A chromatin remodeling complex subunit physically interacts with DELLA proteins and is involved in control of gibberellin biosynthesis and hormonal cross talk.
Abstract
Switch (SWI)/Sucrose Nonfermenting (SNF)-type chromatin-remodeling complexes (CRCs) are involved in regulation of transcription, DNA replication and repair, and cell cycle. Mutations of conserved subunits of plant CRCs severely impair growth and development; however, the underlying causes of these phenotypes are largely unknown. Here, we show that inactivation of SWI3C, the core component of Arabidopsis (Arabidopsis thaliana) SWI/SNF CRCs, interferes with normal functioning of several plant hormone pathways and alters transcriptional regulation of key genes of gibberellin (GA) biosynthesis. The resulting reduction of GA4 causes severe inhibition of hypocotyl and root elongation, which can be rescued by exogenous GA treatment. In addition, the swi3c mutation inhibits DELLA-dependent transcriptional activation of GIBBERELLIN-INSENSITIVE DWARF1 (GID1) GA receptor genes. Down-regulation of GID1a in parallel with the DELLA repressor gene REPRESSOR OF GA1-3 1 in swi3c indicates that lack of SWI3C also leads to defects in GA signaling. Together with the recent demonstration of function of SWI/SNF ATPase BRAHMA in the GA pathway, these results reveal a critical role of SWI/SNF CRC in the regulation of GA biosynthesis and signaling. Moreover, we demonstrate that SWI3C is capable of in vitro binding to, and shows in vivo bimolecular fluorescence complementation interaction in cell nuclei with, the DELLA proteins RGA-LIKE2 and RGA-LIKE3, which affect transcriptional activation of GID1 and GA3ox (GIBBERELLIN 3-OXIDASE) genes controlling GA perception and biosynthesis, respectively. Furthermore, we show that SWI3C also interacts with the O-GlcNAc (O-linked N-acetylglucosamine) transferase SPINDLY required for proper functioning of DELLAs and acts hypostatically to (SPINDLY) in the GA response pathway. These findings suggest that DELLA-mediated effects in GA signaling as well as their role as a hub in hormonal cross talk may be, at least in part, dependent on their direct physical interaction with complexes responsible for modulation of chromatin structure.
The Switch (SWI)/Sucrose Nonfermenting (SNF)-type chromatin remodeling complexes (CRCs) are evolutionary conserved in eukaryotes. They carry a central Snf2-type ATPase in association with several core subunits that correspond to orthologs of SNF5, SWI3, and SWP73 (SWI/SNF-associated protein 73) proteins of the yeast (Saccharomyces cerevisiae) prototype of SWI/SNF CRCs. In mammals, the core noncatalytic subunits of SWI/SNF-type complexes, such as SWI3, directly interact with nuclear hormone receptors and coactivators (DiRenzo et al., 2000; Zraly et al., 2006; John et al., 2008). All known core subunits of SWI/SNF complexes are conserved in plants. The Arabidopsis (Arabidopsis thaliana) genome encodes four SNF2 ATPases and four SWI3-type proteins, which build various SWI/SNF complexes with different subunit composition (Sarnowski et al., 2005). Mutations affecting the Arabidopsis SWI/SNF subunits cause characteristic alterations in plant development and responses to environmental factors. As yet, detailed characterization of knockouts of BRAHMA (BRM) and SPLAYED (SYD) ATPase and four SWI3 genes (SWI3A, SWI3B, SWI3C, and SWI3D) has been reported (Sarnowski et al., 2002, 2005; Farrona et al., 2004; Bezhani et al., 2007; Archacki et al., 2009). However, the exact molecular mechanisms by which these mutations cause complex developmental and physiological defects are so far largely unknown.
Our earlier studies revealed that, in Arabidopsis, the BRM ATPase and SWI3C CRC subunits fulfill most of their functions by acting in a common complex. However, we also found that BRM has additional and specific functions that are independent of SWI3C (Archacki et al., 2009). Transcriptome analysis of brm and syd mutant lines indicated that these mutations modify the expression of genes in several signaling pathways, including the gibberellin (GA) and abscisic acid (ABA) hormone pathways (Bezhani et al., 2007; Saez et al., 2008). GA is responsible for regulation of growth and other basic processes, including germination, shoot and root elongation, flower development, flowering time, seed development and maturation, and aging (Fleet and Sun, 2005). The best-studied downstream elements in the GA pathway are the DELLA proteins that act as general repressors of GA-stimulated processes (Peng et al., 1997, Silverstone et al., 1998). Upon accumulation, GA is perceived by the GIBBERELLIN INSENSITIVE DWARF1 (GID1) nuclear receptors (GID1a, GID1b, and GID1c in Arabidopsis; Ueguchi-Tanaka et al., 2005; Nakajima et al., 2006), and the GA-GID1 complex binds to DELLAs (Griffiths et al., 2006; Willige et al., 2007; Ueguchi-Tanaka et al., 2007). This enables interactions with the F-box protein SLEEPY/GID2 that mediates polyubiquitinylation and subsequent proteasomal degradation of the DELLA repressors (Sasaki et al., 2003; Dill et al., 2004). The activity of DELLAs is likely also regulated by other pathways. The enzyme O-GlcNAc transferase encoded by the SPINDLY (SPY) gene was shown to enhance the repressor activity of DELLAs (Shimada et al., 2006; Silverstone et al., 2007). We have recently demonstrated that BRM affects the expression of a significant number of GA-responsive genes, including GIBBERELLIN 3-OXIDASE (GA3ox1) and that the level of active GA is markedly decreased in the brm null mutant (Archacki et al., 2013).
Here, we show that proper regulation of plant responses to several hormones requires the function of core SWI3C subunit of SWI/SNF CRCs and provides novel clues regarding a possible mechanism underlying SWI/SNF-mediated regulation of the GA hormone response pathway. We show that inactivation of SWI3C results in developmental abnormalities that are characteristic for Arabidopsis mutants impaired in GA biosynthesis and signaling. The swi3c mutation, similarly to the brm mutation, markedly decreases the levels of bioactive GA derivatives by causing pathway-wide alteration in the transcription of genes involved in the biosynthesis and inactivation of GAs. Furthermore, the swi3c mutation also down-regulates the expression of GID1 GA receptor genes, which may affect the GA perception in leaves. Moreover, SWI3C physically interacts in the nucleus with several DELLA proteins and with SPY, which appears to act upstream of SWI3C in the GA response pathway. Physical interactions of SWI3C with DELLAs and SPY suggest that the function of SWI3C-containing SWI/SNF CRCs may be required for some of the DELLA-mediated effects, such as activation of GID1 and GA3ox genes involved in GA perception and biosynthesis, respectively.
RESULTS
The swi3c Mutant Shows Altered Ethylene, ABA, Brassinosteroid, Gravitropic, and GA Responses and Confers GA-Related Growth and Developmental Defects
During initial characterization of the swi3c transfer DNA insertion mutants (Sarnowski et al., 2005), we observed that seedlings carrying either the swi3c-1 or swi3c-2 mutant alleles showed similarly altered phenotypic traits compared with the wild type when germinated on media containing different phytohormones. Subsequently, we used the swi3c-1 mutant to examine in more detail several hormone responses in germination and seedling growth assays. When germinated in the presence of increasing concentrations of ABA, the swi3c mutant displayed reduced germination rate compared with the wild type, indicating that similarly to BRM (Han et al., 2012), inactivation of the SWI3C SWI/SNF subunit results in enhanced sensitivity to ABA (Supplemental Fig. S1A). Dark-grown swi3c seedlings developed short hypocotyls and roots when germinated in the presence of ethephon, which is hydrolyzed to ethylene in the medium, and the ethylene precursor aminocyclopropane-1-carboxylic acid, suggesting an increased ethylene sensitivity (Supplemental Fig. S1B). In response to brassinosteroid treatment, light-grown swi3c-1 plants responded with enhanced hypocotyl elongation compared with the wild type (Supplemental Fig. S1C). Finally, the swi3c-1 mutation showed a defect of root gravitropic response compared with the wild type, indicating that in addition to inhibition of root elongation, the swi3c-1 mutation also prevented auxin-dependent gravitropic root bending (Supplemental Fig. S1D). These preliminary germination and growth assays thus highlighted an alteration of multiple hormonal responses underlying the severe developmental defects observed in the swi3c mutant (Sarnowski et al., 2005).
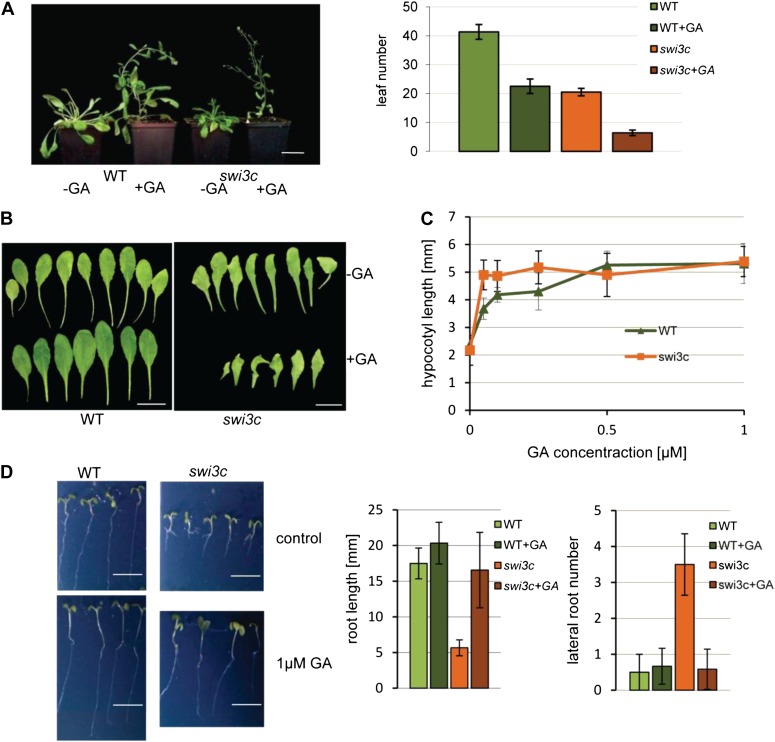
Compared with other hormones, the swi3c mutant showed markedly enhanced growth and flowering responses to external feeding with bioactive GA4+7. Compared with the wild type, GA4+7 treatment resulted in acceleration of flowering of swi3c (counted in number of leaves to flowering), which also flowered earlier than the wild type without GA treatment under short-day (SD) condition (Fig. 1A; Sarnowski et al., 2005). However, unlike the wild type, swi3c plants did not display an increase of leaf blade size upon GA treatment (Fig. 1B). By contrast, treatment with 1µm GA4+7 completely suppressed the defects of hypocotyl and root elongation of swi3c mutant seedlings, which developed like the wild type in the presence of GA (Fig. 1, C and D; Supplemental Fig. S2). At higher (10 µm) concentration, GA4+7 marginally inhibited root but not hypocotyl elongation of both wild-type and swi3c seedlings (Supplemental Fig. S2). In addition, GA treatment abolished characteristic branching of swi3c mutant roots on 0.5% Suc-containing Murashige and Skoog (MS) medium (Fig. 1D). Compared with the wild type, the swi3c seedlings proved to be insensitive to the GA biosynthesis inhibitor paclobutrazol (PAC). Even at very low concentration (2.5 nm), PAC-treatment reduced the hypocotyl length of wild-type seedlings. By contrast, PAC treatment of swi3c seedlings stimulated hypocotyl shortening only when PAC concentration was increased to 1 µm (Supplemental Fig. S2). In summary, several developmental defects observed in the swi3c mutant proved to be similar to those of GA-deficient mutants. Furthermore, suppression of root and hypocotyl elongation defects by GA4+7 indicated that similarly to mutations of the BRM ATPase (Archacki et al., 2013), the hypocotyl and root elongation defects caused by inactivation of the SWI3C SWI/SNF CRC core subunit were due to deficiency of GA biosynthesis.
Figure 1.
The swi3c mutation confers GA-related growth defects. A, Compared the wild type, GA4+7 treatment resulted in acceleration of flowering time in the swi3c mutant that also flowers earlier than the wild type without GA treatment under SD condition. Six-week-old swi3c and wild-type plants grown in SD conditions untreated or treated with 100 μm GA4+7. Bar = 5 cm. B, The leaves of swi3c mutant did not show blade expansion after GA treatment, indicating an organ-specific defect in GA response. Bar = 1 cm. C, Treatment with 1 µm GA4+7 completely suppressed the defect of hypocotyl elongation of swi3c mutant seedlings. D, Treatment with 1 µm GA4+7 suppressed the defects of hypocotyl and root elongation of swi3c mutant seedlings.
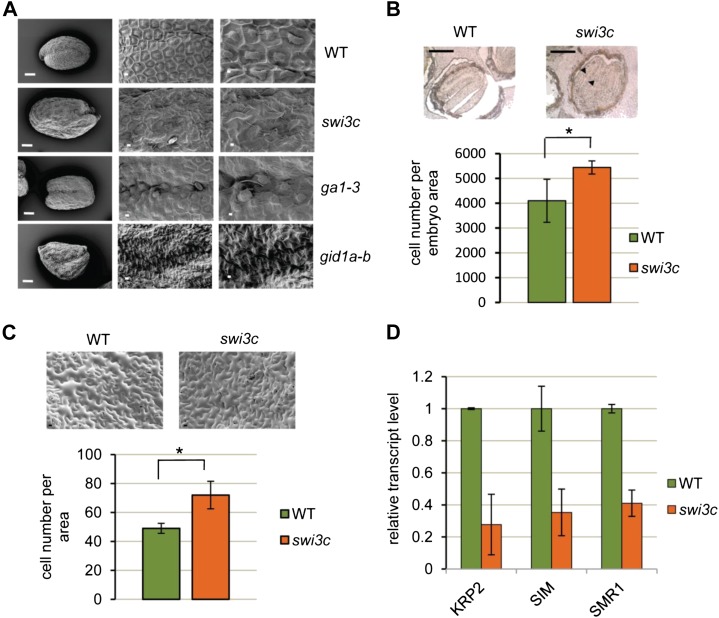
We reported previously that the Arabidopsis swi3c mutation results in complex pleiotropic developmental defects (Sarnowski et al., 2005). Some of these pleiotropic deficiencies, such as enhanced leaf curling and alterations in the development of flower organs, were also identified in the brm mutant and are thus typical for plants deficient in the function of SWI/SNF CRCs (Archacki et al., 2009). By contrast, other phenotypic traits of the swi3c and brm mutants, in particular their dark-green leaf color and semidwarf, stature resemble those of GA-deficient mutants that show reduced GA biosynthesis and accumulation of DELLA proteins (Koornneef and van der Veen, 1980). The Arabidopsis mutants gid1a, gid1b, and ga1-3 deficient in GA perception and biosynthesis, respectively, display reduced germination, abnormal seed shape, and irregular cell division patterns in the seed coat (Iuchi et al., 2007). Using scanning electron microscopic studies, we found that the epidermal cell layer of irregularly shaped swi3c mutant seeds is similarly characterized by highly abnormal patterns of cells, which differ in both size and shape from seed coat epidermal cells of the wild type (Fig. 2A). Next, we examined the structure of mature wild-type and swi3c embryos using cross sections of seeds embedded into wax after 24 h of imbibition and fixation with paraformaldehyde. Cross sections of matured swi3c embryos revealed aberrant development characterized by larger embryo size, increased cell number, and improperly developed cotyledons compared to the wild type. This indicated that swi3c mutation altered normal regulation of cell division not only in seed epidermis, but also during embryogenesis (Fig. 2B). Compared with the wild type, the swi3c mutant had higher density of cells per unit surface area of the leaf epidermis (Fig. 2C). In addition to organ-specific changes in cell number and size, the transcription of genes encoding the cell cycle inhibitors KIP-RELATED PROTEIN2, SIAMESE, and SIAMESE RELATED1 showed a marked reduction in the swi3c mutant (Fig. 2D). Together, these results were consistent with our observations indicating that SWI3C-containing SWI/SNF CRCs are involved in the regulation of multiple hormonal pathways and suggested that at least part of the complex swi3c mutant phenotype resulted from aberrant GA biosynthesis and/or signaling.
Figure 2.
The swi3c mutation confers GA-related developmental defects. A, The scanning electromicroscopic analysis of seed coat structure of swi3c mutant indicates similar changes to those observed in GA pathway mutants ga1-3 and gid1a-b. Bars = 100 μm (left) and 10 μm (middle and right). B, Cross sections of mature swi3c embryos. Arrowheads indicate improperly developed cotyledons. Bar = 500 μm. *P value < 0.05. C, The cell number of 4-week-old LD-grown swi3c mutant leaves is increased. Bar = 10 μm. *P value < 0.05. D, The expression levels of cell cycle inhibitors are markedly reduced in the swi3c mutant.
The swi3c Mutant Has Decreased Level of Bioactive GA4
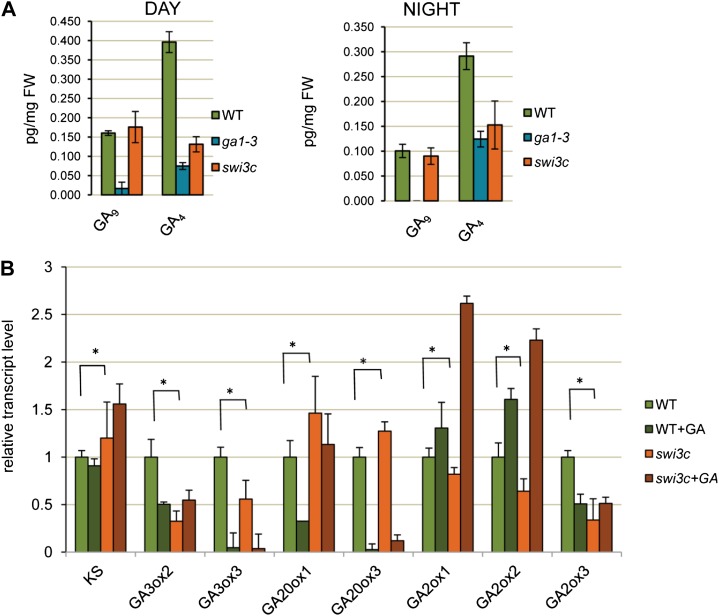
To verify the latter conclusion inferred from physiological assays, we compared the levels of GA biosynthesis intermediates, bioactive GAs, and inactive GA metabolites in swi3c and ga1-3 mutant and wild-type plants, which were collected at the end of day and at the end of night during a diurnal growth period. Quantitative measurements of GAs revealed that similarly to ga1-3, the swi3c mutant contained reduced levels of bioactive GA4, as well as GA34, the inactive metabolite of GA4 (Fig. 3A; Supplemental Fig. S3B; Supplemental Table S1, A and B). We did not observe an accumulation of GA9 but found that the swi3 mutant accumulated higher levels of GA15, GA19, and GA51 compared with the wild type, indicating a shift of GA biosynthesis toward the inactive GA51 derivative rather than active GA4. Consequently, similarly to brm (Archacki et al., 2013), the swi3c mutant appeared to be deficient in the biosynthesis of active GAs.
Figure 3.
swi3c mutant has decreased GA4 content and shows altered transcriptional regulation of GA pathway genes. A, Four-week-old LD-grown wild-type, swi3c, and ga1-3 plants were collected at the end of night and end of day and subjected to GA analysis. Both swi3c and ga1-3 have decreased level of bioactive GA4. B, Transcription of genes acting in GA biosynthesis and metabolism shows coordinate changes in swi3c mutant. Reduction of bioactive GA in swi3c mutant correlates with decreased expression of GA3ox2 and GA3ox3 genes, as well as overexpression of GA2ox1 and GA2ox2. *P value < 0.05.
Transcription of Genes Acting in GA Biosynthesis and Metabolism Show Coordinate Changes in the swi3c Mutant
Next, we examined the abundance of transcripts encoding key enzymes of GA metabolism, including KS (ent-kaurene synthase B), KAO1 and KAO2 (ent-kaurenoic acid hydroxylase), KO (ent-kaurene oxidase), CPS (ent-copalyl diphosphate synthase), GA3ox1-3 (GA 3-β-dioxygenase), GA20ox1 and GA20ox3 (GA 20-oxidase), and GA2ox1-3 (GA 2-β-dioxygenase) in soil-grown swi3c seedlings. Compared with the wild type, the KS (GA2) transcript level was slightly elevated in swi3c mutant in both absence and presence of GA4+7 treatment, whereas the KAO1 transcript was marginally elevated only when swi3c was treated with GA4+7. The KS ent-kaurene synthase B catalyzes a second step in cyclization of geranylgeranyl pyrophosphate to ent-kaurene, whereas the KAO1 ent-kaurenoic acid hydroxylase controls the further three steps in GA biosynthetic pathway from ent-kaurenoic acid to GA12 (Hedden and Phillips, 2000). More importantly, compared with the wild type in the absence of GA treatment, the swi3c mutant showed a 2-fold reduction of transcript level of GA3ox2 encoding GA 3-β-dioxygenase2, which catalyzes the hydroxylation of GA9 and GA20 to bioactive GA4 and GA1, respectively (Curaba et al., 2004). There was also no compensation of GA3ox2 by GA3ox3 gene, which acts in GA-dependent regulation of flower organ development, as the expression of the latter gene was marginally reduced in swi3c. The expression of GA3ox2 was, however, restored to the wild-type level in GA-treated swi3c plants (Fig. 3B). In parallel, the transcript levels of GA2ox1, GA2ox2, and GA2ox3 genes, which code for GA 2-oxidases that inactivate the GA19-derived GAs, including GA9 and GA20 precursors of bioactive GA4 and GA1, were reduced 0.8- to 2.8-fold in swi3c mutant. In comparison, transcript levels of GA20ox1 and GA20ox3 genes, involved in the synthesis of precursors of bioactive GAs, were slightly higher in swi3c compared with the wild type. The treatment with GA4+7 increased the abundance of GA2ox1 and GA2ox2 transcripts over 2-fold to a level 60% to 80% higher than in the wild type, revealing that inactivation of bioactive GAs was enhanced in swi3c mutant when plants were treated with exogenous GA. Together, these observations are consistent with extensive deregulation of the GA-mediated feedback control of GA biosynthesis pathway (Griffiths et al., 2006) in swi3c mutant.
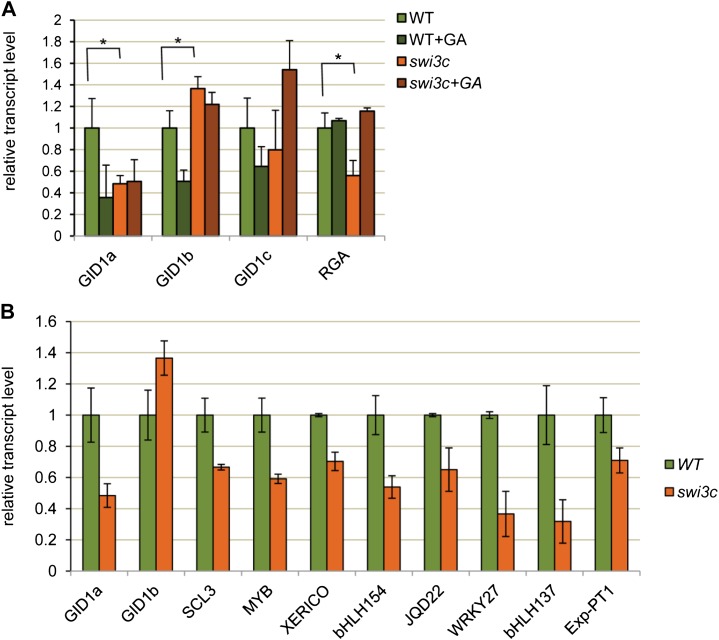
The swi3c Mutation Alters the Regulation of GID1 GA-Receptor Genes and Numerous Known DELLA Target Genes
Although direct measurements of GA levels clearly indicated that the swi3c mutation caused GA deficiency, some of the developmental defects of swi3c plants (e.g. formation of curling leaves and expansion of leaf blades) were not restored to the wild type by GA treatment (Fig. 1B). To check whether this was due to alteration of tissue-specific GA perception in swi3c, we compared the abundance of GID1 GA receptor transcripts in leaves of wild-type and swi3c plants. Transcription of GID1a, encoding the most abundant form of GA receptor expressed at the highest level in all plant organs except roots (Griffiths et al., 2006), showed over 2-fold reduction in swi3c mutant. The transcript level of GID1a in swi3c, both without and with GA treatment, was comparable to that in wild-type plants upon GA-mediated feedback inhibition of GID1a (Fig. 4A). GID1b, which is expressed at higher level than GID1a in roots but similarly inhibited by GA (Griffiths et al., 2006), showed slightly higher transcript levels but no GA inhibition in swi3c. Finally, GID1c, which is expressed at very low level compared with GID1a and GID1b in most plant organs, showed GA-stimulated, rather than -inhibited, transcription in swi3c. In comparison, transcription of REPRESSOR OF GA1-3 1 (RGA), encoding one of the five DELLA repressors, was reduced 2-fold in swi3c leaves but restored to wild-type levels by GA treatment. Additionally, the analysis of expression of genes encoding other DELLA proteins indicated that the RGA-LIKE1 (RGL1), RGL3, and GIBBERELLIC ACID INSENSITIVE (GAI) transcription levels were reduced 1.5- to 2-fold, whereas the RGL2 transcript level was 2-fold elevated in swi3c leaves (Supplemental Fig. S4). Altered transcription of GID1 genes in the absence of GA and a lack of their GA-mediated feedback inhibition in swi3c leaves thus suggested that SWI3C-contaning SWI/SNF chromatin remodeling complexes are required for proper transcriptional regulation of the GA receptors. As the GID1a and GID1b genes are considered to be direct targets of the DELLA repressors, we also tested several known DELLA target genes encoding SCARECROW-LIKE3, a member of the GRAS family of putative transcriptional regulators, the MYB nuclear transcription factor XERICO E3 ubiquitin ligase, IQD22 protein of the IQD (IQ domain) family of calmodulin binding proteins, WRKY27 transcription factor, bHLH137 and bHLH154 basic helix-loop-helix DNA-binding superfamily proteins, and Exp-PT1 (GA marker gene), a protein predicted to be localized in the nucleus. (Zentella et al., 2007). The transcript levels of these known DELLA target genes showed 1.5- to 4-fold reduction in the swi3c mutant (Fig. 4B). Taken together, deregulation of GID1 genes, alteration of expression of all DELLA genes, and altered transcriptional regulation of several known DELLA target genes observed in the swi3c mutant suggested that SWI/SNF CRC complexes play a role in the regulation of DELLA repressors and thereby DELLA-dependent activation and GA-mediated feedback inhibition of transcription of GID1 GA receptor (Griffiths et al., 2006) and other DELLA target genes (Zentella et al., 2007).
Figure 4.
Altered transcriptional regulation of GID1, RGA, and DELLA target genes in the swi3c mutant. A, The swi3c mutation causes altered regulation of GID genes of GA receptors and the RGA gene coding for a DELLA protein. *P value < 0.05. B, The direct target genes for DELLA repressor proteins show altered transcription in the swi3c mutant. *P value < 0.05.
SWI3C Physically Interacts with DELLAs and the O-GlcNAc Transferase SPY in the Nucleus
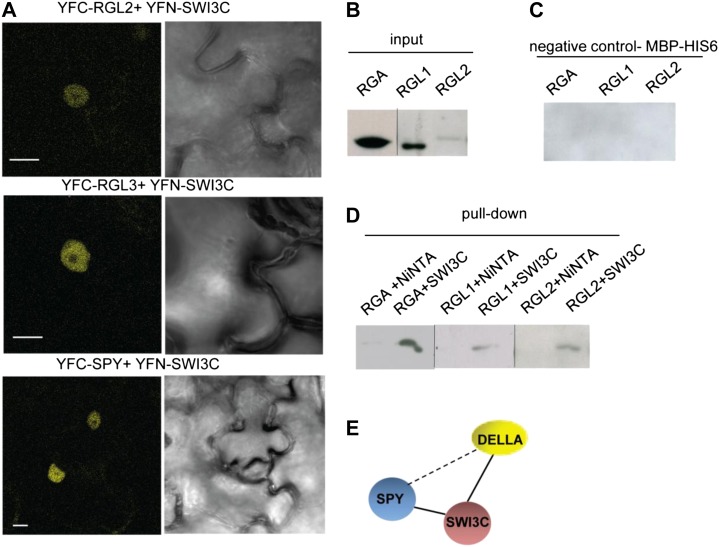
DELLA repressors of GA responses do not bind directly to DNA and are thus thought to regulate the expression of their target genes through interactions with transcription factors, as demonstrated for the PHYTOCHROME INTERACTING FACTOR (PIF) bHLH transcription factors promoting hypocotyl elongation (de Lucas et al., 2008; Feng et al., 2008) and/or chromatin modification complexes (Zentella et al., 2007; Sun, 2010). The failure of proper DELLA-dependent activation and GA-mediated repression of GID1 and GA3ox genes in the swi3c mutant raised the possibility that SWI3C-containing SWI/SNF complexes may somehow mediate the effects of DELLA repressor on these target genes, perhaps analogously to involvement of animal CRCs with nuclear receptors (Zraly et al., 2006). Because both DELLA and SWI3C (Sarnowski et al., 2005 and R. Archacki, unpublished results) proteins self-activate the reporter gene when fused to the GAL4 binding domain in yeast, we were unable to test the interaction using two-hybrid assay. Therefore, we used bimolecular fluorescence complementation (BiFC) assays (Hu et al., 2002), in which SWI3C fused to the N-terminal domain of split Yellow Fluorescent Protein (YFP; YFN-SWI3C) was transiently coexpressed with DELLA repressors in fusion with the C-terminal domain (YFC) of split YFP in epidermal cells of Nicotiana benthamiana. Similarly, we used an YFC fusion of SPY to determine whether this O-GlcNAc transferase required for activation of DELLAs was recruited by SWI3C-containing SWI/SNF CRCs. Using high-resolution confocal microscopy, we detected reconstitution of YFP activity in epidermal cell nuclei revealing in vivo interaction of YFN-SWI3C with the YFC-fused DELLA repressors RGL2 and RGL3 and SPY (Fig. 5A). Subsequently, we performed control BiFC assays, in which YFC fusions of RGL2, RGL3, and SPY were individually coexpressed with YFN fusions of the red fluorescent proteins (YFN-RFP), whereas an YFC-RFP fusion was expressed simultaneously with YFN-SWI3C. The lack of YFP reconstitution in each case and detection of control RFP signal in both cytoplasm and nuclei confirmed the specificity of observed BiFC interactions of SWI3C with RGL2, RGL3, and SPY (Supplemental Fig. S5A). The interaction of SWI3C with SPY was next confirmed by two-hybrid assay (Supplemental Fig. S5B). To test the robustness of observed protein interactions, we performed additional stringent in vitro protein-binding assays. SWI3C was fused to an N-terminal maltose-binding protein-6×His tag (MBP-6×His). Subsequently, equal amounts of purified MBP-6×His-SWI3C and control MBP-6×His proteins were immobilized on nickel-nitrilotriacetic acid agarose (Ni-NTA) resin and used for pull-down assays with total protein extracts from plants expressing one of the 9MYC-tagged DELLA proteins RGA, RGL1, and RGL2, respectively. None of the 9MYC-tagged DELLAs, which were loaded at equal amounts onto the different matrices, were retained on the control MBP-6×His protein (Fig. 5, B and C) and Ni-NTA resins (Fig. 5D). By contrast, anti-c-Myc immunoblotting of proteins eluted from the MBP-6×His-SWI3C matrix detected in vitro binding of all three DELLA proteins, confirming specific interaction of RGA, RGL1, and RGL2 with SWI3C (Fig. 5D). Together with the in vivo BiFC assays, the results of these in vitro pull-down assays indicated that SPY and at least three DELLA proteins interact with core SWI3C subunit of SWI/SNF CRCs. While the observed protein-protein interactions did not resolve whether SPY and DELLAs bind to SWI/SNF together or separately (Fig. 5E), they provided a first mechanistic clue for the observed role of SWI3C in regulation of GA responses.
Figure 5.
SWI3C interacts with the DELLA and SPY proteins. A, BiFC analysis of in vivo interactions between SWI3C and RGL2, RGL3, and SPY. Bar = 10 μm. B, 9MYC-tagged RGA, RGL1, or RGL2 protein levels in the input plant total protein extracts used in pull-down assays. C, Control pull-down assays with Ni-NTA-bound MBP-His-6 protein used as negative control. D, Pull-down assay with recombinant SWI3C protein with MBP-His-6 tag and total protein extracts from plants overexpressing the 9Myc-tagged RGA, RGL1, and RGL2 DELLA proteins. Ni-NTA-protein fraction isolated from bacteria without induction of SWI3C-MBP-His-6 construct was combined with DELLA protein extracts as additional negative control. E, Schematic visualization of DELLA, SPY, and SWI3C interactions. Full lines indicate direct interactions and dashed line indicates functional relation between SPY and DELLA. DIC, Differential interference contrast image.
Genetic Interactions between the swi3c and spy-1 Mutations
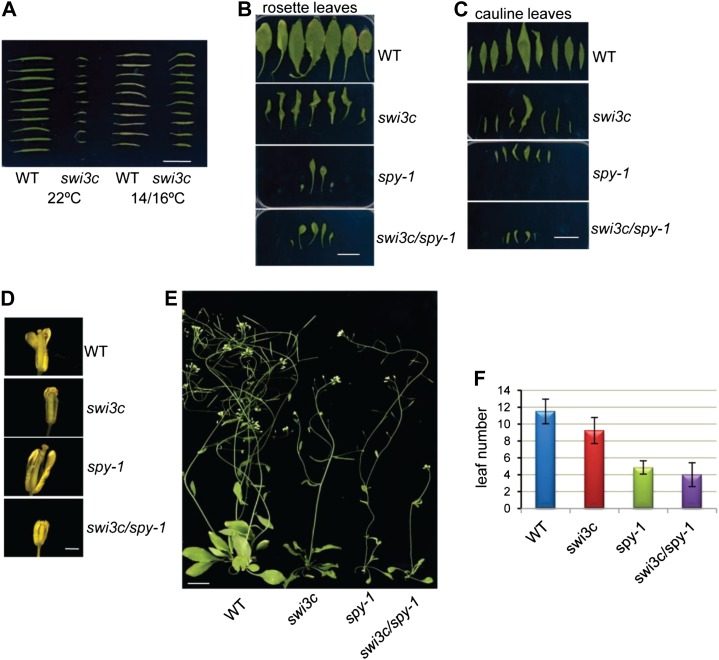
Inactivation of the SPY O-GlcNAc transferase in Arabidopsis dramatically reduces fertility. The spy-1 mutant develops short siliques that produce very low amount of seed at normal temperature. However, seed production is restored to nearly normal when spy-1 is grown at 18°C (Jacobsen and Olszewski, 1993). Given that the swi3c mutant produces very few seeds when grown at normal 22°C day and 18°C night temperature, we tested whether, analogously to spy-1, this defect could be due to temperature sensitivity of the swi3c mutant. When grown at 14°C or 16°C under (16-h/8-h day/night) diurnal cycle, the swi3c mutant produced approximately 2-fold longer siliques containing a higher number of viable seeds (Fig. 6A; Supplemental Fig. S3A). Surprisingly, this suggested that low temperature partially lifted the need for chromatin remodeling for some growth processes connected to seed production in swi3c. Because we found that SWI3C interacts with SPY, it was interesting to determine whether spy-1 mutation alters the phenotypic traits of the swi3c mutant. In fact, swi3c and spy-1 single mutants showed close phenotypic similarity, except that swi3c seedlings developed curling rosette and cauline leaves and had frequent defects of carpel and stamen development in their flowers. Introduction of the spy-1 mutation into swi3c background resulted in the development of spy-1-like noncurling leaves, but the swi3c spy-1 double mutant displayed similar developmental defects of stamens and carpels and even more retarded vegetative growth as the swi3c single mutant (Fig. 6, B–E). In regard to the latter phenotypic traits, the effect of the swi3c mutation on leaf development appeared thus hypostatic to those of spy-1. However, compared with swi3c, the spy-1 single mutant flowered earlier, with about one-half the number of leaves, whereas the swi3c spy-1 double mutant flowered in comparison even earlier (Fig. 6F). Furthermore, the spy-1 mutation synergistically shortened the lengths of flower organs and resulted in complete sterility in combination with swi3c. Thus, SPY turned out necessary for proper execution of flower and seed developmental programs, which were impaired in a temperature-dependent manner by the swi3c mutation.
Figure 6.
Genetic interaction between swi3c and spy-1 mutations. A, swi3c mutant has greatly reduced fertility when grown under optimal conditions, while lower temperature (16°C day/14°C night) stimulates the elongation of swi3c siliques and increases fertility, resembling the behavior of spy-1 mutant. B, swi3c spy-1 double mutant exhibits rosette leaf phenotype similar to spy-1, with no twisting and curling characteristic for swi3c. C, Similarity of cauline leaf phenotype of swi3c spy-1 double mutant and spy-1. D, The flowers of swi3c spy-1 double mutant have similar developmental changes of carpels and stamens as swi3c single mutant, but the double mutant is sterile. E, Twenty-eight-day-old swi3c spy-1 plants have spy-1-like phenotype but show even more retarded vegetative growth and sterility. F, swi3c spy-1 mutant flowers slightly earlier than spy-1. Numbers of leaves were compared at the time of flowering.
DISCUSSION
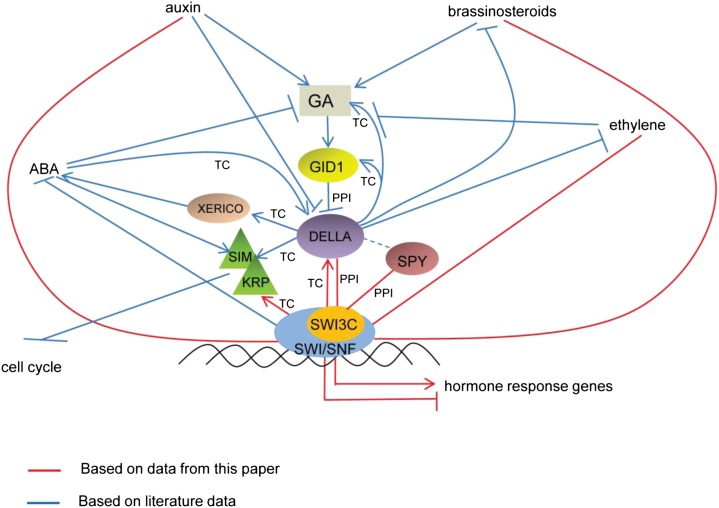
Studies in yeast and animals document that a major function of SWI/SNF complexes is the control of nucleosome dynamics at gene promoters and enhancers (Euskirchen et al., 2011). A particularly well-studied role of SWI/SNF CRCs in animals is their interaction with nuclear receptors. The binding of steroid hormones by nuclear receptor enables their interactions with coactivators, one of which is the SWI/SNF complex. Consistently, genes regulated by steroid hormones are in vivo targets for regulation by SWI/SNF CRCs (Belandia and Parker, 2003; Zraly et al., 2006). As in mammals, SWI/SNF chromatin remodeling complexes in Arabidopsis are involved in transcriptional regulation of genes controlling important developmental and hormonal pathways. Our recent study revealed that BRM, a major SWI/SNF ATPase in Arabidopsis, is involved in regulation of GA signaling (Archacki et al., 2013). The brm mutation was also found to result in derepressed expression of the ABA INSENSITIVE5 gene that encodes a BASIC LEUCINE-ZIPPER transcription factor regulating ABA sensitivity of germinating seeds (Han et al., 2012). Present characterization of phenotypic defects caused by inactivation of the core SWI3C subunit of SWI/SNF CRCs indicates complex alteration of several hormone regulatory pathways. Among these, the swi3c mutation simultaneously affects the ABA, ethylene, brassinosteroid, and GA signaling pathways by differentially modulating plant responses to these hormones (Fig. 7). We reported previously that many, but not all, phenotypic traits of the swi3c mutant overlap with those of brm impaired in the function of the BRAHMA SNF2 ATPase subunit (Sarnowski et al., 2005; Archacki et al., 2009). Therefore, it is not surprising that both brm and swi3c mutations result in similar enhancement of ABA hormone sensitivity, which further supports our notion that these subunits act in the same SWI/SNF CRC. Collectively, the above data implicate the SWI/SNF complexes in cross talk and integration of different hormonal pathways in Arabidopsis. To reveal the possible molecular basis of such a role, we decided to concentrate on the characterization of SWI/SNF subunit mutants to GAs.
Figure 7.
A hypothetical model of regulatory network centered at SWI/ SNF and DELLAs on the basis of published data and the results of this paper. The SWI/SNF complex influences various hormonal pathways either by controlling a response to a hormonal treatment or by direct interaction with elements of hormonal pathways or their target genes modulating hormonal cross talk in Arabidopsis. Red lines correspond to data presented in this paper. Blue lines represent published data (Fu and Harberd, 2003; Achard et al., 2009; Marrocco et al., 2010; Sun, 2010; Han et al., 2012). TC, Transcriptional control; PPI, protein- protein interaction.
The swi3c mutant is characterized by a semidwarf growth habit and other traits, such as altered cell division patterns in embryos, seed coat epidermis, and leaves, which resemble those of GA-deficient mutants. In this study we demonstrate that many of the developmental defects observed in the swi3c mutant, in particular defective elongation of hypocotyls and roots during seedling development, can be suppressed and restored to the wild type by exogenous GA4+7 treatment. Quantitative analysis of precursors, active forms, and inactive derivatives of GAs revealed that similarly as in brm mutant, the amount of bioactive GA4 hormone is largely reduced in swi3c seedlings. Systematic quantitative real-time (qRT)-PCR analysis of transcription of genes involved in GA biosynthesis and inactivation showed that the main reason for reduced biosynthesis of GA4 is the down-regulation of transcription of GA3ox2 and GA3ox3 genes that control the production of bioactive GAs GA4 and GA1 in different organs (Curaba et al., 2004). In addition, exogenous GA treatment enhanced the activation of GA2ox1 and GA2ox2 genes, leading to a conversion of accumulating GA19 precursor toward inactive GAs, such as GA51 in the swi3c mutant.
The biosynthetic GA3ox and GID1a GA receptor genes were reported to show DELLA-dependent activation and GA-dependent feedback inhibition (Griffiths et al., 2006). Because of GA feedback regulation of GID1 transcription, the defect of GA biosynthesis also has consequences for GA signaling via the receptors. In accordance, we found that GID1a, encoding the most abundant GA receptor, displays reduced transcription and lack of apparent GA inhibition in leaves of the swi3c mutant. Down-regulation of the GID1a in mutant leaves may be one of the reasons that swi3c leaves failed to respond to a similar extent as the wild type to externally provided GA by typical leaf blade expansion (Fig. 1B).
The activity and stability of DELLA repressors is negatively controlled by GA binding and activation of GID1 receptors and subsequent formation of stable GID1-DELLA and GID1-DELLA-SLY protein complexes required for DELLA’s inactivation and destruction, respectively (Hartweck, 2008; Murase et al., 2008). Although we observed that the DELLA genes, except RGL2, are also down-regulated in the swi3c mutant, altered transcriptional activation of the GID1a, GA3ox, and several known DELLA target genes suggested that the SWI3C subunit might also be implicated in the control of DELLA’s activity at the protein level. This prompted us to examine whether SWI3C could directly interact with and thus play a role in the binding of DELLAs to the SWI/SNF chromatin remodeling complex. We also included in these studies the SPY gene encoding one of the two Arabidopsis O-GlcNAc transferases shown to act as potent negative regulator of GA signaling (Jacobsen and Olszewski, 1993). While it has been suggested that at the molecular level SPY may act by N-acetyl-glucosamination of DELLAs leading to their activation or stabilization, this has not been demonstrated experimentally, and the real targets of SPY in GA signaling are still largely unknown (Silverstone et al., 2007; for review, see Schwechheimer and Willige, 2009). Because N-acetyl-glucosamination of Ser and Thr residues is now recognized as a highly dynamic posttranslational modification of numerous nuclear and cytoplasmic proteins acting in key signal transduction pathways (Slawson et al., 2006), we decided to examine potential interaction of SPY with SWI3C. Both in vivo BiFC and in vitro protein-protein interaction studies, as well as supplementing yeast two-hybrid tests in the case of SPY, indicated that SWI3C is capable of in vitro and in vivo interactions and therefore may be responsible for forming complexes between SWI/SNF, the DELLA RGA, RGL1, and RGL2 proteins, and SPY. While it is by no means clear whether DELLA, SPY, and SWI/SNF occur in the same complex, the existence of these interactions suggest a potential role of chromatin structural modifications in functioning of both DELLA proteins and SPY.
The stabilization of DELLAs in ga mutants impaired in GA biosynthesis (i.e. as their degradation is inhibited in the absence of GA-mediated activation of GID1 receptors) results in severe retardation of growth. In the spy-1 mutant, the growth inhibitory activities of DELLAs are greatly decreased despite their remarkable stabilization, and therefore the spy-1 mutation alleviates most inhibitory effects of DELLAs also in the GA biosynthesis mutants. Interestingly, the rice (Oryza sativa) homolog of SPY has been shown to function in GA signaling not via changes in the amount or stability of rice DELLA protein Slender Rice-1, but probably through control of suppressive function of Slender Rice-1 (Shimada et al., 2006). The resemblance of spy-1 and swi3c responses to low temperature may therefore suggest that the function of SPY, including possibly the control of DELLA’s growth-suppressive function, can be linked to its role in active chromatin remodeling. It remains an intriguing question whether the association of DELLA proteins and SPY with SWI3C and their possible consequences for chromatin remodeling by the SWI/SNF complexes could underlay SPY effect on DELLA activity. It will be therefore important to further explore whether some of the developmental defects observed in the spy-1 mutant at normal temperature are due to inability to remodel nucleosomes at SWI/SNF-bound nuclear target loci.
The reduced activity of DELLAs could explain decreased transcription of the GID1a GA receptor, other DELLA target genes, and GA3ox2/GA3ox3 genes (i.e. implying altered regulation of their activators and repressors; Richter et al., 2010) as well as defects in the perception and biosynthesis of active GAs in the swi3c mutant. Given that both the availability of bioactive GAs and transcriptional activation of the major GID1a receptor are simultaneously impaired, DELLAs could confer a pronounced growth inhibition in the swi3c mutant. External GA treatment, decreasing DELLA levels by their GID1-mediated inactivation, alleviates the inhibition of hypocotyl and root elongation by the swi3c mutation. It remains to be determined whether the release of PIF bHLH transcription factors from their inactive DELLA complexes plays a role in this process and whether SWI/SNF CRCs also play a role in DELLA-dependent or -independent recruitment or phytochrome B-aided destabilization of PIFs (de Lucas et al., 2008). In contrast to GA-mediated suppression of hypocotyl and root elongation defects, the expansion of leaf blades is not restored and leaf curling is not abolished efficiently by GA treatment of the swi3c plants. This may indicate that low GID1a availability in leaves is not sufficient for GA-induced complete inactivation of DELLAs in this organ, which might reflect a requirement for a functional SWI/SNF complex mediating interaction of GID1s with DELLAs in the chromatin context. It is therefore remarkable that the spy-1 mutation diminishing the activation of DELLAs restores the curling leaf phenotype in the swi3c mutant background. This indicates either a SPY-dependent and DELLA-independent effect or that independently of their simultaneous recruitment by SWI3C, SPY can still control the activation of DELLAs, possibly by interacting with one of the other three SWI3-type SWI/SNF subunits. This might also explain why swi3c mutation shows only partial hypostatic behavior in respect to spy-1 mutation and why the phenotype of the swi3c mutants is milder compared with those in mutants in other swi3 subunits.
The results of this investigation are summarized and placed in the context of current knowledge about hormone cross talk in Figure 7. DELLA growth repressors are known to be under the influence of multiple signals including auxins, ABA, brassinosteroids, and ethylene that arrive and modify at different levels the main GA pathway (Fu and Harberd, 2003; Achard et al., 2009; Marrocco et al., 2010; Sun, 2010; Han et al., 2012). DELLAs also negatively affect the ABA, ethylene, and brassinosteroids pathways, which collectively define them as important hubs in the integration of environmental and developmental signals. All the above-mentioned hormonal pathways were shown here to require functional SWI/SNF for normal activity. The key new element of this work is the discovery that in addition to controlling the transcription of DELLA genes, the SWI3C subunit of SWI/SNF complex also directly interacts with DELLAs, which could explain possible involvement of SWI3C in controlling DELLA’s activity at the protein level. The SWI/SNF chromatin remodelers appear to be uniquely positioned regarding the control of central regulatory hub of DELLAs and therefore emerge as likely important integrators of cross talk between several hormone signaling networks.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
The swi3c-1 mutant (referred further as swi3c) was characterized previously (Sarnowski et al., 2005). Lines overexpressing 9MYC-tagged RGA, GAI, RGL1, RGL2, and RGL3 proteins, respectively, were kindly provided by Xing Wang Deng (Feng et al., 2008). A swi3c spy-1 double mutant was isolated by crossing homozygous swi3c plants with a spy-1 line followed by PCR and phenotype-based screening of mutant alleles in the segregating F2 population. Primers used for genotyping are listed in Supplemental Table S2. Plants were grown under long-day (LD), SD, or darkness conditions (16-h light/8-h dark or 8-h light/16-h dark, respectively) at 18°C to 23°C or using 8-h/16-h night/day conditions at 14°C to 16°C, 70% humidity, and 200 μm m–2 s–1 light intensity. Seedlings were cultivated in medium containing one-half-strength MS salts (Sigma-Aldrich), 0.5% (w/v) Suc, and 0.8% (w/v) agar (pH 5.8) supplemented with various concentrations of GA4+7, PAC, aminocyclopropane-1-carboxylic acid, ethephon, brassinolide, or ABA. In the case of ABA treatment, medium without Suc was used. Wild-type and swi3c seeds were sown on one-half-strength MS plates containing different concentrations of PAC, ABA, or brassinolide and cultivated for 7 d in SD conditions. To test their ethylene response, wild-type and swi3c plants were grown in the darkness for 7 d. After this period, seedlings were harvested and subjected for subsequent analyses. To analyze GA responses, plants were grown in soil and treated with 100 μm GA4+7 by spraying twice a week or were grown on one-half-strength MS medium supplemented with 0.05 μm to 10 μm GA4+7. The gravitropism assays were performed on vertically placed square petri plates. Plants were grown for 6 d, and plates were turned 90° counterclockwise and grown for further 4 d.
qRT-PCR Analyses
Fourteen-day-old LD-grown wild-type and swi3c seedlings were sprayed with 100 μm GA4+7 or with water as control. RNA was extracted from seedlings using the RNeasy plant mini kit (Qiagen), and DNA was removed by DNase treatment using a TURBO DNA-free kit (Ambion). A first-strand complementary DNA synthesis kit (Roche) was used to prepare complementary DNA from 2.5 μg of RNA. Aliquots (3 μL) of 5-fold-diluted complementary DNA samples were used as templates in 20-μL reactions containing SYBR Green Master mix (Bio-Rad) and specific primers for PCR amplification. The final primer concentrations were 0.5 μm, the annealing temperature was set at 56°C, and extension was performed in 72°C. The qRT-PCR data was recorded and analyzed using iQ-PCR (Bio-Rad) or FAST7500 (Applied Biosystems) equipment and software as recommended by the manufacturers. Transcripts of the SERINE/THREONINE PROTEIN PHOSPHATASE2A and UBIQUITIN5 genes were used as normalization controls. Each experiment was performed using at least two independent biological replicates, and the specificity of real-time PCR products was confirmed by melting curve analysis. Specific primers used in quantitative PCR reactions are listed in Supplemental Table S2.
Construction of Vectors Used in BiFC
To obtain YFN-SWI3C and YFC-DELLA (RGA, GAI, RGL1, RGL2, and RGL3) or SPY fusions for BiFC (Hu et al., 2002) analysis, the open reading frames of complementary DNAs encoding SWI3C, SPY, and DELLA proteins were PCR amplified and cloned into the binary vectors pYFN43 or pYFC43 (Belda-Palazón et al., 2012), respectively, using the Gateway (Invitrogen) recombination approach. In vivo interactions between proteins were detected by BiFC using Leica TCS SP2 AOBS, a laser scanning confocal microscope (Leica Microsystems). Excitation of YFP was with the Argon laser line at 514 nm and of RFP with a 563-nm diode laser, and detection of YFP fluorescence was at 518 to 555 nm and of RFP at 568 to 630 nm. The specificity of observed signals was confirmed by measuring the fluorescence emission wavelength (λ scan). Tobacco (Nicotiana benthamiana) epidermal cells were infiltrated Agrobacterium tumefaciens GV3101 (pMP90) strains carrying plasmids encoding SWI3C, DELLA, or SPY fusions and the p19 helper vector (Voinnet et al., 2003) and analyzed by confocal microscopy 3 d later. YFN-RFP and YFC-RFP fusions were used to detect transformed cells in the BiFC assays. At least five nuclei were analyzed in each of three separate experiments.
Overexpression of ATSWI3C and Pull-Down of DELLA Proteins
The coding region of SWI3C gene was cloned into the pDEST-MBP 6×HIS vector (Invitrogen) to express the fusion protein in bacteria. Native SWI3CMBP6×His protein was purified according to protocol 14 (Qiaexpressionist, Qiagen). Nuclear extracts were prepared from 4-week-old Arabidopsis (Arabidopsis thaliana) plants overexpressing the 9MYC-tagged DELLA proteins RGA, RGL1, RGL2, RGL3, and GAI (Feng et al., 2008). One-half gram of plant tissue was ground in liquid nitrogen and resuspended in IP-1 buffer (20 mm HEPES-KOH, pH 8.0, 0.15 m KCl, 0.2% (v/v) Triton, 10% (v/v) glycerol, 0.1 mm phenylmethylsulfonyl fluoride, 5 mm β-mercaptoethanol, and Complete EDTA-free), incubated for 15 min at 4°C, and centrifuged for 15 min at 15,000g to yield a supernatant used in further analyses. For pull-down assays, the SWI3C protein was bound to Ni-NTA agarose beads and incubated with total protein extracts of 9MYC-DELLA-expressing plants in IP-1 buffer for 2 h at 4°C. Beads were washed eight times with IP-1 buffer and boiled, followed by SDS-PAGE (12%) separation and immunoblotting of proteins using a c-Myc primary (dilution, 1:1,500; Covance) and an anti-mouse horseradish peroxidase secondary antibody (dilution, 1:10,000: Sigma).
Seeds Embedding and Tissue Sectioning
Seeds were fixed with paraformaldehyde as described (Porri et al., 2012; Torti et al., 2012). To allow penetration of the fixative, the seeds were vacuum infiltrated, and the samples incubated on ice overnight. The following day, the fixative was replaced with a graded ethanol: water series at 4°C (85% ethanol, 4 h; 95% (v/v) ethanol, 4 h; 100% (v/v) ethanol, overnight; and 100% (v/v) ethanol, fresh). The samples were stored at 4°C in 100% ethanol until embedding. Paraffin embedding in Paraplast Plus (McCormick) was performed using an automated Leica ASP300 tissue processor. Wax blocks were stored at 4°C until sectioning with a rotary microtome (Leica). The Paraplast was removed from the semithin sections with pure Histoclear before images were taken with a light microscope (Leica DMRB), and cell measurements and counting were carried out by using ImageJ software. For scanning electromicroscopic analysis of the seed surface, seeds were mounted on stubs using double-sided adhesive and conductive tabs and sputter coated with platin before imaging with a Zeiss Supra 40VP scanning electron microscope.
Yeast Two-Hybrid Protein Interaction Studies
Yeast (Saccharomyces cerevisiae) two-hybrid assays were performed with the plasmids pGBT9 and pGAD424 containing a full-length complementary DNA of the Arabidopsis SWI3C gene as described previously (Sarnowski et al., 2002). To obtain other pGBT9 and pGAD424 constructs, full-length complementary DNA of SPY was PCR amplified using primers with suitable restriction sites (Supplemental Table S2) and cloned in the pCR-TOPO-TA vector (Invitrogen). After sequencing, the complementary DNAs were excised by restriction endonucleases and cloned into the vectors pGBT9 and pGAD424. Yeast strain Y190 was transformed with the pGBT9 and pGAD424 constructs encoding the protein pairs to be tested and each construct in combination with either empty pGBT9 or pGAD424 as controls. The level of reporter β-galactosidase activity of each yeast strain was monitored using the replica filter lift method described in the Clontech yeast protocols handbook.
Measurement of Endogenous Phytohormones
For the analysis of the endogenous hormone level, the aerial parts of 4-week-old wild-type, swi3c, and ga1-3 plants were collected, flash frozen in liquid nitrogen, and subjected to further analysis. Phytohormones were quantified using a 6410 Triple Quad Liquid chromatography\x{2013}mass spectrometry (Agilent Technologies) with an Agilent 1200 series rapid resolution liquid chromatography system fitted with a ZORBAX Eclipse XDB-C18 column (1.8 µm, 2.1 × 50 mm) as described (Plackett et al., 2012). Isotope-labeled internal standard was obtained from Olchemin, Icon Isotopes, Sigma-Aldrich, and Tokyo Kasei (Seo et al., 2011).
Upon request, all novel materials described in this publication will be made available in a timely manner for noncommercial research purposes, subject to the requisite permission from any third-party owners of all or parts of the material. Obtaining any permissions will be the responsibility of the requestor.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SWI3C (AT1G21700), RGA (AT2G01570), RGL1 (AT1G66350), RGL2 (AT3G03450), RGL3 (AT5G17490), and SPY (AT3G11540).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The swi3c mutant shows altered responses to exogenously applied hormones ethylene, ABA, and brassinosteroids and an altered gravitropic response.
Supplemental Figure S2. The swi3c mutation confers GA-related growth and developmental defects suppressed by the GA4+7 treatment and insensitivity to the GA biosynthesis inhibitor PAC.
Supplemental Figure S3. Low temperature similarly affects fertility of swi3c and spy-1. GA biosynthesis is altered in swi3c.
Supplemental Figure S4. The swi3c mutant exhibits altered expression of genes encoding DELLA repressors.
Supplemental Figure S5. SWI3C interacts with the SPY and DELLA proteins.
Supplemental Table S1. The level of GA intermediates in the swi3c and ga1-3 mutants.
Supplemental Table S2. Primers used in this work.
Acknowledgments
We thank Sabine Schaefer and Ingrid Reinsch for technical assistance, Xing Wang Deng for providing lines overexpressing 9MYC-tagged RGA, GAI, RGL1, RGL2, and RGL3 proteins, respectively (Feng et al., 2008), Tomoe Nose for help with hormone measurements, Neil Olszewski for critical comments to the manuscript, and Janusz Siedlecki for encouragement.
Glossary
- CRC
chromatin-remodeling complex
- ABA
abscisic acid
- MS
Murashige and Skoog
- PAC
paclobutrazol
- BiFC
bimolecular fluorescence complementation
- Ni-NTA
nickel-nitrilotriacetic acid agarose
- qRT
quantitative real-time
- LD
long-day
- SD
short-day
- IP-1
20 mM HEPES-KOH, pH 8.0, 0.15 M KCl, 0.2% (v/v) Triton, 10% (v/v) glycerol, 0.1 mM phenyl-methylsulfonyl fluoride, 5 mM β-mercaptoethanol, and Complete EDTA-free
References
- Achard P, Gusti A, Cheminant S, Alioua M, Dhondt S, Coppens F, Beemster GT, Genschik P. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188–1193 [DOI] [PubMed] [Google Scholar]
- Archacki R, Buszewicz D, Sarnowski TJ, Sarnowska E, Rolicka AT, Tohge T, Fernie AR, Jikumaru Y, Kotlinski M, Iwanicka-Nowicka R, et al. (2013) BRAHMA ATPase of the SWI/SNF chromatin remodeling complex acts as a positive regulator of gibberellin – mediated responses in Arabidopsis. PLoS ONE 8: e58588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archacki R, Sarnowski TJ, Halibart-Puzio JU, Brzeska K, Buszewicz D, Prymakowska-Bosak M, Koncz C, Jerzmanowski A. (2009) Genetic analysis of functional redundancy of BRM ATPase and ATSWI3C subunits of Arabidopsis SWI/SNF chromatin remodelling complexes. Planta 229: 1281–1292 [DOI] [PubMed] [Google Scholar]
- Belandia B, Parker MG. (2003) Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell 114: 277–280 [DOI] [PubMed] [Google Scholar]
- Belda-Palazón B, Ruiz L, Martí E, Tárraga S, Tiburcio AF, Culiáñez F, Farràs R, Carrasco P, Ferrando A. (2012) Aminopropyltransferases involved in polyamine biosynthesis localize preferentially in the nucleus of plant cells. PLoS ONE 7: e46907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezhani S, Winter C, Hershman S, Wagner JD, Kennedy JF, Kwon CS, Pfluger J, Su Y, Wagner D. (2007) Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G. (2004) AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol 136: 3660–3669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Dill AS, Thomas SG, Hu J, Steber CM, Sun TP. (2004) The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16: 1392–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, Kingston R, Brown M. (2000) BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol 20: 7541–7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, Rozowsky J, Bhardwaj N, Gerstein MB, Snyder M. (2011) Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrona S, Hurtado L, Bowman JL, Reyes JC. (2004) The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131: 4965–4975 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. (2008) Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleet CM, Sun TP. (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8: 77–85 [DOI] [PubMed] [Google Scholar]
- Fu X, Harberd NP. (2003) Auxin promotes Arabidopsis root growth by modulating gibberellin response. Nature 421: 740–743 [DOI] [PubMed] [Google Scholar]
- Griffiths J, Murase K, Rieu I, Zentella R, Zhang ZL, Powers SJ, Gong F, Phillips AL, Hedden P, Sun TP, et al. (2006) Genetic characterization and functional analysis of the GID1 gibberellin receptors in Arabidopsis. Plant Cell 18: 3399–3414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Kim D. (2006) AtRTPrimer: database for Arabidopsis genome-wide homogeneous and specific RT-PCR primer-pairs. BMC Bioinformatics 7: 179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SK, Sang Y, Rodrigues A, Wu MF, Rodriguez PL, Wagner D. (2012) The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24: 4892–4906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM. (2008) Gibberellin signaling. Planta 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Hedden P, Phillips AL. (2000) Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Hu CD, Chinenov Y, Kerppola TK. (2002) Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9: 789–798 [DOI] [PubMed] [Google Scholar]
- Iuchi S, Suzuki H, Kim YC, Iuchi A, Kuromori T, Ueguchi-Tanaka M, Asami T, Yamaguchi I, Matsuoka M, Kobayashi M, et al. (2007) Multiple loss-of-function of Arabidopsis gibberellin receptor AtGID1s completely shuts down a gibberellin signal. Plant J 50: 958–966 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. (1993) Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Johnson TA, Sung M-H, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, et al. (2008) Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell 29: 611–624 [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. (1980) Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.). Heynh. Theor. Appl. Genet 58: 257–263 [DOI] [PubMed] [Google Scholar]
- Marrocco K, Bergdoll M, Achard P, Criqui M-C, Genschik P. (2010) Selective proteolysis sets the tempo of the cell cycle. Curr Opin Plant Biol 13: 631–639 [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. (2008) Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456: 459–463 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Iuchi S, Kobayashi M, et al. (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46: 880–889 [DOI] [PubMed] [Google Scholar]
- Plackett AR, Powers SJ, Fernandez-Garcia N, Urbanova T, Takebayashi Y, Seo M, Jikumaru Y, Benlloch R, Nilsson O, Ruiz-Rivero O, et al. (2012) Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP. (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11: 3194–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porri A, Torti S, Romera-Branchat M, Coupland G. (2012) Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Richter R, Behringer C, Müller IK, Schwechheimer C. (2010) The GATA-type transcription factors GNC and GNL/CGA1 repress gibberellin signaling downstream from DELLA proteins and PHYTOCHROME-INTERACTING FACTORS. Genes Dev 24: 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez A, Rodrigues A, Santiago J, Rubio S, Rodriguez PL. (2008) HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20: 2972–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski TJ, Ríos G, Jásik J, Swiezewski S, Kaczanowski S, Li Y, Kwiatkowska A, Pawlikowska K, Koźbiał M, Koźbiał P, et al. (2005) SWI3 subunits of putative SWI/SNF chromatin-remodeling complexes play distinct roles during Arabidopsis development. Plant Cell 17: 2454–2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnowski TJ, Swiezewski S, Pawlikowska K, Kaczanowski S, Jerzmanowski A. (2002) AtSWI3B, an Arabidopsis homolog of SWI3, a core subunit of yeast Swi/Snf chromatin remodeling complex, interacts with FCA, a regulator of flowering time. Nucleic Acids Res 30: 3412–3421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Itoh H, Gomi K, Ueguchi-Tanaka M, Ishiyama K, Kobayashi M, Jeong DH, An G, Kitano H, Ashikari M, et al. (2003) Accumulation of phosphorylated repressor for gibberellin signaling in an F-box mutant. Science 299: 1896–1898 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Willige BC. (2009) Shedding light on gibberellic acid signalling. Curr Opin Plant Biol 12: 57–62 [DOI] [PubMed] [Google Scholar]
- Seo M, Jikumaru Y, Kamiya Y (2011) Profiling of hormones and related metabolites in seed dormancy and germination studies. In AR Kermode, ed, Seed Dormancy: Methods and Protocols. Humana Press, New York, NY, pp 99–111 [DOI] [PubMed] [Google Scholar]
- Shimada A, Ueguchi-Tanaka M, Sakamoto T, Fujioka S, Takatsuto S, Yoshida S, Sazuka T, Ashikari M, Matsuoka M. (2006) The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J 48: 390–402 [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Ciampaglio CN, Sun T-P. (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10: 155–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Tseng TS, Swain SM, Dill A, Jeong SY, Olszewski NE, Sun TP. (2007) Functional analysis of SPINDLY in gibberellin signaling in Arabidopsis. Plant Physiol 143: 987–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Housley MP, Hart GW. (2006) O-GlcNAc cycling: how a single sugar post-translational modification is changing the way we think about signaling networks. J Cell Biochem 97: 71–83 [DOI] [PubMed] [Google Scholar]
- Sun TP. (2010) Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Phys 154: 567–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torti S, Fornara F, Vincent C, Andrés F, Nordström K, Göbel U, Knoll D, Schoof H, Coupland G. (2012) Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell 24: 444–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. (2005) GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437: 693–698 [DOI] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, et al. (2007) Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 19: 2140–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D. (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C. (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19: 1209–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, Thomas SG, Endo A, Murase K, Fleet CM, Jikumaru Y, Nambara E, Kamiya Y, et al. (2007) Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19: 3037–3057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zraly CB, Middleton FA, Dingwall AK. (2006) Hormone-response genes are direct in vivo regulatory targets of Brahma (SWI/SNF) complex function. J Biol Chem 281: 35305–35315 [DOI] [PubMed] [Google Scholar]