Abstract
Background
Ovarian carcinomas consist of at least five distinct diseases: high-grade serous, low-grade serous, clear cell, endometrioid, and mucinous. Biomarker and molecular characterization may represent a more biologically relevant basis for grouping and treating this family of tumors, rather than site of origin. Molecular characteristics have become the new standard for clinical pathology, however development of tailored type-specific therapies is hampered by a failure of basic research to recognize that model systems used to study these diseases must also be stratified. Unrelated model systems do offer value for study of biochemical processes but specific cellular context needs to be applied to assess relevant therapeutic strategies.
Methods
We have focused on the identification of clear cell carcinoma cell line models. A panel of 32 “ovarian cancer” cell lines has been classified into histotypes using a combination of mutation profiles, IHC mutation-surrogates, and a validated immunohistochemical model. All cell lines were identity verified using STR analysis.
Results
Many described ovarian clear cell lines have characteristic mutations (including ARID1A and PIK3CA) and an overall molecular/immuno-profile typical of primary tumors. Mutations in TP53 were present in the majority of high-grade serous cell lines. Advanced genomic analysis of bona-fide clear cell carcinoma cell lines also support copy number changes in typical biomarkers such at MET and HNF1B and a lack of any recurrent expressed re-arrangements.
Conclusions: As with primary ovarian tumors, mutation status of cancer genes like ARID1A and TP53 and a general immuno-profile serve well for establishing histotype of ovarian cancer cell We describe specific biomarkers and molecular features to re-classify generic “ovarian carcinoma” cell lines into type specific categories. Our data supports the use of prototype clear cell lines, such as TOV21G and JHOC-5, and questions the use of SKOV3 and A2780 as models of high-grade serous carcinoma.
Introduction
Ovarian cancer is a diverse set of diseases and amongst the most clinically significant, epithelial ovarian cancers (EOC), at least five distinct entities exist [1]–[9]. At a broad level, the terms type I and type II EOCs are often applied, wherein high-grade serous carcinomas (HGSCs) are type II and all other histologies are type I cancers [8]. However, even within type I, distinct entities exist, namely low-grade serous carcinoma (LGSC), endometrioid carcinoma (ENOCa), clear cell carcinoma (CCC) and mucinous carcinoma (MUC). There is significant data suggesting that a majority of HGSC originate from fallopian tube epithelium [1], [10]–[13], while low-grade serous tumors are generally still thought to arise from the ovarian surface epithelium – though this relationship is being questioned [7], [14]. ENOCa and CCC tumors occur in a background of endometriosis and could represent a spectrum of displaced, malignant endometrium [15]–. Finally, mucinous tumors are exceedingly rare and their true origin is difficult to ascertain with subgroups of distinct histology. Their resemblance to other mucinous epithelial malignancies, most notably gastric cancers, has added to the confusion of their origin [3],[21]–[23].
Clinical responses and epidemiological differences are also apparent between histotypes. High-grade serous cancers show the best initial response rates to the current standard chemotherapy regime of platinum and taxanes [24], [25]. Familial BRCA1/BRCA2 mutations also appear largely restricted to this histology [26]–[28]. Conversely, the minor histotypes tend to occur in younger patient populations and more frequently present at lower stage [29]–[31]. A list of some of the more distinguishing features between histotypes types is given in Table 1.
Table 1. Discriminating Features Of The Five Major Histotypes Of Ovarian Carcinoma.
| Clear Cell Carcinoma | Endometrioid Carcinoma | Mucinous Carcinomas (& Mucinous Borderline Tumors) | Low-Grade Serous Carcinomas (& Serous Borderline tumors) | High-grade serous carcinoma | |
| Presentation | Presents at younger age and low stage (pelvic mass) [4], [29]–[31] | Presents at younger age (than HGSC) [4], [29]–[31] | Presents at younger age (than HGSC) [4], [29]–[31]Histopathological similarity to gastric carcinomas (intestinal type) [1], [8], [31] | Presents at younger age (than HGSC) [4], [29]–[31] | Presents at older age (than other histotypes) and high stage (ascites common) [4], [8], [29]–[31] |
| Precursors | Associated with Endometriosis [1], [8], [16], [82] | Associated with Endometriosis [1], [8], [16], [82] | Potential link to Walthard cell nests [83] | Association between ovarian surface and fallopian tube epithelium is unclear [14] | Significant subset associated with serous tubal intraepithelial carcinoma (STIC) [1], [8], [11], [84] |
| Genetics, Genomics & Biomarkers | TP53 wild-type [4], [15] | TP53 mutations rare [4] | TP53 wild-type (borderline)TP53 mutant (∼1/2 of carcinomas) [4], [8] | TP53 wild-type [4], [8] | TP53 mutant (virtually ubiquitous, >96%) [9], [85] |
| Negligible occurrence of (germline) BRCA1/2 mutations [26]–[28], [86] | Negligible occurrence of (germline) BRCA1/2 mutations [26]–[28], [86] | Negligible occurrence of (germline) BRCA1/2 mutations [26]–[28], [86] | Frequency of BRCA1/2 mutations presumed low | Germline and somatic BRCA dysfunction/high proportion of hereditary (germline) BRCA1/2 mutation carriers [9], [26]–[28], [86] | |
| High frequency of ARID1A and PIK3CA mutations; frequent loss of PTEN expression; near ubiquitous expression of HNF1B [15], [16], [45] | High frequency of ARID1A mutations; Moderate frequency of PIK3CA, CTNNB1, and PTEN (loss/LOH) mutations [16], [45] | High frequency (55–75%, carcinoma-borderline) of KRAS mutations (ras-pathway mutation almost exclusively KRAS); Frequent (19%) of high-level ERBB2 amplification [22] | High frequency mutually exclusive RAS-pathway mutations (KRAS, BRAF, NRAS, or ERBB2) typical of borderline serous tumors [5], [8], [10], [66] | Complex karyotypes suggestive of a period of massive genomic instability [9], [87] | |
| Treatment Response and Outcomes | Higher frequency of thromboembolic complications [15], [88] Low stage outcome better than (stage matched) HGSC; poor initial response to therapy and worse high stage outcomes (vs. HGSC) [15], [89] | Typically longer interval to progression or death than HGSC (confounded by stage). Stage matched analysis (Stage III) suggests little difference in outcome to HGSC [90] | Overall favorable (due to prevalence of low-stage disease), however very poor outcome on recurrence [31], [43] | Poor response to current treatment standards (Platinum/taxane) [91], [92] | Good initial response rates to current treatment standards (Platinum/taxane); relapse and eventual treatment failure is common [4], [24] |
Regardless of origin or histological similarities and differences, biomarker and genomic studies have been successfully used to distinguish each histotype and may represent a far more biologically relevant basis for classifying and subsequently treating EOCs. Although this concept is well-accepted, and gaining traction on becoming a new clinical standard, ambiguous cell line models perpetuated through molecular biology bench research hamper the development of tailored type-specific therapies. Those using bench experiment model systems must recognize that, like primary cancers, the models used to study these diseases must also be stratified. Although biochemical studies can generate useful information from using a variety of unrelated model systems, disease specific studies need to apply cellular context. The vast majority of research employing functional studies on “ovarian cancer” cell lines does not properly ascertain the background of their model systems. Resulting conclusions may be difficult to interpret and the value of potential therapeutic targets may be questionable as is the true relevance to a particular disease.
Cell line studies of ovarian cancer have been severely hampered due to the lack of proper annotation of “ovarian” carcinoma cell lines. Once in culture, cells no longer have easily identifiable morphological traits to aid in histological classification. Additionally, human error, mislabeling and the generic feature of “epithelial-like” cell lines have also led to mix ups of cell lines and contamination which has resulted in un-interpretable data [32], [33]. In the post-genome era, biomarkers and genomic features for ovarian carcinoma subtypes are very well established. Screening techniques to assay biomarkers and verify genomic features are also widely accessible. Here, we present a panel of biomarkers and molecular features across 32 commonly used and in-house derived ovarian carcinoma cell lines. Our initial goal was to establish a bona-fide list of CCC cell lines for our own research program, however we propose establishing type-specificity for these cell lines should became the new standard in planning and executing experiments around any study on epithelial ovarian carcinoma.
Methods
Cell culture
Cells were maintained in a humidified incubator at 37C with 5% CO2. See Table S1 for a list of cell lines, culture conditions and contributing labs and repositories. Some cell lines were derived in-house (labeled with “VOA#”) through continuous in vitro culture of primary patient material obtained through the OVCARE Tumor bank. All patients with tissue deposited in the OVCARE tumor bank provided written consent for experimental studies including sequencing, IHC characterization, and derivation of long-term cell lines from tissue samples. The OVCARE tumor bank study was approved under University of British Columbia and British Columbia Cancer Agency Research Ethics Board H05-60119 protocol.
All cell lines were subjected to identity testing using STR genotyping (AmpFlSTR Identifiler, Applied Biosystems) at the College of American Pathologist's (CAP) accredited Centre For Translational and Applied Genomics (CTAG) as per manufacturer directives. Only lines with profiles matching public repository records, reported STR [32], and/or original patient tumors (in the case of in-house derived cell lines) were retained for further study.
Immunohistochemistry and Calculator of Subtype Prediction (COSP)
Cell lines were scraped from culture plates, washed 2× with PBS and pelleted. Cell pellets were re-suspended in ∼500 μl 10% Neutral Buffered Formalin (NBF) and allowed to fix overnight. Cells were pelleted again and re-suspended in a Histo-gel (Thermo-Fisher) plug prior to embedding in paraffin. A tissue microarray (TMA) was constructed as previously described [4] taking 3×2 mm cores from the cell line plugs. Immunohistochemistry (IHC) was performed on 4 μm sections on a Ventana Discovery XT system as previously described [2], [34], refer to table S2 for details of antibodies used. Histotype prediction was done using the Calculator of Subtype Prediction (COSP) [2] in tumor bank mode. Tumour bank mode was chosen due to the nature of the fixed cell lines and the controlled fixation period similar to the tumor bank process on which this predictor was trained. Scoring criteria for IHC was done visually and followed the exact guidelines proposed in the original COSP paper [2]. IHC for mismatch repair (MMR) proteins (Table S5) was performed as described in [35], a complete absence of staining for any given MMR protein resulted in a score of 0 (negative), and is presumed to result in MMR deficiency.
mRNA transcripts
RNA was extracted from cell lines using Qiazol-miRNeasy kit (Qiagen) protocol and from primary tumors, 12 randomly selected from each histotype, using the miRNeasy FFPE kit (Qiagen). All RNA transcript levels were measured using the NanoString nCounter system [36] and data normalized with nSolver software v1.1 (NanoString Inc.) using endogenous control genes (ACTB, SDHA, RPL19, POLR1B, PGK1) as per manufacturers directives. In the case of TFF3 mRNA levels we considered any sample with detectable transcripts to be positive and substituted a score of “1” in place of TFF3 IHC when using COSP. The detection threshold (DT) for mRNA was considered to be the maximum count from spike-in negative control probes (across all cell line samples) plus 2 standard deviations. Statistical tests were calculated using GraphPad Prism v6.0c software.
Mutation Testing and Genomic Analysis
Genomic DNA was extracted using standard methods (Gentra Puregene kit; Qiagen). Regions encompassing mutations of known significance (Cancer hotspots) were Sanger sequenced using M13-tagged primers. Sequencing of ARID1A was done through a combination of custom hybrid capture and transcriptome sequencing on an Illumina GAII next generation sequencing (NGS) system as described previously [16], [37]. Associated raw data is deposited in the NCBI Sequence Read Archive under BioProjects PRJNA209481, PRJNA209482, and PRJNA209484. All noted variants were either verified by Sanger sequencing or considered validated if recorded in the Cancer Cell Line Encyclopedia (CCLE) [38] and/or the COSMIC database [39]. Expressed re-arrangements were predicted from transcriptome sequencing data for CCC cell lines TOV21G, JHOC-5, JHOC-7, JHOC-9, and RMG-2 using deFuse [40] (Table S3).
Copy Number Analysis
DNA copy number was inferred from Affymetrix SNP 6.0 genome-wide microarrays. Arrays were run as per manufacturers directives and copy number ratio generated from an unpaired reference. Detection of copy number changed regions was done using a segmentation algorithm. All analysis and visualization was executed with Partek Genomics Suite 6.6, raw data is available from NCBI GEO [Accession GSE48351].
Results
Histotype by COSP in ovarian cancer cell lines
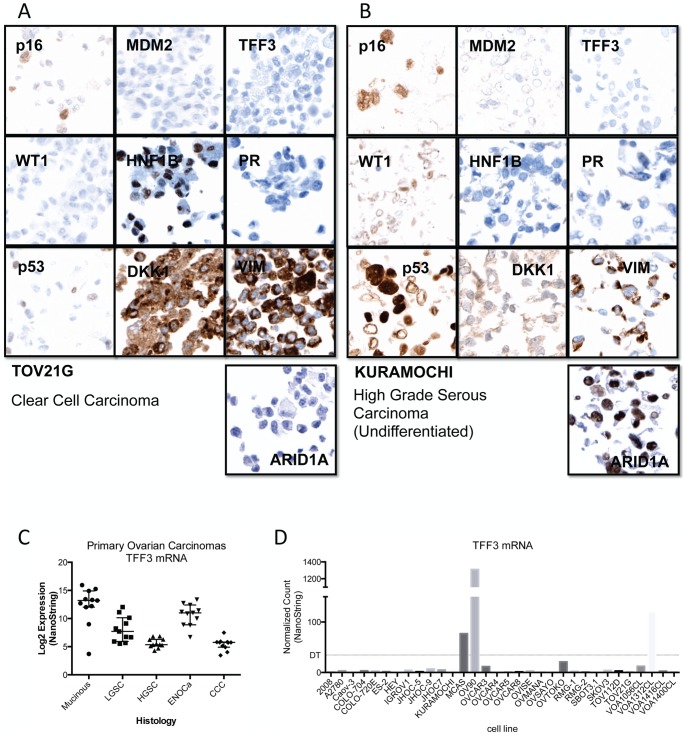
Ovarian cancer cell lines grown in culture do not exhibit the histological phenotypes that are useful for classification into the major disease types. Our group has described a large number of immunohistochemical biomarkers that show specific profiles across these histotypes [4], [41]–[43]. A core panel of 9 IHC markers combined with a predictive algorithm, the Calculator for Ovarian Subtype Prediction (COSP), can be used to reliably distinguish between types [2]. We have previously demonstrated a high level of concordance between our predictive immune-classifier and consensus expert gynecopathological review [2], . Initially, we applied this panel (Fig 1A–B), and the COSP predictive algorithm, to 32 ovarian cancer cell lines of ambiguous histotype to establish if cell lines retained representative characteristics sufficient to classify cell lines to their true disease origins and allow for type-specific ovarian cancer model development. The TFF3 IHC marker, which is normally strongly associated with the mucinous type and seen at moderate frequency in ENOCa and LGSC [2], was negative across all samples (Table S4), suggesting this secreted factor, if expressed at all, may be expelled quickly from the cells and washed away in media. Consequently, TFF3 IHC may not be a reliable biomarker measurement for use with cultured cells. However, the prevalence of TFF3 mRNA in primary samples appeared similar to that reported by IHC [2], [44], with consistently higher expression in mucinous carcinomas (p<0.01; Fig. 1C). We therefore substituted detectable TFF3 mRNA for IHC and scored any cell line with detectable mRNA as “1” in our COSP algorithm (Fig. 1D and Table 2).
Figure 1. Prediction of histotype was in part based on the COSP algorithm using 9 IHC markers [2].
(A–B) representative IHC from a typical high-grade serous ovarian carcinoma cell line, Kuramochi, and a clear cell carcinoma cell line, TOV21G. In addition to the 9-marker COSP panel, IHC for ARID1A (BAF250a) is also shown as a mutation surrogate. (C) TFF3 mRNA expression from 60 ovarian cancer samples (12 of each histotype). As noted previously high expression is most prevalent in MUC, followed by ENOCa and LGSC [2], [4]. Expression in our pilot cohort suggests the highest levels of TFF3 in MUC, which was significantly higher than all other groups (Tukey's adjusted p<0.01); no other pairwise comparisons had p<0.05. (D) TFF3 mRNA detected in ovarian cancer cell lines was used in place of an IHC score as the secreted TFF3 was considered a poor biomarker for cell culture conditions. Any cell line with measurable TFF3 mRNA above the NanoString detection threshold (see methods) was considered positive (score of 1 for use in the COSP algorithm).
Table 2. Validation of the histotype of commonly used ovarian carcinoma cell lines using immunohistochemistry based prediction via COSP and mutational profiling.
| Cell Line | Reported Histotype | COSP Markers | COSP Prediction (Clinical) | Non-COSP Markers | DNA Mutational Profile | Validated Cell Line HistoType | ||||||||||||
| p16 (CDKN2A) | MDM2 | TFF3 [mRNA] | p53 | VIMENTIN | WT1 | HNF1B | PR | DKK1 | CCC | ENOCa | HGSC | MUC | ARID1A (BAF250A) | TP53 | Other* | |||
| JHOC-5 | CCOC | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 85 | 13 | 2 | 0 | 1b | nc | none detected | Clear Cell Carcinoma (CCC) |
| JHOC-7 | CCOC | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 99 | 1 | 0 | 0 | 1b | nc | PIK3CA | |
| JHOC-9 | CCOC | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 85 | 13 | 2 | 0 | 0a | nc | PIK3CA/ARID1A | |
| RMG-2 | CCOC | 0 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 97 | 3 | 0 | 0 | 0a | nc | PPP2R1A/ARID1A | |
| TOV21G | CCOC | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 55 | 41 | 4 | 0 | 0a | nc | KRAS/PTEN/PIK3CA/ARID1A | |
| OVTOKO | CCOC | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 99 | 1 | 0 | 0 | 0b | nc | none detected | |
| OVMANA | CCOC | 1 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 99 | 1 | 0 | 0 | 0a | nc | PIK3CA/ARID1A | |
| A2780 | Adenocarcinoma | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 94 | 6 | 0 | 0a | nc | PTEN/ARID1A | Endometrioid Carcinoma (ENOCa) |
| IGROV1 | Mixed | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 17 | 82 | 1 | 0 | 0a | p.Y126C (het) | ARID1A/PTEN | |
| TOV112D | ENOCa | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 38 | 62 | 0 | 1b | p.R175H (Hm) | CTNNB1 | |
| 2008 | ENOCa | 0 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 98 | 0 | 2 | 0 | 0 | c.572_574 del CTC (het)/c.673-1 G>T (het, splice site) | none detected | Atypical Non-Serous [CCC/ENOCa] cell lines** |
| OVISE | CCOC | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 1 | 20 | 54 | 25 | 0 | 0a | nc | ARID1A | |
| ES-2 | CCOC | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 100 | 0 | 0 | 1b | p.S241F (het) | BRAF | |
| SKOV3 | adenocarcinoma | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 100 | 0 | 1b | nc | PIK3CA/ARID1A | |
| OVSAYO | CCOC | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 100 | 0 | 1 | R249M (Hm) | none detected | High Grade Serous Ovarian Carcinoma (HGSC) |
| CAOV3 | Adenocarcinoma | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 100 | 0 | 1b | p.Q136* (Hm) | none detected | |
| Kuramochi | Undifferentiated | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 97 | 0 | 1 | p.D281Y (Hm) | none detected | |
| OVCAR-3 | Adenocarcinoma | 1 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 1b | p.R248Q (Hm) | none detected | |
| OVCAR-4 | Serous Adenocarc. | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 100 | 0 | 1b | p.L130V (Hm) | none detected | |
| OVCAR-5 | Adenocarcinoma | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 1b | nc | KRAS | |
| OVCAR-8 | Adenocarcinoma | 0 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 0 | 38 | 62 | 0 | 1b | p.Y126_splice (Hm) | none detected | |
| COLO-720E | carcinoma | 1 | 1 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 20 | 80 | 0 | 1 | p.A138V (het)/c.1118delA (het) | PTEN | |
| COLO-704 | carcinoma | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 1 | c.1146delA (het) | PTEN | |
| Hey | carcinoma | 0 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 10 | 90 | 0 | 1 | nc | KRAS | |
| VOA1400_CL | HGSC primary tumour | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 100 | 0 | 1 | E198* (het) | none detected | |
| VOA1416_CL | HGSC ascites | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 1 | 0 | 0 | 100 | 0 | 1 | nc | none detected | |
| VOA1072_CL | HGSC primary tumour | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 1 | R248Q (Hm) | none detected | |
| VOA1312_CL | LGSC ascites | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 100 | 0 | 0 | 1 | nc | KRAS | Low-Grade Serous Carcinoma (LGSC) |
| VOA1056_CL | LGSC primary tumour | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 39 | 61 | 0 | 1 | nc | NRAS | |
| MCAS | mucinous carcinoma | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 100 | 1b | 127bp del (Hm, Ex 4) | KRAS | mucinous carcinoma |
| RMG-1 | CCOC | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 22 | 0 | 76 | 3 | 1 | nc | none detected | unclassified |
| OV90 | Adenocarcinoma | 1 | 1 | 1 | 2 | 0 | 0 | 1 | 1 | 0 | 0 | 53 | 47 | 0 | 1b | p.S215R (Hm) | none detected | |
COSP and AIRD1A markers were scored as positive (1) or negative (0), except for p53: null mutation (0), wildtype (1), mutated (2).
ARID1A IHC: a – corresponding ARID1A nonsense or frameshift mutation detected, b – no ARID1A mutation detected in sequencing data (if no letter code, sequence information was unavailable)
COSP algorithm can be found at http://www.gpec.ubc.ca/index.php?content=papers/ovcasubtype.php
TP53 mutations are noted as heterozygous (het) or Homozygous/Hemizygous (Hm)
Sequencing of BRAF, KRAS, ERBB2, NRAS, CTNNB1, EGFR, PTEN, PIK3CA, PPP2R1A, DICER1 and ARID1A
Many previously described CCC lines showed features characteristic of their expected origins. In addition to the COSP 9-marker panel, we added IHC for ARID1A (BAF250a). Given the strong negative association of mutation status and detectable protein expression [16] we considered this assay as a surrogate mutation test useful in segregating endometriosis associated ovarian cancer from other subtypes, most notably high-grade serous, as ARID1A mutations appear to be exceedingly rare in this subtype [16], [45].
Mutational Profiles: Clear cell specific molecular features
We next tested cell lines for mutations in common ovarian cancer associated genes (Table 2 and Table S5). As some of the cell lines we tested are also part of a larger Cancer Cell Line Encyclopedia (CCLE) repository data set [38], we cross-validated our mutation testing with this database as well as the COSMIC database [39]. We focused on regions of known significance in common cancer genes including hotspots in BRAF, KRAS, NRAS, ERBB2, EGFR, CTNNB1, PIK3CA, PPP2R1A and DICER1. All coding exons of TP53 were verified in all cell lines. ARID1A mutations were tested using a custom NGS gene hybrid capture strategy [37] in RMG-1, RMG-2, JHOC-5, JHOC-7, JHOC-9, TOV21G, and ES-2; for all other cell lines we used ARID1A data from COSMIC and CCLE in addition to IHC as an ARID1A mutation-testing surrogate (Table 2).
As with our IHC data most CCC lines maintained a profile consistent with the CCC histotype including mutations in PIK3CA and ARID1A. Further, loss of ARID1A expression, demonstrated by IHC, showed good concordance with presence of known truncating mutations, as noted for primary tumor specimens [16]. As expected IHC for p53 correlated well with occurrence of mutations. For cell lines with a recorded mutation (at time of submission) in either CCLE or COSMIC all detected mutations matched repository records, except for a homozygous/hemizygous 127-bp deletion of TP53 detected in MCAS. We presume that the 127-bp deletion in MCAS (at the end of exon 4; Fig. S1) may have been overlooked in the CCLE exon sequencing strategy as in our experience false negatives are prevalent in NGS datasets. Overall, the addition of mutation data was particularly helpful in supporting initial classification from COSP (Table 2).
We took note that a number of cell lines often used as high-grade serous models or generically as “ovarian carcinoma” had both ARID1A mutations and immuno-profiles consistent with the endometrioid type, the third most common type accounting for less than 10% of ovarian carcinomas [3], [4]. Along with the immuno-classification the presence of an ARID1A mutation provided compelling evidence of a non-HGSC origin. The incidence of TP53 and ARID1A mutation was near mutually exclusive with the exceptions of IGROV1 and 2008. IGROV1 carries two frame shift mutations in ARID1A (p.M274fs/p.G1847fs) and a mutation of unknown significance in TP53 (p.Y126C (het)), though p53 expression by IHC appeared normal. The 2008 cell line had undetectable ARID1A, suggesting loss of function, and also carried two TP53 mutations (c.572_574 delCTC (het)/c.673-1 G>T (het, splice site)) and corresponding p53 IHC overexpression. These atypical combinations of mutation could plausibly be explained by a propensity to accumulate mutations in cell lines with DNA mismatch repair (MMR) deficiencies, as has been reported for IGROV1, SKOV3, and A2780 [46], [47]. We validated MMR pathway protein expression with IHC for MLH-1, PMS-2, MSH-2, and MSH-6 (Table S5) and observed loss of two or more MMR proteins in IGROV1, SKOV3, A2780 TOV21G, COLO-704 and COLO-720E; no MMR protein deficiency was noted in the 2008 cell line.
Copy Number profiles of clear cell lines
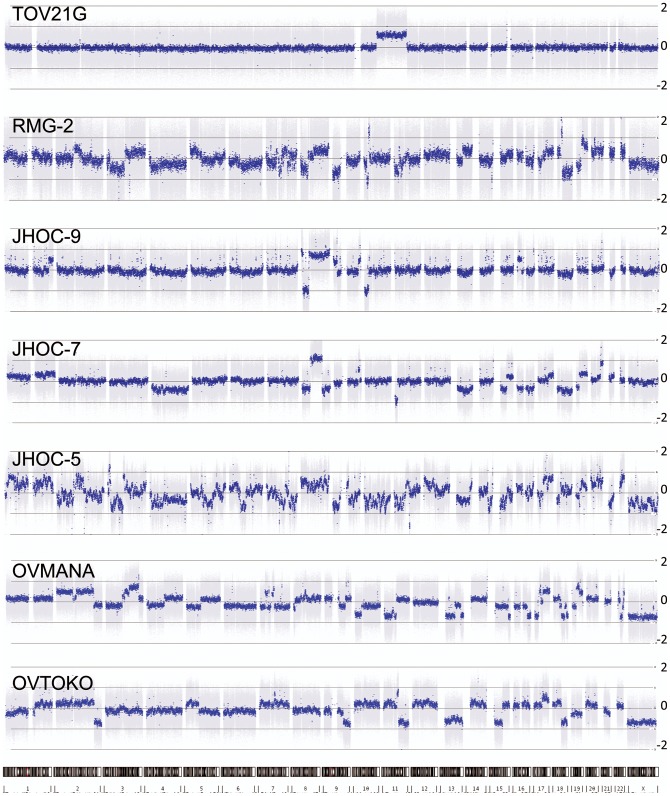
As our primary objective was to describe CCC cell lines we generated copy number profiles of bona-fide CCC cell lines using Affymetrix SNP6.0 microarrays. Consistent with previous reports using primary tumor samples, CCC lines showed a moderate degree of copy number abnormalities, suggesting a genome that has undergone some degree of genomic instability.
A limited number of literature reports have highlighted genes with mutations, overexpression and/or amplification amongst primary CCC, some with a relationship to survival or advanced disease [4], [16], [45], [48]–[59]. As we observed in mutation profiles, our bona-fide CCC cell line panel was representative of clear-cell associated copy number changes (Figure 2, Table 3). Most showed modest copy number gains for HNF1B (5/7) and MET (4/7), including one with high-level amplification (JHOC-5), similar to previous reports for CCC tumors [53], [56], [57]. Although 3/7 CCC lines showed copy number gain of ERBB2, in all cases the amplicon segment also encompassed the nearby CCC biomarker HNF1B, and none were positive for HER2 protein expression by IHC (not shown). Copy number loss around TP53 was observed only in a single CCC cell line (OVMANA; heterozygous loss) and, as noted above, all CCC lines appeared to have a normal-like expression pattern for p53 (IHC score 1).
Figure 2. Genome-wide copy number profiles of bona-fide ovarian CCC cell lines.
A large range of copy number changes are seen including typical Chr8 gains and Chr17 gains surrounding the CCC biomarker HNF1B gene, see also Table 3.
Table 3. Copy number changes across putative CCC oncogenes, tumor suppressors, and biomarkers.
| Segment Copy Number | JHOC-5 | JHOC-7 | JHOC-9 | OVMANA | OVTOKO | RMG-2 | TOV21G | References |
| ARID1A | 3.422 | 2.397 | NC | 2.314 | NC | NC | NC | [16], [45] |
| ERBB2 | 3.061 | NC | NC | NC | 2.514 | 2.382 | NC | [52] |
| HNF1B | 3.061 | 3.109 | 3.533 | NC | 2.514 | 2.382 | NC | [53] |
| MAP1LC3A | NC | NC | NC | NC | NC | NC | NC | [54] |
| MET | 8.465 | NC | NC | 3.451 | 2.346 | 2.667 | NC | [53], [56], [57] |
| PIK3CA | 0.971 | 1.222 | NC | NC | NC | 6.482 | NC | [48], [49] |
| PPM1D | 3.157 | 2.329 | NC | 3.004 | 3.009 | 2.382 | NC | [50], [51] |
| STAT3 | 3.314 | 3.142 | NC | NC | 2.522 | 2.382 | NC | [53] |
| TP53 | NC | NC | NC | 1.320 | 2.410 | NC | NC | [4] |
| YAP1 | NC | NC | NC | NC | 1.264 | NC | NC | [59] |
| ZNF217 | 2.893 | 5.897 | 4.717 | 3.412 | 3.648 | 2.589 | NC | [55], [58] |
| CDKN2A | 0.163 | NC | NC | NC | 0.246 | 1.244 | NC | [58] |
| CDKN2B | 0.454 | NC | NC | NC | 0.602 | 1.244 | NC | [58] |
NC = no change in copy number was detected.
Transcriptome profile of clear cell lines
As with other ovarian carcinoma types, recurrent translocations amongst CCC have not been described, though only a minimal number of studies have been undertaken [16]. Our transcriptome sequencing data on RMG-1, RMG-2, JHOC-5, JHOC-7, JHOC-9, TOV21G, and ES-2 suggests recurrent expressed rearrangements are at least rare and were not detected amongst these cell lines. A moderate number of expressed intra- and inter-chromosomal rearrangements were detectable (Table S4), though all were unique to each respective cell line; some were visible by multi-colored FISH (Figure 3). Both expressed and non-expressed translocations resulting in gene gain/loss of function or promoter exchange may serve to influence pathway activation, and overall expression, profiles of CCC. A systematic analysis was considered beyond the scope of this study and there is currently an absence of an equivalent knowledge base derived from primary CCC tumors for comparison.
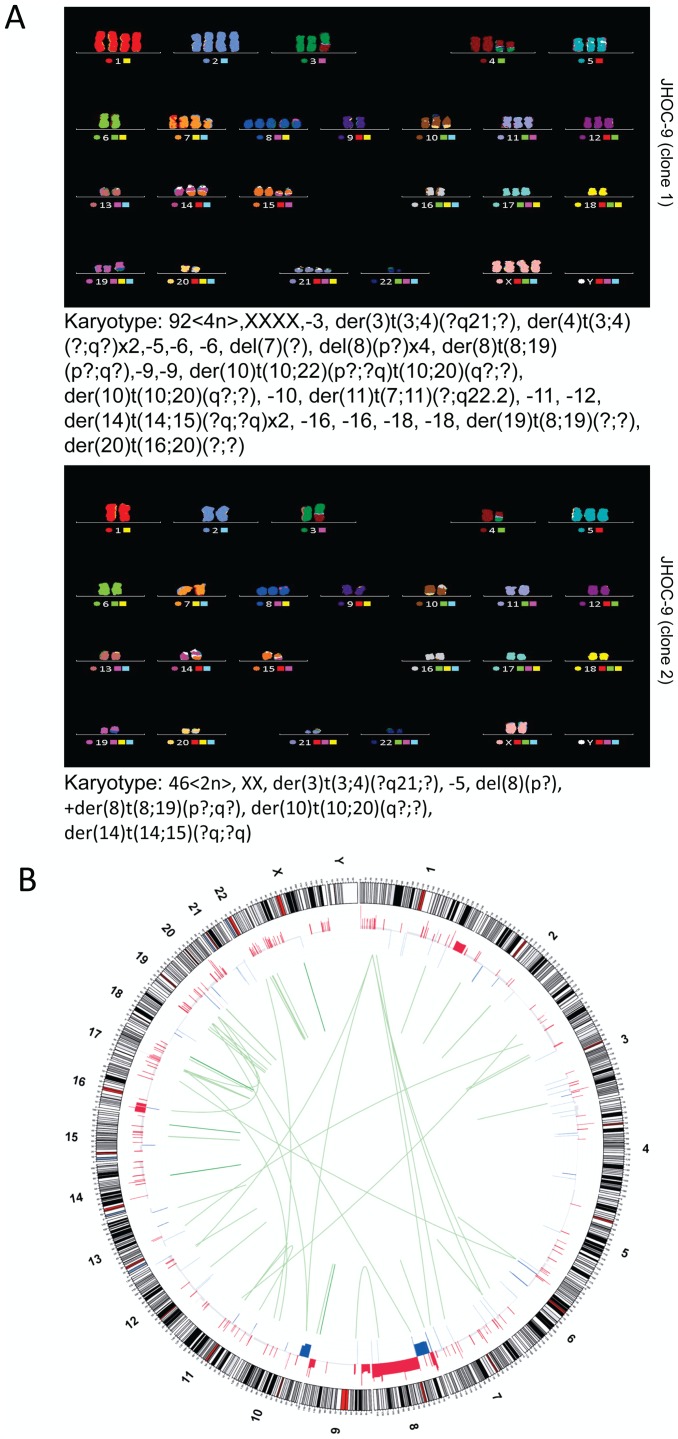
Figure 3. Genomic structure of CCC cell line JHOC-9. (A) 24 color FISH analysis suggested the presence of two dominant clones; one near-diploid and one near-tetraploid in the JHOC-9 CCC cell line.
A number of translocations and rearrangements can be seen in each representative clone. The complex karyotype of each dominant clone is noted below the corresponding 24-colour FISH results. Not all derivative chromosomes were identifiable resulting in a large number of ambiguous translocations and fragments (denoted by question marks in the karyotype notations). (B) Circos plot of RNAseq data and deFuse analysis depicting expressed genomic rearrangements in the JHOC-9 cell line. Translocations seen in the 24-color FISH profile are also visible as expressed transcripts including t(8;19) observed in both 2N and 4N dominant clones. No recurrent translocations were seen across our series (see also Table S3).
Discussion and Conclusions
As our initial goal was identification of bonafide CCC cell lines, we are pleased to report that the majority of reported CCC lines were representative of the primary tumors' molecular and pathological phenotype. Our immuno-classification scheme, COSP, predicted most to be CCC and our own mutation data, as well as that from COSMIC and CCLE, suggested loss of function ARID1A mutations were prevalent in these cell lines. Although three CCC lines did not have identifiable ARID1A mutations, only JHOC-5 cells appeared to have both wild-type sequence and detectable protein expression. The number of ARID1A “normal” CCC lines is lower than might be expected given the frequency of ARID1A mutations (and negative IHC) observed in primary CCC [16], [45] and may indicate some preferential selection for ARID1A null CCC lines to adapt to in vitro culture. However, given the small sample size it may well be a chance occurrence and does not appear to be significant. Other detected mutations (PIK3CA, PTEN, KRAS, PPP2R1A) are all consistent with varying frequencies in CCC. TP53 mutations are notably absent in all of our validated CCC cell lines, as a de-facto defining characteristic, and only a single CCC line had heterozygous copy number loss though still retained normal-like p53 IHC.
Both CCC and ENOCa appear to arise in a background of endometriosis. Atypical endometriosis adjacent to, or contiguous with, either histotype is not unusual for either CCC or ENOCa [16], [60], [61]. Co-occurrence (sometimes contiguous) of both CCC and ENOCa histologies in a mixed-cell type tumor has been reported [62] (and Dr. Blake Gilks, personal communication). Mutational profiles including ARID1A and PIK3CA, are common to both types, overall supporting a related origin and similar route to transformation [16], [45], [48], [49]. We found that both ES2 and OVISE cell lines, reportedly derived from CCC, largely resembled the immuno-profiles of ENOCa. Conversely the 2008 cell line, reportedly derived from serous carcinoma [63], though often referred to as ENOCa [64], appeared more CCC-like from COSP alone. The 2008 line did show mutant p53 staining and has two confirmed TP53 mutations, atypical for true CCC. IHC was negative for ARID1A, supporting a non-serous origin. We favored an assignment of ENOCa base largely on the TP53 mutation though note that this cell line is quite atypical as it may carry loss of function changes for ARID1A, mutation of TP53, and is positive for the CCC biomarker HNF1B. Arguably errors in cell line histotype reports may be explained simply by historically poor reproducibility in cell type assignment, though it is not unforeseeable that the biological relationship between CCC and ENOCa could be influencing these phenotypes. Given the high degree of overlap between the mutational characteristics of CCC and ENOCa, our panel was not able to further segregate or clarify this apparent confusion. SKOV3 is another unique case as it's immuno-phenotype most closely resembles HGSC, yet it caries a truncating mutation for ARID1A, a mutation that has not been observed in HGSC despite widespread testing [16], [45]. Previous studies with SKOV3 have pointed to a clear cell-like histology when grown as xenograft [64] and this may also favor an endometriosis-associated ovarian cancer diagnosis as does the presence of a PIK3CA mutation. Finally, the TOV112D cell line also presents as an exception with a moderately strong prediction of HGSC immuno-phenotype. In spite of this finding we suggest this line is representative of TP53 mutant ENOCa, based on the presence of an ENOCa characteristic CTNNB1 mutation, pathological review of the primary tumor material in the originating laboratory and expression profiling experiments supporting this conclusion [65]. We propose that these atypical CCC/ENOCa may be useful in exploration of some common endometriosis-associated ovarian cancer biology though care should be undertaken to allow proper interpretation of the results.
Unfortunately our COSP tool is unable to differentiate LGSC. Based on expert re-review of primary material we are aware of two cell lines derived from LGSC primary tumors. We therefore confidently favor this classification for VOA1056_CL and VOA1312_CL despite predictions of HGSC or ENOCa obtained from COSP. The VOA1056_CL line carries a Ras-pathway mutation as might be expected of an LGSC tumor, however this is an NRAS Q61R activating mutation. Activating NRAS mutations were recently described in LGSC at the 2012 AACR annual meeting [66] however, this represent the first validated report of an NRAS mutant LGSC tumor and the first validated LGSC derived cell line carrying this mutation. The COSMIC database suggests the cell lines LK-1 (G12D; defined as ovarian carcinoma, type not specified) and TYK-nu (G12D and Q61K; defined as ovarian “serous carcinoma”) also carry activating NRAS mutations, however we were unable to source these cell lines to confirm/reject their histological identity. In the cases of LGSC cell lines derived in-house, mutations of TP53 were not observed, consistent with IHC based literature reports suggesting this is a major molecular discriminator between HGSC and LGSC [67].
Finally only a single cell line in our collection was reported to be of mucinous carcinoma origin. The mutation profile of this cell line is consistent with this diagnosis, including a 127-bp TP53 homozygous deletion, overexpression by IHC, and KRAS G12V mutation.
Cell line records for epithelial ovarian carcinoma have recently come into question with a number contaminated and redundant cell lines acknowledged in a recent study [32]. Most notably 2008 (aka. ov2008) was reported to be frequently contaminated with, or a mislabeled version of, the HPV-positive ME-180 cell line (ATCC HTB-33), the “true” HPV-negative 2008 line defined in the report from Korch et al. [32], is the one used in our study. Maintaining appropriate records, testing and, most importantly, re-testing identity of cell lines in each individual lab's stocks should be paramount even if cell lines are obtained directly from repositories. Here we report only on cell lines that matched the originating repository STR DNA profile or the STR profile of their originating primary tumors (in the case of in-house derived lines). Despite our own best efforts our exercise did yield the discovery of 3 cell lines in our own stocks that were either mislabeled or contaminated, including our original stock of the 2008 cell line noted above. All contaminated lines have since been discarded/replaced. It should be noted that none of our assays were designed or tested to discriminate ovarian from non-ovarian malignancies, and although STR analysis would have eliminated any obviously male cancers (through detection of Chr Y markers), some level of accuracy in repository reported origin of “ovarian” must be assumed. In the case of the more atypical cell lines it is possible these may be of non-ovarian origin, e.g. endometrial carcinomas or other peritoneal cancers of unknown primary, we are currently unable to assess this idea. Further, our analysis may be confounded by dominant expansion of rare tumor sub-clones [68], acquisition of spontaneous mutations during culturing, and MMR deficiency (whether acquired or present in the originating primary tumor). MMR deficiencies have been reported to be prevalent in endometriosis-associated ovarian cancers (CCC and ENOCa) [69]–[72] and the potential acquisition of mutations as a result of MMR deficiency may influence some of the more ambiguous biomarker phenotypes within this group, as well as observed atypical mutation patterns. We noted MMR deficiencies in the non-serous lines TOV21G, SKOV3, A2780, and IGROV1 as well as the HGSC cell lines COLO-704 and COLO-720E (Table S5). MMR-protein deficiencies were not observed in our in-house derived LGSC cell lines (or their corresponding primary tumors) or in the mucinous carcinoma line MCAS.
In the spectrum of ovarian carcinomas, recent evidence strongly supports diagnosis and treatment of the five major histotypes of carcinomas as distinct diseases. Cancer cell lines provide an important intermediate tool for clinically relevant translational science, allowing genomic manipulation and cell biology studies beyond what can be reasonably achieved in clinical trials or animal models of cancer. In order to develop appropriate treatments, translational researchers need to use model systems appropriate to each ovarian carcinoma type. Unfortunately, historical records of ovarian cancer cell lines have rarely included information on histological origin [32], [64]; this is further hampered by a historical lack of reproducibility in histological diagnosis [73]–[75]. Histopathological reproducibility is steadily improving as recognition of the five major histotypes as unique disease entities becomes more widespread [4], [7], [8], [15], [22], [44], [76], [77], histology and grading criteria become unified [78]–[81], and objective biomarker based tools to delineate histotypes are developed [2], [4], [41], [44]. However, cell lines lack morphological features that are recognizable in culture and development of new, well-defined, cell lines is laborious with poor long-term success rates. Assigning histotype to readily available and well-used cell lines will undoubtedly lead to better interpretation of new data and re-interpretation of already published findings.
Supporting Information
127bp homozygous (or hemizygous) deletion affecting TP53 exon 4 in the MCAS mucinous carcinoma cell line. This mutation was apparent by Sanger sequencing though not annotated in the CCLE database. Coding bases are annotated in upper case.
(PDF)
Cell Lines & Sources.
(PDF)
Antibodies and Dilutions.
(PDF)
deFuse predicted expressed re-arrangements from transcriptome sequencing data.
(XLS)
Mutations Found In Ovarian Carcinoma Cell Lines.
(PDF)
Mismatch Repair IHC.
(PDF)
Acknowledgments
The authors wish to thank the Genetic Pathology Evaluation Centre (GPEC) and Centre for Translational and Applied Genomics (CTAG) for technical support, Blake Gilks, Jason Madore, Melissa McConechy, Marcel Bally, Ying Ng, and Jessica McAlpine for resource support and insightful discussions. Some of the cell lines used in this study were donated from the laboratories of James Brenton, Nelly Auersperg, and Ian Campbell, we are grateful for access to these well-maintained and precious resources. Finally, we thank all of the labs that have dedicated significant time and effort to the development of cancer cell lines and the repositories that maintain these resources for the global scientific community.
Funding Statement
Support for this project was provided to the Ovarian Cancer Research Team of BC (OVCARE; http://www.ovcare.ca) through the BC Cancer Foundation, The VGH and UBC Hospitals Foundation and the Canadian Institutes for Health Research (CIHR) Emerging Team Grant: Personalized siRNA-Based Nanomedicines (FRN: 111627). Funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Auersperg N (2011) The origin of ovarian carcinomas: a unifying hypothesis. Int J Gynecol Pathol 30: 12–21. [DOI] [PubMed] [Google Scholar]
- 2. Kalloger SE, Kobel M, Leung S, Mehl E, Gao D, et al. (2011) Calculator for ovarian carcinoma subtype prediction. Mod Pathol 24: 512–521. [DOI] [PubMed] [Google Scholar]
- 3. Soslow RA (2008) Histologic subtypes of ovarian carcinoma: an overview. Int J Gynecol Pathol 27: 161–174. [DOI] [PubMed] [Google Scholar]
- 4. Kobel M, Kalloger SE, Boyd N, McKinney S, Mehl E, et al. (2008) Ovarian carcinoma subtypes are different diseases: implications for biomarker studies. PLoS Med 5: e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anglesio MS, Arnold JM, George J, Tinker AV, Tothill R, et al. (2008) Mutation of ERBB2 provides a novel alternative mechanism for the ubiquitous activation of RAS-MAPK in ovarian serous low malignant potential tumors. Mol Cancer Res 6: 1678–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tothill RW, Tinker AV, George J, Brown R, Fox SB, et al. (2008) Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res 14: 5198–5208. [DOI] [PubMed] [Google Scholar]
- 7. Kurman RJ, Shih Ie M (2011) Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer-Shifting the paradigm. Hum Pathol 42: 918–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kurman RJ, Shih Ie M (2010) The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am J Surg Pathol 34: 433–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. The Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. McCluggage WG (2011) Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis. Pathology 43: 420–432. [DOI] [PubMed] [Google Scholar]
- 11. Levanon K, Ng V, Piao HY, Zhang Y, Chang MC, et al. (2010) Primary ex vivo cultures of human fallopian tube epithelium as a model for serous ovarian carcinogenesis. Oncogene 29: 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carlson JW, Jarboe EA, Kindelberger D, Nucci MR, Hirsch MS, et al. (2010) Serous tubal intraepithelial carcinoma: diagnostic reproducibility and its implications. Int J Gynecol Pathol 29: 310–314. [DOI] [PubMed] [Google Scholar]
- 13. Crum CP, Drapkin R, Kindelberger D, Medeiros F, Miron A, et al. (2007) Lessons from BRCA: the tubal fimbria emerges as an origin for pelvic serous cancer. Clin Med Res 5: 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Laury AR, Ning G, Quick CM, Bijron J, Parast MM, et al. (2011) Fallopian tube correlates of ovarian serous borderline tumors. Am J Surg Pathol 35: 1759–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anglesio MS, Carey MS, Kobel M, Mackay H, Huntsman DG (2011) Clear cell carcinoma of the ovary: a report from the first Ovarian Clear Cell Symposium, June 24th, 2010. Gynecol Oncol 121: 407–415. [DOI] [PubMed] [Google Scholar]
- 16. Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, et al. (2010) ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med 363: 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feeley KM, Wells M (2001) Precursor lesions of ovarian epithelial malignancy. Histopathology 38: 87–95. [DOI] [PubMed] [Google Scholar]
- 18. Thomas EJ, Campbell IG (2000) Evidence that endometriosis behaves in a malignant manner. Gynecol Obstet Invest 50 Suppl 12–10. [DOI] [PubMed] [Google Scholar]
- 19. Thomas EJ, Campbell IG (2000) Molecular genetic defects in endometriosis. Gynecol Obstet Invest 50 Suppl 144–50. [DOI] [PubMed] [Google Scholar]
- 20. Prowse AH, Manek S, Varma R, Liu J, Godwin AK, et al. (2006) Molecular genetic evidence that endometriosis is a precursor of ovarian cancer. Int J Cancer 119: 556–562. [DOI] [PubMed] [Google Scholar]
- 21.Schiavone MB, Herzog TJ, Lewin SN, Deutsch I, Sun X, et al.. (2011) Natural history and outcome of mucinous carcinoma of the ovary. Am J Obstet Gynecol 205: 480 e481–488. [DOI] [PubMed]
- 22. Anglesio MS, Kommoss S, Tolcher MC, Clarke B, Galletta L, et al. (2013) Molecular characterization of mucinous ovarian tumours supports a stratified treatment approach with HER2 targeting in 19% of carcinomas. J Pathol 229: 111–120. [DOI] [PubMed] [Google Scholar]
- 23. Hess V, A'Hern R, Nasiri N, King DM, Blake PR, et al. (2004) Mucinous epithelial ovarian cancer: a separate entity requiring specific treatment. J Clin Oncol 22: 1040–1044. [DOI] [PubMed] [Google Scholar]
- 24. Berns EM, Bowtell DD (2012) The changing view of high-grade serous ovarian cancer. Cancer Res 72: 2701–2704. [DOI] [PubMed] [Google Scholar]
- 25. Vaughan S, Coward JI, Bast RC Jr, Berchuck A, Berek JS, et al. (2011) Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer 11: 719–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. McAlpine JN, Porter H, Kobel M, Nelson BH, Prentice LM, et al. (2012) BRCA1 and BRCA2 mutations correlate with TP53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod Pathol 25: 740–750. [DOI] [PubMed] [Google Scholar]
- 27. Risch HA, McLaughlin JR, Cole DE, Rosen B, Bradley L, et al. (2006) Population BRCA1 and BRCA2 mutation frequencies and cancer penetrances: a kin-cohort study in Ontario, Canada. J Natl Cancer Inst 98: 1694–1706. [DOI] [PubMed] [Google Scholar]
- 28. Alsop K, Fereday S, Meldrum C, deFazio A, Emmanuel C, et al. (2012) BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: a report from the Australian Ovarian Cancer Study Group. J Clin Oncol 30: 2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Quirk JT, Natarajan N (2005) Ovarian cancer incidence in the United States, 1992–1999. Gynecol Oncol 97: 519–523. [DOI] [PubMed] [Google Scholar]
- 30. McGuire V, Jesser CA, Whittemore AS (2002) Survival among U.S. women with invasive epithelial ovarian cancer. Gynecol Oncol 84: 399–403. [DOI] [PubMed] [Google Scholar]
- 31. Gershenson DM (2012) Treatment of ovarian cancer in young women. Clin Obstet Gynecol 55: 65–74. [DOI] [PubMed] [Google Scholar]
- 32. Korch C, Spillman MA, Jackson TA, Jacobsen BM, Murphy SK, et al. (2012) DNA profiling analysis of endometrial and ovarian cell lines reveals misidentification, redundancy and contamination. Gynecol Oncol 127: 241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Capes-Davis A, Theodosopoulos G, Atkin I, Drexler HG, Kohara A, et al. (2010) Check your cultures! A list of cross-contaminated or misidentified cell lines. Int J Cancer 127: 1–8. [DOI] [PubMed] [Google Scholar]
- 34.Kommoss S, Gilks CB, Kommoss F, Anglesio MS, Chow C, et al.. (2013) Accelerating type-specific ovarian carcinoma research: Calculator for Ovarian Subtype Prediction (COSP) is a reliable high-throughput tool for case review. Histopathology (accepted June 2, 2013). [DOI] [PubMed]
- 35. Lu FI, Gilks CB, Mulligan AM, Ryan P, Allo G, et al. (2012) Prevalence of loss of expression of DNA mismatch repair proteins in primary epithelial ovarian tumors. Int J Gynecol Pathol 31: 524–531. [DOI] [PubMed] [Google Scholar]
- 36. Malkov VA, Serikawa KA, Balantac N, Watters J, Geiss G, et al. (2009) Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes 2: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McConechy MK, Ding J, Cheang MC, Wiegand KC, Senz J, et al. (2012) Use of mutation profiles to refine the classification of endometrial carcinomas. J Pathol 228: 20–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, et al. (2012) The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, et al. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39: D945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McPherson A, Hormozdiari F, Zayed A, Giuliany R, Ha G, et al. (2011) deFuse: an algorithm for gene fusion discovery in tumor RNA-Seq data. PLoS Comput Biol 7: e1001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kobel M, Turbin D, Kalloger SE, Gao D, Huntsman DG, et al. (2011) Biomarker expression in pelvic high-grade serous carcinoma: comparison of ovarian and omental sites. Int J Gynecol Pathol 30: 366–371. [DOI] [PubMed] [Google Scholar]
- 42. Kobel M, Reuss A, Bois A, Kommoss S, Kommoss F, et al. (2010) The biological and clinical value of p53 expression in pelvic high-grade serous carcinomas. J Pathol 222: 191–198. [DOI] [PubMed] [Google Scholar]
- 43. McAlpine JN, Wiegand KC, Vang R, Ronnett BM, Adamiak A, et al. (2009) HER2 overexpression and amplification is present in a subset of ovarian mucinous carcinomas and can be targeted with trastuzumab therapy. BMC Cancer 9: 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kobel M, Kalloger SE, Carrick J, Huntsman D, Asad H, et al. (2009) A limited panel of immunomarkers can reliably distinguish between clear cell and high-grade serous carcinoma of the ovary. Am J Surg Pathol 33: 14–21. [DOI] [PubMed] [Google Scholar]
- 45. Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, et al. (2010) Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Taverna P, Liu L, Hanson AJ, Monks A, Gerson SL (2000) Characterization of MLH1 and MSH2 DNA mismatch repair proteins in cell lines of the NCI anticancer drug screen. Cancer Chemother Pharmacol 46: 507–516. [DOI] [PubMed] [Google Scholar]
- 47. Warnick CT, Dabbas B, Ford CD, Strait KA (2001) Identification of a p53 response element in the promoter region of the hMSH2 gene required for expression in A2780 ovarian cancer cells. J Biol Chem 276: 27363–27370. [DOI] [PubMed] [Google Scholar]
- 48. Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, et al. (2004) Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 64: 7678–7681. [DOI] [PubMed] [Google Scholar]
- 49. Kuo KT, Mao TL, Jones S, Veras E, Ayhan A, et al. (2009) Frequent activating mutations of PIK3CA in ovarian clear cell carcinoma. Am J Pathol 174: 1597–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hirasawa A, Saito-Ohara F, Inoue J, Aoki D, Susumu N, et al. (2003) Association of 17q21–q24 gain in ovarian clear cell adenocarcinomas with poor prognosis and identification of PPM1D and APPBP2 as likely amplification targets. Clin Cancer Res 9: 1995–2004. [PubMed] [Google Scholar]
- 51. Tan DS, Lambros MB, Rayter S, Natrajan R, Vatcheva R, et al. (2009) PPM1D is a potential therapeutic target in ovarian clear cell carcinomas. Clin Cancer Res 15: 2269–2280. [DOI] [PubMed] [Google Scholar]
- 52. Tan DS, Iravani M, McCluggage WG, Lambros MB, Milanezi F, et al. (2011) Genomic analysis reveals the molecular heterogeneity of ovarian clear cell carcinomas. Clin Cancer Res 17: 1521–1534. [DOI] [PubMed] [Google Scholar]
- 53. Anglesio MS, George J, Kulbe H, Friedlander M, Rischin D, et al. (2011) IL6-STAT3-HIF signaling and therapeutic response to the angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin Cancer Res 17: 2538–2548. [DOI] [PubMed] [Google Scholar]
- 54.Spowart JE, Townsend KN, Huwait H, Eshragh S, West NR, et al.. (2012) The Autophagy Protein LC3A Correlates with Hypoxia and is a Prognostic Marker of Patient Survival in Clear Cell Ovarian Cancer. J Pathol. [DOI] [PubMed]
- 55. Rahman MT, Nakayama K, Rahman M, Nakayama N, Ishikawa M, et al. (2012) Prognostic and therapeutic impact of the chromosome 20q13.2 ZNF217 locus amplification in ovarian clear cell carcinoma. Cancer 118: 2846–2857. [DOI] [PubMed] [Google Scholar]
- 56. Yamamoto S, Tsuda H, Miyai K, Takano M, Tamai S, et al. (2011) Gene amplification and protein overexpression of MET are common events in ovarian clear-cell adenocarcinoma: their roles in tumor progression and prognostication of the patient. Mod Pathol 24: 1146–1155. [DOI] [PubMed] [Google Scholar]
- 57. Yamamoto S, Tsuda H, Miyai K, Takano M, Tamai S, et al. (2012) Accumulative copy number increase of MET drives tumor development and histological progression in a subset of ovarian clear-cell adenocarcinomas. Mod Pathol 25: 122–130. [DOI] [PubMed] [Google Scholar]
- 58. Kuo KT, Mao TL, Chen X, Feng Y, Nakayama K, et al. (2010) DNA copy numbers profiles in affinity-purified ovarian clear cell carcinoma. Clin Cancer Res 16: 1997–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang X, George J, Deb S, Degoutin JL, Takano EA, et al. (2011) The Hippo pathway transcriptional co-activator, YAP, is an ovarian cancer oncogene. Oncogene 30: 2810–2822. [DOI] [PubMed] [Google Scholar]
- 60. Wei JJ, William J, Bulun S (2011) Endometriosis and ovarian cancer: a review of clinical, pathologic, and molecular aspects. International journal of gynecological pathology: official journal of the International Society of Gynecological Pathologists 30: 553–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. LaGrenade A, Silverberg SG (1988) Ovarian tumors associated with atypical endometriosis. Hum Pathol 19: 1080–1084. [DOI] [PubMed] [Google Scholar]
- 62. Razzouk K, Roman H, Chanavaz-Lacheray I, Scotte M, Verspyck E, et al. (2007) Mixed clear cell and endometrioid carcinoma arising in parietal endometriosis. Gynecol Obstet Invest 63: 140–142. [DOI] [PubMed] [Google Scholar]
- 63. DiSaia PJ, Sinkovics JG, Rutledge FN, Smith JP (1972) Cell-mediated immunity to human malignant cells. A brief review and further studies with two gynecologic tumors. Am J Obstet Gynecol 114: 979–989. [DOI] [PubMed] [Google Scholar]
- 64. Shaw TJ, Senterman MK, Dawson K, Crane CA, Vanderhyden BC (2004) Characterization of intraperitoneal, orthotopic, and metastatic xenograft models of human ovarian cancer. Mol Ther 10: 1032–1042. [DOI] [PubMed] [Google Scholar]
- 65. Madore J, Ren F, Filali-Mouhim A, Sanchez L, Kobel M, et al. (2010) Characterization of the molecular differences between ovarian endometrioid carcinoma and ovarian serous carcinoma. J Pathol 220: 392–400. [DOI] [PubMed] [Google Scholar]
- 66.Farley JH, Brady WE, Birrer MJ, Lankes H, Coleman R, et al.. (2012) A phase II trial of selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum. AACR Annual Meeting 2012, Chicogo, IL, USA. [DOI] [PMC free article] [PubMed]
- 67.Altman AD, Nelson GS, Ghatage P, McIntyre JB, Capper D, et al.. (2013) The diagnostic utility of TP53 and CDKN2A to distinguish ovarian high-grade serous carcinoma from low-grade serous ovarian tumors. Mod Pathol. [DOI] [PubMed]
- 68. Danjoh I, Shirota R, Hiroyama T, Nakamura Y (2013) Dominant expansion of a cryptic subclone with an abnormal karyotype in B lymphoblastoid cell lines during culture. Cytogenet Genome Res 139: 88–96. [DOI] [PubMed] [Google Scholar]
- 69. Cai KQ, Albarracin C, Rosen D, Zhong R, Zheng W, et al. (2004) Microsatellite instability and alteration of the expression of hMLH1 and hMSH2 in ovarian clear cell carcinoma. Hum Pathol 35: 552–559. [DOI] [PubMed] [Google Scholar]
- 70. Liu J, Albarracin CT, Chang KH, Thompson-Lanza JA, Zheng W, et al. (2004) Microsatellite instability and expression of hMLH1 and hMSH2 proteins in ovarian endometrioid cancer. Mod Pathol 17: 75–80. [DOI] [PubMed] [Google Scholar]
- 71. Domanska K, Malander S, Masback A, Nilbert M (2007) Ovarian cancer at young age: the contribution of mismatch-repair defects in a population-based series of epithelial ovarian cancer before age 40. Int J Gynecol Cancer 17: 789–793. [DOI] [PubMed] [Google Scholar]
- 72. Jensen KC, Mariappan MR, Putcha GV, Husain A, Chun N, et al. (2008) Microsatellite instability and mismatch repair protein defects in ovarian epithelial neoplasms in patients 50 years of age and younger. Am J Surg Pathol 32: 1029–1037. [DOI] [PubMed] [Google Scholar]
- 73. Hernandez E, Bhagavan BS, Parmley TH, Rosenshein NB (1984) Interobserver variability in the interpretation of epithelial ovarian cancer. Gynecol Oncol 17: 117–123. [DOI] [PubMed] [Google Scholar]
- 74. Sakamoto A, Sasaki H, Furusato M, Suzuki M, Hirai Y, et al. (1994) Observer disagreement in histological classification of ovarian tumors in Japan. Gynecol Oncol 54: 54–58. [DOI] [PubMed] [Google Scholar]
- 75. Cramer SF, Roth LM, Ulbright TM, Mazur MT, Nunez CA, et al. (1987) Evaluation of the reproducibility of the World Health Organization classification of common ovarian cancers. With emphasis on methodology. Arch Pathol Lab Med 111: 819–829. [PubMed] [Google Scholar]
- 76. Swenerton KD, Santos JL, Gilks CB, Kobel M, Hoskins PJ, et al. (2011) Histotype predicts the curative potential of radiotherapy: the example of ovarian cancers. Ann Oncol 22: 341–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hunter SM, Anglesio MS, Sharma R, Gilks CB, Melnyk N, et al. (2011) Copy number aberrations in benign serous ovarian tumors: a case for reclassification? Clin Cancer Res 17: 7273–7282. [DOI] [PubMed] [Google Scholar]
- 78. Kobel M, Kalloger SE, Santos JL, Huntsman DG, Gilks CB, et al. (2010) Tumor type and substage predict survival in stage I and II ovarian carcinoma: insights and implications. Gynecol Oncol 116: 50–56. [DOI] [PubMed] [Google Scholar]
- 79. Kobel M, Kalloger SE, Huntsman DG, Santos JL, Swenerton KD, et al. (2010) Differences in tumor type in low-stage versus high-stage ovarian carcinomas. Int J Gynecol Pathol 29: 203–211. [DOI] [PubMed] [Google Scholar]
- 80. Kobel M, Kalloger SE, Baker PM, Ewanowich CA, Arseneau J, et al. (2010) Diagnosis of ovarian carcinoma cell type is highly reproducible: a transcanadian study. Am J Surg Pathol 34: 984–993. [DOI] [PubMed] [Google Scholar]
- 81. Gilks CB, Ionescu DN, Kalloger SE, Kobel M, Irving J, et al. (2008) Tumor cell type can be reproducibly diagnosed and is of independent prognostic significance in patients with maximally debulked ovarian carcinoma. Hum Pathol 39: 1239–1251. [DOI] [PubMed] [Google Scholar]
- 82. Campbell IG, Thomas EJ (2001) Endometriosis: candidate genes. Hum Reprod Update 7: 15–20. [DOI] [PubMed] [Google Scholar]
- 83. Seidman JD, Khedmati F (2008) Exploring the histogenesis of ovarian mucinous and transitional cell (Brenner) neoplasms and their relationship with Walthard cell nests: a study of 120 tumors. Arch Pathol Lab Med 132: 1753–1760. [DOI] [PubMed] [Google Scholar]
- 84. Crum CP (2009) Intercepting pelvic cancer in the distal fallopian tube: theories and realities. Mol Oncol 3: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ahmed AA, Etemadmoghadam D, Temple J, Lynch AG, Riad M, et al. (2010) Driver mutations in TP53 are ubiquitous in high grade serous carcinoma of the ovary. J Pathol 221: 49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shaw PA, McLaughlin JR, Zweemer RP, Narod SA, Risch H, et al. (2002) Histopathologic features of genetically determined ovarian cancer. Int J Gynecol Pathol 21: 407–411. [DOI] [PubMed] [Google Scholar]
- 87.McBride DJ, Etemadmoghadam D, Cooke SL, Alsop K, George J, et al.. (2012) Tandem duplication of chromosomal segments is common in ovarian and breast cancer genomes. J Pathol. [DOI] [PMC free article] [PubMed]
- 88. Duska LR, Garrett L, Henretta M, Ferriss JS, Lee L, et al. (2010) When ‘never-events’ occur despite adherence to clinical guidelines: the case of venous thromboembolism in clear cell cancer of the ovary compared with other epithelial histologic subtypes. Gynecol Oncol 116: 374–377. [DOI] [PubMed] [Google Scholar]
- 89. Mackay HJ, Brady MF, Oza AM, Reuss A, Pujade-Lauraine E, et al. (2010) Prognostic relevance of uncommon ovarian histology in women with stage III/IV epithelial ovarian cancer. Int J Gynecol Cancer 20: 945–952. [DOI] [PubMed] [Google Scholar]
- 90. Storey DJ, Rush R, Stewart M, Rye T, Al-Nafussi A, et al. (2008) Endometrioid epithelial ovarian cancer: 20 years of prospectively collected data from a single center. Cancer 112: 2211–2220. [DOI] [PubMed] [Google Scholar]
- 91. Schmeler KM, Sun CC, Bodurka DC, Deavers MT, Malpica A, et al. (2008) Neoadjuvant chemotherapy for low-grade serous carcinoma of the ovary or peritoneum. Gynecol Oncol 108: 510–514. [DOI] [PubMed] [Google Scholar]
- 92. Vang R, Shih Ie M, Kurman RJ (2009) Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Advances in anatomic pathology 16: 267–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
127bp homozygous (or hemizygous) deletion affecting TP53 exon 4 in the MCAS mucinous carcinoma cell line. This mutation was apparent by Sanger sequencing though not annotated in the CCLE database. Coding bases are annotated in upper case.
(PDF)
Cell Lines & Sources.
(PDF)
Antibodies and Dilutions.
(PDF)
deFuse predicted expressed re-arrangements from transcriptome sequencing data.
(XLS)
Mutations Found In Ovarian Carcinoma Cell Lines.
(PDF)
Mismatch Repair IHC.
(PDF)