Light signaling genes are dynamically regulated during shade avoidance with adaptation to a low red/far-red light environment mediated by enhancing the activity of HY5, a master regulator of seedling deetiolation.
Abstract
Shade-intolerant plants perceive the reduction in the ratio of red light (R) to far-red light (FR) as a warning of competition with neighboring vegetation and display a suite of developmental responses known as shade avoidance. In recent years, major progress has been made in understanding the molecular mechanisms underlying shade avoidance. Despite this, little is known about the dynamics of this response and the cascade of molecular events leading to plant adaptation to a low-R/FR environment. By combining genome-wide expression profiling and computational analyses, we show highly significant overlap between shade avoidance and deetiolation transcript profiles in Arabidopsis (Arabidopsis thaliana). The direction of the response was dissimilar at the early stages of shade avoidance and congruent at the late ones. This latter regulation requires LONG HYPOCOTYL IN FAR RED1/SLENDER IN CANOPY SHADE1 and phytochrome A, which function largely independently to negatively control shade avoidance. Gene network analysis highlights a subnetwork containing ELONGATED HYPOCOTYL5 (HY5), a master regulator of deetiolation, in the wild type and not in phytochrome A mutant upon prolonged low R/FR. Network analysis also highlights a direct connection between HY5 and HY5 HOMOLOG (HYH), a gene functionally implicated in the inhibition of hypocotyl elongation and known to be a direct target of the HY5 transcription factor. Kinetics analysis show that the HYH gene is indeed late induced by low R/FR and that its up-regulation depends on the action of HY5, since it does not occur in hy5 mutant. Therefore, we propose that one way plants adapt to a low-R/FR environment is by enhancing HY5 function.
Plants have evolved two opposing strategies in response to competition for light: shade tolerance and shade avoidance. Angiosperms have an impressive capacity to avoid shade. Daylight contains roughly equal proportions of red light (R) and far-red light (FR), but within a vegetation community, that ratio is lowered as a result of R absorption by photosynthetic pigments. The reduction in the R/FR ratio is perceived as an early signal of neighbor proximity, resulting in a suite of developmental responses known as shade avoidance. The most dramatic response to low R/FR is the stimulation of elongation growth that is remarkably rapid, with a lag phase of a few minutes. In dicotyledonous plants, elongation growth induced by low R/FR is often associated with a reduction of leaf development. In the long term, low R/FR exposure leads to early flowering with a reduced seed set, which is considered an escape mechanism because it shortens generation time. All of these responses occur both in natural dense communities and in shade simulations (low R/FR). Furthermore, similar responses are induced by exposing plants to horizontal FR radiation with white light from above. This is because shade-avoiding plants are able to perceive light reflected by neighboring plants as partially depleted of the R wavelengths, and they can activate responses to avoid shade even before canopy closure and actual shading occurs (Franklin, 2008; Ruberti et al., 2012; Casal, 2013). However, at high canopy density, multiple light signals control the shade-avoidance response (Ballaré, 1999). There is evidence that both low R/FR and reduced blue light are required for full expression of shade avoidance in plant canopies. Interestingly, the blue light responses seem to be mediated through pathways that showed only limited overlap with those activated by low R/FR (Keller et al., 2011; Keuskamp et al., 2011).
Changes in the R/FR ratio of light occurring within a vegetation community are detected by the phytochrome (phy) family of R/FR photoreceptors (phyA–phyE in Arabidopsis [Arabidopsis thaliana]). Phytochromes are photochromic biliproteins that exist in two photoconvertible isoforms: Pr and Pfr. They are synthesized in their inactive Pr form; upon absorption of R, Pr is converted into the biologically active Pfr form, which can absorb FR and switch back to Pr, resulting in a dynamic photoequilibrium between the two forms of phytochrome. Following conversion to the Pfr form, phytochromes translocate to the nucleus. phyA is the only phytochrome that is rapidly degraded in its Pfr form and can signal during rapid photoconversion between the Pr and Pfr forms. All the other phytochromes are relatively stable in the Pfr form (Bae and Choi, 2008; Franklin and Quail, 2010; Casal, 2013).
Among the light-stable phytochromes, phyB plays a key role in shade avoidance. Arabidopsis phyB mutants constitutively display shade-avoidance traits such as elongated hypocotyl, stem, petioles, and leaves, acceleration of flowering, and higher apical dominance under high R/FR (Reed et al., 1993). However, other light-stable phytochromes (phyD and phyE) also contribute to shade avoidance (Smith and Whitelam, 1997; Devlin et al., 1998, 1999). By contrast, phyA seems to attenuate the elongation response induced by low R/FR (Johnson et al., 1994; Devlin et al., 2003; Wang et al., 2011).
In the nucleus, phytochromes physically interact with a subfamily of basic helix-loop-helix (bHLH) proteins, the PHYTOCHROME-INTERACTING FACTORS (PIFs), controlling several aspects of photomorphogenesis (Castillon et al., 2007; Jiao et al., 2007; Leivar and Quail, 2011). This interaction in turn leads to PIF’s phosphorylation, ubiquitination, and degradation via the 26S proteasome, providing an elegant mechanism for the rapid regulation of gene expression in response to changes in the light environment (Bauer et al., 2004; Park et al., 2004; Shen et al., 2005, 2007; Al-Sady et al., 2006; Nozue et al., 2007).
PIF1, PIF3, PIF4, PIF5, and PIF7 have been demonstrated to directly contribute to shade avoidance (Lorrain et al., 2008; Hornitschek et al., 2012; Leivar et al., 2012; Li et al., 2012). They all interact physically with phyB through the conserved N-terminal sequence, called the active phyB-binding motif (Leivar and Quail, 2011). As a result, PIF1, PIF3, PIF4, and PIF5 become phosphorylated and degraded via the ubiquitin-proteasome system, with degradation half-times in the range of 5 to 20 min (Leivar and Quail, 2011). PIF3, PIF4, and PIF5 protein levels increase rapidly in photoautotrophic seedlings upon exposure to low R/FR (Lorrain et al., 2008; Leivar et al., 2012). Unlike its close relatives, PIF7 is not rapidly degraded in high R/FR (Leivar et al., 2008a). However, this PIF protein accumulates in its dephosphorylated form in shade, suggesting the existence of a protein phosphatase and a protein kinase whose activities or availability are regulated by light quality changes (Li et al., 2012). Shade-induced elongation response is significantly attenuated in pif4 pif5 and, to an even greater extent, in pif1 pif3 pif4 pif5 quadruple (pifq) and pif7 mutants (Lorrain et al., 2008; Leivar et al., 2012; Li et al., 2012). Conversely, PIF4- and PIF5-overexpressing seedlings have constitutively long hypocotyls and petioles (Lorrain et al., 2008).
Consistent with the rapidity of the elongation growth response to low R/FR and its reversibility upon perception of high R/FR, changes in gene expression are very rapid and reversible (Carabelli et al., 1996; Salter et al., 2003). The transcript levels of the homeodomain-leucine zipper (HD-Zip) II ARABIDOPSIS THALIANA HOMEOBOX2 (ATHB2) and bHLH PIF3-LIKE1 (PIL1) transcription factor genes, functionally implicated in the elongation response provoked by neighbor detection (Steindler et al., 1999; Salter et al., 2003), increase within a few minutes of low R/FR exposure (Carabelli et al., 1996; Salter et al., 2003). Significantly, ATHB2 and PIL1 transcript levels fall very rapidly after transfer from low R/FR to high R/FR (Carabelli et al., 1996; Salter et al., 2003). ATHB2 and PIL1 induction by low R/FR does not require de novo protein synthesis (Roig-Villanova et al., 2006) and is significantly reduced in loss-of-function pif mutants (pif1 pif3, pif4 pif5, and pif7; Lorrain et al., 2008; Hornitschek et al., 2009; Leivar et al., 2012; Li et al., 2012). There is evidence that PIL1 and ATHB2 are recognized in vivo by PIF4 and PIF5 (de Lucas et al., 2008; Hornitschek et al., 2009, 2012), and physical interaction between the PIL1 promoter and PIF7 has also been reported (Li et al., 2012).
An ever-increasing body of evidence shows that auxin biosynthesis, signaling, and transport are all critical for shade avoidance (Morelli and Ruberti, 2000; Kanyuka et al., 2003; Carabelli et al., 2007; Tao et al., 2008; Keuskamp et al., 2010; Sassi et al., 2013), and links between this hormone and transcription factors triggering plant responses to low R/FR (PIF and HD-Zip II proteins) have been established (Steindler et al., 1999; Hornitschek et al., 2012; Li et al., 2012; Turchi et al., 2013). PIF4 and PIF5 physically interact with the promoter of YUCCA8 (YUC8), which encodes a rate-limiting enzyme in auxin synthesis (Hornitschek et al., 2012), and PIF7 in its dephosphorylated form binds G-boxes of the auxin biosynthetic genes YUC5, YUC8, and YUC9 and increases their expression, thus directly linking the perception of a low R/FR signal to changes in free INDOLE-3-ACETIC ACID (IAA) required for shade-induced growth (Li et al., 2012). By contrast, how HD-Zip II proteins promote auxin response and transport remains to be investigated.
By exploiting mutant analysis in combination with genome-wide expression profiling, Sessa et al. (2005) uncovered a negative regulatory mechanism active in low R/FR that involves HYPOCOTYL FAR RED1/SLENDER IN CANOPY SHADE1 (HFR1/SICS1). The HFR1/SICS1 gene is rapidly induced by low R/FR, and there is evidence that its promoter is recognized in vivo by PIF5 (Sessa et al., 2005; Hornitschek et al., 2009). HFR1/SICS1 encodes an atypical bHLH protein and acts as a helix-loop-helix inhibitor. Upon prolonged exposure to low R/FR, HFR1/SICS1 accumulates and interacts with PIF4 and PIF5, forming non-DNA-binding heterodimers, thus limiting PIF-mediated gene expression (Hornitschek et al., 2009). Consistent with this, several genes rapidly and transiently induced by low R/FR are significantly up-regulated in hfr1/sics1 loss-of-function mutants upon prolonged exposure to simulated shade (Sessa et al., 2005). Another atypical bHLH protein gene, HELIX LOOP HELIX1/PHYTOCHROME RAPIDLY REGULATED1 (HLH1/PAR1; Sessa et al., 2005; Roig-Villanova et al., 2006), is also rapidly regulated by low R/FR, and its induction does not require de novo protein synthesis (Roig-Villanova et al., 2006). HLH1/PAR1 has also been involved in the negative regulation of shade-induced elongation and proposed to act as a dominant negative antagonist of conventional bHLH transcription factors (Roig-Villanova et al., 2007; Galstyan et al., 2011; Hao et al., 2012).
Despite the significant advances over the last decade in understanding shade avoidance, little is known about the dynamics of this response. Here, the shade-avoidance response was examined by genome-wide expression profiling in wild-type and genetically altered plants exposed to low R/FR for different times. Functional associations by response overlap (FARO) between shade avoidance transcript profiles and available microarray gene expression data revealed highly significant overlap with genes regulated in deetiolation experiments. Remarkably, the direction of the response was dissimilar at the early stages of shade avoidance and congruent at the late ones. This latter regulation involves both HFR1/SICS1 and phyA, which function largely independently to control the shade-avoidance response. Computational gene network analysis identified a subnetwork containing ELONGATED HYPOCOTYL5 (HY5), a key regulator of the transcriptional cascades promoting seedling photomorphogenesis (Lau and Deng, 2010), in wild-type seedlings and not in phyA mutants upon prolonged exposure to low R/FR. Based on these data, we propose that one way in which plants adapt to a low R/FR environment is by modulating the HY5 pathway.
RESULTS
Simulated Shade Environment
Several laboratories use white light supplemented with FR to study early events of the shade-avoidance response (neighbor detection). The light conditions utilized are quite different one from the other both in terms of photosynthetic active radiation (PAR) and R/FR ratio, and PAR is usually quite low (Tao et al., 2008; Hornitschek et al., 2009; Wang et al., 2011). For several years, we have been using a different experimental setup to simulate a shade environment (Sessa et al., 2005; Carabelli et al., 2007; Ciarbelli et al., 2008; Ruberti et al., 2012). To study changes in gene expression underlying early and late responses to a low-R/FR environment, we set up simulated shade conditions in which R is reduced and FR is increased, maintaining total light quantity (400–800 nm) constant (Sessa et al., 2005) and thus making entirely negligible the well-known influence of this parameter on the stability of several transcription factors involved in photomorphogenesis (Henriques et al., 2009). Varying R/FR while maintaining total light quantity constant inevitably implies a nonconstant supply of PAR (400–700 nm). Therefore, control experiments were performed lowering PAR without changing the ratio between R and FR. Columbia (Col-0) seedlings were grown for 7 d in a light/dark (L/D) cycle (16/8 h) in high R/FRhigh PAR (PAR 111) and then either maintained in the same regimen or exposed to high R/FRlow PAR (PAR 27) or to low R/FRlow PAR (PAR 27) for 1, 2, 4, 8, and 12 h. No significant difference was observed in the expression of several genes rapidly induced by low R/FR, such as ATHB2 (Carabelli et al., 1996), PIL1 (Salter et al., 2003), and HFR1/SICS1 (Sessa et al., 2005), between high R/FRhigh PAR and high R/FRlow PAR, thus demonstrating that these genes are specifically regulated at the transcriptional level by light quality changes under our simulated shade environment (Supplemental Fig. S1A). Hypocotyl elongation rate upon exposure to high R/FRlow PAR was also measured. To this end, Col-0 seedlings were grown for 4 d in a L/D cycle in high R/FRhigh PAR and then either maintained in the same regimen or exposed to high R/FRlow PAR or to low R/FRlow PAR for 1 d. No significant difference was observed in hypocotyl elongation in high R/FRlow PAR relative to high R/FRhigh PAR, indicating that the increase in elongation rate observed upon exposure to low R/FRlow PAR is induced by the change in the ratio of R to FR (Supplemental Fig. S1B).
Genome-Wide Expression Profiling during the Shade-Avoidance Response
To analyze the dynamics of the shade-avoidance response at a genome-wide scale, we inspected gene expression global profiles upon brief (1 and 4 h) and prolonged (1 d) exposure to low R/FR by means of Affymetrix Arabidopsis Genome GeneChip array (ATH1) analyses on 8-d-old Arabidopsis seedlings. A total of 399 genes were identified as differentially regulated by low R/FR (Supplemental Tables S1–S3). Among them, we could find a number of genes whose FR-rich light regulation has been previously demonstrated. To this group, among others, belong the genes encoding the HD-Zip transcription factors ATHB2 and ATHB4, the atypical bHLH HLH1/PAR1, the auxin efflux carrier PIN3, the IAA proteins IAA1 and IAA3, and the floral inducer FLOWERING LOCUS T (FT; Carabelli et al., 1993, 1996; Devlin et al., 2003; Sessa et al., 2005; Roig-Villanova et al., 2006; Tao et al., 2008; Keuskamp et al., 2010; Leivar et al., 2012). Moreover, although we could not observe PIL1 transcriptional regulation in our experiments because the Affymetrix ATH1 GeneChip lacks a probe set for this gene (Salter et al., 2003), its homolog PIL2, previously shown to be regulated by light quality changes (Salter et al., 2003), was found to be significantly induced upon 4 h of exposure to low R/FR (Supplemental Tables S1–S3).
To gain insights into the large number of low-R/FR-regulated genes, we proceeded to cluster them into different functional groups according to the putative or established gene functions in the plant, taking advantage of the Database for Annotation, Visualization, and Integrated Discovery (DAVID) functional annotation clustering (Huang et al., 2009). For each time point, the most enriched groups (top 10 ranked with EASE score [a modified Fisher Exact P-Value] greater than 1) were identified and described by means of a Gene Ontology (GO) term (descriptor). Intriguingly, the relative abundance of each functional class varies significantly between genes early and late regulated by low R/FR (Supplemental Tables S4–S6).
Among the very early-regulated genes (1 h of low R/FR), transcription factor- and hormone-related genes were significantly more abundant than other functional classes. In agreement with the major role of auxin in shade avoidance (Steindler at al., 1999; Carabelli et al., 2007; Tao et al., 2008; Keuskamp et al., 2010; Li et al., 2012), DAVID analysis evidenced a highly significant overrepresentation of “response to auxin stimulus” and “auxin-mediated signaling pathway” functional classes, whereas the importance of transcriptional regulation at the early stages of shade avoidance (Carabelli et al., 1993, 1996; Salter et al., 2003; Sessa et al., 2005; Roig-Villanova et al., 2006; Sorin et al., 2009) was highlighted by the strong enrichment of “transcription regulator activity” and “basic helix-loop-helix dimerization region” class descriptors. Furthermore, consistent with evidence on the existence of cross talk between auxin and brassinosteroid signal transduction pathways active in the modulation of plant growth and tropic responses (Bao et al., 2004; Nemhauser et al., 2004; Nakamoto et al., 2006; Keuskamp et al., 2011), besides R and FR signaling pathway overrepresentation, we found that “brassinosteroid metabolic process” and “developmental growth involved in morphogenesis” classes were also enriched among genes rapidly regulated by simulated shade (Supplemental Table S4).
After 4 h of low R/FR, DAVID analysis revealed a behavior partially similar to that observed at the very early stages of shade avoidance, further highlighting the involvement of transcription factors (“basic helix-loop-helix dimerization region”) and auxin pathways (“response to auxin stimulus” and “auxin-mediated signaling pathway”) in plant responses to light quality changes. However, overrepresentation of these functional classes among low-R/FR-regulated genes was weaker, and some differences emerged (Supplemental Table S5). Among them is a significant overrepresentation of ethylene signaling genes (“regulation of ethylene-mediated signaling pathway”), supporting recent evidences on the function of this hormone in the shade-avoidance response (Pierik et al., 2009) and suggesting a strict temporal coordination of hormone interplay. Moreover, DAVID analysis highlighted a role for sulfur metabolism, probably related to secondary pathways (“regulation of glycosinolate biosynthetic process” and “sulfotransferase activity”; Supplemental Table S5).
Among the late-regulated genes (1 d of low R/FR), class enrichment scores appeared to be, on average, lower than among the early ones. The most overrepresented functional classes were related to light signaling (“response to red or far-red light” and “phototransduction”; Supplemental Table S6). Moreover, in agreement with the down-regulation of the jasmonic acid response by prolonged exposure to low R/FR (Moreno et al., 2009; Robson et al., 2010), the “response to jasmonic acid stimulus” class descriptor was also overrepresented. Finally, at late stages of shade avoidance, DAVID analysis highlighted a rearrangement of cellular metabolism and circadian rhythm pathways (Supplemental Table S6).
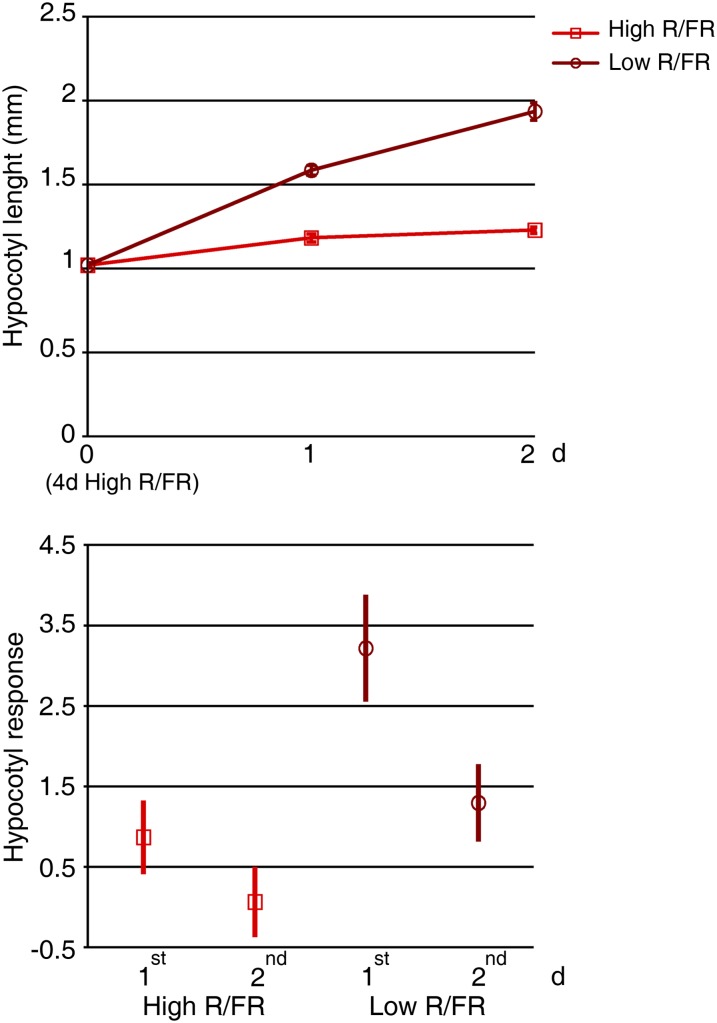
The finding that the gene expression pattern is dynamic in a low-R/FR environment suggests that the elongation growth response to simulated shade might change over time. To investigate whether this is the case, the effect of low R/FR on hypocotyl growth during days 1 and 2 of exposure to light enriched in FR was measured. Elongation occurs during both days 1 and 2. However, the growth response is promoted significantly more during day 1 than day 2 of exposure to low R/FR (Fig. 1).
Figure 1.
The growth elongation response to low R/FR decreases over time. Col-0 seedlings were grown in a L/D cycle (16/8 h) for 4 d in high R/FR (control) and then maintained in high R/FR or transferred to low R/FR for 1 and 2 d under the same light regimen. The top graph shows hypocotyl lengths (means ± se) of wild-type seedlings grown as described. At least 30 seedlings were analyzed for each condition. The bottom graph shows the strength of the hypocotyl elongation response of wild-type seedlings during days 1 and 2 in low R/FR. High-R/FR controls are also shown. Symbols (squares and circles) and vertical lines depict the standardized effect size values and 95% confidence intervals, respectively (Hedges and Olkin, 1985).
Deriving Functional Associations between Shade-Avoidance Transcript Profiles and Available Microarray Gene Expression Data
The DAVID functional analysis of the genes regulated by low R/FR indicated that shade-driven transcriptional alterations are highly dynamic, involving profound changes in the regulation of gene expression over time. However, the analysis of some functional classes (e.g. “response to red or far-red light”) suggests that DAVID may be not entirely effective in describing the dynamics of light-regulated genes. To overcome this limitation, we inspected in more detail the genes regulated by low R/FR by means of the FARO Web tool, which compares genome-wide expression profiles of a query (our experiments) with a large number of studies (“factors”) from a microarray data repository (“response compendium”) to find a significant functional overlap (Nielsen et al., 2007). For each time point (1 h, 4 h, and 1 d), we defined significantly overlapping groups as the top 15 ranked ones with more than 25% of genes shared with repository experiments. A large majority of top-ranked associated factors by FARO turned out to be composed of light- and hormone-related experiments, which were selected for further analysis (Supplemental Tables S7–S9). Remarkably, we found an opposite regulation of genes regulated in deetiolation experiments with respect to early shade-regulated genes and consistent regulation for late-regulated ones (Fig. 2; Supplemental Table S7). In fact, among the 12 factors showing significant overlap with genes regulated in the wild type upon 1 h of exposure to low R/FR, 10 of them pertain to deetiolation experiments and always display a congruence lower than 30% (Fig. 2; Supplemental Table S7). A similar result was observed for genes regulated upon 4 h of exposure to low R/FR (Fig. 2; Supplemental Table S8). By contrast, FARO analyses highlighted a strong congruence between genes late regulated by low R/FR (1 d) and deetiolation experiments (Fig. 2; Supplemental Table S9). Moreover, a clear positive correlation of an early shade-avoidance response with auxin- and ethylene-responsive genes was also shown by FARO (Fig. 2).
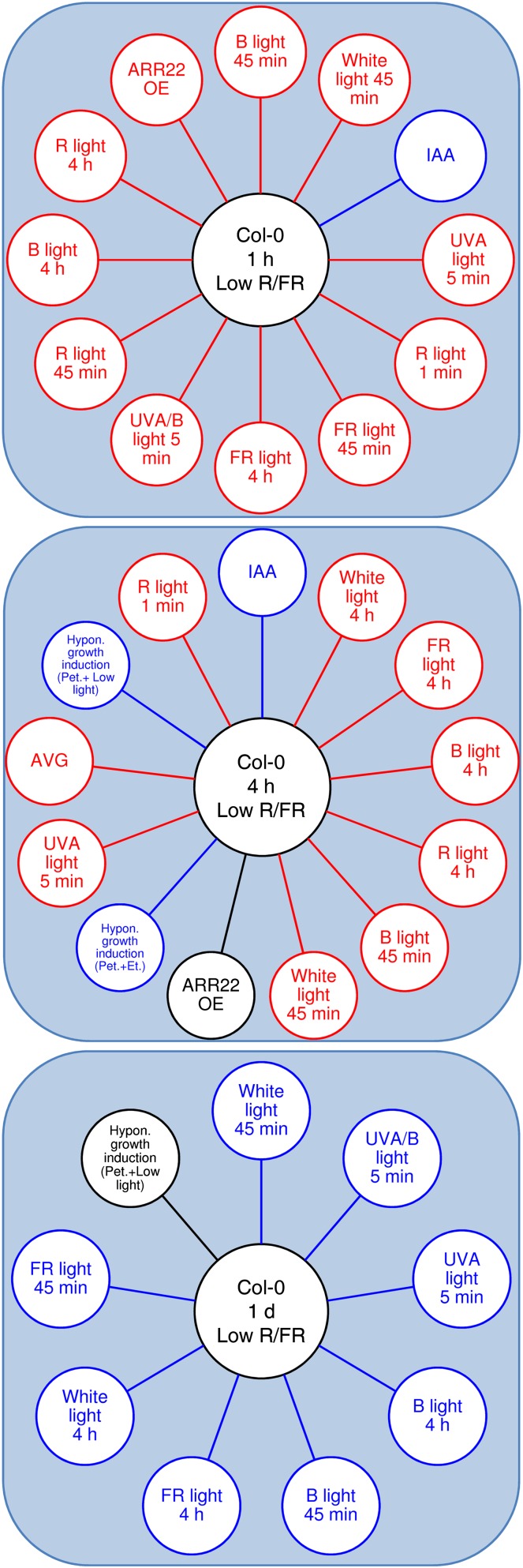
Figure 2.
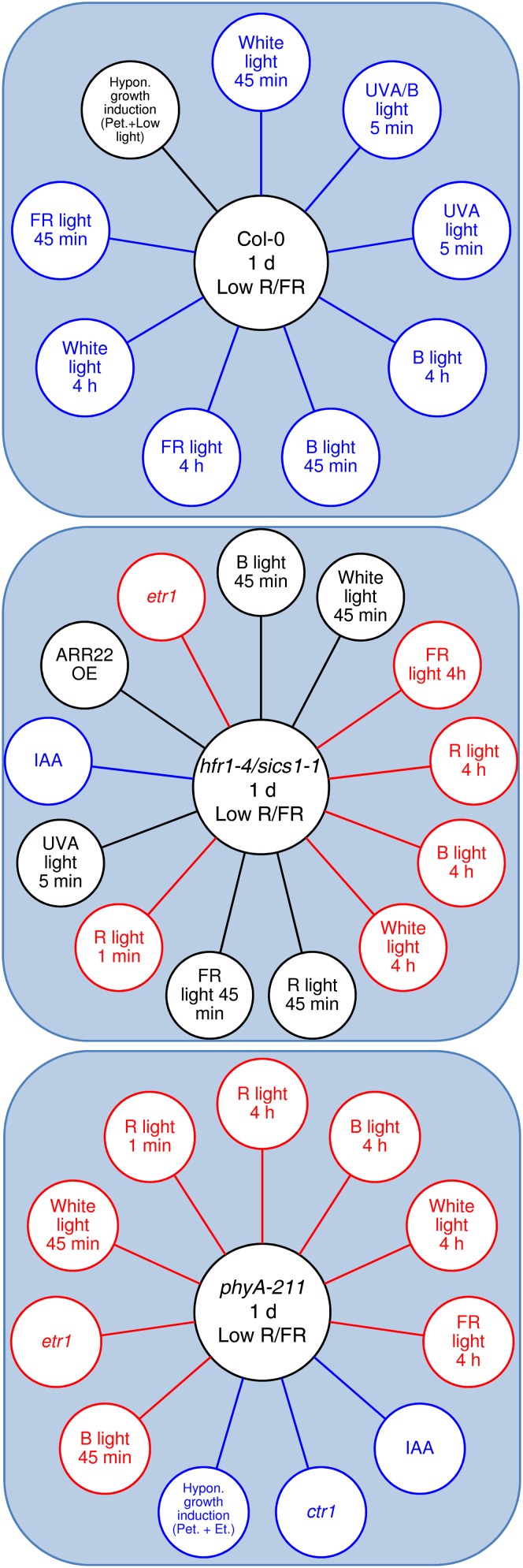
FARO of Arabidopsis seedlings exposed to low R/FR for different times. Low-R/FR-regulated genes involved in light and hormone pathways are visualized by means of the FARO Web tool, which compares genome-wide expression profiles of a query (our microarray experiments) with a large number of studies (factors) from a microarray data repository (response compendium) to find a significant functional overlap. The large majority of top-ranked associated factors by FARO were composed of experiments related to light and hormone signaling, which were selected for functional analysis (for details, see “Materials and Methods”). Genes differentially regulated in Col-0 after 1 h, 4 h, or 1 d of low-R/FR exposure are displayed in the central circles. In the outer circles, experimental factors with strong associations to the wild type exposed to low R/FR are shown. The factors showing overlap with the opposite direction (i.e. congruence less than 30%) with respect to our experiments are depicted in red; those showing the highest congruence (greater than 70%) are depicted in blue.
Inference of Functional Gene Networks in the Shade-Avoidance Response
For each functional cluster obtained by DAVID analysis of the genes differentially expressed in the wild type after exposure to low R/FR for different times, the genes associated with corresponding functional annotations were extracted to infer gene regulatory networks by means of the GeneMANIA prediction server, utilizing a large set of available genomics and proteomics data (protein, physical and genetic interactions, coexpression, and colocalization data) stored in its database, and assigning an equal weight to all different data sets. Each cluster was extended with associated genes considering the 10 top-scored ones by GeneMANIA. Then, we removed the genes poorly connected (e.g. networks composed of less than three nodes) to reduce prediction bias. Hence, starting from these extended clusters, we ran a further GeneMANIA algorithm on a single time-point level. This strategy globally resulted in three different gene networks depicting the shade-avoidance response during time (Figs. 3 and 4; Supplemental Fig. S2).
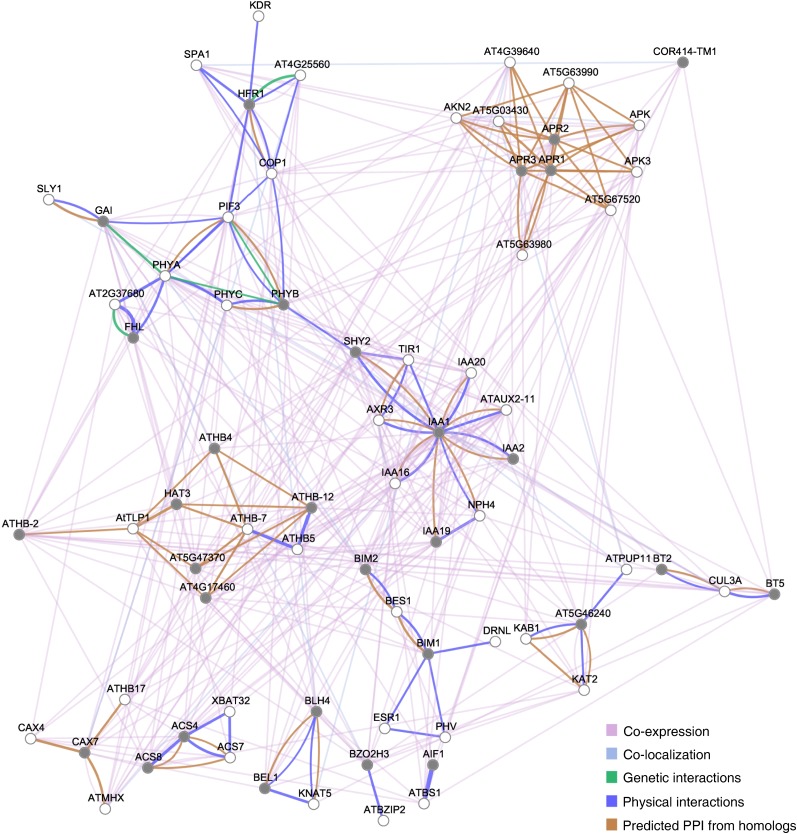
Figure 3.
Network analysis of genes regulated very early in the shade-avoidance response. DAVID functional clusters of genes regulated upon 1 h of exposure to low R/FR were utilized to infer gene networks by means of the GeneMANIA server using available genomics and proteomics data. Networks are represented as graphs, in which the nodes correspond to genes and the edges correspond to interactions between them. Nodes colored with gray indicate significant differential gene expression with respect to the control in our array data (Supplemental Table S1; for details, see “Materials and Methods”). PPI, Protein-protein interactions.
Figure 4.
Network analysis of genes regulated late in the shade-avoidance response. DAVID functional clusters of genes regulated upon 1 d of exposure to low R/FR were utilized to infer gene networks by means of the GeneMANIA server using available genomics and proteomics data. Networks are represented as graphs, in which the nodes correspond to genes and the edges correspond to interactions between them. Nodes colored with gray indicate significant differential gene expression with respect to the control in our array data (Supplemental Table S3; for details, see “Materials and Methods”). PPI, Protein-protein interactions.
In the very early stages of shade avoidance, gene networks underscore the connections among the phy-PIF signaling pathway (Bae and Choi, 2008; Leivar and Quail, 2011), transcription factors (e.g. HD-Zip II; Ruberti et al., 2012), and hormones (Fig. 3). The genes involved in phytochrome transduction pathways form a clear subnetwork, connected also with GIBBERELLIN INSENSITIVE (GAI) and SLEEPY1 (SLY1), key gibberellin signaling components (Hartweck, 2008; Harberd et al., 2009). Moreover, this subnetwork is mostly related to the auxin signaling pathway (Chapman and Estelle, 2009), further supporting the central role of this hormone during early phases of the shade-avoidance response. Another subnetwork is composed of HD-Zip genes and highlights the interaction among family II members with a documented role in the shade-avoidance response (ATHB2, ATHB4, HOMEOBOX ARABIDOPSIS THALIANA1 [HAT1], HAT2, HAT3; Carabelli et al., 1996; Steindler et al., 1999; Sessa et al., 2005; Ciarbelli et al., 2008, Sorin et al., 2009) and family I members (ATHB5, ATHB7, and ATHB12; Henriksson et al., 2005). Finally, GeneMANIA evidenced a tight cluster of genes involved predominantly in sulfur primary metabolism (ADENOSINE-5′-PHOSPHOSULFATE REDUCTASES [APRs] and ADENOSINE-5′-PHOSPHOSULFATE KINASES [APKs]; Kopriva et al., 2009; Mugford et al., 2011; Fig. 3).
After 4 h of low R/FR, networks show a higher number of genes linked in two major interwoven subnetworks. The first one is related to phytochrome signaling pathways similar to 1 h; however, beyond the connection with GAI and SLY1 emerges the association with ethylene signaling (Lin et al., 2009; Yoo et al., 2009). The second one is constituted of genes known to be involved in sulfate assimilation pathways, with a prominent role of secondary metabolism (APRs-APKs-SULFOTRANSFERASES; Kopriva et al., 2009; Mugford et al., 2011; Supplemental Fig. S2).
Interestingly, upon prolonged exposure to low R/FR (1 d), GeneMANIA analysis describes a different scenario with respect to the early phases of the shade-avoidance response: the phytochrome transduction pathways do not involve PIFs, whereas HY5, a crucial regulator of photomorphogenesis (Bae and Choi, 2008), gains a central position in this subnetwork (Fig. 4). The results also suggest casein kinases (CKA1 and CKBs) and central circadian oscillator (CCA1) as points of convergence of different cues (Portolés and Más, 2010; Fig. 4). Furthermore, the analysis highlights smaller subnetworks containing genes belonging to metabolic pathways, mainly in sulfur primary metabolism (APRs-APKs; Kopriva et al., 2009; Mugford et al., 2011; Fig. 4).
Promoter cis-Element Analysis of Genes Regulated at Different Times during Shade Avoidance
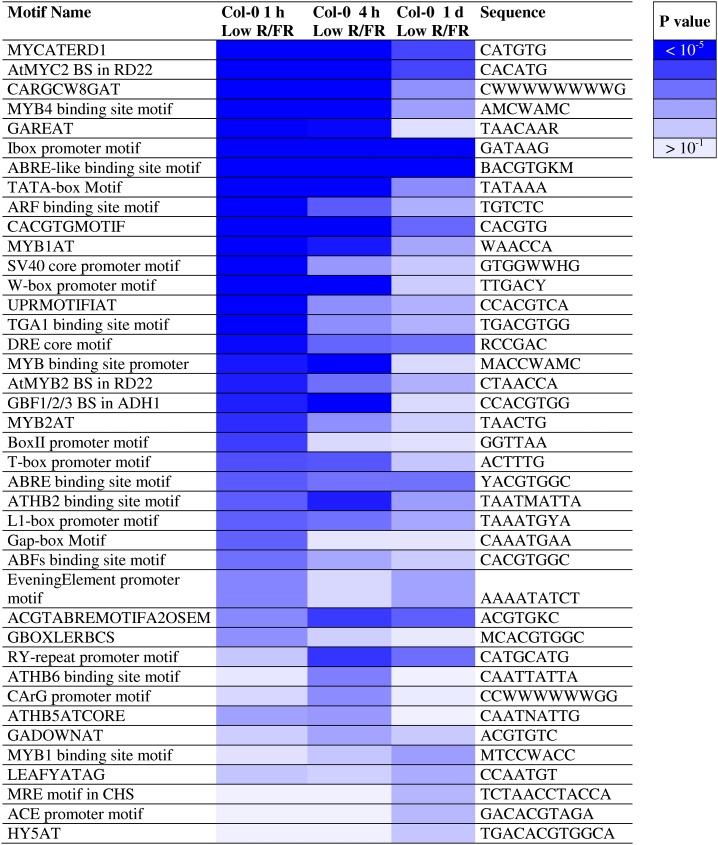
To gain further insights on transcriptional gene regulation during the shade-avoidance response, cis-acting element enrichment analysis was carried out by means of the Athena Web tool (O’Connor et al., 2005). We focused on the promoter regions (−2,000 bp, truncated if they overlap with an upstream gene) of the genes regulated by low R/FR upon 1 h, 4 h, or 1 d. For each promoter, we identified the top 30 enriched motifs with P < 0.05: upon score calculation, we displayed cis-elements nonredundantly by means of a heat map to highlight their overrepresentation at different time points (Fig. 5).
Figure 5.
Overrepresented DNA motifs on promoters of genes regulated in Col-0 seedlings upon exposure to low R/FR. The heat map displays the P values of significantly overrepresented cis-elements (top-ranked 30 motifs with P < 0.05, calculated using a hypergeometric distribution) in the wild type upon low-R/FR exposure for different times, identified by means of the Athena Web tool (http://www.bioinformatics2.wsu.edu/Athena).
Promoter analyses further highlighted the complex, dynamic transcriptional regulation taking place during the shade-avoidance response. In fact, cis-elements related to hormonal pathways, mainly those of auxin (“ARF [for auxin response factor] binding site motif”; Liu et al., 1994; Ulmasov et al., 1995) and gibberellin (“GAREAT”; Ogawa et al., 2003), display a rapid and transient enrichment (Fig. 5). A similar behavior is depicted for HD-Zip II transcription factor-binding sites (e.g. “ATHB2 binding site motif,” “ATHB6 binding site motif,” and “ATHB5ATCORE”; Sessa et al., 1993, 1997), whereas the opposite is true for the DNA elements recognized by the key regulator of photomorphogenesis, HY5 (“HY5AT”; Chattopadhyay et al., 1998), and CHALCONE SYNTHASE (CHS; “MRE motif in CHS” and “ACE promoter motif”; Hartmann et al., 1998), which are significantly enriched only in late-regulated genes (Fig. 5). Moreover, consistent with DAVID analyses, cis-elements involved in phytochrome and light signaling, such as E-box/G-box (Toledo-Ortiz et al., 2003) or I-box/GATA (Giuliano et al., 1988; Terzaghi and Cashmore, 1995), display an overrepresentation both upon brief and prolonged low R/FR treatment (Fig. 5).
Genome-Wide Functional Analyses of Negative Regulators of the Shade-Avoidance Response
To further explore, at a genome-wide scale, the dynamics of the plant response to light quality changes, we took advantage of loss-of-function mutants in negative regulators of shade avoidance (hfr1/sics1 and phyA).
HFR1/SICS1, originally described as a downstream component of phyA and CRYPTOCHROME1 in the deetiolation process (Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000; Duek and Fankhauser 2003; Bae and Choi, 2008), has been subsequently demonstrated to be specifically regulated by light quality changes and to act as a master negative regulator of the shade-avoidance response (Sessa et al., 2005; Hornitschek et al., 2009). The rapid induction of HFR1/SICS1 by low R/FR as well as its reversibility by high R/FR have strongly suggested the involvement of type II phytochromes in the regulation of this gene during shade avoidance (Sessa et al., 2005). Consistent with this hypothesis, HFR1/SICS1 is significantly up-regulated in phyB-9 seedlings in high R/FR (Lorrain et al., 2008; Supplemental Fig. S3A); the expression level of HFR1/SICS1 in phyB-9 in high R/FR is comparable to that observed in the wild type briefly exposed to low R/FR (Supplemental Fig. S3A), implying a major role for phyB in the regulation of HFR1/SICS1 expression by light quality changes. To investigate whether the up-regulation of HFR1/SICS1 in the phyB-9 mutant is functionally relevant, we constructed the double mutant hfr1-4/sics1-1 phyB-9 and analyzed its phenotype in high R/FR. As expected, phyB-9 is extremely elongated relative to the wild type (Reed et al., 1993; Supplemental Fig. S3B). However, in accordance with HFR1/SICS1 acting as a negative regulator of elongation growth, the hypocotyls of hfr1-4/sics1-1 phyB-9 seedlings are significantly longer than those of phyB-9 (Supplemental Fig. S3B). Furthermore, several genes early induced by low R/FR (Carabelli et al., 1996; Devlin et al., 2003; Salter et al., 2003; Sessa et al., 2005; Roig-Villanova et al., 2006) are up-regulated in hfr1-4/sics1-1 phyB-9 relative to phyB-9 in high R/FR (Supplemental Fig. S3C).
Several findings have indicated the involvement of phyA in attenuating the elongation response to low R/FR (Johnson et al., 1994; Devlin et al., 2003; Wang et al., 2011). Consistently, phyA-211 mutants display an exaggerated elongation response upon 1 and 2 d of exposure to our simulated shade environment relative to the wild type (Supplemental Fig. S4).
To verify that the elongated phenotype of hfr1-4/sics1-1 and phyA-211 under simulated shade (Sessa et al., 2005; Supplemental Fig. S4) does indeed depend on low R/FR, control experiments were performed lowering PAR without changing the ratio between R and FR. Col-0, hfr1-4/sics1-1, and phyA-211 seedlings were grown for 4 d in a L/D cycle in high R/FRhigh PAR and then either maintained in the same regimen or exposed to high R/FRlow PAR for 1 and 2 d. No significant difference was observed in the hypocotyl elongation of hfr1-4/sics1-1 and phyA-211 relative to the wild type upon exposure to high R/FRlow PAR, indicating that the increase in elongation rate observed upon exposure to low R/FRlow PAR is induced by the change in the ratio of R to FR (Supplemental Fig. S4). Interestingly, whereas the growth response to low R/FR is reduced over time in the wild type, that of both hfr1-4/sics1-1 and phyA-211 occurs with little variation between days 1 and 2 (Supplemental Fig. S4).
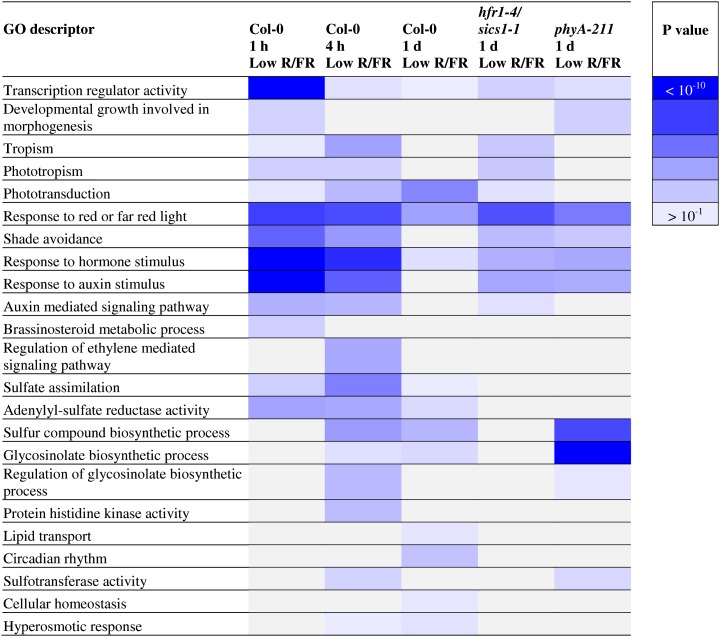
To gain further genome-wide insights into the dynamics of shade avoidance, we analyzed global expression profiles of hfr1-4/sics1-1 (Sessa et al., 2005; Supplemental Table S10) and phyA-211 (Supplemental Table S11) mutants upon prolonged exposure to low R/FR (1 d) by means of the DAVID functional classification and selected the enriched GO terms corresponding to the ones present in the most enriched functional annotation clusters in the wild type upon 1 h, 4 h, and 1 d of low R/FR (top 10 ranked with EASE score greater than 1; Supplemental Tables S4–S6). We then used a heat map to evaluate the enrichment of each GO descriptor in the wild type at different times during shade avoidance and in the mutants upon prolonged exposure to low R/FR (Fig. 6). The “shade avoidance” functional class is enriched in both hfr1-4/sics1-1 and phyA-211 upon prolonged exposure to FR-rich light, similar to the wild type upon brief low R/FR treatment. Furthermore, transcription factors (“transcription regulator activity”) and auxin (“response to hormone stimulus” and “response to auxin stimulus”) classes, which are the ones most enriched in wild-type seedlings briefly exposed to FR-rich light, are overrepresented in both hfr1-4/sics1-1 and phyA-211 mutants upon prolonged exposure to low R/FR (Fig. 6).
Figure 6.
Heat map of GO descriptor enrichment in Col-0 and mutants upon exposure to low R/FR. The heat map displays the P values of enriched GO descriptors identified by means of DAVID in the wild type upon low-R/FR exposure for different times (10 leading ranks with EASE score greater than 1 for each time; Supplemental Tables S4–S6). The P values of the same GO descriptors in hfr1-4/sics1-1 and phyA-211 seedlings upon prolonged exposure to low R/FR are also shown.
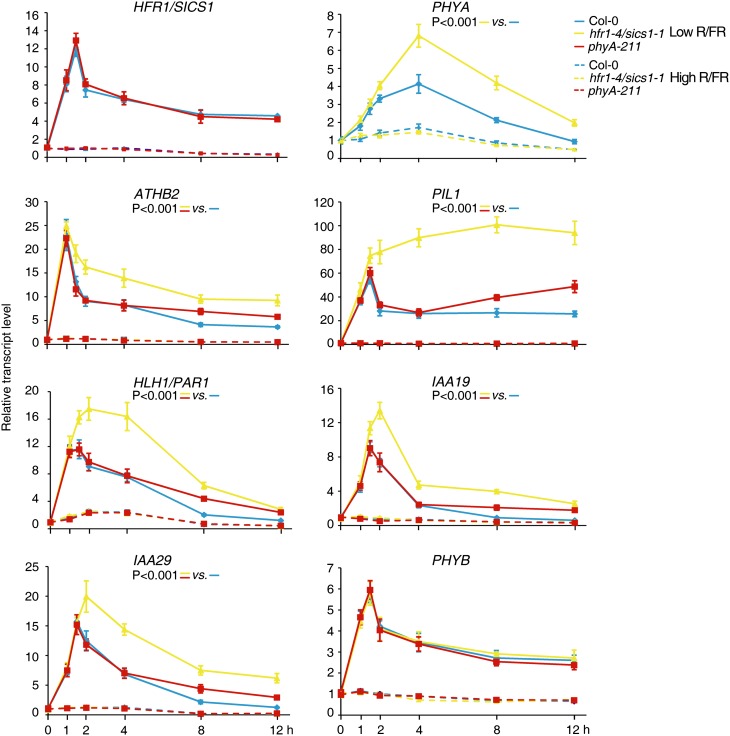
To understand the temporal dynamics of gene expression changes in seedlings lacking functional HFR1/SICS1 and PHYA proteins with respect to the wild type upon exposure to the simulated shade environment, we performed low-R/FR kinetics experiments in Col-0, hfr1-4/sics1-1, and phyA-211 and analyzed the expression of key regulators of plant responses to FR-rich light. No significant difference was observed in the induction of several genes early regulated by low R/FR in hfr1-4/sics1-1 and phyA-211 relative to the wild type. By contrast, the transcript levels of all of these genes are significantly higher in both hfr1-4/sics1-1 and phyA-211 mutants than the wild type at later times of exposure to low R/FR (Fig. 7; Sessa et al., 2005). Interestingly, the expression profile of genes early induced by light quality changes is different in hfr1-4/sics1-1 and phyA-211 mutants. Several transcription factor genes, such as ATHB2 and PIL1, functionally implicated in the shade-avoidance response (Steindler et al., 1999; Salter et al., 2003), as well as Aux/IAA genes, rapidly induced by FR-rich light, are significantly up-regulated upon 2 and 8 h of low R/FR exposure in hfr1-4/sics1-1 and phyA-211 mutant seedlings, respectively (Fig. 7). At the later times (8 and 12 h), transcript levels of genes rapidly regulated by low R/FR are significantly higher in both mutants (Fig. 7). Consistent with HFR1/SICS1 being largely regulated through phyB in photoautotrophic seedlings (Supplemental Fig. S3), no significant difference was observed in the HFR1/SICS1 transcript levels in phyA-211 seedlings relative to the wild type upon exposure to low R/FR (Fig. 7).
Figure 7.
phyA is required to down-regulate genes early induced by low R/FR. Col-0, hfr1-4/sics1-1, and phyA-211 seedlings were grown for 7 d in a L/D cycle (16/8 h) in high R/FR and then maintained in high R/FR or transferred to low R/FR for different times. The graphs show the relative expression levels in high (dashed lines) and low (solid lines) R/FR of HFR1/SICS1, PHYA, ATHB2, PIL1, HLH1/PAR1, IAA19, and IAA29 in the different genotypes. Each value is the mean ± sd of three biological replicates normalized to EF1α expression. PHYB is a light-regulated gene not differentially expressed in mutant seedlings relative to the wild type. Statistical significances between different backgrounds were assessed by means of two-way ANOVA followed by the Bonferroni posthoc test.
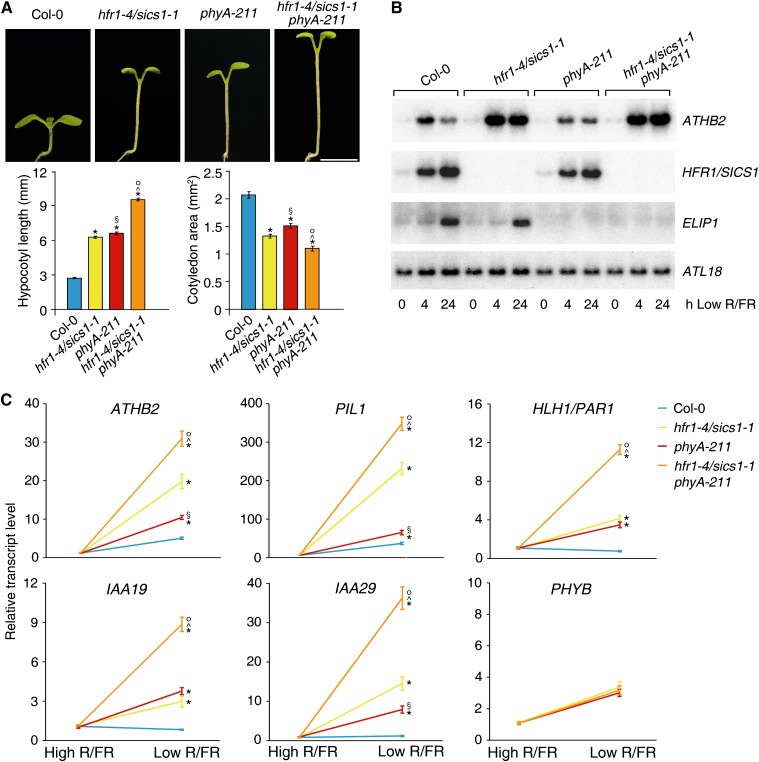
To investigate whether phyA indeed acts independently of HFR1/SICS1 in low R/FR, we constructed the double mutant hfr1-4/sics1-1 phyA-211 and analyzed its phenotype. No major difference was observed in hfr1-4/sics1-1 phyA-211 relative to hfr1-4/sics1-1 and phyA-211 in high R/FR (Supplemental Table S12). By contrast, the phenotype of the double mutant is extremely more severe than those of hfr1-4/sics1-1 and phyA-211 single mutants in low R/FR (Fig. 8A; Supplemental Fig. S5). Particularly striking is the appearance of elongated internodes such that under low R/FR, hfr1-4/sics1-1 phyA-211 plants no longer display a rosette habit (Supplemental Fig. S6). Consistent with the extremely exaggerated phenotype of the double mutant in low R/FR, the expression of several genes rapidly and transiently induced by low R/FR is significantly higher in hfr1-4/sics1-1 phyA-211 relative to both hfr1-4/sics1-1 and phyA-211 upon prolonged exposure to FR-rich light (Fig. 8, B and C).
Figure 8.
HFR1/SICS1 and phyA act largely independently in the regulation of the shade-avoidance response. A, Col-0, hfr1-4/sics1-1, phyA-211, and hfr1-4/sics1-1 phyA-211 seedlings grown for 4 d in a L/D cycle (16/8 h) in high R/FR and then transferred to low R/FR for 4 d under the same L/D regimen. The histograms depict the mean ± se hypocotyl lengths (left) and the mean ± se cotyledon areas (right) of seedlings grown as described. At least 30 seedlings were analyzed for each line. Statistical significance was assessed by means of one-way ANOVA followed by Tukey’s test. *P < 0.01 for hfr1-4/sics1-1, phyA-211, and hfr1-4/sics1-1 phyA-211 versus Col-0; §P < 0.01 for phyA-211 versus hfr1-4/sics1-1; ^P < 0.01 for hfr1-4/sics1-1 phyA-211 versus hfr1-4/sics1-1; °P < 0.01 for hfr1-4/sics1-1 phyA-211 versus phyA-211. Bar = 3 mm. B, Northern-blot analyses of ATHB2 and HFR1/SICS1 in Col-0, hfr1-4/sics1-1, phyA-211, and hfr1-4/sics1-1 phyA-211. Plants were grown for 7 d in a L/D cycle (16/8 h) in high R/FR (0) and then transferred to low R/FR under the same L/D regimen. Plant transfer to low R/FR was performed 4 h after the beginning of the light period, and seedlings were harvested 4 and 24 h later for northern-blot analyses. ELIP1 is a gene not differentially regulated by low R/FR in hfr1/sics1 mutants relative to the wild type. ATL18 was used to monitor equal loading. C, RT-qPCR analyses of ATHB2, PIL1, HLH1/PAR1, IAA19, and IAA29 in Col-0, hfr1-4/sics1-1, phyA-211, and hfr1-4/sics1-1 phyA-211. Plants were grown for 7 d in a L/D cycle (16/8 h) in high R/FR and then either maintained in high R/FR or transferred to low R/FR under the same L/D regimen for 1 d. Plant transfer to low R/FR was performed 4 h after the beginning of the light period. The graphs show the relative expression levels in high and low R/FR of ATHB2, PIL1, HLH1/PAR1, IAA19, and IAA29 in the different genotypes. PHYB is a gene not differentially regulated by low R/FR in mutant seedlings relative to the wild type. Each value is the mean ± sd of five biological replicates normalized to EF1α expression. Statistical significance was assessed by means of one-way ANOVA followed by Tukey’s test. *P < 0.001 for hfr1-4/sics1-1, phyA-211, and hfr1-4/sics1-1 phyA-211 versus Col-0; §P < 0.001 for phyA-211 versus hfr1-4/sics1-1; ^P < 0.001 for hfr1-4/sics1-1 phyA-211 versus hfr1-4/sics1-1; °P < 0.001 for hfr1-4/sics1-1 phyA-211 versus phyA-211.
Previous work has shown that hfr1/sics1 mutants display an early-flowering phenotype in low R/FR (Sessa et al., 2005; Supplemental Fig. S7, A and B). Consistent with this, it has also been shown that the expression of FT, encoding a key integrator of different floral induction signals including low R/FR (Cerdán and Chory, 2003; Casal et al., 2004; Kim et al., 2008; Wollenberg et al., 2008; Adams et al., 2009), was substantially higher in hfr1/sics1 relative to the wild type (Sessa et al., 2005; Supplemental Fig. S7C). Recent work has demonstrated that the HFR1/SICS1 protein interacts with PIF transcription factors forming non-DNA-binding heterodimers, thus limiting PIF-mediated gene expression (Hornitschek et al., 2009). Furthermore, it has been reported that PIF4 directly regulates FT at high temperature (Franklin et al., 2011; Kumar et al., 2012). It seems likely, therefore, that the higher FT transcript levels observed in hfr1/sics1 in low R/FR might result from the increased PIF protein activity in this mutant. By contrast, phyA mutants are late flowering in low R/FR (Johnson et al., 1994; Supplemental Fig. S7), and a role of phyA in enhancing the stability of CONSTANS (CO), a positive regulator of FT expression, has been demonstrated (Valverde et al., 2004). Consistent with HFR1/SICS1 and phyA acting largely independently of one another, hfr1/sics1 phyA-211 plants flower significantly earlier than phyA-211 under low R/FR (Supplemental Fig. S7).
FARO of hfr1/sics1 and phyA Mutants upon Prolonged Exposure to Low R/FR
FARO analyses of hfr1-1/sics1-4 and phyA-211 showed that both mutants still have a strong association with IAA profiles upon prolonged exposure to low R/FR (Fig. 9, IAA; Supplemental Tables S13 and S14), further supporting the functional relevance of auxin-related pathway enrichment highlighted by DAVID during early phases of the shade-avoidance response, followed later by the attenuation of this process through the action of HFR1/SICS1 and phyA (Fig. 6). FARO results also revealed a highly significant overlap of hfr1-1/sics1-4 and phyA-211 mutants exposed to low R/FR for a prolonged time with deetiolation experiments (Fig. 9). However, strikingly, the direction of the response was dissimilar, as observed in the wild type at the early stages of shade avoidance (Figs. 2 and 9).
Figure 9.
FARO of hfr1-4/sics1-1 and phyA-211 in low R/FR. Low-R/FR-regulated genes involved in light and hormone pathways are visualized by means of the FARO Web tool. The large majority of top-ranked associated factors by FARO to hfr1-4/sics1-1 and phyA-211 upon prolonged exposure to low R/FR were composed of genes involved in light and hormone pathways, which were consequently selected for functional analysis. Genes differentially regulated in Col-0, hfr1-4/sics1-1, and phyA-211 upon 1 d of low-R/FR treatment are displayed in the central circles. In the outer circles, experimental factors with strong associations with the wild type and the mutants exposed to low R/FR are shown. The factors showing overlap with the opposite direction (i.e. congruence less than 30%) with respect to our experiments are depicted in red; those showing the highest congruence (greater than 70%) are depicted in blue.
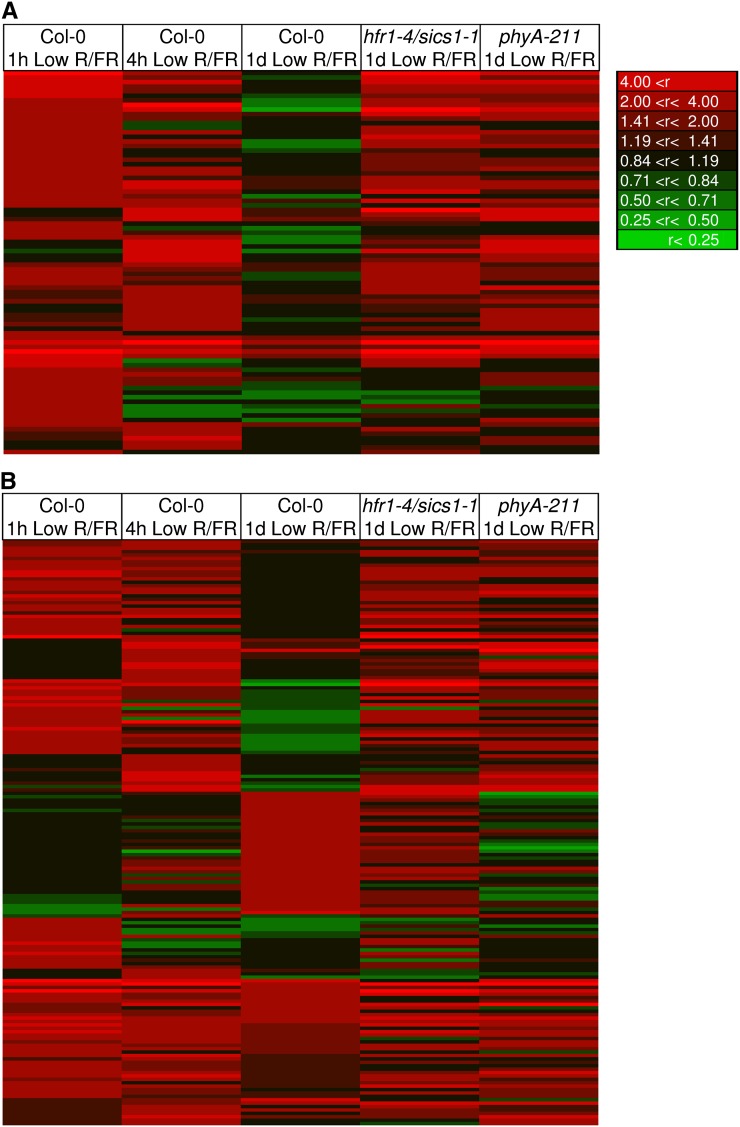
To examine further the effects of the hfr1-1/sics1-4 and phyA-211 mutations, a visual inspection of gene expression profiles during the shade-avoidance response in wild-type and mutant seedlings was conducted by means of a heat map (Fig. 10). This analysis indicated that the great majority of early shade-induced genes (Supplemental Tables S1 and S2), which FARO depicts as auxin responsive (Fig. 9), were still up-regulated upon prolonged low R/FR in both hfr1-4/sics1-1 and phyA-211 mutants (Fig. 10A). By contrast, genes induced by simulated shade (Supplemental Tables S1–S3), which FARO identifies as regulated during deetiolation (Fig. 9), displayed a different expression profile between the two mutants (Fig. 10B). The genes rapidly and transiently induced by light quality changes in wild-type seedlings (e.g. ATHB2) were still up-regulated in both mutants upon prolonged exposure to low R/FR, whereas those late induced by low R/FR were mostly up-regulated in hfr1/sics1 but not in phyA-211 (Fig. 10B).
Figure 10.
Expression profiles of auxin and light pathway genes in Col-0, hfr1-4/sics1-1, and phyA-211 exposed to low R/FR. Genes significantly up-regulated by brief or prolonged exposure to low R/FR in wild-type seedlings, and indicated by FARO as involved in auxin (A) and light (B) pathways, are visualized by means of heat maps. The expression profiles of the same genes in hfr1/sics1 and phyA-211 exposed to low R/FR for a prolonged time are also shown. Red and green depict up- and down-regulation relative to control (high R/FR), respectively. r, Gene expression ratio.
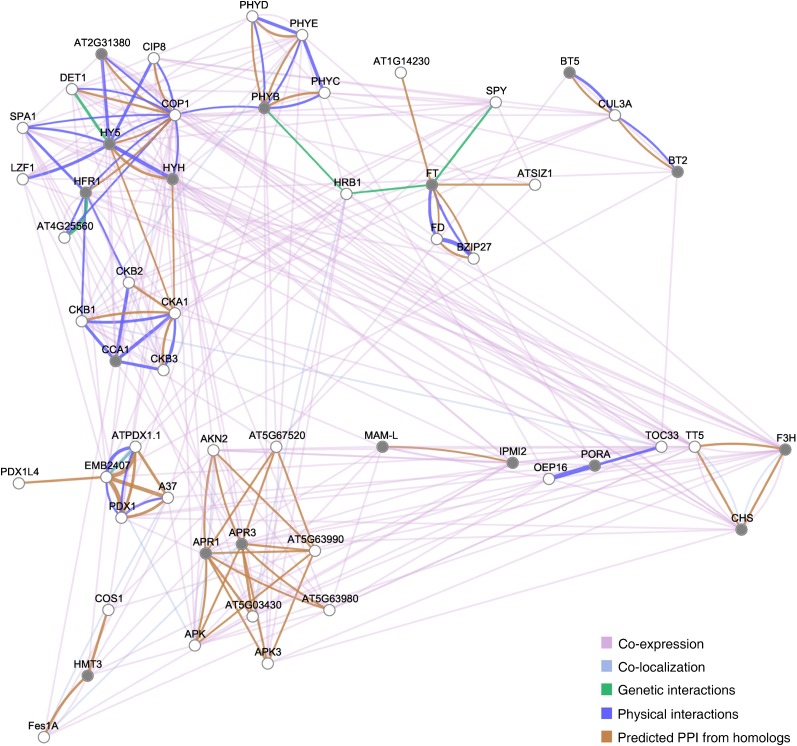
Light pathway genes regulated in the wild type and not in phyA-211 upon prolonged exposure to low R/FR, extracted from the cluster that emerged from heat map analysis (Fig. 10B), were utilized to infer gene networks by means of GeneMANIA. Network analysis identified a subnetwork involving HY5 (Fig. 11), thus suggesting that phyA might play a major role in the regulation of this transcription factor gene during the shade-avoidance response. Reverse transcription (RT)-quantitative PCR (qPCR) analyses demonstrated that HY5 is indeed significantly induced by low R/FR in a phyA-dependent manner (Fig. 12A). The PHYA gene is itself induced by low R/FR (Devlin et al., 2003; Fig. 12A), and its up-regulation precedes that of HY5 during shade avoidance (Fig. 12A). Moreover, no significant changes in HY5 transcript levels were observed in low R/FR relative to high R/FR in phyA-211 seedlings, thus demonstrating that HY5 induction by light quality changes does depend on the action of phyA (Fig. 12A). Consistent with HFR1/SICS1 and phyA acting largely independently one from the other, no significant difference was observed in HY5 induction by low R/FR in hfr1-4/sics1-1 with respect to the wild type (Supplemental Fig. S8). In addition to HY5, among the genes generating the congruence between shade avoidance and deetiolation are also HY5 HOMOLOG (HYH) and UNFERTILIZED EMBRYO SAC10 (UNE10; Fig. 11). There is evidence that HYH, a close homolog of HY5, has a role in the inhibition of hypocotyl elongation (Holm et al., 2002). The phenotype of the hyh mutant is evident only in blue light; however, increased levels of HYH can suppress the elongated phenotype of hy5 in white light (Holm et al., 2002; Sibout et al., 2006). Furthermore, hy5 hyh double mutants have longer hypocotyls than hy5 in white light (Sibout et al., 2006). hy5 hyh seedlings also display shoot phenotypes that are absent from the single mutants, including delayed leaf development and reduced vasculature (Sibout et al., 2006). Interestingly, based on both morphological and molecular phenotypes of hy5 and hy5 hyh seedlings, it was proposed that HY5 and HYH act as negative regulators of auxin signaling (Sibout et al., 2006; Lau and Deng, 2010). UNE10 encodes a bHLH transcription factor similar to PIF7 not yet characterized at the functional level (Leivar et al., 2008a). However, UNE10 has been described as a gene rapidly induced in response to continuous FR during seedling deetiolation (Tepperman et al., 2004). Moreover, UNE10 was found to be up-regulated in phyB but not in phyA phyB seedlings upon prolonged exposure to low R/FR (Devlin et al., 2003). RT-qPCR analyses demonstrated that HYH and UNE10 are both significantly induced by low R/FR in a phyA-dependent manner (Fig. 12B). Furthermore, HY5, HYH, and UNE10 are all up-regulated upon prolonged exposure to low R/FR (1 d) compared with high R/FR. This up-regulation occurs in Col-0 and hfr1-4/sics1-1 and not in phyA-211 (Fig. 12C).
Figure 11.
Network analysis of light pathway genes regulated in Col-0 but not in phyA-211 upon prolonged exposure to low R/FR. Light pathway genes regulated in the wild type but not in phyA-211 upon prolonged exposure to low R/FR (1 d), extracted from the cluster that emerged in the heat map (Fig. 10B), were utilized to infer gene networks by means of the GeneMANIA server using available genomics and proteomics data. Networks are represented as graphs, in which the nodes correspond to genes and the edges correspond to interactions between them. Nodes colored with gray show significant differential gene expression with respect to the control in our array data (Supplemental Tables S3 and S11; for details, see “Materials and Methods”). PPI, Protein-protein interactions.
Figure 12.
HY5, HYH, and UNE10 are late induced by low R/FR largely through the action of phyA. A, RT-qPCR analyses of PHYA in Col-0 and HY5 in Col-0 and phyA-211 upon exposure to low R/FR for different times. Col-0 and phyA-211 seedlings were grown for 7 d in a L/D cycle (16/8 h) in high R/FR (control) and then maintained in high R/FR or transferred to low R/FR for different times. The graphs show the relative expression levels in high (dashed lines) and low (solid lines) R/FR of PHYA in Col-0 and HY5 in Col-0 and phyA-211. Each value is the mean ± sd of three biological replicates normalized to EF1α expression. Statistical significance was assessed by means of one-way ANOVA followed by Dunnett’s test. ◊P < 0.001 for low R/FR versus control (0) in Col-0. Statistical significance between different backgrounds was assessed by means of two-way ANOVA followed by the Bonferroni posthoc test. B, RT-qPCR analyses of HYH and UNE10 in Col-0 and phyA-211 upon exposure to low R/FR for different times. Col-0 and phyA-211 seedlings were grown and treated as described in A. The graphs show the relative expression levels in high (dashed lines) and low (solid lines) R/FR of HYH and UNE10 in Col-0 and phyA-211. Each value is the mean ± sd of three biological replicates normalized to EF1α expression. Statistical significance between different treatments and backgrounds was assessed by means of two-way ANOVA followed by the Bonferroni posthoc test. C, RT-qPCR analyses of HY5, HYH, and UNE10 upon prolonged exposure to low R/FR in Col-0, hfr1-4/sics1-1, and phyA-211. Col-0, hfr1-4/sics1-1, and phyA-211 seedlings were grown for 7 d in a L/D cycle (16/8 h) in high R/FR and then either maintained in high R/FR or transferred to low R/FR under the same L/D regimen for 1 d. Plant transfer to low R/FR was performed 4 h after the beginning of the light period. The graphs show the relative expression levels in high and low R/FR of HY5, HYH, and UNE10 in the different genotypes. PHYB is a gene not differentially regulated by low R/FR in mutant seedlings relative to the wild type. Each value is the mean ± sd of five biological replicates normalized to EF1α expression. Statistical significance was assessed by means of one-way ANOVA followed by Tukey’s test. *P < 0.001 for Col-0 low R/FR versus Col-0 high R/FR; §P < 0.001 for hfr1-4/sics1-1 low R/FR versus hfr1-4/sics1-1 high R/FR; ^P < 0.001 for phyA-211 low R/FR versus phyA-211 high R/FR.
To exclude that the observed effects were caused by the higher level of FR under our simulated shade conditions (see “Simulated Shade Environment”), phenotype and gene expression experiments were performed keeping FR constant and reducing R (low R/FR*; see “Materials and Methods”). phyA-211 mutant seedlings display an exaggerated elongation response upon 1 and 2 d of exposure to low R/FR* relative to the wild type (Supplemental Fig. S9A). The PHYA gene is induced by low R/FR* (Supplemental Fig. S9B). HY5, HYH, and UNE10 are all significantly up-regulated by low R/FR* in Col-0 and not in phyA-211 (Supplemental Fig. S9C).
Interestingly, HYH and UNE10 are among the genes known to be bound in vivo by HY5 (Lee et al., 2007). To assess whether HY5 does regulate HYH and UNE10 expression under shade conditions, we performed low R/FR kinetics experiments in the loss-of-function hy5-215 mutant (Oyama et al., 1997). The up-regulation of HYH and UNE10 expression by low R/FR depends on the action of HY5, since it does not occur in the hy5 mutant (Fig. 13). No significant difference was observed in HYH and UNE10 induction by low R/FR in hfr1-4/sics1-1 with respect to the wild type (Fig. 13).
Figure 13.
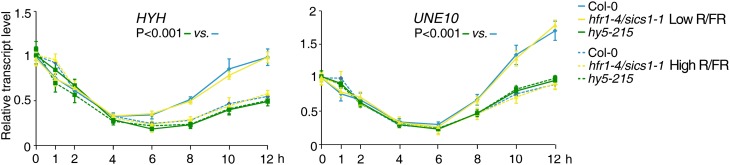
HY5 is required to up-regulate the HYH and UNE10 transcription factor genes in low R/FR. Col-0, hfr1-4/sics1-1, and hy5-215 seedlings were grown for 7 d in a L/D cycle (16/8 h) in high R/FR and then maintained in high R/FR or transferred to low R/FR for different times. The graphs show the relative expression levels in high (dashed lines) and low (solid lines) R/FR of HYH and UNE10 in the different genotypes. Each value is the mean ± sd of three biological replicates normalized to EF1α expression. Statistical significance between different backgrounds was assessed by means of two-way ANOVA followed by the Bonferroni posthoc test.
DISCUSSION
Plant responses to light quality changes are regulated by a balance of positive (PIFs) and negative (HFR1/SICS1) regulators of gene expression, which ensures a fast reshaping of the plant body toward an environment optimal for growth while at the same time avoiding an exaggerated reaction to low R/FR (Sessa et al., 2005; Lorrain et al., 2008; Hornitschek et al., 2009). Here, by combining genome-wide expression profiling and computational analyses, we show highly significant overlap between shade avoidance and deetiolation transcript profiles. Strikingly, the direction of the response is dissimilar at the early stages of shade avoidance and congruent at the late ones. Down-regulation of genes early induced by light quality changes upon prolonged exposure to low R/FR depends not only on HFR1/SICS1 (Sessa et al., 2005), which interacts with PIF transcription factors forming non-DNA-binding heterodimers, thus limiting PIF-mediated gene expression (Hornitschek et al., 2009), but also on phyA. By contrast, phyA and not HFR1/SICS1 is required for the up-regulation of genes late induced by low R/FR. Remarkably, among them is the HY5 transcription factor gene, known to act as the master regulator of seedling deetiolation (Lau and Deng, 2010).
Light Signaling Genes Are Dynamically Regulated during Shade Avoidance
Signaling downstream of the photoreceptors involves two main pathways: CONSTITUTIVE PHOTOMORPHOGENIC1 (COP1)-HY5 and PIFs. COP/DEETIOLATED (DET)/FUSCA (FUS) are central repressors of photomorphogenesis, which, in the dark, function in concert to target positive regulators of photomorphogenesis (e.g. HY5) for degradation through the 26S proteasome, thus preventing deetiolation. In daylight, the activity of COP/DET/FUS proteins is reduced, resulting in the accumulation of transcription factors required for photomorphogenesis. COP1, one of the COP/DET/FUS proteins, is an E3 ligase that interacts with several transcription factors and promotes their ubiquitination together with the SUPPRESSOR OF PHYA105 proteins (Lau and Deng, 2010). By contrast, a pifq mutant displays a cop-like phenotype in darkness, demonstrating that these PIF transcription factors function in the dark to promote skotomorphogenesis (Leivar et al., 2008b; Shin et al., 2009). Upon light exposure, photoactivated phytochromes interact with PIFs and promote their degradation via the ubiquitin-proteasome pathway (Leivar and Quail, 2011). The rapid light-induced degradation of PIF molecules does not lead to their disappearance; rather, it results in a lower steady-state level of these proteins in daylight (Leivar and Quail, 2011).
Genome-wide expression profiling of wild-type etiolated seedlings briefly exposed to light and dark-grown pifq mutants identified a set of genes that are potential direct targets of these PIF molecules (Leivar et al., 2009). More recently, by comparing these transcriptome profiles with those of wild-type and pifq seedlings briefly exposed to low R/FR, Leivar and coauthors (2012) identified a number of genes rapidly repressed and induced by light and shade signals, respectively. Among them are ATHB2, previously shown to be down-regulated in etiolated seedlings briefly exposed to R and FR and strongly and rapidly induced in photoautotrophic plants exposed to low R/FR (Carabelli et al., 1996), IAA19, and IAA29 (Leivar et al., 2012). Here, by taking advantage of the FARO Web tool, we extended and broadened these results, showing an opposite regulation of genes regulated in deetiolation experiments with respect to early shade-regulated genes and consistent regulation for late-regulated ones. Consistent with this, whereas at the very early stages of shade avoidance, gene network analysis highlights the connections among phy-PIF signaling, HD-Zip transcription factors, and auxin, later, HY5 seems to gain a central role in light signal transduction.
HFR1/SICS1 and phyA Function Largely Independently in Negatively Regulating the Shade-Avoidance Response
phyA mutants elongate more than the wild type in low R/FR (Johnson et al., 1994; Devlin et al., 2003; Wang et al., 2011; this work). Our analysis of seedlings lacking a functional phyA demonstrates a major role of this phytochrome in regulating other aspects of the shade-avoidance response as well. phyA function is complex: it negatively regulates the growth responses of the hypocotyl, cotyledons, and leaves induced by light quality changes, thus preventing an exaggerated plant reaction to low R/FR, and positively regulates flowering, shortening the generation time and, therefore, ensuring species survival in an unfavorable environment. A significant number of genes rapidly and transiently induced by light quality changes are up-regulated in phyA with respect to the wild type upon prolonged exposure to low R/FR. Interestingly, several of these genes are also up-regulated under the same conditions in seedlings lacking a functional HFR1/SICS1 (Sessa et al., 2005; Fig. 7), a negative regulator of shade avoidance (Sessa et al., 2005; Hornitschek et al., 2009), also described as a downstream component of phyA signaling during deetiolation (Fairchild et al., 2000; Fankhauser and Chory, 2000; Soh et al., 2000; Lorrain et al., 2009). However, hfr1/sics1 phyA seedlings display a phenotype more severe than those of hfr1/sics1 and phyA single mutants in the hypocotyl, cotyledons, and leaves in low R/FR. Moreover, the expression of several genes early induced by low R/FR is significantly higher in the double mutant relative to both hfr1/sics1 and phyA upon prolonged exposure to FR-rich light. In agreement with HFR1/SICS1 and phyA acting largely independently from one another, we found that HFR1/SICS1 is regulated through phyB in photoautotrophic seedlings. Finally, we also found that hfr1/sics1 phyA plants flower significantly earlier than phyA but later than hfr1/sics1 under low R/FR.
Consistent with HFR1/SICS1 and phyA acting in the negative control of shade avoidance, FARO analyses of hfr1/sics1 and phyA upon prolonged exposure to low R/FR revealed, in both mutants, a response dissimilar to that of deetiolation as observed in the wild type at the early stages of shade avoidance. However, differences emerged with respect to gene expression profiles between hfr1/sics1 and phyA. In fact, whereas the great majority of early shade-induced genes that FARO depicts as regulated during deetiolation were still up-regulated in both mutants upon prolonged exposure to low R/FR, those late induced by low R/FR were mostly up-regulated in hfr1/sics1 but not in phyA-211.
Taken together, the data indicate that phyA largely signals through transcription factors other than HFR1/SICS1 in low R/FR.
phyA Is Required for Late Induction by Low R/FR of the HY5 Transcription Factor Gene
Network analysis of the genes late induced by low R/FR in the wild type and not in the phyA mutant identified a subnetwork involving the HY5 transcription factor gene. Low-R/FR kinetics experiments demonstrated that HY5 up-regulation is preceded by the rapid induction of PHYA, and it is indeed impaired in phyA mutant seedlings. Furthermore, network analysis also highlighted a direct connection between HY5 and HYH, a gene functionally implicated in the inhibition of hypocotyl elongation (Holm et al., 2002) and known to be a direct target of the HY5 transcription factor (Lee et al., 2007). Expression studies in wild-type and mutant seedlings exposed to FR-rich light for different times revealed that the HYH gene is indeed late induced by low R/FR, and its up-regulation does depend on the action of HY5, since it does not occur in the hy5 mutant.
The role of HY5 has been most extensively studied at the early stages of seedling development. Originally identified as a negative regulator of cell elongation acting downstream of multiple families of the photoreceptors (Oyama et al., 1997; Osterlund et al., 2000), it has been shown subsequently to function as a key controller of the transcriptional cascades promoting seedling photomorphogenesis (Lau and Deng, 2010). More recently, HY5 has also been implicated in the inhibition of hypocotyl elongation induced in shaded plants by brief exposure to direct sunlight perceived primarily by phyB (Sellaro et al., 2011). HY5 is 15 to 20 times more abundant in seedlings grown in the light than in the dark (Osterlund et al., 2000). Consistent with its prominent role in the commitment to photomorphogenic development, HY5 abundance exhibits its highest level 2 to 3 d after germination and then drastically decreases at later times of seedling development (Hardtke et al., 2000). Multiple photoreceptors are involved in regulating the abundance of HY5. phyB and phyA are primarily responsible for HY5 accumulation in continuous R and FR, respectively. CRY1 and CRY2 are largely redundant and involved in the accumulation of HY5 in continuous B light. In all cases, the abundance of HY5 directly correlates with the degree of photomorphogenic development (Osterlund et al., 2000). Thus, it seems likely that increased HY5 expression upon prolonged exposure to low R/FR may be a mechanism through which phyA exerts its regulatory role in the shade-avoidance response.
A recent study combining genome-wide analysis of HY5 binding sites under continuous white light and during the light-to-dark transition and gene expression profiling of wild-type and hy5 seedlings identified more than 1,000 target genes positively or negatively regulated by HY5 (Zhang et al., 2011). The positive and negative functions of this transcription factor in regulating gene expression have been further strengthened by the finding that HY5 acts as a repressor of FAR-RED ELONGATED HYPOCOTYL1 (FHY1) and FHY1-LIKE (FHL) expression by modulating the transcriptional activities of FHY3 and its homolog FAR-RED IMPAIRED RESPONSE1 (FAR1), two crucial components in phyA signaling (Li et al., 2010), whereas it acts as a transcriptional activator of EARLY FLOWERING4 (ELF4), a key player of the central oscillator of the circadian clock, in concert with FHY3/FAR1 (Li et al., 2011). Interestingly, the cis-elements of HY5 and FHY3/FAR1 are very close to each other in the FHY1/FHL promoters, whereas they are around 20 bp away in the ELF4 promoter (Li et al., 2010, 2011). Comparison of FHY3 (Ouyang et al., 2011) and HY5 direct targets (Lee et al., 2007; Zhang et al., 2011) identified a subset of genes coregulated by these two transcription factors, including FHY1 and ELF4, suggesting that HY5 is likely to work with other transcription factors to coregulate the expression of genes other than those targets of FHY3/HY5 (Ouyang et al., 2011). Low-R/FR kinetics experiments indicated that phyA is also involved in the negative regulation of genes early induced by light quality changes upon prolonged exposure to low R/FR. Remarkably, among them is ATHB2, functionally involved in the shade-avoidance response and identified as a gene directly repressed by HY5 (Zhang et al., 2011). IAA19, previously implicated in the regulation of cell elongation in the phototropic response (Tatematsu et al., 2004), and HLH1/PAR1, both identified as genes recognized in vivo by HY5 (Lee et al., 2007; Zhang et al., 2011), are also significantly up-regulated upon prolonged low-R/FR treatment in seedlings lacking phyA. Therefore, it is tempting to speculate that phyA through HY5 not only positively regulates photomorphogenesis-promoting genes upon prolonged exposure to low R/FR but also down-regulates genes early induced by light quality changes. Future work will be needed to identify the transcription factor(s) that may work in concert with HY5 to both activate and repress gene expression in low R/FR and to understand their regulatory mode of action on promoters of common target genes.
MATERIALS AND METHODS
Plant Lines and Growth Conditions
The wild-type strain used was Arabidopsis (Arabidopsis thaliana) Col-0. Other lines used were hfr1-4/sics1-1 (Sessa et al., 2005; SALK_037727), phyB-9 (Reed et al., 1993; NASC#N6217), phyA-211 (Reed et al., 1994; NASC#N6223), and hy5-215 (Oyama et al., 1997). The lines hfr1-4/sics1-1 phyB-9 and hfr1-4/sics1-1 phyA-211 were generated by crossing. The F2 progeny of the cross hfr1-4/sics1-1 × phyB-9 was grown for 7 d in high R/FR together with phyB-9. Seedlings displaying a hypocotyl similar to or longer than that shown by phyB-9 mutants were further analyzed for the presence of the hfr1-4/sics1-1 mutation by PCR using the primers detailed below. The F2 progeny of the cross hfr1-4/sics1-1 × phyA-211 was grown for 4 d in continuous FR (41.2 μmol m−2 s−1) together with phyA-211 and hfr1-4/sics1-1. Under these conditions, phyA-211 is extremely elongated whereas hfr1-4/sics1-1 displays only a mild phenotype (Fairchild et al., 2000). Seedlings displaying an elongated phenotype, analogous to that shown by phyA-211 mutants, were then analyzed for the presence of the hfr1-4/sics1-1 mutation by PCR. The primers used for hfr1-4/sics1-1 genotyping were as follows: HFR1/SICS15′, 5′-TGGAATTGGGATGGAGAAACGAC-3′; HFR1/SICS13′, 5′-CGAGAACCGAAACCTTGTCCGT-3′; LBb1, 5′-GCGTGGACCGCTTGCTGCAACT-3′.
Plants were grown as described previously (Sessa et al., 2005) in a light-emitting diode growth chamber (E30-LED; Percival Scientific) at 21°C temperature, 75% humidity, 16-h-light/8-h-dark cycles. Light outputs in high R/FRhigh PAR were as follows: 670 nm (R), 96 μmol m−2 s−1; 735 nm (FR), 21 μmol m−2 s−1; 470 nm (blue light), 15 μmol m−2 s−1. Light outputs in high R/FRlow PAR were as follows: 670 nm, 12 μmol m−2 s−1; 735 nm, 2 μmol m−2 s−1; 470 nm, 15 μmol m−2 s−1. Light outputs in low R/FRlow PAR were as follows: 670 nm, 12 μmol m−2 s−1; 735 nm, 105 μmol m−2 s−1; 470 nm, 15 μmol m−2 s−1. Light outputs in low R/FR* were as follows: 670 nm, 12 μmol m−2 s−1; 735 nm, 21 μmol m−2 s−1; 470 nm, 15 μmol m−2 s−1.
Phenotypic Analyses
Images of whole seedlings and green leaves were taken with an MZ 12 binocular microscope (Leica) using ProgRes C10 plus (Jenoptik) or captured with a Coolpix 990 digital camera (Nikon). For hypocotyl, cotyledon, leaf, and internode measurements, images were taken with the same devices and subsequently analyzed with NIH Image analysis software (http://rsb.info.nih.gov/ij), as described previously (Sessa et al., 2005; Carabelli et al., 2007). Statistical significance was assessed by means of one-way ANOVA followed by Tukey’s or Games-Howell tests (Prism 5 [GraphPad Software] and PASW Statistics 18 [SPSS]). The 95% confidence intervals for the ratio of two means were computed by the method of Fieller (1940). The strength of the hypocotyl response during growth in different light regimes was measured by means of standardized effect size as described by Hedges and Olkin (1985).
Microarray Data Analyses
For gene expression analyses, 8-d-old seedlings were harvested after the designated treatments for the indicated periods of time. Affymetrix Arabidopsis Genome GeneChip array (ATH1) experiments were performed as described by Sessa et al. (2005). Two biological replicates were performed for each time point and each line. Pixel-level files from scanned arrays (.dat files) were analyzed with Affymetrix MAS 5.0 software to obtain text format files with intensity values for perfect match and mismatch features (.cel files) and absent/present calls (.chp files). All data were submitted to the ArrayExpress database (accession nos. E-MEXP-443, E-MEXP-444, E-MEXP-3266, and E-MEXP-3267) in compliance with minimum information about a microarray experiment guidelines. All .cel files were imported into GeneSpring 7.1 (Agilent Technologies) and normalized in two steps. First, the robust multiarray average algorithm (Irizarry et al., 2003) was used to convert the probe-level expression data into probe-set (or gene-level) expression data and to ensure that the distribution of the expression values was comparable across the different chips or samples; second, to account for the difference in detection efficiency between spots and to compare the relative change in gene expression levels, the expression value for one gene across the different conditions was centered on 1 by dividing the expression value by the median value of the expression values for that gene across the conditions. To identify low-R/FR-regulated genes, a rank-product analysis was carried out by means of R software, imposing false discovery rate < 0.05 (Breitling et al., 2005; Supplemental Tables S1–S3, S10, and S11). Heat maps obtained by means of FiRe 2.2 (Garcion et al., 2006) were used to compare gene expression patterns in multiple microarray experiments: fold change values were computed after robust multiarray average normalization of all the experiments together using GeneSpring 7.1 (Agilent Technologies).
DAVID Analyses
We used DAVID (version 6.7; http://david.abcc.ncifcrf.gov/home.jsp) functional annotation clustering to cluster the genes differentially regulated by low R/FR in the wild type (1 h, 4 h, or 1 d) and hfr1-4/sics1-1 and phyA-211 mutants (1 d) into different functional groups. DAVID uses a fuzzy clustering concept by measuring relationships among the annotation terms on the basis of the degree of their coassociation with genes within the user’s list in order to cluster somewhat heterogenous, yet highly similar, annotation into functional annotation groups (Huang et al., 2009). For each condition, we identified the significantly enriched functional groups as the top 10 ranked with EASE score greater than 1. Heat maps were used to compare P values of GO descriptors obtained from each DAVID analysis in the different genotypes.
Network Analyses
DAVID analyses of the genes differentially expressed in the wild type after exposure to low R/FR for 1 h, 4 h, and 1 d identified 29 significantly enriched functional clusters. For each cluster, the genes associated with corresponding functional annotations were extracted to feed them into the GeneMANIA server to infer network topology (version 2.1; http://www.genemania.org) using available genomics and proteomics data (protein, physical, and genetic interactions, coexpression, and colocalization data). For each cluster of genes, we ran the network analysis by weighting every individual data source equally (“equal-by-network” option) and displaying a fixed number of related genes as computed by the GeneMANIA algorithm: 10 genes in Figures 3 and 4 and Supplemental Figure S2 and 20 genes in Figure 11. After this step, we manually removed genes that were not structured in well-connected networks (e.g. composed of less than three nodes) and used the remaining ones to run the GeneMANIA Web tool on a single time point level (utilizing again the equal-by-network option but displaying no other related genes) to obtain global networks (Figs. 3, 4, and 11; Supplemental Fig. S2).
Promoter Analyses
Putative promoter regions upstream of low-R/FR-regulated genes were analyzed to identify overrepresented cis-regulatory elements by means of the Athena Web tool (http://www.bioinformatics2.wsu.edu/Athena; O’Connor et al., 2005), inspecting −2,000 bp from the 5′ untranslated region, if known (otherwise from ATG), and truncating promoter sequences that overlap with upstream genes. For each single time point, significantly overrepresented cis-elements were defined as the top-ranked 30 motifs with P < 0.05, calculated using a hypergeometric probability distribution.
FARO Analyses
FARO analyses were performed as described by Nielsen et al. (2007) by means of the appropriate Web tool (http://www.cbs.dtu.dk/services/faro/). The top-ranked 15 “associated factors” with more than 25% overlap were chosen for subsequent analysis. Following manual inspection, only factors related to light/hormone pathways were selected for functional analyses.
Northern-Blot Analyses
Northern-blot experiments were performed using 10 μg of total RNA as described previously (Carabelli et al., 1996). The ATHB2, RIBOSOMAL PROTEIN L18 (ATL18), EARLY LIGHT-INDUCIBLE PROTEIN1 (ELIP1), and HFR1/SICS1 probes were described previously (Carabelli et al., 1993, 1996; Sessa et al., 2005).
RT-qPCR
For RT-qPCR experiments, total RNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions except for step 6, for which the incubation time was extended to 1 h. qPCR was performed with the LightCycler 480 instrument (Roche) using LightCycler 480 Probes Master (Roche) and Universal ProbeLibrary (UPL) probes (Roche), 5′ labeled with fluorescein and 3′ labeled with a dark quencher dye, according to the manufacturer’s instructions, as described previously (Ciarbelli et al., 2008). Quantification of target gene expression was expressed in comparison with the reference gene TRANSLATION ELONGATION FACTOR1α (EF1α; Sibout et al., 2006), and relative expression ratio was calculated based on the qPCR efficiency for each gene and the crossing point deviation of target genes versus control (Pfaffl, 2001). Specific UPL probes and primers for RT-qPCR analyses are listed in Supplemental Table S15. Statistical analyses were performed on log-transformed relative expression ratio values as described by Rieu and Powers (2009). Relative transcript abundance of each gene was normalized to Col-0 level in high R/FR. Subsequent to data standardization (Willems et al., 2008), one-way ANOVA followed by Tukey’s posthoc test was used to assess differences among means (Prism 5; GraphPad Software). Time-course experiments were analyzed by mean of one-way ANOVA followed by Dunnett’s posthoc test; experiments with different backgrounds were evaluated by mean of two-way ANOVA followed by Bonferroni posthoc tests (Prism 5; GraphPad Software).
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ATHB2 (At4g16780), ATL18 (At3g05590), CO (At5g15840), EF1α (At1g18070.3), ELIP1 (At3g22840), FT (At1g65480), HFR1/SICS1 (At1g02340), HLH1/PAR1 (At2g42870), HY5 (At5g11260), HYH (At3g17609), IAA19 (At3g15540), IAA29 (At4g32280), PHYA (At1g09570), PHYB (At2g18790), PIL1 (At2g46970), and UNE10 (At4g00050). All array data were submitted to the ArrayExpress database (accession nos. E-MEXP-3266 and E-MEXP-3267) in compliance with minimum information about a microarray experiment guidelines.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Arabidopsis seedling responses to low R/FRlow PAR versus high R/FRlow PAR.
Supplemental Figure S2. Network analysis of genes regulated early in the shade-avoidance response.
Supplemental Figure S3. HFR1/SICS1 is regulated through phyB in photoautotrophic seedlings.
Supplemental Figure S4. Hypocotyl elongation of Col-0, hfr1-4/sics1-1, and phyA-211 seedlings in high R/FRhigh PAR, high R/FRlow PAR, and low R/FRlow PAR.
Supplemental Figure S5. Leaf phenotype of hfr1-4/sics1-1 phyA-211 in low R/FR.
Supplemental Figure S6. hfr1-4/sics1-1 phyA-211 plants exhibit internode elongation in low R/FR.
Supplemental Figure S7. hfr1-4/sics1-1 phyA-211 plants are early flowering relative to phyA-211 in low R/FR.
Supplemental Figure S8. Kinetics of HY5 induction by low R/FR in Col-0 and hfr1-4/sics1-1 seedlings.
Supplemental Figure S9. Arabidopsis seedling responses to low R/FR*.
Supplemental Table S1. Genes regulated in Col-0 after 1 h of exposure to low R/FR.
Supplemental Table S2. Genes regulated in Col-0 after 4 h of exposure to low R/FR.
Supplemental Table S3. Genes regulated in Col-0 after 1 d of exposure to low R/FR.
Supplemental Table S4. Significantly enriched cluster descriptors in Col-0 after 1 h of exposure to low R/FR.
Supplemental Table S5. Significantly enriched cluster descriptors in Col-0 after 4 h of exposure to low R/FR.
Supplemental Table S6. Significantly enriched cluster descriptors in Col-0 after 1 d of exposure to low R/FR.
Supplemental Table S7. List of experiments functionally associated with Col-0 at 1 h of low R/FR according to FARO.
Supplemental Table S8. List of experiments functionally associated with Col-0 at 4 h of low R/FR according to FARO.
Supplemental Table S9. List of experiments functionally associated with Col-0 at 1 d of low R/FR according to FARO.
Supplemental Table S10. Genes regulated in hfr1-4/sics1-1 upon 1 d of exposure to low R/FR.
Supplemental Table S11. Genes regulated in phyA-211 upon 1 d of exposure to low R/FR.
Supplemental Table S12. Phenotypes of Col-0, hfr1-4/sics1-1, phyA-211, and hfr1-4/sics1-1 phyA-211 seedlings in high R/FR.
Supplemental Table S13. List of experiments functionally associated with hfr1-4/sics1-1 at 1 d of low R/FR according to FARO.
Supplemental Table S14. List of experiments functionally associated with phyA-211 at 1 d of low R/FR according to FARO.
Supplemental Table S15. Primers and UPL probes for RT-qPCR analyses.
Acknowledgments
We thank Jorg Becker (Affymetrix Core Facility, Instituto Gulbenkian de Ciencia) for microarray experiments. We also thank the European Arabidopsis Stock Centre for N6217 and N6223 seeds. We are grateful to Daniela Bongiorno (Institute of Molecular Biology and Pathology, National Research Council) for skilled technical assistance.
Glossary
- R
red light
- FR
far-red light
- bHLH
basic helix-loop-helix
- FARO
functional associations by response overlap
- PAR
photosynthetic active radiation
- Col-0
Columbia
- L/D
light/dark
- DAVID
Database for Annotation, Visualization, and Integrated Discovery
- GO
Gene Ontology
- RT
reverse transcription
- qPCR
quantitative PCR
- low R/FR*
far-red light constant and reduced red light
- UPL
Universal ProbeLibrary
- EASE score
a modified Fisher Exact P-Value
References
- Adams S, Allen T, Whitelam GC. (2009) Interaction between the light quality and flowering time pathways in Arabidopsis. Plant J 60: 257–267 [DOI] [PubMed] [Google Scholar]
- Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. (2006) Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell 23: 439–446 [DOI] [PubMed] [Google Scholar]
- Bae G, Choi G. (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59: 281–311 [DOI] [PubMed] [Google Scholar]
- Ballaré CL. (1999) Keeping up with the neighbours: phytochrome sensing and other signalling mechanisms. Trends Plant Sci 4: 97–102 [DOI] [PubMed] [Google Scholar]
- Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z. (2004) Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134: 1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, et al. (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R, Armengaud P, Amtmann A. (2005) Vector analysis as a fast and easy method to compare gene expression responses between different experimental backgrounds. BMC Bioinformatics 6: 181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Morelli G, Whitelam G, Ruberti I. (1996) Twilight-zone and canopy shade induction of the Athb-2 homeobox gene in green plants. Proc Natl Acad Sci USA 93: 3530–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Possenti M, Sessa G, Ciolfi A, Sassi M, Morelli G, Ruberti I. (2007) Canopy shade causes a rapid and transient arrest in leaf development through auxin-induced cytokinin oxidase activity. Genes Dev 21: 1863–1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli M, Sessa G, Baima S, Morelli G, Ruberti I. (1993) The Arabidopsis Athb-2 and -4 genes are strongly induced by far-red-rich light. Plant J 4: 469–479 [DOI] [PubMed] [Google Scholar]
- Casal JJ. (2013) Photoreceptor signaling networks in plant responses to shade. Annu Rev Plant Biol 64: 403–427 [DOI] [PubMed] [Google Scholar]
- Casal JJ, Fankhauser C, Coupland G, Blázquez MA. (2004) Signalling for developmental plasticity. Trends Plant Sci 9: 309–314 [DOI] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12: 514–521 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Chory J. (2003) Regulation of flowering time by light quality. Nature 423: 881–885 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N. (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, Fruscalzo A, Sessa G, Morelli G, Ruberti I. (2008) The Arabidopsis homeodomain-leucine zipper II gene family: diversity and redundancy. Plant Mol Biol 68: 465–478 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière J-M, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blázquez MA, Titarenko E, Prat S. (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC. (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Robson PRH, Patel SR, Goosey L, Sharrock RA, Whitelam GC. (1999) Phytochrome D acts in the shade-avoidance syndrome in Arabidopsis by controlling elongation growth and flowering time. Plant Physiol 119: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA. (2003) A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133: 1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duek PD, Fankhauser C. (2003) HFR1, a putative bHLH transcription factor, mediates both phytochrome A and cryptochrome signalling. Plant J 34: 827–836 [DOI] [PubMed] [Google Scholar]
- Fairchild CD, Schumaker MA, Quail PH. (2000) HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev 14: 2377–2391 [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. (2000) RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol 124: 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fieller EC. (1940) The biological standardization of insulin. Suppl J R Statist Soc 7: 1–64 [Google Scholar]
- Franklin KA. (2008) Shade avoidance. New Phytol 179: 930–944 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Quail PH. (2010) Phytochrome functions in Arabidopsis development. J Exp Bot 61: 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galstyan A, Cifuentes-Esquivel N, Bou-Torrent J, Martinez-Garcia JF. (2011) The shade avoidance syndrome in Arabidopsis: a fundamental role for atypical basic helix-loop-helix proteins as transcriptional cofactors. Plant J 66: 258–267 [DOI] [PubMed] [Google Scholar]
- Garcion C, Baltensperger R, Fournier T, Pasquier J, Schnetzer MA, Gabriel JP, Métraux JP. (2006) FiRe and microarrays: a fast answer to burning questions. Trends Plant Sci 11: 320–322 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR. (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Oh E, Choi G, Liang Z, Wang ZY. (2012) Interactions between HLH and bHLH factors modulate light-regulated plant development. Mol Plant 5: 688–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harberd NP, Belfield E, Yasumura Y. (2009) The angiosperm gibberellin-GID1-DELLA growth regulatory mechanism: how an “inhibitor of an inhibitor” enables flexible response to fluctuating environments. Plant Cell 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW. (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B. (1998) Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol 36: 741–754 [DOI] [PubMed] [Google Scholar]
- Hartweck LM. (2008) Gibberellin signaling. Planta 229: 1–13 [DOI] [PubMed] [Google Scholar]
- Hedges LV, Olkin I (1985) Statistical Methods for Meta-Analysis. Academic Press, New York [Google Scholar]
- Henriksson E, Olsson AS, Johannesson H, Johansson H, Hanson J, Engström P, Söderman E. (2005) Homeodomain leucine zipper class I genes in Arabidopsis: expression patterns and phylogenetic relationships. Plant Physiol 139: 509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Jang IC, Chua NH. (2009) Regulated proteolysis in light-related signaling pathways. Curr Opin Plant Biol 12: 49–56 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma L-G, Qu L-J, Deng X-W. (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Hornitschek P, Lorrain S, Zoete V, Michielin O, Fankhauser C. (2009) Inhibition of the shade avoidance response by formation of non-DNA binding bHLH heterodimers. EMBO J 28: 3893–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Lau OS, Deng XW. (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8: 217–230 [DOI] [PubMed] [Google Scholar]
- Johnson E, Bradley M, Harberd NP, Whitelam GC. (1994) Photoresponses of light-grown phyA mutants of Arabidopsis: phytochrome A is required for the perception of daylength extensions. Plant Physiol 105: 141–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanyuka K, Praekelt U, Franklin KA, Billingham OE, Hooley R, Whitelam GC, Halliday KJ. (2003) Mutations in the huge Arabidopsis gene BIG affect a range of hormone and light responses. Plant J 35: 57–70 [DOI] [PubMed] [Google Scholar]
- Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, Ballaré CL. (2011) Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J 67: 195–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Pollmann S, Voesenek LA, Peeters AJ, Pierik R. (2010) Auxin transport through PIN-FORMED 3 (PIN3) controls shade avoidance and fitness during competition. Proc Natl Acad Sci USA 107: 22740–22744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keuskamp DH, Sasidharan R, Vos I, Peeters AJ, Voesenek LA, Pierik R. (2011) Blue-light-mediated shade avoidance requires combined auxin and brassinosteroid action in Arabidopsis seedlings. Plant J 67: 208–217 [DOI] [PubMed] [Google Scholar]
- Kim SY, Yu X, Michaels SD. (2008) Regulation of CONSTANS and FLOWERING LOCUS T expression in response to changing light quality. Plant Physiol 148: 269–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopriva S, Mugford SG, Matthewman C, Koprivova A. (2009) Plant sulfate assimilation genes: redundancy versus specialization. Plant Cell Rep 28: 1769–1780 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alós E, Alvey E, Harberd NP, Wigge PA. (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13: 571–577 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW. (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19: 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Al-Sady B, Carle C, Storer A, Alonso JM, Ecker JR, Quail PH. (2008a) The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell 20: 337–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Monte E, Oka Y, Liu T, Carle C, Castillon A, Huq E, Quail PH. (2008b) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18: 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Cohn MM, Monte E, Al-Sady B, Erickson E, Quail PH. (2012) Dynamic antagonism between phytochromes and PIF family basic helix-loop-helix factors induces selective reciprocal responses to light and shade in a rapidly responsive transcriptional network in Arabidopsis. Plant Cell 24: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH. (2009) Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Siddiqui H, Teng Y, Lin R, Wan X-Y, Li J, Lau O-S, Ouyang X, Dai M, Wan J, et al. (2011) Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol 13: 616–622 [DOI] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee J-H, Zhang H, Shen Y, et al. (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22: 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung H-S, et al. (2012) Linking photoreceptor excitation to changes in plant architecture. Genes Dev 26: 785–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhong S, Grierson D. (2009) Recent advances in ethylene research. J Exp Bot 60: 3311–3336 [DOI] [PubMed] [Google Scholar]
- Liu Z-B, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ. (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C. (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Trevisan M, Pradervand S, Fankhauser C. (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de-etiolation in continuous far-red light. Plant J 60: 449–461 [DOI] [PubMed] [Google Scholar]
- Morelli G, Ruberti I. (2000) Shade avoidance responses: driving auxin along lateral routes. Plant Physiol 122: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno JE, Tao Y, Chory J, Ballaré CL. (2009) Ecological modulation of plant defense via phytochrome control of jasmonate sensitivity. Proc Natl Acad Sci USA 106: 4935–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford SG, Lee BR, Koprivova A, Matthewman C, Kopriva S. (2011) Control of sulfur partitioning between primary and secondary metabolism. Plant J 65: 96–105 [DOI] [PubMed] [Google Scholar]
- Nakamoto D, Ikeura A, Asami T, Yamamoto KT. (2006) Inhibition of brassinosteroid biosynthesis by either a dwarf4 mutation or a brassinosteroid biosynthesis inhibitor rescues defects in tropic responses of hypocotyls in the Arabidopsis mutant nonphototropic hypocotyl 4. Plant Physiol 141: 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2: E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen HB, Mundy J, Willenbrock H. (2007) Functional associations by response overlap (FARO), a functional genomics approach matching gene expression phenotypes. PLoS ONE 2: e676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN. (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- O’Connor TR, Dyreson C, Wyrick JJ. (2005) Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Ogawa M, Hanada A, Yamauchi Y, Kuwahara A, Kamiya Y, Yamaguchi S. (2003) Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW. (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Ouyang X, Li J, Li G, Li B, Chen B, Shen H, Huang X, Mo X, Wan X, Lin R, et al. (2011) Genome-wide binding site analysis of FAR-RED ELONGATED HYPOCOTYL3 reveals its novel function in Arabidopsis development. Plant Cell 23: 2514–2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11: 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Kim J, Lee Y, Shin J, Oh E, Chung WI, Liu JR, Choi G. (2004) Degradation of phytochrome interacting factor 3 in phytochrome-mediated light signaling. Plant Cell Physiol 45: 968–975 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierik R, Djakovic-Petrovic T, Keuskamp DH, de Wit M, Voesenek LA. (2009) Auxin and ethylene regulate elongation responses to neighbor proximity signals independent of gibberellin and della proteins in Arabidopsis. Plant Physiol 149: 1701–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés S, Más P. (2010) The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. (1994) Phytochrome A and phytochrome B have overlapping but distinct functions in Arabidopsis development. Plant Physiol 104: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagpal P, Poole DS, Furuya M, Chory J. (1993) Mutations in the gene for the red/far-red light receptor phytochrome B alter cell elongation and physiological responses throughout Arabidopsis development. Plant Cell 5: 147–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieu I, Powers SJ. (2009) Real-time quantitative RT-PCR: design, calculations, and statistics. Plant Cell 21: 1031–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, Harris S-R, Wasternack C, Brearley C, Turner JG. (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou J, Sorin C, Devlin PF, Martínez-García JF. (2006) Identification of primary target genes of phytochrome signaling: early transcriptional control during shade avoidance responses in Arabidopsis. Plant Physiol 141: 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF. (2007) Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 26: 4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruberti I, Sessa G, Ciolfi A, Possenti M, Carabelli M, Morelli G. (2012) Plant adaptation to dynamically changing environment: the shade avoidance response. Biotechnol Adv 30: 1047–1058 [DOI] [PubMed] [Google Scholar]
- Salter MG, Franklin KA, Whitelam GC. (2003) Gating of the rapid shade-avoidance response by the circadian clock in plants. Nature 426: 680–683 [DOI] [PubMed] [Google Scholar]
- Sassi M, Wang J, Ruberti I, Vernoux T, Xu J. (2013) Shedding light on auxin movement: light-regulation of polar auxin transport in the photocontrol of plant development. Plant Signal Behav 8: e23355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellaro R, Yanovsky MJ, Casal JJ. (2011) Repression of shade-avoidance reactions by sunfleck induction of HY5 expression in Arabidopsis. Plant J 68: 919–928 [DOI] [PubMed] [Google Scholar]
- Sessa G, Carabelli M, Sassi M, Ciolfi A, Possenti M, Mittempergher F, Becker J, Morelli G, Ruberti I. (2005) A dynamic balance between gene activation and repression regulates the shade avoidance response in Arabidopsis. Genes Dev 19: 2811–2815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Morelli G, Ruberti I. (1993) The Athb-1 and -2 HD-Zip domains homodimerize forming complexes of different DNA binding specificities. EMBO J 12: 3507–3517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa G, Morelli G, Ruberti I. (1997) DNA-binding specificity of the homeodomain-leucine zipper domain. J Mol Biol 274: 303–309 [DOI] [PubMed] [Google Scholar]
- Shen H, Moon J, Huq E. (2005) PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J 44: 1023–1035 [DOI] [PubMed] [Google Scholar]
- Shen Y, Khanna R, Carle CM, Quail PH. (2007) Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol 145: 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov IS, Bae G, Lee CH, Lee D, Choi G. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS. (2006) Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet 2: e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H, Whitelam GC. (1997) The shade avoidance syndrome: multiple responses mediated by multiple phytochromes. Plant Cell Environ 20: 840–844 [Google Scholar]
- Soh M-S, Kim Y-M, Han S-J, Song P-S. (2000) REP1, a basic helix-loop-helix protein, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12: 2061–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martínez-García JF. (2009) ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 59: 266–277 [DOI] [PubMed] [Google Scholar]
- Steindler C, Matteucci A, Sessa G, Weimar T, Ohgishi M, Aoyama T, Morelli G, Ruberti I. (1999) Shade avoidance responses are mediated by the ATHB-2 HD-zip protein, a negative regulator of gene expression. Development 126: 4235–4245 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. (2008) Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133: 164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT. (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepperman JM, Hudson ME, Khanna R, Zhu T, Chang SH, Wang X, Quail PH. (2004) Expression profiling of phyB mutant demonstrates substantial contribution of other phytochromes to red-light-regulated gene expression during seedling de-etiolation. Plant J 38: 725–739 [DOI] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR. (1995) Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol 46: 445–474 [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15: 1749–1770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchi L, Carabelli M, Ruzza V, Possenti M, Sassi M, Peñalosa A, Sessa G, Salvi S, Forte V, Morelli G, et al. (2013) Arabidopsis HD-Zip II transcription factors control apical embryo development and meristem function. Development 140: 2118–2129 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Liu Z-B, Hagen G, Guilfoyle TJ. (1995) Composite structure of auxin response elements. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang X, Roig-Villanova I, Khan S, Shanahan H, Quail PH, Martinez-Garcia JF, Devlin PF. (2011) A novel high-throughput in vivo molecular screen for shade avoidance mutants identifies a novel phyA mutation. J Exp Bot 62: 2973–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willems E, Leyns L, Vandesompele J. (2008) Standardization of real-time PCR gene expression data from independent biological replicates. Anal Biochem 379: 127–129 [DOI] [PubMed] [Google Scholar]
- Wollenberg AC, Strasser B, Cerdán PD, Amasino RM. (2008) Acceleration of flowering during shade avoidance in Arabidopsis alters the balance between FLOWERING LOCUS C-mediated repression and photoperiodic induction of flowering. Plant Physiol 148: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y, Sheen J. (2009) Emerging connections in the ethylene signaling network. Trends Plant Sci 14: 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, He H, Wang X, Wang X, Yang X, Li L, Deng XW. (2011) Genome-wide mapping of the HY5-mediated gene networks in Arabidopsis that involve both transcriptional and post-transcriptional regulation. Plant J 65: 346–358 [DOI] [PubMed] [Google Scholar]