Abstract
Myeloid/lymphoid or mixed-lineage AF4 acute lymphoblastic leukemia (MLL-AF4 ALL) is a pediatric leukemia that occurs rarely in adults. MLL-AF4 ALL is typically characterized by the presence of chromosomal translocation (t(4;1l)(q21;q23)), leading to expression of MLL-AF4 fusion protein. Although MLL-AF4 fusion protein triggers a molecular pathogenesis and hematological presentations that are unique to leukemias, the precise role of this oncogene in leukemogenesis remains unclear. Previous studies have indicated that microRNAs (miRs) might modulate the expression of MLL-AF4 ALL fusion protein, thereby suggesting the involvement of miR in progression or suppression of MLL-AF4 ALL. We have previously demonstrated that miR-205 negatively regulates transcription of an MLL-AF4 luciferase reporter. Here, we report that exogenous expression of miR-205 in MLL-AF4 human cell lines (RS4;11 and MV4-11) inversely regulates the expression of MLL-AF4 at both messenger RNA (mRNA) and protein level. Furthermore, miR-205 significantly induced apoptosis in MLL-AF4 cells as evidenced by Annex in V staining using fluorescence-activated cell sorting (FACS) analysis. The proliferative capacity of leukemic cells was suppressed by miR-205. The addition of an miR-205 inhibitor was able to restore the observed effects. In conclusion, these findings demonstrate that miR-205 may have potential value as a novel therapeutic agent in the treatment of MLL-AF4 ALL.
Keywords: miR-205, MLL-AF4, leukemia, microRNA, oncogene expression, untranslated regions, proliferation
Introduction
MLL-AF4 is an acute lymphoblastic leukemia (ALL) that results from a balanced translocation between the myeloid/lymphoid or mixed lineage (MLL) and AF4 genes. This translocation (t(4;1l)(q21;q23)) is present in 50% of ALL cases in infants and 2% in children; however, it can also be found in 5%–6% of ALL cases in adults.1 Even when treated with stem cell transplantation, MLL-AF4 ALL is still associated with a high relapse rate and poor prognosis.2 Understanding of MLL-AF4 at the molecular level is necessary to improve current therapeutic approaches and to devise novel treatment strategies.
The MLL-AF4 oncogene or fusion protein is important for leukemic clonogenicity and for engraftment of this highly aggressive leukemia. Overexpression of the MLL-AF4 gene in lymphoid cells induces resistance to etoposide-mediated cytotoxicity. Reduction of MLL-AF4 transcript levels induces apoptosis and impairs cell proliferation.3,4 Therefore, targeted inhibition of MLL-AF4 expression may lead to an effective and highly specific treatment for this therapy-resistant leukemia.
MicroRNAs (miRs) are small, noncoding RNAs that can downregulate specific genes. miRs translationally repress genes when partially complementary sequences are present in the 3′-untranslated regions (UTRs) of target mRNAs (messenger RNAs), or by directing mRNA degradation.5 Mammalian miRs can control a number of cellular functions, including cellular proliferation and differentiation, and are also involved in tumorigenesis. Furthermore, the downregulation of subsets of miRs has been described in the initiation and progression of leukemia.6–10 Therefore, the identification of miRs that downregulate MLL-AF4 will provide a better understanding of leukemogenesis and may represent a novel targeted therapy for MLL-AF4 ALL.
A previous study reported that the MLL-AF4 fusion gene is downregulated by miR-128b. Downregulation of these specific miR is implicated in glucocorticoid resistance, and restoration of their levels may represent a potential therapeutic approach to the treatment of MLL-AF4 ALL.11 Using a luciferase reporter assay, we have earlier demonstrated that overexpression of miR-205 and miR-143 significantly reduced the activity of a reporter containing the MLL-AF4 3′-UTR. Conversely, mutations at the miR-205 and miR-143 target sites in the MLL-AF4 3′-UTR luciferase reporter rescued the activity.1
In this study, we explored the regulatory role of miR-205 on MLL-AF4 in ALL cells. Our results suggest that miR-205 downregulate s the expression of MLL-AF4 at both mRNA and protein levels. Evidence is also presented that the restoration of miR-205 expression in MLL-AF4 ALL cells leads to suppression of cell proliferation and induces apoptotic cell death.
These results reveal a regulatory role for miR-205 in the tumorigenesis of MLL-AF4 ALL. We propose that upregulation of miR-205 may present a novel therapeutic approach for treatment of MLL-AF4 ALL.
Materials and methods
Cell culture
The human leukemia cell line RS4;11 and MV4-11 (American Type Culture Collection [ATCC], Manassas, VA, USA) carrying the chromosomal translocation t(4;ll) (q21;q23) were used to express different MLL-AF4 variants. The human leukemia cell line THP-1 (ATCC) harboring MLL-AF9 fusion protein was used as a control.
Transfection
On the day before transfection, RS4;11 cells were seeded in six-well plates at a density of 1–2 × 105 cells per well. Cells were transfected using Hiperfect (Qiagen, Hilden, Germany) according to the manufacturer’s instructions with synthetic miR-205 at a concentration of 100 nM. The mature miR-205 sequence was UCCUUCAUUCCACCGGAGUCUG. Transfection efficiency in RS4;11 cells was assessed using fluorescent duplex small interfering RNA (siRNA)-FAM (Genepharma, Shanghai, People’s Republic of China) as a positive transfection control, and scrambled oligonucleotides were used as controls for nonspecific effects (sense 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense 5′-ACGUGACACGUUCGGAGAATT-3′). miR-205 inhibitors are synthetic RNA that inhibit miR-205 expression (Genepharma, Shanghai, People’s Republic of China). Cells were transfected using Hiperfect according to the manufacturer’s instructions with miR-205 inhibitors at a concentration of lOO nM.
Real-time reverse transcriptase polymerase chain reaction
Total RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA, USA) as instructed by the manufacturer. Quantitative real-time polymerase chain reaction (qRT-PCR) analyses were performed as described. The primers were designed using PRIMER-EXPRESS software (Applied Biosystems Ine, Foster City, CA, USA): MLL-AF4 (sense 5′-ACAGAAAAAAGTGGCTCCCCG-3′; antisense 5′-TATTGCTGTCAAAGGAGGCGG-3′); HOXA9 (sense 5′-CCACCATCCCCGCACA-3′; antisense 5′-AACAGGGTTTGCCTTGGAAA-3′); HOXA7 (sense 5′-CGCCAGACCTACACGCG-3′; antisense 5′-CAGGTAGCGGTTGAAGTGGAA-3′); β-Actin (sense 5′-CCGACAGGATGCAGAAGGAG-3′; antisense 5′-CAAGAAAGGGTGTAACGCAACT-3′). All qRT-PCR data include at least three independent experiments, with three replicates per experiment. The expression of mature miR-205 was analyzed using the TaqMan miR Assay (Applied Biosystems Inc, Foster City, CA, USA).
Western blot analysis
Protein extracts were prepared with a radioimmunoprecipitation assay (RIPA) buffer containing the protease inhibitor phenylmethanesulfonylfluoride (PMSF). Standard techniques were used for Polyacrylamide gel electrophoresis, tank-based transfer to Immobilon Hybond-C membranes (Amersham Biosciences, Piscataway, NJ, USA), and immunodetection. Western blot analysis using antibodies against AF4 (Abcam, Cambridge, MA, USA) was performed in accordance with the manufacturer’s instructions. Transferred proteins were visualized with SuperSignal West Pico Chemoluminescent Substrate (Pierce, Rockford, IL, USA).
Cell cycle analysis, apoptosis assay, and fluorescence-activated cell sorting analysis
Cell cycle analysis was performed as described.1 The program ModFit (Verity Software House, Topsham, ME, USA) was used to analyze and evaluate the data. Apoptosis was measured with the human Annexin V-FITC (fluorescein isothiocyanate)/7AAD kit (BD Biosciences, Franklin Lakes, NJ, USA) according to the manufacturer’s instructions. Briefly, 2–5 × 105 cells were washed with phosphate-buffered saline (PBS) at indicated time points after transfection and were incubated with Annexin V-FITC solution for 10 minutes at room temperature. A fluorescence-activated cell sorting (FACS) Calibur flow cytometry instrument (Beeton Dickinson, Biosciences, San Jose, CA, USA) was used to analyze the cells immediately after cell preparation. A cell counting kit (CCK)-8 assay (Dojindo Laboratories, Kumamoto, Japan) was used to monitor cell growth. A phycoerythrin-conjugated cluster of differentiation (CD)-133 antibody (eBioscience, San Diego, CA, USA) was used to monitor cell surface expression of CD133 using FACS analysis.
Statistical analysis
The statistical significance of differences in miR expression was assessed using the Wilcoxon rank sum test and the modified t-test. Analysis of variance (ANOVA) or t-tests were used to compare data in apoptosis assays and qRT-PCR, and P-values < 0.05 were considered statistically significant. All analyses were performed with Statistical Package for the Social Sciences version 11.0 (SPSS Inc., Chicago, IL, USA).
Results
miR-205 downregulates MLL-AF4 expression in human MLL-AF4 leukemia cell lines
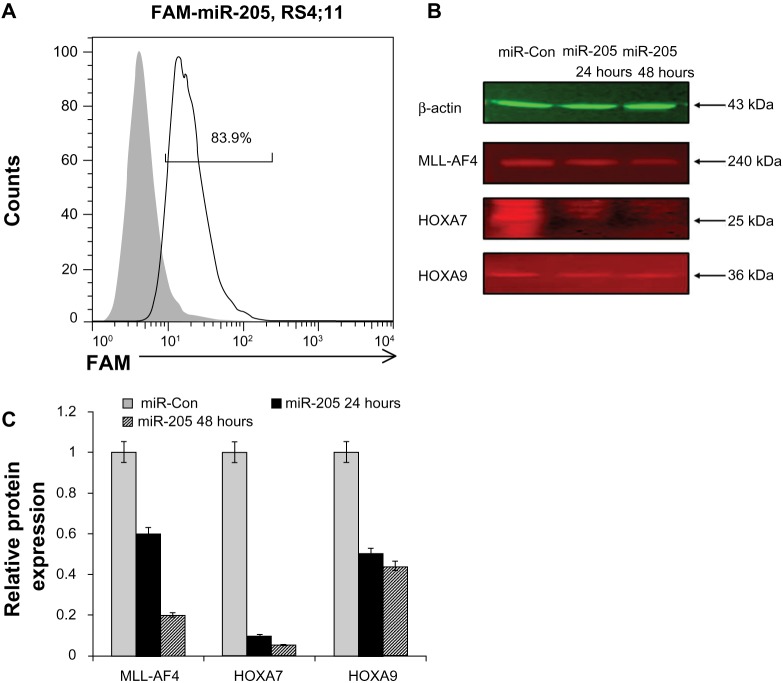
We have previously demonstrated that miR-205 represses transcription of a luciferase reporter containing the MLL-AF43′-UTR. To confirm that miR-205 could downregulate endogenous MLL-AF4 expression in leukemic cells, we transfected synthetic miR-205 or scrambled oligonucleotides into the human MLL-AF4 leukemia cell line RS4;11, which contains the chromosomal translocation t(4;ll)(q21;q23). MLL-AF4 mRNA levels and MLL-AF4 protein levels were measured by qRT-PCR and Western blot, respectively. The RS4;11 cell line expresses MLL-AF4 and very low levels of miR-205. The trans feetion efficiency was determined using fluorescent FAM-labeled miR, and FACS showed that approximately 84% of RS4;11 cells were FAM-positive 24 hours after transfection, indicating that the synthetic miRs were efficiently transfected into the cells (Figure 1A). In addition, transfection efficiencies were also monitored with glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-positive control, and we found that GAPDH-positive control could downregulate the expression of GAPDH (data not shown). Overexpression of miR-205 resulted in significantly reduced (60%–70%) MLL-AF4 mRNA expression levels, and the strongest effect was observed 48 hours after transfection, while overexpression of a scrambled control miR had little effect (Figure 1B). Western blot analysis showed that MLL-AF4 protein levels were also reduced in miR-205-overexpressing cells, indicating that miR-205 downregulates MLL-AF4 expression in a human leukemia cell line at both the RNA and the protein levels (Figure 1C).
Figure 1.
Expression of miR-205 in RS4;11 cells results in the downregulation of MLL-AF4, H0XA7, and H0XA9 mRNA and protein levels. (A) FACS of cells transfected with FAM-conjugated siRNA to determine transfection efficiency. The percentage of FAM-positive cells 48 hours after transfection was approximately 84%. (B) Expression of endogenous MLL-AF4, HOXA7, and HOXA9 proteins in MLL-AF4 ALL transfected with miR-205. Protein expression was determined using Western blot analysis. (C) Protein fold change of MLL-AF4, HOXA7, and HOXA9 proteins in MLL-AF4 ALL transfected with miR-205 detected with Western blot.
Abbreviations: ALL, acute lymphoblastic leukemia; miR-Con, microRNA control; FACS, fluorescence-activated cell sorting; miR, microRNA; MLL, mixed lineage; mRNA, messenger RNA; siRNA, small interfering RNA.
miR-205 inhibits MLL-AF4 downstream target genes HOXA7 and HOXA9 in a human MLL-AF4 leukemia cell line
To determine if miR-205 regulates genes downstream of MLL-AF4, we examined HOXA7 and HOXA9 mRNA and protein levels in RS4;11 cells transfected with miR-205 (Figure 2). Synthetic miR-205 or scrambled oligonucleotides were transfected into RS4;11 cells, and significant levels of exogenous mature miR-205 could be detected 48 hours post-transfeetion (Figure 2A). These levels of miR-205 were capable of causing the repression of HOXA7 and HOXA9, the mRNA levels of which decreased by 63.0% and 45.5%, respectively, 48 hours post-transfection with miR-205 (Figure 2C). Western blot analysis showed that HOXA7 and HOXA9 protein levels were also reduced in miR-205-transfected cells (Figure 1C). Therefore, apart from its regulatory effect on MLL-AF4, miR-205 also appears to downregulate HOXA7 and HOXA9 expression, at least in part by targeting the MLL-AF4 gene. Surface expression of the differentiation marker CD133 did not change after miR-205 transfection (data not shown).
Figure 2.
Expression of miR-205 in the RS4;11 cell line inhibits MLL-AF4 downstream target genes HOXA7, HOXA9 and HOXA10. (A) miR-205 mRNA levels in RS4;11 cells transfected with synthetic miR-205 were measured by qRT-PCR at 24 hours and 48 hours. (B) The level of miR-205 mRNA was measured by qRT-PCR at 24 hours and 48 hours following transfection with synthetic miR-205 inhibitors. (C) mRNA levels of MLL-AF4, HOXA7, HOXA9 and HOXAIO in RS4;11 cells transfected with miR-205 as measured by qRT-PCR. (D) mRNA levels of MLL-AF4, HOXA7, and HOXA9 in RS4;11 cells transfected with miR-205 inhibitor as measured by qRT-PCR.
Note: P-values <0.05 were considered statistically significant.
Abbreviations: Con, control; miR, microRNA; MLL, mixed lineage; mRNA, messenger RNA; qRT-PCR, quantitative real-time polymerase chain reaction.
Transfection of RS4;11 cells with an miR-205 inhibitor did not significantly change the levels of miR-205 (Figure 2B), but the inhibitor was capable of inhibiting endogenous miR-205 sufficiently to restore mRNA levels of the downstream target genes H0XA7 and H0XA9 (Figure 2D).
miR-205 induces apoptosis in the RS4;11 cell lines and inhibits leukemic proliferation
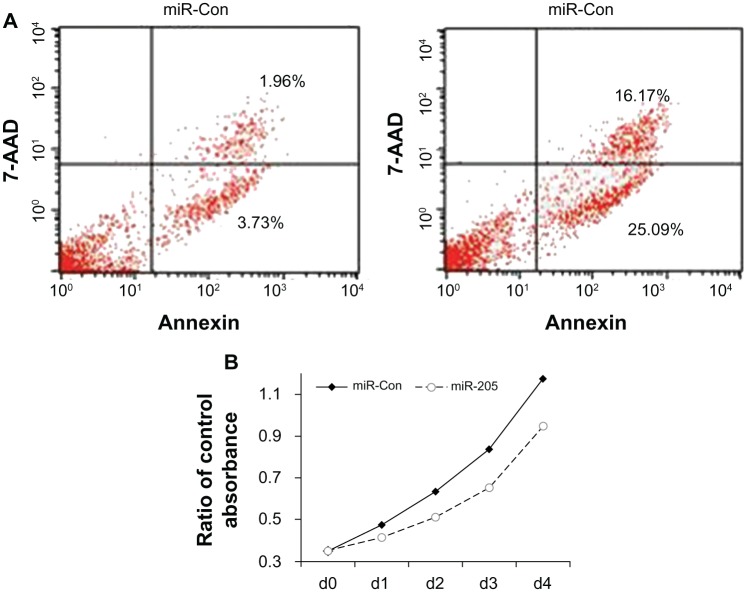
MLL-AF4 is important for engraftment of this ALL, and downregulation of MLL-AF4 expression induces apoptosis and impairs proliferation. We hypothesized that if miR-205 could inhibit MLL-AF4 expression, cell proliferation and apoptosis might also be impaired. To examine the biological effects of miR-205 expression in ALL cells harboring the MLL-AF4 translocation, we overexpressed miR-205 in the RS4;11 human leukemia cell line, and examined its effects on cell cycle, differentiation, growth, and apoptosis. The RS4;11 cell line was used in our experiments because it is known to contain endogenous miR-205 at low levels only. Cells that were transfected with synthetic miR-205 exhibited a 7-fold increase (P < 0.05) in apoptosis 96 hours post-transfection, as measured by Annexin V staining using FACS analysis (Figure 3A). The overexpression of miR-205 resulted in the inhibition of cell growth when compared with controls, as seen in the difference in viable cells between the treatment groups (Figure 3B). Surface expression of CD133 and cell cycle did not change after miR-205 transfection (data not shown). These observations could be explained by the fact that miR-205 induces apoptosis in almost half of RS4;11 cells, therefore making it virtually impossible to study the effect on differentiation marker and cell cycle. Although transfection of the miR-205 duplex upregulated miR-205 levels up to 100-fold in MLL-AF9-positive THP-1 cells, it failed to induce any apoptotic cell death (data not shown).
Figure 3.
miR-205 expression inhibits proliferation and induces apoptosis in RS4;11 cells. (A) FACS analysis to determine Annexin and V/7AAD staining in RS4;11 cells transfected with miR-205 or the negative control. (B) Growth of RS4;11 cells following transfection with miR-205 or the negative control. Viable cells were measured using a CCK-8 assay. X-axis shows the different time points. Cell proliferation analysis was performed in three independent experiments.
Abbreviations: CCK, cell counting kit; FACS, fluorescence-activated cell sorting; miR, microRNA; MLL, mixed lineage; d, day; Con, control.
We further validated these findings using a different MLL-AF4-positive cell line, MV4-11. Our results demonstrate that miR-205 downregulates MLL-AF4 protein expression (Figure 4A), and inhibits cell growth (Figure 4B) effectively in the MV4-11 cell line.
Figure 4.
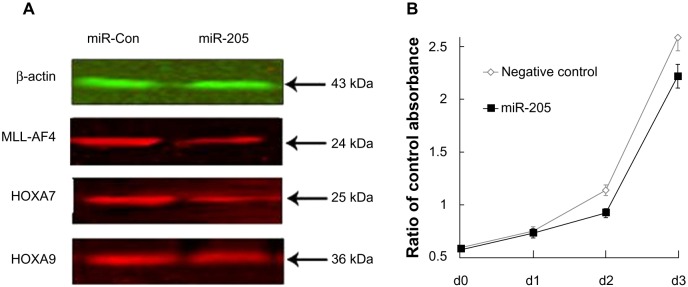
MiR-205 expression inhibits proliferation and MLL-AF4 expression in MV4-11 cells. (A) Expression of endogenous MLL-AF4, HOXA7, and HOXA9 proteins in MV4-11 cells transfected with miR-205. Protein expression was determined using Western blot analysis. (B) Growth of MV4-11 cells following transfection with miR-205 or the negative control.Viable cells were measured using a CCK-8 assay. X-axis shows the different time points. Cell proliferation analysis was performed in three independent experiments.
Abbreviations: CCK, cell counting kit; miR, microRNA; MLL, mixed lineage; d, day; Con, control.
Discussion
MLL-AF4 ALL is an aggressive cancer that is difficult to treat. Current treatment strategies involving intensive chemotherapy and/or stem cell transplantation are associated with a high relapse rate and poor prognosis; therefore, novel treatment approaches are required. Several recent studies have proposed that miRs are directly involved in regulating MLL-AF4 oncogene expression and therefore have become the focus for therapeutic applications. For instance, miR-128b and miR-221 are two miRs that are downregulated in MLL-AF4 ALL. Ectopic expression of both miRs restores glucocorticoid resistance in these cells through downregulation of the MLL-AF4 gene.12 Another recent report has demonstrated that miR-143 is a negative regulator of MLL-AF4 expression, and acts as a tumor suppressor in MLL-AF4 B-cell ALL.1
It has been reported that the miR-205 can function as a tumor suppressor in renal cancer, prostate cancer, head and neck squamous cell carcinoma, and breast cancer. 13–16 miR-205 is reported as a novel regulator of the anti-apoptotic protein Bcl2, and is downregulated in prostate cancer. Consistent with its anti-apoptotic target Bc12, miR-205 promoted apoptosis in prostate cancer cells. To the best of our knowledge, this is the first report to establish that miR-205 regulates MLL-AF4 oncogene expression in the human MLL-AF4 ALL cell lines. Overexpression of miR-205 not only downregulates the MLL-AF4 expression directly at both the mRNA and the protein levels, but also negatively regulates the expression of downstream MLL-AF4-targeted genes HOXA7 and HOXA9. Although surface expression of the differentiation marker CD133 did not alter after miR-205 transfection, this observation could be because miR-205 induced apoptosis in the majority of RS4;11 cells, hence rendering the study of miR-205 on the CD 133 differentiation marker difficult.
The MLL-AF4 fusion protein plays an important role in leukemic clonogenicity and engraftment of this highly aggressive leukemia. Decreased expression of the MLL-AF4 oncogene led to induced apoptosis and impaired cellular proliferation, and our findings provide further support to these observations. The restoration of miR-205 expression in MLL-AF4 ALL cells inhibited MLL-AF4 expression, resulting in a marked reduction of cell growth and induction of apoptosis. However, this effect may also have been indirectly caused by the downregulation of MLL-AF4-targeted genes HOXA7 and HOXA9. However, our results indicated that the miR-205 inhibitor slightly altered expression of miR-205 (but this was not statistically significant). Interestingly, the inhibitor was capable of inhibiting endogenous miR-205 sufficiently to restore mRNA levels of the downstream target genes HOXA7 and HOXA9 (Figure 2D). Taking the above into account, no differences in mRNA expression of miR-205 could be explained by the fact that although miR inhibitors may act as a modulator of suppression of endogenous miR, they may fail to alter the expression at the miR level. There are now multiple other examples where silencing by an miR is observed, with either no change in the mRNA level, or with a significantly smaller decrease in mRNA levels than observed for protein.17–19 Expression of an miR-205 inhibitor increased MLL-AF4 oncogene expression and its downstream target genes, further confirming the regulatory effect of miR-205 on MLL-AF4.
This study adds to our current understanding of the mechanisms involved in MLL-AF4 ALL development and tumorigenesis. In addition, it is highly possible that low endogenous miR-205 levels in the RS4;11 cell line are contributing to the upregulation of ML.L-AF4 expression, a critical mediator of malignant cell proliferation. Overall, our findings provide strong evidence that dysregulation of miR-205 plays an important role in the development of MLL-AF4 ALL. Therefore, strategies regulating miR-205 expression might offer a potential novel therapy for MLL-AF4 ALL.
Acknowledgments
This work was supported by the Beijing Nova Program (2011114), National Natural Science Foundation of China (Nos 30800482, 30971297, 81102242, 81000221, 81270610, and 90919044), the Beijing Natural Science Foundation of China (No 7102147 and 7132217), High and New Technology Program of the PLA (2010gxjs091), Capital Medical Development Scientific Research Fund (No 2007–2040), National Public Health Grand Research Foundation (No 201202017), the funding of the public health project (Zl11107067311070), and the National 973 Project of China (No 2005CB522400).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Dou L, Zheng D, Li J, et al. Methylation-mediated repression of microRNA-143 enhances MLL-AF4 oncogene expression. Oncogene. 2012;31:507–517. doi: 10.1038/onc.2011.248. [DOI] [PubMed] [Google Scholar]
- 2.Bueno C, Montes R, Catalina P, Rodriguez R, Menendez P. Insights into the cellular origin and etiology of the infant pro-B acute lymphoblastic leukemia with MLL-AF4 rearrangement. Leukemia. 2011;25:400–10. doi: 10.1038/leu.2010.284. [DOI] [PubMed] [Google Scholar]
- 3.Thomas M, Gessner A, Vornlocher HP, Hadwiger P, Greil J, Heidenreich O. Targeting MLL-AF4 with short interfering RNAs inhibits clonogenicity and engraftment of t(4;11)-positive human leukemic cells. Blood. 2005;106:3559–3566. doi: 10.1182/blood-2005-03-1283. [DOI] [PubMed] [Google Scholar]
- 4.Starn RW. MLL-AF4 driven leukemogenesis: what are we missing? Cell Res. 2012;22:948–949. doi: 10.1038/cr.2012.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alemdehy MF, Erkeland SJ. MicroRNAs: key players of normal and malignant myelopoiesis. Curr Opin Hematol. 2012;19:261–267. doi: 10.1097/MOH.0b013e328353d4e9. [DOI] [PubMed] [Google Scholar]
- 6.Mendier JH, Maharry K, Radmacher MD, et al. RUNX1 mutations are associated with poor outcome in younger and older patients with cytogenetically normal acute myeloid leukemia and with distinct gene and Micro RNA expression signatures. J Clin Oncol. 2012;30:3109–3118. doi: 10.1200/JCO.2011.40.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye H, Liu X, Lv M, et al. MicroRNA and transcription factor co-regulatory network analysis reveals miR-19 inhibits CYLD in T-cell acute lymphoblastic leukemia. Nucleic Acids Res. 2012;40:5201–5214. doi: 10.1093/nar/gks175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baer C, Claus R, Frenzel LP, et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012;72:3775–3785. doi: 10.1158/0008-5472.CAN-12-0803. [DOI] [PubMed] [Google Scholar]
- 9.Marcucci G, Mrózek K, Radmacher MD, Garzon R, Bloomfield CD. The prognostic and functional role of microRNAs in acute myeloid leukemia. Blood. 2011;117:1121–1129. doi: 10.1182/blood-2010-09-191312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabbri M, Croce CM. Role of microRNAs in lymphoid biology and disease. Curr Opin Hematol. 2011;18:266–272. doi: 10.1097/MOH.0b013e3283476012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kotani A, Ha D, Hsieh J, et al. miR-128b is a potent glucocorticoid sensitizer in MLL-AF4 acute lymphocytic leukemia cells and exerts cooperative effects with miR-221. Blood. 2009;114:4169–4178. doi: 10.1182/blood-2008-12-191619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotani A, Ha D, Schotte D, den Boer ML, Armstrong SA, Lodish HF. A novel mutation in the miR-128b gene reduces miRNA processing and leads to glucocorticoid resistance of MLL-AF4 acute lymphocytic leukemia cells. Cell Cycle. 2010;9:1037–1042. doi: 10.4161/cc.9.6.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majid S, Saini S, Dar AA, et al. MicroRNA-205 inhibits Src-mediated oncogenic pathways in renal cancer. Cancer Res. 2011;71:2611–2621. doi: 10.1158/0008-5472.CAN-10-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majid S, Dar AA, Saini S, et al. MicroRNA-205-directed transcriptional activation of tumor suppressor genes in prostate cancer. Cancer. 2010;116:5637–5649. doi: 10.1002/cncr.25488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Childs G, Fazzari M, Kung G, et al. Low-level expression of microRNAs let-7d and miR-205 are prognostic markers of head and neck squamous cell carcinoma. Am J Pathol. 2009;174:736–745. doi: 10.2353/ajpath.2009.080731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H, Zhu S, Mo YY. Suppression of cell growth and invasion by miR-205 in breast cancer. Cell Res. 2009;19:439–448. doi: 10.1038/cr.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 18.Chen X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science. 2004;303:2022–2025. doi: 10.1126/science.1088060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimmino A, Calin GA, Fabbri M et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sei USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]