Abstract
Microcystins (MCs) are the most commonly-reported hepatotoxins produced by various cyanobacterial taxa in fresh waters to constitute a potential threat to human and animal health. The biological role of MCs in the producer organisms is not known, and it would be very useful to understand the driving force behind the toxin production. Recent studies have suggested that MCs may have a protective function in cells facing environmental stress. Following this starting premise, we speculate that under adverse conditions the expression of stress-related genes coding for Heat Shock Proteins (Hsp) might be different in an MC-producing strain and its MC-deficient mutant. We therefore used RT-qPCR to compare the expression of 13 hsp genes of an MC-producing strain of Planktothrix agardhii (CYA126/8) and its MC-deficient ΔmcyD mutant over different periods of exposure to high light stress (HL). Three reference genes (RGs) were selected from six candidates to normalize the RT-qPCR data. Of these three RGs (rsh, rpoD, and gltA), gltA is used here for the first time as an RG in prokaryotes. Under HL stress, five genes were found to be strongly up-regulated in both strains (htpG, dnaK, hspA, groES, and groEL). Unexpectedly, we found that the MC-producing wild type strain accumulated higher levels of htpG and dnaK transcripts in response to HL stress than the MC-deficient mutant. In addition, a significant increase in the mcyE transcript was detected in the mutant, suggesting that MCs are required under HL conditions. We discuss several possible roles of MCs in the response to HL stress through their possible involvement in the protective mechanisms of the cells.
Introduction
Cyanobacterial bloom-forming species are a worldwide problem, because of the toxins they produce. The situation has become a cause of increased concern in recent decades as the frequency and intensity of bloom occurrence have increased, due in part to anthropic activities [1]. One of the diverse groups of cyanotoxins, that of the hepatotoxin microcystins (MCs) which includes up to 90 variants [2], is widespread and frequently reported. Microcystins [3] are becoming a real threat to human and animal health due to the contamination of freshwater [4]. However, the ecological significance and biological function of the MCs for the producer cells, which could elucidate the forces underlying toxin production, remain elusive. MCs are typically intracellular components, which are usually released into the environment after the cells die [5]. It has been shown that they are not essential for growth, but are probably involved in intracellular processes [6]. Various different hypotheses for the possible role of cyanotoxins have been proposed, these include a role as: grazer deterrent compounds [7] iron scavenging molecules [8], allelopathic compounds [9], growth regulators permitting successful adaptation [10], light harvesting and chromatic adaptation [11], and infochemicals [11]–[13]. However, the findings of the various studies are contradictory in many respects, and difficult to understand [14].
More recently, emerging investigations have suggested that MCs may have a protective role in the response to unfavorable conditions. Significantly greater growth rates have been observed in an MC-producing strain Planktothrix agardhii, than an MC-free strain; both strains had previously been collected from environmental samples (i.e. had different genomes) [15]. It has been also reported that transcription of the genes responsible for MCs production in Microcystis increased in response to strong illumination or iron starvation suggesting that MCs may play a protective role under various stressful conditions, including oxidative stress [11], [16]. To date, most of the data about the physiological roles of MCs has been reported by Neilan and colleagues in Microcystis [17], where they show that MCs production has a complex and deep effect on the proteome. Furthermore, MCs may be involved in the carbon-nitrogen metabolism, in redox control, in the perception of redox changes, and in providing protection against oxidative stress [17]–[19]. However, cyanobacterial responses to abiotic stresses are complex, and several mechanisms usually act in concert and synergistically to prevent cell damage and to re-establish cellular homeostasis [20]–[21]. The activities of heat shock proteins (hsps) constitute an important component of the cell’s response to stress. Depending on their size, Hsps are divided into 5 main classes: Hsp100, Hsp90, Hsp70, Hsp60 and small Hsp; representatives of each class are found in cyanobacteria [21]. Under normal conditions, Hsps play an important role in the folding, assembly, and trafficking of newly-synthesized polypeptides and in the degradation of denatured or aggregated proteins. Under stressful conditions, as a result of increased levels of aberrant proteins, the importance of Hsps increases, and this is usually reflected in their up-regulation [20]. However, much remains to be learned about the “in vivo” function of the Hsps proteins in cyanobacteria, as their biological role may extend to stress responses in general (known as HSRs for Heat shock responses) [22]–[23], and to multiple molecular cell defenses. Indeed, the protective effects of Hsps can be attributed to the network of the chaperone machinery, in which many Hsps play complementary and sometimes overlapping roles [20]. These major studies are initiated at the transcriptional level, using RT-qPCR (real-time quantitative polymerase chain reaction) analysis, which is one of the most powerful tools available for investigating quantitative differences in gene expression responses under experimental conditions [24]. However it is also a demanding tool, and calls for preliminary evaluation of the stability of a panel of reference genes (RGs) to provide accurate normalization of the gene expression analysis.
A few studies have identified the intracellular function of MCs in cyanobacteria against abiotic stressors at the transcriptomic level, and they involved the use of two strains sharing identical genomes, but with one gene engineered to inhibit MCs production. Unicellular Microcystis strains were investigated [19], but no study has yet been carried out of two identical clones of filamentous Planktothrix agardhii (differing solely by a single insertional mutation in one mcy gene), despite its hazardous impact on ecosystem functioning and the current increase of blooms in waterbodies. We selected P. agardhii for our study, due to its ecological preference for low light intensities [25], as HL (high light) could be expected to induce more explicit responses. We attempted to i) validate that the RGs were stable under our experimental conditions (i.e. control vs stressed conditions) in order to optimize RT-qPCR accuracy; ii) determine the gene expression profiles of 13 hsps and one mcy gene (mcyE, which is involved in the synthesis of Adda, and the incorporation of D-glu during MCs production); iii) determine the fold changes in the expression levels of the hsps and mcyE in the MC-producing strain (CYA126/8) and its MC-free mutant (ΔmcyD) when exposed to HL stress, using RT-qPCR analysis. To the best of our knowledge, this is the first study to report the transcriptional shift of a panel of 13 stress related genes (hsps) in MC-producing cells during a short period of exposure to stress (5–24 h).
Materials and Methods
Strains and Culture Conditions
Two strains of P. agardhii were used in this study. The MC-producing strain CYA126/8 (i.e. the wild type: WT) and its MC-deficient mutant (ΔmcyD) were kindly provided by Dr Kurmayer (University of Vienna). Both strains were monoclonal and not axenic and are maintained in the PMC (Paris Museum Collection, Paris). They shared an identical genome except that in the ΔmcyD mutant; a chloramphenicol cartridge was inserted into the mcyD gene to inhibit MC biosynthesis. We used LC/MS to confirm that CYA126/8 WT was an MC-producer, and that the mutant was free of MCs (Combes, unpublished data). Both cultures were maintained in Z8 liquid medium [26] at 20°C under white light (Osram white FM 11W/730 universal white) at 22±2 µmol.m−2.s−1 and with a light/dark cycle of 16/8. Cultures of mutant (ΔmcyD) cells were maintained under a constant selective pressure with chloramphenicol (1 µg/ml Z8 medium) to avoid potential wild type copies to grow (as the cyanobacteria may contain several genome copies), which may lead to the restoration of MC-producing cells. To avoid any effect that chloramphenicol may have had on physiological processes, two weeks before each experiment, the mutant culture was transferred to chloramphenicol-free Z8 medium.
Experimental RT-qPCR Conditions
The cultures under optimal conditions (i.e. control) were obtained during the exponential growth phase under a continuous light intensity of 22 µmol m−2 s−1. For the HL treatment, the cultures in the exponential phase were shifted from control conditions to an intensity of 600 µmol m−2 s−1 during 24 h. For both conditions, the temperature was maintained at 20°C using a Binder phytotron. Experiments were performed using equivalent culture densities (OD750 nm = 0.3) of both strains. Samples were taken at 0 h (control), and 1 h, 2 h, 5 h and 24 h after transferring to HL conditions, and were used for subsequent analysis. Two independent replicates were performed.
RNA Extraction and cDNA Synthesis
For each sample, 40 ml of the culture suspension (OD750 nm = 0.3) was centrifuged at 4°C, for 15 min, at 4000 rpm. Total RNA extraction was carried out using Trizol reagent (Invitrogen, USA) followed by purification using PureLink™ RNA Mini Kit (Invitrogen), according to the Manufacturer’s instructions. The pellet was mixed with 3 ml of Trizol, and then immediately frozen in liquid nitrogen and conserved at −80°C until extraction. Phase separation was obtained by adding 600 µl of chloroform to the cell lysate and shaking vigorously by hand for 15 seconds (8 times), storing at room temperature for 5 minutes, and then centrifuging at 12 000 g for 15 minutes at 4°C. RNA purification was performed using PureLink™ RNA Mini Kit (Invitrogen). Purified RNA, previously treated with a DNA-free Kit (Ambion), was quantified using a NanoDrop 2000 Spectrophotometer (Thermo Scientific); and its integrity was checked on 1.5% agarose gel (data not shown). Genomic DNA contamination was checked by PCR on a total RNA template using primers targeting the citrate synthetase sequence (data not shown).
The A260/A280 ratio of the RNA samples was 2.089±0.017 (mean ± SD), indicating the absence of protein and the purity of all the total RNA samples required for an accurate qRT-PCR analysis. First-strand cDNA was synthesized from 0.8 µg total RNA using SuperScript III First-Strand Synthesis SuperMix (Invitrogen, Carlsbad, USA), with 1 µl of random hexamers in a 20-µl reaction mixture, according to the Manufacturer’s instructions. cDNA samples were stored at −20°C.
Genes Investigated in the Study
Two series of genes were used in this study (Table 1). As the relevance of RT-qPCR analysis greatly depends on transcript normalization with stably-expressed reference genes (RGs), 6 candidates were selected from different functional classes. Four conventional candidate RGs were tested: 16S rRNA [27], rpoD [28], GAPDH [27], rsh [27], plus two RGs genes that had never so far been tested in prokaryotes: gltA and rpsL. The sequence of 16S rRNA is available in a database (GeneBank FJ184435.1), but the other 5 RGs candidates were all isolated in this study.
Table 1. Information about the genes investigated in this study.
| Gene | Name | Description/function | Accession number |
| Rsh * | (p)ppGpp synthase/hydrolase | Control of metabolism of (p)ppGpp thereby involved in responses to nutritionaldeprivation | KF275118 |
| rpoD * | RNA polymerase sigma factor | Primary RNA polymerase sigma factor | KF275120 |
| gltA * | Citrate synthase | Citric acid cycle | KF275124 |
| GAPDH * | Glyceraldehyde 3-phosphatedehydrogenase | Glycolysis | KF275123 |
| rpsL * | 30S ribosomal protein S12 | Structural constituent of ribosome | KF275122 |
| 16S rRNA * | 16S ribosomal RNA | Structural constituent of ribosome, acting as scaffold defining the positions ofribosomal proteins | FJ184435.1 |
| hspA ** | Small heat shock protein | Prevent irreversible protein aggregation during stress | KF294790 |
| hslO ** | 33 kDa heat shock protein | Chaperon holdase, functioning as a first line of defense during oxidative stressconditions that cause protein unfolding. | KF294782 |
| hsp40 ** | 40 kDa heat shock protein | Co-chaperone of Hsp70, regulating complex formation between Hsp70 and client proteins. | KF294789 |
| grpE ** | Nucleotide exchange factorfor DnaK | Stimulate the release of ADP from Hsp70, fostering substrate dissociation,thereby ‘recycling’ Hsp70 molecule. | KF294788 |
| dnaK ** | 70 kDa heat shock protein | Help the folding of nascent proteins under normal conditions, prevent the aggregationof unfolding proteins and assist in refold aggregated proteins under stress conditions. | KF294783 |
| hsp70(1) ** | KF294784 | ||
| hsp70(2) ** | KF294785 | ||
| hsp70(3) ** | KF294786 | ||
| hsp70(4) ** | KF294787 | ||
| clpC ** | 100 kDa heat shock protein | Regulatory ATPase/chaperone subunit of Clp protease, involved in the efficientdegradation of irreversibly damaged proteins. | KF275115 |
| htpG ** | 90 kDa heat shock protein | Recognize and bind non-native proteins to prevent their nonspecific aggregation | KF275116 |
| groEL ** | 60 kDa heat shock protein | Bind to partially folded/unfolded protein and enable them to fold in a protectedenvironment where they do not interact with any other proteins. | KF275121 |
| groES ** | 10 kDa heat shock protein | Co-chaperone of GroEL | KF275119 |
Reference gene candidates;
Target genes.
The second series were the target genes. They included nine hsp genes: hspA, hslO, hsp40, grpE, dnaK, hsp70 (1), hsp70(2), hsp70(3), and hsp70(4), which had previously been sequenced, and were kindly provided by Dr Quiblier (MNHN, Paris) plus four other genes: clpC (hsp100), htpG (hsp90), groEL (hsp60) and groES (hsp10)), which were isolated in this study. The full-length sequences of the genes obtained here are available in the EMBL database under the following accession numbers: KF275115 to KF275124, and KF294782 to KF294790. We also included mcyE, which is involved in Adda synthesis and the incorporation of D-Glu in the biosynthesis of MCs [29] as a target gene.
Primer Design and qPCR Conditions
All primer sets except that of mcyE (Table 2) were manually designed and then analyzed using NetPrimer algorithm, PREMIER Biosoft International (http://www.premierbiosoft.com/netprimer/). Furthermore, all primer pairs were checked for specificity using Primer-BLAST [30]. For mcyE, the primers mcyE-plaR3 [31] and mcyE-F2 [32] were used.
Table 2. Real-time PCR primers used in this study.
| Name | Primer sequence (5′ –3′) | Amplicon size (bp) | |
| Forward | Reverse | ||
| HspA | GCGATGTCCCTCTTTCCTCC | CCTTTTTCTTCGGTTTGGTTG | 167 |
| HslO | CCACATCCAGAGTCAATATCCG | CCATAACCAACATCTCGCACC | 202 |
| Hsp40 | ACCTGCGTTTAGAGTTCAAAGAAG | CGGACAAACGGAAACCTGAG | 206 |
| GrpE | GCGAATAATCCTGATGAACTAACG | CGCTTGATTCTATTTCCTGACC | 181 |
| DnaK | GAACGCATTGAACGCAAAAAC | GCTTGTTGTAAATCCGTAGTTAGGG | 191 |
| Hsp70(1) | CTGCTAAACGGGGTATTCCTC | CATCTTCATCGGCATAAACTTCTG | 189 |
| Hsp70(2) | CATTGGCATAGACTTAGGGACAAC | GTATTTTCCGCATTGGTAACGAC | 183 |
| Hsp70(3) | CCCGTTGTGATTGCTAACTCTG | GTGTAGGGAACCCGTTTTGAG | 200 |
| Hsp70(4) | GTAACGGCAGAGGATAACACCC | CCCCTAACCAAACGGAAAGAC | 259 |
| ClpC | GTTTCCCGTGCCATTCGTC | GTTGTCCGCCTTCGTTGTATC | 247 |
| HtpG | GAACGCAATAAAGAACGCCAC | GTCATCTAGTTCCGCATCCACC | 201 |
| GroEL | GCTCAAGTCGGTTCTATCTCTGC | CTTCCATCCGTTCGGTATCG | 205 |
| GroES | CTGTATCTCTAAGCGTATCAACCG | CATCATTGCGTTTGCCAGG | 168 |
| Rsh | CCTCACCTACCTTCCTATTCTCAAC | CGAATCTTTCTCCCTCCACG | 208 |
| RpoD | GACTCGCAACCCTTCCACTG | CTTTGTTCTCGTCATCTTCCTCC | 168 |
| GltA | CCACCAAAGATGAGTTAGCAGAC | GGATTATCTAAAGCCCGACGAG | 175 |
| GAPDH | GAAAGGGTGAAGGCGTGG | GTGGGTTGTGGTCATTGTGC | 173 |
| RpsL | GCTAACCTCTGGCTATGAAGTGAC | CTCCCGCCGTATCTAATGTTC | 152 |
| 16S rRNA | GGAGTACGCACGCAAGTGTG | GATGGCAACTAACGACGAGG | 246 |
| mcyE | GAAATTTGTGTAGAAGGTGC | CTCAATCTGAGGATAACGAT | 250 |
The specificity of the primer sets were tested by Real-time PCR using cDNA of P. agardhii CYA 128/6 and confirmed by melting curve (Fig. S1) and gel electrophoresis (Fig. S2). The identities of all PCR products were further confirmed by TA cloning using pGEM-T Easy vector (Promega, WI, USA) and subsequent sequencing.
The melting curve analysis indicated that all the primer pairs produced a single peak (Fig.S1), and only one band of the expected size was obtained on 2.5% agarose gel electrophoresis, which confirmed the specificity of all the primer pairs (Fig. S2).
Real time PCR was performed on a LightCycler 2.0 (Roche) using 32-capillary carousel combined with the LightCycler FastStart DNA Master SYBR Green I (Roche). Each 20-µl capillary contained a total volume reaction of 10 µl including: 1 µl of ready-to-use hot start PCR reaction mix; 1 µl of primer mix (Table S1); 4 µl of 1∶64 diluted cDNA sample; and 4 µl of MgCl2. Each run included a non-template control (NTC). Real-time amplification reactions for each gene of the WT and mutant strains from one biological replicate were performed in a technical duplicate and in the same PCR run. The cycling conditions were: 1 cycle at 95°C for 10 min, followed by 40 cycles at 95°C for 10 s, 62–65°C (depending on the target – Table S1) for 4 s, and 72°C for 10 s. In this study, the CT was automatically identified using the “Second Derivative Maximum Method” [33]. At the end of the amplification, the melting temperature of the product was also determined using the melting curve program: 65–95°C, with a heating rate of 0.1°C per s and continuous fluorescence measurement.
Expression Stability of the Candidate Reference gene and Data Analysis
In order to determine the true gene-specific variation, at least one stably-expressed RG is required to normalize the expression level of the target genes. The expression levels of the six candidate RGs were determined by RT-qPCR under the same experimental conditions as for the target genes (i.e. control condition+HL stress).
To identify the genes most stably expressed during HL treatment, the CT values of these six candidate RGs were analyzed using three different mathematical algorithms: geNorm [34], Normfinder [35] and BestKeeper [36]. In brief, GeNorm is the one most commonly used in the literature and it relies on the transformation of raw CT values (using the delta-CT method). The gene expression stability measure (M) for a candidate RG is computed by averaging pairwise variations of that gene versus all the other candidates tested. A decrease in the M value reflects an increase in expression stability. NormFinder is a model-based algorithm used to identify the optimum RGs from a group of candidates. This algorithm required the transformation of CT values to linear scale expression quantities. The genes with the lowest stability values have the most stable expression. BestKeeper uses the raw CT as the input for calculation. A Pearson correlation coefficient was calculated for each candidate pair as well as the probability that the correlation was significant. An index value was calculated as the geometric mean of the CT values of all highly-correlated candidate RGs. Stable RGs show a strong correlation with the BestKeeper index.
The gene-specific PCR efficiency was determined for each pair of primers using a 5-fold serial dilution of cDNA as template. The standard curve was obtained by plotting CT values against a logarithm of serial dilutions of the target nucleic acid. The efficiency of the reaction (E) was calculated from the slope value of a standard curve, as follows: E = 10(−1/sl°pe) –1.
The relative quantity of each gene (Q), which was used in geNorm and NormFinder, was calculated as: Q = E(min CT − sample C T) where Q = sample quantity relative to the sample with the highest expression; E = amplification efficiency; min CT = lowest CT value = CT value of the sample with the highest expression.
To calculate the normalized relative gene expression levels, data were analyzed using Relative Expression Software Tool (REST) (http://gene-quantification.com/rest.html) [37].
Data of transcript expression levels were analyzed using one-way analysis of variance (ANOVA) at a confidence level of p<0.05; followed by Tukey’s test on GraphPad Prism 5.0 software.
Results
Selection and Validation of Reference genes for RT-qPCR Normalization
In order to compare the expression levels of hsp target genes in the WT and ΔmcyD mutant of P. agardhii CYA 126/8 under HL conditions, we normalized all the samples using the same RGs. The CT values were obtained for each candidate gene across all the samples (Fig. 1) and revealed the differences in transcript levels. The 16S rRNA gene gave the lowest CT (8.81), corresponding to the highest expression level, whereas gltA and rsh showed the lowest expression levels, with mean CT values of 25.66 and 25.99, respectively (Fig. 1). The expression stability of the candidate RGs were analyzed using geNorm, NormFinder and BestKeeper, which provided complementary measures of the cDNA samples. Both geNorm and NormFinder classified 16S rRNA as the least stable gene (Table 3), regardless of the data series used (combined or single WT and mutant).The transcript level of this candidate was also much higher than that of the others (Fig. 1). Because it is crucial to use RGs with ranges of expression similar to those of the target genes in the samples for analysis [38], we excluded the 16S rRNA gene from the BestKeeper analysis. The three most stable genes identified by all three programs were similar, especially for the first two in all the data sets (Table 3). Rsh, rpoD and gltA were identified as the best performing genes, whereas rpsL and 16S rRNA were always identified as the least stable genes. The optimum number of RGs required for an accurate normalization was provided by the pairwise variation (Vn/n+1) calculation using the GeNorm program. The closest value to 0.15 (the cut-off value for validation - [35]) in our analyses, was found for V2/3 (0.12–0.14 according to data series); the value for V3/4 was even lower (0.09), indicating that three reference genes were the optimal number for accurately normalized gene expression. Consequently, we validated the 3 most stable reference genes (rsh, rpoD and gltA) for the normalization of all RT-qPCR data.
Figure 1. Real-time PCR CT values in the samples collected.
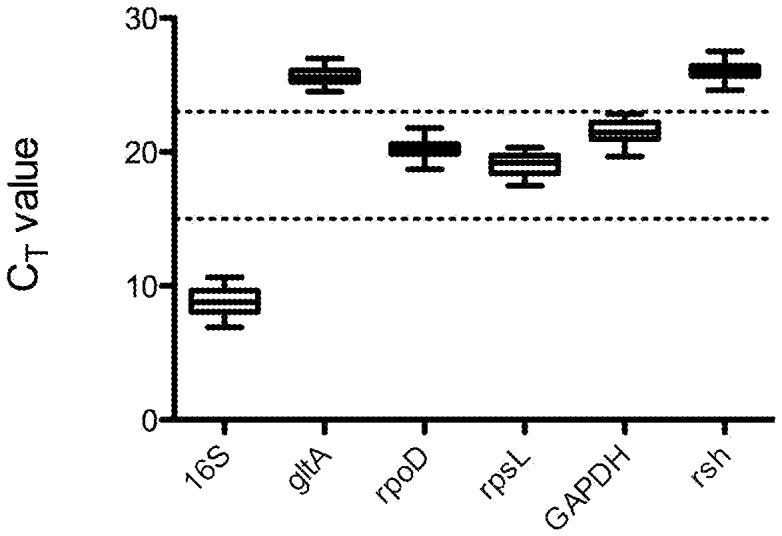
The distribution of the expression levels of candidate reference genes is shown by the median (lines), the lower and upper quartiles (boxes), and ranges (whiskers) (n = 20). The genes were divided into three groups by the arbitrary lines at CT 15 and 23, on the basis of their different expression levels.
Table 3. Ranking of candidate reference genes by three different algorithms.
| Gene name | Ranking order | ||||||||||||
| geNorm | NormFinder | BestKeeper | |||||||||||
| Combined | WT | M | No group | 2-group | Combined | WT | M | ||||||
| rsh | 1/2 | 1/2 | 1/2 | 1 | 1 | 1 | 2 | 1 | |||||
| rpoD | 1/2 | 1/2 | 3 | 2 | 3 | 2 | 1 | 3 | |||||
| gltA | 3 | 3 | 1/2 | 4/5 | 2 | 3 | 4 | 2 | |||||
| GAPDH | 4 | 4 | 4 | 3 | 4 | 4 | 3 | 4 | |||||
| rpsL | 5 | 5 | 5 | 4/5 | 5 | 5 | 5 | 5 | |||||
| 16S rRNA | 6 | 6 | 6 | 6 | 6 | ||||||||
GeNorm (75), NormFinder (2) and BestKeeper (51) used to identify the most stably expressed genes in control and HL conditions. Wild type (WT) and mutant (M) strains of Planktothrix agardhii.
Expression Levels of hsp genes of P. agardhii under Control Condition
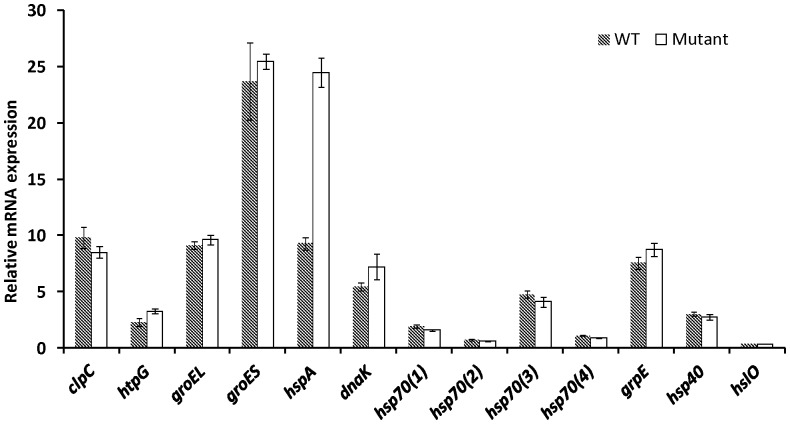
The expression level of the 13 target hsp genes (Fig. 2) was determined under control conditions versus the geometric mean of the transcription levels of the three selected reference genes. They could be divided into four groups (with no significant differences within mRNA abundance (p>0.05)) listed in decreasing order of mRNA abundance: (i) groES (WT and ΔmcyD) and hspA (ΔmcyD strain); (ii) groEL, clpC, grpE (WT and ΔmcyD) and hspA (WT); (iii) htpG, dnaK, hsp70(3), hsp40 (WT and ΔmcyD), and (iv) hsp70(1), hsp70(2), hsp70(4), hslO (WT and ΔmcyD).
Figure 2. Relative mRNA expression levels of P. agardhii hsp genes under optimal conditions.
Normalization against three references genes: rsh, rpoD and gltA. Error bars correspond to the standard deviation, including two technical replicates for two independent biological samples. Asterisks indicate a significant difference in the expression levels of the WT and the mutant strain; ***: p<0.001.
Under control conditions, the groES gene was the one most highly expressed in both the WT and ΔmcyD (23.72±3.45 and 25.47±0.7, respectively). Except for hspA, no significant difference was found under control conditions between the WT and ΔmcyD (p>0.05). The hspA expression level was significantly higher (p<0.001) in ΔmcyD than in WT (24.5±1.26 and 9.27±0.52, respectively).
Effects of HL on the Expression Levels of hsp genes in P. agardhii WT and ΔmcyD Mutant
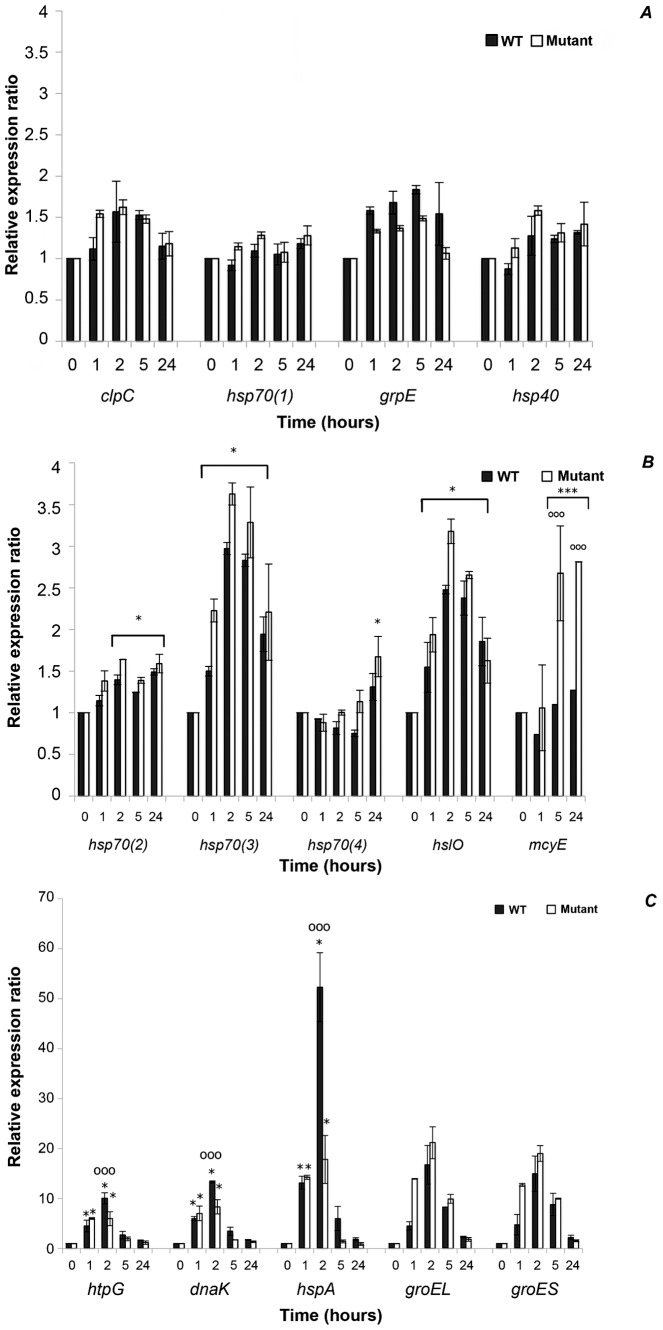
The transcriptional response of 13 hsp genes of P. agardhii (WT and ΔmcyD) was compared at different times (0, 1 h, 2 h, 5 h and 24 h) under HL conditions (Fig. 3 and Table 4). Based on the relative expression levels (shown as the fold change in gene expression versus control at time T0), three different expression profiles were obtained: (i) clpC, hsp70(1), grpE and hsp40 (Fig. 3.A) showed no significant difference in expression level (p>0.05) under control and HL conditions; (ii) hsp70(2), hsp70(3), hsp70(4) and hslO (Fig. 3.B) showed a slight (<4 fold) but significant change (p<0.05) in expression levels under HL and (iii) htpG, dnaK, hspA, groEL and groES (Fig. 3.C) showed a strong (>4 fold) and significant (p<0.05) increase in expression level under HL conditions.
Figure 3. Relative expression levels of the hsp genes and mcyE gene of P. agardhii from control (T0) to high light stress (1 h to 24 h).
Relative mRNA expression of hsp genes was normalized against three RGs: rsh, rpoD and gltA. (A): unchanged expression; (B): <4 fold up-regulated; (C): >4 fold up-regulated. Error bars correspond to the standard deviation, including two technical replicates for two independent biological samples. An asterisk indicates a significant difference versus control (T0) *: p<0.05. Circles indicate a significant difference in the expression level between the WT and the mutant strains; °°: p<0.01; °°°: p<0.001.
Table 4. Relative fold change of transcript of the genes obtained by qRT-PCR.
| Gene | WT | M | Statistical difference between WT and Mutant | |
| Group 1 | clpC | 1.57±0.37 | 1.62±0.09 | NS |
| hsp70(1) | 1.1±0.08 | 1.28±0.04 | NS | |
| grpE | 1.68±0.14 | 1.37±0.03 | NS | |
| hsp40 | 1.28±0.24 | 1.58±0.06 | NS. | |
| Group 2 | hsp70(2) | 1.40±0.06 | 1.64±0.001 | * |
| hsp70(3) | 2.97±0.07 | 3.63±0.13 | NS | |
| hsp70(4) | 0.82±0.08 | 1.00±0.03 | NS | |
| hslO | 2.48±0.05 | 3.18±0.15 | NS | |
| mcyE | 1.1±0.13 | 2.7±0.57 | *** | |
| Group 3 | htpG | 10.06±1.09 | 5.98±1.40 | *** |
| dnaK | 13.34±0.22 | 8.34±1.44 | ** | |
| hspA | 52.31±6.90 | 17.82±4.80 | *** | |
| groEL | 16.77±3.86 | 21.19±3.21 | NS | |
| groES | 14.98±3.53 | 19.02±1.59 | NS |
In the WT and mutant (M) strains of P. agardhii after exposure to HL stress for 2 h (for hsp genes) and 5 h (for mcyE). Data normalization was done using the three most stable RGs (rsh, rpoD and gltA). Group 1: genes which showed no significant change under HL conditions; Group 2: genes which were slightly up-regulated (<4 fold) under HL conditions; and Group 3: genes which were strongly up-regulated (>4 fold) under HL conditions. Values are reported as mean ± SD; NS: p>0.05;
p<0.05;
p<0.01;
p<0.001.
The expression of the following genes was significantly higher under HL than under control conditions (T0) (Fig. 3.B): Hsp70(3) and hslO showed a significant increase at 1 h, with a peak after 2 h under HL (≈3.5 fold for both strains) (p<0.05). Hsp70(4) exhibited a significant (p<0.01) increase after 24 hours for both the WT and the ΔmcyD (1.31±0.16 and 1.67±0.24, respectively). For hsp70(2), a slight but significant increase (p<0.05) was observed in the WT and the mutant after 2 hours under HL (1.38±0.12 and 1.64±0.001, respectively) (p<0.05).
Five genes were strongly up-regulated under the HL conditions (Fig. 3.C) and they displayed a similar expression pattern: a significant increase of expression level (p<0.001) at 1 hour, a peak reached after 1 to 2 hours under HL, and then a decrease to the background level. The expression profiles of htpG, dnaK, and hspA were quite similar in both strains. Interestingly, quantitatively significant differences were found between the WT and the ΔmcyD for these three genes (Fig. 3.C). In the WT, the expressions of htpG, dnaK, and hspA were up-regulated to a greater extent after 2 h than in ΔmcyD (Fig. 3.C). The fold change in dnaK expression induced by HL was 13.34 in the WT versus 8.34 in the ΔmcyD (p<0.01). For htpG, the expression fold change reached its maximum of 10.06±1.08 after 2 h of exposure to HL in the WT, which was significantly higher (p<0.001) than that in the mutant (5.98±1.39).
The relative expression of hspA after 2 h under HL was 52.31±6.9 in the WT strain compared to 17.82±4.8 in the ΔmcyD (p<0.001). However, it is noteworthy that under control conditions, the level of hspA in WT was significantly lower than in the ΔmcyD strain (9.27±0.52 and 24.5±1.26, respectively) (Fig. 2). Therefore, even though the maximum level of change in WT reached 52.31±6.9 fold versus 17.82±4.8 fold in the mutant, there was no significant difference between the amounts of hspA mRNA under HL in the two strains (p>0.05).
For groEL and groES genes, in both strains, the mRNA reached the greatest level 2 hours after being exposed to HL stress and then decreased (Fig. 3.C). However, the pattern of change differs in timing with a significant increase (p<0.001) right after 1 h in the mutant, in contrast to the unchanged value in the WT (Fig. 3.C). The maximum level was reached after 2 h in both strains. For groEL and groES expression in HL, no significant differences were observed between the maximum levels of the WT and ΔmcyD.
Effects of HL on Expression Levels of the mcyE gene in P. agardhii WT and ΔmcyD Mutant
The transcription level of mcyE of the WT strain was not affected by HL during the test period (0–24 h) (Fig. 3.B). The main finding was that the insertion of the CmR gene into mcyD did not disrupt the expression of another mcy gene (mcyE) in P. agardhii. Under HL, a significant increase in the abundance of mcyE mRNA was observed in the mutant (2.7 fold 5 h after the shift from control conditions to HL) (p<0.001). This mRNA level remained high till 24 h.
Discussion
There is a general consensus that the selection of suitable reference genes for normalization of the target genes is a prerequisite for RT-qPCR [24]. In this study, six candidate genes were chosen from independent pathways to avoid possible effects due to co-regulation. Four genes had previously been used as conventional RGs in prokaryotes (16SrRNA, rpoD, GAPDH, rsh) [27]–[28]. To the best of our knowledge, the other two candidate genes (gltA, rpsL) had never been tested in prokaryotes. A combination of three computational programs (GeNorm, NormFinder and BestKeeper) was used to provide accurate validation of the most stable genes for normalization [39]. The same three genes (rsh, rpoD and gltA) were identified as being the most stable genes, despite some slight differences in ranking order (Table 3). Since these three algorithms rely on different mathematical approaches to calculate stability (see Mat & Meth), these minor differences between their outputs were not unexpected [36], [40]. Such discrepancies have been reported in several studies as minor changes in gene stability rankings [41]. According to the geNorm analysis, V2/3 was below 0.15, indicating that the minimum number of RGs required for reliable normalization in this study would be two. However, it should be pointed out that using additional genes is usually an option [40]–[41], and using the three best RGs is a valid normalization strategy in most cases [35]. Moreover, in our study, including the third most stable candidate gene gave a significantly lower V-value. We therefore decided to use the three most stable genes as RGs for normalization of the RT-qPCR data. These included gltA, which will be listed as a newly-recommended RG for further RT-qPCR data normalization, because validation of the suitability of RGs for normalization in prokaryotes is still lacking [42].
The most unstable genes were also the same in all the sample sets (Table 3). We showed here that 16SrRNA and GAPDH, two RGs used in previous studies [28], [43] were not appropriate for normalization under our experimental conditions for the two Planktothrix strains we used. The suitability of 16SrRNA gene for use as an RG is currently disputed [44] due to its high abundance compared to target gene transcripts (resulting in too large a difference in CT values), which can bias interpretations [17], [35]. This once more showed that appropriate RGs can be very different in different organisms as well as under different experimental conditions and the more commonly used RGs are not necessarily always suitable. Each RG needs to be validated under the same experimental set-up as the target gene. The validation of three RGs that are stably expressed under our experimental settings allowed us to evaluate the relative expression levels of 13 hsp genes of the MC-producing strain and of its MC-deficient mutant under optimum and HL conditions over a period of 24 h.
Under control conditions, all the hsp genes were found to be constitutively expressed. Among them, groES showed the highest expression level, which was at least double that of the others (Fig. 2). This was not surprising, as most Hsps are constantly produced to assist the proper folding of nascent proteins, and to prevent protein aggregation throughout the lifetime of cells [21], [45]. Furthermore, groES is a part of the groESL operon, a major chaperone system in bacteria, which plays an important role in the conformational homeostasis of cell proteins [38]. However, under control conditions, a higher expression level (about 2.5-fold greater) of a small hsp (hspA) was observed in the mutant strain than in the WT strain (Fig. 2), something that we cannot explain.
Under HL stress, among the 13 genes under investigation, only five hsps were strongly up-regulated in both strains, with a 6 to 52-fold change relative to control conditions, four genes were slightly up-regulated, and the other four were unchanged (Fig. 3, Table 4). It has been observed that the Hsps stress response varies considerably in some organisms depending on the species, Hsp family, developmental stage, and stressor [46]–[47]. Diverse Hsp isotypes in different species may have different roles and modes of action. Therefore, the true significance and role of Hsps in difference species must be confirmed using methods appropriate for each species.
However, it had previously been reported that HL stress may induce changes in gene transcription within as little as 15 minutes, and the expression level may also return to the basal level very quickly [48]. In this study, over our experimental time course (1–24 h) the five genes were all strongly up-regulated and displayed a similar kinetic pattern. The expression level increased 1 h after the shift to high light; it then reached its maximum level after 1 to 2 h, and thereafter declined (Fig. 3). This pattern of change is typical of how gene expression usually responds to stress [48]. These changes are usually transient and, even with persistent stress, gene expression fairly soon reaches a new homeostasis, in which the physiology of the cell has adjusted to new conditions [48]–[50].
The pronounced inductions of the five hsp genes (htpG, dnaK, hspA, groEL, groES) are consistent with those reported for Synechocystis PCC6803, where htpG, dnaK2, groESL, and hspA were also conspicuously up-regulated in response to a shift from low light to HL [48], [51]. This may suggest that these genes play a significant physiological role in protecting cells against this specific abiotic stress, regardless of the cell’s ability to produce MCs.
The most important finding of our study was detecting some transcriptional differences between the WT and its MC-deficient mutant. Indeed, two hsps genes (htpG and dnaK) were ≈ 1.7-fold more highly expressed under HL stress in WT than in ΔmcyD (Table 4 and Fig. 3). HtpG had previously been reported to play a role in the ability of cyanobacteria to tolerate various stresses [52]–[54], including providing effective protection against the oxidative stress caused by HL in unicellular cyanobacteria [50]. Some studies have suggested that HtpG is involved in regulating the biosynthesis of tetrapyrrole [55] and that it interacts with the linker polypeptides of phycobilisome in cyanobacteria to prevent their thermal aggregation [56]. These activities may endow HtpG with an effective photoprotective role in response to HL stress.
For the dnaK gene, it has been shown that cyanobacteria contain multiple dnaK homologs, the expressions of which are differently regulated [57]. However, the alignment of our sequence (Accession no.: KF294788) revealed the greatest similarity to dnaK2 of Synechocystis PCC6803 (91% similarity- data not shown). dnaK2 has been reported to be induced by various abiotic stresses [50], [58]–[59], including HL [50], although its essential function is still elusive [60].
Even if their multiple functions are still unclear, one would expect that higher expression levels of dnaK and htpG may contribute to better protection of macromolecular complexes, such as the photosynthetic apparatus, and thus enable the WT strain to tolerate HL better.
Finally, an unexpected finding was about the expression profile of the mcyE in both strains when they were transferred from control conditions to HL stress (Fig. 3.B, Table 4). In the WT strain, stable expression of the mcyE gene was observed after the transition from control to HL stress conditions for a short period of time (0–24 h), in contrast to some previous findings in Microcystis strains. An up-regulation of mcyB and mcyD was found as a result of HL intensities [11]. Using RT-qPCR we showed that the expression level remained constant, which was corroborated by constant MCs production by the P. agardhii WT cells during the first 24 h (450 ng eq. MC-LR per mL of culture normalized to OD750 nm = 1, unpublished data). The mcy operon seems to be expressed at a basic level corresponding to the intracellular-MC present in the cell. Up-regulation in the WT seems to be unnecessary during the first 24 h under this level of HL. Unexpectedly, a basal level of the mcyE transcript was observed under control conditions in the MC-deficient mutant that was similar to that in the WT. The disruption of mcyD by CmR has no effect on the expression of mcyE gene expression in P. agardhii. This absence of any polar effect of the mutation of one mcy gene on the others had previously been reported in Microcystis [61]. In the MC-deficient mutant, HL induced a significant increase of the mcyE transcript that reached its maximum level from 5 to 24 h (Fig. 3.B). The enhancement of the mcyE transcript induced by HL conditions in the ΔmcyD mutant strain might reflect a requirement for MCs production under such stress. HL is known to cause direct severe damage of the photosynthetic apparatus, and an indirect increase in ROS production (which induces oxidative stress). As a consequence, many different mechanisms and substances may act as cellular defenses in different ways. MCs may be one of them, as suggested by Zilliges and colleagues [19], as MCs bind to cysteine-residues and to specific protein targets involved in photosynthesis processes and against oxidative stress conditions. The possibility cannot be excluded that the depletion or the absence of MCs in deficient cells could increase the damage caused, and may thus contribute to an increase in the susceptibility to environmental stress. This might explain the sporadic changes seen within populations, where MC-producing genotypes can replace non-producing strains in the field under unfavorable conditions [15], [62].
In conclusion, our findings support the hypothesis that MCs have an intracellular function in Planktothrix agardhii related to the transcriptional variations of mRNA, and that this could be attributed to the intracellular presence of MCs in the producer cells (related to HL stress). However, further investigations are needed to identify the nature of the interactions between MCs and Hsps modulated-responses (if any), and finally, to define a possible connection between MCs and the primary metabolism of cyanobacteria that produce this “secondary” metabolite.
Supporting Information
Examples of melting curve profile of 19 genes investigated in the study. RG candidates : A – Rsh; B – RpoD; C – GltA; D – GAPDH; E – RpsL; F –16S rRNA; GOIs : G – ClpC; H – HtpG; I – GroEL; K – GroES; L – HspA; M – dnaK; N – Hsp70 (1); O – Hsp70 (2); P – Hsp70 (3); Q – Hsp 70(4); R – GrpE; S– Hsp 40; T – HslO;
(DOC)
Agarose gel electrophoresis showing specific RT PCR products of the expected size for each gene. A: Ultra Low Range DNA Ladder (lane 1); 16S rRNA (246 bp) (lane 2); Clp (247 bp) (lane 3); gltA (175 bp) (lane 4); GroES (168 bp) (lane 5); GroEL (205 bp) (lane 6); DnaK (191 bp) (lane 7); HspA (167 bp) (lane 8); HslO (202 bp) (lane 9); Hsp70(1) (189 bp) (lane 10); Hsp70(2) (183 bp) (lane 11); Hsp70(4) (200 bp) (lane 12); Hsp70(5) (259 bp) (lane 13); hsp40 (206 bp) (lane 14); rpoD (168 bp) (lane 15); NTC (lane 16) B: Ultra Low Range DNA Ladder (lane 1); mcyE (250 bp) (lane 2,3); GAPDH (173 bp) (lane 4,5); GrpE (181 bp) (lane 6,7); HK (186 bp) (lane 8,9) C: Ultra Low Range DNA Ladder (lane 1); rspL (152 bp) (lane 2); NTC (lane 3); rsh (208 bp) (lane 4)
(TIFF)
Optimal parameters obtained for each transcript and its amplification efficiency.
(DOC)
Acknowledgments
We are very grateful to Dr. Rainer Kurmayer for providing us with the P. agardhii CYA 126/8 strain and its ΔmcyD mutant used in this study. We thank Dr. Valérie Barbe from the Genomic Institut – Genoscope (http://www.genoscope.cns.fr), Evry, France for giving us 9 gene sequences. Many thanks to Monika Gosh for improving the English version of the manuscript.
Funding Statement
TDCT was a PhD scholarship recipient of Vietnam. Financial support was provided through ATM (“Biodiversité et rôle des micro-organismes dans les écosystèmes actuels et passés”) fellowship from the National Natural History Museum (MNHN) from 2010 to 2011 and an annual grant from Yves Rocher foundation (grant number 660/09). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. Paerl HW, Huisman J (2009) Climate change: a catalyst for global expansion of harmful cyanobacterial blooms. Env Microbiol Rep 1: 27–37. [DOI] [PubMed] [Google Scholar]
- 2. Krüger T, Christian B, Luckas B (2009) Development of an analytical method for the unambiguous structure elucidation of cyclic peptides with special appliance for hepatotoxic desmethylated microcystins. Toxicon 54: 302–312. [DOI] [PubMed] [Google Scholar]
- 3. Tillett D, Dittmann E, Erhard M, von Dohren H, Borner T, et al. (2000) Structural organization of microcystin biosynthesis in Microcystis aeruginosa PCC7806: an integrated peptide-polyketide synthetase system Chem Biol. 7: 753–64. [DOI] [PubMed] [Google Scholar]
- 4.Chorus I, Bartram J (1999) In Toxic Cyanobacteria in Water - A Guide to their Public Health Consequences, Monitoring and Management. London: E & FN Spon Press. 595 p.
- 5. Rapala J, Sivonen K, Lyra C, Niemela SI (1997) Variation of microcystins, cyanobacterial hepatotoxins, in Anabaena spp. as a function of growth stimuli. Appl Environ Microb 63: 2206–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hesse K, Kohl JG (2001) Effects of light and nutrient supply on growth and microcystin content of different strains of Microcystis aeruginosa. In: Chorus Editor. Cyanotoxins: Occurrence Causes Consequences. 104–15.
- 7. Demott WR, Zhang QX, Carmichael WW (1991) Effects of toxic cyanobacteria and purified toxins on the survival and feeding of a copepod and 3 species of Daphnia . Limnol Oceanogr 36: 1346–57. [Google Scholar]
- 8. Utkilen H, Gjolme N (1995) Iron-stimulated toxin production in Microcystis aeruginosa . Appl Environ Microb 61: 797–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Christoffersen K (1996) Ecological implications of cyanobacterial toxins in aquatic food webs. Phycologia 35: 42–50. [Google Scholar]
- 10. Sedmak B, Kosi G (1998) The role of microcystins in heavy cyanobacterial bloom formation. J Plankton Res 20: 691–708. [Google Scholar]
- 11. Kaebernick M, Neilan BA, Borner T, Dittmann E (2000) Light and the transcriptional response of the microcystin biosynthesis gene cluster. Appl Environ Microb 66: 3387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dittmann E, Erhard M, Kaebernick M, Scheler C, Neilan BA, et al. (2001) Altered expression of two light-dependent genes in a microcystin-lacking mutant of Microcystis aeruginosa PCC 7806. Microbiology 147: 3113–9. [DOI] [PubMed] [Google Scholar]
- 13. Schatz D, Keren Y, Vardi A, Sukenik A, Carmeli S, et al. (2007) Towards clarification of the biological role of microcystins, a family of cyanobacterial toxins. Environ Microbiol 9: 965–70. [DOI] [PubMed] [Google Scholar]
- 14. Babica P, Blaha L, Marsalek B (2006) Exploring the natural role of microcystins - A review of effects on photoautotrophic organisms. J Phycol 42: 9–20. [Google Scholar]
- 15. Briand E, Yepremian C, Humbert JF, Quiblier C (2008) Competition between microcystin- and non-microcystin-producing Planktothrix agardhii (cyanobacteria) strains under different environmental conditions. Environ Microbiol 10: 3337–48. [DOI] [PubMed] [Google Scholar]
- 16. Sevilla E, Martin-Luna B, Vela L, Bes MT, Fillat MF, et al. (2008) Iron availability affects mcyD expression and microcystin-LR synthesis in Microcystis aeruginosa PCC7806. Environ Microbiol 10: 2476–83. [DOI] [PubMed] [Google Scholar]
- 17.Alexova R, Haynes PA, Ferrari BC, Neilan BA (2011) Comparative protein expression in different strains of the bloom-forming cyanobacterium Microcystis aeruginosa. Mol Cell Proteomics 10: DOI: 10.1074/mcp 003749. [DOI] [PMC free article] [PubMed]
- 18.Dziallas C, Grossart HP (2011) Increasing Oxygen Radicals and Water Temperature Select for Toxic Microcystis sp. PloS ONE 6:DOI: 10–1371/journal.pone 0025569. [DOI] [PMC free article] [PubMed]
- 19.Zilliges Y, Kehr JC, Meissner S, Ishida K, Mikkat S, et al.. (2011) The Cyanobacterial Hepatotoxin Microcystin Binds to Proteins and Increases the Fitness of Microcystis under Oxidative Stress Conditions. PloS ONE 6DOI: 10–1371/journal.pone 0017615. [DOI] [PMC free article] [PubMed]
- 20. Wang WX, Vinocur B, Shoseyov O, Altman A (2004) Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 9: 244–52. [DOI] [PubMed] [Google Scholar]
- 21.Wase NV, Yen SO, Wright PC (2013) A global understanding of light stress in cyanobacteria: Environmental and bioproducts perspectives. In: Srivastava AK, Rai AN, Neilan BA, editors. Stress Biology of Cyanobacteria: Molecular Mechanisms to Cellular Responses: CRC Press; 394 p.
- 22. Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu Rev Physiol 61: 243–82. [DOI] [PubMed] [Google Scholar]
- 23. Lindquist S (1986) The heat-shock response. Annual Review of Biochemistry 55: 1151–91. [DOI] [PubMed] [Google Scholar]
- 24. Bustin SA, Benes V, Garson JA, Hellemans J (2010) The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55: 611–622. [DOI] [PubMed] [Google Scholar]
- 25. Reynolds CS, Huszar V, Naselli-Flores L, Melo S (2002) Towards a functional classification of the freshwater phytoplankton. J Plankton Res 24: 417–28. [Google Scholar]
- 26. Rippka R (1988) Isolation and purification of cyanobacteria. Method Enzymol 167: 3–27. [DOI] [PubMed] [Google Scholar]
- 27. Zhao W, Li Y, Gao P, Sun Z, Sun T, et al. (2011) Validation of reference genes for real-time quantitative PCR studies in gene expression levels of Lactobacillus casei Zhang. J Ind Microbiol Biot 38: 1279–86. [DOI] [PubMed] [Google Scholar]
- 28. Botteldoorn N, Van Coillie E, Grijspeerdt K, Werbrouck H, Haesebrouck F, et al. (2006) Real-time reverse transcription PCR for the quantification of the mntH expression of Salmonella enterica as a function of growth phase and phagosome-like conditions. J Microbiol Meth 66: 125–35. [DOI] [PubMed] [Google Scholar]
- 29. Dittmann E, Borner T (2005) Genetic contributions to the risk assessment of microcystin in the environment. Toxicol Appl Pharm 203: 192–200. [DOI] [PubMed] [Google Scholar]
- 30.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, et al.. (2012) Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13. DOI: 10–1186/1471–2105–13–134. [DOI] [PMC free article] [PubMed]
- 31. Rantala A, Rajaniemi-Wacklin P, Lyra C, Lepisto L, Rintala J, et al. (2006) Detection of microcystin-producing cyanobacteria in Finnish lakes with genus-specific microcystin synthetase gene E (mcyE) PCR and associations with environmental factors. Appl EnvironMicrob 72: 6101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vaitomaa J, Rantala A, Halinen K, Rouhiainen L, Tallberg P, et al. (2003) Quantitative real-time PCR for determination of microcystin synthetase e copy numbers for microcystis and anabaena in lakes. Appl EnvironMicrob 69: 7289–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rasmussen R (2001) Quantification on the LightCycler. In: Meuer S, Wittwer C, Nakagawara K, editors. Rapid Cycle Real-time PCR, Methods and Applications. Heidelberg: Springer Press 21–34.
- 34.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, et al.. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3 (7)Research0034. [DOI] [PMC free article] [PubMed]
- 35. Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–50. [DOI] [PubMed] [Google Scholar]
- 36. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper - Excel-based tool using pair-wise correlations. Biotechnol Lett 26: 509–15. [DOI] [PubMed] [Google Scholar]
- 37. Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30 (9): e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chaurasia AK, Apte SK (2009) Overexpression of the groESL Operon Enhances the Heat and Salinity Stress Tolerance of the Nitrogen-Fixing Cyanobacterium Anabaena sp Strain PCC7120. Appl EnvironMicrob 75: 6008–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayers D, Clements DN, Salway F, Day PJR (2007) Expression stability of commonly used reference genes in canine articular connective tissues. BMC Vet Res 3DOI: 10–1186/1746–6148–3-7. [DOI] [PMC free article] [PubMed]
- 40.Lee JM, Roche JR, Donaghy DJ, Thrush A, Sathish P (2010) Validation of reference genes for quantitative RT-PCR studies of gene expression in perennial ryegrass (Lolium perenne L.). BMC Mol Biol 11DOI: 10–1186/1471–2199–11–8. [DOI] [PMC free article] [PubMed]
- 41. Cruz F, Kalaoun S, Nobile P, Colombo C, Almeida J, et al. (2009) Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol Breeding. 23: 607–16. [Google Scholar]
- 42. Huggett J, Dheda K, Bustin S, Zumla A (2005) Real-time RT-PCR normalisation; strategies and considerations. Genes Immun 6: 279–84. [DOI] [PubMed] [Google Scholar]
- 43. Venkatesh B, Babujee L, Liu H, Hedley P, Fujikawa T, et al. (2006) The Erwinia chrysanthemi 3937 PhoQ sensor kinase regulates several virulence determinants. J Bacteriol 188: 3088–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ritz M, Garenaux A, Berge M, Federighi M (2009) Determination of rpoA as the most suitable internal control to study stress response in C-jejuni by RT-qPCR and application to oxidative stress. J Microbiol Meth 76: 196–200. [DOI] [PubMed] [Google Scholar]
- 45.Mayer MP (2010) Gymnastics of molecular chaperones. Mol Cell 39: DOI: 10.1016/j.molcel.2010.07.012. [DOI] [PubMed]
- 46. Clark MS, Peck LS (2009) HSP70 heat shock proteins and environmental stress in Antarctic marine organisms: A mini-review. Mar Genom 2: 11–8. [DOI] [PubMed] [Google Scholar]
- 47. Iwama GK, Afonso LOB, Todgham A, Ackerman P, Nakano K (2004) Are hsps suitable for indicating stressed states in fish? J Exp Biol 207: 15–9. [DOI] [PubMed] [Google Scholar]
- 48. Hihara Y, Kamei A, Kanehisa M, Kaplan A, Ikeuchi M (2001) DNA microarray analysis of cyanobacterial gene expression during acclimation to high light. Plant Cell 13: 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lopez-Maury L, Marguerat S, Bahler J (2008) Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat Rev Genet 9: 583–93. [DOI] [PubMed] [Google Scholar]
- 50. Mary I, Tu CJ, Grossman A, Vaulot D (2004) Effects of high light on transcripts of stress-associated genes for the cyanobacteria Synechocystis sp. PCC 6803 and Prochlorococcus MED4 and MIT9313. Microbiology 150: 1271–81. [DOI] [PubMed] [Google Scholar]
- 51. Huang LX, McCluskey MP, Ni H, LaRossa RA (2002) Global gene expression profiles of the cyanobacterium Synechocystis sp strain PCC 6803 in response to irradiation with UV-B and white light. J Bacteriol 184: 6845–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fang F, Barnum SR (2003) The heat shock gene, htpG, and thermotolerance in the cyanobacterium, Synechocystis sp PCC 6803. Curr Microbiol 47: 341–6. [DOI] [PubMed] [Google Scholar]
- 53. Hossain MM, Nakamoto H (2002) HtpG plays a role in cold acclimation in cyanobacteria. Curr Microbiol 44: 291–6. [DOI] [PubMed] [Google Scholar]
- 54. Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp PCC 6803. Bioch Bioph Res Co 290: 339–48. [DOI] [PubMed] [Google Scholar]
- 55. Watanabe S, Kobayashi T, Saito M, Sato M, Nimura-Matsune K, et al. (2007) Studies on the role of HtpG in the tetrapyrrole biosynthesis pathway of the cyanobacterium Synechococcus elongatus PCC 7942. Bioch Bioph Res Co 352: 36–41. [DOI] [PubMed] [Google Scholar]
- 56. Sato T, Minagawa S, Kojima E, Okamoto N, Nakamoto H (2010) HtpG, the prokaryotic homologue of Hsp90, stabilizes a phycobilisome protein in the cyanobacterium Synechococcus elongatus PCC 7942. Mol Microbiol 76: 576–89. [DOI] [PubMed] [Google Scholar]
- 57. Lee BH, Hibino T, Jo J, Viale AM, Takabe T (1997) Isolation and characterization of a dnaK genomic locus in a halotolerant cyanobacterium Aphanothece halophytica . Plant Mol Biol 35: 763–75. [DOI] [PubMed] [Google Scholar]
- 58. Chitnis PR, Nelson N (1991) Molecular cloning of the genes encoding 2 chaperone proteins of the cyanobacterium Synechocystis sp PCC 6803. J Biol Chem 266: 58–65. [PubMed] [Google Scholar]
- 59. Fulda S, Mikkat S, Huang F, Huckauf J, Marin K, et al. (2006) Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6: 2733–45. [DOI] [PubMed] [Google Scholar]
- 60. Katano Y, Nimura-Matsune K, Yoshikawa H (2006) Involvement of DnaK3, one of the three DnaK proteins of cyanobacterium Synechococcus sp PCC7942, in translational process on the surface of the thylakoid membrane. Biosc Biotech Bioch 70: 1592–8. [DOI] [PubMed] [Google Scholar]
- 61. Pearson LA, Hisbergues M, Borner T, Dittmann E, Neilan BA (2004) Inactivation of an ABC transporter gene, mcyH, results in loss of microcystin production in the cyanobacterium Microcystis aeruginosa PCC 7806. Appl Environ Microb 70: 6370–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. KurmayerR, Dittman E, Fastner J, Chorus I (2001) Diversity of microcystin genes within a population of the toxic cyanobacterium Microcystis spp. in lake Wannsee (Berlin, germany). Microb Ecol 43: 107–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Examples of melting curve profile of 19 genes investigated in the study. RG candidates : A – Rsh; B – RpoD; C – GltA; D – GAPDH; E – RpsL; F –16S rRNA; GOIs : G – ClpC; H – HtpG; I – GroEL; K – GroES; L – HspA; M – dnaK; N – Hsp70 (1); O – Hsp70 (2); P – Hsp70 (3); Q – Hsp 70(4); R – GrpE; S– Hsp 40; T – HslO;
(DOC)
Agarose gel electrophoresis showing specific RT PCR products of the expected size for each gene. A: Ultra Low Range DNA Ladder (lane 1); 16S rRNA (246 bp) (lane 2); Clp (247 bp) (lane 3); gltA (175 bp) (lane 4); GroES (168 bp) (lane 5); GroEL (205 bp) (lane 6); DnaK (191 bp) (lane 7); HspA (167 bp) (lane 8); HslO (202 bp) (lane 9); Hsp70(1) (189 bp) (lane 10); Hsp70(2) (183 bp) (lane 11); Hsp70(4) (200 bp) (lane 12); Hsp70(5) (259 bp) (lane 13); hsp40 (206 bp) (lane 14); rpoD (168 bp) (lane 15); NTC (lane 16) B: Ultra Low Range DNA Ladder (lane 1); mcyE (250 bp) (lane 2,3); GAPDH (173 bp) (lane 4,5); GrpE (181 bp) (lane 6,7); HK (186 bp) (lane 8,9) C: Ultra Low Range DNA Ladder (lane 1); rspL (152 bp) (lane 2); NTC (lane 3); rsh (208 bp) (lane 4)
(TIFF)
Optimal parameters obtained for each transcript and its amplification efficiency.
(DOC)