Multiple herbicide resistance in waterhemp is due to distinct biochemical mechanisms for herbicide detoxification.
Abstract
Previous research reported the first case of resistance to mesotrione and other 4-hydroxyphenylpyruvate dioxygenase (HPPD) herbicides in a waterhemp (Amaranthus tuberculatus) population designated MCR (for McLean County mesotrione- and atrazine-resistant). Herein, experiments were conducted to determine if target site or nontarget site mechanisms confer mesotrione resistance in MCR. Additionally, the basis for atrazine resistance was investigated in MCR and an atrazine-resistant but mesotrione-sensitive population (ACR for Adams County mesotrione-sensitive but atrazine-resistant). A standard sensitive population (WCS for Wayne County herbicide-sensitive) was also used for comparison. Mesotrione resistance was not due to an alteration in HPPD sequence, HPPD expression, or reduced herbicide absorption. Metabolism studies using whole plants and excised leaves revealed that the time for 50% of absorbed mesotrione to degrade in MCR was significantly shorter than in ACR and WCS, which correlated with previous phenotypic responses to mesotrione and the quantity of the metabolite 4-hydroxy-mesotrione in excised leaves. The cytochrome P450 monooxygenase inhibitors malathion and tetcyclacis significantly reduced mesotrione metabolism in MCR and corn (Zea mays) excised leaves but not in ACR. Furthermore, malathion increased mesotrione activity in MCR seedlings in greenhouse studies. These results indicate that enhanced oxidative metabolism contributes significantly to mesotrione resistance in MCR. Sequence analysis of atrazine-resistant (MCR and ACR) and atrazine-sensitive (WCS) waterhemp populations detected no differences in the psbA gene. The times for 50% of absorbed atrazine to degrade in corn, MCR, and ACR leaves were shorter than in WCS, and a polar metabolite of atrazine was detected in corn, MCR, and ACR that cochromatographed with a synthetic atrazine-glutathione conjugate. Thus, elevated rates of metabolism via distinct detoxification mechanisms contribute to mesotrione and atrazine resistance within the MCR population.
Waterhemp (Amaranthus tuberculatus) is a troublesome annual weed species in midwestern U.S. corn (Zea mays) and soybean (Glycine max) production. The change to production systems with limited tillage has favored waterhemp germination and growth (Hager et al., 2002). Waterhemp seeds are small, and one female plant can produce up to one million seeds (Steckel et al., 2003), which endow waterhemp with an effective short-distance dispersal mechanism. In addition, multiple herbicide resistance mechanisms in waterhemp are facilitated by its dioecious biology and wind-pollinated flowers (Steckel, 2007). The long-distance flow of pollen may be one of the main reasons that multiple herbicide resistance in waterhemp has become widespread in the United States (Liu et al., 2012).
Mesotrione (2-[4-(methylsulfonyl)-2-nitrobenzoyl]-1,3-cyclohexanedione) belongs to the triketone class of 4-hydroxyphenylpyruvate dioxygenase (HPPD)-inhibiting herbicides (Beaudegnies et al., 2009). Molecular information regarding plant HPPD gene sequences and expression patterns is limited (for review, see Pallett, 2000; Kim and Petersen, 2002; Riechers and Stanford, 2002; Matringe et al., 2005), and only a single expressed HPPD gene was detected in waterhemp (Riggins et al., 2010). Herbicidal activity of mesotrione in sensitive plants is due to competitive inhibition of the HPPD enzyme, which is a key enzyme in the biosynthesis of tocopherols and plastoquinone. Plastoquinone is an electron acceptor for the phytoene desaturase reaction in the pathway of carotenoid biosynthesis and also serves as an electron acceptor in PSII (Hess, 2000). Tocopherols and carotenoids are responsible for the detoxification of reactive oxygen species and scavenging of free radicals in plant tissues (Maeda and DellaPenna, 2007; Triantaphylidès and Havaux, 2009; Mène-Saffrané and DellaPenna, 2010), and carotenoids also protect chlorophyll from photooxidation (Cazzonelli and Pogson, 2010). Following mesotrione treatment, carotenoid biosynthesis is inhibited in sensitive plants, resulting in bleaching and necrosis. In particular, new leaves and meristems are primarily affected due to the need for protective carotenoids and tocopherols in photosynthetic tissues (Triantaphylidès and Havaux, 2009) and the systemic nature of mesotrione, which is translocated in the phloem (Mitchell et al., 2001; Beaudegnies et al., 2009).
There are two main mechanisms of herbicide resistance in plants: (1) target site alterations, such as mutations that affect herbicide-binding kinetics or amplification of the target site gene (Powles and Yu, 2010), and (2) nontarget site mechanisms, including metabolism, translocation, and sequestration (Yuan et al., 2007; Powles and Yu, 2010). Metabolic detoxification is a common nontarget-based mechanism for herbicide resistance, which typically may result from elevated levels of cytochrome P450 monooxygenase (P450) or glutathione S-transferase (GST) activity (Powles and Yu, 2010; Délye et al., 2011). In addition to conferring resistance in weeds, these enzymes also confer natural tolerance in crops (Kreuz et al., 1996; Riechers et al., 2010). Similar to tolerant sorghum (Sorghum bicolor) lines (Abit and Al-Khatib, 2009), corn is tolerant to mesotrione via rapid metabolism (i.e. ring hydroxylation catalyzed by P450 activity) in combination with slower uptake relative to sensitive weeds and a less sensitive form of the HPPD enzyme in grasses relative to dicots (Hawkes et al., 2001; Mitchell et al., 2001).
Atrazine (2-chloro-4-(ethylamino)-6-(isopropylamino)-S-triazine) is a symmetrical triazine herbicide commonly used in corn to selectively control annual dicot weeds. Atrazine disrupts electron transport by competing with plastoquinone for the secondary electron-accepting plastoquinone-binding site on the D1 protein of PSII in chloroplasts (Hess, 2000). Atrazine resistance in weeds can be due to a mutation in the psbA gene that causes a Ser-Gly substitution at amino acid position 264 of the D1 protein (Hirschberg and McIntosh, 1983; Devine and Preston, 2000). Corn and grain sorghum are naturally tolerant to atrazine via the rapid metabolism of atrazine through conjugation with reduced glutathione (GSH; Frear and Swanson, 1970; Lamoureux et al., 1973), which is catalyzed by GST activities (Shimabukuro et al., 1971). Enhanced metabolism of atrazine and simazine in weedy species has been reported in Abutilon theophrasti, Lolium rigidum, and Alopecurus myosuroides due to either GST- or P450-mediated detoxification mechanisms (Burnet et al., 1993; Gray et al., 1996; Cummins et al., 1999; Délye et al., 2011).
A population of waterhemp (designated MCR for McLean County mesotrione- and atrazine-resistant) from Illinois is resistant to HPPD inhibitors (Hausman et al., 2011) and atrazine as well as to acetolactate synthase (ALS)-inhibiting herbicides. A different population of waterhemp (designated ACR for Adams County mesotrione-sensitive but atrazine-resistant; Patzoldt et al., 2005) that is atrazine resistant but sensitive to mesotrione (Hausman et al., 2011) and a waterhemp population (designated WCS for Wayne County herbicide-sensitive; Patzoldt et al., 2005) that is sensitive to both mesotrione and atrazine (Hausman et al., 2011) were used in comparison with MCR in this research. MCR displayed 10- and 35-fold resistance to mesotrione in comparison with ACR and WCS, respectively, in greenhouse studies (Hausman et al., 2011). In addition, waterhemp populations with similar patterns of multiple resistance have recently been identified (Hausman et al., 2011; McMullan and Green, 2011; Heap, 2012). However, the mechanisms of resistance to mesotrione and atrazine in these waterhemp populations are currently unknown. Therefore, the objective of this study was to determine if the multiple-herbicide-resistant phenotype of MCR (in regard to mesotrione and atrazine resistance) is due to either target site or nontarget site mechanisms.
RESULTS AND DISCUSSION
HPPD-resistant plants have been genetically engineered (Matringe et al., 2005; Dufourmantel et al., 2007), but evolved HPPD resistance in weeds had never been documented until 2009 (Heap, 2012). Interestingly, the recently discovered HPPD-resistant weed populations are also resistant to S-triazine herbicides. These findings are particularly significant because mesotrione and atrazine display synergistic foliar activity on atrazine-resistant weeds regardless of the triazine resistance mechanism (Hugie et al., 2008; Woodyard et al., 2009; Walsh et al., 2012), yet they did not completely control the MCR population in previous research (Hausman et al., 2011). Our results here indicate that enhanced herbicide detoxification by different enzymatic mechanisms contributes significantly to mesotrione and atrazine resistance within the MCR population.
HPPD Target Site Analyses
The MCR population displayed resistance to multiple HPPD-inhibiting herbicides (Hausman et al., 2011). An altered target site was hypothesized to confer resistance in MCR as one of several potential mechanisms. In accord with our previous transcriptome study (Riggins et al., 2010), a single expressed HPPD gene was detected in waterhemp. The WCS HPPD coding sequence, which is 1,305 bp in length, is deposited in GenBank as accession JX259255. The HPPD coding sequences from four MCR plants confirmed to be resistant to mesotrione, as well as from an ACR plant (mesotrione sensitive), were obtained and aligned to the WCS (mesotrione sensitive) coding sequence (Supplemental Fig. S1). Although nucleotide polymorphisms were identified among the sequences, a total of six amino acid polymorphisms were inferred, but none were associated with resistance. Among the six amino acid polymorphisms, one was between the two mesotrione-sensitive plants (WCS and ACR), two were present in only two of the four mesotrione-resistant MCR plants, and three were consistent across all four MCR plants but were also observed in the ACR plant (Supplemental Fig. S1). Therefore, it was concluded that mesotrione resistance in the MCR population was not due to an HPPD mutation conferring an insensitive target site.
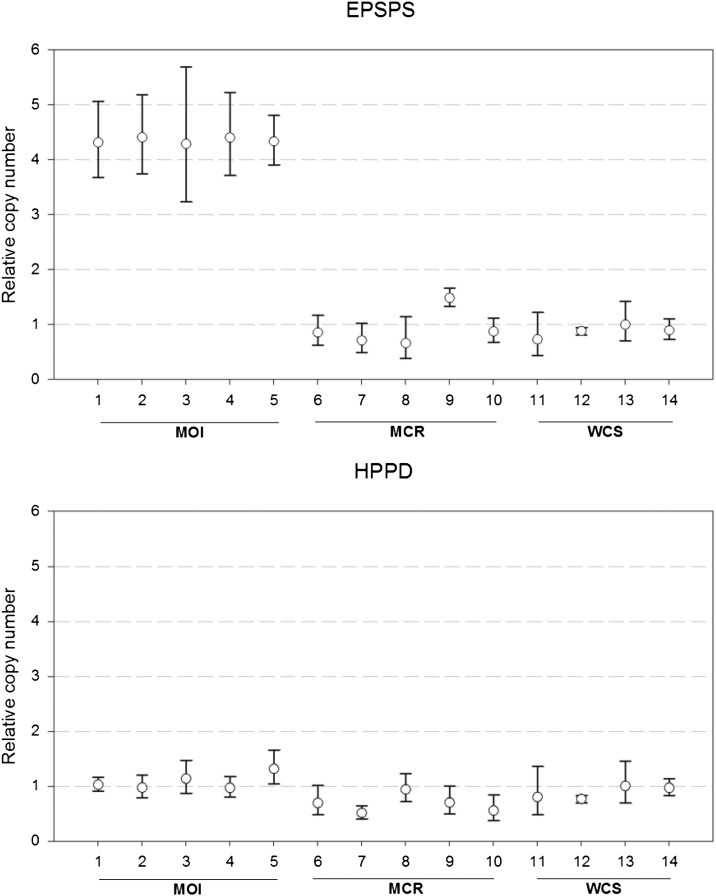
Gene amplification has recently been reported as a mechanism of herbicide resistance (Gaines et al., 2010); as a result, this mechanism was investigated in MCR. However, evidence for HPPD gene amplification was not detected in resistant and sensitive plants (Fig. 1). Additionally, preliminary results from quantitative reverse transcription-PCR experiments (data not shown) were consistent with genomic analyses and suggest that resistance is not due to a change in target site expression. Thus, it was concluded that the mechanism of mesotrione resistance in MCR is nontarget site based.
Figure 1.
Relative EPSP and HPPD gene amplification in Missouri glyphosate-resistant (MO1), MCR, and WCS waterhemp populations. The experiment measuring increased EPSP relative amplification in MO1 was included as a positive control to test the robustness of the procedure. MO1 individuals, which have demonstrated EPSP gene amplification (Tranel et al., 2011), were used to test the robustness of the qPCR assay. Relative gene copy number was determined by real-time qPCR using a modified protocol from a previous study (Gaines et al., 2010).
Mesotrione Uptake and Metabolism in Whole Plants
Decreased mesotrione uptake was initially investigated as a potential nontarget site resistance mechanism in MCR. Differences in uptake between HPPD-sensitive and -resistant populations were not time dependent (data not shown; P = 0.368). The mean uptake of applied radiolabeled mesotrione across all time points for MCR, ACR, and WCS was 84.0%, 83.8%, and 78.1%, respectively. The uptake of mesotrione was similar in MCR and ACR during the entire time course, which was significantly higher than in WCS (P < 0.0001). These minor differences in uptake may have resulted from genetic variation among populations. Alternatively, higher rates of detoxification may have resulted in a greater driving force for absorption from more rapid herbicide metabolism within underlying leaf tissues (Devine et al., 1993; Riechers et al., 1996). However, since the mesotrione-resistant MCR population had an equal or greater amount of herbicide uptake at all time points (relative to both sensitive populations), this indicates that differential uptake of mesotrione is not directly involved as a primary resistance mechanism.
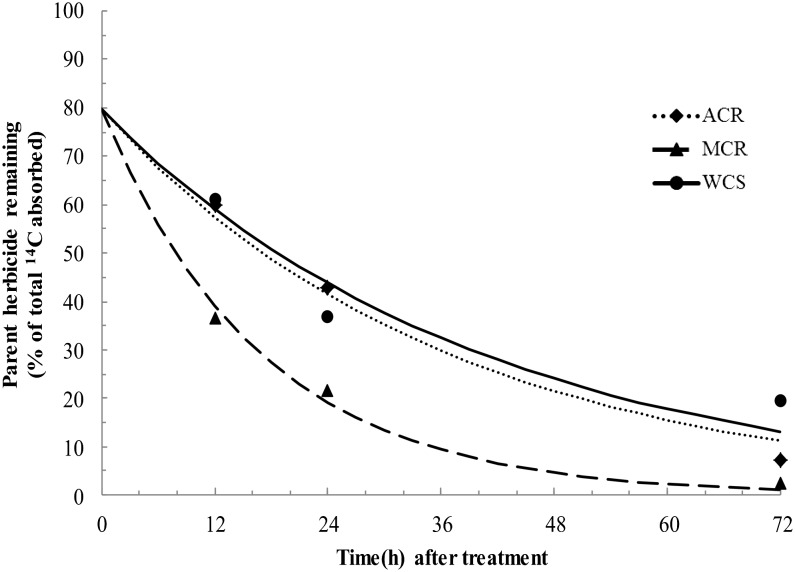
In order to determine if differences in metabolism existed among the three waterhemp populations, the amount of parent herbicide remaining (clearly resolved from polar metabolites as described below) as a percentage of total extractable radioactivity was quantified by HPLC. The amount of parent herbicide remaining was analyzed by nonlinear least-squares regression analysis and fit with a simple first-order curve in order to estimate the time for 50% of absorbed herbicide to degrade (DT50; Fig. 2). Based on the regression analyses, the DT50 values determined for MCR, ACR, and WCS were 11.7, 25.4, and 27.8 h, respectively. The significantly shorter DT50 for mesotrione in MCR is consistent with dose-response analyses reported previously for each population (Hausman et al., 2011) and phenotypes following mesotrione treatment in that all three populations of waterhemp display initial bleaching in new leaves and meristems, but the degree of bleaching in MCR is less severe and MCR plants recover more quickly compared with WCS and ACR. The results of the whole-plant metabolism study are thus consistent with the hypothesis that elevated rates of herbicide metabolism may contribute to mesotrione resistance in MCR.
Figure 2.
Time course of mesotrione metabolism in whole plants of MCR, ACR, and WCS waterhemp populations. All waterhemp plants (10–12 cm tall) were treated with unlabeled mesotrione (105 g ai ha−1) including 1% (v/v) COC and 2.5% (v/v) liquid ammonium sulfate as adjuvants, followed immediately by 701 Bq of [U-14C]mesotrione (with the same adjuvants) applied as 33- × 0.3-μL droplets to the upper surface of the fourth youngest leaf for analyses of metabolism. Data were analyzed by nonlinear least-squares regression analysis and fit with a simple first-order curve to estimate a DT50 separately for each population. The DT50 values determined for MCR, ACR, and WCS were 11.7 h (95% confidence interval of 9.3–14.1), 25.4 h (95% confidence interval of 19.2–31.7), and 27.8 h (95% confidence interval of 20.9–34.7), respectively.
Mesotrione Metabolism in Excised Leaves
In addition to analyzing mesotrione metabolism in waterhemp seedlings, a different experimental approach was utilized to determine rates of metabolism in excised leaves. This method has been used previously in the study of primisulfuron metabolism in corn leaves (Kreuz and Fonne-Pfister, 1992). An advantage of this technique is that it is independent of whole-plant translocation patterns of mesotrione (Mitchell et al., 2001). In addition to employing an excised leaf assay, a vegetative cloning strategy was utilized in this study. Due to the dioecious biology of waterhemp and the large degree of genetic diversity within Amaranthus spp. populations (Steckel, 2007), the use of vegetatively cloned plants ensured that identical waterhemp genotypes are analyzed within a time-course experiment and that accurate mesotrione DT50 values could be determined for each waterhemp population. Additionally, corn leaves (grown from seed) were used as a positive control in this study as an example of a mesotrione-tolerant crop (Hawkes et al., 2001).
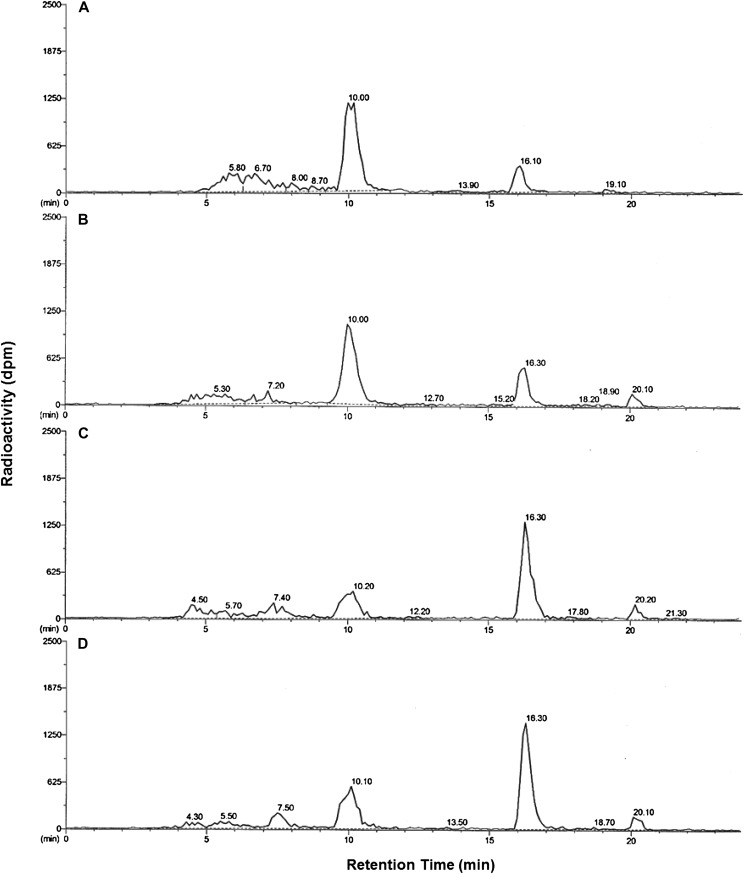
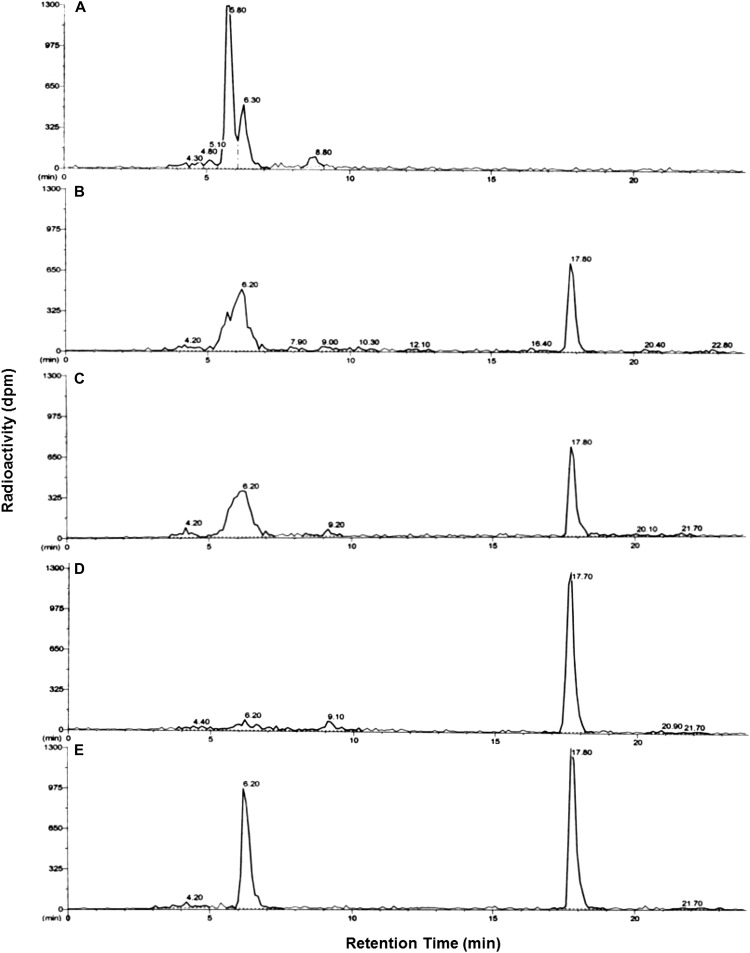
Parent mesotrione was clearly resolved from several polar metabolites under our conditions for reverse-phase HPLC (Fig. 3). The peak areas of parent mesotrione by HPLC (Fig. 3; retention time of about 16.3 min) were smaller in corn and MCR than in sensitive waterhemp populations (ACR and WCS) at 24 h after treatment (HAT). Additionally, a prominent peak with a retention time of about 10.1 min was more abundant in corn and MCR than in ACR and WCS (Fig. 3). The accumulation of polar metabolite(s) within this peak correlates with the reduction in parent compound in all samples. According to the HPLC results in Figure 3, unique polar metabolites were not detected in corn and MCR compared with ACR and WCS under our conditions. Thus, the rate of mesotrione metabolism and the formation of polar metabolites in MCR are quantitatively higher but not qualitatively different from those in ACR and WCS. An additional nonpolar metabolite (retention time of 20.1 min; Fig. 3) was detected in all three waterhemp populations but not in corn. Its chemical structure or route of enzymatic/nonenzymatic formation is not known; however, it is clearly not related to mesotrione resistance in MCR.
Figure 3.
Mesotrione-resistant waterhemp (MCR) and corn accumulated higher levels of polar metabolites than mesotrione-sensitive populations at 24 HAT. Representative reverse-phase HPLC results are depicted for plant samples (24 HAT) supplied with 150 μm [U-14C]mesotrione and extracted from corn (A) and MCR (B), ACR (C), and WCS (D) waterhemp excised leaves. Peak retention time around 10.1 min, 4-OH and possibly other hydroxylated forms of mesotrione; peak at retention time 16.3 min, mesotrione; peak at retention time 20.1 min (waterhemp only), unidentified metabolite.
The median DT50 values of mesotrione in corn and MCR in the excised leaf assay were 11.9 and 12.0 h, respectively (Table I). Thus, MCR is similar to corn in its metabolic capacity for mesotrione detoxification (Hawkes et al., 2001). By contrast, in mesotrione-sensitive ACR and WCS populations, median DT50 values of mesotrione were 29.8 h and greater than 45.2 h, respectively, which are not significantly different from each other (Table I). The significantly longer DT50 values of mesotrione in ACR and WCS, relative to MCR (and tolerant corn), correlate with the phenotypic responses to foliar-applied mesotrione in corn and in these waterhemp populations (Hausman et al., 2011) and to the data shown in Figure 3.
Table I. DT50 in excised leaves from vegetatively cloned MCR, ACR, and WCS waterhemp populations.
Excised leaves (third youngest leaf, 2–3 cm in length) from vegetatively cloned waterhemp (10–12 cm) or corn seedlings were placed in 0.1 m Tris-HCl buffer (pH 6.0) for 1 h, followed by 0.1 m Tris-HCl (pH 6.0) plus 150 μm [U-14C]mesotrione for 1 h, then one-quarter-strength Murashige and Skoog salts solution for 0, 5, 11, 23, or 35 h. DT50 values for waterhemp were estimated for each individual plant from a simple first-order degradation curve fit by nonlinear least squares with a common intercept parameter. A single DT50 for corn was estimated separately using the same model. Population medians were compared using the Wilcoxon rank-sum test through SAS Release 9.2. Pairwise comparisons are as follows: ACR versus MCR, P = 0.005; ACR versus WCS, P = 0.126; MCR versus WCS, P = 0.005. For comparison, the DT50 of mesotrione in excised corn leaves was 11.9 h.
Liquid chromatography-tandem mass spectrometry [LC-MS/MS] multiple reaction monitoring (MRM) was conducted to identify and quantify mesotrione and its known metabolites (Hawkes et al., 2001; Armel et al., 2005) using analytical standards. MCR6, which demonstrated the shortest DT50 (9.5 min) among the six MCR clonal lines (Table I), was utilized to identify mesotrione metabolites. Consistent with our previous metabolic data, the concentration of parent mesotrione (Mr 339.1) in MCR6 extracts (8.9 pmol mL−1) was significantly lower than in ACR (17.1 pmol mL−1) and WCS (23.6 pmol mL−1) extracts at 24 HAT (Table II). Additionally, the concentration of 4-hydroxy-mesotrione (4-OH; Mr 355.0) in MCR6 extracts was significantly higher than in ACR and WCS extracts (Table II). The accumulation of 4-OH, which was the main mesotrione metabolite identified in corn (Hawkes et al., 2001), is likely related to enhanced P450 activity in MCR and correlates with the concomitant reduction in parent mesotrione. Accordingly, the ratio of 4-OH to mesotrione in MCR6 was 2.1, which is significantly higher than in ACR (0.9) and WCS (0.6; Table II). The significantly lower level of mesotrione in MCR6 extracts (Table II) is in accord with the reductions of parent mesotrione quantified in Figure 2 (whole plants) and Table I (excised leaves) and as illustrated in Figure 3 (HPLC scan of excised leaves).
Table II. Concentrations of mesotrione and 4-OH in excised leaf extracts of mesotrione-resistant clonal line MCR6 and mesotrione-sensitive (ACR and WCS) waterhemp populations at 24 h following 150 µm mesotrione (1.86 mCi mmol−1) treatment.
The two reverse-phase LC-MS/MS (MRM) transitions utilized and peak retention time (RT) are listed per compound. The same quantities of extracted samples were compared by normalizing the amounts of U-14C-labeled compounds by LSS prior to MRM analysis. In addition, the same amount of prometryne (250 ng mL−1) was added to each sample as an internal standard to normalize peak areas obtained from each run. MRM data acquisition consisted of monitoring analytical standards and plant samples in the positive ion mode (M+H)+. The concentrations (pmol mL−1) of mesotrione and 4-OH were obtained according to standard curves (Supplemental Table S2) using Proc REG (SAS Release 9.2). Waterhemp populations were compared (P) to determine if the quantity of parent herbicide in MCR6 is lower, or if the 4-OH metabolite is higher, than in ACR or WCS utilizing Proc GLM one-sided contrasts (SAS Release 9.2). m/z, Mass-to-charge ratio.
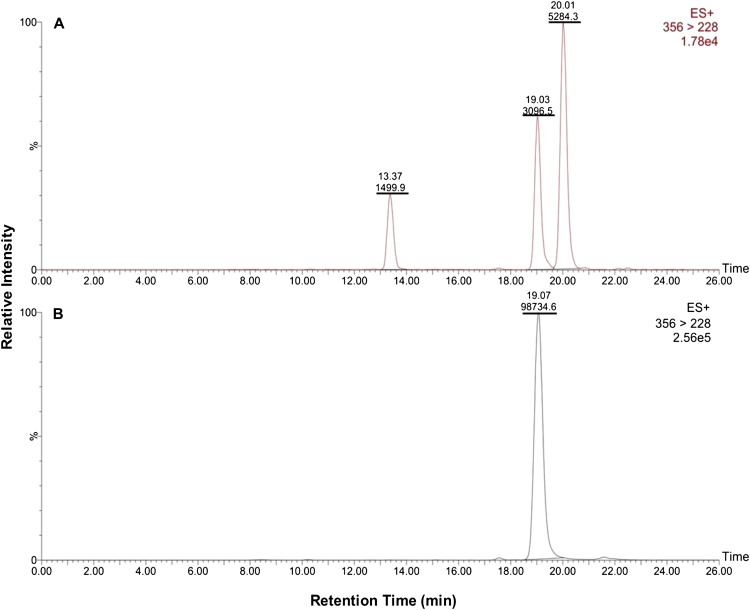
When comparing the MCR6 extract at 24 HAT with the analytical 4-OH standard via LC-MS/MS (MRM), one extracted compound had identical retention time (19.0 min) and Mr (355.0) as the standard. However, two additional peaks were identified with the same Mr but different retention times as the 4-OH standard (Fig. 4). These additional compounds were typically observed in ACR and WCS extracts as well (data not shown), but always at lower levels than in MCR6 extracts (in addition to 4-OH). The identities of these two additional compounds are not known, however, but could possibly represent structural isomers or stereoisomers of hydroxylated mesotrione (Urlacher, 2012).
Figure 4.
Evidence that mesotrione is metabolized to 4-OH in waterhemp clonal line MCR6. Representative LC-MS/MS (MRM) results are depicted for plant samples (24 HAT) supplied with 150 µm mesotrione and extracted (as described in “Materials and Methods”) from MCR6 (A) and the 4-OH analytical standard (Mr 355.0; B). MRM data acquisition consisted of monitoring the analytical standard and plant sample in positive mode (M+H)+ using the transition channel of parent ion (356) → fragment ion (228). Peaks at mass-to-charge ratio of (M+H)+ 356.0 identified from MCR extracts are displayed in A. Underlined numbers indicate peak areas, and their retention times are listed above the peak area. [See online article for color version of this figure.]
Additionally, a minor polar metabolite, 2-amino-4-(methyl-sulfonyl)benzoic acid (AMBA), was more abundant in MCR6 than in ACR and WCS extracts but was approximately 50-fold less abundant than 4-OH in MCR6 at 24 HAT (data not shown). AMBA formation was also detected in a previous mesotrione metabolism study in Cirsium arvense (Armel et al., 2005). Whether AMBA is an enzymatically formed catabolite of mesotrione and/or 4-OH or is nonenzymatically formed has not been determined.
Metabolism results from the excised leaf study using vegetatively cloned plants (Tables I and II; Fig. 3) and the whole-plant study (Fig. 2) demonstrated more rapid mesotrione metabolism in MCR than in ACR and WCS, although mesotrione metabolism in WCS in the whole-plant study was faster than expected (Fig. 2). This finding may be due to the different leaf stages used between studies (fourth leaf versus third leaf in the excised leaf study), pretreatment with unlabeled mesotrione in the whole-plant study, or the different types of experimental methods used to investigate mesotrione metabolism. However, in spite of the different rates of mesotrione metabolism in WCS between studies (Fig. 2; Table I), it is noteworthy that significant differences in mesotrione DT50 values between mesotrione-resistant (MCR) and mesotrione-sensitive (ACR and WCS) waterhemp populations were consistently observed in both studies (as well as in Fig. 3), which provide further evidence that increased metabolism is related to the mechanism of mesotrione resistance in MCR.
Influence of P450 Inhibitors on Mesotrione Metabolism in Excised Leaves
The metabolism of mesotrione in corn is due to a relatively rapid rate of P450-catalyzed ring hydroxylation, which was supported by increased corn sensitivity to mesotrione following addition of the P450 inhibitor malathion (Hawkes et al., 2001). Similarly, malathion inhibited metabolism of the ALS inhibitor primisulfuron in corn (Kreuz and Fonne-Pfister, 1992). Malathion (an organophosphate insecticide) acts as a “suicide substrate” for P450s via the release of reactive sulfur species during its metabolism that covalently bind to certain P450 enzymes (Correia and Ortiz de Montellano, 2005). As a result, potential herbicide substrates cannot be metabolized by these P450s, leading to crop injury (Kreuz and Fonne-Pfister, 1992; Barrett, 1997, 2000; Hawkes et al., 2001). Tetcyclacis is also a broad-range inhibitor of plant P450s (Leah et al., 1991). It belongs to the nitrogen-containing family of inhibitors that strongly bind to the prosthetic heme iron of P450 enzymes, which is related to sterol biosynthesis inhibition by certain fungicides (Werck-Reichhart et al., 1990).
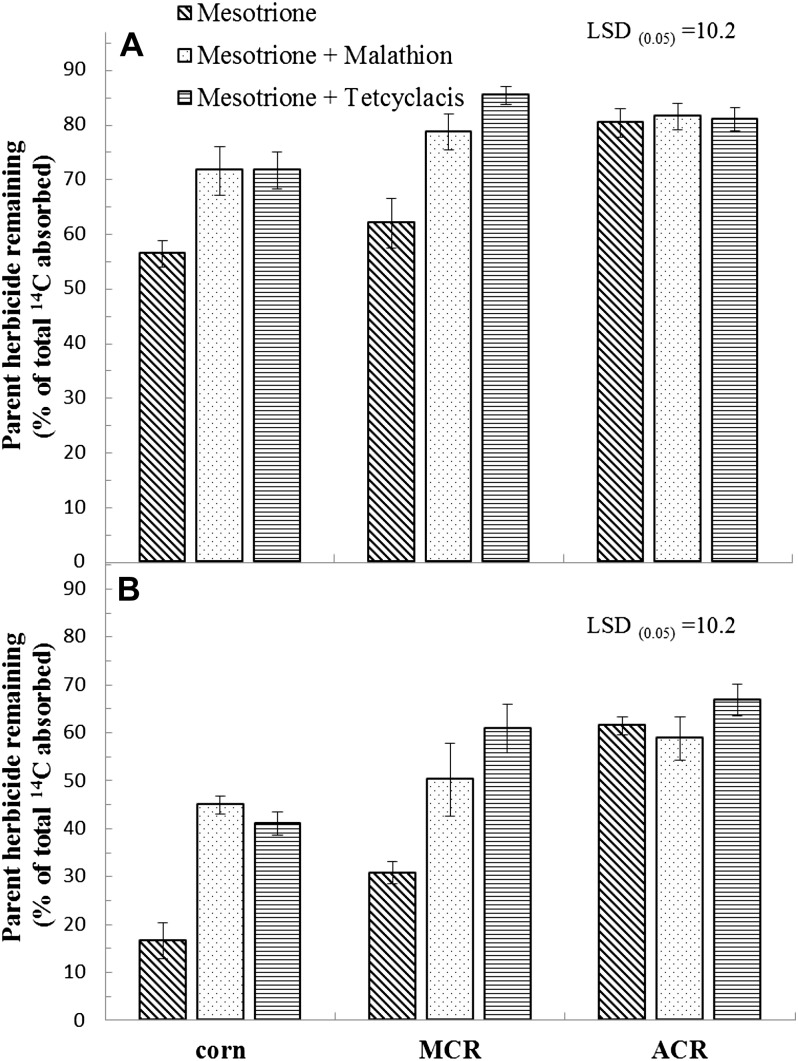
To determine if the biochemical basis for enhanced mesotrione metabolism in MCR is based on P450 activity, as has been shown previously in corn (Hawkes et al., 2001), each of these P450 inhibitors was applied in combination with mesotrione in a separate excised leaf study (Fig. 5). At 6 HAT, 72%, 72%, and 52% of parent mesotrione remained in corn samples treated with malathion, tetcyclacis, and the minus-P450 inhibitor control, respectively (Fig. 5A). At this same time point, 79% and 85% of parent mesotrione remained in the MCR leaves with malathion and tetcyclacis treatments, respectively, but only 62% of parent mesotrione remained in the absence of P450 inhibitor treatment (Fig. 5A). A decrease of parent mesotrione remaining in corn and waterhemp leaves was observed with malathion or tetcyclacis treatments at 24 HAT relative to 6 HAT (Fig. 5). For example, there was less nonmetabolized mesotrione remaining in the leaves of corn and MCR with P450 inhibitor treatments at 24 HAT compared with the same treatments at 6 HAT, which may indicate a partial metabolic degradation or inactivation of the P450 inhibitors or other metabolic activities that are not inhibited by malathion or tetcyclacis (Kreuz and Fonne-Pfister, 1992; Hidayat and Preston, 2001). Importantly, however, treatments with P450 inhibitors in corn and MCR at 24 HAT showed higher amounts of parent mesotrione compared with treatments without a P450 inhibitor (Fig. 5B).
Figure 5.
Influence of P450 inhibitors on mesotrione metabolism in excised leaves of corn and MCR and ACR waterhemp populations. Excised leaves (third youngest leaf; 2–3 cm in length) from vegetatively cloned waterhemp (10–12 cm) or corn seedlings were placed in 0.1 m Tris-HCl buffer (pH 6.0) with or without (control) the P450 inhibitors malathion or tetcyclacis (100 μm) for 2 h, followed by 0.1 m Tris-HCl (pH 6.0) plus 150 μm [U-14C]mesotrione with or without (control) 100 μm P450 inhibitor for 1 h, then one-quarter-strength Murashige and Skoog salts solution for 5 h (A) or 23 h (B). Mean comparisons were performed by lsd(P = 0.05) = 10.2, with error bars displaying se of treatment means.
By contrast, all treatments in ACR leaves were not significantly different in regard to parent mesotrione remaining, with or without malathion or tetcyclacis treatment, during the same time course (Fig. 5). This suggests a lower amount of P450-based metabolism of mesotrione in ACR relative to MCR, different P450 enzymes that are not inhibited, or other forms of mesotrione detoxification enzymes in ACR. Higher amounts of nonmetabolized mesotrione in corn and MCR leaves at each time point indicate that both malathion and tetcyclacis inhibited the metabolism of mesotrione, suggesting a similar detoxification mechanism in corn and MCR. In addition, the HPLC metabolite profile of MCR extracts was qualitatively similar to that in corn extracts (Fig. 3). These results confirmed that the specific P450(s) inhibited by malathion and tetcyclacis are also related to the metabolism of mesotrione in corn and MCR (Table I; Fig. 5). The overall number or specific P450(s) involved with mesotrione metabolism in MCR or other waterhemp populations are not yet known. However, cross resistance to other HPPD-inhibiting herbicides (Hausman et al., 2011) suggests that either a single P450 enzyme with broad substrate specificity (Nordby et al., 2008) or multiple P450s with low substrate specificity (Urlacher, 2012) may be involved with resistance in MCR, but these remain to be determined.
Increased Mesotrione Efficacy with Malathion in Waterhemp Seedlings
In addition to quantifying mesotrione metabolism with P450 inhibitors in excised leaves, a whole-plant study of MCR treated with malathion plus either mesotrione or atrazine was conducted in the greenhouse to verify the results from the previous excised leaf study (Fig. 5). The biomass of MCR seedlings treated with only malathion was not significantly different from the untreated controls (Table III). Since tolerant plants typically detoxify atrazine via GST-catalyzed GSH conjugation (Shimabukuro et al., 1971), it was hypothesized that malathion would not affect foliar atrazine activity in MCR. As expected, the biomass of MCR following treatment with malathion plus atrazine was not reduced significantly relative to atrazine alone (Table III). The biomass of MCR following treatment with malathion plus mesotrione was moderately reduced compared with mesotrione alone (Table III). These results are consistent with the excised leaf study (Fig. 5) and indicate that malathion increased mesotrione activity in MCR through the inhibition of P450s (but not atrazine; Table III), although the moderate level of biomass reduction suggests that other resistance mechanism(s) to mesotrione may also exist in MCR. These data implicate distinct detoxification mechanisms contributing to multiple herbicide resistance in the MCR population.
Table III. Foliar activity of malathion plus either mesotrione or atrazine applied to MCR waterhemp populations.
MCR waterhemp seedlings (10–12 cm tall) were treated with malathion at 2,000 g ai ha−1, including 0.25% (v/v) nonionic surfactant as an adjuvant, 1 h before a postemergence application of either mesotrione (105 g ai ha−1) with 1% (v/v) COC and 2.5% liquid ammonium sulfate (v/v) as adjuvants or atrazine (1,681 g ai ha−1) with 1% (v/v) COC as the adjuvant, followed by a soil drench of 5 mm malathion solution (50 mL pot−1). The comparison of the mixture of malathion plus each herbicide with the herbicide tested alone was analyzed by ANOVA using PROC MIXED in SAS Release 9.2.
| Treatment | Mean Biomass | Percentage of Control | Difference | P | Conclusion |
|---|---|---|---|---|---|
| g | g | ||||
| Control (experiments. 1, 2, and 3) | 3.28 | ||||
| Malathion | 2.83 | 86 | |||
| Mesotrione | 1.65 | 50 | |||
| Malathion + mesotrione | 1.16 | 35 | −0.49 | 0.118 | Moderate evidence of increased efficacy |
| Control (experiments 1 and 2) | 3.64 | ||||
| Malathion | 3.04 | 83 | |||
| Atrazine | 3.01 | 83 | |||
| Malathion + atrazine | 2.47 | 68 | −0.54 | 0.160 | No evidence of increased efficacy |
Atrazine Resistance in ACR and MCR
In previous research, the target site for atrazine (the psbA gene) encoding the D1 protein in PSII (Hess, 2000) was partially sequenced in an atrazine-resistant waterhemp population (SegR), but SegR did not contain the Ser-Gly-264 substitution that is commonly found in atrazine-resistant plants (Devine and Preston, 2000; Patzoldt et al., 2003). The waterhemp population ACR was subsequently identified with several similar characteristics to SegR and thus was predicted to possess a nontarget site mechanism for atrazine resistance (Patzoldt et al., 2005). Similarly, in this study, MCR did not possess an altered psbA sequence (data not shown). Since ACR showed a similar phenotype to MCR following atrazine treatment (Hausman et al., 2011), rates of atrazine metabolism were investigated in MCR, ACR, and atrazine-sensitive (WCS) waterhemp as well as in tolerant corn using the excised leaf assay described previously (Figs. 3–5; Tables I and II).
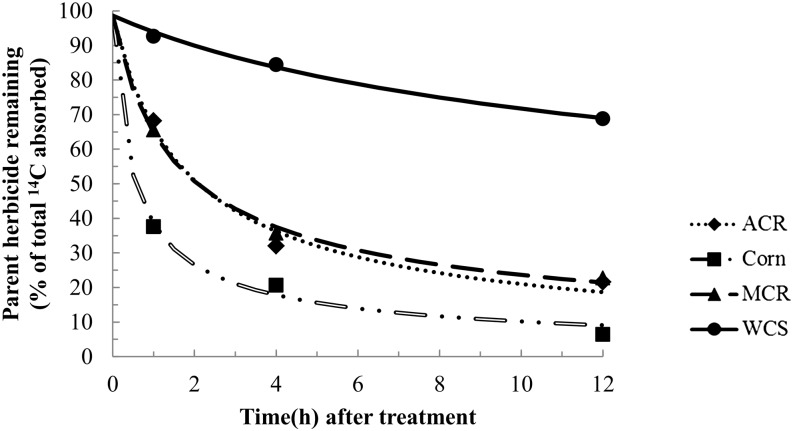
In the atrazine metabolism study, the amount of parent herbicide remaining was analyzed by nonlinear least-squares regression analysis and fit with a first-order multicompartment (FOMC) model (Gustafson and Holden, 1990) in order to estimate the time for 50% of absorbed atrazine to degrade (Fig. 6). The results showed a large difference in rates of atrazine metabolism between WCS and either MCR or ACR (Fig. 6). Furthermore, the DT50 values of atrazine determined by regression analysis were 0.6 h in corn, 2.2 h in MCR, 2.2 h in ACR, and greater than 12 h in WCS. The DT50 values of atrazine were significantly lower in MCR and ACR when compared with WCS, but the DT50 values of atrazine in MCR and ACR were significantly higher than in corn (Fig. 6).
Figure 6.
Time course of atrazine metabolism in excised leaves of corn and MCR, ACR, and WCS waterhemp populations. Excised leaves (third youngest leaf; 2–3 cm in length) from waterhemp (10–12 cm) or corn seedlings were placed in 0.1 m Tris-HCl buffer (pH 6.0) for 1 h, followed by 0.1 m Tris-HCl (pH 6.0) plus 150 μm [U-14C]atrazine for 1 h, then one-quarter-strength Murashige and Skoog salts solution for 0, 3, or 11 h. Data were analyzed by nonlinear least-squares regression analysis and fit with a FOMC model (Gustafson and Holden, 1990) to estimate a DT50 separately for each waterhemp population and corn. The DT50 values of atrazine determined by regression analysis were 0.6 h (95% confidence interval of 0.3–0.9) in corn, 2.2 h (95% confidence interval of 1.1–3.2) in MCR, 2.2 h (95% confidence interval of 1.1–3.2) in ACR, and greater than 12 h in WCS.
Rapid metabolism of atrazine in corn is due to high activity of GSTs that detoxify atrazine through carbon-chlorine bond displacement and subsequent formation of an atrazine-GSH conjugate (Frear and Swanson, 1970; Shimabukuro et al., 1971). To determine if the same GST-mediated detoxification mechanism exists in MCR and ACR, leading to rapid atrazine metabolism (Fig. 6) and atrazine resistance (Hausman et al., 2011), a synthetic standard of an atrazine-GSH conjugate was synthesized in vitro for comparative analysis of atrazine metabolism in corn and waterhemp by reverse-phase HPLC. The peak areas of atrazine in the HPLC scan (Fig. 7; retention time of about 17.8 min) were much smaller in MCR and ACR than in WCS at 4 HAT. This same peak (i.e. parent atrazine) was too small to detect in the corn sample at the same time point (Fig. 7). Importantly, a predominant polar metabolite with a retention time of about 6.2 min was extracted from excised MCR and ACR leaves and cochromatographed with the synthetic atrazine-GSH conjugate (Fig. 7).
Figure 7.
Atrazine-resistant waterhemp populations (MCR and ACR) and corn accumulated higher levels of polar metabolites than the atrazine-sensitive population (WCS) at 4 HAT. Representative reverse-phase HPLC results are depicted for plant samples (4 HAT) supplied with 150 μm [U-14C]atrazine and extracted from corn (A) and MCR (B), ACR (C), and WCS (D) waterhemp excised leaves and for the in vitro conjugation reaction of GSH with atrazine as an analytical standard (E). Peak retention time around 6.2 min, atrazine-GSH conjugate (E) as well as possible catabolite(s) of the atrazine-GSH conjugate (A–D); peak at retention time 17.8 min, atrazine.
Although several additional polar metabolites were extracted from corn, MCR, and ACR leaves at 4 HAT (Fig. 7), these compounds most likely represent catabolites of the atrazine-GSH conjugate, such as dipeptide and Cys conjugates of atrazine (Lamoureux et al., 1973). Catabolites of herbicide-GSH conjugates are typically formed rapidly in plants via the activities of vacuolar peptidases or other catabolic enzymes (Kreuz et al., 1996; Riechers et al., 1996, 2010). As a result, the rapid initial metabolism of atrazine in MCR and ACR is likely related to increased GST activity (relative to atrazine-sensitive WCS), although the number of GST isozymes involved in atrazine detoxification in resistant waterhemp populations is currently unknown. GST-mediated metabolism of atrazine in MCR is consistent with the greenhouse study in that the biomass following treatment with malathion plus atrazine was not reduced significantly relative to atrazine alone (Table III).
CONCLUSION
Previous greenhouse studies suggested that mesotrione plus atrazine might not provide acceptable control of the MCR population in the field (Hausman et al., 2011). Our findings here indicate that this may be due to enhanced metabolism of mesotrione via P450 activity and atrazine via GST activity in MCR. Increased rates of metabolism of both herbicides may also limit the potential for achieving synergistic activity of mesotrione and atrazine applied postemergence (Hugie et al., 2008; Woodyard et al., 2009; Walsh et al., 2012), which had previously been documented in an A. theophrasti biotype displaying atrazine resistance due to increased GST-catalyzed metabolism (Gray et al., 1996). Importantly, nontarget site mechanisms can confer plants with forms of multiple herbicide resistance that are more genetically complex, resulting in patterns of inheritance that are more difficult to determine (Délye et al., 2011). In addition to multiple resistance in MCR toward existing herbicides for weed management in corn, it is also of great concern that detoxification-based resistance mechanisms may have already been selected for herbicides that are not yet commercially available due to increased metabolism by P450 or GST activities. This scenario is particularly salient in regard to other cross-pollinating weed species, such as Lolium spp. and A. myosuroides, which also possess biotypes with distinct nontarget site mechanisms for herbicide resistance (Burnet et al., 1993; Délye et al., 2011).
While different detoxification mechanisms appear to be a contributing factor for mesotrione and atrazine resistance in the MCR population, other distinct nontarget site resistance mechanisms for mesotrione (such as altered translocation patterns or subcellular sequestration) cannot be discounted at this stage and will be investigated in future research. This will require the use of HPPD inhibitors that are metabolically blocked on the cyclohexane and/or phenyl ring (Beaudegnies et al., 2009). For example, less-metabolized HPPD inhibitors in weeds of the genus Amaranthus will be used in whole-plant studies to confirm whether the mechanism of mesotrione resistance in MCR is solely due to enhanced herbicide metabolism, and the possible induction of metabolism by mesotrione pretreatment in MCR will also be investigated. Genetics and inheritance studies are currently being conducted to determine the number of resistance genes or alleles within the MCR population. Whole-plant translocation studies will determine if MCR sequesters mesotrione from moving to apical and axillary meristems, since increased metabolism to polar metabolites may also affect whole-plant translocation patterns of radiolabeled mesotrione or its metabolites (Devine et al., 1993; Mitchell et al., 2001). The working hypothesis that enhanced oxidative metabolism accounts for resistance to mesotrione in MCR will continue to direct our future mechanistic work toward a more comprehensive understanding of multiple herbicide resistance within the MCR population.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Three waterhemp (Amaranthus tuberculatus) populations (mesotrione and atrazine resistant [MCR], mesotrione sensitive but atrazine resistant [ACR], and mesotrione and atrazine sensitive [WCS]) were investigated in this research (Patzoldt et al., 2005; Hausman et al., 2011). Seeds were collected and suspended in 0.1 g L−1 agar-water solution at 4°C for at least 30 d to enhance germination. Seeds of all three waterhemp populations were germinated in 12- × 12-cm trays with a commercial potting medium (Sun Gro Horticulture) in the greenhouse. Emerged seedlings (2 cm tall) were then transplanted into 80-cm3 pots in the greenhouse. When the seedlings were 4 cm tall, they were transplanted into 950-cm3 pots containing a 3:1:1:1 mixture of potting mix:soil:peat:sand. Slow-release fertilizer (Nutricote; Agrivert) was added to this mixture. Corn (Zea mays) seeds (hybrid DKC 63-14 RR) were planted 2 cm deep in the same soil mixture. When plants were 10 to 12 cm tall, they were transferred to a growth chamber and utilized for herbicide absorption and whole-plant metabolism studies. Greenhouse and growth chamber (Controlled Environments) conditions were maintained at 28°C/22°C day/night with a 16/8-h photoperiod. Natural sunlight was supplemented with mercury halide lamps, providing a minimum of 500 µmol m−2 s−1 photon flux at plant canopy level in the greenhouse. Light in the growth chamber was provided by incandescent and fluorescent bulbs delivering 550 µmol m−2 s−1 photon flux at plant canopy level.
HPPD and psbA Gene Identification
A nearly full-length HPPD sequence was first identified from the waterhemp transcriptome data set (Riggins et al., 2010). However, the 5′ end was missing from this data set, so a full-length wild-type sequence from a mesotrione-sensitive waterhemp individual was obtained by creating and screening a complementary DNA library using the Creator SMART cDNA library construction kit from Clontech. Subsequently, forward and reverse primers (Supplemental Table S1) encompassing the entire HPPD coding region were designed to amplify the HPPD gene in a number of sensitive and mesotrione-resistant plants. Primers for obtaining the full-length chloroplastic psbA gene in waterhemp were designed from shotgun genomic sequence data (Lee et al., 2009) and are listed in Supplemental Table S1. PCR protocols for amplifying and sequencing both genes are the same as provided by Riggins et al. (2010).
HPPD Gene Amplification
Genomic DNA was extracted from fresh leaf tissue using the modified cetyl-trimethyl-ammonium bromide method (Doyle and Doyle, 1990). Multiple plants from three waterhemp populations were analyzed: MCR, WCS, and MO1 (a glyphosate-resistant population from Missouri). MO1 individuals were included to test the robustness of the quantitative PCR (qPCR) assay, since this population has demonstrated EPSP gene amplification. Relative HPPD gene amplification was determined by real-time qPCR using a modified protocol from a previous study (Gaines et al., 2010). Quantitations of HPPD and EPSP were normalized to the endogenous single-copy control gene CPS, which encodes the large subunit of carbamoylphosphate synthetase (EC 6.3.5.5). To our knowledge, CPS is not associated with herbicide resistance and was selected as an alternative reference gene to the ALS gene previously used by Gaines et al. (2010), which confers resistance to a variety of ALS-inhibiting herbicides. CPS was inferred to be a single-copy gene based on BLAST searches using the waterhemp transcriptome (Riggins et al., 2010) against single-copy orthologous gene databases (Wu et al., 2006). In addition, qPCR experiments (data not shown) comparing the relative copy number of the CPS gene with the single-copy waterhemp ALS gene demonstrated copy number stability in both mesotrione-resistant and mesotrione-susceptible individuals. Each biological sample was analyzed in triplicate using the ABI Prism 7900 Detection System. Primers were anchored in continuous exons for each gene and were as follows: CPS (forward, 5′-ATTGATGCTGCCGAGGATAG-3′, and reverse, 5′-GATGCCTCCCTTAGGTTGTTC-3′); HPPD (forward, 5′-CTGTCGAAGTAGAAGACGCAG-3′, and reverse, 5′-TACATACCGAAGCACAACATCC-3′); and EPSP (forward, 5′-GGTTGTGGTGGTCTGTTTCC-3′, and reverse, 5′-CATCGCTGTTCCTGCATTTC-3′). The following parameters were used for qPCR: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 30 s and 60°C for 1 min.
Estimations of gene amplification were made using the comparative cycle threshold (Ct; 2-ΔΔCt) method (Pfaffl, 2001). Relative standard curves for each gene were generated from a 5× dilution series (100, 20, 4, 0.8, and 0.16 ng) using DNA from WCS. Amplification efficiencies were determined for each gene using the equation:
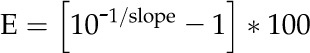 |
(1) |
where the slope is obtained from the linear regression of Ct values plotted against template concentration. Standard curve plots had high correlation coefficients (r2 ≥ 0.99) and slopes within the acceptable range of −3.6 and −3.1 (90%–110% efficiency). Average Ct values for each biological sample (run in triplicate) were calculated with the formula:
| (2) |
Using data from the standard curves, a validation experiment was performed by plotting ΔCt values against the log-transformed concentration of genomic DNA. The absolute value of the slope was 0.1 or less, which demonstrates equal PCR efficiencies of the three genes (Livak and Schmittgen, 2001). The ΔΔCt value was calculated by subtracting the ΔCt of the herbicide-sensitive population WCS (i.e. the calibrator sample) from the ΔCt of each biological sample. The fold difference and range in HPPD or EPSP amplification relative to WCS were calculated by 2-ΔΔCt with ΔΔCt ± sd, where sd is the sd of the ΔΔCt value. Two different experimental runs including all samples were performed to verify the results.
Mesotrione Uptake in Whole Plants
Seedlings of mesotrione-resistant (MCR) and mesotrione-sensitive (ACR and WCS) populations of waterhemp (10–12 cm tall) were moved from the greenhouse to the growth chamber 1 d before herbicide treatment to allow plants to acclimate. All waterhemp plants were treated with mesotrione at a rate of 105 g active ingredient (ai) ha−1, including 1% (v/v) crop oil concentrate (COC; Herbimax; Loveland Products) and 2.5% (v/v) liquid ammonium sulfate (AMS Liquid N-PaK; Agriliance) as adjuvants. Herbicide treatments were applied using a compressed air research sprayer (DeVries Manufacturing) equipped with a TeeJet 80015 EVS nozzle (Spraying Systems) calibrated to deliver 185 L ha−1 at 275 kPa. After 30 min, plants were returned to the growth chamber and 701 Bq of [U-14C]mesotrione (8 μm final concentration; specific activity of 19.7 mCi mmol−1) including the same adjuvants as before was applied as 33- × 0.3-μL droplets to the top surface of the fourth youngest leaf, which was marked with a black dot for clear recognition when harvesting. At 4, 12, 24, 72, and 120 HAT, the treated leaves were washed with 20% methanol for 30 s and total radioactivity absorbed in each leaf was quantified with a biological oxidizer (RJ Harvey Instrument) and liquid scintillation spectrometry (LSS; Packard Instrument). To determine the average recovery and calculate mass balance for uptake studies, radioactivity from all aboveground plant tissues (14CO2 trapped during plant tissue oxidation) plus [U-14C]mesotrione recovered from leaf washes was expressed as a percentage (98%) of the total radiolabeled herbicide applied to the treated leaf in all treatments and samples.
Mesotrione Metabolism in Whole Plants
Similar to the herbicide uptake study, all three populations of waterhemp were moved to the growth chamber 1 d before herbicide application and sprayed with unlabeled mesotrione (105 g ai ha−1 plus adjuvants) followed by 701 Bq of [U-14C]mesotrione (8 μm plus adjuvants) applied 30 min later as 33- × 0.3-μL droplets to the top surface of the fourthyoungest leaf. At 12, 24, and 72 HAT, each treated leaf was harvested, washed with 20% methanol for 30 s, and ground in liquid nitrogen. [U-14C]Mesotrione and its metabolites were extracted with 14 mL of 90% acetone at −4°C for 16 h. Following 90% acetone extraction of treated leaves, extractable radioactivity was approximately 99% of radiolabeled compounds that remained in the treated leaf. Nonextractable radioactivity (bound residue) in treated leaves slightly increased in each population during the study but averaged only 0.3% across all time points. Samples were centrifuged at 5,000g for 10 min, and supernatants were concentrated at 40°C until a final volume of 0.5 mL was reached with a rotary evaporator. Acetonitrile:water (50:50, v/v) was added to adjust the final volume of the extracts to 1.25 mL, and extracts were centrifuged at 10,000g for 10 min. Total radioactivity in each sample was measured by LSS. The same quantities of extracted samples (8,000 dpm) were compared by normalizing amounts of U-14C-labeled compounds by LSS prior to analysis. Total extractable radioactivity was resolved into parent mesotrione and its polar metabolites by reverse-phase HPLC (Hewlett-Packard) as described below. Parent [U-14C]mesotrione remaining in each sample as a percentage of total radioactivity detected by the HPLC Flow Scintillation Analyzer was recorded to determine the rates of mesotrione metabolism in each waterhemp population.
Reverse-phase HPLC was performed with a Hypersil Gold C18 column (4.6 × 250 mm, 5-µm particle size; Thermo Scientific) at a flow rate of 1 mL min−1. Eluent A was 0.1% (v/v) formic acid in water and eluent B was acetonitrile. The elution profile was as follows: step 1, A:B (4:1, v/v) to A:B (3:2, v/v) linear gradient for 12 min; step 2, A:B (3:2, v/v) to A:B (3:7, v/v) linear gradient for 5 min; step 3, A:B (3:7, v/v) to A:B (1:9, v/v) linear gradient for 2 min (19 min total). This gradient was immediately followed by A:B (1:9, v/v) to A:B (4:1, v/v) in a linear gradient for 3 min and A:B (4:1, v/v) isocratic hold for 2 min to reequilibrate the column before injecting the next sample. [U-14C]Mesotrione displayed a retention time of 16.3 min. Radiolabeled compounds were detected with a 500 TR Series Flow Scintillation Analyzer (Packard) and Ultima-Flo M cocktail (Perkin-Elmer). For whole-plant mesotrione metabolism studies, the average amount of radioactivity extracted from the treated leaf plus nonextractable radioactivity combined with radioactivity recovered from leaf washes was 95% of radiolabeled compounds remaining in the treated leaf or on the leaf surface at each time point.
Vegetative Cloning of Waterhemp Seedlings for Mesotrione DT50 Analysis
Due to the large amount of genetic variability within waterhemp populations (Steckel, 2007), a vegetative cloning procedure was devised for metabolism studies using intact leaves. Five axillary shoots (3 cm in length) were excised from all three waterhemp populations. Most of the leaves on these shoots were removed to decrease water evaporation upon further handling, but the two youngest leaves were left. The shoots were transplanted into 80-cm3 pots (one seedling per pot) with commercial potting medium and were placed in the growth chamber for 7 d to establish roots. When the cloned plants were 4 cm tall, they were transplanted into 950-cm3 pots containing a 3:1:1:1 mixture of potting mix:soil:peat:sand including slow-release fertilizer and were moved to the greenhouse. When these seedlings again displayed 3-cm axillary shoots, the plants were cloned for a second time using the same method. Six independent parental lines of waterhemp were cloned for each population of waterhemp and were recorded as lines M1 to M6, A1 to A6, and W1 to W6.
When cloned waterhemp plants were 10 to 12 cm tall, the third youngest leaves (2–3 cm in length) were excised for mesotrione metabolism and DT50 analysis. Leaves from distinct lines of vegetatively cloned waterhemp plants (as well as corn seedlings) were then excised a second time under water, and cut ends were placed into 1.5-mL plastic vials (one leaf per vial; based on methods described by Kreuz and Fonne-Pfister, 1992) containing 200 µL of 0.1 m Tris-HCl buffer (pH 6.0) for 1 h, then 200 µL of 0.1 m Tris-HCl (pH 6.0) with 150 μm [U-14C]mesotrione for 1 h, then washed with deionized water and transferred to a one-quarter-strength Murashige and Skoog salts solution (500 µL) for 0, 5, 11, 23, or 35 h in the growth chamber as a time-course study. Mean absorption of mesotrione was approximately 9% of the supplied radioactivity during the 1-h incubation period. [U-14C]Mesotrione and its metabolites were extracted, resolved, and detected using the same methods and chromatographic conditions as described previously with the mesotrione metabolism study in whole plants.
LC-MS/MS (MRM) Analysis of Mesotrione Metabolism in Excised Leaves
For LC-MS/MS (MRM) analysis, excised leaves from clonal line MCR6, ACR, and WCS waterhemp plants were collected at 24 HAT and extracted as described above for excised leaf studies (Fig. 3; Table I), except that 150 µm mesotrione with a lower specific activity (1.86 mCi mmol−1) was utilized. MCR6, which demonstrated the shortest DT50 (9.5 min) among the six MCR clonal lines (Table I), was utilized to identify mesotrione metabolites (Table II; Fig. 4). To increase the concentration of mesotrione and its metabolites in samples for LC-MS/MS (MRM) analysis, 16 leaf samples derived from different plants from each waterhemp population were pooled and concentrated. Mesotrione and its metabolites in each plant extract were normalized by quantifying the amount of U-14C-labeled compounds via LSS prior to MRM analysis. Analytical standards of mesotrione, AMBA, and 4-OH (provided by Syngenta) were analyzed by LC-MS/MS (MRM) to establish standard curves (Supplemental Table S2) with concentrations of 1, 5, 10, 25, and 100 ng mL−1.
LC-MS/MS (MRM) was employed for the quantitation of 4-OH and mesotrione parent ions using their corresponding fragmented ions, with the assistance of an internal standard (prometryne, a nonpolar triazine herbicide), at 250 ng mL−1 per sample. The LC-MS/MS system consisted of an analytical HPLC separation module (Waters Alliance 2795) coupled with an electrospray ionization mass spectrometer (Waters QuattroUltima). Samples were analyzed using a reverse-phase Hypersil Gold C18 column (4.6 × 250 mm, 5-µm particle size; Thermo Scientific). The mobile phase was composed of two solutions: eluent A was 0.1% (v/v) formic acid in water and eluent B was acetonitrile with 0.1% (v/v) formic acid. The elution profile was as follows: step 1, A:B (4:1, v/v) to A:B (3:2, v/v) linear gradient for 24 min; step 2, A:B (3:2, v/v) to A:B (3:7, v/v) linear gradient for 10 min; step 3, A:B (3:7, v/v) to A:B (1:9, v/v) linear gradient for 6 min. This gradient was immediately followed by A:B (1:9, v/v) to A:B (4:1, v/v) in a linear gradient for 6 min and A:B (4:1, v/v) isocratic hold for 40 min to reequilibrate the column before injecting the next sample. The longer gradient used for LC-MS/MS (MRM) analysis increased the retention times of 4-OH (19.0 min) and parent mesotrione (33.2 min) relative to previous reverse-phase HPLC methods (Fig. 3).
MRM data acquisition consisted of monitoring the following analytes in the positive ion mode (M+H)+ using the following transitions (parent ion → fragment ion, cone voltage, collision voltage): AMBA (215.9 → 136.0, 20 eV, 20 eV and 215.9 → 198.0, 20 eV, 10 eV); 4-OH (356.0 → 55.2, 35 eV, 25 eV and 356.0 → 228.0, 30 eV, 17 eV); mesotrione (340.1 → 104.0, 30 eV, 30 eV and 340.1 → 228.0, 35 eV, 15 eV); and the internal standard prometryne (242.0 → 158.0, 30 eV, 20 eV and 242.0 → 200.0, 30 eV, 20 eV), all with 100-ms dwell time. Quantitation of each analyte was determined using the peak areas normalized to the internal standard (prometryne at 250 ng mL−1) and a previously calculated standard curve. Analytical data were processed using Waters Mass Lynx software (version 4.1).
Influence of P450 Inhibitors on Mesotrione Metabolism in Excised Leaves
To determine the effect of P450 inhibitors on mesotrione metabolism, excised leaves from MCR and ACR cloned waterhemp lines and corn were used in a second metabolism study. The P450 inhibitors malathion or tetcyclacis (100 μm) were added to 200 µL of 0.1 m Tris-HCl buffer (pH 6.0) and supplied to excised leaves for 2 h, followed by 200 µL of 0.1 m Tris-HCl (pH 6.0) with 150 μm [U-14C]mesotrione and 100 μm P450 inhibitor for 1 h, then 500 µL of Murashige and Skoog salts solution as described previously for 5 or 23 h in the growth chamber. [U-14C]Mesotrione and its metabolites were extracted, resolved, and detected using the same methods and chromatographic conditions as described previously for the mesotrione metabolism study in whole plants (Fig. 2).
Malathion-Herbicide Studies in the Greenhouse
To investigate the activity of either mesotrione or atrazine plus malathion, MCR seedlings (10–12 cm tall) were treated with malathion at a rate of 2,000 g ai ha−1, including 0.25% nonionic surfactant (Agriliance) as an adjuvant, at 1 h before a foliar application of mesotrione or atrazine, followed by a soil drench of 5 mm malathion solution (50 mL pot−1) at 2 d after herbicide treatment. Mesotrione treatments at 105 g ai ha−1 included 1% (v/v) COC (Herbimax) and 2.5% (v/v) liquid ammonium sulfate (AMS Liquid N-PaK; Agriliance) as adjuvants. This mesotrione rate is known to reduce MCR dry biomass by approximately 60% (Hausman et al., 2011) and allowed for the detection of significant reductions in biomass relative to mesotrione alone. The rate of atrazine was 1,681 g ai ha−1 (plus 1% [v/v] COC as an adjuvant), which can discriminate among the three waterhemp populations under greenhouse conditions. Herbicides were applied individually or in combination with malathion by using spray methods similar to those described previously in the mesotrione uptake and whole-plant metabolism studies. All aboveground plant tissue was harvested at 17 d after herbicide treatment and dried at 65°C for 7 d. The dry weight of all plants was recorded and converted to a percentage of the untreated control.
Atrazine Metabolism in Excised Leaves of Waterhemp and Corn
Excised leaves from atrazine-resistant (MCR and ACR) or atrazine-sensitive (WCS) waterhemp and corn seedlings were supplied with [U-14C]atrazine (150 μm; specific activity of 16.5 mCi mmol−1) to determine the rates of metabolism using methods similar to those described previously in the mesotrione metabolism study with excised leaves, although a different time course (1, 4, and 12 HAT) was examined. Mean absorption of atrazine was approximately 11% of the supplied radioactivity during the 1-h incubation period. As a standard for HPLC analysis of polar metabolites, a synthetic glutathione (GSH) conjugate of atrazine was generated in vitro using a base-catalyzed method described previously for preparing a dimethenamid-GSH conjugate (Riechers et al., 1996). Briefly, the protocol consisted of incubating 0.1 mm [U-14C]atrazine with 20 mm GSH in 60 mm 3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid (TAPS) buffer (adjusted to a final pH of 9.5 with NaOH; final reaction volume was 300 μL) at 35°C for 24 h. As a negative control, 0.1 mm [U-14C]atrazine incubated at 35°C for 24 h in 60 mm TAPS buffer (adjusted to a final pH of 9.5 with NaOH) without sulfhydryl showed no conjugation product or alteration in parent atrazine. [U-14C]Atrazine (17.8 min) and the [U-14C]atrazine-GSH conjugate (6.2 min) were used as standards for cochromatography in HPLC analysis and the determination of atrazine metabolism in waterhemp and corn leaves. HPLC conditions were identical to those described previously for mesotrione metabolism in whole plants and excised leaves.
Statistical Procedures
Treatments were arranged in a completely randomized design (unless otherwise noted), and data from each independent experiment were combined and analyzed using the statistical procedures described below. For experiments involving mesotrione absorption in whole plants, mesotrione metabolism in excised leaves, and atrazine metabolism in excised leaves, two identical independent experiments were carried out, with each treatment comprising three biological replicates. For the experiment investigating the effect of P450 inhibitors on mesotrione metabolism in excised leaves, the two independent experiments were composed of three replicates in the first and two in the second. For the malathion-herbicide study in the greenhouse, three independent experiments were carried out with three, four, and five replicates, respectively. For experiments regarding mesotrione absorption in whole plants and the effect of P450 inhibitors on mesotrione metabolism in excised leaves, the data were analyzed by ANOVA and individual treatments were compared using Fisher’s lsd(P = 0.05). For the experiments of mesotrione metabolism in whole plants and mesotrione metabolism in excised leaves from cloned plants, the data were analyzed by nonlinear least-squares regression analysis and fit with a simple first-order curve in order to estimate DT50 values. The model was described by the equation:
| (3) |
For the analysis of atrazine metabolism in excised leaves, the FOMC model (Gustafson and Holden, 1990) described the data more accurately. The model was described by the equation:
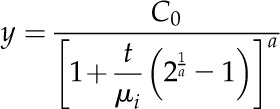 |
(4) |
where, in both models, y represents the percentage of the parent herbicide remaining at time t, µi is the DT50 for each biotype i, and the parameter C0 is the estimated amount of parent herbicide present at t = 0. In the FOMC model, a represents a shape parameter. In the case of cloned plants, DT50 values were estimated separately for lines derived from each parent waterhemp plant, and biotypes were compared using the nonparametric Wilcoxon rank-sum test. Greenhouse biomass reduction data involving malathion plus mesotrione or atrazine were analyzed by ANOVA using PROC MIXED to compare the mixture with the herbicide tested alone. All statistical analyses were conducted in SAS Release 9.2.
For the LC-MS/MS (MRM) analyses, four independent experiments were conducted with one replicate per treatment arranged in a randomized complete block design. Each waterhemp population sample (treatment) consisted of a pooled extract of 16 leaf samples derived from different plants for mass spectrometry analyses. Concentrations (pmol mL−1) of mesotrione and 4-OH were obtained according to standard curves using Proc REG (SAS Release 9.2) and used to calculate 4-OH:mesotrione for each population. Waterhemp populations were compared to determine if the quantity of mesotrione in MCR is lower, if 4-OH is higher, and if 4-OH:mesotrione is higher than in ACR or WCS by utilizing Proc GLM one-sided contrasts (SAS Release 9.2).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JX259255.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amino acid alignment of HPPD sequences from MCR, ACR, and WCS waterhemp plants.
Supplemental Table S1. Sequences and combinations of primers used for PCR amplification and sequencing of the HPPD and psbA genes from waterhemp plants.
Supplemental Table S2. Mesotrione and 4-OH standards used to generate a standard curve.
Acknowledgments
We thank Lisa Gonzini, Douglas Maxwell, and Nicholas Hausman for technical assistance, Dr. Germán Bollero and Dr. Maria Villamil for helpful suggestions concerning statistical analyses, and Dr. Kevin Tucker for assistance with LC-MS/MS (MRM) analysis. Appreciation is also extended to Wendy Zhang, Austin Tom, Jacquie Janney, and Caleb Shearrow for their assistance in plant extractions and to Dr. Anatoli Lygin for helpful discussions regarding LC-MS/MS (MRM).
Glossary
- HPPD
4-hydroxyphenylpyruvate dioxygenase
- MOI
Missouri glyphosate-resistant
- MCR
McLean County mesotrione- and atrazine-resistant
- ACR
Adams County mesotrione-sensitive but atrazine-resistant
- WCS
Wayne County herbicide-sensitive
- P450
cytochrome P450 monooxygenase
- GST
glutathione S-transferase
- GSH
reduced glutathione
- ALS
acetolactate synthase
- DT50
time for 50% of absorbed mesotrione to degrade
- HAT
hours after treatment
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- 4-OH
4-hydroxy-mesotrione
- AMBA
2-amino-4-(methyl-sulfonyl)benzoic acid
- FOMC
first-order multicompartment
- qPCR
quantitative PCR
- Ct
cycle threshold
- COC
crop oil concentrate
- LSS
liquid scintillation spectrometry
- TAPS
3-{[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]amino}-1-propanesulfonic acid
- MRM
multiple reaction monitoring
- ai
active ingredient
References
- Abit MJM, Al-Khatib K. (2009) Absorption, translocation, and metabolism of mesotrione in grain sorghum. Weed Sci 57: 563–566 [Google Scholar]
- Armel GR, Hall GJ, Wilson HP, Cullen N. (2005) Mesotrione plus atrazine mixtures for control of Canada thistle (Cirsium arvense). Weed Sci 53: 202–211 [Google Scholar]
- Barrett M (1997) Regulation of xenobiotic degrading enzymes with insecticides and other synergists. In KK Hatzios, ed, Regulation of Enzymatic Systems Detoxifying Xenobiotics in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 289–304 [Google Scholar]
- Barrett M (2000) The role of cytochrome P450 enzymes in herbicide metabolism. In AH Cobb, RC Kirkwood, eds, Herbicides and Their Mechanisms of Action. CRC Press, Boca Raton, FL, pp 25–37 [Google Scholar]
- Beaudegnies R, Edmunds AJF, Fraser TEM, Hall RG, Hawkes TR, Mitchell G, Schaetzer J, Wendeborn S, Wibley J. (2009) Herbicidal 4-hydroxyphenylpyruvate dioxygenase inhibitors—a review of the triketone chemistry story from a Syngenta perspective. Bioorg Med Chem 17: 4134–4152 [DOI] [PubMed] [Google Scholar]
- Burnet MWM, Loveys BR, Holtum JAM, Powles SB. (1993) Increased detoxification is a mechanism of simazine resistance in Lolium rigidum. Pestic Biochem Physiol 46: 207–218 [Google Scholar]
- Cazzonelli CI, Pogson BJ. (2010) Source to sink: regulation of carotenoid biosynthesis in plants. Trends Plant Sci 15: 266–274 [DOI] [PubMed] [Google Scholar]
- Correia MA, Ortiz de Montellano PR (2005) Inhibition of cytochrome P450 enzymes. In PR Ortiz de Montellano, ed, Cytochrome P450: Structure, Mechanism, and Biochemistry, Ed 3. Kluwer Academic/Plenum Publishers, New York, pp 247–322 [Google Scholar]
- Cummins I, Cole DJ, Edwards R. (1999) A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. Plant J 18: 285–292 [DOI] [PubMed] [Google Scholar]
- Délye C, Gardin JAC, Boucansaud K, Chauvel B, Petit C. (2011) Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Res 51: 433–437 [Google Scholar]
- Devine MD, Duke SO, Fedtke C (1993) Herbicide translocation. In Physiology of Herbicide Action. PTR Prentice Hall, Englewood Cliffs, NJ, pp 67–94 [Google Scholar]
- Devine MD, Preston C (2000) The molecular basis of herbicide resistance. In AH Cobb, RC Kirkwood, eds, Herbicides and Their Mechanisms of Action. Sheffield Academic, Sheffield, UK, pp 72–104 [Google Scholar]
- Doyle JJ, Doyle JL. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15 [Google Scholar]
- Dufourmantel N, Dubald M, Matringe M, Canard H, Garcon F, Job C, Kay E, Wisniewski J-P, Ferullo J-M, Pelissier B, Sailland A, Tissot G. (2007) Generation and characterization of soybean and marker-free tobacco plastid transformants over-expressing a bacterial 4-hydroxyphenylpyruvate dioxygenase which provides strong herbicide tolerance. Plant Biotechnol J 5: 118–133 [DOI] [PubMed] [Google Scholar]
- Frear DS, Swanson HR. (1970) Biosynthesis of S-(4-ethylamino-6-isopropylamino-2-S-triazino) glutathione: partial purification and properties of a glutathione S-transferase from corn. Phytochemistry 9: 2123–2132 [Google Scholar]
- Gaines TA, Zhang W, Wang D, Bukun B, Chisholm ST, Shaner DL, Nissen SJ, Patzoldt WL, Tranel PJ, Culpepper AS, Grey TL, Webster TM, et al. (2010) Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc Natl Acad Sci USA 107: 1029–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JA, Balke NE, Stoltenberg DE. (1996) Increased glutathione conjugation of atrazine confers resistance in a Wisconsin velvetleaf (Abutilon theophrasti) biotype. Pestic Biochem Physiol 55: 157–171 [Google Scholar]
- Gustafson DI, Holden LR. (1990) Nonlinear pesticide dissipation in soil: a new model based on spatial variability. Environ Sci Technol 24: 1032–1038 [Google Scholar]
- Hager AG, Wax LM, Bollero GA, Simmons FW. (2002) Common waterhemp (Amaranthus rudis Sauer) management with soil-applied herbicides in soybean (Glycine max (L.) Merr.). Crop Prot 21: 277–283 [Google Scholar]
- Hausman NE, Singh S, Tranel PJ, Riechers DE, Kaundun SS, Polge ND, Thomas DA, Hager AG. (2011) Resistance to HPPD-inhibiting herbicides in a population of waterhemp (Amaranthus tuberculatus) from Illinois, United States. Pest Manag Sci 67: 258–261 [DOI] [PubMed] [Google Scholar]
- Hawkes TR, Holt DC, Andrews CJ, Thomas PG, Langford MP, Hollingworth S, Mitchell G (2001) Mesotrione: mechanism of herbicidal activity and selectivity in corn. In The BCPC Proceedings - Weeds, British Crop Protection Council, Brighton, UK, Vol 2. pp 563–568 [Google Scholar]
- Heap I (2012) The International Survey of Herbicide Resistant Weeds. http://www.weedscience.org (April 29, 2012)
- Hess FD. (2000) Light-dependent herbicides: an overview. Weed Sci 48: 160–170 [Google Scholar]
- Hidayat I, Preston C. (2001) Cross-resistance to imazethapyr in a fluazifop-P-butyl-resistant population of Digitaria sanguinalis. Pestic Biochem Physiol 71: 190–195 [Google Scholar]
- Hirschberg J, McIntosh L. (1983) Molecular basis of herbicide resistance in Amaranthus hybridus. Science 222: 1346–1349 [DOI] [PubMed] [Google Scholar]
- Hugie JA, Bollero G, Tranel PJ, Riechers DE. (2008) Defining the rate requirements for synergism between mesotrione and atrazine in redroot pigweed (Amaranthus retroflexus). Weed Sci 56: 265–270 [Google Scholar]
- Kim KH, Petersen M. (2002) cDNA cloning and functional expression of hydroxyphenylpyruvate dioxygenase from cell suspension cultures of Coleus blumei. Plant Sci 163: 1001–1009 [Google Scholar]
- Kreuz K, Fonne-Pfister R. (1992) Herbicide-insecticide interaction in maize: malathion inhibits cytochrome P450-dependent primisulfuron metabolism. Pestic Biochem Physiol 43: 232–240 [Google Scholar]
- Kreuz K, Tommasini R, Martinoia E. (1996) Old enzymes for a new job: herbicide detoxification in plants. Plant Physiol 111: 349–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoureux GL, Stafford LE, Shimabukuro RH, Zaylskie RG. (1973) Atrazine metabolism in sorghum: catabolism of the glutathione conjugate of atrazine. J Agric Food Chem 21: 1020–1030 [DOI] [PubMed] [Google Scholar]
- Leah JM, Worrall TL, Cobb AH. (1991) A study of bentazon uptake and metabolism in the presence and the absence of cytochrome P-450 and acetyl-coenzyme A carboxylase inhibitors. Pestic Biochem Physiol 39: 232–239 [Google Scholar]
- Lee RM, Thimmapuram J, Thinglum KA, Gong G, Hernandez AG, Wright CL, Kim RW, Mikel MA, Tranel PJ. (2009) Sampling the waterhemp (Amaranthus tuberculatus) genome using pyrosequencing technology. Weed Sci 57: 463–469 [Google Scholar]
- Liu J, Davis AS, Tranel PJ. (2012) Pollen biology and dispersal dynamics in waterhemp (Amaranthus tuberculatus). Weed Sci 60: 416–422 [Google Scholar]
- Livak KJ, Schmittgen TD. (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Δ Δ C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Maeda H, DellaPenna D. (2007) Tocopherol functions in photosynthetic organisms. Curr Opin Plant Biol 10: 260–265 [DOI] [PubMed] [Google Scholar]
- Matringe M, Sailland A, Pelissier B, Rolland A, Zink O. (2005) p-Hydroxyphenylpyruvate dioxygenase inhibitor-resistant plants. Pest Manag Sci 61: 269–276 [DOI] [PubMed] [Google Scholar]
- McMullan PM, Green JM. (2011) Identification of a tall waterhemp (Amaranthus tuberculatus) biotype resistant to HPPD-inhibiting herbicides, atrazine and thifensulfuron in Iowa. Weed Technol 25: 514–518 [Google Scholar]
- Mène-Saffrané L, DellaPenna D. (2010) Biosynthesis, regulation and functions of tocochromanols in plants. Plant Physiol Biochem 48: 301–309 [DOI] [PubMed] [Google Scholar]
- Mitchell G, Bartlett DW, Fraser TEM, Hawkes TR, Holt DC, Townson JK, Wichert RA. (2001) Mesotrione: a new selective herbicide for use in maize. Pest Manag Sci 57: 120–128 [DOI] [PubMed] [Google Scholar]
- Nordby JN, Williams MM, II, Pataky JK, Riechers DE, Lutz JD. (2008) A common genetic basis in sweet corn inbred Cr1 for cross-sensitivity to multiple cytochrome P450-metabolized herbicides. Weed Sci 56: 376–382 [Google Scholar]
- Pallett KE (2000) The mode of action of isoxaflutole: a case study of an emerging target site. In AH Cobb, RC Kirkwood, eds, Herbicides and Their Mechanisms of Action. Sheffield Academic, Sheffield, UK, pp 215–238 [Google Scholar]
- Patzoldt WL, Dixon BS, Tranel PJ. (2003) Triazine resistance in Amaranthus tuberculatus (Moq) Sauer that is not site-of-action mediated. Pest Manag Sci 59: 1134–1142 [DOI] [PubMed] [Google Scholar]
- Patzoldt WL, Tranel PJ, Hager AG. (2005) A waterhemp (Amaranthus tuberculatus) biotype with multiple resistance across three herbicide sites of action. Weed Sci 53: 30–36 [Google Scholar]
- Pfaffl MW. (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: 2002–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powles SB, Yu Q. (2010) Evolution in action: plants resistant to herbicides. Annu Rev Plant Biol 61: 317–347 [DOI] [PubMed] [Google Scholar]
- Riechers DE, Fuerst EP, Miller KD. (1996) Initial metabolism of dimethenamid in safened and unsafened wheat shoots. J Agric Food Chem 44: 1558–1564 [Google Scholar]
- Riechers DE, Kreuz K, Zhang Q. (2010) Detoxification without intoxication: herbicide safeners activate plant defense gene expression. Plant Physiol 153: 3–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechers DE, Stanford MM. (2002) Molecular characterization of the HPPD gene from velvetleaf. Proc North Central Weed Sci Soc 57: 38 [Google Scholar]
- Riggins CW, Peng Y, Stewart CN, Jr, Tranel PJ. (2010) Characterization of de novo transcriptome for waterhemp (Amaranthus tuberculatus) using GS-FLX 454 pyrosequencing and its application for studies of herbicide target-site genes. Pest Manag Sci 66: 1042–1052 [DOI] [PubMed] [Google Scholar]
- Shimabukuro RH, Frear DS, Swanson HR, Walsh WC. (1971) Glutathione conjugation. An enzymatic basis for atrazine resistance in corn. Plant Physiol 47: 10–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckel LE. (2007) The dioecious Amaranthus spp.: here to stay. Weed Technol 21: 567–570 [Google Scholar]
- Steckel LE, Sprague CL, Hager AG, Simmons FW, Bollero GA. (2003) Effects of shading on common waterhemp (Amaranthus rudis) growth and development. Weed Sci 51: 898–903 [Google Scholar]
- Tranel PJ, Riggins CW, Bell MS, Hager AG. (2011) Herbicide resistances in Amaranthus tuberculatus: a call for new options. J Agric Food Chem 59: 5808–5812 [DOI] [PubMed] [Google Scholar]
- Triantaphylidès C, Havaux M. (2009) Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci 14: 219–228 [DOI] [PubMed] [Google Scholar]
- Urlacher VB (2012) Oxidation: stereoselective oxidations with cytochrome P450 monooxygenases. In EM Carreira, H Yamamoto, eds, Comprehensive Chirality. Oxford Elsevier, Amsterdam, The Netherlands, pp 275–294 [Google Scholar]
- Walsh MJ, Stratford K, Stone K, Powles SB. (2012) Synergistic effects of atrazine and mesotrione on susceptible and resistant wild radish (Raphanus raphanistrum) populations and the potential for overcoming resistance to triazine herbicides. Weed Technol 26: 341–347 [Google Scholar]
- Werck-Reichhart D, Gabriac B, Teutsch H, Durst F. (1990) Two cytochrome P-450 isoforms catalysing O-de-ethylation of ethoxycoumarin and ethoxyresorufin in higher plants. Biochem J 270: 729–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodyard AJ, Hugie JA, Riechers DE. (2009) Interactions of mesotrione and atrazine in two weed species with different mechanisms for atrazine resistance. Weed Sci 57: 369–378 [Google Scholar]
- Wu F, Mueller LA, Crouzillat D, Pétiard V, Tanksley SD. (2006) Combining bioinformatics and phylogenetics to identify large sets of single-copy orthologous genes (COSII) for comparative, evolutionary and systematic studies: a test case in the euasterid plant clade. Genetics 174: 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan JS, Tranel PJ, Stewart CN., Jr (2007) Non-target-site herbicide resistance: a family business. Trends Plant Sci 12: 6–13 [DOI] [PubMed] [Google Scholar]