Recent progress in the biochemistry and molecular biology of cuticle synthesis and function highlights major questions that will drive future research in this field.
Abstract
The plant cuticle is an extracellular hydrophobic layer that covers the aerial epidermis of all land plants, providing protection against desiccation and external environmental stresses. The past decade has seen considerable progress in assembling models for the biosynthesis of its two major components, the polymer cutin and cuticular waxes. Most recently, two breakthroughs in the long-sought molecular bases of alkane formation and polyester synthesis have allowed construction of nearly complete biosynthetic pathways for both waxes and cutin. Concurrently, a complex regulatory network controlling the synthesis of the cuticle is emerging. It has also become clear that the physiological role of the cuticle extends well beyond its primary function as a transpiration barrier, playing important roles in processes ranging from development to interaction with microbes. Here, we review recent progress in the biochemistry and molecular biology of cuticle synthesis and function and highlight some of the major questions that will drive future research in this field.
The first plant colonizers of land, approximately 450 million years ago in the mid-Paleozoic era, faced a daunting set of challenges associated with their new terrestrial environment, including desiccation, temperature extremes, gravity, and increased exposure to UV radiation (Waters, 2003; Leliaert et al., 2011). The transition from an exclusively aquatic to a terrestrial life style, therefore, would have necessitated the evolution of a toolbox of morphological and physiological features, some of which are apparent through studies of the fossil record or by examining extant plant lineages. For example, the development of architecturally complex cell walls for biomechanical support and structural protection, which typify modern land plants, can be traced back to divergence and radiation within the Charophycean green algae, their immediate ancestors (Sørensen et al., 2011). However, the most critical adaptive trait for survival during terrestrialization would have been the ability to retain water in increasingly dehydrating habitats. Consequently, the capacity to synthesize, deposit, and maintain a hydrophobic surface layer, or cuticle, over the surfaces of aerial organs was arguably one of the most important innovations in the history of plant evolution. This idea is borne out by both fossil evidence (Edwards, 1993) and the ubiquity of cuticles among all extant embryophytes, from bryophytes (Budke et al., 2012) to angiosperms.
Armed with a protective skin, together with a range of adaptive strategies for acquiring and conserving water, as well as for avoiding or tolerating water stress, embryophytes now thrive in a wide range of desiccating environments (Ogburn and Edwards, 2010; Aroca et al., 2012; Delaux et al., 2012; Jones and Dolan, 2012; Obata and Fernie, 2012; Gaff and Oliver, 2013). Accordingly, cuticles from a broad range of species, and in various ecological and agricultural contexts, have been studied from the perspective of their role as the primary barrier to transpirational water loss. However, it is now clear that cuticles play numerous other roles in plant development, physiology, and interactions with the abiotic environment and other organisms. Indeed, in recent years, there have been many instances of unexpected associations between the cuticle and diverse aspects of plant biology. In parallel, the past decade has seen considerable progress in understanding the biosynthesis of the major cuticle components and the complex regulatory networks that control cuticle synthesis and assembly.
This review summarizes recent progress in elucidating the biochemistry and molecular biology of cuticle synthesis and function and highlights some of the connections to other aspects of plant biology, including signaling, pathogen defense, and development. Given the broad scope and space limitation, not every aspect of cuticle biosynthesis is covered in depth, and recent specialized reviews focusing on cuticle biomechanical properties (Domínguez et al., 2011), defensive functions (Reina-Pinto and Yephremov, 2009), and transport barrier properties (Burghardt and Riederer, 2006) may be of further interest. In addition, key ongoing questions in the field are discussed, and potential future approaches to resolving those questions are suggested.
CUTICLE STRUCTURE, BIOSYNTHESIS, AND ASSEMBLY
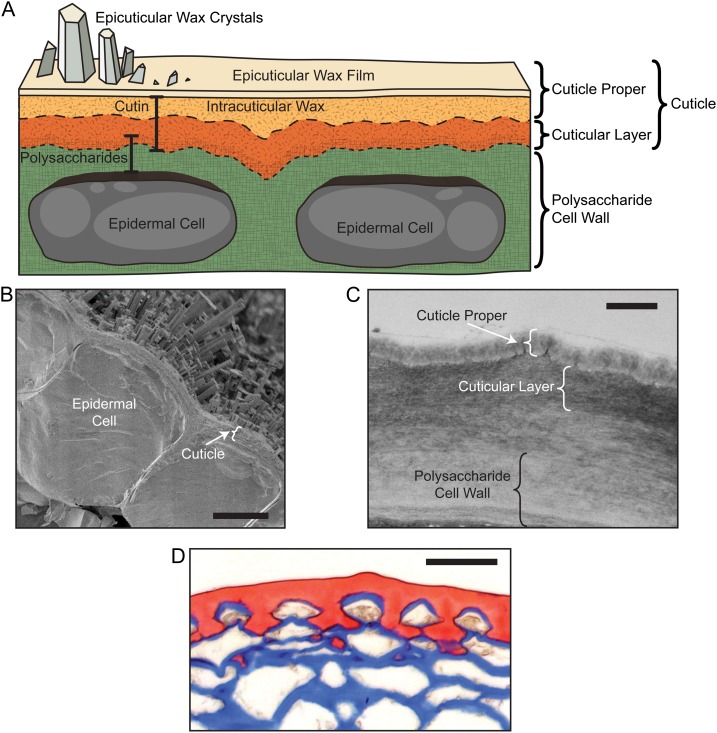
Plant cuticles are composite structures, composed of a covalently linked macromolecular scaffold of cutin and a variety of organic solvent-soluble lipids that are collectively termed waxes. Although the cuticle is usually considered independently from the underlying polysaccharide cell wall of the epidermis, the two structures are physically associated and have some overlapping functions. Indeed, the cuticle can be considered a specialized lipidic modification of the cell wall, just as lignification is a common modification of plant secondary cell walls. The microscopic structure of the cuticle is often divided into two domains based on histochemical staining and their presumed chemical composition: a cutin-rich domain with embedded polysaccharides, which is referred to as the “cuticular layer,” and an overlying layer that is less abundant in polysaccharides but enriched in waxes, referred to as the “cuticle proper” (Fig. 1A). The waxes are either deposited within the cutin matrix (intracuticular wax) or accumulate on its surface as epicuticular wax crystals, or films. These epicuticular waxes can confer distinct macroscopic surface properties: epicuticular films are responsible for the glossy appearance common to many leaves and fruits, while epicuticular wax crystals account for the dull, glaucous appearance of broccoli (Brassica oleracea) leaves and Arabidopsis (Arabidopsis thaliana) stems. Cuticle architectural organization can be discerned using a number of microscopic techniques. Scanning electron microscopy can reveal the elaborate and diverse morphologies of epicuticular wax crystals (Fig. 1B; Jeffree, 2006), while transmission electron microscopy shows the distinct patterning of interior layers of the cuticle, although this approach does not allow the visualization of wax structures (Fig. 1C). Cuticles vary considerably in their architecture and, depending on species and ontogeny, differ dramatically in hickness, from the nanometer to the micrometer scale (Jeffree, 2006). In the latter case, light microscopy can be used to elucidate the fine structures of the cuticle and epidermal cell wall (Fig. 1D), while histochemical staining coupled with confocal microscopy can further resolve three-dimensional cuticle architecture (Buda et al., 2009).
Figure 1.
Plant cuticle structure. A, Schematic diagram highlighting the major structural features of the cuticle and underlying epidermal cell layer (not drawn to scale). B, Scanning electron micrograph image of an Arabidopsis leaf epidermis and overlying cuticle, seen in cross section. Bar = 5 μm. (Image courtesy of Dr. Lacey Samuels.) C, Transmission electron micrograph image of an Arabidopsis stem epidermal cell wall and cuticle. Bar = 500 nm. (Image courtesy of Dr. Christiane Nawrath.) D, Light microscopy image showing the cuticle of a mature green-stage tomato fruit stained with Sudan Red and the polysaccharide cell walls stained with Alcian Blue. Bar = 50 μm.
Wax Biosynthesis
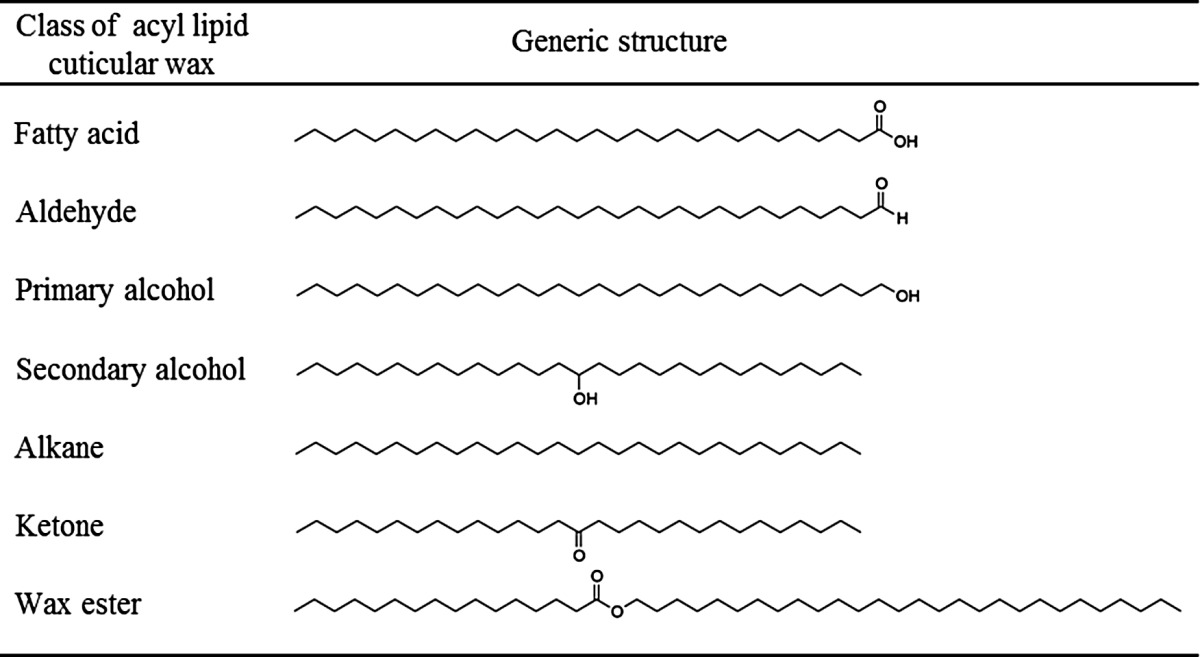
Wax composition can vary substantially with species, ontogeny, and environmental growth conditions (Jenks and Ashworth, 1999). In most cases, the majority of compounds comprising the cuticular wax are derived from very-long-chain fatty acids (VLCFAs; C20–C34), including alkanes, aldehydes, primary and secondary alcohols, ketones, and esters (Table I). In some species, various lipophilic secondary metabolites, such as pentacyclic triterpenoids, flavonoids, and tocopherols, can also be substantial components (Jetter et al., 2006). There has been impressive progress in revealing the molecular biology underlying VLCFA-derived wax biosynthesis, and to this end, Arabidopsis has provided an excellent experimental model (Bernard and Joubès, 2013). In addition to its well-known advantages as a genetic system, the presence of stem epicuticular wax crystals, which impart a glaucous appearance in the wild type, has enabled an easy screen for wax-deficient mutants. Such mutants, termed eceriferum (cer; Koornneef et al., 1989), typically exhibit a glossy stem phenotype, and it has primarily been through molecular analyses of these and other wax mutants that an increasingly complete pathway for acyl wax biosynthesis has been established.
Table I. Major acyl-lipid classes found in cuticular waxes.
Most classes occur as homologous series with broad distributions of chain lengths, compounds with typical average chain lengths are shown.
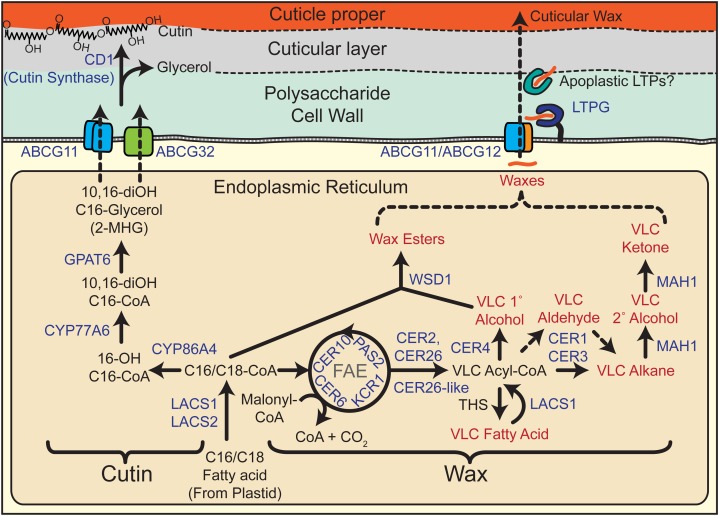
Wax biosynthesis begins with de novo C16 or C18 fatty acid biosynthesis in the plastid of epidermal cells (Fig. 2). These long-chain fatty acid compounds are converted to CoA thioesters by a long-chain acyl-coenzyme A synthase (LACS) isozyme and are ultimately transferred to the endoplasmic reticulum (ER). The mechanism of intracellular trafficking of fatty acid from the chloroplast to the ER remains unknown, although heterologous expression of Arabidopsis LACS1, LACS2, and LACS3 facilitates fatty acid uptake in yeast, suggesting that this class of enzymes may play dual roles in fatty acid trafficking and activation (Pulsifer et al., 2012). For reference, Table II provides a list of the corresponding genes, as well as others discussed in this review. The C16 acyl-CoA is then a substrate for the fatty acid elongase (FAE) complex. Through successive addition of two carbons per cycle derived from malonyl-CoA, the ultimate products of this complex are VLCFAs. The complex consists of four core subunits: β-ketoacyl-CoA synthase, β-ketoacyl-CoA reductase, β-hydroxyacyl-CoA dehydratase, and enoyl-CoA reductase. In Arabidopsis, 21 genes are predicted to encode β-ketoacyl-CoA synthase, and for wax biosynthesis, the most important gene, based on the mutant phenotype, is CER6 (Fiebig et al., 2000). Genes encoding the remaining subunits of the FAE complex, represented by KCR1, PAS2, and CER10, respectively, are less redundant, and their pleiotropic mutant phenotypes underscore the shared importance of the FAE in generating VLCFA precursors for sphingolipid biosynthesis (Zheng et al., 2005; Bach et al., 2008; Beaudoin et al., 2009). An additional family of proteins, composed of CER2, CER26, and CER26-like, appears to be required for the elongation of fatty acids to lengths greater than 28C (Haslam et al., 2012; Pascal et al., 2013). Curiously, these enzymes have sequence homology to BAHD acyltransferases, but conserved catalytic amino acid residues of this family of enzymes are dispensable for the elongation-promoting activity of CER2 (Haslam et al., 2012). The elongation cycles can be terminated by a thioesterase to form free VLCFAs, or the VLCFA-CoA esters can undergo further modifications.
Figure 2.
Cutin and wax biosynthetic pathways. Genes (blue text) are described in the review. Red text denotes compound classes that are typically observed in cuticular wax mixtures.
Table II. Cuticle-associated genes discussed in this review.
| Gene Symbol | Gene Name | Species | Locus Identifier | Description |
|---|---|---|---|---|
| ABCG11 | ATP-BINDING CASSETTE G11 | Arabidopsis | At1G17840 | ABC half transporter |
| ABCG13 | ATP-BINDING CASSETTE G13 | Arabidopsis | At1G51460 | ABC half transporter |
| ABCG32 | ATP-BINDING CASSETTE G32 | Arabidopsis | At2G26910 | ABC full transporter |
| BDG | BODYGUARD | Arabidopsis | At1G64670 | α/β-Hydrolase family protein |
| BDG3 | BODYGUARD3 | Arabidopsis | At4G24140 | Homolog of BDG |
| CD1 | CUTIN DEFICIENT1 | Tomato | Solyc11G006250 | Cutin synthase/hydroxyacylglycerol transesterase |
| CD2 | CUTIN DEFICIENT2 | Tomato | Solyc01G091630 | Homeodomain-Leu zipper IV transcription factor |
| CER1 | ECERIFERUM1 | Arabidopsis | At1G02205 | Involved in alkane formation |
| CER10 | ECERIFERUM10 | Arabidopsis | At3G55360 | Enoyl-CoA reductase |
| CER2 | ECERIFERUM2 | Arabidopsis | At4G24510 | Required for C28 to C30 elongation of fatty acids |
| CER26 | ECERIFERUM26 | Arabidopsis | At4G13840 | Homolog of CER2, required for elongation of fatty acids greater than C30 |
| CER26-like | ECERIFERUM26-like | Arabidopsis | At3G23840 | Homolog of CER2 and CER26 |
| CER3 | ECERIFERUM3 | Arabidopsis | At5G57800 | Involved in alkane formation |
| CER4 | ECERIFERUM1 | Arabidopsis | At4G33790 | VLCFA-CoA by fatty acyl-CoA |
| CER5/ABCG12 | ECERIFERUM5/ATP-BINDING CASSETTE G12 | Arabidopsis | At1G51500 | ABC half transporter |
| CER6 | ECERIFERUM6 | Arabidopsis | At1G68530 | β-Ketoacyl-CoA synthase |
| CER7 | ECERIFERUM7 | Arabidopsis | At3G60500 | Exosomal exoribonuclease |
| CER9 | ECERIFERUM9 | Arabidopsis | At4G34100 | Putative E3 ubiquitin ligase |
| CFL1 | CURLY FLAG LEAF1 | Rice | Os02G31140 | WW domain-containing protein |
| CYP77A6 | CYP77A6 | Arabidopsis | At3G10570 | CYP77A subfamily of cytochrome P450 |
| CYP86A4 | CYP86A4 | Arabidopsis | At1G01600 | CYP86A subfamily of cytochrome P450 |
| DCR | DEFECTIVE IN CUTICULAR RIDGES | Arabidopsis | At5G23940 | BAHD acyltransferase |
| FDH | FIDDLEHEAD | Arabidopsis | At2G26250 | β-Ketoacyl-CoA synthase |
| GPAT6 | GLYCEROL-3-PHOSPHATE SN-2-ACYLTRANSFERASE6 | Arabidopsis | At2G38110 | Bifunctional glycerol-3-phosphate sn-2-acyltransferase/phosphatase |
| HDG1 | HOMEODOMAIN GLABROUS1 | Arabidopsis | At3G61150 | Homeodomain-Leu zipper IV transcription factor |
| HTH | HOTHEAD | Arabidopsis | At1G72970 | Glc-methanol-choline oxidoreductase family protein |
| IRG1 | INHIBITOR OF RUST GERM TUBE DIFFERENTIATION1 | M. truncatula | Medtr5G014400 | C2H2 zinc finger transcription factor |
| KCR1 | β-KETOACYL-COENZYME A REDUCTASE1 | Arabidopsis | At1G67730 | β-Ketoacyl-CoA reductase |
| LACS1 | LONG-CHAIN ACYL-COENZYME A SYNTHASE1 | Arabidopsis | At2G47240 | Long-chain acyl-CoA synthase |
| LACS2 | LONG-CHAIN ACYL-COENZYME A SYNTHASE2 | Arabidopsis | At1G49430 | Long-chain acyl-CoA synthase |
| LACS3 | LONG-CHAIN ACYL-COENZYME A SYNTHASE3 | Arabidopsis | At1G64400 | Long-chain acyl-CoA synthase |
| LCR | LACERATA | Arabidopsis | At2G45970 | CYP86A subfamily of cytochrome P450 |
| LTL1 | LI-TOLERANT LIPASE1 | Arabidopsis | At3G04290 | Homolog of CD1 |
| LTPG1 | GPI-ANCHORED LIPID TRANSFER PROTEIN1 | Arabidopsis | At1G27950 | GPI-anchored lipid transfer protein |
| LTPG2 | GPI-ANCHORED LIPID TRANSFER PROTEIN2 | Arabidopsis | At3G43720 | GPI-anchored lipid transfer protein |
| MAH1 | MIDCHAIN ALKANE HYDROXYLASE1 | Arabidopsis | At1G57750 | CYP96A subfamily of cytochrome P450 |
| MYB106 | Myb DOMAIN PROTEIN106 | Arabidopsis | At3G01140 | Myb transcription factor |
| MYB16 | Myb DOMAIN PROTEIN16 | Arabidopsis | At5G15310 | Myb transcription factor |
| MYB30 | Myb DOMAIN PROTEIN30 | Arabidopsis | At3G28910 | Myb transcription factor |
| MYB41 | Myb DOMAIN PROTEIN41 | Arabidopsis | At4G28110 | Myb transcription factor |
| MYB96 | Myb DOMAIN PROTEIN96 | Arabidopsis | At5G62470 | Myb transcription factor |
| OCL1 | OUTER CELL LAYER1 | Maize | GRMZM2G026643 | Homeodomain-Leu zipper IV transcription factor |
| PAS2 | PASTICCINO2 | Arabidopsis | At5G10480 | β-Hydroxyacyl-CoA dehydratase |
| SHN2 | SHINE2 | Arabidopsis | At5G11190 | Homolog of WIN1/SHN1 |
| WIN1/ SHN1 | WAX INDUCER1/SHINE1 | Arabidopsis | At1G15360 | AP2 domain-containing transcription factor |
| WSD1 | WAX SYNTHASE/ACYL-COENZYME A:DIACYLGLYCEROL ACYLTRANSFERASE1 | Arabidopsis | At5G37300 | Wax synthase/acyl-CoA:diacylglycerol acyltransferase family protein |
| WXP1 | WAX PRODUCTION1 | M. truncatula | Medtr5G062700 | AP2 domain-containing transcription factor |
| – | – | Arabidopsis | At5G33370 | Homolog of CD1 |
Primary alcohols can be produced from VLCFA-CoA by fatty acyl-CoA reductase, an enzyme encoded by CER4 in Arabidopsis (Rowland et al., 2006). Free primary alcohols can occur in the wax mixture, or they can be esterified to a fatty acid in order to form wax esters. In this case, the alcohol is coupled to an acyl group derived from fatty acyl-CoA. The Arabidopsis enzyme responsible for this is WSD1, an enzyme of the wax synthase/diacylglycerol acyltransferase family (Li et al., 2008).
A second branch of acyl wax biosynthesis leads to the formation of aldehydes and, ultimately, alkanes. Interestingly, in Arabidopsis, LACS1, which is also required for C16 cutin monomer biosynthesis, appears to have an additional specificity for C30 VLCFA and is required for the normal accumulation of downstream wax compounds (Lü et al., 2009). This suggests that conversion of an intracellular pool of free VLCFA back to VLCFA-CoA is an important route to aldehyde and alkane biosynthesis, rather than VLCFA-CoA directly derived from FAE. A long unresolved question in wax biosynthesis is the enzymatic basis of alkane synthesis. Classical biochemistry, using crude extracts from pea (Pisum sativum), indicated that the reaction likely occurs via the reduction of VLCFA-CoA to an aldehyde intermediate followed by decarbonylation, yielding an alkane that is 1C shorter (Cheesbrough and Kolattukudy, 1984; Schneider-Belhaddad and Kolattukudy, 2000). Although this enzyme was not purified and identified, compelling evidence was recently obtained, through studies of Arabidopsis, that CER1 and CER3 in complex act together to catalyze the formation of alkanes from VLCFA-CoA. It was shown by a split ubiquitin yeast two-hybrid assay and an Arabidopsis split luciferase assay that CER1 interacts with CER3 as well as several isoforms of cytochrome b5. Furthermore, heterologous expression of the combination of CER1, CER3, a cytochrome b5, and LACS1 in yeast resulted in the formation of very-long-chain alkanes (Bernard et al., 2012). This strongly suggests that a complex including CER1 and CER3 with cytochrome b5 as an electron donor catalyzes the reduction and decarbonylation of VLCFA-CoA in order to form cuticular alkanes. Aside from being a major component of the wax mixture, alkanes can undergo further modification to form secondary alcohols and ketones. In Arabidopsis, both of these oxidations are performed by the cytochrome P450 enzyme midchain alkane hydroxylase (MAH1; Greer et al., 2007).
Synthesis of Cutin Precursors
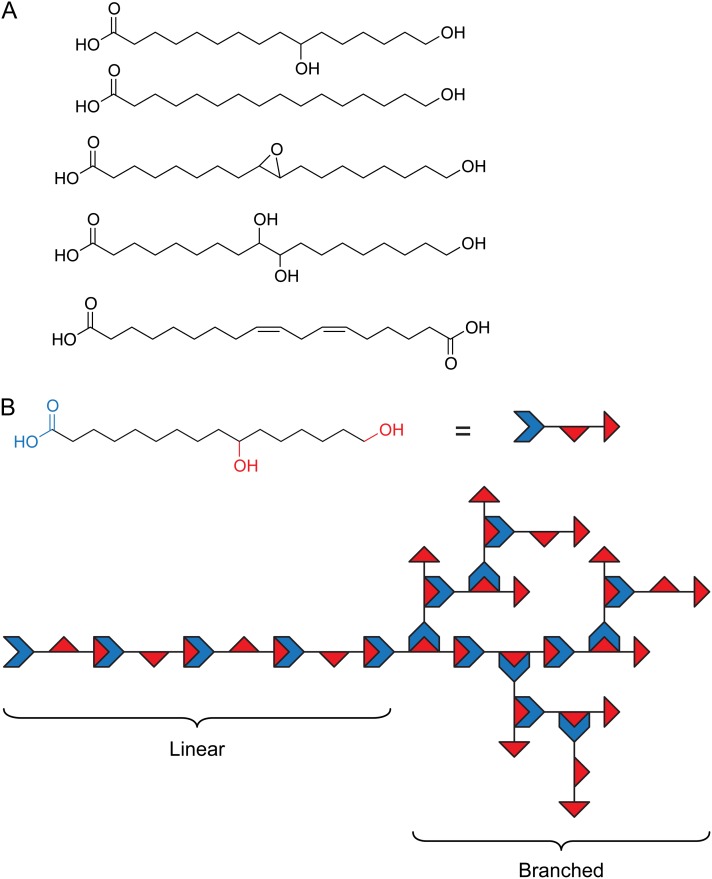
Cutin is typically composed of interesterified hydroxy fatty acids, with lesser amounts of glycerol, phenylpropanoids, and dicarboxylic acids (Kolattukudy, 2001). Chemical processes that cleave ester bonds, such as saponification, readily release these monomeric constituents, although in some species an additional lipidic polymer, referred to as cutan, remains recalcitrant to such treatments. Cutan is rich in ether and C-C bonds, but its structure is otherwise unknown, and it appears to be restricted to relatively few extant species (Gupta et al., 2006). The hydroxy fatty acids of cutin are typically ω-hydroxy fatty acids, usually with one or two additional midchain hydroxyl groups or an epoxy group (Fig. 3A). Despite extensive surveys of the chemical composition of plant cutins in the 1970s and 1980s (Kolattukudy, 2001), the composition of Arabidopsis cutin was not determined until relatively recently (Bonaventure et al., 2004; Franke et al., 2005). It is important to note that, in this important model species, the cutin of stems and leaves is atypical in that its major component is a dicarboxylic acid (Fig. 3A), implying that the predominant structural motif must be a copolymer with an unknown polyhydroxy compound, presumably glycerol (Pollard et al., 2008). However, despite the atypical composition of its cutin, Arabidopsis has proven to be an important model for deciphering the pathway of cutin biosynthesis, and more recently, it was discovered that the cutin of its floral organs is more typical, in that it is composed primarily of 10,16-dihydroxyhexadecanoic acid (Li-Beisson et al., 2009).
Figure 3.
Typical cutin monomers and polymeric structure. A, Some typical C16 and C18 fatty acid-derived cutin monomers. From top to bottom: 10,16-dihydroxyhexadecanoic acid, 16-hydroxyhexadecanoic acid, 9,10-epoxyoctadecanoic acid, 9,10,18-trihydroxyoctadecanoic acid, and octadeca-cis-6,cis-9-diene-1,18-dioate, the major cutin monomer of Arabidopsis stems and leaves. B, Linear and branched domains made possible by different ester linkages of 10,16-dihydroxyhexadecanoic acid, depicted schematically as indicated.
While there is considerable diversity in the structure of cutin monomers, the pathway for the biosynthesis of 10,16-dihydroxyhexadecanoic acid-based cutin is the most complete, and the major themes of cutin biosynthesis are likely shared for other cutin monomers. Here, we summarize this pathway based on recent molecular genetic and biochemical studies using Arabidopsis and tomato (Solanum lycopersicum).
Intracellular Acyltransferases and Hydroxylases
The biosynthesis of cutin begins with de novo fatty acid synthesis in the plastid of epidermal cells (Fig. 2). The next three steps occur in the ER and consist of ω-hydroxylation and midchain hydroxylation and the synthesis of an acyl-CoA intermediate. The relative order of these steps is not known, although it has been shown that the ω-hydroxylation precedes the midchain hydroxylation and that the final product of these steps is most likely a dihydroxyhexadecanoic acid-CoA ester (Li-Beisson et al., 2009). The ω-hydroxylase is encoded by members of the CYP86 subfamily of cytochrome P450s (CYP86A4 in Arabidopsis flowers; Li-Beisson et al., 2009), while the midchain hydroxylase is encoded by the CYP77 subfamily (CYP77A6 in Arabidopsis flowers; Li-Beisson et al., 2009). The acyltransferases that synthesize acyl-CoA are encoded by the LACS family, which consists of nine members in Arabidopsis, and both LACS1 and LACS2 appear to be responsible for C16 cutin monomer biosynthesis (Lü et al., 2009).
An additional intracellular acyltransferase required for the synthesis of cutin polyester is a glycerol 3-phosphate acyltransferase (GPAT). Recently, it was shown that plants possess a unique subfamily of bifunctional GPATs encoding enzymes with both sn-2-specific glycerol-3-phosphate:acyl-CoA acyltransferase activity as well as phosphatase activity, yielding a 2-monoacylglyceryl ester (Yang et al., 2010). In the case of Arabidopsis floral cutin, this activity is encoded by GPAT6 (Li-Beisson et al., 2009). Although the specific sequence of all intracellular biosynthetic steps will require additional characterization of the substrate specificity of each enzyme, biochemical characterization of Arabidopsis bifunctional GPATs indicates that they have a strong preference for ω-hydroxylated acyl-CoA, suggesting that hydroxylation precedes the transfer to glycerol (Yang et al., 2012). In any case, the ultimate product of the intracellular steps of cutin biosynthesis is likely to be 2-monoacylglyceryl esters of cutin monomers. In the case of 10,16-dihydroxyhexadecanoic acid-based cutin, this is 2-mono(10,16)-dihydroxyhexadecanoyl glycerol (2-MHG).
Transport of Cuticle Precursors
After the synthesis of wax and cutin precursors, they are exported from the ER, across the plasma membrane, through the polysaccharide cell wall, and to the nascent cuticular membrane. Most of these transport processes are poorly understood, although trafficking of both wax and cutin precursors across the plasma membrane has been shown to depend on ATP-binding cassette (ABC) transporters. In Arabidopsis, CER5/ABCG12 (Pighin et al., 2004) and ABCG11 (Bird et al., 2007) are required for wax export. Both of these encode half transporters, and based on double mutant analysis and bimolecular fluorescence complementation analyses, it has been suggested that an ABCG11/ABCG12 heterodimer is required for wax secretion (McFarlane et al., 2010). ABCG11 is also required for cutin accumulation, and since it is also able to dimerize with itself, it has been proposed that this homodimer is the functional complex responsible for cutin export (McFarlane et al., 2010). Additionally, a third Arabidopsis half transporter, ABCG13, was shown to be required for cutin deposition in flowers (Panikashvili et al., 2011).
More recently, full transporters required for cutin deposition were identified in Arabidopsis (ABCG32; Bessire et al., 2011) as well as wild barley (Hordeum spontaneum) and rice (Oryza sativa; Chen et al., 2011). Despite the clear genetic evidence supporting a role for ABC transporters in cuticular lipid export, the substrate specificity of these transporters has not yet been demonstrated in vitro. However, all the ABC transporters that have been implicated in cuticle biosynthesis to date are members of the ABCG subfamily, which has been associated with the transport of lipids and hydrophobic compounds in other systems (Moitra et al., 2011). Moreover, in several cases, intracellular lipidic inclusions were observed in ABC transporter mutants, further supporting their direct involvement in cuticular lipid export (Pighin et al., 2004; Bird et al., 2007; Bessire et al., 2011).
Export of some wax compounds also appears to be facilitated by glycosylphosphatidylinositol (GPI)-anchored lipid-transfer proteins (LTPs), LTPG1 and LTPG2, which are bound to the extracellular side of the plasma membrane (Debono et al., 2009; Lee et al., 2009; Kim et al., 2012). These proteins represent a unique class of LTPs, a family of small and typically soluble proteins that bind a variety of lipid substrates in vitro (Yeats and Rose, 2008). A major remaining question is how hydrophobic cuticle precursors are transported across the hydrophilic environment of the polysaccharide cell wall to the cuticle. Apoplastic LTPs have been proposed to play a role, although genetic or biochemical evidence for their involvement in transport is generally lacking (Yeats and Rose, 2008). In the case of the dihydroxyacyl cutin precursor 2-MHG, the glycerol moiety imparts sufficient polarity to allow aqueous solubility at low millimolar concentrations (Yeats et al., 2012b). This suggests that lipid-binding proteins or other factors are not necessary in order to facilitate the transport of this major precursor of cutin biosynthesis. However, the solubility of glyceryl esters of less polar cutin monomers has not been investigated, and they, along with waxes, may require additional factors to increase their solubility in the apoplast.
Cutin Polymerization
The final step of cutin synthesis is incorporation of the hydroxyacyl monomer into the polymer, but the molecular mechanism of cutin polymerization has been a longstanding enigma. Recent progress in this area was achieved by studying the tomato mutant cutin deficient1 (cd1) and transgenic tomato plants in which CD1 expression was suppressed using an RNA interference strategy (Girard et al., 2012; Yeats et al., 2012b). The cd1 mutant exhibits a severe reduction in the amount of polymerized cutin in the fruit cuticle (Isaacson et al., 2009), although chemical analysis indicated that, unlike wild-type fruit, those of the mutant accumulate nonpolymerized 2-MHG (Yeats et al., 2012b). Cloning of the mutated gene revealed that it encodes a protein of the GDSL-motif lipase/hydrolase (GDSL) family, which localizes to the developing cuticle (Girard et al., 2012; Yeats et al., 2012b). Despite its similarity to lipolytic enzymes, the recombinant protein acts as an acyltransferase in vitro, forming polyester oligomers from 2-MHG (Yeats et al., 2012b).
The identification of CD1 as the first known cutin synthase raises several questions about the specificity and generality of the reaction that it catalyzes. Phylogenetic analysis of CD1 and homologous genes indicates that despite belonging to a very large gene family, the subfamily of GDSLs represented by CD1 is relatively small and well conserved, with sequences represented across diverse taxa of land plants (Volokita et al., 2011). In Arabidopsis, its putative orthologs form a five-member gene family, and silencing of the expression of two of these (LTL1 and At5g33370) resulted in plants exhibiting floral organ fusions and lacking nanoridges on the petal surface, phenotypes that are consistent with a cutin deficiency (Shi et al., 2011). An additional putative ortholog of CD1 from Agave americana exhibited similar localization and expression, further supporting a conserved mechanism of CD1-like enzymes acting as cutin synthases (Reina et al., 2007). Despite the presence of a null allele, the cd1 mutant is not completely deficient in cutin, so the identity of additional cutin synthases, or perhaps nonenzymatic mechanisms of cutin synthesis, represents an intriguing line of future research.
The polymeric structure of cutin is not well understood. Monomeric composition can provide a “parts list,” but the relative abundance of possible linkages in the polymer is difficult to determine, largely due to the difficulty of solubilizing intact cutin (Serra et al., 2012). Nevertheless, the multiple functionalities present in many cutin monomers suggests that native cutin polymers can range from linear to branched or cross-linked structures (Pollard et al., 2008). For example, in an idealized cutin polymer composed exclusively of 10,16-dihydroxyhexadecanoic acid, the monomers can be joined by esterification of either the terminal or midchain hydroxyl group. Esterification of a single hydroxyl would result in a linear polymer, while esterification of both hydroxyl groups would generate branched structures (Fig. 3B). The identification of the hydroxyl groups that are esterified by CD1 and other cutin synthases should indicate whether the regiospecificity of cutin polymerization is enzymatically controlled and whether specific cutin synthases catalyze the formation of linear or branched domains of the cutin polymer. Moreover, it is not known how branching or cross linking of cutin affects cuticle function, and the identification of additional cutin synthases will allow this to be investigated using genetic approaches.
REGULATION OF CUTICLE BIOSYNTHESIS
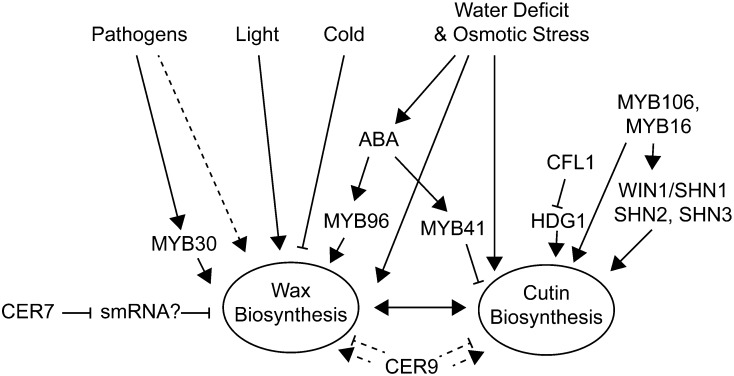
The regulation of cuticle biosynthesis is complex and involves interacting signaling networks associated with environmental stress responses, pathogen responses, and feedback regulation based on the structure and integrity of the cuticle itself. Furthermore, as the cuticle is exclusively synthesized by epidermal cells, the regulation of epidermis identity during development can also be considered to play a regulatory role in cuticle development. This is covered in more depth in an excellent review by Javelle et al. (2011), and we focus here only on direct regulators of cutin and wax biosynthesis (Fig. 4). Even within this restricted context, the analysis of regulatory mutants is complicated by compensatory mechanisms between cutin and wax biosynthesis and other pleiotropic phenotypes. Nevertheless, a complex regulatory network that responds to developmental and environmental cues, mediated by hormones, transcription factors, and posttranscriptional regulation, is beginning to emerge.
Figure 4.
Regulation of cuticle biosynthesis. A summary of the interaction of environmental factors and regulatory genes that are known to influence cutin or wax biosynthesis is shown.
Environment and Hormones
A systematic analysis of both cuticle composition and gene expression in Arabidopsis indicates that wax synthesis is induced by water deficit, sodium chloride, and abscisic acid (ABA) treatments (Kosma et al., 2009). In contrast, cutin biosynthesis was reported only to be induced by water deficit and not ABA or sodium chloride, suggesting that, at least in Arabidopsis, the detection of various osmotic stresses is complex and only partially dependent on ABA (Kosma et al., 2009). However, given that ABA is already well established as a regulator of plant responses to water deficit through the regulation of stomatal aperture (Lee and Luan, 2012), ABA regulation of cuticle biosynthesis is an intriguing area for further research aimed at understanding and engineering drought tolerance in crops.
In addition, dark and cold treatments have been shown to reduce the expression of several components of the FAE complex (Hooker et al., 2002; Joubès et al., 2008). Several wax biosynthetic genes have been shown to be induced by bacterial pathogens (Raffaele et al., 2008) and during infestation of wheat (Triticum aestivum) by the Hessian fly (Mayetiola destructor; Kosma et al., 2010), but in general, the relevance of the induction of cuticle synthesis to pest or pathogen resistance is poorly understood.
Transcription Factors and Cuticle Biosynthesis
The first transcription factor gene identified as having a role in regulating cuticle biosynthesis was the AP2 domain-containing WAX INDUCER1/SHINE1 (WIN1/SHN1; Aharoni et al., 2004; Broun et al., 2004). Overexpression of this gene led to glossy leaves with a greater wax load than the wild type and lower transpiration, although this was likely due to a reduced density of stomata rather than the wax phenotype (Aharoni et al., 2004). Later studies indicated that cutin levels are also increased in WIN1/SHN1-overexpressing plants and that the up-regulation of genes encoding cutin biosynthetic enzymes precedes the induction of wax biosynthetic genes (Kannangara et al., 2007). WIN1/SHN1 is part of a three-member gene family in Arabidopsis, and silencing of all three genes led to a reduction in the amount of cutin but not waxes (Shi et al., 2011). These authors also demonstrated that these transcription factors directly activate promoters of several cutin biosynthetic genes, further supporting a primary role in cutin regulation with a downstream effect on wax biosynthesis (Shi et al., 2011). In addition to regulating cutin biosynthesis, the SHN transcription factors also induced the expression of several pectin-modifying enzymes, suggesting a coordination of the synthesis of the cuticle with the polysaccharide cell wall (Shi et al., 2011). This second function of SHN transcription factors in regulating the polysaccharide cell wall is further suggested by experiments in which the overexpression of Arabidopsis SHN2 in rice resulted in a significant increase in the amount of cellulose and a concomitant decrease in lignin (Ambavaram et al., 2011). On the other hand, a general role of WIN1/SHN1-related transcription factors in the regulation of cutin synthesis is indicated by studies of orthologous genes in barley (Hordeum vulgare; Taketa et al., 2008) and tomato (Shi et al., 2013). The balance of evidence thus suggests that SHN transcription factors coordinate not just the synthesis of cutin but also the polysaccharide cell wall of the epidermis. This ultimately highlights the fact that the cuticle is a specialized modification of the cell wall, and like other modifications, such as lignification or suberization, it should be considered within the context of polysaccharide cell wall components. Aside from the SHN family, other AP2 domain transcription factors from different clades may also play a role in cuticle regulation. For example, overexpression of WXP1 from Medicago truncatula in alfalfa (Medicago sativa) induced wax production (Zhang et al., 2005).
Recently, two transcription factors, MYB106 and MYB16, were identified as regulators of cuticle biosynthesis that function in a similar manner to WIN1/SHN1 (Oshima et al., 2013). They both appear to act upstream of, and directly activate, WIN1/SHN1 but also some cuticle biosynthetic genes (Oshima et al., 2013). Several other transcription factors of the MYB family have also been implicated in the regulation of wax and cutin biosynthesis in response to environmental stresses. MYB30 is induced during infection by bacterial pathogens, leading to the up-regulation of several genes of the FAE complex, and ectopic overexpression of MYB30 leads to an increased wax load (Raffaele et al., 2008). MYB96 was identified as an ABA-inducible transcription factor that mediates drought tolerance (Seo et al., 2009), in part due to an induction of wax biosynthesis resulting from MYB96 directly activating the promoters of several wax synthesis genes (Seo et al., 2011). While MYB96 positively regulates wax production in response to stress, MYB41 mediates the negative regulation of cutin biosynthesis in response to similar stresses. MYB41 is induced by ABA, drought, and osmotic stress, leading to the down-regulation of cutin biosynthesis genes and the disruption of cuticle structure (Cominelli et al., 2008).
Another regulatory factor was identified through characterization of the rice CURLY FLAG LEAF1 (CFL1) gene, which encodes a WW domain-containing protein that negatively regulates cuticle biosynthesis. Studies of the orthologous CFL1 gene in Arabidopsis indicated that it down-regulates cutin biosynthesis by suppressing the function of HDG1, a homeodomain-leucine zipper IV transcription factor (HD-ZIP IV), which has been shown to induce the expression of several cutin biosynthesis genes (Wu et al., 2011). A more general role of HD-ZIP IV proteins in regulating cutin synthesis is further suggested by the homologous tomato gene CD2, which is required for the biosynthesis of cutin in the fruit and other organs (Isaacson et al., 2009; Nadakuduti et al., 2012). In maize (Zea mays), the HD-ZIP IV gene OUTER CELL LAYER1 (OCL1) was shown to be an epidermis-specific positive regulator of wax biosynthesis, although cutin was not quantified in plants overexpressing this gene (Javelle et al., 2010). Interestingly, HD-ZIP IV proteins have also been implicated in regulating other epidermis-specific processes, such as trichome differentiation and the formation of root hairs and stomatal guard cells (Masucci et al., 1996; Nakamura et al., 2006; Takada et al., 2013). Given their additional association with cuticle biosynthesis, it appears that a common feature of members of this protein family is playing key roles in the biology of the plant epidermis and the determination of epidermal cell fate.
Beyond Transcription Factors
In addition to the network of transcription factors that regulate cuticle biosynthesis, regulatory mechanisms that do not involve direct transcriptional activation or repression by promoter binding have recently been discovered. A recent example resulted from studies of the Arabidopsis cer9 mutant, which exhibits alterations in the amount and composition of leaf and stem waxes. Cloning of the CER9 gene revealed it to encode a protein with sequence similarity to yeast Doa10, an E3 ubiquitin ligase involved in ER-associated degradation of misfolded proteins (Lü et al., 2012). Given the ER localization of wax and cutin biosynthetic processes, the authors proposed a role for CER9 in the homeostasis of key cuticle biosynthetic enzyme levels. Experiments further addressing this hypothesis will be particularly interesting, given the surprising finding that the cer9 mutant actually exhibits enhanced drought tolerance and water use efficiency (Lü et al., 2012).
One of the most intriguing mechanisms of cuticle regulation resulted from characterization of the cer7 mutant. CER7 encodes an exosomal exoribonuclease, and the cer7 mutant exhibits reductions in stem wax and transcription of CER3, a major wax biosynthetic enzyme (Hooker et al., 2007). Recently, two suppressors of cer7 that restore the CER3 transcript and stem wax levels were identified, and cloning of the respective genes identified RDR1 and SGS3, two conserved components of the RNA-mediated gene-silencing pathway (Lam et al., 2012). A model was proposed wherein CER7 is involved in the degradation of a small RNA species that negatively regulates the CER3 transcript. Future work involving the identification of such a small RNA species and other components of this pathway will be especially intriguing, since no known plant small RNA species mapped to the CER7-dependent region of the CER3 promoter (Lam et al., 2012).
ENIGMATIC FACTORS IN CUTICLE BIOSYNTHESIS
In addition to the characterized components of cuticle biosynthesis that can be incorporated into a coherent model, as discussed above, several genes/proteins have been identified that are required for cuticle formation but that lack a clear associated biochemical function that would place them in a specific point in the pathways. One example is HOTHEAD (HTH), a Glc-methanol-choline oxidoreductase family protein that is required for proper cuticle organization (Krolikowski et al., 2003). Chemical analysis indicated that the Arabidopsis hth mutant has wild-type wax levels but abnormal cutin quantity and composition. Specifically, it has decreased levels of dicarboxylic acids and increased amounts of ω-hydroxy acids, leading the authors to suggest that HTH may have a role in the oxidation of ω-hydroxy fatty acids to the dicarboxylic acid cutin monomers that are characteristic of Arabidopsis stem and leaf cuticles (Kurdyukov et al., 2006b). As dicarboxylic acid cutin monomers are unusually abundant in Arabidopsis, it will be interesting to see whether HTH-related proteins are as essential to cuticle formation in other species where this class of monomers is scarce.
Another example of an “orphan” cuticle-associated protein resulted from analysis of the Arabidopsis bodyguard (bdg) mutant, which exhibits a microscopically disorganized cuticle with increased permeability but significantly increased levels of wax and cutin (Kurdyukov et al., 2006a). The BDG protein has sequence similarity to the α/β-hydrolase family of proteins, but no enzymatic activity has been reported. The protein is localized in the outer cell wall of the epidermis below the cuticle, which led the authors to propose that BDG may be involved in cutin polymerization, although the increased amounts of polymeric cutin in the mutant would argue against this (Kurdyukov et al., 2006a). Mutation of BDG3, a close homolog of BDG, resulted in the disorganization of floral nanoridges, petal epidermis structures that are composed of cutin (Shi et al., 2011). Moreover, the key cutin regulatory transcription factors SHN1, SHN2, and SHN3 were shown to activate the BDG3 promoter (Shi et al., 2011). Taken together, these results strongly indicate that BDG proteins are closely linked to cutin polymer formation, although their mode of action remains mysterious.
Lastly, a defect in the formation of floral nanoridges was also identified in the Arabidopsis mutant defective in cuticular ridges (dcr), which showed a substantial deficiency in floral cutin but a less drastic alteration of leaf and stem cutin (Panikashvili et al., 2009). DCR encodes a protein of the BAHD acyltransferase family that localizes to the cytoplasm, and it has been proposed that it may be involved in acyl transfer of cutin monomers to form precursor intermediates or oligomeric structures (Panikashvili et al., 2009). However, DCR was later biochemically characterized and shown to possess in vitro diacylglycerol acyltransferase activity, leading to the formation of triacylglycerol (Rani et al., 2010). A role for cytoplasmic triacylglycerol intermediates in cutin biosynthesis is not consistent with any known steps in this pathway, yet DCR is clearly required for cutin biosynthesis in Arabidopsis floral organs. Further work will be needed in order to determine the native substrate and product of DCR in order for its role in cutin biosynthesis to be elucidated.
FUNCTIONS OF THE CUTICLE
The plant cuticle is most typically associated with providing a fixed barrier to excessive transpirational water loss, allowing gas exchange and transpiration to be dynamically controlled by stomata. However, it has evolved a number of secondary functions that are consistent with its place as the outermost layer of primary aerial organs: it forms a physical barrier that is the first line of defense against pests and pathogens; in many species, elaborate epicuticular crystals help to form a self-cleaning surface, preventing dust and other debris from blocking sunlight; in some cases, it can act to screen excessive UV light; finally, as a defining feature of the epidermis, it plays a central role in development by physically establishing organ boundaries.
Cuticle Structure and Water Barrier Properties
A common perception is that a thick cuticle is associated with a lower water permeability and thus increased tolerance to water stress. However, comparative studies of the water permeability of cuticles from diverse species have indicated that there is no correlation with either the thickness of the cuticle or the amount of wax (Riederer and Schreiber, 2001). Similarly, the amount of cutin is not necessarily an indication of cuticular water permeability (CWP). For example, studies of three tomato mutants (cd1–cd3), each of which has a greater than 95% reduction in fruit cutin levels, revealed only minor increases in the rate of water loss, and even among the mutants there was no clear correlation between cutin amount and susceptibility to desiccation (Isaacson et al., 2009). However, cutin deficiency that leads to organizational defects can be detrimental to the cuticle permeability (Bessire et al., 2011). In contrast to the lack of association with cutin, extensive removal of wax from tomato fruit, accomplished by brief immersion of the fruit in an organic solvent, indicates that waxes contribute approximately 95% of the cuticle-mediated resistance to water diffusion, at least in tomato fruit (Leide et al., 2007).
Specific compound classes appear to be associated with water barrier properties of the cuticle; notably, the more nonpolar components, such as alkanes, tend to be associated with decreased CWP, while nonaliphatic wax compounds, such as triterpenoids, are likely a less effective water barrier (Leide et al., 2007; Buschhaus and Jetter, 2012). This is consistent with a model in which cuticular waxes localize within either crystalline or amorphous domains of the cuticle, with aliphatic compounds forming crystallite “rafts” that are impervious to water, forcing water, and other polar metabolites, to diffuse by a circuitous route through the amorphous domains that are formed by more polar and cyclic waxes (Riederer and Schreiber, 1995). The idea that the proportion of alkanes and not the total wax amount has the most significant effect on CWP was illustrated by a recent study with a backcrossed population of Capsicum annum and Capsicum chinense, two pepper species with high and low postharvest water loss rates, respectively. In 20 backcrossed families, CWP was inversely correlated with the amount of alkanes in the wax but not the total amount of wax, and the more rapidly desiccating parent had three times the wax coverage as the parent that exhibited low postharvest water loss (Parsons et al., 2012). In summary, resistance to water loss is primarily attributed to wax and not cutin, but there is not a direct correlation between the amount of either component and CWP. Rather, it appears that CWP is primarily determined by the particular mixture of intracuticular and epicuticular waxes and by their packing and organization within the cuticle architecture.
The Lotus Effect
A striking feature of many plant leaves is that water tends to bead into drops and roll to the ground, collecting and washing particles and debris from the leaf surface. The efficiency of this self-cleaning mechanism, termed the “lotus effect,” varies between species and during organ ontogeny, but it has been correlated with the abundance of epicuticular wax crystals that repel water and allow a pocket of air to form beneath the droplets (Barthlott and Neinhuis, 1997). It is thought that this self-cleaning surface helps to prevent the buildup of dust that would block sunlight and slow photosynthesis and that this could also play an important role in washing away pathogen spores before they germinate. Despite the apparent advantages of a self-cleaning surface, there is not a clear example of this trait conferring an adaptive advantage. In terms of photosynthesis, there is likely a tradeoff between a self-cleaning surface and the increased dispersion of light by epicuticular wax crystals, as discussed below. Nevertheless, based on the discovery of this effect, surfaces with high degrees of hydrophobicity and microscopic texture have been employed as effective biomimetic technical materials (Bhushan, 2012), and improved self-cleaning surfaces in agricultural crops may be a productive avenue of research.
The Cuticle as a Barrier against Pests and Pathogens
The plant cuticle presents a physical barrier to pathogens that do not otherwise enter the plant by way of the stomata, wounds, or vectors. However, fungal pathogens have been shown to breach the cuticle using a combination of enzymatic degradation and mechanical rupture. The latter is often accomplished by the formation of a swollen appressorium structure that extends an infectious peg via turgor pressure (Deising et al., 2000). While mechanical rupture may be sufficient for cuticle penetration, particularly of thinner cuticles (Tenberge, 2007), most fungal pathogens also secrete cutinases, a class of small, nonspecific esterases that hydrolyze the cutin polyester and release free cutin monomers (Longhi and Cambillau, 1999). The cutin monomers that are released during polymeric cutin hydrolysis can act as elicitors of plant defense responses and are thus classified as damage-associated molecular patterns. At micromolar concentrations, these compounds induce the production of hydrogen peroxide and other defense responses (Schweizer et al., 1996; Kauss et al., 1999). However, the mechanism of plant perception of free cutin monomers is currently unknown (Boller and Felix, 2009).
Cutin appears to be more important than wax for forming a barrier to pathogen entry, although there is not a consistent correlation between cutin amount and pathogen resistance. In tomato fruit, severely decreased cutin levels in three cd mutants was associated with increased susceptibility to infection by Botrytis cinerea surface inoculation and also to opportunistic microbes (Isaacson et al., 2009). However, in Arabidopsis, a number of cutin-deficient mutants and plants that ectopically overexpress fungal cutinases exhibit enhanced resistance to B. cinerea (Bessire et al., 2007, 2011; Chassot et al., 2007; Tang et al., 2007). In this case, increased cuticular permeability appears to enhance the diffusion of inoculum-derived elicitors that induce the production of small, polar antifungal compounds, which in turn inhibit B. cinerea growth (Bessire et al., 2007). Conversely, the Arabidopsis lacs2 mutant and cutinase overexpressers exhibited no alteration in their susceptibility to a range of other fungal pathogens (Bessire et al., 2007), and the lacs2 mutation also increased susceptibility to a normally avirulent strain of Pseudomonas syringae (Tang et al., 2007). Thus, cutin plays an important role as a physical barrier to many pathogens, yet extreme deficiencies in Arabidopsis can result in increased resistance to some pathogens by way of a secondary, but not well understood, mechanism that involves the induction of plant defenses. An additional layer of complexity was suggested by the observation that cutin can induce gene expression in plant pathogens and has been shown to induce appressorium expression in Colletotrichum trifolii (Dickman et al., 2003). This highlights the competing selective pressures to generate and breach cuticle barriers at the frontier of the plant surface (Chassot and Metraux, 2005).
Despite the importance of cutin in plant-pathogen interactions, the first surface encountered by foliar pathogens is formed by epicuticular wax crystals and films. In addition to the lotus effect that promotes the washing of spores from the plant surface before germination, there are several indications that the epicuticular wax structures and composition are important in determining fungal pathogen development and, thus, pathogenicity. The C26 aldehyde n-hexacosanyl, a component of cuticular wax in many species of the Poaceae, can induce in vitro appressorium formation by the powdery mildew Blumeria graminis (Tsuba et al., 2002; Ringelmann et al., 2009; Hansjakob et al., 2010). This observation is further corroborated by studies of the maize mutant glossy1, which does not accumulate aldehydes in its wax complement. B. graminis appressorium formation is substantially reduced on the leaf surface of the glossy1 mutant but can be restored to normal levels by the application of n-hexacosanyl (Hansjakob et al., 2011). Another example of the influence of waxes on pathogenicity is provided by the inhibitor of rust tube germination1 (irg1) mutant of M. truncatula, which exhibits decreased amounts of epicuticular wax crystals on the abaxial leaf surface, corresponding to a substantial decrease in wax primary alcohol groups. This surface alteration was shown to reduce spore differentiation of the rust fungal pathogens Phakopsora pachyrhizi and Puccinia emaculata and the anthracnose fungus C. trifolii, resulting in nonhost resistance (Uppalapati et al., 2012). The IRG1 gene was found to encode a C2H2 zinc finger transcription factor that had previously been identified as a regulator of dissected leaf morphology (Chen et al., 2010). Reduced transcript levels of putative MYB96 and CER4 orthologs were also observed in the irg1 mutant, which is consistent with the wax phenotype. The significance of waxes and cutin in pathogen resistance, therefore, is suggested in a general sense, but, as with cuticle permeability, little is known about the relative importance of specific molecular classes or their intermolecular associations and packing within the architecture of the cuticle.
Epicuticular waxes may also play an important role in plant-insect interactions; indeed, epicuticular wax crystals can form an unstable surface that prevents insect attachment or locomotion on plant surfaces (Borodich et al., 2010). A striking example of this is seen in the carnivorous pitcher plants (Nepenthes spp.), which catch insects by way of a slippery interior surface that is coated with epicuticular wax crystals (Riedel et al., 2007). For a more detailed review of cuticle chemical ecology, see Müller and Riederer (2005).
The Cuticle and Development
In addition to providing physical barriers to water and microbes, the cuticle appears to play an important role in defining organ boundaries during development, since plants with cuticles showing increased permeability and structural defects often exhibit numerous ectopic organ fusions. This phenomenon has been observed in a wax-deficient tomato mutant (Smirnova et al., 2013), a range of Arabidopsis mutants with abnormal cuticles (Yephremov et al., 1999; Wellesen et al., 2001; Kurdyukov et al., 2006a; Bird et al., 2007), and transgenic Arabidopsis plants overexpressing a secreted fungal cutinase (Sieber et al., 2000). The fusion zones are often marked by two adjacent polysaccharide cell walls with no visible cuticle separating the two organs, although the fused epidermal layers maintain their identity, as indicated by the differentiation of internal nonfunctional stomata within fusion zones (Sieber et al., 2000). In each of three Arabidopsis mutants exhibiting organ fusions, lacerata, bodyguard, and fiddlehead, ectopic organ fusions and cuticular permeability defects could be partially suppressed by a second mutation in SERRATE (Voisin et al., 2009). SERRATE is a C2H2 zinc finger protein that is required for microRNA biogenesis, and hypomorphic alleles exhibit numerous developmental defects, including serrated leaf margins (Dong et al., 2008). While the mechanism of SERRATE action as a suppressor of cuticle fusions remains unclear, this result suggests the existence of a cuticle integrity pathway that is integrated with epidermal developmental programs. The identification of additional suppressors of cuticle mutant-associated developmental phenotypes should be informative in elucidating the cuticle integrity pathway.
Protection against UV Radiation
UV light in the UV-B spectrum is a considerable portion of the daylight that reaches the terrestrial surface, and it can threaten plant life by damaging DNA, the photosynthetic apparatus, and membrane lipids (Rozema et al., 1997). As a result, plants have evolved a number of strategies for screening UV-B radiation. These include a variety of soluble flavonoid pigments that are typically localized within the vacuoles of epidermal cells, phenolic compounds present in the polysaccharide cell wall, and lipophilic phenolic molecules that are covalently bound to cutin or associated with waxes (Pfündel et al., 2006). A survey of isolated cuticles from a range of species indicated generally effective screening of the UV-B spectrum but consistently high transmittance in the higher wavelengths that are photosynthetically active (Krauss et al., 1997). In addition to absorbing light, the plant cuticle can reflect light to some degree, presumably depending on the abundance of epicuticular wax crystals. For example, Dudleya brittonnii can reflect up to 83% of UV-B, but this value is substantially reduced when epicuticular waxes are removed (Mulroy, 1979). Smooth, glossy “glabrous” cuticles typically reflect only small amounts of light (less than 10%), but glaucous plant surfaces are moderately reflective and generally show approximately 20% to 30% reflectance in the UV and visible spectra (Pfündel et al., 2006). Waxes reflect both UV and visible light, but not necessarily to the same extent, and the reflectance of UV has been reported to be greater in some cases (Holmes and Keiller, 2002). While light reflection provides an important protective mechanism, especially by limiting damaging UV radiation, there is likely a tradeoff with photosynthetic efficiency under conditions when light intensity is limiting (Pfündel et al., 2006). In this regard, an interesting area of future research might to determine whether relative proportions of UV and visible light reflection can be predictively changed by altering the composition of epicuticular waxes.
CONCLUSION AND PERSPECTIVES
As described above, several key areas of cuticle biogenesis remain poorly understood. First, the mechanism of intracellular and extracellular transport of wax and cutin precursors remains unknown, although key ABC transporters required for their export across the plasma membrane have been identified (Pighin et al., 2004; Bird et al., 2007; Chen et al., 2011). The first cutin synthase has been identified (Girard et al., 2012; Yeats et al., 2012b), but there are certainly additional cutin synthases, and whether they are closely related to CD1 or belong to distinct protein families remains to be discovered. After cutin is polymerized, is modification of the polymeric structure required to accommodate organ expansion? If so, which enzymes are involved in this process?
While our understanding of cuticle biosynthesis at the molecular level remains incomplete, recent progress in deciphering these pathways is bringing us closer than ever to an ability to selectively modify cuticle properties in order to improve agricultural productivity. However, the ability to make such modifications rationally will require an understanding of the complexity of cuticle function at the molecular level, and far less progress has been made in this regard. To this end, further work aimed at understanding the ecophysiological functions of the cuticle in defined mutant backgrounds, as well as in genetically tractable wild species, will provide a framework for understanding the complex interaction of structure, composition, and function of cuticles (Yeats et al., 2012a). While the past decade has seen unprecedented progress in the molecular biology of cuticle biogenesis, many studies have revealed complexities in cuticle function that underscore the fact that the cuticle is much more than just a preformed barrier to water loss.
Acknowledgments
We thank Drs. Gregory Buda, Christiane Nawrath, and Lacey Samuels for generously providing microscopy images and Eric Fich, Laetitia Martin, and Dr. Iben Sørensen for helpful comments and discussion.
Glossary
- VLCFA
very-long-chain fatty acid
- ER
endoplasmic reticulum
- FAE
fatty acid elongase
- ABC
ATP-binding cassette
- LTP
lipid-transfer protein
- GPI
glycosylphosphatidylinositol
- ABA
abscisic acid
- CWP
cuticular water permeability
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. (2004) The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 16: 2463–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambavaram MM, Krishnan A, Trijatmiko KR, Pereira A. (2011) Coordinated activation of cellulose and repression of lignin biosynthesis pathways in rice. Plant Physiol 155: 916–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aroca R, Porcel R, Ruiz-Lozano JM. (2012) Regulation of root water uptake under abiotic stress conditions. J Exp Bot 63: 43–57 [DOI] [PubMed] [Google Scholar]
- Bach L, Michaelson LV, Haslam R, Bellec Y, Gissot L, Marion J, Da Costa M, Boutin JP, Miquel M, Tellier F, et al. (2008) The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc Natl Acad Sci USA 105: 14727–14731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthlott W, Neinhuis C. (1997) Purity of the sacred lotus, or escape from contamination in biological surfaces. Planta 202: 1–8 [Google Scholar]
- Beaudoin F, Wu X, Li F, Haslam RP, Markham JE, Zheng H, Napier JA, Kunst L. (2009) Functional characterization of the Arabidopsis β-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol 150: 1174–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Domergue F, Pascal S, Jetter R, Renne C, Faure JD, Haslam RP, Napier JA, Lessire R, Joubès J. (2012) Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 24: 3106–3118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard A, Joubès J. (2013) Arabidopsis cuticular waxes: advances in synthesis, export and regulation. Prog Lipid Res 52: 110–129 [DOI] [PubMed] [Google Scholar]
- Bessire M, Borel S, Fabre G, Carraça L, Efremova N, Yephremov A, Cao Y, Jetter R, Jacquat AC, Métraux JP, et al. (2011) A member of the PLEIOTROPIC DRUG RESISTANCE family of ATP binding cassette transporters is required for the formation of a functional cuticle in Arabidopsis. Plant Cell 23: 1958–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessire M, Chassot C, Jacquat AC, Humphry M, Borel S, Petétot JM, Métraux JP, Nawrath C. (2007) A permeable cuticle in Arabidopsis leads to a strong resistance to Botrytis cinerea. EMBO J 26: 2158–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhushan B. (2012) Bioinspired structured surfaces. Langmuir 28: 1698–1714 [DOI] [PubMed] [Google Scholar]
- Bird D, Beisson F, Brigham A, Shin J, Greer S, Jetter R, Kunst L, Wu X, Yephremov A, Samuels L. (2007) Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J 52: 485–498 [DOI] [PubMed] [Google Scholar]
- Boller T, Felix G. (2009) A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60: 379–406 [DOI] [PubMed] [Google Scholar]
- Bonaventure G, Beisson F, Ohlrogge J, Pollard M. (2004) Analysis of the aliphatic monomer composition of polyesters associated with Arabidopsis epidermis: occurrence of octadeca-cis-6,cis-9-diene-1,18-dioate as the major component. Plant J 40: 920–930 [DOI] [PubMed] [Google Scholar]
- Borodich FM, Gorb EV, Gorb SN. (2010) Fracture behaviour of plant epicuticular wax crystals and its role in preventing insect attachment: a theoretical approach. Appl Phys A Mater Sci Process 100: 63–71 [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. (2004) WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA 101: 4706–4711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buda GJ, Isaacson T, Matas AJ, Paolillo DJ, Rose JKC. (2009) Three-dimensional imaging of plant cuticle architecture using confocal scanning laser microscopy. Plant J 60: 378–385 [DOI] [PubMed] [Google Scholar]
- Budke JM, Goffinet B, Jones CS. (2012) The cuticle on the gametophyte calyptra matures before the sporophyte cuticle in the moss Funaria hygrometrica (Funariaceae). Am J Bot 99: 14–22 [DOI] [PubMed] [Google Scholar]
- Burghardt M, Riederer M (2006) Cuticular transpiration. In M Riederer, C Müller, eds, Biology of the Plant Cuticle. Blackwell, Oxford, pp 292–311 [Google Scholar]
- Buschhaus C, Jetter R. (2012) Composition and physiological function of the wax layers coating Arabidopsis leaves: β-amyrin negatively affects the intracuticular water barrier. Plant Physiol 160: 1120–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot C, Metraux JP. (2005) The cuticle as source of signals for plant defense. Plant Biosyst 139: 28–31 [Google Scholar]
- Chassot C, Nawrath C, Métraux JP. (2007) Cuticular defects lead to full immunity to a major plant pathogen. Plant J 49: 972–980 [DOI] [PubMed] [Google Scholar]
- Cheesbrough TM, Kolattukudy PE. (1984) Alkane biosynthesis by decarbonylation of aldehydes catalyzed by a particulate preparation from Pisum sativum. Proc Natl Acad Sci USA 81: 6613–6617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Komatsuda T, Ma JF, Nawrath C, Pourkheirandish M, Tagiri A, Hu YG, Sameri M, Li X, Zhao X, et al. (2011) An ATP-binding cassette subfamily G full transporter is essential for the retention of leaf water in both wild barley and rice. Proc Natl Acad Sci USA 108: 12354–12359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Yu J, Ge L, Wang H, Berbel A, Liu Y, Chen Y, Li G, Tadege M, Wen J, et al. (2010) Control of dissected leaf morphology by a Cys(2)His(2) zinc finger transcription factor in the model legume Medicago truncatula. Proc Natl Acad Sci USA 107: 10754–10759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cominelli E, Sala T, Calvi D, Gusmaroli G, Tonelli C. (2008) Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J 53: 53–64 [DOI] [PubMed] [Google Scholar]
- Debono A, Yeats TH, Rose JKC, Bird D, Jetter R, Kunst L, Samuels L. (2009) Arabidopsis LTPG is a glycosylphosphatidylinositol-anchored lipid transfer protein required for export of lipids to the plant surface. Plant Cell 21: 1230–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deising HB, Werner S, Wernitz M. (2000) The role of fungal appressoria in plant infection. Microbes Infect 2: 1631–1641 [DOI] [PubMed] [Google Scholar]
- Delaux P-M, Nanda AK, Mathé C, Sejalon-Delmas N, Dunand C. (2012) Molecular and biochemical aspects of plant terrestrialization. Perspect Plant Ecol Evol Syst 14: 49–59 [Google Scholar]
- Dickman MB, Ha YS, Yang Z, Adams B, Huang C. (2003) A protein kinase from Colletotrichum trifolii is induced by plant cutin and is required for appressorium formation. Mol Plant Microbe Interact 16: 411–421 [DOI] [PubMed] [Google Scholar]
- Domínguez E, Heredia-Guerrero JA, Heredia A. (2011) The biophysical design of plant cuticles: an overview. New Phytol 189: 938–949 [DOI] [PubMed] [Google Scholar]
- Dong Z, Han MH, Fedoroff N. (2008) The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA 105: 9970–9975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D. (1993) Cells and tissues in the vegetative sporophytes of early land plants. New Phytol 125: 225–247 [DOI] [PubMed] [Google Scholar]
- Fiebig A, Mayfield JA, Miley NL, Chau S, Fischer RL, Preuss D. (2000) Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 12: 2001–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R, Briesen I, Wojciechowski T, Faust A, Yephremov A, Nawrath C, Schreiber L. (2005) Apoplastic polyesters in Arabidopsis surface tissues: a typical suberin and a particular cutin. Phytochemistry 66: 2643–2658 [DOI] [PubMed] [Google Scholar]
- Gaff DF, Oliver M. (2013) The evolution of desiccation tolerance in angiosperm plants: a rare yet common phenomenon. Funct Plant Biol 40: 315–328 [DOI] [PubMed] [Google Scholar]
- Girard AL, Mounet F, Lemaire-Chamley M, Gaillard C, Elmorjani K, Vivancos J, Runavot JL, Quemener B, Petit J, Germain V, et al. (2012) Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell 24: 3119–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer S, Wen M, Bird D, Wu X, Samuels L, Kunst L, Jetter R. (2007) The cytochrome P450 enzyme CYP96A15 is the midchain alkane hydroxylase responsible for formation of secondary alcohols and ketones in stem cuticular wax of Arabidopsis. Plant Physiol 145: 653–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta NS, Collinson ME, Briggs DEG, Evershed RP, Pancost RD. (2006) Reinvestigation of the occurrence of cutan in plants: implications for the leaf fossil record. Paleobiology 32: 432–449 [Google Scholar]
- Hansjakob A, Bischof S, Bringmann G, Riederer M, Hildebrandt U. (2010) Very-long-chain aldehydes promote in vitro prepenetration processes of Blumeria graminis in a dose- and chain length-dependent manner. New Phytol 188: 1039–1054 [DOI] [PubMed] [Google Scholar]
- Hansjakob A, Riederer M, Hildebrandt U. (2011) Wax matters: absence of very-long-chain aldehydes from the leaf cuticular wax of the glossy11 mutant of maize compromises the prepenetration processes of Blumeria graminis. Plant Pathol 60: 1151–1161 [Google Scholar]
- Haslam TM, Mañas-Fernández A, Zhao L, Kunst L. (2012) Arabidopsis ECERIFERUM2 is a component of the fatty acid elongation machinery required for fatty acid extension to exceptional lengths. Plant Physiol 160: 1164–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes MG, Keiller DR. (2002) Effects of pubescence and waxes on the reflectance of leaves in the ultraviolet and photosynthetic wavebands: a comparison of a range of species. Plant Cell Environ 25: 85–93 [Google Scholar]
- Hooker TS, Lam P, Zheng H, Kunst L. (2007) A core subunit of the RNA-processing/degrading exosome specifically influences cuticular wax biosynthesis in Arabidopsis. Plant Cell 19: 904–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooker TS, Millar AA, Kunst L. (2002) Significance of the expression of the CER6 condensing enzyme for cuticular wax production in Arabidopsis. Plant Physiol 129: 1568–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson T, Kosma DK, Matas AJ, Buda GJ, He Y, Yu B, Pravitasari A, Batteas JD, Stark RE, Jenks MA, et al. (2009) Cutin deficiency in the tomato fruit cuticle consistently affects resistance to microbial infection and biomechanical properties, but not transpirational water loss. Plant J 60: 363–377 [DOI] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Depège-Fargeix N, Arnould C, Oursel D, Domergue F, Sarda X, Rogowsky PM. (2010) Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor Outer Cell Layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol 154: 273–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javelle M, Vernoud V, Rogowsky PM, Ingram GC. (2011) Epidermis: the formation and functions of a fundamental plant tissue. New Phytol 189: 17–39 [DOI] [PubMed] [Google Scholar]
- Jeffree CE (2006) The fine structure of the plant cuticle. In M Riederer, C Müller, eds, Biology of the Plant Cuticle. Blackwell, Oxford, pp 11–125 [Google Scholar]
- Jenks MA, Ashworth EN. (1999) Plant epicuticular waxes: function, production and genetics. Hortic Rev 23: 1–68 [Google Scholar]
- Jetter R, Kunst L, Samuels AL (2006) Composition of plant cuticular waxes. In M Riederer, C Müller, eds, Biology of the Plant Cuticle. Blackwell, Oxford, pp 145–181 [Google Scholar]
- Jones VAS, Dolan L. (2012) The evolution of root hairs and rhizoids. Ann Bot (Lond) 110: 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joubès J, Raffaele S, Bourdenx B, Garcia C, Laroche-Traineau J, Moreau P, Domergue F, Lessire R. (2008) The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol Biol 67: 547–566 [DOI] [PubMed] [Google Scholar]
- Kannangara R, Branigan C, Liu Y, Penfield T, Rao V, Mouille G, Höfte H, Pauly M, Riechmann JL, Broun P. (2007) The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 19: 1278–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H, Fauth M, Merten A, Jeblick W. (1999) Cucumber hypocotyls respond to cutin monomers via both an inducible and a constitutive H2O2-generating system. Plant Physiol 120: 1175–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Lee SB, Kim HJ, Min MK, Hwang I, Suh MC. (2012) Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol 53: 1391–1403 [DOI] [PubMed] [Google Scholar]
- Kolattukudy PE. (2001) Polyesters in higher plants. Adv Biochem Eng Biotechnol 71: 1–49 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Hanhart CJ, Thiel F. (1989) A genetic and phenotypic description of eceriferum (cer) mutants in Arabidopsis thaliana. J Hered 80: 118–122 [Google Scholar]
- Kosma DK, Bourdenx B, Bernard A, Parsons EP, Lü S, Joubès J, Jenks MA. (2009) The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol 151: 1918–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosma DK, Nemacheck JA, Jenks MA, Williams CE. (2010) Changes in properties of wheat leaf cuticle during interactions with Hessian fly. Plant J 63: 31–43 [DOI] [PubMed] [Google Scholar]
- Krauss P, Markstädter C, Riederer M. (1997) Attenuation of UV radiation by plant cuticles from woody species. Plant Cell Environ 20: 1079–1085 [Google Scholar]
- Krolikowski KA, Victor JL, Wagler TN, Lolle SJ, Pruitt RE. (2003) Isolation and characterization of the Arabidopsis organ fusion gene HOTHEAD. Plant J 35: 501–511 [DOI] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Nawrath C, Bär S, Voisin D, Efremova N, Franke R, Schreiber L, Saedler H, Métraux JP, et al. (2006a) The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 18: 321–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdyukov S, Faust A, Trenkamp S, Bär S, Franke R, Efremova N, Tietjen K, Schreiber L, Saedler H, Yephremov A. (2006b) Genetic and biochemical evidence for involvement of HOTHEAD in the biosynthesis of long-chain alpha-,omega-dicarboxylic fatty acids and formation of extracellular matrix. Planta 224: 315–329 [DOI] [PubMed] [Google Scholar]
- Lam P, Zhao L, McFarlane HE, Aiga M, Lam V, Hooker TS, Kunst L. (2012) RDR1 and SGS3, components of RNA-mediated gene silencing, are required for the regulation of cuticular wax biosynthesis in developing inflorescence stems of Arabidopsis. Plant Physiol 159: 1385–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Go YS, Bae HJ, Park JH, Cho SH, Cho HJ, Lee DS, Park OK, Hwang I, Suh MC. (2009) Disruption of glycosylphosphatidylinositol-anchored lipid transfer protein gene altered cuticular lipid composition, increased plastoglobules, and enhanced susceptibility to infection by the fungal pathogen Alternaria brassicicola. Plant Physiol 150: 42–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SC, Luan S. (2012) ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ 35: 53–60 [DOI] [PubMed] [Google Scholar]
- Leide J, Hildebrandt U, Reussing K, Riederer M, Vogg G. (2007) The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: effects of a deficiency in a β-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol 144: 1667–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, Verbruggen H, Zechman FW. (2011) Into the deep: new discoveries at the base of the green plant phylogeny. Bioessays 33: 683–692 [DOI] [PubMed] [Google Scholar]
- Li F, Wu X, Lam P, Bird D, Zheng H, Samuels L, Jetter R, Kunst L. (2008) Identification of the wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase WSD1 required for stem wax ester biosynthesis in Arabidopsis. Plant Physiol 148: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Pollard M, Sauveplane V, Pinot F, Ohlrogge J, Beisson F. (2009) Nanoridges that characterize the surface morphology of flowers require the synthesis of cutin polyester. Proc Natl Acad Sci USA 106: 22008–22013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longhi S, Cambillau C. (1999) Structure-activity of cutinase, a small lipolytic enzyme. Biochim Biophys Acta 1441: 185–196 [DOI] [PubMed] [Google Scholar]
- Lü S, Song T, Kosma DK, Parsons EP, Rowland O, Jenks MA. (2009) Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J 59: 553–564 [DOI] [PubMed] [Google Scholar]
- Lü S, Zhao H, Des Marais DL, Parsons EP, Wen X, Xu X, Bangarusamy DK, Wang G, Rowland O, Juenger T, et al. (2012) Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol 159: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci JD, Rerie WG, Foreman DR, Zhang M, Galway ME, Marks MD, Schiefelbein JW. (1996) The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122: 1253–1260 [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Shin JJ, Bird DA, Samuels AL. (2010) Arabidopsis ABCG transporters, which are required for export of diverse cuticular lipids, dimerize in different combinations. Plant Cell 22: 3066–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moitra K, Silverton L, Limpert K, Im K, Dean M. (2011) Moving out: from sterol transport to drug resistance. The ABCG subfamily of efflux pumps. Drug Metabol Drug Interact 26: 105–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C, Riederer M. (2005) Plant surface properties in chemical ecology. J Chem Ecol 31: 2621–2651 [DOI] [PubMed] [Google Scholar]
- Mulroy TW. (1979) Spectral properties of heavily glaucous and non-glaucous leaves of a succulent rosette-plant. Oecologia 38: 349–357 [DOI] [PubMed] [Google Scholar]
- Nadakuduti SS, Pollard M, Kosma DK, Allen CJ, Jr, Ohlrogge JB, Barry CS. (2012) Pleiotropic phenotypes of the sticky peel mutant provide new insight into the role of CUTIN DEFICIENT2 in epidermal cell function in tomato. Plant Physiol 159: 945–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T. (2006) Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol 141: 1363–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obata T, Fernie AR. (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69: 3225–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogburn RM, Edwards EJ. (2010) The ecological water-use strategies of succulent plants. Adv Bot Res 55: 179–225 [Google Scholar]
- Oshima Y, Shikata M, Koyama T, Ohtsubo N, Mitsuda N, Ohme-Takagi M. (2013) MIXTA-like transcription factors and WAX INDUCER1/SHINE1 coordinately regulate cuticle development in Arabidopsis and Torenia fournieri. Plant Cell 25: 1609–1624 10.1105/tpc.113.110783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A. (2009) The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol 151: 1773–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panikashvili D, Shi JX, Schreiber L, Aharoni A. (2011) The Arabidopsis ABCG13 transporter is required for flower cuticle secretion and patterning of the petal epidermis. New Phytol 190: 113–124 [DOI] [PubMed] [Google Scholar]
- Parsons EP, Popopvsky S, Lohrey GT, Lü S, Alkalai-Tuvia S, Perzelan Y, Paran I, Fallik E, Jenks MA. (2012) Fruit cuticle lipid composition and fruit post-harvest water loss in an advanced backcross generation of pepper (Capsicum sp.). Physiol Plant 146: 15–25 [DOI] [PubMed] [Google Scholar]
- Pascal S, Bernard A, Sorel M, Pervent M, Vile D, Haslam RP, Napier JA, Lessire R, Domergue F, Joubès J. (2013) The Arabidopsis cer26 mutant, like the cer2 mutant, is specifically affected in the very long chain fatty acid elongation process. Plant J 73: 733–746 [DOI] [PubMed] [Google Scholar]
- Pfündel EE, Agati G, Cerovic ZG (2006) Optical properties of plant surfaces. In M Riederer, C Müller, eds, Biology of the Plant Cuticle. Blackwell, Oxford, pp 216–249 [Google Scholar]
- Pighin JA, Zheng H, Balakshin LJ, Goodman IP, Western TL, Jetter R, Kunst L, Samuels AL. (2004) Plant cuticular lipid export requires an ABC transporter. Science 306: 702–704 [DOI] [PubMed] [Google Scholar]
- Pollard M, Beisson F, Li Y, Ohlrogge JB. (2008) Building lipid barriers: biosynthesis of cutin and suberin. Trends Plant Sci 13: 236–246 [DOI] [PubMed] [Google Scholar]
- Pulsifer IP, Kluge S, Rowland O. (2012) Arabidopsis long-chain acyl-CoA synthetase 1 (LACS1), LACS2, and LACS3 facilitate fatty acid uptake in yeast. Plant Physiol Biochem 51: 31–39 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Léger A, Joubès J, Miersch O, Huard C, Blée E, Mongrand S, Domergue F, Roby D. (2008) A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20: 752–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani SH, Krishna TH, Saha S, Negi AS, Rajasekharan R. (2010) Defective in cuticular ridges (DCR) of Arabidopsis thaliana, a gene associated with surface cutin formation, encodes a soluble diacylglycerol acyltransferase. J Biol Chem 285: 38337–38347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina JJ, Guerrero C, Heredia A. (2007) Isolation, characterization, and localization of AgaSGNH cDNA: a new SGNH-motif plant hydrolase specific to Agave americana L. leaf epidermis. J Exp Bot 58: 2717–2731 [DOI] [PubMed] [Google Scholar]
- Reina-Pinto JJ, Yephremov A. (2009) Surface lipids and plant defenses. Plant Physiol Biochem 47: 540–549 [DOI] [PubMed] [Google Scholar]
- Riedel M, Eichner A, Meimberg H, Jetter R. (2007) Chemical composition of epicuticular wax crystals on the slippery zone in pitchers of five Nepenthes species and hybrids. Planta 225: 1517–1534 [DOI] [PubMed] [Google Scholar]
- Riederer M, Schreiber L (1995) Waxes: the transport barriers of plant cuticles. In RJ Hamilton, ed, Waxes: Chemistry, Molecular Biology and Functions. Oily Press, Dundee, UK, pp 131–156 [Google Scholar]
- Riederer M, Schreiber L. (2001) Protecting against water loss: analysis of the barrier properties of plant cuticles. J Exp Bot 52: 2023–2032 [DOI] [PubMed] [Google Scholar]
- Ringelmann A, Riedel M, Riederer M, Hildebrandt U. (2009) Two sides of a leaf blade: Blumeria graminis needs chemical cues in cuticular waxes of Lolium perenne for germination and differentiation. Planta 230: 95–105 [DOI] [PubMed] [Google Scholar]
- Rowland O, Zheng H, Hepworth SR, Lam P, Jetter R, Kunst L. (2006) CER4 encodes an alcohol-forming fatty acyl-coenzyme A reductase involved in cuticular wax production in Arabidopsis. Plant Physiol 142: 866–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozema J, van de Staaij J, Björn LO, Caldwell M. (1997) UV-B as an environmental factor in plant life: stress and regulation. Trends Ecol Evol 12: 22–28 [DOI] [PubMed] [Google Scholar]
- Schneider-Belhaddad F, Kolattukudy P. (2000) Solubilization, partial purification, and characterization of a fatty aldehyde decarbonylase from a higher plant, Pisum sativum. Arch Biochem Biophys 377: 341–349 [DOI] [PubMed] [Google Scholar]
- Schweizer P, Felix G, Buchala A, Müller C, Métraux JP. (1996) Perception of free cutin monomers by plant cells. Plant J 10: 331–341 [Google Scholar]
- Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM. (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23: 1138–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151: 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra O, Chatterjee S, Huang W, Stark RE. (2012) Mini-review: what nuclear magnetic resonance can tell us about protective tissues. Plant Sci 195: 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi JX, Adato A, Alkan N, He Y, Lashbrooke J, Matas AJ, Meir S, Malitsky S, Isaacson T, Prusky D, et al. (2013) The tomato SlSHINE3 transcription factor regulates fruit cuticle formation and epidermal patterning. New Phytol 197: 468–480 [DOI] [PubMed] [Google Scholar]
- Shi JX, Malitsky S, De Oliveira S, Branigan C, Franke RB, Schreiber L, Aharoni A. (2011) SHINE transcription factors act redundantly to pattern the archetypal surface of Arabidopsis flower organs. PLoS Genet 7: e1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Métraux JP, Nawrath C. (2000) Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusions. Plant Cell 12: 721–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnova A, Leide J, Riederer M. (2013) Deficiency in a very-long-chain fatty acid β-ketoacyl-coenzyme A synthase of tomato impairs microgametogenesis and causes floral organ fusion. Plant Physiol 161: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen I, Pettolino FA, Bacic A, Ralph J, Lu F, O’Neill MA, Fei Z, Rose JKC, Domozych DS, Willats WG. (2011) The charophycean green algae provide insights into the early origins of plant cell walls. Plant J 68: 201–211 [DOI] [PubMed] [Google Scholar]
- Takada S, Takada N, Yoshida A. (2013) ATML1 promotes epidermal cell differentiation in Arabidopsis shoots. Development 140: 1919–1923 [DOI] [PubMed] [Google Scholar]
- Taketa S, Amano S, Tsujino Y, Sato T, Saisho D, Kakeda K, Nomura M, Suzuki T, Matsumoto T, Sato K, et al. (2008) Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proc Natl Acad Sci USA 105: 4062–4067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang D, Simonich MT, Innes RW. (2007) Mutations in LACS2, a long-chain acyl-coenzyme A synthetase, enhance susceptibility to avirulent Pseudomonas syringae but confer resistance to Botrytis cinerea in Arabidopsis. Plant Physiol 144: 1093–1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenberge KB (2007) Morphology and cellular organisation in Botrytis interactions with plants. In Y Elad, B Williamson, P Tudzynski, N Delen, eds, Botrytis: Biology, Pathology and Control. Springer, Dordrecht, The Netherlands, pp 67–84 [Google Scholar]
- Tsuba M, Katagiri C, Takeuchi Y, Takada Y, Yamaoka N. (2002) Chemical factors of the leaf surface involved in the morphogenesis of Blumeria graminis. Physiol Mol Plant Pathol 60: 51–57 [Google Scholar]
- Uppalapati SR, Ishiga Y, Doraiswamy V, Bedair M, Mittal S, Chen J, Nakashima J, Tang Y, Tadege M, Ratet P, et al. (2012) Loss of abaxial leaf epicuticular wax in Medicago truncatula irg1/palm1 mutants results in reduced spore differentiation of anthracnose and nonhost rust pathogens. Plant Cell 24: 353–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voisin D, Nawrath C, Kurdyukov S, Franke RB, Reina-Pinto JJ, Efremova N, Will I, Schreiber L, Yephremov A. (2009) Dissection of the complex phenotype in cuticular mutants of Arabidopsis reveals a role of SERRATE as a mediator. PLoS Genet 5: e1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volokita M, Rosilio-Brami T, Rivkin N, Zik M. (2011) Combining comparative sequence and genomic data to ascertain phylogenetic relationships and explore the evolution of the large GDSL-lipase family in land plants. Mol Biol Evol 28: 551–565 [DOI] [PubMed] [Google Scholar]
- Waters ER. (2003) Molecular adaptation and the origin of land plants. Mol Phylogenet Evol 29: 456–463 [DOI] [PubMed] [Google Scholar]
- Wellesen K, Durst F, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A. (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid omega-hydroxylation in development. Proc Natl Acad Sci USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R, Li S, He S, Wassmann F, Yu C, Qin G, Schreiber L, Qu LJ, Gu H. (2011) CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 23: 3392–3411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Pollard M, Li-Beisson Y, Beisson F, Feig M, Ohlrogge J. (2010) A distinct type of glycerol-3-phosphate acyltransferase with sn-2 preference and phosphatase activity producing 2-monoacylglycerol. Proc Natl Acad Sci USA 107: 12040–12045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Simpson JP, Li-Beisson Y, Beisson F, Pollard M, Ohlrogge JB. (2012) A land-plant-specific glycerol-3-phosphate acyltransferase family in Arabidopsis: substrate specificity, sn-2 preference, and evolution. Plant Physiol 160: 638–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Buda GJ, Wang Z, Chehanovsky N, Moyle LC, Jetter R, Schaffer AA, Rose JKC. (2012a) The fruit cuticles of wild tomato species exhibit architectural and chemical diversity, providing a new model for studying the evolution of cuticle function. Plant J 69: 655–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Martin LB, Viart HM, Isaacson T, He Y, Zhao L, Matas AJ, Buda GJ, Domozych DS, Clausen MH, et al. (2012b) The identification of cutin synthase: formation of the plant polyester cutin. Nat Chem Biol 8: 609–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeats TH, Rose JKC. (2008) The biochemistry and biology of extracellular plant lipid-transfer proteins (LTPs). Protein Sci 17: 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. (1999) Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 11: 2187–2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY. (2005) Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J 42: 689–707 [DOI] [PubMed] [Google Scholar]
- Zheng H, Rowland O, Kunst L. (2005) Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 17: 1467–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]