Arabidopsis RIBOSOMAL PROTEIN L10 family members are nonredundant and contribute differentially to male gametophyte functionality and UV-B stress responses.
Abstract
The RIBOSOMAL PROTEIN L10 (RPL10) is an integral component of the eukaryotic ribosome large subunit. Besides being a constituent of ribosomes and participating in protein translation, additional extraribosomal functions in the nucleus have been described for RPL10 in different organisms. Previously, we demonstrated that Arabidopsis (Arabidopsis thaliana) RPL10 genes are involved in development and translation under ultraviolet B (UV-B) stress. In this work, transgenic plants expressing ProRPL10:β-glucuronidase fusions show that, while AtRPL10A and AtRPL10B are expressed both in the female and male reproductive organs, AtRPL10C expression is restricted to pollen grains. Moreover, the characterization of double rpl10 mutants indicates that the three AtRPL10s differentially contribute to the total RPL10 activity in the male gametophyte. All three AtRPL10 proteins mainly accumulate in the cytosol but also in the nucleus, suggesting extraribosomal functions. After UV-B treatment, only AtRPL10B localization increases in the nuclei. We also here demonstrate that the three AtRPL10 genes can complement a yeast RPL10 mutant. Finally, the involvement of RPL10B and RPL10C in UV-B responses was analyzed by two-dimensional gels followed by mass spectrometry. Overall, our data provide new evidence about the nonredundant roles of RPL10 proteins in Arabidopsis.
In eukaryotes, the cytosolic ribosomes consist of large 60S and small 40S subunits. In Arabidopsis (Arabidopsis thaliana), ribosomal protein genes exist as families composed of two to seven members that could be differentially incorporated into the cytosolic ribosome under specific situations (Schmid et al., 2005; Byrne, 2009). In this way, ribosomal heterogeneity would allow selective translation of specific mRNAs under particular cell conditions (Barakat et al., 2001; Szick-Miranda and Bailey-Serres, 2001; Giavalisco et al., 2005; Carroll et al., 2008; Carroll, 2013). Arabidopsis mutants in ribosomal proteins exhibit a large range of developmental phenotypes with extreme abnormalities, including embryonic lethality, suggesting that ribosomes also have specific functions regulating the expression of developmental genes (Van Lijsebettens et al., 1994; Degenhardt and Bonham-Smith, 2008; Byrne, 2009; Horiguchi et al., 2011, 2012; Szakonyi and Byrne, 2011). Furthermore, it has been recently demonstrated that ribosomal proteins control auxin-mediated developmental programs by translational regulation of auxin response factors (Rosado et al., 2012). In addition, the characterization of single, double, and, in certain cases, triple mutants as well as complementation by paralog genes have demonstrated full, partial, and no redundancy between members of ribosomal protein families (Briggs et al., 2006; Guo and Chen, 2008; Guo et al., 2011; Horiguchi et al., 2011; Stirnberg et al., 2012).
RIBOSOMAL PROTEIN L10 (RPL10) was initially identified in humans as a putative suppressor of Wilms’ tumor (Dowdy et al., 1991). Since then, RPL10 has been studied in different organisms from archaea and bacteria to eukaryotes such as mammals, insects, yeast, and plants (Marty et al., 1993; Mills et al., 1999; Hwang et al., 2000; Zhang et al., 2004; Wen et al., 2005; Singh et al., 2009). A remarkable property of this protein is its high degree of amino acid conservation, suggesting fundamental and critical conserved functions of RPL10 in different organisms (Farmer et al., 1994; Eisinger et al., 1997; Hofer et al., 2007; Nishimura et al., 2008). Likewise, the crystallographic structural similarity observed among RPL10 orthologs in eukaryotes, bacteria, and archaea (called L16) established the conservation of this universal ribosomal protein family and provided evidence of the inalterability of the ribosome during evolution (Spahn et al., 2001; Nishimura et al., 2008). Nevertheless, besides being a constituent of ribosomes and participating in protein translation, additional extraribosomal functions have been described for RPL10 (Mills et al., 1999; Hwang et al., 2000; Chávez-Rios et al., 2003; Zhang et al., 2004; Singh et al., 2009). In yeast, RPL10 is essential for viability, organizes the union site of the aminoacyl-tRNA, and its incorporation into the 60S subunit is a prerequisite for subunit joining and the initiation of translation (West et al., 2005; Hofer et al., 2007). Extensive analysis of the in vivo assembly of ribosomes revealed that RPL10 is loaded to the ribosome in the cytosol with the assistance of its chaperone suppressor of QSR1 truncations (Hedges et al., 2005; West et al., 2005).
Arabidopsis has three genes encoding RPL10 proteins, AtRPL10A, AtRPL10B, and, AtRPL10C. Recently, we demonstrated that Arabidopsis RPL10 genes are differentially regulated by UV-B radiation: RPL10B is down-regulated, RPL10C is up-regulated, while RPL10A is not UV-B regulated. Arabidopsis single mutants showed that RPL10 genes are not functionally equivalent. Heterozygous rpl10a mutant plants are translation deficient under UV-B conditions, knockout rpl10A mutants are not viable, and knockdown homozygous rpl10B mutants show abnormal growth. Conversely, knockout homozygous rpl10C mutants do not exhibit any visible phenotype. Overall, RPL10 genes are involved in development and translation under UV-B stress (Falcone Ferreyra et al., 2010b). Furthermore, coimmunoprecipitation studies showed an association of RPL10 with nuclear proteins, suggesting that at least one of the RPL10 isoforms could have an extraribosomal function in the nucleus (Falcone Ferreyra et al., 2010a).
The aim of this work was to further investigate the contribution of each Arabidopsis RPL10 to plant development and UV-B responses. We examined the spatiotemporal expression of each AtRPL10 using transgenic plants expressing ProRPL10:GUS fusions. By AtRPL10-GFP fusions, we analyzed the subcellular localization of each RPL10, demonstrating that the three isoforms are mainly localized in the cytosol but also in the nucleus. In order to investigate the functional redundancy between AtRPL10 genes in more detail, we generated and characterized double rpl10 mutants. We also here demonstrate that the three AtRPL10 genes can complement a yeast RPL10 mutant. Finally, the involvement of RPL10B and RPL10C in UV-B responses was analyzed by two-dimensional (2D) gels followed by mass spectrometry. Overall, our data provide new insights into the role of each RPL10 in Arabidopsis.
RESULTS
AtRPL10s Are Expressed in Specific Tissues during Plant Development
We have previously shown, by reverse transcription-quantitative PCR (RT-qPCR) analysis, that AtRPL10s are expressed in leaves, roots, stems, and flowers (Falcone Ferreyra et al., 2010b). In order to analyze the spatiotemporal expression of AtRPL10s in more detail, we generated transgenic plants expressing the GUS gene under the control of the RPL10A, RPL10B, and RPL10C promoters (ProAtRPL10s:GUS), as described in “Materials and Methods.”
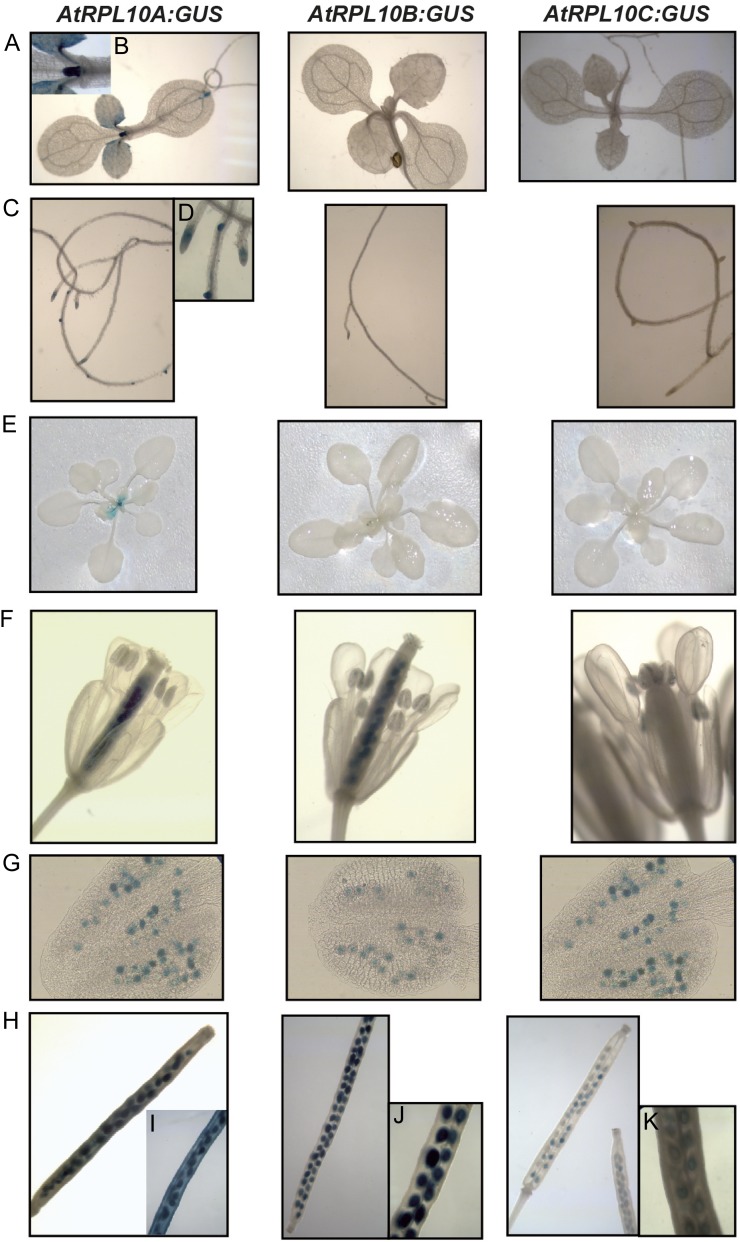
Histochemical GUS staining of 7-d-old seedlings reveals a high expression of RPL10A in young leaves, mostly in dividing cells and in the hydathodes (Fig. 1, A and B), while RPL10B and RPL10C expression was not detected in this organ (Fig. 1A). Seven-day-old roots also showed RPL10A expression, mainly in the root tips and lateral root primordia, while expression of RPL10B and RPL10C was not detected (Fig. 1, C and D). In 21-d-old plants, similar expression patterns to 7-d-old seedlings were detected, with GUS staining being present only for RPL10A (Fig. 1E).
Figure 1.
Histochemical localization of GUS activity in transgenic plants carrying the AtRPL10 promoters. A, Seven-day-old seedlings. B, Magnification of A (RPL10A). C, Roots from 7-d-old seedlings. D, Magnification of C (RPL10A). E, Twenty-day-old plants. F, Open flowers. G, Anthers from open flowers shown in F. H Siliques. I to K, Magnifications of H. [See online article for color version of this figure.]
AtRPL10 genes also show expression differences in the female and male reproductive organs. Expression of AtRPL10A and AtRPL10B was detected in pistils, anthers, and pollen grains (Fig. 1, F and G), while AtRPL10C was only detected in pollen grains (Fig. 1G).
In immature siliques, the expression of the three AtRPL10 genes was different. For AtRPL10A, GUS staining was present in the valves, septum, and developing seeds (Fig. 1, H and I), while AtRPL10B expression was limited to developing seeds (Fig. 1, H and J). AtRPL10C also showed expression in developing seeds, but with a weaker intensity than AtRPL10A and AtRPL10B (Fig. 1, H and K).
AtRPL10C Contributes with AtRPL10A to Plant Viability
We have previously shown that RPL10A is essential for the female gametophyte, as the mutant alleles cause female gametophyte lethality, while RPL10B has an important role during plant development (Imai et al., 2008; Falcone Ferreyra et al., 2010b). On the contrary, under standard growth conditions, rpl10C single mutants showed a phenotype that is indistinguishable from wild-type plants both at the vegetative and reproductive stages, suggesting functional redundancy within the RPL10 gene family; alternatively, it could have a role under different stress conditions. In order to analyze the functional redundancy between AtRPL10 genes in more detail, we generated double mutants by crossing the single mutants, rpl10B and rpl10C, with rpl10A heterozygous plants (rpl10A/+) as the male parent. The resultant F1 population of the rpl10A/+ with rpl10B (knockdown homozygous for RPL10B) cross produced rpl10A/+ rpl10B/+ (heterozygous for both genes) and RPL10A rpl10B/+ (wild type for RPL10A, heterozygous for RPL10B) at a 1:2 ratio (Supplemental Table S1), indicating a distortion in the transmission of the rpl10A allele through the male gametophyte when the rpl10B alleles are present. Similarly, when rpl10A/+ plants were crossed with rpl10C mutants (knockout homozygous for RPL10C), 67% of progeny had the RPL10A rpl10C/+ genotype, while 33% was rpl10A/+ rpl10C/+ (Supplemental Table S2). This deviation from the expected 1:1 ratio in both crosses indicates that rpl10A has an effect in the transmission through the male gametophyte when the RPL10B or RPL10C gene is mutated. On the contrary, when rpl10B was crossed with rpl10C, the resultant F1 population was 100% heterozygous for RPL10B and RPL10C. We further screened the resultant selfed F2 progeny for rpl10A/+ rpl10B/+, rpl10A/+ rpl10C/+, and rpl10B/+ rpl10C/+ double mutants. Again, the rpl10A allele was transmitted to the progeny at a frequency lower than expected: for rpl10A/+ rpl10B/+ plants, 34% of the progeny included rpl10A/+ RPL10B, rpl10A/+ rpl10B/+, and rpl10A/+ rpl10B mutant plants, while 66% had RPL10A RPL10B, RPL10A rpl10B/+, and RPL10A rpl10B genotypes (Supplemental Table S1). Similarly, for rpl10A/+ rpl10C/+ plants, rpl10A/+ RPL10C, rpl10A/+ rpl10C/+, and rpl10A/+ rpl10C mutants constituted 38% of the total, whereas 62% was represented by RPL10A RPL10C, RPL10A rpl10C/+, and RPL10A rpl10C (Supplemental Table S2). In contrast, the expected frequencies were found for the selfed F2 progeny from the rpl10B/+ rpl10C/+ double mutants (Supplemental Table S3). Moreover, reciprocal crosses of rpl10B/+ rpl10C/+ mutants showed unaffected transmission of the mutant alleles through the female and male gametes, while reciprocal crosses using rpl10A/+ rpl10C/+ and rpl10A/+ rpl10B plants as a male parent yielded unexpected ratios in F1 progeny, confirming the impaired transmission of the rpl10A allele through the male gametophyte (Supplemental Table S4).
After genotyping more than 150 F2 plants, rpl10A/+ rpl10C plants were not found, indicating that the presence of at least one functional RPL10C allele is essential for plant viability when one rpl10A allele is present. Moreover, to generate the rpl10 triple mutant (rpl10A/+ rpl10B rpl10C), rpl10A/+ rpl10B mutants were crossed with rpl10B rpl10C mutants as male and female parents, respectively. Siliques did not develop and all had aborted embryos, indicating that the triple mutant is not viable.
The rpl10A/+ rpl10B and rpl10B rpl10C double mutants were indistinguishable from their parent rpl10B single mutants. Likewise, rpl10A/+ and rpl10C single mutants and rpl10A/+ rpl10C/+ double mutants looked like wild-type Columbia (Col-0) plants (Supplemental Fig. S1).
To confirm the absence of functional compensation in the RPL10 family by variations in their transcript levels, we performed RT-qPCR analysis using RNA from the three double mutants (rpl10A/+ rpl10B, rpl10B rpl10C, and rpl10A/+ rpl10C/+), and similar RPL10 transcript levels were detected in single mutants, double mutants, and wild-type plants (Supplemental Fig. S2).
Arabidopsis RPL10 Proteins Complement Saccharomyces cerevisiae Conditional Lethal rpl10 Mutants
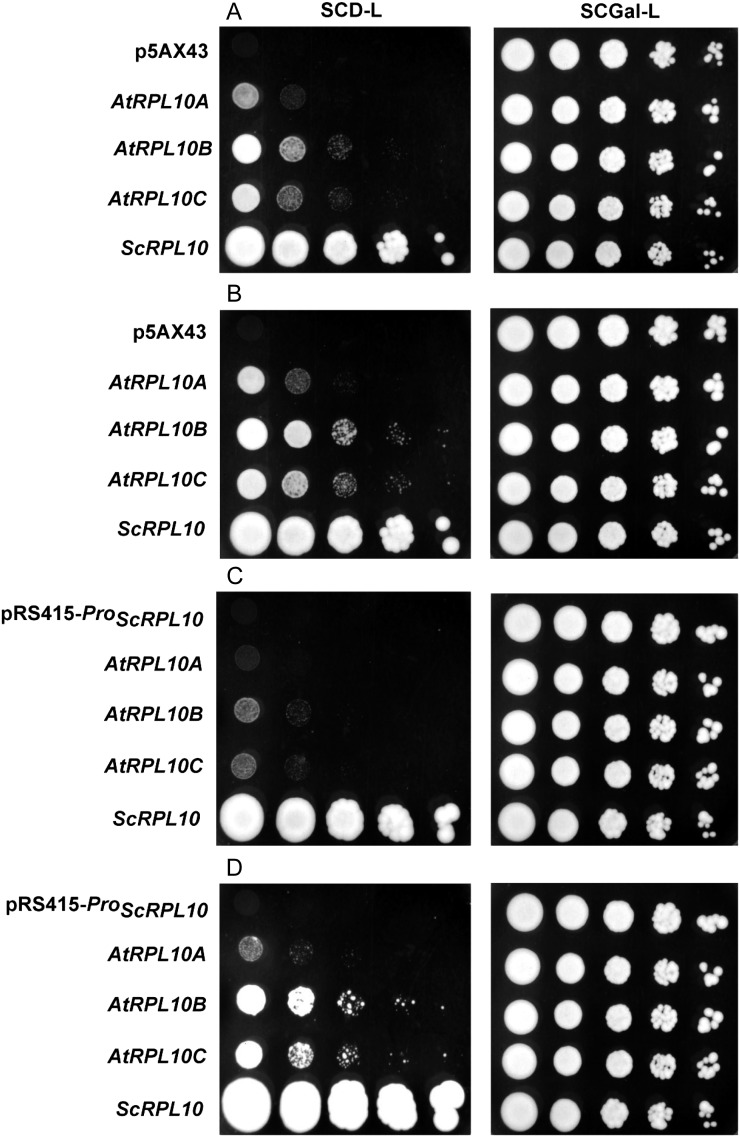
Arabidopsis RPL10 proteins exhibit 94% to 97% similarity and also share a high degree of primary sequence conservation with other eukaryotic orthologs; for example, they show 62% similarity with S. cerevisiae RPL10 (Supplemental Fig. S3). Based on this, we investigated whether Arabidopsis RPL10 proteins can complement an S. cerevisiae conditional lethal mutant in the RPL10 gene. The AJY2104 strain was used for these experiments, in which the RPL10 gene is expressed from its genomic locus but under the control of the GAL1 promoter; consequently, this strain is unviable under repressing conditions in the presence of Glc, while it grows similarly to the wild-type strain with Gal (Hofer et al., 2007). We expressed the coding region of each AtRPL10 under the control of the constitutive GDP promoter using the expression vector p5AX43 as well as under the control of the endogenous RPL10 promoter using the centromeric vector pRS415. In both cases, the endogenous RPL10 gene was expressed from the same plasmids as positive controls. Serial dilutions of transformed yeast were plated on selective medium with Glc, and simultaneously in the same medium with Gal, to verify the homogenous growth in all cases. The expression of Arabidopsis RPL10 proteins, as well as the endogenous RPL10 protein in transformed strains, was verified by immunoblot assays showing an immunoreactive band in all cases (Supplemental Fig. S4). Figure 2, A and B, shows yeast growth using the p5AX43 expression vector. After incubation for 3 and 4 d at 30°C, complementation was observed with either of the AtRPL10 proteins; however, the rate of growth was lower than the one reached with the endogenous RPL10 protein.
Figure 2.
Complementation of conditional mutant yeast with RPL10s from Arabidopsis. AJY2104 was transformed with plasmids expressing AtRPL10A to AtRPL10C under the control of the GDP promoter (A and B) or the RPL10 promoter from S. cerevisiae (C and D). Empty vectors (p5AX43 and pRS415-ProScRPL10) and ScRPL10 were assayed as negative and positive controls, respectively. Ten-fold serial dilutions of cultures grown in selective medium with Gal were spotted on a selective medium containing Glc or Gal (SCD-L and SCGal-L) and incubated at 30°C for 3 d (A), 4 d (B and C), and 7 d (D).
When each AtRPL10 was expressed under the control of the endogenous RPL10 promoter, transformed yeast showed the formation of colonies with all the constructs, but the rate of growth was slower than that observed using the GAL1 promoter (Fig. 2, C and D). Taken together, these results demonstrate that Arabidopsis RPL10 proteins are functionally equivalent to the S. cerevisiae RPL10; the three AtRPL10 proteins can rescue the lethality due to the absence of the endogenous protein.
Arabidopsis RPL10 Proteins Localize in the Cytosol But Also in the Nucleus
By coimmunoprecipitation studies using Arabidopsis leaves, we have previously demonstrated that RPL10 proteins associate with proteins localized in the nucleus and with proteins involved in cytoplasm-nuclei traffic, suggesting that at least one of the AtRPL10 isoforms may have extraribosomal functions in the nuclei (Falcone Ferreyra et al., 2010b). To examine the subcellular localization of each AtRPL10 isoform, we generated Arabidopsis transgenic plants that express each AtRPL10 as a fusion protein to the N terminus of the GFP driven by the cauliflower mosaic virus 35S promoter (Pro35S:AtRPL10s-GFP constructs). Transgenic plants were selected and examined by RT-qPCR as described in “Materials and Methods” (Supplemental Fig. S5). It should be mentioned that it has been reported that GFP-tagged ribosomal proteins are correctly incorporated into the ribosomes (Mustroph et al., 2009). Here, to verify that AtRPL10-GFP fusion proteins are functional in Arabidopsis plants, we transformed the Arabidopsis rpl10B mutants with a construct expressing the AtRPL10B-GFP driven by the 35S promoter. As previously published, these mutants exhibit an abnormal growth phenotype (Falcone Ferreyra et al., 2010b). F1 progeny were indistinguishable from wild-type (Col-0) plants and rpl10B/+ mutants, indicating the ability of the AtRPL10B-GFP fusion protein to complement mutant plants in the RPL10B gene and, consequently, the functionality of the AtRPL10B-GFP fusion protein in Arabidopsis. Thus, given the high identity between RPL10 proteins from Arabidopsis, we assume that all AtRPL10-GFP proteins are functional in Arabidopsis.
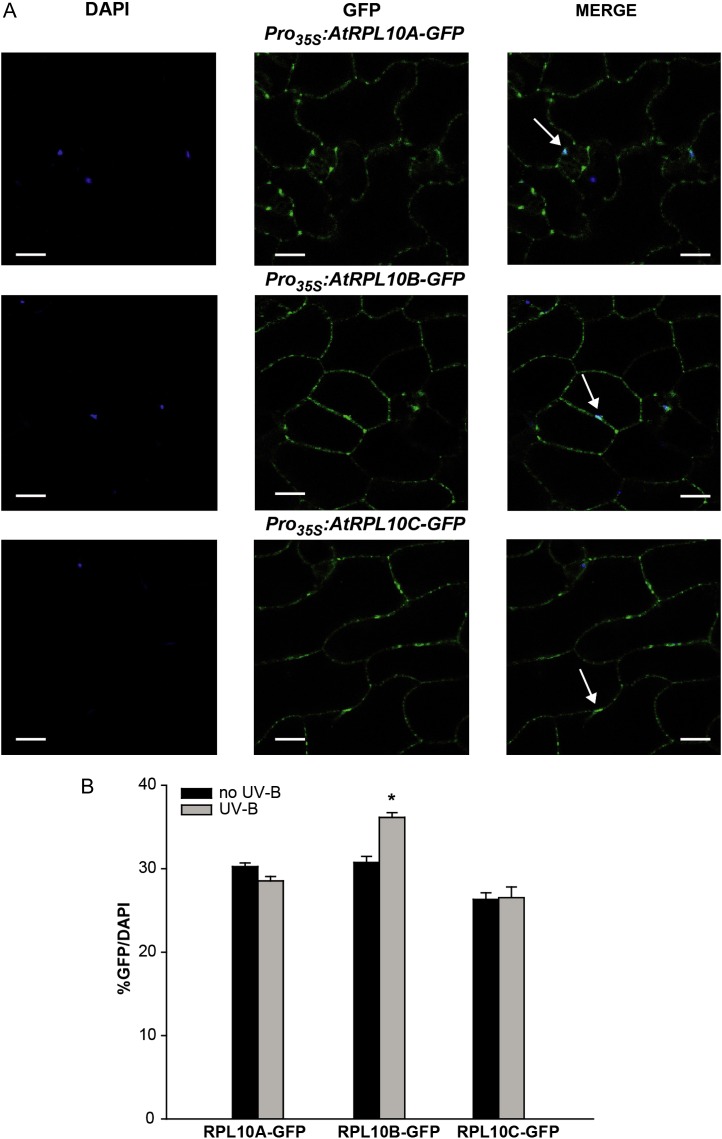
AtRPL10-GFP fusions were mainly detected in the cytosol, but GFP fluorescence was also detected in the nuclei (stained with 4′,6-diamino-phenylindole [DAPI]; Fig. 3A). All three fusion proteins exhibit a necklace appearance, suggesting their association with rough endoplasmic reticulum ribosomes, as described for the human RPL10 (Loftus et al., 1997). In order to quantify the AtRPL10s-GFP localization in the nucleus, we measured the percentage of nuclei stained with DAPI that also showed detectable GFP fluorescence, as described by Kaiserli and Jenkins (2007). For all three proteins, approximately 30% of the nuclei had detectable GFP fluorescence, without significant differences between the three RPL10 proteins (Fig. 3B). We also analyzed GFP localization in transgenic plants expressing GFP driven by the 35S promoter (Pro35S:GFP) in order to discard the presence of GFP fluorescence in the nucleus as a result from the cleavage of GFP fusion proteins. In these transgenic plants, GFP was only detected in the cytosol (Supplemental Fig. S6).
Figure 3.
Subcellular localization of RPL10s from Arabidopsis. A, Confocal images show DAPI and GFP fluorescence in epidermal tissues of leaves from Pro35S:AtRPL10s-GFP transgenic plants. DAPI staining indicates the position of the nuclei in cells; GFP staining indicates the localization of AtRPL10s in the cytoplasm and the nuclei of epidermal cells of transgenic plants. Images were merged to show signal overlap. Arrows indicate the colocalization of DAPI and GFP. Bars = 50 μm. B, Percentage of GFP/DAPI for each AtRPL10-GFP fusion protein in its respective transgenic plant. Error bars indicate se of the samples. The asterisk indicates a statistically significant difference between samples (P < 0.05; one-way ANOVA). [See online article for color version of this figure.]
Previously, we demonstrated that RPL10s were UV-B regulated and RPL10s were involved in translation under UV-B stress (Falcone Ferreyra et al., 2010b). Therefore, we examined whether UV-B radiation regulates RPL10 subcellular localization. After 4 h of exposure with UV-B radiation, AtRPL10A and AtRPL10C did not show changes in their subcellular distribution; however, AtRPL10B-GFP fluorescence increased by about 20% in the nuclei after the treatment (Fig. 3B; Supplemental Fig. S7).
Proteome Analysis of Knockout rpl10C and Knockdown rpl10B Single Mutants
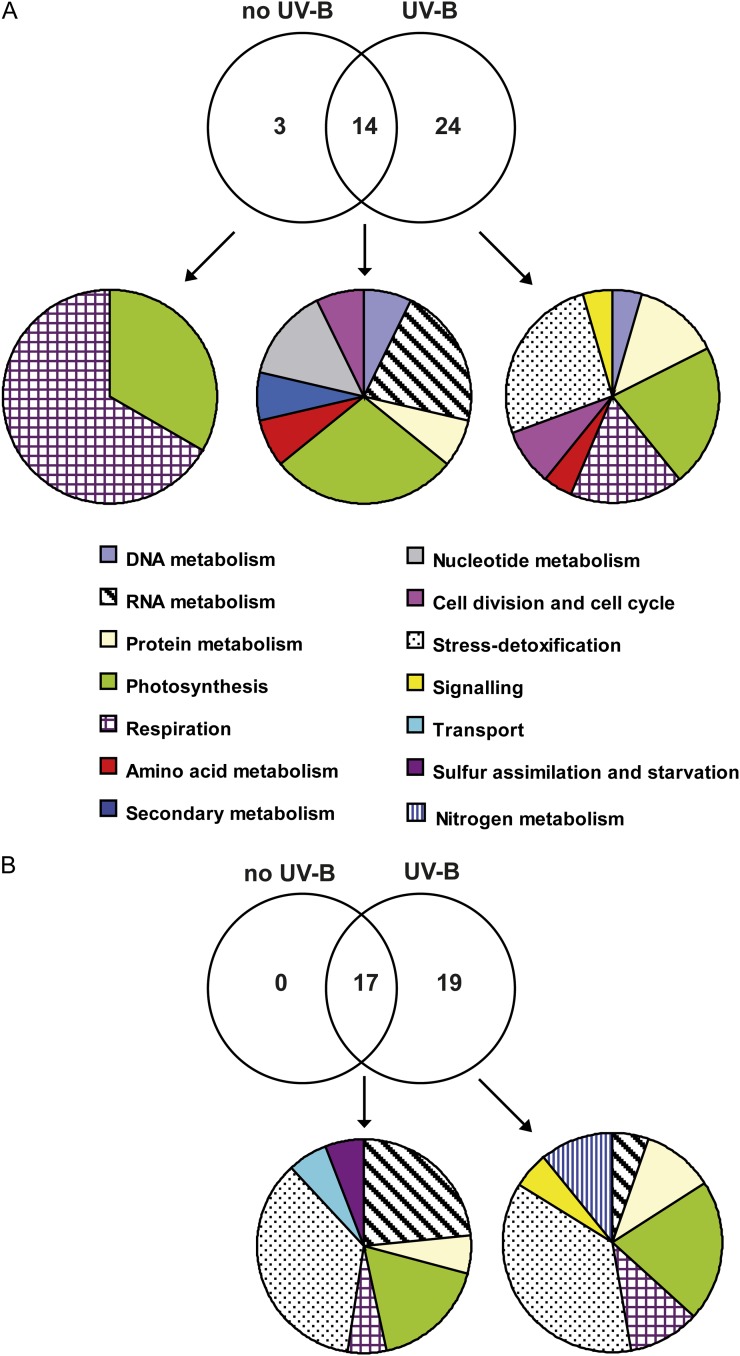
Both AtRPL10B and AtRPL10C are regulated by UV-B radiation: AtRPL10B is repressed, while AtRPL10C is induced after a 4-h UV-B radiation treatment. In addition, we here show that nuclear localization of AtRPL10B is increased after UV-B irradiation (Fig. 3B). Thus, in order to investigate metabolic processes in which AtRPL10B and AtRPL10C isoforms could be involved and their participation in responses under UV-B stress, we carried out 2D gel electrophoresis to compare changes in the leaf proteome of rpl10C and rpl10B homozygous plants under control conditions in the absence of UV-B and after UV-B irradiation with those in wild-type plants, as reported previously for rpl10A heterozygous mutant plants (Falcone Ferreyra et al., 2010b). The differential proteomes were analyzed in two different ways: (1) the proteomes from the mutant plant leaves were compared with those from wild-type plants under the same radiation conditions (no UV-B and UV-B); (2) for each plant type (wild type and mutant), we compared their proteomes under control conditions (no UV-B) and after UV-B irradiation, and only proteins that were differentially UV-B regulated in the wild-type and mutant plants were selected. For rpl10C mutants, a total of 45 protein spots showed differential levels (increased or decreased 1.5-fold or more) in at least one of the comparisons, and interpretable tandem mass spectrometry spectra were obtained for 41 of the 45 spots (Supplemental Table S5). Of these 41 spots, 25 were similarly changed as observed previously in the rpl10A mutants (Falcone Ferreyra et al., 2010b), while 16 spots were exclusively differential for the rpl10C mutants. Under control conditions in the absence of UV-B, nine protein spots were increased in rpl10C mutants, while eight protein spots were decreased compared with wild-type plants; however, after a 4-h UV-B treatment, the number of proteins that changed in rpl10C mutants was increased to 38 (12 increased and 26 decreased), indicating that the rpl10C mutation more severely affects protein expression patterns after a UV-B exposure than under control conditions (Fig. 4A). Similarly, when we compared the proteomes of both plants after the UV-B treatment, rpl10C mutant plants exhibited a higher number of proteins that were decreased by UV-B (12 protein spots) than those increased (six protein spots). Also, some proteins that were changed by UV-B in wild-type plants did not show any difference in the mutants after the same treatment (nine protein spots).
Figure 4.
Venn diagrams comparing proteome changes between rpl10C and rpl10B mutants and wild-type plants. The intersections of proteins with differential levels in rpl10C (A) and rpl10B (B) mutant plants in comparison with the wild type under control conditions without UV-B and after a 4-h UV-B treatment are shown. Each mutant plant was compared with the wild type under the same light conditions. For each group of proteins, the classification based on their cell functions is shown. Proteins were identified by 2D gel electrophoresis followed by mass spectrometry, and those showing changes in abundances of at least 1.5-fold were included. [See online article for color version of this figure.]
Total identified proteins (41) were classified according to their functions (Fig. 4A). The functional groups with the largest number of proteins differentially changed in the rpl10C mutants in comparison with the wild-type plants after the UV-B treatment are involved in photosynthesis, stress and detoxification, respiration, protein biosynthesis, and posttranslational mechanisms. Other groups include proteins implicated in DNA metabolism (DNA repair), RNA metabolism (transcription and regulation), signaling, and cell division and cycle. In summary, general processes in plant metabolism and growth are affected in the rpl10C mutant after UV-B exposure, as was observed previously for rpl10A heterozygous mutant plants. Thus, although rpl10C mutants are not more sensitive to UV-B radiation than wild-type plants, these results indicate that RPL10C may also be involved in translation under UV-B conditions.
For rpl10B mutants, a total of 43 protein spots showed differential levels (increased or decreased 1.5-fold or more) in at least one of the comparisons. Supplemental Figure S8 shows a representative 2D gel of the proteome of rpl10B mutant leaves after a 4-h UV-B treatment used in one set of comparisons. Interpretable tandem mass spectrometry spectra were obtained for 36 of the 43 spots (Supplemental Table S6). These differential spots were not identified in the same analysis using the rpl10A and rpl10C mutants. Under control conditions in the absence of UV-B, nine protein spots were increased, while eight protein spots were decreased in rpl10B mutants compared with wild-type plants; however, after a 4-h UV-B treatment, the number of proteins that changed in rpl10B mutants was increased to 36 (18 increased and 18 decreased), suggesting, as observed for the rpl10A and rpl10C mutants, that the rpl10B mutation more severely disrupts the protein expression pattern after a UV-B exposure than under control conditions (Fig. 4B). In addition, when we compared the proteomes of wild-type and rpl10B plants after the UV-B treatment, some proteins that changed by UV-B in the wild-type plants did not show any difference in the mutants after the same treatment (17 protein spots).
Identified proteins were classified according to their functions (Fig. 4B; Supplemental Fig. S8B). The major group of identified proteins corresponds to proteins involved in stress responses (41% and 36% in control and UV-B conditions, respectively), mainly biotic stress, representing 35% and 23% of the total in control and UV-B conditions, respectively (Fig. 4B). Some proteins can be grouped in more than one process; for example, spot 38 is involved in RNA metabolism and biotic stress, while spot 39 can be grouped with proteins involved in cell division and biotic stress (Supplemental Table S6). Other important groups include proteins implicated in RNA metabolism (transcription and regulation), protein metabolism, photosynthesis, and signaling. It is noteworthy that a high percentage of proteins identified are predicted to be localized in the chloroplast, with a lower proportion in the nucleus.
Networks of associations were generated for proteins that showed different levels in rpl10C and rpl10B mutants in comparison with wild-type plants under UV-B stress (Supplemental Fig. S9). In particular, when analyzing the enrichment of any particular biological process in each network, rpl10B mutants showed a significantly larger number of proteins involved in stress responses (Supplemental Fig. S9A). By contrast, rpl10C mutants showed proteins involved in general biological and metabolic processes and a lower percentage of proteins involved in stress responses than rpl10B mutants (Supplemental Fig. S9B). In general, predicted interactions were found between proteins belonging to the same functional group, although some proteins did not show any interaction with proteins in the networks for both mutants.
DISCUSSION
RPL10 is an integral component of the large subunit of eukaryotic ribosomes. In order to gain knowledge about the participation of the three AtRPL10 proteins in translation, we investigated the conservation of this function between AtRPL10 proteins and its ortholog in S. cerevisiae by testing their ability to complement an S. cerevisiae RPL10 mutant (Hofer et al., 2007). By using two different constructs containing the coding regions of each AtRPL10, we demonstrated the complementation of a conditional lethal yeast mutant with their Arabidopsis orthologs, suggesting that heterologous proteins can be assembled into a hybrid ribosome (Fig. 2). Proteomic studies of the 80S cytosolic ribosome using different Arabidopsis tissues by 2D electrophoresis followed by spectrometric mass showed that both RPL10A and RPL10C were components of the 60S subunit, while RPL10B was not identified (Chang et al., 2005; Carroll et al., 2008; Turkina et al., 2011). However, our complementation studies in yeast suggest that RPL10B, as well as RPL10A and RPL10C, may fulfill the function of yeast RPL10 and, consequently, are all probably involved in translation. Thus, Arabidopsis RPL10 proteins are functionally equivalent to the RPL10 from S. cerevisiae and, therefore, these proteins are interchangeable. Similarly, the three Arabidopsis RACK1 proteins, which are components of the 40S ribosome subunit, complement S. cerevisiae cross pathway control2/rack1 mutants, as was shown previously for the mammalian RACK1 protein (Gerbasi et al., 2004; Guo et al., 2011). However, although all three Arabidopsis genes are functional in rescuing yeast lethality, slower growth rates were observed when compared with that obtained using the endogenous yeast protein, indicating differences in the complementation efficiency. Previous studies demonstrated the complementation of RPL10 yeast mutants with its maize (Zea mays) and human orthologs (Dick and Trumpower, 1998). Similarly, a decreased growth rate was observed in complemented mutants. Western-blot assays of the 80S ribosome showed that although both maize and human RPL10s are incorporated into the yeast ribosome, hybrid ribosomes are defective in subunit assembly and display a partial reduction of their translational activity (Dick and Trumpower, 1998). Therefore, a similar situation may occur in our complementation assays. Extensive mutational analysis of ScRPL10 revealed amino acids that are essential for viability and others involved in interactions with the ribosome and partners (Hofer et al., 2007). Thus, amino acid changes between Arabidopsis and S. cerevisiae RPL10 sequences may be a cause of growth deficiencies in complemented yeast. Moreover, it is worth mentioning that no suppressor of QSR1 truncations ortholog gene is present in the Arabidopsis genome, suggesting that the binding of RPL10 into the ribosomes would be different in plants. Future site-directed mutagenesis experiments will be necessary to determine whether differences in particular amino acids between RPL10 proteins are responsible for the poor binding of AtRPL10s into the ribosome or the decreased activity of the hybrid ribosomes.
By using transgenic plants expressing ProRPL10:GUS fusions, we found that RPL10A has the widest expression of the three AtRPL10 proteins. In addition, rpl10A/+ rpl10C/+ double mutants are phenotypically undistinguishable from wild-type plants and rpl10A/+ and rpl10C single mutants. However, rpl10A/+ rpl10C plants are not viable. Also, the progeny of rpl10A/+ rpl10C/+ and rpl10A/+ rpl10B/+ double mutants show a deviation from the expected frequency of inheritance of the rpl10A allele (Supplemental Tables S1, S2, and S4). These results demonstrate that RPL10A is also important for the male gametophyte, in addition to its reported role in the female gametophyte (Imai et al., 2008; Ferreyra et al., 2010b). Furthermore, these results agree with the expression pattern of RPL10A in both reproductive organs as detected by GUS staining (Fig. 1).
On the other hand, RPL10B showed a very specific pattern of expression only in reproductive organs. In addition, we here show that mutations in RPL10A or RPL10C do not increase the developmental defects of rpl10B single mutants (Supplemental Fig. S1). RPL10B represents only 10% of all AtRPL10 transcripts (Falcone Ferreyra et al., 2010b). Taken together, these data suggest that RPL10B could have a very specific role that is not shared with RPL10A or RPL10C. Our complementation studies in yeast suggest that RPL10B may be involved in translation. Then, taking into account the ribosome heterogeneity model (Horiguchi et al., 2012) and assuming that RPL10B is a constituent of ribosomes, one possible explanation is that this protein is essential for the translation of some specific transcripts during plant development and in both reproductive organs, and that function cannot be supplied by RPL10A or RPL10C. Additionally, it is possible that this protein is required for translation only under certain environmental conditions.
Regarding RPL10C, as reported by Falcone Ferreyra et al. (2010b), mutant plants in RPL10C do not show any abnormal phenotype. Furthermore, when crossing rpl10C single mutants with the rpl10B mutants, the resultant F1 population was 100% rpl10B/+ rpl10C/+; however, rpl10A/+ rpl10C plants were not found, indicating that the presence of at least one functional RPL10C allele is essential for plant viability when one rpl10A allele is present. Transgenic plants expressing ProRPL10C:GUS showed a marked expression of RPL10C only in the male gametophyte, as did RPL10A and RPL10B. Johnson et al. (2004) reported that the Arabidopsis hap3 mutant, which has a transfer DNA (T-DNA) insertion 1,814 bp upstream the translational start codon of RPL10C, exhibits defective pollen tube growth at the stigma, indicating that RPL10C may be important for pollen development and germination. Thus, according to AtRPL10 expression levels during pollen development (Arabidopsis eFP Browser; Supplemental Fig. S10A) and our data, the three AtRPL10 genes would independently contribute, and in different proportions, to AtRPL10 pool activity required in male gametophyte development. Interestingly, AtRPL10A contributes similarly in all microgametogenesis stages, while AtRPL10B seems to be more important during the first stages and AtRPL10C in mature pollen (Winter et al., 2007; Supplemental Fig. S10A). As mentioned before, RPL10A is also important for pollen development, so one possibility is that RPL10C may partially supply RPL10A function during this stage, because rpl10A/+ rpl10C/+ plants are viable while rpl10A/+ rpl10C are not. Thus, we hypothesize that a threshold level of AtRPL10 activity is required for proper pollen development. In concordance with this hypothesis, microarray data of mature pollen indicated that the mature pollen grains contain transcripts to be translated during tube germination (Becker et al., 2003; Honys and Twell, 2003). Hence, RPL10s could have important roles as constituents of ribosomes in protein biosynthesis. Nevertheless, we cannot rule out that RPL10 proteins may also play unknown extraribosomal functions in these tissues. On the contrary, during pollen germination (Arabidopsis eFP Browser; Supplemental Fig. S10B), the AtRPL10C transcript mainly contributes to total AtRPL10 transcript levels (10- and 50-fold higher than RPL10A and RPL10B, respectively), in concordance with the fact that hap3 mutants show defective pollen tube growth at the stigma (Johnson et al., 2004). Thus, while the three AtRPL10 proteins are important for pollen development, AtRPL10C seems to be the most important AtRPL10 protein during pollen germination.
Previous RT-qPCR analysis showed high RPL10C and low RPL10B expression levels in 1- and 3-week-old leaves and roots (Falcone Ferreyra et al., 2010b); however, GUS staining was not observed in these tissues in this work (Fig. 1). Although it is known that RT-qPCR is a more sensitive technique than GUS staining, these different observations could also be a consequence of the promoter length used for the GUS fusions. Different cis-regulatory elements may be present in other regions of the chromosome surrounding the RPL10C locus. Nevertheless, the length of the AtRPL10 promoters used in the experiments was enough to analyze expression in other tissues not previously studied by RT-qPCR due to the difficulty of isolation, such as both reproductive organs and developing seeds, which was one of the objectives of this work.
We have previously demonstrated that RPL10A has an important role in translation under UV-B stress (Falcone Ferreyra et al., 2010b). Nevertheless, RPL10B and RPL10C transcripts are UV-B regulated. To gain knowledge about the role of the AtRPL10s in the UV-B response, we generated transgenic plants expressing each AtRPL10 gene fused to GFP and showed that the three AtRPL10 proteins are mainly localized in the cytosol, in agreement with being constituents of cytosolic ribosomes (Chang et al., 2005; Carroll et al., 2008; Turkina et al., 2011). Interestingly, all AtRPL10 proteins were also detected in the nuclei in similar proportions, suggesting that the three isoforms may have extraribosomal roles. Moreover, after a 4-h UV-B irradiation treatment, AtRPL10B showed increased nuclear localization while AtRPL10A and AtRPL10C did not (Fig. 3B). This observation supports the hypothesis that AtRPL10B has a different role from the other two AtRPL10 proteins. Also, previous coimmunoprecipitation experiments carried out after a UV-B treatment revealed a higher number of nuclear proteins associated to AtRPL10 with respect to those in control conditions in the absence of UV-B (Falcone Ferreyra et al., 2010b). One possibility is that the increased number of nuclear proteins associated to AtRPL10 arises from increased translocation of AtRPL10B to the nucleus after the UV-B treatment. Carvalho et al. (2008) previously demonstrated that, after viral infection, AtRPL10A is phosphorylated by the NSP-INTERACTING KINASE1 (NIK1), redirecting the protein to the nucleus, whereas when NIK1 is mutated, RPL10A fails to accumulate in the nucleus. Therefore, a similar situation may occur after UV-B exposure with AtRPL10B, where it may fulfill specific functions in response to this radiation.
Additionally, we investigated the participation of AtRPL10B and AtRPL10C in the response to UV-B radiation by analyzing the proteome changes in rpl10B and rpl10C single mutants after a UV-B treatment. A large number of proteins (61%) that showed differential levels in the rpl10C mutant in comparison with wild-type plants were also previously identified in the same comparisons using rpl10A mutants (Falcone Ferreyra et al., 2010b). According to the functions of all differential proteins, general processes for plant metabolism and growth are affected in the rpl10C mutant after a 4-h UV-B exposure. Consequently, RPL10C would also be involved in translation after UV-B exposure, as reported previously for RPL10A, and the up-regulation of RPL10C expression by UV-B is needed for its translational function under this stressful condition.
The analysis of differential changes in rpl10B in comparison with the wild-type proteome under control conditions and after UV-B exposure also shows a larger number of proteins that change under UV-B stress than under control conditions in the absence of UV-B, indicating that RPL10B could be important under UV-B conditions. However, the identified proteins in this analysis are different from those identified in the same comparisons using rpl10A and rpl10C mutants. The largest functional group corresponds to proteins involved in stress responses, mainly biotic stress, suggesting that RPL10B could have a role in the response to pathogens, and these responses may be common for UV-B responses. Diverse responses to UV-B radiation are shared with other stress responses, for example, against pathogens (Bilgin et al., 2010; Mukherjee et al., 2010; Fraire-Velázquez et al., 2011). Many proteins identified in the rpl10B mutant were also identified in Arabidopsis by proteome analysis after infection with Alternaria brassicicola (Mukherjee et al., 2010). Interestingly, by coimmunoprecipitation studies after UV-B treatment followed by the identification of proteins associated to RPL10 in Arabidopsis, we demonstrated that RPL10s associate with proteins involved in the response to pathogens, including transcription factors, suggesting that at least one RPL10 isoform may also have a role in biotic stress (Falcone Ferreyra et al., 2010a). Moreover, RPL10B has also been involved in cold acclimation in Arabidopsis (Kawamura and Uemura, 2003). Collectively, these findings indicate that RPL10B may participate in responses against different stress conditions. Additionally, a large number of proteins that change in rpl10B mutants are localized in the chloroplast, indicating that RPL10B could also have some roles in chloroplast homeostasis; for example, RPL10B may be involved in the translation of specific chloroplast proteins. These observations support our hypothesis that RPL10B has specific roles different from the roles of RPL10A and RPL10C.
AtRPL10 gene expression is differentially regulated by UV-B radiation (Falcone Ferreyra et al., 2010b). In knockout rpl10C mutant plants, the regulation of the expression of RPL10 genes is affected when plants are irradiated with UV-B: mainly AtRPL10A transcript levels increase, suggesting that high levels of RPL10 are necessary for translation under UV-B stress. Sormani et al. (2011) proposed a model of ribosome biogenesis by three transcriptional pathways, where one of the pathways involves cytosolic ribosomal protein genes that are down-regulated under stress conditions. This mechanism results in a decrease in general translation or a reduction in the translation of particular mRNAs. Based on this model and our results, AtRPL10C may contribute with AtRPL10A to general translation under UV-B stress. Furthermore, the decrease in AtRPL10B expression by UV-B and its accumulation in the nuclei may lower the heterogeneity of ribosomes; therefore, only specific and necessary mRNAs could be translated, avoiding excessive energy consumption. Additionally or alternatively, AtRPL10B may also have extraribosomal activities as a regulator of transcription in the nucleus, as described in different organisms (Monteclaro and Vogt, 1993; Chávez-Rios et al., 2003).
In conclusion, we here demonstrate that the three AtRPL10 proteins complement a yeast mutant in RPL10, suggesting that they could all be involved in translation. Also, we present data demonstrating that AtRPL10B has a role different from the one of AtRPL10A and AtRPL10C in control and UV-B conditions. Finally, we show that RPL10C could participate with RPL10A in general translation. Together, our data demonstrate that AtRPL10s all contribute differently to plant development and stress responses.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and UV-B Treatment
Following a cold treatment (72 h at 4°C in the dark), Arabidopsis (Arabidopsis thaliana) ecotype Col-0 plants, transgenic plants, and the T-DNA insertion lines were grown in a growth chamber under light (100 μE m−2 s−1) with a 16-h-light/8-h-dark photoperiod at 22°C. Arabidopsis plants (wild-type Col-0 and transgenic plants) were also germinated and grown in Murashige and Skoog (MS) salt-agar medium. Arabidopsis seeds were sterilized and germinated on MS medium, and they were grown after vernalization for 3 d at 4°C. For subcellular localization analysis, plants were grown for 20 d on MS plates supplemented with kanamycin (50 mg L−1).
UV-B treatments were done in a growth chamber as described previously (Falcone Ferreyra et al., 2010b). Arabidopsis plants (Col-0 and rpl10B and rpl10C mutants) were treated for 4 h with a UV-B intensity of 2 W m−2 and a UV-A intensity of 0.65 W m−2. The UV-B bulbs were covered with cellulose acetate filters (100-mm extra-clear cellulose acetate plastic; Tap Plastics); the cellulose acetate sheeting does not remove any UV-B radiation from the spectrum but excludes wavelengths lower than 300 nm. Control plants without UV-B were exposed for the same period of time under the same UV-B lamps but covered with polyester filters (100-μm clear polyester plastic; Tap Plastics), which absorb both UV-B and wavelengths lower than 280 nm (UV-B, 0.04 W m−2, UV-A, 0.4 W m−2). Samples were collected immediately after irradiation and stored at −80°C. The experiments were repeated at least three times. For subcellular localization studies using Arabidopsis transgenic plants (Pro35S:AtRPL10s-GFP), plants were irradiated as described above. Samples were analyzed immediately after irradiation. The experiments were repeated at least three times.
Protein Extraction under Denaturing Conditions, Labeling with Alexa 610 and Alexa 532 Dyes, 2D Gel Electrophoresis, and Gel Image Analysis
All these experiments were performed as described previously (Falcone Ferreyra et al., 2010b).
In-Gel Digestion, Mass Spectrometry, and Database Searching
Gel spots of interest were manually excised from the gels and sent to CEBIQUIEM facilities (Facultad de Ciencias Exactas y Naturales, Universidad de Buenos Aires) for further analyses. Spots were subjected to in-gel digestion (donatello.ucsf.edu/ingel.html) with trypsin according to Casati et al. (2005). The mass spectrometric data were obtained using a matrix-assisted laser-desorption ionization time of flight/time of flight spectrometer (Ultraflex II; Bruker). The spectra obtained were submitted for National Center for Biotechnology Information database searching using MASCOT (www.matrixscience.com; Perkins et al., 1999) and analyzed as described previously (Casati et al., 2005). Protein functional classification was carried out according to literature data (Usadel et al., 2006).
Generation and Identification of Double Insertional T-DNA Mutants
rpl10A heterozygous (rpl10A/+ and rpl10A-1), rpl10B homozygous (rpl10B-1), and rpl10C homozygous (rpl10C-1) single mutants (Falcone Ferreyra et al., 2010b) were crossed, and their resultant F2 populations were screened for rpl10A/+ rpl10B, rpl10A/+ rpl10C, and rpl10B rpl10C double mutants. In order to obtain the triple mutant (rpl10A/+ rpl10B rpl10C), rpl10A/+ rpl10B mutants were crossed with rpl10B rpl10C mutants. The genotypes were determined by PCR on genomic DNA using combinations of specific primers for each gene and one primer that hybridizes with the left border of the T-DNA, as described previously (Falcone Ferreyra et al., 2010b).
Generation of Arabidopsis Transgenic Plants Expressing AtRPL10s-GFP and GFP
Full-length open reading frames for each AtRPL10 were amplified from complementary DNA (cDNA) obtained from leaf tissues of wild-type Arabidopsis plants (Col-0). The primers F-KpnI-RPL10 and R-SalI-RPL10 with the KpnI and SalI restriction sites, respectively, were used for further cloning (Supplemental Table S7). PCR was performed with GoTaq (Promega) and Pfu polymerases (Invitrogen; 10:1) under the following conditions: 1× GoTaq buffer, 1.5 mm MgCl2, 0.5 µm of each primer, and 0.5 mm of each deoxyribonucleotide triphosphate (dNTP), in a final volume of 25 μL. The amplified products were purified, cloned into pGEM-T-Easy vector (Promega), and sequenced. The KpnI-SalI fragments were further subcloned into pCS052_GFP_pCHF3 (a modified version of pCHF3; GFP coding sequence without the start codon is inserted into SalI-PstI sites), generating Pro35S:AtRPL10s-GFP constructs.
Full-length open reading frames for GFP were reamplified by PCR using pCS052_GFP_pCHF3 vector as a template under the same conditions described above using the primers F-SalI-GFP and R-PstI-GFP with the SalI and PstI restriction sites, respectively, for further cloning (Supplemental Table S7). PCR product was purified, cut with SalI and PstI restriction enzymes, purified again, and cloned into pCS052_GFP_pCHF3 that had been cut with the same enzyme and purified.
The Pro35S:GFP and Pro35S:AtRPL10s-GFP constructs were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation, and the transformation of Arabidopsis (Col-0) and rpl10B single mutants with the resulting bacteria was performed by the floral dip method (Clough and Bent, 1998). Transformed seedlings (T1) were identified by selection on solid MS medium containing kanamycin (50 mg L−1), and then the plants were transferred to soil. The presence of Pro35S:GFP and Pro35S:AtRPL10s-GFP transgenes in transformed plants was analyzed by PCR on the genomic DNA using the following combinations of primers: F-35Sprom and a reverse primer specific for each AtRPL10, and forward primers specific for each AtRPL10 and R-PstI-GFP (Supplemental Table S7; Supplemental Fig. S5, A and B). The expression of the AtRPL10s-GFP transgenes in transformed plants was analyzed by RT-qPCR using forward primers specific for each AtRPL10 and R-PstI-GFP (Supplemental Table S7; Supplemental Fig. S5C). PCR conditions were as follows: 1× GoTaq buffer, 3 mm MgCl2, 0.2 mm dNTP, 0.25 μm of each primer, 0.625 units of GoTaq (Promega), and sterile water added to obtain a volume of 25 μL. Cycling conditions were as follows: 2 min of denaturation at 95°C, followed by 35 cycles of 15 s of denaturation at 95°C, 20 s of annealing at 52°C, 1 min of amplification at 72°C, and finally, 7 min of amplification at 72°C. PCR products were separated on a 1% (w/v) agarose gel and stained with SYBR Green (Invitrogen). In each of these transgenic plants, the expression of the other AtRPL10 endogenous genes was analyzed by quantitative PCR as described below, with each transgenic plant showing no significant difference in transcript levels in comparison with wild-type plants (Supplemental Table S7; Supplemental Fig. S5D).
Subcellular Localization Analysis
Arabidopsis transgenic plants (Pro35S:AtRPL10s-GFP; T2 lines) were grown on MS agar plates for 20 d as described above. To visualize the nuclei, leaves were incubated with 2 µg mL−1 DAPI (Sigma) in phosphate-buffered saline buffer (10 mm sodium phosphate, 130 mm NaCl, pH 7.2) plus 0.1% (v/v) Tween for 15 min. The fluorescence of GFP and DAPI was visualized by confocal laser-scanning microscopy (Nikon C1) under water with a 40× objective. GFP and DAPI were excited using an argon laser at 488 nm and a UV laser at 395 nm, respectively. GFP emission was collected between 515 and 530 nm to avoid cross talk with chloroplast autofluorescence. To analyze the percentage of GFP/DAPI colocalization, each nucleus identified by DAPI fluorescence was examined for GFP fluorescence, and whenever GFP fluorescence was detected, the nucleus was scored as showing colocalization. Thus, the relative intensity of nuclear GFP fluorescence was not considered in these measurements. The percentage of GFP/DAPI was calculated by the relation between the number of nuclei showing GFP localization and the total number of nuclei identified by DAPI fluorescence. For each condition, more than 100 nuclei were analyzed (20 images) from five different plants using two independent T2 transgenic lines. The experiments were repeated at least three times.
Yeast Strains and Transformation
Saccharomyces cerevisiae AJY2104 strain (MATα kanMX6:GAL1:RPL10 ura-3 leu-2 ade-2 ade-3) has been described previously (Hofer et al., 2007). Yeast were grown in 1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) Gal, and 20 mg L−1 adenine. Yeast transformation was made following the Trafo method (Gietz and Woods, 2002), and transformants were selected on synthetic complete medium agar plates (Sherman et al., 1974) containing 2% (w/v) Gal without Leu (SCGal-L). Transformed colonies appeared within 3 to 4 d at 30°C.
Vector Constructs for Complementation Experiments in Yeast
For complementation studies, yeast expression plasmids were constructed in pRS415 (LEU-marked centromeric vector) and p5AX43 (LEU-marked vector) as follows. Full-length cDNAs for each AtRPL10 were amplified by PCR as described above using specific primers for each gene; the forward primers included the start codon and BamHI sites, while the reverse primers included the stop codons and the XhoI restriction site (Supplemental Table S7). The amplified products were purified, cloned into pGEM-T-Easy vector (Promega), generating pGEMT-AtRPL10s constructs, and sequenced. The BamHI-XhoI fragments were subcloned into p5AX43 plasmid, generating the p5AX43-ProGDP:AtRPL10s plasmids. The p5AX43 vector corresponds to a modified version of plasmid YEplac181 (Gietz and Sugino, 1988), in which the glyceraldehyde 3-phosphate dehydrogenase promoter (GDP) was inserted at the HindIII site. The S. cerevisiae RPL10 promoter was amplified by PCR using pAJ2522 plasmid (Hofer et al., 2007) as template and the F-SacI-ScRPL10prom and R-BamHI-ScRPL10prom oligonucleotides (Supplemental Table S7). PCR was performed with GoTaq (Promega) and Pfu polymerases (Invitrogen; 10:1) under the following conditions: 1× GoTaq buffer, 1.5 mm MgCl2, 0.5 µm of each primer, and 0.5 mm of each dNTP in a final volume of 25 μL. Cycling conditions were as follows: 5 min of denaturation at 95°C, followed by 35 cycles of 20 s of denaturation at 95°C, 30 s of annealing at 57°C, 45 s of amplification at 72°C, and finally, 7 min of amplification at 72°C. The PCR product was purified, cut with the corresponding restriction enzymes, cloned into the pRS415 vector, generating the pRS415-ProScRPL10 construct, and sequenced. Then, the AtRPL10s were inserted into this plasmid downstream of the ScRPL10 promoter as follows. pGEMT-AtRPL10s constructs were digested with BamHI and EcoRI restriction enzymes, then BamHI-EcoRI fragments were purified from the gels and ligated into the pRS415-ProScRPL10 construct cut with BamHI and EcoRI restriction enzymes. The resulting constructs, named pRS415-ProScRPL10:AtRPL10s, contain AtRPL10 coding sequences downstream of the ScRPL10 promoter. The full-length open reading frame for RPL10 from S. cerevisiae was amplified by PCR using the pAJ2522 plasmid (Hofer et al., 2007) as template and the F-BamHI-ScRPL10 and R-KpnI-ScRPL10 oligonucleotides as primers (Supplemental Table S7). PCR conditions were as described above for the ScRPL10 promoter amplification, with the following modifications: the annealing was at 54°C and the amplification was for 1 min at 72°C. The PCR products were purified, cut with BamHI and KpnI restriction enzymes, purified, and cloned into pRS415-ProScRPL10 vector, generating the pRS415-ProScRPL10:ScRPL10 plasmid.
Complementation Analysis
Complementation of the AJY2104 conditional lethal rpl10 mutant yeast with RPL10s from Arabidopsis was evaluated by replica plating to Leu− Glc plates. Single colonies were grown in liquid SCGal-L medium for 48 h at 30°C. An aliquot of the culture was harvested, washed three times with distilled water to remove the Gal medium, and the cells were then inoculated into water to give an initial optical density at 600 nm of 0.5 (this initial solution was considered as 10−1). Then, serial dilutions (10−2, 10−3, 10−4, and 10−5) were prepared, and 10 µL of each dilution was plated as drops on selective medium containing glucose (SCD)-L and SCGal-L plates. Colonies were examined for 7 d and photographed.
Yeast Extract Preparation and Protein Concentration Determination
Cells (2–3 × 108) were resuspended in 0.1 mL of buffer containing 40 mm Tris-HCl, pH 6.8, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 0.1% (v/v) β-mercaptoethanol, and 5% (w/v) SDS. Glass beads (425–600 μm; Sigma) were added, and six series of 1 min of vortex mixing alternating with 1 min of incubation on ice, were performed. Samples were centrifuged to pellet glass beads and cell debris. The supernatants were used for immunoblot analysis. Protein concentration was determined using Bradford reagent (Bio-Rad).
Immunoblot Analysis
For immunodetection, yeast extracts were subjected to 12% (w/v) SDS-PAGE, and proteins were electroblotted onto a nitrocellulose membrane for immunoblotting according to Burnette (1981). Affinity-purified rabbit polyclonal antibody raised against an N-terminal peptide of human QM was used for the detection of RPL10 (Santa Cruz Biotechnology). Bound antibody was visualized by linking to alkaline phosphatase-conjugated goat anti-rabbit IgG according to the manufacturer’s instructions (Bio-Rad).
AtRPL10 Promoter:GUS Expression in Transgenic Arabidopsis Plants
To make the AtRPL10s promoter:GUS constructs (ProAtRPL10s:GUS), the 5′ flanking DNAs of AtRPL10 coding regions were amplified by PCR with specific primers for each gene with SalI and BamHI restriction sites in the forward and reverse primers, respectively (Supplemental Table S7). The PCR fragments (1,401, 1,267, and 1,506 bp for AtRPL10A, AtRPL10B, and AtRPL10C, respectively) were cloned into the pGEM-T-Easy vector and sequenced. Then, these constructs were digested with SalI and BamHI restriction sites, and the products were purified from the gels and cloned into pBI101 binary vector, generating pBI101-ProAtRPL10s:GUS constructs. These plasmids were transformed into Col-0 plants as described above. Transgenic plants were selected on solid MS medium containing kanamycin (50 mg L−1). Three independent T3 transgenic lines were used for histochemical analysis. Samples were stained with 5-bromo-4-chloro-3-indolyl-d-glucuronide at 37°C for 24 h, followed by washes with ethanol, and kept in 50% (v/v) ethanol and 5% (v/v) acetic acid before being photographed. The experiments were repeated at least three times with similar results.
RT-qPCR
Tissues from three independent biological replicates were frozen in liquid nitrogen and stored at −80°C. Total RNA was isolated from 100 mg of tissue using the TRIzol reagent (Invitrogen). RNA was converted into first-strand cDNA using SuperScript II reverse transcriptase (Invitrogen) with oligo(dT) as a primer in previous treatment with DNase (Promega). The resultant cDNAswere used as a template for quantitative PCR amplification in a MiniOPTICON2 apparatus (Bio-Rad), using the intercalation dye SYBR Green I (Invitrogen) as a fluorescent reporter and Platinum Taq Polymerase (Invitrogen). Primers were designed to generate unique 150- to 250-bp fragments using the PRIMER3 software (Rozen and Skaletsky, 2000). Primers for CBP20 were used for normalization. All primer sequences are listed in Supplemental Table S7. Amplification conditions were as follows: 2 min of denaturation at 94°C; 40 to 45 cycles at 94°C for 15 s, 57°C for 20 s, and 72°C for 20 s; followed by 10 min at 72°C. Melting curves for each PCR were determined by measuring the decrease of fluorescence with increasing temperature (from 65°C to 98°C).To confirm the size of the PCR products and to check that they corresponded to unique and expected products, the final products were separated on a 2% (w/v) agarose gel.
Statistical Analysis
The data presented were analyzed using one-way ANOVA. Minimum significant differences were calculated by the Bonferroni, Holm-Sidak, Dunett, and Duncan tests (P < 0.05) using the Statgraphics Plus 5.0 Software.
Sequence data from this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: RPL10A, At1g14320; RPL10B, At1g26910; RPL10C, At1g66580; CAP-BINDING PROTEIN20, At5g44200.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Double mutants in RPL10 genes do not exhibit phenotypic differences with respect to rpl10 single mutants.
Supplemental Figure S2. Analysis of RPL10 transcript levels in single and double mutants with respect to wild-type plants analyzed by RT-qPCR.
Supplemental Figure S3. Alignment of amino acid sequences of Arabidopsis and S. cerevisiae RPL10 proteins.
Supplemental Figure S4. Immunoblot analysis of RPL10 proteins in complemented yeast.
Supplemental Figure S5. Presence and expression of the Pro35S:AtRPL10s-GFP transgenes in Arabidopsis transgenic plants.
Supplemental Figure S6. Subcellular localization of GFP in Arabidopsis.
Supplemental Figure S7. Subcellular localization of RPL10s from Arabidopsis after a 4-h UV-B treatment.
Supplemental Figure S8. Typical 2D gel of proteins from rpl10B mutant leaves after a 4-h UV-B treatment.
Supplemental Figure S9. Networks of associations for proteins that showed different levels in the rpl10B and rpl10C mutants in comparison with wild-type plants under UV-B stress.
Supplemental Figure S10. Relative expression levels of RPL10 transcripts during pollen development and germination.
Supplemental Table S1. Inheritance and segregation of rpl10A and rpl10B alleles.
Supplemental Table S2. Inheritance and segregation of rpl10A and rpl10C alleles.
Supplemental Table S3. Segregation analysis of selfed F2 progeny from rpl10B/+ rpl10C/+.
Supplemental Table S4. Transmission of mutant alleles through the male gamete analyzed by reciprocal crosses.
Supplemental Table S5. Proteins showing different levels in rpl10C mutant plants in comparison with wild-type plants under control conditions and after a 4-h UV-B treatment identified by mass spectrometry.
Supplemental Table S6. Proteins showing different levels in rpl10B mutant plants in comparison with wild-type plants under control conditions and after a 4-h UV-B treatment identified by mass spectrometry.
Supplemental Table S7. Primer sequences used for PCR.
Acknowledgments
We are grateful to Dr. Arlen W. Johnson (University of Texas) for providing the AJY2104 strain and the AJ2522 plasmid. We thank Dr. Julia Cricco (Instituto de Biología Molecular y Celular de Rosario-Consejo Nacional de Investigaciones Científicas y Técnicas) for the pRS415 vector, Dr. Javier Palatnik (Instituto de Biología Molecular y Celular de Rosario-Consejo Nacional de Investigaciones Científicas y Técnicas) for the pCS052_GFP_pCHF3 binary vector, Dr. Erich Grotewold (The Ohio State University) for the p5AX43 plasmid, Dr. Rodrigo Gomez (Centro de Estudios Fotosintéticos y Bioquímicos-Consejo Nacional de Investigaciones Científicas y Técnicas) for his excellent technical assistance with complementation analysis in yeast, and Virginia Fernandez (The Ohio State University) for generating two double mutants.
Glossary
- 2D
two-dimensional
- RT-qPCR
reverse transcription-quantitative PCR
- Col-0
Columbia
- DAPI
4′,6-diamino-phenylindole
- T-DNA
transfer DNA
- MS
Murashige and Skoog
- cDNA
complementary DNA
- dNTP
deoxyribonucleotide triphosphate
- SCGal-L
synthetic complete medium agar plates containing 2% (w/v) Gal without Leu
References
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J. (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415 [PMC free article] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA. (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgin DD, Zavala JA, Zhu J, Clough SJ, Ort DR, DeLucia EH. (2010) Biotic stress globally downregulates photosynthesis genes. Plant Cell Environ 33: 1597–1613 [DOI] [PubMed] [Google Scholar]
- Briggs GC, Osmont KS, Shindo C, Sibout R, Hardtke CS. (2006) Unequal genetic redundancies in Arabidopsis: a neglected phenomenon? Trends Plant Sci 11: 492–498 [DOI] [PubMed] [Google Scholar]
- Burnette WN. (1981) “Western blotting”: electrophoretic transfer of proteins from sodium dodecyl sulfate-polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem 112: 195–203 [DOI] [PubMed] [Google Scholar]
- Byrne ME. (2009) A role for the ribosome in development. Trends Plant Sci 14: 512–519 [DOI] [PubMed] [Google Scholar]
- Carroll AJ. (2013) The Arabidopsis cytosolic ribosomal proteome: from form to function. Front Plant Sci 4: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AJ, Heazlewood JL, Ito J, Millar AH. (2008) Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol Cell Proteomics 7: 347–369 [DOI] [PubMed] [Google Scholar]
- Carvalho CM, Santos AA, Pires SR, Rocha CS, Saraiva DI, Machado JPB, Mattos EC, Fietto LG, Fontes EP. (2008) Regulated nuclear trafficking of rpL10A mediated by NIK1 represents a defense strategy of plant cells against virus. PLoS Pathog 4: e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Zhang X, Burlingame AL, Walbot V. (2005) Analysis of leaf proteome after UV-B irradiation in maize lines differing in sensitivity. Mol Cell Proteomics 4: 1673–1685 [DOI] [PubMed] [Google Scholar]
- Chang IF, Szick-Miranda K, Pan S, Bailey-Serres J. (2005) Proteomic characterization of evolutionarily conserved and variable proteins of Arabidopsis cytosolic ribosomes. Plant Physiol 137: 848–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez-Rios R, Arias-Romero LE, Almaraz-Barrera MdJ, Hernández-Rivas R, Guillén N, Vargas M. (2003) L10 ribosomal protein from Entamoeba histolytica share structural and functional homologies with QM/Jif-1: proteins with extraribosomal functions. Mol Biochem Parasitol 127: 151–160 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Degenhardt RF, Bonham-Smith PC. (2008) Arabidopsis ribosomal proteins RPL23aA and RPL23aB are differentially targeted to the nucleolus and are disparately required for normal development. Plant Physiol 147: 128–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick FA, Trumpower BL. (1998) Heterologous complementation reveals that mutant alleles of QSR1 render 60S ribosomal subunits unstable and translationally inactive. Nucleic Acids Res 26: 2442–2448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdy SF, Lai KM, Weissman BE, Matsui Y, Hogan BL, Stanbridge EJ. (1991) The isolation and characterization of a novel cDNA demonstrating an altered mRNA level in nontumorigenic Wilms’ microcell hybrid cells. Nucleic Acids Res 19: 5763–5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisinger DP, Dick FA, Trumpower BL. (1997) Qsr1p, a 60S ribosomal subunit protein, is required for joining of 40S and 60S subunits. Mol Cell Biol 17: 5136–5145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone Ferreyra ML, Biarc J, Burlingame AL, Casati P. (2010a) Arabidopsis L10 ribosomal proteins in UV-B responses. Plant Signal Behav 5: 1222–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcone, Ferreyra MLF, Pezza A, Biarc J, Burlingame AL, Casati P. (2010b) Plant L10 ribosomal proteins have different roles during development and translation under ultraviolet-B stress. Plant Physiol 153: 1878–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer AA, Loftus TM, Mills AA, Sato KY, Neill JD, Tron T, Yang M, Trumpower BL, Stanbridge EJ. (1994) Extreme evolutionary conservation of QM, a novel c-Jun associated transcription factor. Hum Mol Genet 3: 723–728 [DOI] [PubMed] [Google Scholar]
- Fraire-Velázquez S, Rodríguez-Guerra R, Sánchez-Calderón L (2011) Abiotic and biotic stress response crosstalk in plants. In A Shanker, B Venkateswarlu, eds, Abiotic Stress Response in Plants: Physiological, Biochemical and Genetic Perspectives. Intech, Rijeka, Croatia, pp 3–26 [Google Scholar]
- Gerbasi VR, Weaver CM, Hill S, Friedman DB, Link AJ. (2004) Yeast Asc1p and mammalian RACK1 are functionally orthologous core 40S ribosomal proteins that repress gene expression. Mol Cell Biol 24: 8276–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P, Wilson D, Kreitler T, Lehrach H, Klose J, Gobom J, Fucini P. (2005) High heterogeneity within the ribosomal proteins of the Arabidopsis thaliana 80S ribosome. Plant Mol Biol 57: 577–591 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. (1988) New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74: 527–534 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Woods RA. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol 350: 87–96 [DOI] [PubMed] [Google Scholar]
- Guo J, Chen J-G. (2008) RACK1 genes regulate plant development with unequal genetic redundancy in Arabidopsis. BMC Plant Biol 8: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang S, Valerius O, Hall H, Zeng Q, Li JF, Weston DJ, Ellis BE, Chen JG. (2011) Involvement of Arabidopsis RACK1 in protein translation and its regulation by abscisic acid. Plant Physiol 155: 370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, West M, Johnson AW. (2005) Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J 24: 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer A, Bussiere C, Johnson AW. (2007) Mutational analysis of the ribosomal protein Rpl10 from yeast. J Biol Chem 282: 32630–32639 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D. (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiguchi G, Mollá-Morales A, Pérez-Pérez JM, Kojima K, Robles P, Ponce MR, Micol JL, Tsukaya H. (2011) Differential contributions of ribosomal protein genes to Arabidopsis thaliana leaf development. Plant J 65: 724–736 [DOI] [PubMed] [Google Scholar]
- Horiguchi G, Van Lijsebettens M, Candela H, Micol JL, Tsukaya H. (2012) Ribosomes and translation in plant developmental control. Plant Sci 191-192: 24–34 [DOI] [PubMed] [Google Scholar]
- Hwang JS, Goo TW, Yun EY, Lee JH, Kang SW, Kim KY, Kwon OY. (2000) Tissue-/stage-dependent expression of a cloned Bombyx mandarina QM homologue. Biomol Eng 16: 211–215 [DOI] [PubMed] [Google Scholar]
- Imai A, Komura M, Kawano E, Kuwashiro Y, Takahashi T. (2008) A semi-dominant mutation in the ribosomal protein L10 gene suppresses the dwarf phenotype of the acl5 mutant in Arabidopsis thaliana. Plant J 56: 881–890 [DOI] [PubMed] [Google Scholar]
- Johnson MA, von Besser K, Zhou Q, Smith E, Aux G, Patton D, Levin JZ, Preuss D. (2004) Arabidopsis hapless mutations define essential gametophytic functions. Genetics 168: 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiserli E, Jenkins GI. (2007) UV-B promotes rapid nuclear translocation of the Arabidopsis UV-B specific signaling component UVR8 and activates its function in the nucleus. Plant Cell 19: 2662–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Uemura M. (2003) Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36: 141–154 [DOI] [PubMed] [Google Scholar]
- Loftus TM, Nguyen YH, Stanbridge EJ. (1997) The QM protein associates with ribosomes in the rough endoplasmic reticulum. Biochemistry 36: 8224–8230 [DOI] [PubMed] [Google Scholar]
- Marty I, Brugidou C, Chartier Y, Meyer Y. (1993) Growth-related gene expression in Nicotiana tabacum mesophyll protoplasts. Plant J 4: 265–278 [DOI] [PubMed] [Google Scholar]
- Mills AA, Mills MJ, Gardiner DM, Bryant SV, Stanbridge EJ. (1999) Analysis of the pattern of QM expression during mouse development. Differentiation 64: 161–171 [DOI] [PubMed] [Google Scholar]
- Monteclaro FS, Vogt PK. (1993) A Jun-binding protein related to a putative tumor suppressor. Proc Natl Acad Sci USA 90: 6726–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee AK, Carp MJ, Zuchman R, Ziv T, Horwitz BA, Gepstein S. (2010) Proteomics of the response of Arabidopsis thaliana to infection with Alternaria brassicicola. J Proteomics 73: 709–720 [DOI] [PubMed] [Google Scholar]
- Mustroph A, Zanetti ME, Jang CJH, Holtan HE, Repetti PP, Galbraith DW, Girke T, Bailey-Serres J. (2009) Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. Proc Natl Acad Sci USA 106: 18843–18848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Kaminishi T, Takemoto C, Kawazoe M, Yoshida T, Tanaka A, Sugano S, Shirouzu M, Ohkubo T, Yokoyama S, et al. (2008) Crystal structure of human ribosomal protein L10 core domain reveals eukaryote-specific motifs in addition to the conserved fold. J Mol Biol 377: 421–430 [DOI] [PubMed] [Google Scholar]
- Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551–3567 [DOI] [PubMed] [Google Scholar]
- Rosado A, Li R, van de Ven W, Hsu E, Raikhel NV. (2012) Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc Natl Acad Sci USA 109: 19537–19544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In SA Krawetz, S Misener, eds, Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann JU. (2005) A gene expression map of Arabidopsis thaliana development. Nat Genet 37: 501–506 [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C (1974) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Singh K, Paul A, Kumar S, Ahuja PS. (2009) Cloning and differential expression of QM like protein homologue from tea [Camellia sinensis (L.) O. Kuntze]. Mol Biol Rep 36: 921–927 [DOI] [PubMed] [Google Scholar]
- Sormani R, Masclaux-Daubresse C, Daniel-Vedele F, Chardon F. (2011) Transcriptional regulation of ribosome components are determined by stress according to cellular compartments in Arabidopsis thaliana. PLoS ONE 6: e28070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Beckmann R, Eswar N, Penczek PA, Sali A, Blobel G, Frank J. (2001) Structure of the 80S ribosome from Saccharomyces cerevisiae: tRNA-ribosome and subunit-subunit interactions. Cell 107: 373–386 [DOI] [PubMed] [Google Scholar]
- Stirnberg P, Liu JP, Ward S, Kendall SL, Leyser O. (2012) Mutation of the cytosolic ribosomal protein-encoding RPS10B gene affects shoot meristematic function in Arabidopsis. BMC Plant Biol 12: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szakonyi D, Byrne ME. (2011) Ribosomal protein L27a is required for growth and patterning in Arabidopsis thaliana. Plant J 65: 269–281 [DOI] [PubMed] [Google Scholar]
- Szick-Miranda K, Bailey-Serres J. (2001) Regulated heterogeneity in 12-kDa P-protein phosphorylation and composition of ribosomes in maize (Zea mays L.). J Biol Chem 276: 10921–10928 [DOI] [PubMed] [Google Scholar]
- Turkina MV, Klang Årstrand H, Vener AV. (2011) Differential phosphorylation of ribosomal proteins in Arabidopsis thaliana plants during day and night. PLoS ONE 6: e29307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Lijsebettens M, Vanderhaeghen R, De Block M, Bauw G, Villarroel R, Van Montagu M. (1994) An S18 ribosomal protein gene copy at the Arabidopsis PFL locus affects plant development by its specific expression in meristems. EMBO J 13: 3378–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y, Shao J-Z, Pan X-X, Xiang L-X. (2005) Molecular cloning, characterization and expression analysis of QM gene from grass carp (Ctenopharyngodon idellus) homologous to Wilms’ tumor suppressor. Comp Biochem Physiol B Biochem Mol Biol 141: 356–365 [DOI] [PubMed] [Google Scholar]
- West M, Hedges JB, Chen A, Johnson AW. (2005) Defining the order in which Nmd3p and Rpl10p load onto nascent 60S ribosomal subunits. Mol Cell Biol 25: 3802–3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ. (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang J, Meng Q, Jiang T, Xie L, Wang Z, Zhang R. (2004) Molecular cloning and expression of a pearl oyster (Pinctada fucata) homologue of mammalian putative tumor suppressor QM. Mar Biotechnol (NY) 6: 8–16 [DOI] [PubMed] [Google Scholar]