Abstract
Spectral fingerprints were acquired for Rio Red grapefruit using flow injection electrospray ionization with ion trap and time-of-flight mass spectrometry (FI-ESI-IT-MS and FI-ESI-TOF-MS). Rio Red grapefruits were harvested 3 times a year (early, mid, and late harvests) in 2005 and 2006 from conventionally and organically grown trees. Data analysis using analysis of variance principal component analysis (ANOVA-PCA) demonstrated that, for both MS systems, the chemical patterns were different as a function of farming mode (conventional vs organic), as well as growing year and time of harvest. This was visually obvious with PCA and was shown to be statistically significant using ANOVA. The spectral fingerprints provided a more inclusive view of the chemical composition of the grapefruit and extended previous conclusions regarding the chemical differences between conventionally and organically grown Rio Red grapefruit.
Keywords: Grapefruit (Citrus paradisi), mass spectral fingerprints, analysis of variance principal component analysis
INTRODUCTION
The demand for organic food is growing rapidly in the U.S. as sales of organic food and beverages have increased from$1 billion in 1990 to an estimated $20 billion in 2007 (1). Organic food sales are anticipated to increase an average of 18% each year from2007 to 2010(1). This growth is primarily due to consumers’ perception that organic foods are safer, more nutritious, and better-tasting than conventionally grown foods (2).
Many studies have been conducted to determine whether significant differences in yield and quality truly exist between organic and conventionally grown foods (3–11). However, few have exercised the necessary controls to account for the basic environmental factors affecting plant development and yield, such as soil physical, biological and chemical properties, rainfall, radiation, water management practices, pest and disease management, and crop varieties, etc. (12). Additionally, most were based on the analysis of only a few chemical components. According to a British review of studies done over the past 50 years, organic and conventionally produced foods have about the same nutrient content, suggesting that neither is better in terms of health benefits (13).
Evaluating the “nutritional” quality of a plant material is difficult. Every plant contains compounds that are nutritional or potentially health-promoting for humans. Since the presence and concentration of these compounds vary considerably, an evaluation of total “nutritional quality” is difficult. From a chemist’s point of view, the simplest approach is to first determine whether the chemical contents of two plant materials are the same. Obviously, if the chemical content is the same, the “nutritional quality” must be the same. If the chemical content is different, then further tests are needed to determine which compounds differentiate the two materials.
In the field of metabolomics, comparison of “global” chemical patterns of plant materials is accomplished using spectral fingerprints or chromatographic profiles. Spectral fingerprints usually do not employ chromatographic separation and require pattern recognition software to obtain useful information. Fingerprints acquired by mass spectrometry (MS) can provide information for specific ions, but identification of specific compounds requires expensive, high resolution systems. Chromatographic profiles provide more information but are more time-consuming, and retention times must be aligned prior to pattern recognition analysis.
A recent study compared organically and conventionally grown Rio Red grapefruit (Citrus paradisi) with respect to market quality, nutritional and bioactive compounds, and consumer taste intensity and acceptance (12). They found that conventional fruits were better colored, less tart, higher in lycopene, lower in naringin (a bitter flavonoid), and better accepted by consumers. The organic fruit had a thinner peel and was higher in ascorbic acid and sugars and lower in nitrate and furanocoumarins (known for adverse drug interaction). This study was unique since the fruits were grown under carefully matched conditions to eliminate as many variables as possible. Consequently, these grapefruit samples represent a unique opportunity to investigate the chemical patterns resulting from conventional and organic farming.
In the present study, MS fingerprints were obtained for the Rio Red grapefruit described above using flow injection electrospray ionization with ion trap and time-of-flight mass spectrometry (FI-ESI-IT-MS and FI-ESI-TOF-MS). The chemical patterns represented by the spectral fingerprints were analyzed using analysis of variance principal component analysis (ANOVAPCA) to deconvolute variance introduced by growing year, harvest, and farming mode. While the main goal of the research was to compare the farming mode (conventional versus organic), the other experimental factors, year (2005 versus 2006) and harvest (early, mid, and late season), were included for statistical accuracy.
MATERIALS AND METHODS
Standard Compounds and Other Chemicals
Optima grade water and acetonitrile were purchased from Fisher Scientific (Pittsburgh, PA). MS grade formic acid and reserpine were purchased from Sigma/Aldrich (St. Louis, MO).
Plant Materials
A total of 120 of Rio Red grapefruit (Citrus paradisi) were grown by using conventional and organic cultivation methods were harvested at three distinct growing phases (early, mid, and late season harvest) over two years (2005 and 2006) (12). Grapefruit is usually harvested 3 times during the year since all the fruit do not ripen simultaneously.
The selected fruits were juiced, and the juice was flash frozen with liquid nitrogen and stored at −80 °C until analyzed. As shown in Figure 1, 10 samples were collected for each of 3 harvests for both conventional and organic farming modes and for each of 2 years (10×3×2×2= 120).
Figure 1.
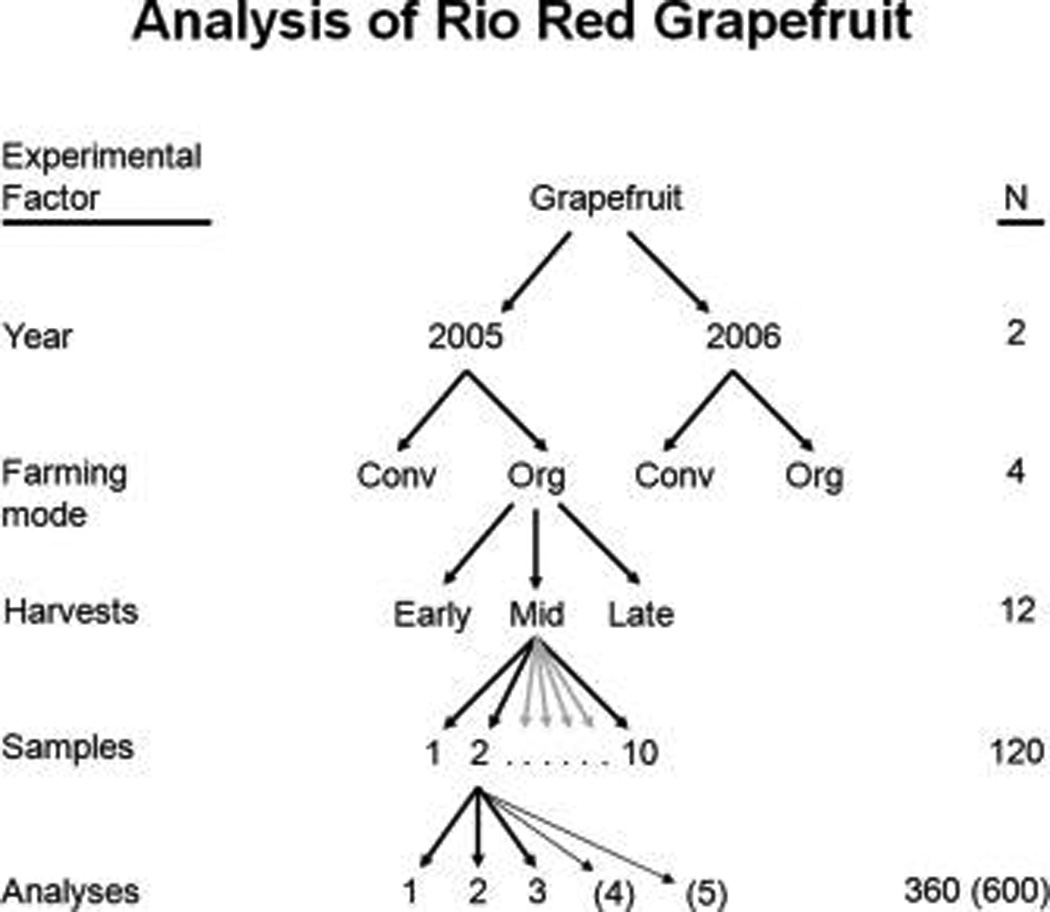
Grapefruit sample analysis scheme. Extra repeat analyses for IT-MS are shown in parentheses.
MS Conditions
Two MS systems (FI-ESI-IT-MS and FI-ESI-TOFMS) were used for this study. The first system consisted of a binary pump with a vacuum degasser, a thermostated column compartment, and an autosampler (Agilent 1100, Agilent Technologies, Palo Alto, CA) in combination with a ion-trap mass spectrometer (LCQ Classic, Thermo Fisher Scientific Inc., Waltham, MA). The second system consisted of a binary pump with a vacuum degasser, a thermostated column compartment, and an autosampler (Agilent 1100, Agilent Technologies, Palo Alto, CA) in combination with a time-of-flight mass spectrometer (TOF-MS, Unique, Leco, St. Joseph, MI). Both spectrometers used electrospray ionization (ESI) in positive ion mode and were optimized daily using reserpine with electrospray ionization.
The conditions for IT-MS were as follows: sheath gas flow rate, 80 (arbitrary units); aux gas flow rate, 10 (arbitrary units); spray voltage, 4.50 kV; heated capillary temperature, 200 °C; capillary voltage, −4.0 V; tube lens offset, 20 V. The conditions for the TOF-MS were as follows: detector voltage, 2700 V; electrospray voltage, 3500 V; desolvation gas, 6 L/min; desolvation temperature, 250 °C; interface temperature, 75 °C; nozzle voltage, 240 V; skimmer voltage, 100 V.
Samples were introduced into the spectrometers using an abbreviated liquid chromatographic system that consisted of only a guard column (Adsorbosphere All-Guard Cartridge, C18, 5 µm, 4.6 × 7.5 mm, Alltech Associates, Inc., Deerfield, IL). The mobile phase consisted of 0.1% formic acid in H2O (A) and 0.1% formic acid in acetonitrile (B) with isocratic elution at 60:40 (v/v) at a flow rate of 0.5 mL/min. Flash frozen grapefruit juice samples were thawed, diluted 10 times with H2O, and filtered through a 17 mm (0.45 µm) PVDF syringe filter (VWR Scientific, Seattle, WA) before 5 µL was injected. Spectra were collected across the bolus of the sample for 1 min (from 0.5 to 1.5 min for IT-MS and from 0.2 to 1.2 min for TOF-MS.
Data were collected from 150 to 1000 m/z for IT-MS and from 0.1 to 1000 m/z for TOF-MS. The lower limit for IT-MS was programmable, but there is no lower limit for the TOF-MS. The lower threshold for IT-MS was programmed at 2%. There was no programmable threshold for the TOF-MS. The threshold is set by the manufacturer’s software.
All samples were analyzed once a day for 5 days by IT-MS and once a day for 3 days by TOF-MS, producing 600 and 360 analyses (spectra), respectively. Samples were initially analyzed by TOF-MS in triplicate. These results suggested that more repeats might be beneficial, so analyses by IT-MS were repeated 5 times. More repeats had little effect on the quality of the data, so more repeats for TOF-MS were not made.
Data Processing
MS fingerprints were obtained as one-dimensional spectra; counts versus m/z. Five repeat analyses of the 120 different grapefruit samples IT-MS provided 600 spectra. Three repeat analyses of the 120 different grapefruit samples extracts TOF-MS provided a second set of data with 360 spectra. The data were analyzed using the analysis of variance principal components analysis (ANOVA-PCA) method originally described by Harrington et al. (14) and modified by Harnly et al. (15). The ANOVA portion of the evaluation was performed using Excel (Microsoft, Redmond, WA) and the PCA portion was performed using Solo (Eigenvector Research, Inc., Wenatchee, WA).
The raw data were imported into Excel and truncated to 150 m/z at the lower end. Prior to the ANOVA, the common masses of all the spectra were aligned using an Excel macro developed in-house to provide a uniform data matrix. Zero values were inserted for the masses for which no intensity was observed. The data were then processed, or “filtered”, in 3 ways to produce 3 data sets. In the first data set, all the data were accepted without modification (labeled “unfiltered”). In the second data set, intensity values were listed for a mass only if the intensities for all the repeats of a single sample were nonzero (“5of5” filter for IT-MS and “3of3” filter for the TOF-MS). If one of the repeats was zero, all 5 (or 3) repeats of that mass were listed as zero. In the third data set, intensity values were listed for a mass only if the intensities for all the repeats for all 10 samples from a single harvest were nonzero (“50of50” filter for IT-MS and “30of30” filter for TOF-MS).If a single analysis had zero intensity, all 50 (or 30) repeats for that mass were listed as zero. After filtering, if a mass contained a column of zeros (i.e., not a single analyses had a nonzero value) the mass was deleted from the data set.
Each data set was analyzed by ANOVA-PCA. The calculation of the ANOVA matrices in Excel has been described previously (15, 16). Briefly, for each data set, individual spectra were normalized to a unit vector (the sum of the squares of all the intensities is set equal to 1.0). The data for each mass were then averaged and mean-centered (the average is subtracted from each individual data point). The normalized, mean-centered matrix was then used to construct 10 submatrices, the means and residuals matrix for each experimental variable: year (Y), farming mode (M), harvest (H), analytical run (R), and interaction of the four factors (Y × M × H × R). The sum of the squares of the matrices was used for ANOVA calculations. The residuals for the factor interaction (Y × M × H × R), corresponding to the variability between fruit (biological variability), were added to the means matrices for each variable prior to PCA.
RESULTS AND DISCUSSION
MS Fingerprints
Figures 2 and 3 show typical fingerprints for the conventionally and organically grown grapefruit samples acquired by IT-MS and TOF-MS, respectively, prior to any data processing. Visual inspection provides two impressions: first, there is a considerable difference between the spectra of the two spectrometers, and, second, there appear to be no differences between farming modes. With respect to the spectrometers, considerably more ions were detected using IT-MS than with TOF-MS. In addition, no significant ions were detected above m/z 600 using TOF-MS. Common ions can be seen at m/z 175, 365, 381, 525, 557 in both spectra (Figures 2 and 3). IT-MS had prominent ions at m/z 175, 365 (its highest ion) and 696, while TOF-MS featured prominent ions at m/z 365, 381, and 557. The differences in the spectra between the two mass spectrometers arise from the completely different operating principals and the software of the instruments. The ESI sources, the ion optics, the separation mechanisms (IT versus TOF), the ion detectors, and the software used to processing the raw data for the two spectrometers are different.
Figure 2.
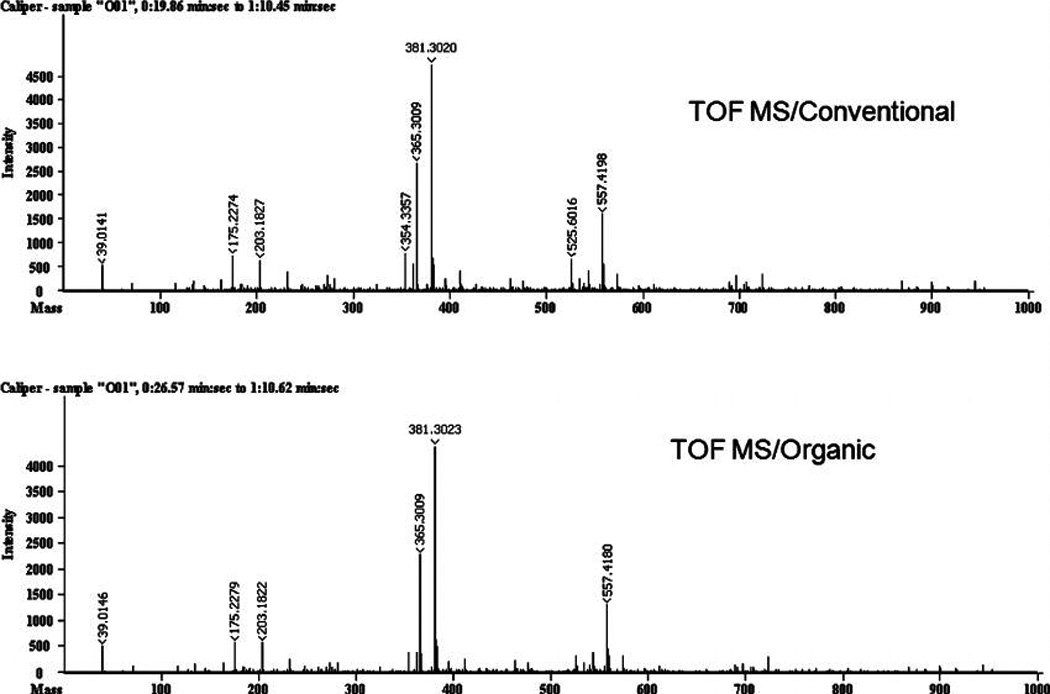
MS fingerprint for IT-MS; conventionally and organically grown.
Figure 3.
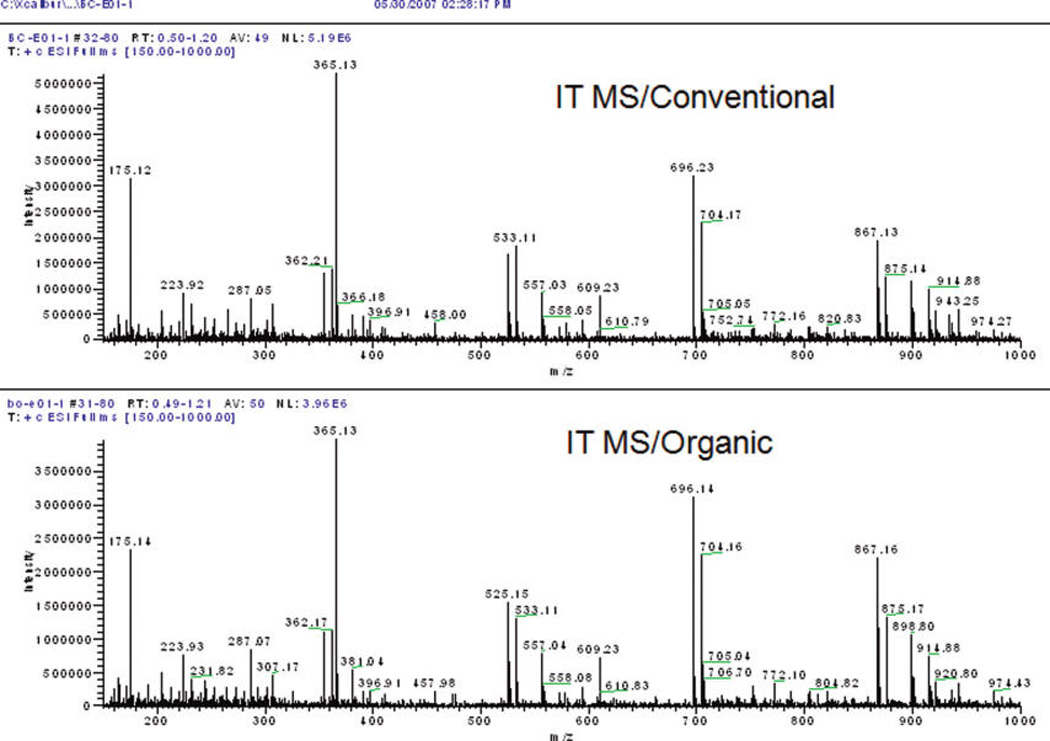
MS fingerprint for TOF-MS; conventionally and organically grown.
The similar appearance of spectra for conventional and organic grapefruit fingerprints is consistent with the previous results reported by Lester et al. (12) that there was no apparent difference between the framing modes with respect to the concentrations of many of the fruit components. This is not too surprising since basic metabolic functions associated with growth and energy metabolism would be expected to be similar. This is especially true in this study where the whole juice is analyzed rather than an extract of compounds of a specific polarity. However, changes in environmentally sensitive compounds are expected. Hence, the use of ANOVA and PCA to detect less obvious changes in the patterns of ion intensities.
ANOVA
A raw data matrix was constructed for each MS. For IT-MS, the raw data matrix was 600 × 831 (600 analyses by 831 masses). Application of a “5of5” filter to the data (as described in Materials and Methods) reduced the matrix size to 600 × 631. Use of a “50of50” filter further reduced the matrix to 600 × 345. For TOF-MS, the raw data matrix was 360 × 84 which was reduced to 360 × 26 and 360 × 14 for “3of3” and “30of30” filtering, respectively.
We chose to filter the data with respect to their presence for repeat analyses rather than use a signal-to-noise ratio (S/N) or threshold filter for the following reasons. First, so few repeats (3 or 5) makes the S/N highly irreproducible. Second, as discussed in the previous paragraph, we anticipated that some of the most important compounds for discriminating between the experimental conditions would be the environmentally sensitive compounds which would most likely not provide ions with the highest count rates. Thus, we did not want to set a threshold that would exclude less intense peaks that were present for all repeat analyses. This point is made clearer in a later discussion on significant masses.
The ANOVA process, as described in Materials and Methods, produced 10 submatrices; means and residuals for year (Y), farming mode (F), harvest (H), analytical run (R), and factor interaction (Y × F × H × R). The residuals matrix for the factor interaction provided the uncertainty between samples, or the biological uncertainty. The biological uncertainty matrix was added to each of the mean matrices (year, farming mode, and harvest) before submitting the data to PCA.
The sums of the squared data in the submatrices can be used to compute the total variance and the variance associated with the means and residuals of each experimental factor and to construct an ANOVA table. The 4-way ANOVA data computed for IT-MS (Table 1) shows that year, farming mode, harvest, and run accounted for 22.3%, 5.2%, 4.2%, and 3.0%, of the total variance. Factor interaction accounted for 62.5% of the total variance and indicates that the interaction between the experimental factors was not constant; e.g. the harvest means was different for different years and different farming modes. The biological variance was 2.8% of the total variance.
Table 1.
Analysis of Variance for IT-MS and TOF-MS Data
| n | df | variance | percent | mean variance | F value | |
|---|---|---|---|---|---|---|
| IT-MS | ||||||
| grand means residuals (G) | 87.890 | |||||
| between years (Y) | 2 | 1 | 19.613 | 22.3% | 19.6130 | 4312.30 |
| between farming modes (M) | 2 | 1 | 4.597 | 5.2% | 4.5970 | 1010.74 |
| between harvests (H) | 3 | 2 | 3.697 | 4.2% | 1.8485 | 406.43 |
| between runs (R) | 5 | 4 | 2.602 | 3.0% | 0.6505 | 143.03 |
| between Y × M × H × R | 10 | 8 | 54.924 | 62.5% | 6.8655 | 1509.52 |
| within Y × M ×H × R (i.e. biological) | 540 | 2.456 | 2.8% | 0.0045 | ||
| 87.889 | 100.0% | |||||
| TOF-MS | ||||||
| grand means residuals (G) | 3.686 | |||||
| between years (Y) | 2 | 1 | 0.384 | 10.4% | 0.3840 | 407.92 |
| between farming modes (M) | 2 | 1 | 0.200 | 5.4% | 0.2000 | 212.46 |
| between harvests (H) | 3 | 2 | 0.844 | 22.9% | 0.4220 | 448.29 |
| between runs (R) | 3 | 2 | 0.023 | 0.6% | 0.0115 | 12.22 |
| between Y × M × H × R | 10 | 4 | 1.929 | 52.3% | 0.4823 | 512.29 |
| within Y × M ×H × R (i.e. biological) | 324 | 0.305 | 8.3% | 0.0009 | ||
| 3.685 | 100.0% | |||||
The percentage of variance associated with year and harvest were considerably different for TOF-MS (“30of30” filter). The 4-way ANOVA data computed for TOF-MS, shows that the variance for year, farming mode, harvest, run, and factor interaction was 10.4%, 5.4%, 22.9%, 0.6%, and 52.3%. Since a considerably different family of masses were measured by the two spectrometers (345 for IT-MS versus 14 for TOF-MS, also see Figures 2 and 3), these differences were not unexpected. It should be noted, however, that in every case the F values were very high indicating a low statistical probability that the means for growing year, farming mode, harvest, and analytical run were equal.
Use of less stringent filters (“5of5” or “3of3” instead of “50of50” or “30of30”) resulted in the incorporation of more masses in the calculation (631 for IT-MS and 87 for TOF-MS), decreased percent variance associated with the growing year, farming mode, and harvest, and increased percent variance associated with the biological uncertainty (data not shown). As a result, the F values were lower and the differences in the means for growing year, farming mode, and harvest were not statistically significant. The F values were even lower when all the data (“unfiltered”) were used. These results suggest that the additional masses contributed little with respect to discriminating between the experimental factors and only contributed to the total variance.
Initially, a 3-way ANOVA was used with the data; only year, farming mode, and harvest were considered. However, the residuals for the factor interaction (Y × M × H), providing the analytical and biological uncertainty, gave PCA score plots that were structured (data not shown). Although the clusters were clearly separated, they lacked the random distribution expected for unbiased analyses. The nonrandom distribution suggested a systematic bias between the analytical runs on different days despite the normalization of the data. This may be due to the fact that the ambient temperature of the laboratory is not well controlled. Consequently, a 4-way ANOVA was used that included the analytical run as one of the experimental factors. The factor interaction residual gave a PCA score plot that was unstructured. Interestingly, the variance for between runs was only 2.8% and 0.6% for IT-MS and TOF-MS, respectively.
PCA
Figures 4 and 5 show the PCA score and loading plots acquired for IT-MS (“50of50” filter), and Figure 6 shows the PCA score plot for TOF-MS (“30of30” filter). Despite the difference in the spectral fingerprints (Figure 2 versus Figure 3) both spectrometers provide excellent discrimination between grapefruit grown in 2005 and 2006. The means for grapefruit from the two years are statistically different at a very high confidence level. These results visually and statistically confirm the results obtained by ANOVA (Table 1). Similar agreements of the PCA score plots between the two spectrometers were observed for farming mode and harvest. Consequently, only the score and loading plots for IT-MS are shown.
Figure 4.
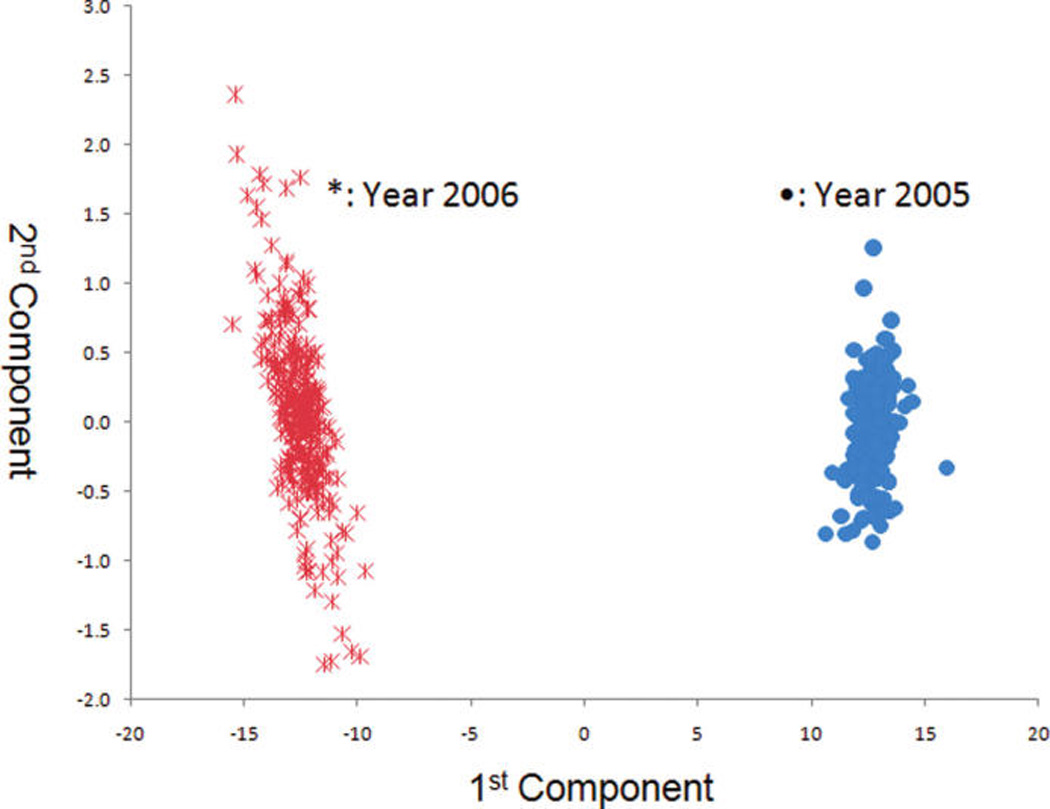
PCA scores plot for year for the IT-MS (n = 600).
Figure 5.
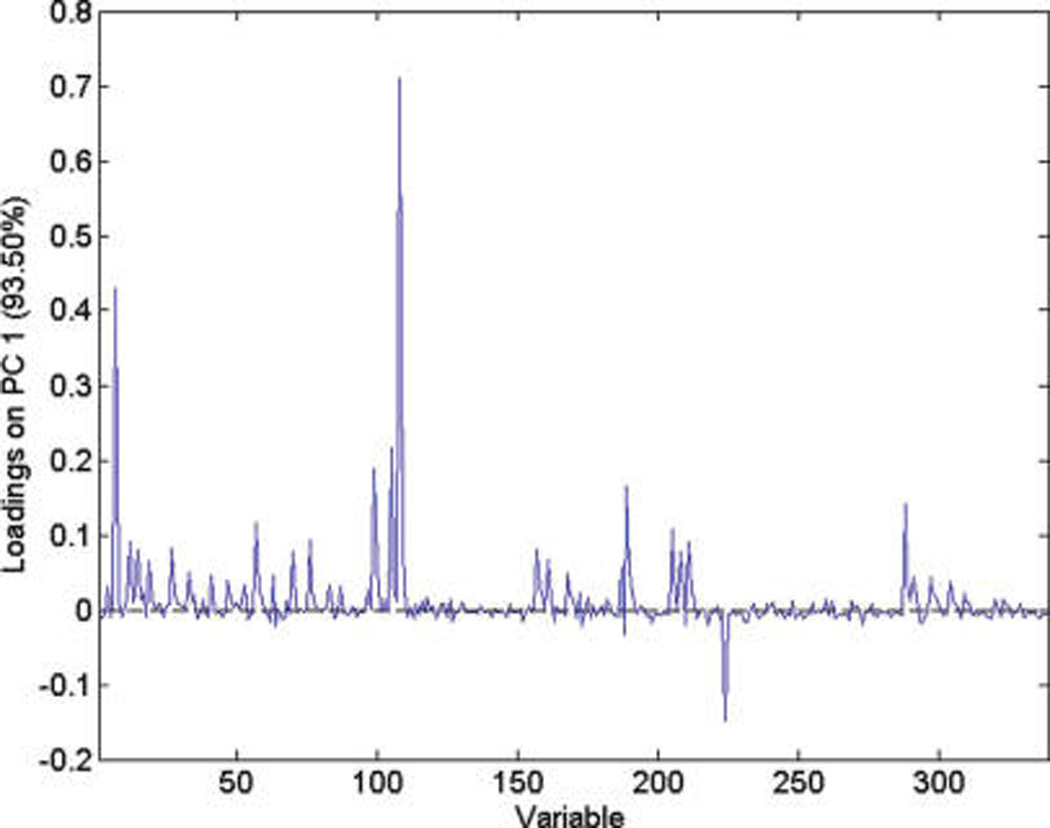
PCA loading plot for year for the IT-MS.
Figure 6.
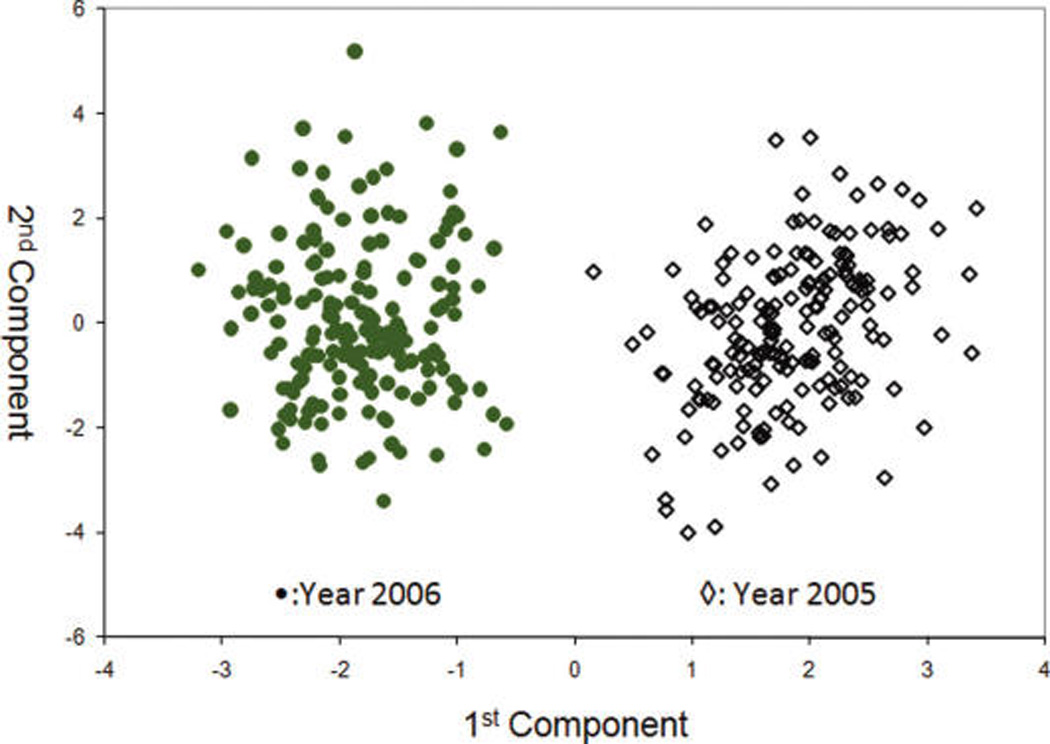
PCA scores plot for year for the TOF-MS (n = 360).
Figures 7 and 8 show the score plot and loading plot obtained from the IT-MS for the farming mode (organic vs conventional). There is excellent discrimination between samples grown with conventional and organic methods. Figures 9 and 10 shows the score and loading plots for early, mid, and late harvests. In this case, the scores for all three harvests are plotted together rather than a comparison of each pair. In either case, there is excellent separation of the three harvests. Again, the data in these figures visually and statistically confirm the results obtained in the previous section using ANOVA.
Figure 7.
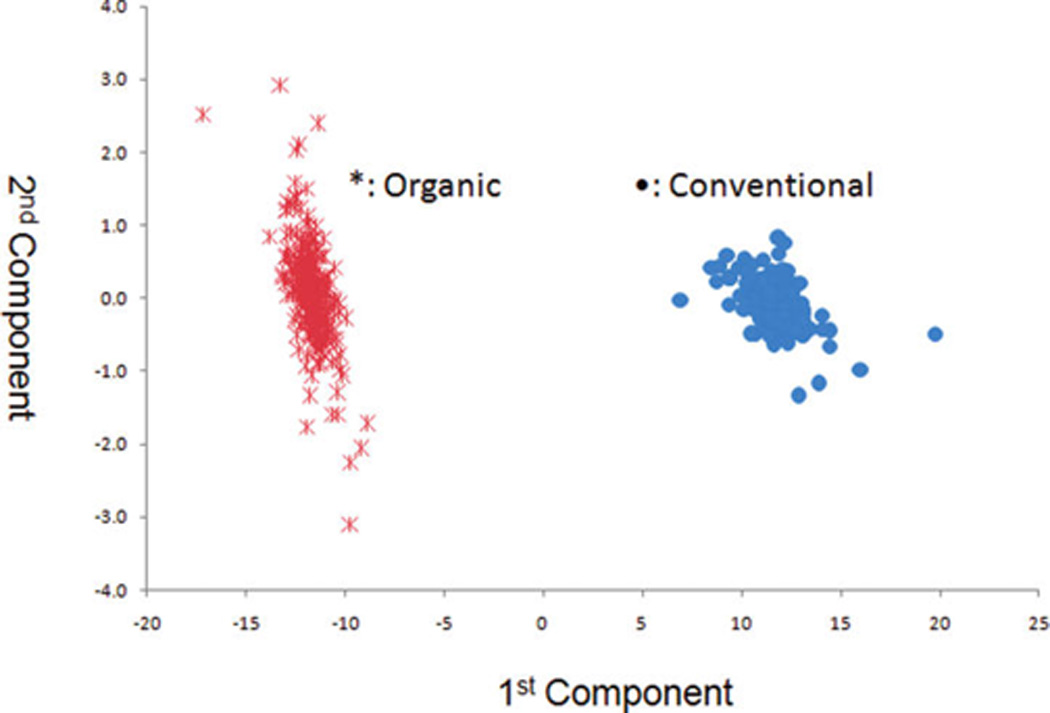
PCA scores plot for farming mode for the IT-MS MS (n = 600).
Figure 8.
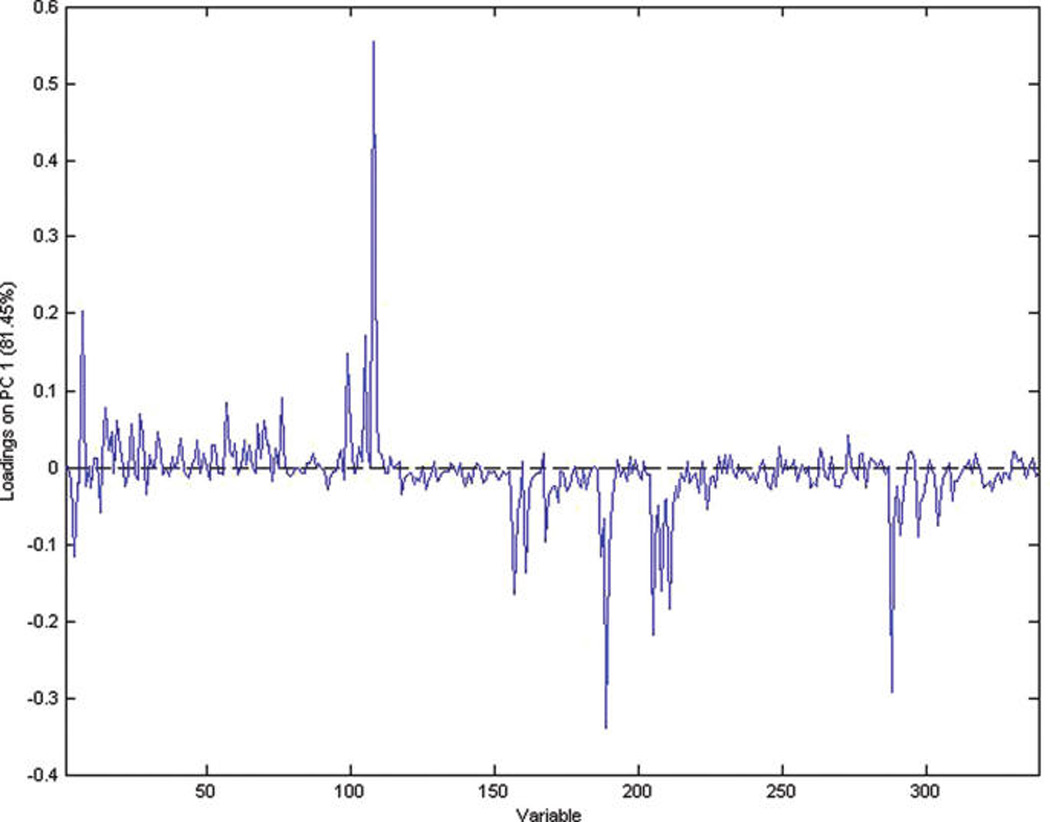
PCA loading plot for farming mode for the IT-MS.
Figure 9.
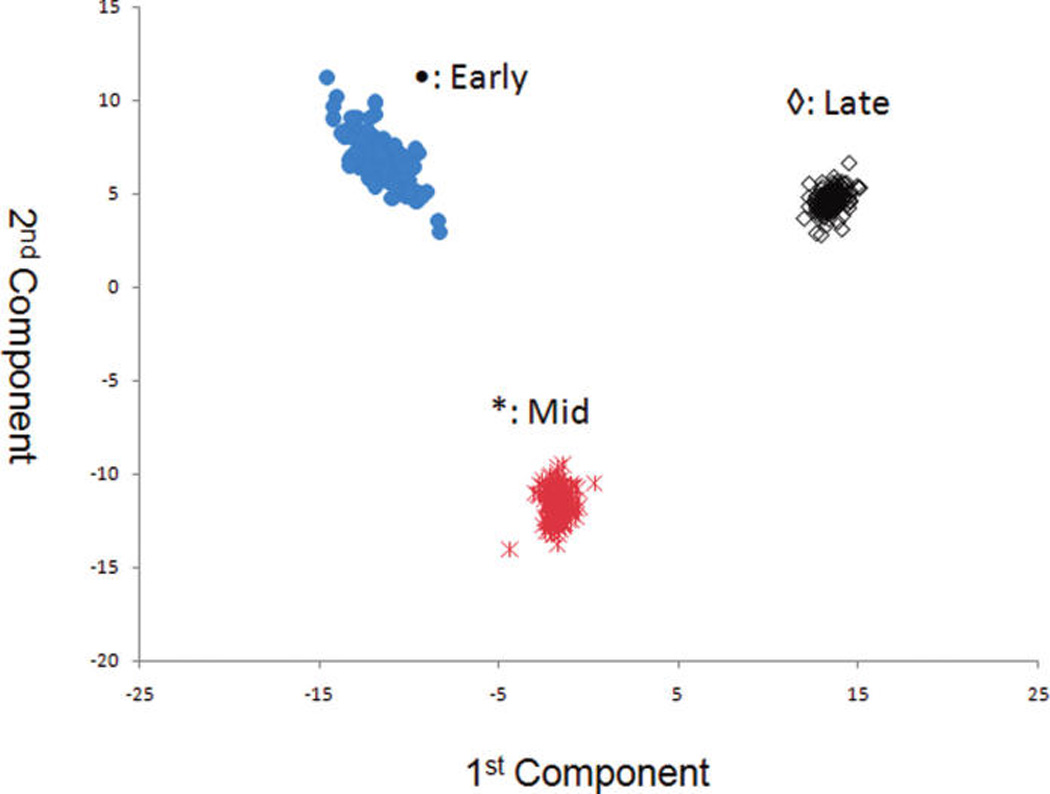
PCA scores plot for harvest for the IT-MS MS (n = 600).
Figure 10.
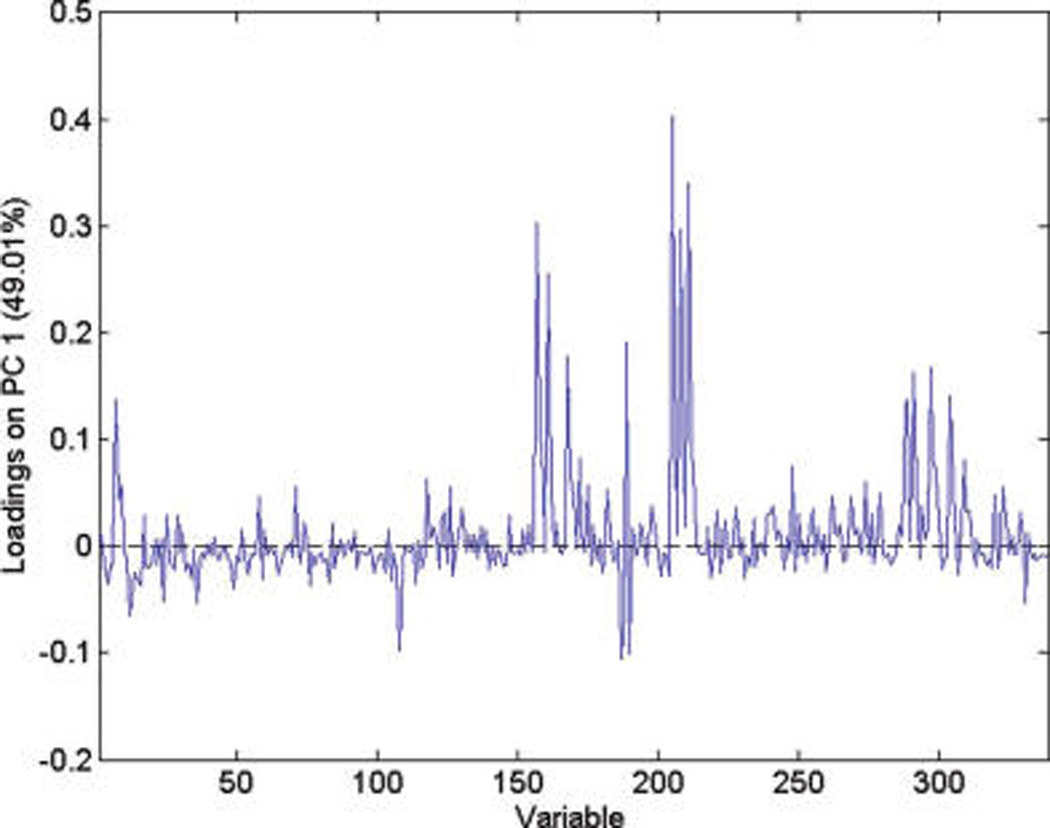
PCA loading plot for harvest for the IT-MS.
Significant Masses
The PCA loadings calculated for each variable (m/z in this case) correspond to the weighting (or usefulness) of that variable in discriminating between the two populations. With ANOVA, the variance can be calculated for each variable in each matrix. The variance for each variable (Table 2) clearly indicates its usefulness in separating the populations. The 33 ions in Table 2 for IT-MS represent approximately one tenth of the ions used in the ANOVA calculation and contribute 86.9% of the total variance. The remaining 312 ions contribute 13.1% of the total variance. Only those ions that contribute at least 0.1% have been listed.
Table 2.
Contribution of Ions and Experimental Factors to Total Variance for IT-MS
| % of mass total variance |
|||||||
|---|---|---|---|---|---|---|---|
| mass | total | year | farming mode |
harvest | run | Y×F×H×R | residuals |
| 163 | 0.2% | 9% | 29% | 3% | 12% | 44% | 4% |
| 175 | 18.8% | 21% | 1% | 1% | 6% | 66% | 4% |
| 204 | 0.7% | 26% | 0% | 5% | 13% | 51% | 5% |
| 212 | 0.1% | 2% | 15% | 4% | 11% | 56% | 13% |
| 224 | 0.7% | 21% | 4% | 0% | 8% | 65% | 2% |
| 232 | 0.4% | 24% | 5% | 0% | 6% | 63% | 2% |
| 244 | 0.5% | 28% | 5% | 0% | 2% | 61% | 4% |
| 252 | 0.2% | 24% | 5% | 0% | 5% | 61% | 5% |
| 265 | 0.2% | 23% | 4% | 0% | 6% | 64% | 3% |
| 273 | 0.1% | 29% | 5% | 0% | 1% | 60% | 3% |
| 287 | 1.5% | 20% | 3% | 0% | 8% | 64% | G% |
| 293 | 0.2% | 26% | 4% | 0% | 3% | 60% | 6% |
| 301 | 0.4% | 30% | 4% | 0% | 1% | 61% | 3% |
| 307 | 0.7% | 27% | 6% | 1% | 3% | 61% | 3% |
| 354 | 2.5% | 31% | 5% | 0% | 1% | 62% | 1% |
| 362 | 3.4% | 30% | 4% | 0% | 2% | 62% | 2% |
| 365 | 36.2% | 31% | 4% | 0% | 1% | 62% | 1% |
| 525 | 1.7% | 8% | 8% | 17% | 1% | 65% | 1% |
| 533 | 1.2% | 9% | 8% | 17% | 0% | 63% | 3% |
| 557 | 0.6% | 8% | 8% | 15% | 1% | 65% | 3% |
| 573 | 0.2% | 6% | 6% | 11% | 3% | 72% | 3% |
| 607 | 1.0% | 8% | 7% | 15% | 1% | 62% | 7% |
| 608 | 0.1% | 17% | 17% | 8% | 2% | 53% | 3% |
| 696 | 2.9% | 9% | 9% | 17% | 0% | 64% | 1% |
| 704 | 1.7% | 8% | 8% | 16% | 0% | 63% | 4% |
| 707 | 4.0% | 4% | 4% | 9% | 4% | 77% | 1% |
| 723 | 2.2% | 22% | 1% | 6% | 23% | 44% | 4% |
| 772 | 0.3% | 1% | 1% | 10% | 0% | 32% | 5% |
| 867 | 2.4% | 19% | 18% | 9% | 1% | 50% | 3% |
| 875 | 0.5% | 8% | 8% | 17% | 0% | 62% | 4% |
| 899 | 0.5% | 9% | 9% | 18% | 0% | 63% | 2% |
| 915 | 0.4% | 9% | 8% | 17% | 0% | 63% | 2% |
| 921 | 0.1% | 9% | 8% | 17% | 0% | 63% | 3% |
| 86.9% | |||||||
The data in Table 2 present the percent variance (column 2) each ion (m/z) contributes to the total variance and the percent that each experimental factor (columns 3–8) contributes to the ion variance (column 2). Thus, the sum of columns 3–8 is 100%. For example, consider m/z 175. The variance contributed by this ion makes up 18.8% of the total variance of the data. Of this variance, 21% comes from variance between years, i.e. from variance arising from the differences in the means for the two years, not random variance around the means. Little variance comes from farming mode or harvest; variance between run means accounts for 6%, variance between samples means accounts for 4%, and factor interaction accounts for 66%. Thus, m/z 175 would be useful for distinguishing between grapefruit from different years but would contribute far less toward other experimental factors.
It can be seen that most of the low mass ions (m/z 365 or lower) are useful for distinguishing between the growing years. To distinguish between farming modes, m/z 163, 212, 608, and 867 are useful. To distinguish between harvests, most of the high mass ions (m/z 525 and higher) are useful and m/z 723 can be used to distinguish between runs. The identification of the compounds associated with these ions is desirable but was not possible with any degree of certainty with the IT-MS and unit mass resolution. Use of TOF-MS with 1000 times better resolution still provided ambiguity in identification of the peaks when using online chemical identification programs. Since the spectra were obtained using direct injection, it was not possible to identify molecular and fragment ions and establish their association. HPLC—MS remains the best method for identification of individual compounds.
All of the intense peaks (100,000 counts, or more) in Figure 1 are found in Table 2. The high total counts of these ions means that any correlation with year, farming, mode, or run, no matter how small, will be statistically significant. Conversely, more than half the ions in Table 2 have less than 40,000 counts (less than 10% of the most intense ion). In some cases, they have counts at the 5% level. Since their total counts are low, their contribution is significant because they are highly correlated with one of the experimental factors. Consequently, care should be taken not to throw out ions because of their low count rate.
Conclusion
MS fingerprints, when combined with ANOVA-PCA, clearly established that there are chemical differences between grapefruit samples with respect to growing year, farming mode (conventional versus organic), and harvest. Statistical results for both the IT-MS and TFO-MS agreed despite the significant differences in their spectra. This data does not make it possible to identify specific compounds or to determine whether the difference in chemical composition affects the nutritional quality of the grapefruit.
Contributor Information
Pei Chen, Beltsville Human Nutrition Research Center, Agricultural Research Service, U.S. Department of Agriculture, Beltsville, Maryland 20705.
James M. Harnly, Beltsville Human Nutrition Research Center, Agricultural Research Service, U.S. Department of Agriculture, Beltsville, Maryland 20705
Gene E. Lester, Kika de la Garza Subtropical Agricultural Research Center, Agricultural Research Service, U.S. Department of Agriculture, Weslaco, Texas 78596
LITERATURE CITED
- 1.Executive Summary of Organic Trade Association’s 2007 Manufacturer Survey.
- 2.Magkos F, Arvaniti F, Zampelas A. Organic food: Nutritious food or food for thought? A review of evidence. Int. J. Food Sci. 2003;54:357–371. doi: 10.1080/09637480120092071. [DOI] [PubMed] [Google Scholar]
- 3.Asami DK, Hong YJ, Barrett DM, Mitchell AE. Comparison of the total phenolic and ascorbic acid content of freeze-dried and air dried marionberry, strawberry, and corn grown using conventional, organic and sustainable agricultural practices. J. Agric. Food Chem. 2003;51:1237–1241. doi: 10.1021/jf020635c. [DOI] [PubMed] [Google Scholar]
- 4.Blatt CR, McRae KB. Comparison of four organic amendments with a chemical fertilizer applied to three vegetables in rotation. Can. J. Plant Sci. 1998;78:641–646. [Google Scholar]
- 5.Meier-Ploeger A, Duden R, Vogtmann H. Quality of food plants grown with compost from biogenic waste. Agrie, Ecosyst. Environ. 1989;27:483–491. [Google Scholar]
- 6.Rembialkowska E. Organic agriculture and food quality. NATO Sci. Ser. Sci. Technol. Policy. 2004;44:185–204. [Google Scholar]
- 7.Roinila P, Vaisanen J, Granstedt A. Effects of different organic fertilization practices and mineral fertilization on potato quality. Biol. Agric. Hortic. 2003;21:165–194. [Google Scholar]
- 8.Warman PR, Havard KA. Yield, vitamin and mineral contents of organically and conventionally grown carrots and cabbage. Agrie, Ecosyst. Environ. 1997;61:155–162. [Google Scholar]
- 9.Weibel FP, Bickel R, Luthold ST, Alfoeldi T, Niffli U. Are organically grown apples tastier and healthier? A comparative field study using conventional and alternative methods to measure fruit quality. Acta Hortic. 2000;517:417–426. [Google Scholar]
- 10.Worthington V. Nutritional quality of organic versus conventional fruits, vegetables and grains. J. Ahem. Complementary Med. 2002;7:161–173. doi: 10.1089/107555301750164244. [DOI] [PubMed] [Google Scholar]
- 11.Woese K, Lang D, Boess C, Bogl KW. A comparison of organically and conventionally grown foods—Results of a review of the relevant literature. J. Sci. Food Agric. 1997;74:281–293. [Google Scholar]
- 12.Lester GE, Manthey JA, Buslig BS. Organic vs. conventionally grown Rio Red whole grapefruit and juice: comparison of production inputs, market quality, consumer acceptance, and human health-bioactive compounds. J. Agric. Food Chem. 2007;55:4474–4480. doi: 10.1021/jf070901s. [DOI] [PubMed] [Google Scholar]
- 13.Dangour AD, Dodhia SK, Hayter A, Allen E, Lock K, Uauy R. Nutritional quality of organic foods: a systematic review. Am. J. Clin. Nutr. 2009;90:680–685. doi: 10.3945/ajcn.2009.28041. [DOI] [PubMed] [Google Scholar]
- 14.Harrington DB, Vieira NE, Espinoza J, Kien JK, Romero R, Yergey AL. Analysis of variance-principal component analysis: a soft tool for proteomic discovery. Anal. Chim. Acta. 2005;544:118–127. [Google Scholar]
- 15.Luthria DL, Mukhopadhyay S, Robbins RJ, Finley JW, Banuelos GS, Harnly JM. UV spectral fingerprinting and analysis of variance-principal component analysis: a useful tool for characterizing sources of variance in plant materials. J. Agric. Food Chem. 2008;56:5457–5462. doi: 10.1021/jf0734572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harnly JM, Pastor-Corralesb MS, Devanand LL. Variance in the chemical composition of dry beans determined from UV spectral fingerprints. J. Agric. Food Chem. 2008;56:9819–9827. doi: 10.1021/jf900852y. [DOI] [PubMed] [Google Scholar]


