 plants showed higher accumulation of anthocyanin (Supplemental Fig. S8), indicating that JAMs negatively regulate not only JA-dependent but also Suc-dependent anthocyanin biosynthesis.
To estimate the effect of Suc on gene expression, we extracted RNAs from MJ-treated seedlings grown on GM containing 0.5% Suc and tested the expression profiles of PAP1, DFR, and VSP2 by qRT-PCR analysis. In this experimental condition, the expression of PAP1, DFR, and VSP2 in jamx3 was higher than in the wild type, and these phenotypes were attenuated by myc2 (Supplemental Fig. S9). Furthermore, by contrast to 1% Suc in the medium, 0.5% Suc resulted in significantly lower DFR expression levels in the mock-treated jam1jam2 and jamx3, indicating that DFR is induced also by Suc only, probably via PAP1 (Fig. 7; Supplemental Fig. S9).
DISCUSSION
Based on network analysis combined with genetic studies, we show that JAM1 and its close homologs JAM2 and JAM3 play an important role in regulating JA-responsive genes. The expression of JAM1 and JAM2 was induced not only by MJ treatments but also under stress conditions such as wounding, pathogen infection, and drought stress, while JAM3 expression was almost unchanged in any condition (Genevestigator [https://www.genevestigator.com/gv/]; Hruz et al., 2008). As JAM1, JAM2, and JAM3 transcript levels were all reduced in coi1-1 and myc2 (Fig. 2), we concluded that JAM1, JAM2, and JAM3 are involved in JA signaling. MYC2 expression was also COI1 dependent but JAM independent in MJ-treated plants, suggesting that MYC2 is a regulator of JAMs but JAMs are not regulators of MYC2 (Fig. 2C; Supplemental Fig. S7). Although JAM genes encode MYC2-like bHLH transcription factors, their functions appear to be antagonistic to those of MYC2. For example, root lengths were shorter in jamx3 but longer in myc2 than in the wild type on the MJ-containing medium (Fig. 3). The root phenotype in jamx3 was attenuated by myc2, confirming that MYC2 and JAM genes function antagonistically.
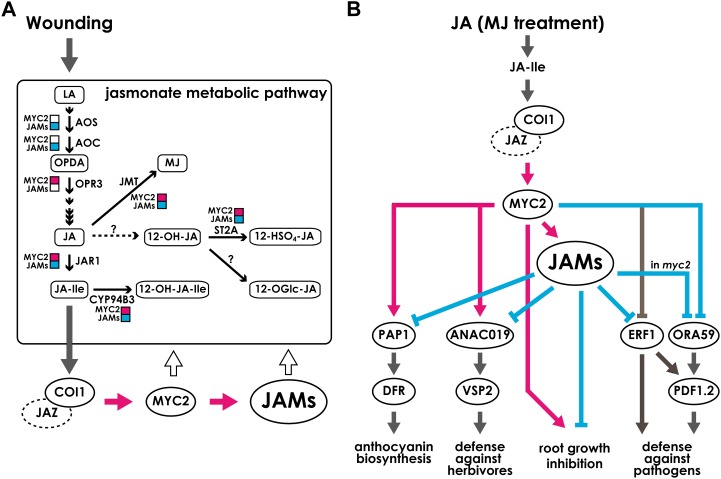
The antagonistic functionality between JAM genes and MYC2 is also apparent in gene regulation in the jasmonate metabolic pathway. For example, JAR1, CYP94B3, JMT, and ST2A expression levels in wounded leaves were significantly higher in jamx3 but lower in myc2 than in the wild type (Figs. 5A and 8A). However, the effects of JAMs and MYC2 on the accumulation of the jasmonates are rather complicated. For example, the negative regulation by JAMs is evident in the accumulation profiles of OPDA and JA, while we did not observe a positive effect of MYC2 on OPDA and JA contents other than 1 h after wounding for JA. Furthermore, for reasons that are not clear, MYC2 did not attenuate the effect of JAMs in the case of OPDA and JA contents (Fig. 5B). We found that the JA-Ile level in jamx3 was about one-half of the wild-type level at 1 h after wounding, while 12-OH-JA was more accumulated in the mutant (Fig. 5B). The content of active jasmonates is determined by the combination of substrate amounts and biosynthetic and catabolic activities. Therefore, the level of an active jasmonate may not directly correlate with the expression levels of jasmonate biosynthetic and catabolic genes. JAMs are the negative regulators of jasmonate biosynthetic and catabolic genes, but it is likely that a mechanism other than transcriptional regulation is more critical in determining the jasmonate contents. As 12-OH-JA normally accumulates higher than JA-Ile upon wounding (Glauser et al., 2008; Kitaoka et al., 2011), it is plausible that increased accumulation of 12-OH-JA may have a negative effect on JA-Ile synthesis and/or accumulation. Alternatively, there is a possibility that a yet unknown gene(s) encoding an enzyme that catalyzes JA to 12-OH-JA is induced in jamx3, favoring 12-OH-JA accumulation rather than JA-Ile (Fig. 8A).
Figure 8.
Proposed models for the regulation of JA-responsive pathways by JAMs and MYC2. A, After wounding, synthesized JA-Ile activates jasmonate signaling. JAMs and MYC2 antagonistically regulated the jasmonate metabolic pathway in wounded leaves. It should be noted that effects on the expression of jasmonate biosynthetic and catabolic genes are shown, but those do not correlate with oxylipin levels. B, model of JAM-regulated transcription factors and metabolic enzymes during JA responses. After the recognition of JA-Ile by the COI1-JAZ complex, MYC2 is activated and regulates other transcription factors. JAMs also are targets of MYC2 and function in jasmonate signaling. JAMs negatively regulate the targets of MYC2. Note that the arrows show transcriptional regulation except for the arrows pointing to “root growth inhibition.” Pink arrows and squares represent positive effects shown experimentally in this paper. Pale blue arrows and squares represent negative effects shown experimentally in this paper. Black thin arrows represent metabolic reactions. Dark gray arrows are drawn based on previous reports. White arrows represent the feedback regulation by transcription factors. Note that the negative effect of JAMs on ORA59 expression was observed in the myc2 background. For abbreviations, see Figure 1.
We observed no inhibitory effect of exogenous 12-OH-JA on the root lengths of the wild type, jamx3, myc2, and myc2jamx3, consistent with a previous study (Supplemental Fig. S6; Miersch et al., 2008). However, the negative regulation of 12-OH-JA metabolism by JAMs may still imply a biological role of this compound. For instance, after wounding, 12-OH-JA but not JA and JA-Ile was detected even in the midveins of unwounded leaves because of the rapid diffusion to the distil part (Glauser et al., 2008). Thus, it is possible that 12-OH-JA may be a metabolic form to transmit JA signaling to the distal parts of plants. In fact, 12-OH-JA induces potato (Solanum tuberosum) tuber formation (Yoshihara et al., 1989). Furthermore, 12-hydroxyjasmonic acid glucoside, a Glc conjugate of 12-OH-JA, was recently shown to specifically cause the leaf closure of albizzia (Samanea saman) leaves, and that process is independent from the COI1-JAZ module (Nakamura et al., 2011). Thus, we cannot exclude the possibility that 12-OH-JA may be an intermediate to generate other JA metabolites that have specific roles in plants.
We observed the antagonistic effects of JAMs and MYC2 not only on the regulation of jasmonate metabolic genes in wounded plants but also on anthocyanin-related genes in MJ-treated plants. Consistent with the anthocyanin levels, PAP1 and DFR expression levels in MJ-treated jamx3 were significantly higher than in the wild type, indicating negative effects of JAMs on the expression of those genes (Figs. 6D, 7, and 8B). We found that the antagonistic regulation of PAP1 and DFR by JAMs and MYC2 in MJ-treated plants was more apparent when we used the plants grown on GM with a lower concentration of Suc (0.5%; Supplemental Fig. S9), indicating that Suc effects may partially mask the JA-dependent antagonistic regulation by JAMs and MYC2. Previous reports showed that the expression of PAP1 and anthocyanin biosynthetic genes is induced by Suc treatments in a dose-dependent manner (Solfanelli et al., 2006; Shan et al., 2009). Thus, it is possible that JAMs negatively regulate the Suc-dependent anthocyanin accumulation through the transcriptional regulation of PAP1 and anthocyanin biosynthetic genes, including DFR. Most importantly, the anthocyanin phenotypes, including higher accumulation of anthocyanin and transcriptional levels of PAP1 and DFR, in jamx3 are attenuated by myc2 (Fig. 7; Supplemental Fig. S9), indicating that JAM genes and MYC2 function antagonistically.
Our data also show that JAMs and MYC2 antagonistically regulate the expression of ANAC019 and VSP2, as the enhanced expression phenotype in jamx3 is attenuated by myc2 (Figs. 4A and 6A). The JAM-MYC2 antagonistic relationship on VSP2 expression is again more evident when plants are grown on GM with a lower concentration of Suc (Supplemental Fig. S9). Both MJ and Suc treatments affect the expression level of VSP in Arabidopsis (Berger et al., 1995). Thus, Suc effects also mask the JAM-MYC2 antagonism on VSP2 expression (Fig. 8B). Unlike on the expression of PAP1 and ANAC019, the JAM-MYC2 antagonism is not seen on ERF1 expression (Fig. 6B). Previous reports showed that MYC2 negatively regulates ERF1 expression, and such an effect is more apparent 3 h after JA treatments (Dombrecht et al., 2007; Çevik et al., 2012; Chen et al., 2012). The negative effect of MYC2 on ERF1 expression was not so clear in our experimental conditions, probably because we sampled at an earlier time point (Fig. 6B). This is consistent with the previous finding that ERF1 shows a relatively late response against JA treatments (Lorenzo et al., 2003). Thus, it is likely that both JAMs and MYC2 have negative effects on ERF1 expression, but their timing is distinct (Fig. 8B). Like on ERF1, MYC2 has a negative effect on PDF1.2 expression as well as its upstream regulator ORA59 (Figs. 4B and 6C; Lorenzo et al., 2004; Çevik et al., 2012). However, in jamx3, the expression levels of PDF1.2 and ORA59 were comparable to those in the wild type (Figs. 4B and 6C). Notably, the expression levels of PDF1.2 and ORA59 in myc2jamx3 were significantly higher than those in myc2 (Figs. 4B, 6C, and 8B), indicating that the negative effect by JAMs on these two genes is only apparent in the myc2 background, for yet unknown reasons.
MYC2 partially regulates the expression of JAM genes, but JAMs have little effect on the expression of MYC2 (Fig. 2; Supplemental Fig. S7). In this sense, MYC2 is upstream of JAM genes. However, the regulation of downstream genes by JAM and MYC2 proteins is more complicated, as the effects of these proteins are in some cases antagonistic (for PAP1 and ANAC019) or additive (for ERF1 and ORA59; Fig. 6; Supplemental Fig. S9). Therefore, JAM and MYC2 proteins do not represent a typical downstream-upstream relationship. Previous reports showed that MYC2 preferentially binds to the G-box-related sequences (Abe et al., 1997; Yadav et al., 2005; Dombrecht et al., 2007). As MYC2 and JAM proteins share significant amino acid identities around the bHLH motif, it is tempting to speculate that JAMs, as repressors, compete with MYC2, as an activator, for binding to the G-box-related sequence as an explanation for the antagonistic regulation of downstream genes. Very recently, Nakata et al. (2013) showed that JAM1 is a transcriptional repressor and that it can bind to the G-box sequence. Interestingly, although MYC2 binds to the G-box sequence on the MYC2 promoter (Dombrecht et al., 2007) and JAM1 can bind to the G-box sequence, JAMs seem to have little effect on MYC2 expression (Supplemental Fig. S7; Nakata et al., 2013). Therefore, it is likely that MYC2 and JAMs compete for binding to specific variants of the G-box that are not present in the MYC2 promoter.
Alternatively, the formation of distinct transcriptional complexes is another possible mechanism. For example, MEDIATOR25 (MED25; previously known as PHYTOCHROME AND FLOWERING TIME1 [PFT1]) is a subunit of the Arabidopsis Mediator complex and provides a bridge between DNA-bound transcriptional activators and the RNA polymerase II complex (Bäckström et al., 2007). Recently, MED25 was characterized as an important component to regulate the expression of JA-responsive genes by directly binding to MYC2, modulating the expression of transcription factors such as ERF1 and ORA59 (Çevik et al., 2012; Chen et al., 2012). In addition, pft1 mutants and MED25/PFT1-overexpressed plants accumulated less and more anthocyanin compared with the wild type, respectively (Kidd et al., 2009), implying that, like MYC2, the Mediator complex functions antagonistically to JAMs. Thus, it is plausible that JAMs could affect the status of MED25 or other component(s) of the Mediator complex on the promoter region of the JAM-regulated genes that are also regulated by MYC2. Identification of the target cis-elements and the protein interaction network on JAMs, MYC2, and the Mediator complex in the near future will help to build a unified view on how JA signals are incorporated and the fine-tuning of plant responses against environmental stresses.
MATERIALS AND METHODS
Coexpression Gene Network Analysis
Using our previously published Arabidopsis (Arabidopsis thaliana) two oligomicroarray data sets (Taki et al., 2005), we selected 2,220 genes whose expression was increased or decreased by JA, MJ, OPDA, or wounding more than 2-fold compared with the control. Then, we constructed coexpression gene networks of the 2,220 genes by using data sets from our work (Taki et al., 2005; 12 points) and from AtGenExpress (Kilian et al., 2007; stress data, 298 points). To normalize the two data sets, weighted Pearson’s correlation coefficient was calculated using an in-house perl script with the following weights: 30 for JA samples and one for AtGenExpress stress samples, which were manually adjusted. When we set the threshold of Pearson’s correlation coefficient as 0.9, 233 genes were clustered into 11 groups (Supplemental Table S1). Hierarchical clustering was performed by a clustering tool on the ATTED-II Web site (http://atted.jp/; Supplemental Fig. S1; Obayashi et al., 2011).
jam T-DNA Lines
Arabidopsis ecotype Col-0 was used as the wild type in this study. The seeds of T-DNA knockout mutants of JAM1 (jam1-1 [SAIL_536_F09] and jam1-2 [GK-285E09-015277]), JAM2 (jam2-1 [GK-696A04-024559]), and JAM3 (jam3-1 [SALK_050954] and jam3-2 [GK-301G05-015558]) were obtained from the Nottingham Arabidopsis Stock Centre, Genomanalyse im Biologischen System Pflanz, and Arabidopsis Biological Resource Center collections. Homozygous plants were isolated by PCR. The primer sequences for genotyping are shown in Supplemental Table S3. Expression of the coding sequence in each T-DNA insertion line was tested by reverse transcription (RT)-PCR and qRT-PCR. We initially observed reduced anthocyanin contents in the original line from GK-285E09-015277; however, the phenotype was segregated out by back-crossing to the wild type. Note that JAM1 was previously designated AtAIB (for ABA-inducible bHLH protein) by (Li et al., 2007). Because Takagi’s group (Nakata et al., 2013), who recently published JAM1 function, and our group identified that JAM1 has an essential function in JA signaling rather than abscisic acid signaling, both of us codesignated the MYC2-related bHLH-type transcription factor (At2g46510) as JAM1.
Plant Growth Conditions
For gene expression analyses and measurements of anthocyanin content, we used 7-d-old seedlings. Seeds were incubated on a 12-well plate with liquid GM containing 1% Suc under continuous light (96–112 μmol m−2 s−1). Plants were treated with 50 μm MJ (Wako) and harvested at the indicated time points. Plant materials were frozen in liquid nitrogen and stored at −80°C until they were used. For the measurement of root lengths and COI1-dependent gene expression analyses, we used 2-week-old seedlings grown on GM containing 50 μm MJ, 0.8% agar, and 1% Suc under 16-h-light/8-h-dark conditions at a light intensity of 58 to 79 μmol m−2 s−1. We also used the same experimental conditions for the root length measurement of 50 or 100 μm (±)-12-OH-JA (Olchemim)-treated plants. For root length measurement, we placed the plates in the vertical orientation. We took photographs of roots and measured root lengths by ImageJ software (http://rsbweb.nih.gov/ij/; Abràmoff et al., 2004). For COI1-dependent gene expression analyses, we placed the plates with 50 μm MJ-containing medium in the horizontal orientation. The coi1-1 plants were selected for the MJ-insensitive phenotype, and aerial parts of plants were used to extract RNAs for qRT-PCR analysis. For the wounding assay, plants were grown for 3 weeks under 16-h-light/8-h-dark conditions at an intensity of 58 to 79 μmol m−2 s−1 on GM containing 0.8% agar and 1% Suc, and then rosette leaves were wounded by crushing across the midrib with a hemostat. At the indicated time points after wounding, damaged leaves and undamaged leaves from unwounded plants were harvested. For anthocyanin measurement after Suc treatment, plant materials were grown in liquid GM without Suc for 3 d and then harvested 5 d after Suc treatment as described (Saijo et al., 2009).
RT-PCR and qRT-PCR Analyses
Total RNA was isolated from the indicated plant materials, converted to cDNA using RevaTraAce (TOYOBO), and used as a template for RT-PCR and qRT-PCR analyses. For RT-PCR and qRT-PCR analyses, we used KOD FX (TOYOBO) and THUNDERBIRD SYBR qPCR Mix (TOYOBO), respectively, according to the manufacturer’s instructions. For the primer sequences used for RT-PCR and qRT-PCR analyses, see Supplemental Table S3.
GeneChip Analysis
RNAs extracted from Col-0 and jamx3 seedlings treated with ethanol (mock) or 50 μm MJ for 1 h were reverse transcribed and labeled using the 3′ IVT Express Kit (Affymetrix). Labeled samples were used to hybridize Affymetrix GeneChip ATH1 arrays according to the manufacturer’s instructions. GeneSpring version 12 was used for normalization by the GeneChip Robust Multiarray Averaging method. Then, we averaged the normalized intensities that were derived from two biologically independent RNAs. We designated these values as averaged intensity and used them for further analysis. No probe set was removed before the averaging step.
Gene Network Analysis
For gene network analysis, we calculated the MR (Obayashi and Kinoshita, 2009) of the 98 JAM-regulated genes using ATTED-II and visualized the gene network based on the MR values by Cytoscape (http://www.cytoscape.org/; Smoot et al., 2011). We selected correlations whose MR values were less than 50. We also calculated the jamx3-wild type ratio in MJ-treated plants and included those values on the gene network as a node color.
Measurement of Anthocyanin Content
Anthocyanin extraction was carried out in triplicate as described with slight modification (Tohge et al., 2005). Frozen seedlings were homogenized and extracted in 10 μL of extraction solvent (methanol:acetate:water = 9:1:10) per 1 mg fresh weight of tissues. After centrifugation at 12,000g, cell debris was discarded. Absorbance of the extracted solution was measured at 530 nm.
Measurement of Jasmonate Contents
Plant materials were lyophilized, and their dry weights were measured. JA-related molecules were extracted and quantified as described in previous reports with slight modification (Yoshimoto et al., 2009; Ohkama-Ohtsu et al., 2011). After grinding with 10-mm zirconia beads, 1 mL of acetonitrile:acetic acid:water (80:1:19) solution containing D5-OPDA (1 ng; Olchemim), D2-JA (100 pg; Kanto Chemicals), D2-12-OH-JA (1 ng; Olchemim), [13C6]JA-Ile (10 pg; Jikumaru et al., 2004), and [13C6]12-OH-JA-Ile (1 ng; Kitaoka et al., 2011) was added to 1 mg dry weight of unwounded Arabidopsis aerial part and extracted for 1 h. A 100 times larger amount of each stable isotope-labeled chemical was added for wounded plant materials. Supernatants were collected after centrifugation, and residue was extracted again with 1 mL of acetonitrile:acetic acid:water (80:1:19) solution for 10 min. The supernatant was collected after centrifugation and combined to supernatant prepared previously. Acetonitrile in the combined supernatant was removed in a SpeedVac (Thermo Scientific), and acidic water extract was loaded onto an Oasis HLB column cartridge (30 mg, 1 mL; Waters). After washing with 1 mL of water:acetic acid (99:1) solution, JA-related molecules were extracted two times with 1 mL of acetonitrile:acetic acid:water (80:1:19) solution. Acetonitrile in the remaining extract was removed in a SpeedVac, and acidic water extract was loaded onto an Oasis WAX column cartridge (30 mg, 1 mL; Waters). After washing with 1 mL of water:acetic acid (99:1) solution and 1 mL of acetonitrile:water (80:20) solution, JA-related molecules were extracted two times with 1 mL of acetonitrile:acetic acid:water (80:1:19) solution.
Purified extract was dried and dissolved in 30 μL of water:acetic acid (99:1) solution, and 15 μL was subjected to liquid chromatography-tandem mass spectrometry (Agilent 1200-Agilent 6410) equipped with an ODS column (ZORBAX Eclipse XDB-C18, 2.1 × 50 mm, 1.8 μm; Agilent). For quantification of JA-related molecules, separation was performed using a gradient of water containing 0.01% acetic acid as solvent A and acetonitrile containing 0.05% acetic acid as solvent B. Concentration of solvent B was increased from 3% to 70% over 30 min with a flow rate of 0.2 mL min−1. After washing with 98% solvent B for 5 min, the initial condition was restored and allowed to equilibrate for 5 min. Retention time was 24.6 min for OPDA and D5-OPDA, 15.5 min for JA and D2-JA, 18.6 min for JA-Ile and [13C6]JA-Ile, 8.4 min for 12-OH-JA and D2-12-OH-JA, and 12.2 min for 12-OH-JA-Ile and [13C6]12-OH-JA-Ile. Tandem mass spectrometry conditions were as follows. Tandem mass spectrometry transitions for quantification were m/z 291→165 for OPDA, 296→170 for D5-OPDA, 209→59 for JA, 211→59 for D2-JA, 322→130 for JA-Ile, 328→135 for [13C6]JA-Ile, 225→59 for 12-OH-JA, 227→61 for D2-12-OH-JA, 338→130 for 12-OH-JA-Ile, and 344→136 for [13C6]12-OH-JA-Ile. The collision energy was 12 for OPDA and D5-OPDA, 10 for JA and D2-JA, 14 for JA-Ile and [13C6]JA-Ile, 8 for 12-OH-JA and D2-12-OH-JA, and 20 for 12-OH-JA-Ile and [13C6]12-OH-JA-Ile. The fragmentor was 160 for OPDA and D5-OPDA, 150 for JA and D2-JA, 160 for JA-Ile and [13C6]JA-Ile, 120 for 12-OH-JA and D2-12-OH-JA, and 150 for 12-OH-JA-Ile and [13C6]12-OH-JA-Ile.
Arabidopsis Genome Initiative numbers for the sequences described in this article are as follows: MYC2 (At1g32640), COI1 (At2g39940), JAZ1 (At1g19180), VSP2 (At5g24770), PDF1.2 (At5g44420), AOC2 (At3g25770), OPR3 (At2g06050), JAR1 (At2g46370), and ORA59 (At1g06160).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Hierarchical clustering of the 128 genes in cluster 7.
Supplemental Figure S2. Identification of single jam knockout plants.
Supplemental Figure S3. MJ sensitivities of the single jam knockout plants were comparable to Col-0.
Supplemental Figure S4. Scatterplots of averaged intensities from Col-0 and jamx3 treated with 50 μm MJ for 1 h.
Supplemental Figure S5. Gene expression analysis of jasmonate metabolic genes after MJ treatment.
Supplemental Figure S6. Root lengths of Col-0, jamx3, myc2, and myc2jamx3 seedlings after 12-OH-JA treatment.
Supplemental Figure S7. Correlated expression of JAM-regulated transcription factors and metabolic genes.
Supplemental Figure S8. Anthocyanin levels after Suc treatment.
Supplemental Figure S9. Gene expression analysis of PAP1, DFR, and VSP2 after MJ treatment.
Supplemental Table S1. Cluster analysis of jasmonate and wound-responsive genes.
Supplemental Table S2. Genes that show enhanced expression after MJ treatment (50 μm treatment 1 h).
Supplemental Table S3. Primer sequences.
Supplemental Data Set S1. Microsoft Excel file with Arabidopsis Genome Initiative numbers, normalized intensities, averaged intensities, P values, and fold-change information of the probe set.
Acknowledgments
We thank the Nottingham Arabidopsis Stock Centre, Genomanalyse im Biologischen System Pflanz, and the Arabidopsis Biological Resource Center for providing seeds of Arabidopsis mutants and T-DNA insertion lines. We also thank Salome Prat for kindly providing myc2-2 seeds, Hideyuki Matsuura for kindly providing labeled 12-OH-JA-Ile, Sachiko Ohyama for her contribution to GeneChip analyses, Kaori Takizawa, Akiko Ueno, and Reiko Sakamoto for their assistance, and all the members of Ken Shirasu and Hirofumi Nakagami laboratories for their helpful discussions and technical support.
Glossary
- JA
jasmonic acid
- JA-Ile
jasmonoyl-l-isoleucine
- 12-OH-JA-Ile
12-hydroxyjasmonoyl-l-isoleucine
- 12-OH-JA
12-hydroxyjasmonic acid
- bHLH
basic helix-loop-helix
- MJ
methyl jasmonate
- Col-0
Columbia
- T-DNA
transfer DNA
- qRT
quantitative reverse transcription
- OPDA
12-oxo-phytodienoic acid
- MR
mutual rank of the Pearson’s correlation coefficient
- qPCR
quantitative PCR
- RT
reverse transcription
- T-DNA
transferred DNA
- qRT
quantitative real-time
- GM
germination medium
- Col-0
Columbia-0
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. (1997) Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell 9: 1859–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abràmoff MD, Hospitals I, Magalhães PJ, Abràmoff M. (2004) Image processing with ImageJ. Biophotonics International 11: 36–42 [Google Scholar]
- Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol Cell 26: 717–729 [DOI] [PubMed] [Google Scholar]
- Bender J, Fink GR. (1998) A Myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc Natl Acad Sci USA 95: 5655–5660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE. (1995) Arabidopsis thaliana Atvsp is homologous to soybean VspA and VspB, genes encoding vegetative storage protein acid phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol 27: 933–942 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S. (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18: 1577–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browse J. (2009) Jasmonate passes muster: a receptor and targets for the defense hormone. Annu Rev Plant Biol 60: 183–205 [DOI] [PubMed] [Google Scholar]
- Browse J, Howe GA. (2008) New weapons and a rapid response against insect attack. Plant Physiol 146: 832–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li C-B, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C. (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18: 756–767 [DOI] [PubMed] [Google Scholar]
- Çevik V, Kidd BN, Zhang P, Hill C, Kiddle S, Denby KJ, Holub EB, Cahill DM, Manners JM, Schenk PM, et al. (2012) MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol 160: 541–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. (2012) The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell 24: 2898–2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. (2011) The bHLH transcription factor MYC3 interacts with the jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol Plant 4: 279–288 [DOI] [PubMed] [Google Scholar]
- Chico JM, Chini A, Fonseca S, Solano R. (2008) JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 11: 486–494 [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. (2007) The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671 [DOI] [PubMed] [Google Scholar]
- Devoto A, Ellis C, Magusin A, Chang H-S, Chilcott C, Zhu T, Turner JG. (2005) Expression profiling reveals COI1 to be a key regulator of genes involved in wound- and methyl jasmonate-induced secondary metabolism, defence, and hormone interactions. Plant Mol Biol 58: 497–513 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. (2007) MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19: 2225–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. (2011) The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23: 701–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gfeller A, Dubugnon L, Liechti R, Farmer EE. (2010) Jasmonate biochemical pathway. Sci Signal 3: cm3. [DOI] [PubMed] [Google Scholar]
- Gidda SK, Miersch O, Levitin A, Schmidt J, Wasternack C, Varin L. (2003) Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J Biol Chem 278: 17895–17900 [DOI] [PubMed] [Google Scholar]
- Glauser G, Grata E, Dubugnon L, Rudaz S, Farmer EE, Wolfender J-L. (2008) Spatial and temporal dynamics of jasmonate synthesis and accumulation in Arabidopsis in response to wounding. J Biol Chem 283: 16400–16407 [DOI] [PubMed] [Google Scholar]
- Heim MA, Jakoby M, Werber M, Martin C, Weisshaar B, Bailey PC. (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20: 735–747 [DOI] [PubMed] [Google Scholar]
- Heitz T, Widemann E, Lugan R, Miesch L, Ullmann P, Désaubry L, Holder E, Grausem B, Kandel S, Miesch M, et al. (2012) Cytochromes P450 CYP94C1 and CYP94B3 catalyze two successive oxidation steps of plant hormone jasmonoyl-isoleucine for catabolic turnover. J Biol Chem 287: 6296–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Sugiyama K, Sawada Y, Tohge T, Obayashi T, Suzuki A, Araki R, Sakurai N, Suzuki H, Aoki K, et al. (2007) Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA 104: 6478–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. (2008) Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics 2008: 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jikumaru Y, Asami T, Seto H, Yoshida S, Yokoyama T, Obara N, Hasegawa M, Kodama O, Nishiyama M, Okada K, et al. (2004) Preparation and biological activity of molecular probes to identify and analyze jasmonic acid-binding proteins. Biosci Biotechnol Biochem 68: 1461–1466 [DOI] [PubMed] [Google Scholar]
- Katsir L, Chung HS, Koo AJK, Howe GA. (2008) Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol 11: 428–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21: 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D’Angelo C, Bornberg-Bauer E, Kudla J, Harter K. (2007) The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Kitaoka N, Matsubara T, Sato M, Takahashi K, Wakuta S, Kawaide H, Matsui H, Nabeta K, Matsuura H. (2011) Arabidopsis CYP94B3 encodes jasmonyl-L-isoleucine 12-hydroxylase, a key enzyme in the oxidative catabolism of jasmonate. Plant Cell Physiol 52: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Koo AJK, Chung HS, Kobayashi Y, Howe GA. (2006) Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J Biol Chem 281: 33511–33520 [DOI] [PubMed] [Google Scholar]
- Koo AJK, Cooke TF, Howe GA. (2011) Cytochrome P450 CYP94B3 mediates catabolism and inactivation of the plant hormone jasmonoyl-L-isoleucine. Proc Natl Acad Sci USA 108: 9298–9303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnaswamy S, Verma S, Rahman MH, Kav NNV. (2011) Functional characterization of four APETALA2-family genes (RAP2.6, RAP2.6L, DREB19 and DREB26) in Arabidopsis. Plant Mol Biol 75: 107–127 [DOI] [PubMed] [Google Scholar]
- Li H, Sun J, Xu Y, Jiang H, Wu X, Li C. (2007) The bHLH-type transcription factor AtAIB positively regulates ABA response in Arabidopsis. Plant Mol Biol 65: 655–665 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. (2004) JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16: 1938–1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. (2003) ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreti E, Povero G, Novi G, Solfanelli C, Alpi A, Perata P. (2008) Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol 179: 1004–1016 [DOI] [PubMed] [Google Scholar]
- Miersch O, Neumerkel J, Dippe M, Stenzel I, Wasternack C. (2008) Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol 177: 114–127 [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Mithöfer A, Kombrink E, Boland W, Hamamoto S, Uozumi N, Tohma K, Ueda M. (2011) 12-Hydroxyjasmonic acid glucoside is a COI1-JAZ-independent activator of leaf-closing movement in Samanea saman. Plant Physiol 155: 1226–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. (2013) A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell 25: 1641–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. (2011) Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot 62: 2143–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Kinoshita K. (2009) Rank of correlation coefficient as a comparable measure for biological significance of gene coexpression. DNA Res 16: 249–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Nishida K, Kasahara K, Kinoshita K. (2011) ATTED-II updates: condition-specific gene coexpression to extend coexpression analyses and applications to a broad range of flowering plants. Plant Cell Physiol 52: 213–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Sasaki-Sekimoto Y, Oikawa A, Jikumaru Y, Shinoda S, Inoue E, Kamide Y, Yokoyama T, Hirai MY, Shirasu K, et al. (2011) 12-Oxo-phytodienoic acid-glutathione conjugate is transported into the vacuole in Arabidopsis. Plant Cell Physiol 52: 205–209 [DOI] [PubMed] [Google Scholar]
- Park J-H, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. (2002) A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J 31: 1–12 [DOI] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, Tilleman S, Grunewald W, Pérez AC, Chico JM, Bossche RV, Sewell J, Gil E, et al. (2010) NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464: 788–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Morreel K, De Witte E, Lammertyn F, Van Montagu M, Boerjan W, Inzé D, Goossens A. (2008) Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc Natl Acad Sci USA 105: 1380–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. (2008) The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol 147: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. (2011) The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 23: 1795–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe C, Springer A, Samol I, Reinbothe S. (2009) Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J 276: 4666–4681 [DOI] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Häweker H, Dong X, Robatzek S, Schulze-Lefert P. (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Asamizu E, Shibata D, Nakamura Y, Kaneko T, Awai K, Amagai M, Kuwata C, Tsugane T, Masuda T, et al. (2001) Monitoring of methyl jasmonate-responsive genes in Arabidopsis by cDNA macroarray: self-activation of jasmonic acid biosynthesis and crosstalk with other phytohormone signaling pathways. DNA Res 8: 153–161 [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Taki N, Obayashi T, Aono M, Matsumoto F, Sakurai N, Suzuki H, Hirai MY, Noji M, Saito K, et al. (2005) Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 44: 653–668 [DOI] [PubMed] [Google Scholar]
- Schilmiller AL, Howe GA. (2005) Systemic signaling in the wound response. Curr Opin Plant Biol 8: 369–377 [DOI] [PubMed] [Google Scholar]
- Seo HS, Song JT, Cheong JJ, Lee YH, Lee YW, Hwang I, Lee JS, Choi YD. (2001) Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc Natl Acad Sci USA 98: 4788–4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X, Zhang Y, Peng W, Wang Z, Xie D. (2009) Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J Exp Bot 60: 3849–3860 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu F-F, Sharon M, Browse J, et al. (2010) Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468: 400–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. (2006) The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol 24: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley BW, Kubasek WL, Storz G, Bruggemann E, Koornneef M, Ausubel FM, Goodman HM. (1995) Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J 8: 659–671 [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. (2011) Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solfanelli C, Poggi A, Loreti E, Alpi A, Perata P. (2006) Sucrose-specific induction of the anthocyanin biosynthetic pathway in Arabidopsis. Plant Physiol 140: 637–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE. (2008) JAZing up jasmonate signaling. Trends Plant Sci 13: 66–71 [DOI] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, Kikuta A, Kobayashi K, Ainai T, Yagi K, Sakurai N, Suzuki H, Masuda T, et al. (2005) 12-Oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiol 139: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. (2005) Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol 139: 1840–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. (2007) JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448: 661–665 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Wasternack C. (2007) Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann Bot (Lond) 100: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG. (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay S. (2005) A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17: 1953–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara T, Omer E-SA, Koshino H, Sakamura S, Kikuta Y, Koda Y. (1989) Structure of a tuber-inducing stimulus from potato leaves (Solanum tuberosum L.). Agric Biol Chem 53: 2835–2837 [Google Scholar]
- Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K. (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21: 2914–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarei A, Körbes AP, Younessi P, Montiel G, Champion A, Memelink J. (2011) Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol Biol 75: 321–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. (2003) A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development 130: 4859–4869 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Turner JG. (2008) Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3: e3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-Y, Spivey NW, Zeng W, Liu P-P, Fu ZQ, Klessig DF, He SY, Dong X. (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H. (2010) The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene 457: 1–12 [DOI] [PubMed] [Google Scholar]