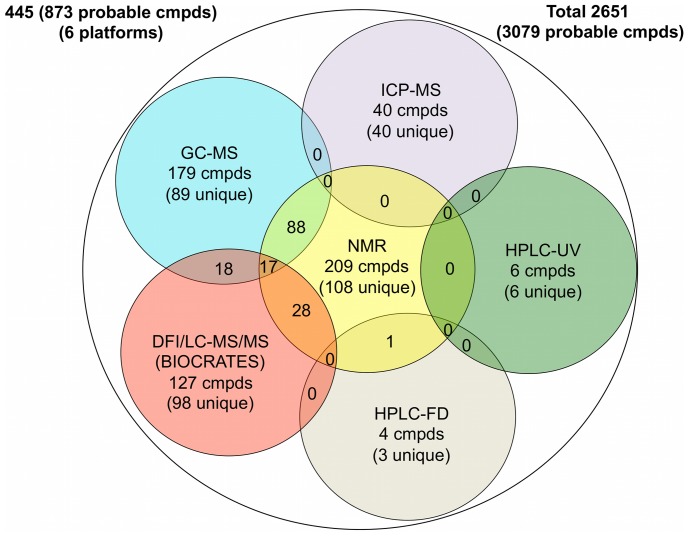
Abstract
Urine has long been a “favored” biofluid among metabolomics researchers. It is sterile, easy-to-obtain in large volumes, largely free from interfering proteins or lipids and chemically complex. However, this chemical complexity has also made urine a particularly difficult substrate to fully understand. As a biological waste material, urine typically contains metabolic breakdown products from a wide range of foods, drinks, drugs, environmental contaminants, endogenous waste metabolites and bacterial by-products. Many of these compounds are poorly characterized and poorly understood. In an effort to improve our understanding of this biofluid we have undertaken a comprehensive, quantitative, metabolome-wide characterization of human urine. This involved both computer-aided literature mining and comprehensive, quantitative experimental assessment/validation. The experimental portion employed NMR spectroscopy, gas chromatography mass spectrometry (GC-MS), direct flow injection mass spectrometry (DFI/LC-MS/MS), inductively coupled plasma mass spectrometry (ICP-MS) and high performance liquid chromatography (HPLC) experiments performed on multiple human urine samples. This multi-platform metabolomic analysis allowed us to identify 445 and quantify 378 unique urine metabolites or metabolite species. The different analytical platforms were able to identify (quantify) a total of: 209 (209) by NMR, 179 (85) by GC-MS, 127 (127) by DFI/LC-MS/MS, 40 (40) by ICP-MS and 10 (10) by HPLC. Our use of multiple metabolomics platforms and technologies allowed us to identify several previously unknown urine metabolites and to substantially enhance the level of metabolome coverage. It also allowed us to critically assess the relative strengths and weaknesses of different platforms or technologies. The literature review led to the identification and annotation of another 2206 urinary compounds and was used to help guide the subsequent experimental studies. An online database containing the complete set of 2651 confirmed human urine metabolite species, their structures (3079 in total), concentrations, related literature references and links to their known disease associations are freely available at http://www.urinemetabolome.ca.
Introduction
Metabolomics is a relatively young branch of “omics” science concerned with the systematic study of the chemical products or metabolites that cells and organisms generate. Because metabolites are the downstream products of numerous genome-wide or proteome-wide interactions, the metabolome (the sum of all metabolites in an organism) can be a very sensitive measure of an organism’s phenotype. This fact has made metabolomics particularly useful in the study of environment-gene interactions [1], [2], [3], [4], the identification of disease biomarkers [5], [6], [7], [8], [9] and the discovery of drugs [10]. Unlike its older “omics” cousins, where complete or near-complete coverage of the genome or proteome is fairly routine, metabolomics still struggles to cover even a tiny fraction of the metabolome. Indeed, most human metabolomic studies published today, even those exploiting the latest and most sensitive LC-MS/MS technologies, typically succeed in identifying or characterizing fewer than 100 compounds [11], [12], [13], [14]. This corresponds to less than 1% of the known human metabolome [15], [16]. In an effort to help improve this situation, we (and others) have started to undertake the systematic characterization of various human biofluid metabolomes. This includes the human cerebrospinal fluid metabolome [17], [18], the human saliva metabolome [19], and the human serum metabolome [20]. We have now turned our attention to characterizing the human urine metabolome.
Urine, as produced by mammals, is a transparent, sterile, amber-colored fluid generated by the kidneys. The kidneys extract the soluble wastes from the bloodstream, as well as excess water, sugars, and a variety of other compounds. The resulting urine contains high concentrations of urea (from amino acid metabolism), inorganic salts (chloride, sodium, and potassium), creatinine, ammonia, organic acids, various water-soluble toxins and pigmented products of hemoglobin breakdown, including urobilin, which gives urine its characteristic color. Urination is the primary route by which the body eliminates water-soluble waste products. The average adult generates between 1.5–2.0 liters of urine per day, which over the course of their lifetime would be enough to fill a small backyard swimming pool (5 X 8 X 1.5 m).
While largely viewed as a waste product, urine has considerable value as a diagnostic biofluid. Indeed the analysis of urine for medical purposes dates back to ancient Egypt [21], [22], [23], [24]. Hippocrates largely legitimized the medical practice of uroscopy (the study of urine for medical diagnostics) where examination of the color, cloudiness, smell and even the taste of urine was used to identify a variety of diseases. Throughout the Byzantine era and well into the Middle Ages, urine color wheels (a diagram that linked the color of urine to a particular disease) were commonly used by physicians to make diagnoses [21], [25]. A brownish color would indicate jaundice, a red hue (blood) might indicate urinary tract tumors, absence of color would be indicative of diabetes and foamy urine would indicate proteinuria. With the advent of modern clinical techniques in the middle of the 19th century, uroscopy largely disappeared. However, urine has continued to be an important cornerstone to modern medical practice. In fact, it was the first biofluid to be used to clinically diagnose a human genetic disease - alkaptonuria [26]. Even today urine analysis is routinely performed with dipstick tests that can readily measure urinary glucose, bilirubin, ketone bodies, nitrates, leukocyte esterase, specific gravity, hemoglobin, urobilinogen and protein. More detailed urinalysis can be also used to study a variety of renal conditions, such as bladder, ovarian and kidney diseases [27], [28], [29], [30].
Being an important and easily accessible biological fluid, urine has been the subject of detailed chemical analysis for more than 100 years [31], [32]. Extensive tables of normal reference ranges have been published for more than 100 urine ions and metabolites [33], [34], [35], [36], [37]. In addition to these referential clinical chemistry studies, several groups have applied various “global” metabolomic or metabolite profiling methods to urine, such as high resolution nuclear magnetic resolution (NMR) spectroscopy [38], [39], [40], high performance liquid chromatography (HPLC) [41], high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) [42], ultra high performance liquid chromatography-high resolution orbitrap mass spectrometry (UHPLC-HRMS) [43], ultra high performance liquid chromatography (UPLC-RP) [31], [44], fast atom bombardment ionization coupled with mass spectroscopy (FAB-MS) [35], two-dimensional gas chromatography coupled to quadrupole mass spectrometry (GCxGC-MS) [32] or time-of-flight mass spectrometry (GCxGC-TOF-MS) [37], nanoflow liquid chromatography-electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS) [33] and liquid chromatography coupled with electrospray quadruple time-of-flight mass spectrometry (LC/QTOF-MS) [36]. A combination of two or more profiling methods has also been used by many research groups to help improve metabolite coverage [34], [40].
A comprehensive list of methods used to analyze urine and the numbers of metabolites identified and/or quantified by these methods (along with references) is provided in Table 1. As seen from this table, it is possible to (tentatively) identify up to 294 different metabolites in human urine. However, quantification is somewhat more difficult, with the largest number of quantified metabolites ever reported in human urine being slightly less than 100. In addition to these global metabolomic studies, hundreds of other “targeted” or single-metabolite studies have been conducted on human urine that have led to the identification and quantification of hundreds of other urine metabolites. Unfortunately, this information is not located in any central repository. Instead it is highly dispersed across numerous textbooks and periodicals [16].
Table 1. Historic data on metabolite characterization of human urine by different platforms.
| Method(s) | # Identified | # Quantified | Year | Reference |
| Multiple | 51 | 51 | 1971 | Putnam [105] |
| GC-MS | 52 | 0 | 1976 | Lawson [85] |
| GC-MS | 72 | 72 | 1978 | Gates [106] |
| GC-MS | 36 | 36 | 1989 | Hoffman [107] |
| GC-MS | 137 | 0 | 1990 | Liebich [108] |
| GC-MS | 131 | 95 | 1991 | Shoemaker [84] |
| GC-MS | 69 | 69 | 1994 | Guneral [56] |
| GC-MS | 75 | 75 | 1996 | Matsumoto [109] |
| ICP-MS | 33 | 33 | 2005 | Goulle [110] |
| SPE/NMR | 73 | 0 | 2008 | Yang [40] |
| SPE/LC/NMR | 72 | 0 | 2008 | Rezzi [78] |
| LC-MS/NMR | 50 | 0 | 2008 | Law [34] |
| LC-MS | 93 | 93 | 2009 | Guo [111] |
| NMR | 50 | 50 | 2009 | Shaykhutdinov [112] |
| GC-MS-MS | 48 | 48 | 2011 | Kvitvang [113] |
| SPE-GC-MS | 79 | 0 | 2011 | Silva [114] |
| LC-MS | 167 | 0 | 2011 | Guo [115] |
| NMR | 42 | 42 | 2011 | Saude [116] |
| NMR/GC-MS | 77 | 0 | 2012 | Zhang [117] |
| LC-MS | 219 (25 listed) | 0 | 2012 | Van der Kloet [118] |
| GC-TOF/LC-MS | 261 | 0 | 2012 | Cheng [119] |
| GC-MS | 258 | 0 | 2012 | Roux [83] |
| GCxGC-TOF-MS | 294 | 0 | 2012 | Rocha [120] |
To facilitate future research into urine chemistry and urine metabolomics, we believe it is critical to establish a comprehensive, electronically accessible database of the detectable metabolites in human urine. This document describes just such a database, one that contains the metabolites that can, with today’s technology, be detected in human urine along with their respective concentrations and disease associations. This resource was assembled using a combination of both our own experimental and literature-based research. Experimentally, we used high-resolution NMR spectroscopy, gas chromatography mass spectrometry (GC-MS), direct flow injection tandem mass spectrometry (DFI/LC-MS/MS), inductively coupled plasma mass spectrometry (ICP-MS) and high performance liquid chromatography (HPLC) with ultraviolet (UV) or fluorescence detection (FD) techniques performed on multiple human urine samples to identify 445 metabolites or metabolite species and quantify 378 of these compounds. To complement these “global” metabolic profiling efforts, our team also surveyed and extracted metabolite and disease-association data from more than 3000 books and journal articles that had been identified through computer-aided literature and in-house developed text-mining software. This “bibliomic” effort yielded data for another 2206 metabolites. The resulting Urine Metabolome Database (UMDB - http://www.urinemetabolome.ca) is a comprehensive, web-accessible resource containing a total of 2651 confirmed urine metabolites or metabolite species (corresponding to 3079 defined structures), their corresponding concentrations and their disease associations that were revealed or identified from these combined experimental and literature mining efforts.
Results and Discussion
The Content of the Human Urine Metabolome – The Urine Metabolome Database
The Urine Metabolome Database (UMDB: http://www.urinemetabolome.ca) contains a complete list of all (to the best of our knowledge) possible metabolites that have been detected in human urine using current technologies. The UMDB is freely available, easily queried, web-enabled database which provides a list of the metabolite names, level of verification (confirmed or probable), normal and disease-associated concentration ranges, associated diseases and corresponding literature references for all human urine metabolites that have ever been detected and/or quantified in the literature. The UMDB also contains concentration data compiled from the experimental studies described here. Each urine metabolite entry in this database is linked to a MetaboCard button [15], [16] that, when clicked, brings up detailed information about that particular entry. This detailed information includes nomenclature, chemical, clinical and molecular/biochemical data. Each MetaboCard entry contains up to 120 data fields many of which are hyperlinked to other databases (KEGG [45], PubChem [46], MetaCyc [47], ChEBI [48], PDB [49], UniProt [50], and GenBank [51] as well as to GeneCard IDs [52], GeneAtlas IDs [53] and HGNC IDs [54] for each of the corresponding enzymes or proteins known to act on that metabolite). Additionally, the UMDB through its MetaboCard/HMDB links includes nearly 450 hand-drawn, zoomable and fully hyperlinked human metabolic pathway maps (SMPDB: http://www.smpdb.ca/). These maps are intended to help users visualize the chemical structures on metabolic pathways and to get detailed information about metabolic processes [55]. These UMDB pathway maps are quite specific to human metabolism and explicitly show the subcellular compartments where specific reactions are known to take place.
The UMDB’s simple text query (TextQuery) supports general text queries including names, synonyms, conditions and disorders. Clicking on the Browse button (on the UMDB navigation panel) generates a tabular view that allows users to casually scroll through the database or re-sort its contents by compound name or by concentration. Users can choose either the “Metabolite View”, “Concentration View” or “Diseases View” to facilitate their browsing or searching. Clicking on a given MetaboCard button brings up the full data content (from the HMDB) for the corresponding metabolite. Users may also search the database using a variety of options listed uner the “Search” menu. For instance, the ChemQuery button allows users to draw or write (using a SMILES string) a chemical compound to search the UMDB for chemicals similar or identical to the query compound. ChemQuery also supports chemical formula and molecular weight searches. The Sequence Search button allows users to conduct BLAST sequence searches of the 4075 protein sequences contained in the UMDB. Both single and multiple sequence BLAST queries are supported. “Advanced Search” which is also located under the “Search” menu is the most sophisticated search tool in the UMDB and opens an easy-to-use query search tool that allows users to select or search over various combinations of subfields. The UMDB’s “MS Search” allows users to submit mass spectral peak lists that will be searched against the Human Metabolome Database (HMDB)’s library of MS/MS spectra. This potentially allows facile identification of urine metabolites from mixtures via MS/MS spectroscopy. UMDB’s NMR Search allows users to submit peak lists from 1H or 13C NMR spectra (both pure and mixtures) and have these peak lists compared to the NMR libraries contained in the HMDB. This allows the identification of metabolites from mixtures via NMR spectral data. The Download button provides links to collected sequence, image and text files associated with the UMDB. In the About menu, the “Data Fields Explained” button lists source data used to assemble the UMDB.
Currently the UMDB contains information on 2651 detectable metabolites or metabolite species (which corresponds to 3079 metabolites with precisely defined structures) and 3832 concentration ranges or values associated with 220 different conditions, diseases and disorders. The number of metabolites in the UMDB is not a number that will remain unchanged. Rather it reflects the total number of metabolites – most of which are endogenous - that have ever been detected and/or quantified by ourselves and others. Certainly as technology improves, we anticipate this number will increase as other, lower abundance, metabolites are detected and added to future versions of the UMDB. Likewise, if the list was expanded to include intermittent, exogenous compounds such as all possible drugs or drug metabolites or rare food additives and food-derived phytochemicals, the database could be substantially larger.
Inspection of the on-line tables in UMDB generally shows that human urine contains a substantial number of hydrophilic molecules. This is further reiterated in Table 2, which provides a listing of the chemical “superclasses” (using the HMDB definitions) in human urine and the number of representative compounds that can be found in this biofluid. Excluding lipids (which are in very low concentration), human urine is dominated by amino acids and derivatives, carbohydrates and carbohydrate conjugates. This simply reinforces the fact that urine is a key carrier of hydrophilic waste products. Other small molecule components found in high abundance in urine include hydroxy acids and derivatives (such as citric acid), urea, ammonia, creatinine and hippuric acid. A more detailed description of both our literature and experimental findings is given in the following 7 sections covering: 1) Literature Review/Text Mining; 2) NMR; 3) DFI/LC-MS/MS; 4) GC-MS; 5) ICP-MS; 6) HPLC/UV and 7) HPLC/FD.
Table 2. Chemical superclasses in the urine metabolome database.
| Chemical superclass | # Compounds |
| Aliphatic Acyclic Compounds | 93 |
| Aliphatic Heteromonocyclic Compounds | 43 |
| Aliphatic Heteropolycyclic Compounds | 40 |
| Aliphatic Homomonocyclic Compounds | 18 |
| Aliphatic Homopolycyclic Compounds | 5 |
| Alkaloids and Derivatives | 45 |
| Amino Acids, Peptides, and Analogues | 286 |
| Aromatic Heteromonocyclic Compounds | 67 |
| Aromatic Heteropolycyclic Compounds | 728 |
| Aromatic Homomonocyclic Compounds | 432 |
| Aromatic Homopolycyclic Compounds | 6 |
| Carbohydrates and Carbohydrate Conjugates | 116 |
| Homogeneous Metal Compounds | 45 |
| Homogeneous Non-metal Compounds | 15 |
| Inorganic compounds | 1 |
| Lignans and Norlignans | 12 |
| Lipids | 866 |
| Mixed Metal/Non-metal Compounds | 7 |
| Nucleosides, Nucleotides, and Analogues | 49 |
| Organic Acids and Derivatives | 108 |
| Organic Halides | 3 |
| Organometallic Compounds | 1 |
| Organophosphorus Compounds | 17 |
| Polyketides | 74 |
| Tannins | 2 |
Metabolite Concentration in Urine – Literature Survey
In addition to the experimentally derived values obtained for this study, the urine metabolome database (UMDB) also presents literature-derived concentrations of urine metabolites with references to either PubMed IDs or clinical textbooks. In many cases, multiple concentration values are given for “normal” conditions. This is done to provide users/readers with a better estimate of the potential concentration variations that different technologies or laboratories may measure. As a general rule, there is good agreement between most laboratories and methods. However, the literature results presented in the UMDB do not reflect the true state of the raw literature. A number of literature-derived concentration values were eliminated through the curation process after being deemed mistaken, disproven (by subsequent published studies), mis-typed or physiologically impossible. Much of the curation process involved having multiple curators carefully reading and re-reading the primary literature to check for unit type, unit conversion and typographical inconsistencies.
Other than the inorganic ions and gases such as sodium (14.7 ± 9.0 mM/mM creatinine – average value), chlorine (8.8 ± 6.2 mM/mM creatinine), potassium (4.6 ± 0.1 mM/mM creatinine), ammonia (2.8 ± 0.9 mM/mM creatinine), the 4 most abundant organic metabolites found in urine (based on average values) are urea (22.5 ± 4.4 mM/mM creatinine), creatinine (10.4 ± 2.0 mM), hippuric acid (298.5 ± 276.8 µM/mM creatinine) and citric acid (280.6 ± 115.2 µM/mM creatinine). The least abundant (detectable) metabolites in urine include oxytocin (0.9 ± 0.1 pM/mM creatinine), angiotensin II (1.2 ± 0.2 pM/mM creatinine), 15-deoxy-d-12,14-PGJ2 (2.3 ± 1.0 pM/mM creatinine) and melatonin (3.3 ± 2.7 pM/mM creatinine). This shows that the current lower limit of detection for metabolites in urine is in the low pM/mM creatinine range and that the concentration range of analytes in urine spans nearly 11 orders of magnitude.
One point that is particularly interesting is the fact that the concentration (scaled to creatinine) of the average metabolite in normal urine varies by about ± 50%, with some metabolites varying by as much as ± 350% (such as normetanephrine (0.00085 ± 0.00317 µM/mM creatinine), pipecolic acid (0.03 ± 0.07 µM/mM creatinine), enterodiol (0.032 ± 0.072 µM/mM creatinine), tungsten (0.010 ± 0.022 µM/mM creatinine) and chlorogenic acid (0.0014 ± 0.0029 µM/mM creatinine). Therefore, drawing conclusions about potential disease biomarkers without properly taking into account this variation would be ill-advised. We believe that these relatively large metabolite concentration ranges are due to a number of factors, including age, gender, genetic background, diurnal variation, health status, activity level and diet [56], [57], [58], [59]. Indeed, some UMDB entries explicitly show such variations based on the populations (age, gender) from which these metabolite concentrations were derived. Clearly more study on the contributions to the observed variations in urine is warranted, although with thousands of metabolites to measure for dozens of conditions, these studies will obviously require significant technical and human resources.
Experimental Quantification and Identification of Urine Metabolites – NMR
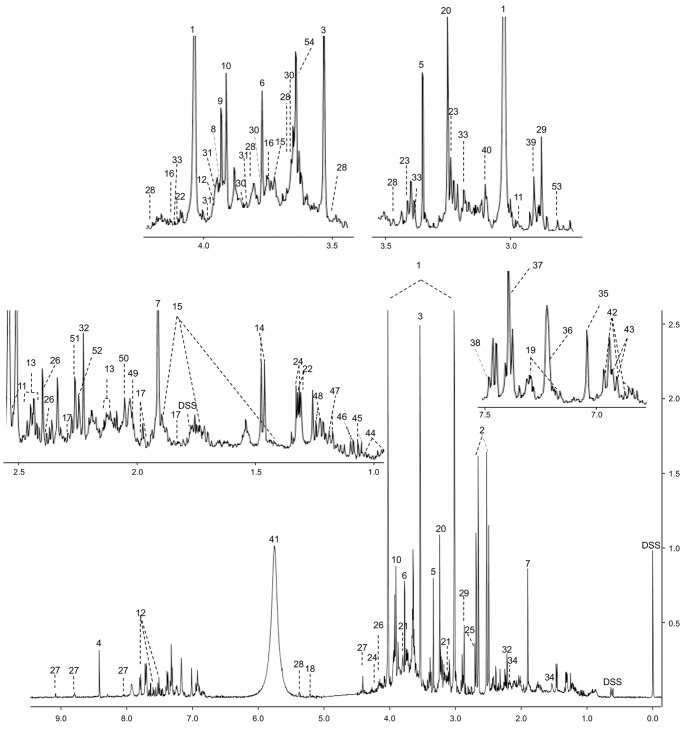
A representative high-resolution NMR spectrum of urine from a healthy individual is shown in Figure 1. As can be readily seen from this figure, urine NMR spectra are very information-rich and surprisingly complex, with thousands of resolved peaks. From the 22 healthy control urine samples analyzed, we could identify a total of 209 unique compounds with an average of 167 ± 19 compounds being identified per sample. Every compound was unequivocally identified and quantified using spectral fitting (via Chenomx) and/or spike-in experiments with authentic standards. The concentration of each metabolite was normalized to each urine sample’s corresponding creatinine value to compensate for variations in urine volume (the concentration of metabolites is expressed as µM/mM creatinine).
Figure 1. Typical 500 MHz 1H-NMR spectra of urine from human urine.
Numbers indicates the following metabolites: 1: creatinine; 2: citric acid; 3: glycine; 4: formic acid; 5: methanol; 6: guanidoacetic acid; 7: acetic acid; 8: L-cysteine; 9: glycolic acid; 10: creatine; 11: isocitric acid; 12: hippuric acid; 13: L-glutamine; 14: L-alanine; 15: L-lysine; 16: gluconic acid; 17∶2- hydroxyglutaric acid; 18: D-glucose; 19: indoxyl sulfate; 20: trimethyl-N-oxide; 21: ethanolamine; 22: L-lactic acid; 23: taurine; 24: L-threonine; 25: dimethylamine; 26: pyroglutamic acid; 27: trigonelline; 28: sucrose; 29: trimethylamine; 30: mannitol; 31: L-serine; 32: acetone; 33: L-cystine; 34: adipic acid; 35: L-histidine; 36: L-tyrosine; 37: imidazole; 38: mandelic acid; 39: dimethylglycine; 40: Cis-aconitic acid; 41: urea; 42∶3-(3-hydroxyphenyl)-3-hydroxypropanoic acid (HPHPA); 43: phenol; 45: isobutyric acid; 46: methylsuccinic acid; 47∶3-aminoisobutyric acid; 48: L-fucose; 49: N-acetylaspartic acid; 50: N-acetylneuraminic acid; 51: acetoacetic acid; 52: Alpha-aminoadipic acid; 53: methylguanidine; 54: phenylacetylglutamine.
The 209 compounds identified and quantified from these NMR studies represent a “high-water” mark for NMR-based metabolomics. Previous studies have reported up to 70 compounds being identified and/or quantified in human urine [40]. Indeed, compared to other platforms previously used to analyze human urine (Table 1), it appears that NMR may currently be the most comprehensive and certainly the most quantitative approach to characterizing this biofluid. Based on the fitted area under each urinary NMR spectrum and the number of unidentified peaks we estimate that more than 96% of the spectral area and more than 92% of all NMR-detectable compounds in our human urine samples are listed in Table 3. In other words, for NMR-based metabolomics, human urine is essentially “solved”. The same “solved” status has already been achieved for human serum (with 49 definitive compounds, [20]) and for human cerebrospinal fluid (with 53 definitive compounds, [17]). Knowing the expected or detectable composition of these biofluids should open the door to automated NMR-based metabolomics [60].
Table 3. Concentrations [Mean (range)] and % occurrence of urine metabolites as determined by NMR.
| Compounds | HMDB ID | Concentration (µM/mM creatinine) | Oc (%) | Literature Values (µM/mM creatinine) |
| 1,3-Diaminopropane | HMDB00002 | 1.2 | 5 | 2.55 (0.18–5.85) |
| 1,3-Dimethyluric acid | HMDB01857 | 3.1 (1.3–6.8) | 68 | 1.5 (1.3–2.3) |
| 1-Methyladenosine | HMDB03331 | 1.7 (0.9–3.6) | 82 | 2.03±0.20 |
| 1-Methylhistidine | HMDB00001 | 8.3 (2.4–28.4) | 100 | 15.9±19.5 |
| 1-Methylnicotinamide | HMDB00699 | 5.8 (1.2–15.0) | 77 | 6.1 (0.2–12.0) |
| 2-Furoylglycine | HMDB00439 | 4.0 (0.9–8.4) | 100 | 9.95 (2.0–18.66) |
| 2-Hydroxy-3-methylpentanoic acid | HMDB00317 | 3.9 (1.7–7.9) | 100 | NA |
| 2-Hydroxybutyric acid | HMDB00008 | 2.8 (1.2–6.9) | 77 | 2.4 (0.0–4.9) |
| 2-Hydroxyglutaric acid | HMDB00694 | 33.0 (13.3–77.9) | 100 | 33.0 (20.0–46.0) |
| 2-Ketobutyric acid | HMDB00005 | 2.9 (0.7–5.4) | 100 | 2.6 (1.1–4.1) |
| 2-Methyl-3-hydroxybutyric acid | HMDB00354 | 4.2 (1.6–6.7) | 95 | 5.5 (0.0–11.0) |
| 2-Methyl-3-ketovaleric acid | HMDB00408 | 1.1 (0.8–1.3) | 18 | NA |
| 2-Methylbutyrylglycine | HMDB00339 | 2.0 (1.2–3.3) | 32 | 1 (0–2) |
| 2-Methylerythritol | HMDB11659 | 25.3 (17.0–49.0) | 14 | NA |
| 2-Methylglutaric acid | HMDB00422 | 0.8 (0.3–1.7) | 45 | NA** |
| 3-Aminoisobutanoic acid | HMDB03911 | 26.0 (2.2–140.0) | 100 | 2.91–116.43 |
| 3-Hydroxybutyric acid | HMDB00357 | 3.6 (1.3–6.4) | 77 | 1.4±1.3 |
| 3-Hydroxyhippuric acid | HMDB06116 | 7.4 (5.7–9.1) | 32 | 2.5 (0.2–5.0) |
| 3-Hydroxyisovaleric acid | HMDB00754 | 6.8 (3.2–21.8) | 100 | 8.5±3.2 |
| 3-Hydroxymethylglutaric acid | HMDB00355 | 2.8 (1.1–5.8) | 68 | 3 (0–10) |
| 3-Hydroxyphenylacetic acid | HMDB00440 | 7.7 (1.4–22.1) | 91 | 6.1±4.1 |
| 3-Methyl-2-oxovaleric acid | HMDB00491 | 2.2 (0.4–4.8) | 91 | 2.0±2.0 |
| 3-Methyladipic acid | HMDB00555 | 2.9 (1.1–7.2) | 86 | 2.9 (0.7–10.5) |
| 3-Methylglutaconic acid | HMDB00522 | 7.5 (3.4–9.5) | 22 | 4.5 (0.0–9.0) |
| 3-Methylhistidine | HMDB00479 | 16.5 (2.8–59.8) | 100 | 15.1 (3.9–26.3) |
| 3-Methylxanthine | HMDB01886 | 6.2 (1.4–17.8) | 100 | 4.0 (2.3–6.3) |
| 4-Aminobutyric acid | HMDB00112 | 2.9 (2.7–3.3) | 9 | 0.75 (0.0–1.50) |
| 4-Aminohippuric acid | HMDB01867 | 3.3 (1.0–7.2) | 86 | 0.3 |
| 4-Deoxythreonic acid | HMDB02453 | 12.3 (4.4–19.8) | 100 | 25±10 |
| 4-Ethylphenol | HMDB29306 | 0.9 (0.6–1.2) | 18 | 0.409±0.174 |
| 4-Hydroxybenzoic acid | HMDB00500 | 1.7 (0.9–3.0) | 77 | 1.8±1.8 |
| 4-Hydroxybutyric acid | HMDB00710 | 7.2 (2.7–22.3) | 100 | 3.3 (0.0–10.8) |
| 4-Hydroxyhippuric acid | HMDB13678 | 2.8 (1.4–6.0) | 100 | 0–14 |
| 4-Hydroxyphenyllactic acid | HMDB00755 | 3.2 (1.5–3.9) | 50 | 1.1 (0.2–2.6) |
| 4-Pyridoxic acid | HMDB00017 | 3.1 (0.4–7.5) | 95 | 0.78±0.45a |
| 5,6-Dihydrouracil | HMDB00076 | 3.8 (2.1–8.3) | 68 | 3.0±2.7 |
| 5-Aminolevulinic acid | HMDB01149 | 2.9 (1.2–4.4) | 95 | 1.45±0.72 |
| 5-Aminopentanoic acid | HMDB03355 | 1.1 | 5 | 2 |
| 7-Methylxanthine | HMDB01991 | 10.0 (3.9–16.1) | 9 | 10.0 (6.7–14.6) |
| Acetaldehyde | HMDB00990 | 2.6 (0.8–4.2) | 32 | NA** |
| Acetaminophen | HMDB01859 | 11.1 | 5 | 5.3 |
| Acetaminophen glucuronide | HMDB10316 | 33.4 | 5 | NA** |
| Acetaminophen sulfate | HMDB59911 | 138.6 | 5 | NA** |
| Acetic acid | HMDB00042 | 13.0 (2.5–106.0) | 100 | 45.3±53.3 |
| Acetoacetic acid | HMDB00060 | 11.1 (2.2–24.9) | 100 | 33 (0–67) |
| Acetone | HMDB01659 | 3.9 (0.8–17.6) | 100 | 2.24±4.13 |
| Acetyl-L-carnitine | HMDB00201 | 2.8 (0.6–7.5) | 100 | 1.9±0.3 |
| Adenine | HMDB00034 | 2.9 (1.4–5.1) | 45 | 1.17 |
| Adenosine | HMDB00050 | 1.4 (0.9–2.3) | 45 | 0.50 (0.09–0.92) |
| Adipic acid | HMDB00448 | 4.4 (1.6–14.3) | 91 | 5.1 (0.8–35.0) |
| ADP | HMDB01341 | 1.8 (1.0–3.4) | 77 | NA** |
| Allantoin | HMDB00462 | 15.4 (4.9–29.3) | 100 | 19.7±10.5 |
| Alpha-Aminoadipic acid | HMDB00510 | 7.3 (3.4–11.2) | 73 | 4.6 |
| Alpha-Aminobutyric acid | HMDB00452 | 2.2 (1.1–4.4) | 95 | 1.37±1.05 |
| Alpha-Hydroxyisobutyric acid | HMDB00729 | 3.8 (1.7–5.9) | 100 | 3.8 |
| Alpha-Ketoisovaleric acid | HMDB00019 | 0.5 (0.2–1.1) | 50 | 1 (0–2) |
| Anserine | HMDB00194 | 7.0 (3.3–20.0) | 100 | 16.16 |
| Arabinitol | HMDB01851 | 31.8 (10.2–64.3) | 100 | 19.28±6.32 |
| Ascorbic acid | HMDB00044 | 32.5 (4.6–78.0) | 14 | 1.70–22.72 |
| Asymmetric dimethylarginine | HMDB01539 | 3.0 (0.2–6.7) | 100 | 4.6 (3.0–7.0) |
| Azelaic acid | HMDB00784 | 2.8 (1.8–4.8) | 64 | 4.8 (1.3–15.0) |
| Benzoic acid | HMDB01870 | 4.7 (1.2–10.5) | 86 | 4.2 (1.9–6.5) |
| Beta-Alanine | HMDB00056 | 5.9 (3.4–13.0) | 68 | 5 (0–10) |
| Betaine | HMDB00043 | 11.5 (2.7–24.7) | 100 | 4.14 |
| Butyric acid | HMDB00039 | 1.6 (0.5–4.0) | 77 | 0.131±0.013 |
| Caffeine | HMDB01847 | 1.2 | 5 | 0.33 (0.0–1.01) |
| Carnosine | HMDB00033 | 5.5 (1.4–12.6) | 91 | 4.0±2.5 |
| Choline | HMDB00097 | 3.5 (1.4–6.1) | 100 | 5.27 |
| Cinnamic acid | HMDB00567 | 1.6 (0.9–2.2) | 9 | 0.77 |
| Cis-Aconitic acid | HMDB00072 | 20.9 (3.8–95.3) | 100 | 13.0 (2.7–44.0) |
| Citraconic acid | HMDB00634 | 2.0 (1.2–2.6) | 73 | 2.2 |
| Citramalic acid | HMDB00426 | 2.4 (1.0–4.8) | 91 | 3.1 |
| Citric acid | HMDB00094 | 203 (49–600) | 100 | 242.0±129.6 |
| Creatine | HMDB00064 | 46 (3–448) | 100 | 46 (9–135) |
| Creatinine* | HMDB00562 | 14,743±9,797 | 100 | 10,090±310 |
| Cytosine | HMDB00630 | 4.1 (1.1–10.7) | 36 | NA** |
| Dehydroascorbic acid | HMDB01264 | 9.1 (5.8–7.1) | 32 | 1.72–7.47 |
| D-Galactose | HMDB00143 | 11.9 (5.4–25.2) | 100 | 4.4 (0.0–31.0) |
| D-Glucose | HMDB00122 | 37.5 (12.5–58.4) | 100 | 31.12 |
| Dihydrothymine | HMDB00079 | 2.4 (1.3–4.4) | 64 | 5 (0–10) |
| Dimethyl sulfone | HMDB04983 | 8.0 (1.3–49.0) | 100 | 4.6 (2.5–7.5) |
| Dimethylamine | HMDB00087 | 30.8 (20.3–59.2) | 100 | 39.3±8.3 |
| Dimethylglycine | HMDB00092 | 4.4 (1.6–10.4) | 100 | 6.2 (0.8–11.2) |
| D-Threitol | HMDB04136 | 19.3 (5.3–32.7) | 100 | 10.0 (5.6–15.0) |
| D-Xylitol | HMDB02917 | 8.4 (6.0–13.8) | 95 | 3.50±2.37 |
| D-Xylose | HMDB00098 | 20 (3.0–102) | 91 | 51.19 |
| Erythritol | HMDB02994 | 33.4 (6.8–64.0) | 100 | 51±18 |
| Ethanol | HMDB00108 | 3.1 | 5 | 5–500b |
| Ethanolamine | HMDB00149 | 37.0 (24.8–56.2) | 100 | 21.4 (6.6–36.2) |
| Ethyl glucuronide | HMDB10325 | 8.2 | 5 | 26.38±15.26 |
| Ethylmalonic acid | HMDB00622 | 3.0 (1.2–5.8) | 95 | 2.0 (0.5–3.9) |
| Formic acid | HMDB00142 | 26.8 (6.9–120.9) | 100 | 20.39±11.84 |
| Fumaric acid | HMDB00134 | 0.7 (0.2–1.7) | 50 | 1 (0–2) |
| Glucaric acid | HMDB00663 | 8.4 (5.8–12.3) | 82 | 2.9±0.2 |
| Gluconic acid | HMDB00625 | 21.5 (8.1–38.8) | 100 | NA** |
| Glucuronic acid | HMDB00127 | 9.7 (3.7–20.6) | 77 | NA |
| Glutaconic acid | HMDB00620 | 3.1 (1.2–3.1) | 68 | 1 (0–2) |
| Glutaric acid | HMDB00661 | 2.1 (0.7–3.6) | 18 | 1.3 (0.6–2.6) |
| Glyceric acid | HMDB00139 | 15.9 (7.8–31.1) | 100 | 4.5 (0.0–9.0) |
| Glycerol | HMDB00131 | 13 (4–19) | 100 | 20 (0–40) |
| Glycine | HMDB00123 | 106 (44–300) | 100 | 151 (233–248) |
| Glycolic acid | HMDB00115 | 42.0 (3.7–122.0) | 100 | 35 (18–55) |
| Guanidoacetic acid | HMDB00128 | 41.8 (10.6–97.3) | 100 | 89.0 (11.0–124.0) |
| Hippuric acid | HMDB00714 | 229 (19–622) | 100 | 257 (20–770) |
| Homocitrulline | HMDB00679 | 7.5 (3.8–13.8) | 91 | 6.0 (0.0–8.8) |
| Homogentisic acid | HMDB00130 | 1.70 (0.55–2.8) | 45 | 1 (0–2) |
| Homovanillic acid | HMDB00118 | 6.2 (1.8–12.7) | 100 | 4.9±3.4 |
| HPHPA | HMDB02643 | 5.9 (1.4–22.1) | 100 | 0.55–54.93 |
| Hydroxyproline | HMDB00725 | 6.5 (2.4–12.5) | 55 | 2 |
| Hydroxypropionic acid | HMDB00700 | 12.3 (4.4–17.9) | 82 | 6.5 (3.0–10.0) |
| Hypoxanthine | HMDB00157 | 7.2 (1.8–24.1) | 100 | 4.67 (2.80–6.38) |
| Indoleacetic acid | HMDB00197 | 3.4 (1.8–6.2) | 100 | 2.0±1.0 |
| Indoxyl sulfate | HMDB00682 | 22.4 (6.0–64.8) | 100 | 19.74±5.26 |
| Inosine | HMDB00195 | 1.4 (0.4–6.5) | 50 | 1.04±0.56 |
| Isobutyric acid | HMDB01873 | 3.5 (1.2–9.0) | 95 | 3.9 |
| Isocitric acid | HMDB00193 | 46.9 (20.4–89.2) | 68 | 58 (36–84) |
| Isovaleric acid | HMDB00718 | 1.4 (0.8–2.5) | 86 | 0.42±0.02 |
| Isovalerylglycine | HMDB00678 | 2.0 (0.4–4.0) | 91 | 5.5 (1.0–10.0) |
| Kynurenic acid | HMDB00715 | 2.1 (0.8–4.2) | 64 | 1.53 (0.66–4.13) |
| Kynurenine | HMDB00684 | 1.7 (1.1–2.5) | 23 | 2.85±1.94 |
| Lactose | HMDB00186 | 11.8 (1.0–24.2) | 91 | 36.31±6.6c |
| L-Alanine | HMDB00161 | 21.8 (7.1–43.0) | 100 | 22.0±11.0 |
| L-Arabinose | HMDB00646 | 13.3 (1.3–22.0) | 100 | 8.75 (6.12–11.84) |
| L-Arginine | HMDB00517 | 8.6 (3.8–18.6) | 95 | 6.6 (2.5–10.8) |
| L-Asparagine | HMDB00168 | 10.0 (4.6–17.8) | 95 | 10.0 (4.6–16.3) |
| L-Aspartic acid | HMDB00191 | 10.9 (3.5–21.8) | 95 | 4.63 (1.71–7.56) |
| L-Carnitine | HMDB00062 | 5.0 (0.7–16.4) | 100 | 5.7±4.8 |
| L-Cystathionine | HMDB00099 | 14.7 (3.9–20.7) | 82 | 5.3 (1.0–9.6) |
| L-Cysteine | HMDB00574 | 65.8 (23.1–134.5) | 100 | 33.4±15.5 |
| L-Cystine | HMDB00192 | 12.5 (5.0–24.6) | 100 | 9.0±3.0 |
| Levoglucosan | HMDB00640 | 10.0 (2.4–29.0) | 27 | NA** |
| Levulinic acid | HMDB00720 | 1.3 (0.4–2.0) | 64 | 1.28 (0.33–2.75) |
| L-Fucose | HMDB00174 | 11.8 (6.7–26.0) | 91 | 9.6 (6.4–16.0) |
| L-Glutamic acid | HMDB00148 | 8.5 (3.3–18.4) | 100 | 10.0±3.7 |
| L-Glutamine | HMDB00641 | 37.2 (19.1–77.9) | 100 | 33.32±10.78 |
| L-Histidine | HMDB00177 | 43 (17–90) | 100 | 60.75±27.45 |
| L-Isoleucine | HMDB00172 | 1.3 (0.4–2.6) | 100 | 1.64 |
| L-Lactic acid | HMDB00190 | 11.6 (3.5–29.3) | 100 | 12.3±6.2 |
| L-Leucine | HMDB00687 | 2.8 (1.6–5.4) | 100 | 2.5±1.3 |
| L-Lysine | HMDB00182 | 17.2 (3.7–51.3) | 100 | 18 (7–29) |
| L-Methionine | HMDB00696 | 1.4 (0.5–2.5) | 41 | 1.3±0.97 |
| L-Phenylalanine | HMDB00159 | 6.4 (3.5–11.2) | 100 | 4.50±1.15 |
| L-Serine | HMDB00187 | 25.3 (11.6–53.4) | 100 | 26.0±10.0 |
| L-Threonine | HMDB00167 | 13.3 (6.4–25.2) | 100 | 5.17–24.59 |
| L-Tryptophan | HMDB00929 | 6.3 (3.4–11.1) | 100 | 2.45–29.40 |
| L-Tyrosine | HMDB00158 | 9.5 (4.1–23.5) | 100 | 8.8±4.5 |
| L-Valine | HMDB00883 | 3.4 (2.0–7.7) | 100 | 3.0±1.0 |
| Maleic acid | HMDB00176 | 0.4 (0.3–0.5) | 14 | 0.5 |
| Malonic acid | HMDB00691 | 2.9 (2.0–3.5) | 77 | 1 (0–2) |
| Maltose | HMDB00163 | 6.0 (1.3–21.4) | 73 | NA** |
| Mandelic acid | HMDB00703 | 1.4 (1.1–1.7) | 9 | 0.966±0.669 |
| Mannitol | HMDB00765 | 32.4 (5.2–85.1) | 95 | 10.26±9.14 |
| Methanol | HMDB01875 | 37 (10–117) | 100 | NA** |
| Methylamine | HMDB00164 | 4.0 (1.5–11.9) | 100 | 10.5 (9.8–12.5) |
| Methylglutaric acid | HMDB00752 | 3.1 (2.6–3.6) | 9 | 3.5 (0.0–7.0) |
| Methylguanidine | HMDB01522 | 2.7 (1.2–6.0) | 95 | 1.25±0.72 |
| Methylmalonic acid | HMDB00202 | 1.9 (0.7–3.5) | 95 | 1.6 (0.0–3.6) |
| Methylsuccinic acid | HMDB01844 | 3.2 (0.7–9.9) | 100 | 2.3 (0.8–10.8) |
| Monomethyl glutaric acid | HMDB00858 | 2.3 (0.6–4.4) | 68 | NA |
| m-Thymol | HMDB01878 | 2.2 (0.7–4.0) | 86 | NA** |
| Myoinositol | HMDB00211 | 22.4 (7.9–36.1) | 100 | 18.8±2.9 |
| N-Acetylaspartic acid | HMDB00812 | 4.0 (1.3–7.0) | 100 | 4.66±1.14 |
| N-Acetylneuraminic acid | HMDB00230 | 5.4 (2.5–8.6) | 100 | 8.60±1.58 |
| N-Acetylputrescine | HMDB02064 | 0.5 (0.4–0.7) | 32 | 1.18 (0.58–2.25) |
| N-Methylhydantoin | HMDB03646 | 4.9 (1.0–11.0) | 55 | NA |
| o-Hydroxyphenylacetic acid | HMDB00669 | 2.0 (0.9–4.5) | 91 | 7.71±3.65d |
| Oxoglutaric acid | HMDB00208 | 4.8 (2.0–17.0) | 100 | 2.98 (0.42–18.30) |
| Oxypurinol | HMDB00786 | 13.3 (5.1–29.3) | 50 | 3.29 |
| Pantothenic acid | HMDB00210 | 1.9 (0.6–4.4) | 100 | 2.7±0.9 |
| p-Cresol sulfate | HMDB11635 | 1.3 (0.3–5.5) | 91 | NA** |
| Phenol | HMDB00228 | 4.8 (0.6–12.8) | 95 | 7.0 (5.6–9.2) |
| Phenylacetic acid | HMDB00209 | 1.9 | 5 | 4.16±0.91 |
| Phenylacetylglutamine | HMDB06344 | 34.0 (4.5–70.0) | 100 | 47.03 (3.84–85.51) |
| Phenylglyoxylic acid | HMDB01587 | 7.34 | 5 | 36.6±0.48e |
| Phosphorylcholine | HMDB01565 | 1.1 (0.7–3.0) | 41 | NA |
| p-Hydroxyphenylacetic acid | HMDB00020 | 5.5 (1.4–14.6) | 100 | 9.68 |
| Picolinic acid | HMDB02243 | 19.4 (7.2–37.6) | 18 | 1.65–10.28 |
| Pimelic acid | HMDB00857 | 2.2 (0.7–4.0) | 77 | NA** |
| Proline-betaine | HMDB04827 | 2.3 (0.8–4.0) | 100 | 12.7 (1.6–23.7) |
| Propylene glycol | HMDB01881 | 6.7 (1.4–44.3) | 100 | NA** |
| Pseudouridine | HMDB00767 | 28.9 (13.3–41.3) | 100 | 26.02±4.62 |
| Pyrocatechol | HMDB00957 | 4.7 (2.0–8.5) | 100 | 6.0±4.3 |
| Pyroglutamic acid | HMDB00267 | 20.7 (10.2–32.6) | 100 | 28.8 (3.4–54.2) |
| Pyruvic acid | HMDB00243 | 2.7 (0.8–6.4) | 100 | 2.13 (0.54–8.67) |
| Quinolinic acid | HMDB00232 | 5.2 (1.0–17.5) | 100 | 2.5±1.1 |
| Sarcosine | HMDB00271 | 2.9 (0.5–5.4) | 77 | 2.8 (0.0–5.6) |
| Scyllitol | HMDB06088 | 4.2 (2.2–8.1) | 68 | NA |
| Sebacic acid | HMDB00792 | 1.9 | 5 | 1 (0–2) |
| Sorbitol | HMDB00247 | 9.9 (2.5–18.7) | 100 | 3.50±2.24 |
| Suberic acid | HMDB00893 | 1.6 (0.9–2.4) | 32 | 0.5 (0.0–2.9) |
| Succinic acid | HMDB00254 | 6.0 (0.3–33.3) | 91 | 5.6±3.8 |
| Succinylacetone | HMDB00635 | 2.8 (0.6–4.7) | 36 | 1 (0–2) |
| Sucrose | HMDB00258 | 7.4 (1.4–19.5) | 100 | 7.12±3.23 |
| Sumiki’s acid | HMDB02432 | 1.7 | 5 | NA** |
| Symmetric dimethylarginine | HMDB03334 | 2.7 (1.5–5.2) | 100 | 3.72±0.35 |
| Syringic acid | HMDB02085 | 1.5 | 5 | 0.9±0.2 |
| Tartaric acid | HMDB00956 | 11.8 (2.6–64.4) | 82 | 12.0 (1.3–46.0) |
| Taurine | HMDB00251 | 81 (13–251) | 100 | 4.00–159.98 |
| Threonic acid | HMDB00943 | 20.8 (10.2–39.3) | 100 | 10.0±7.0 |
| Thymidine | HMDB00273 | 2.2 (0.7–3.9) | 36 | 6.87±4.86 |
| Trans-Aconitic acid | HMDB00958 | 6.6 (1.8–25.1) | 55 | 3.38 |
| Trans-Ferulic acid | HMDB00954 | 1.2 | 5 | 36.3±11.2f |
| Trigonelline | HMDB00875 | 31.1 (5.5–109.3) | 100 | 16.08 |
| Trimethylamine | HMDB00906 | 2.5 (0.3–19.4) | 86 | 7.7±7.4 |
| Trimethylamine N-oxide | HMDB00925 | 91.0 (4.8–509.0) | 100 | 118.7 (35.5–202.1) |
| Uracil | HMDB00300 | 9.5 (2.6–22.8) | 100 | 12 (2–22) |
| Urea | HMDB00294 | 12,285 (174–49,097) | 100 | 22,566±4,407 |
| Vanillic acid | HMDB00484 | 1.6 (0.6–3.3) | 95 | 1.0 (0.0–2.5) |
| Vanillylmandelic acid | HMDB00291 | 2.3 (1.0–3.4) | 9 | 1.2±0.5 |
Concentration of metabolite (Mean ± SD) is expressed by µM.
The mean daily vitamin B6 intake was well below the recommended dietary allowance for men and woman;
depends greatly on amount of alcohol consumed;
after administration of lactose;
after administration of 1 g of coumarin;
After styrene exposure; and
after consumption of chocolate.
NA: not available; NA**: not available for healthy adults; ADP: Adenosine-5-diphosphate; HPHPA: 3-(3-hydroxyphenyl)-3-hydroxypropanoic acid; Oc: occurrence.
Based on these NMR studies, the most abundant constituents (based on average values) of urine from a healthy individual are urea (12.3 ± 14.5 mM/mM creatinine), creatinine (14.7 ± 9.8 mM), hippuric acid (229 ± 160 µM/mM creatinine) and citric acid (203 ± 129 µM/mM creatinine). The least abundant compounds were trans-ferulic acid (1.2 µM/mM creatinine), phosphorylcholine (1.1 ± 0.7 µM/mM creatinine), 4-ethylphenol (0.9 ± 0.1 µM/mM creatinine), 2-methylglutaric acid (0.8 ± 0.4 µM/mM creatinine). The lowest concentrations that could be reliably detected using NMR were 0.4 ± 0.1 µM/mM creatinine (for maleic acid), 0.5 ± 0.2 µM/mM creatinine (for alpha-ketoisovaleric acid and N-acetylputrescine) and 0.7 ± 0.4 µM/mM creatinine (for fumaric acid). The complete list of average compound concentrations (including their range), literature-reported concentrations (for healthy adult) and the frequency of their occurrence is shown in Table 3. In general, there is good agreement between the NMR-measured concentrations and those reported in the literature (often obtained by other analytical techniques).
However, not all of the NMR-derived urine concentrations agree with literature-derived values. A total of 34 compounds had average concentrations somewhat higher (>1 SD) than previously reported values (for example, 1, 3-dimethyluric acid, glucaric acid, L-aspartic acid, mannitol), while 21 compounds had average concentrations lower (<1 SD) than previously reported (for example, 1-methylhistidine, phenol, dihydrothymine, thymidine). Another 7 metabolites also exhibited somewhat greater range in concentrations than those previously reported in the literature. These included: acetic acid, butyric acid, isovaleric acid, lactose, phenylglyoxylic acid, proline-betaine and trans-ferulic acid. Some of these discrepancies are likely due to differences in diet, physiological status, pharmacological effects and the age of the different cohorts that were analyzed. Other differences may be due to storage effects, sample preparation methods and the analytical methods being used. As a rule, NMR concentration determinations are very accurate since they involve direct measurement of the compound, as opposed to an indirect measurement of a derivative compound. Therefore we are quite confident in the NMR concentration values reported in Table 3 and would tend to view these as more reliable than those measured via other technologies.
A number of the compounds exhibiting higher-than or lower-than-reported concentrations appear to be associated with dietary intake. For example, mannitol is a sugar alcohol that is poorly absorbed by humans but its presence in urine can be explained by its occurrence in commonly consumed foods such as apples, pineapples, asparagus and carrots. Likewise, the urinary excretion of trans-ferulic acid (a polyphenolic derivative) increases after the ingestion of breakfast cereals [61] and chocolate. The relatively low value we measured is likely due to the fact that the literature value of trans-ferulic acid reported in Table 3 was measured for people on a special diet [62]. Also, the low level of proline-betaine we measured in urine may be due to a lower frequency of exposure to dietary citrus fruits in our population sample [63]. Proline-betaine is an osmoprotectant found in citrus fruit and urinary excretion of this metabolite is increased after consumption of fruits such as orange juice [64]. Similarly the higher levels of dimethyl sulfone we detected in urine could be attributed to dietary sources that contain DMSO [65]. For example, onions contain many sulfoxides including DMSO which can be oxidized in the liver and kidneys to produce dimethyl sulfone [66], [67]. The consumption of meat could also significantly increase the concentration of some metabolites in urine as reported in the case of 1-methylhistidine [68]. 1-methylhistidine is produced from the metabolism of anserine (a dipeptide) which is commonly found in meats [69], [70]. In addition to diet, metabolite levels in urine can also be affected by physiological status. For instance, the level of 3-hydroxybutyric acid in urine increases during fasting and can range from 0 to 200 µM/mM creatinine, with the maximum level reported in the literature (200 µM/mM creatinine) corresponding to healthy male after 35 h of fasting [50].
Some of the metabolites we measured by NMR did not have any previously reported literature values. For example, glucuronic acid is usually reported as total glucuronic acid (the free acid plus glucuronide conjugates) after hydrolysis [71], [72]. Here we report the concentration of free glucuronic acid, as indicated in Table 3. Another example, 2-methylerythritol, was previously detected in human urine but no concentration was reported [73]. The urinary excretion of 2-methylerythritol is most likely a result of dietary consumption of fruits or vegetables containing 2-methylerythritol and/or 2-methylerythritol-4-phosphate. 2-methylerythritol-4-phosphate is an intermediate in isoprenoid biosynthesis [74] and has been found to be quite abundant in certain plants [75].
A number of compounds we measured by NMR appear to be normal constituents of human urine but seem not to have been previously reported as being detectable by NMR (a total of 42 compounds) or reported as detected but not-quantified by any other method (a total of 8 compounds). The identification of these “NMR-novel” compounds was aided by their prior identification by GC-MS and DI-MS (see following sections) and through a careful literature analysis of compounds that had previously been detected in human urine via other methods. The list of detected but not previously quantified by NMR compounds includes: 2-hydroxy-3-methylpentanoic acid, 2-methyl-3-ketovaleric acid, 2-methylerythritol, glucuronic acid, monomethyl glutaric acid, N-methylhydantoin, phosphorylcholine and scyllitol. All of these compounds were confirmed using authentic standards.
This NMR study also revealed a number of common identification errors made in previously published NMR-based human urine metabolomic studies. In particular, several earlier reports identified phenylacetylglycine [76], N-acetylglutamic acid [77], cresol [78], isonicotinic acid [78], yellow 7.1 [79], meta-hydroxyphenylpropionic acid [80], 2-oxoisocaproic acid [81], urocanic acid, glycylproline and ornithine [80] as being detectable by NMR in human urine. Using our NMR instrument and the samples available to us, we were unable to detect any of these compounds, even after performing multiple spike-in experiments using authentic compounds. While some of these metabolites have been previously reported to be in human urine, they were reported at concentrations far below the lower limit of detection of modern NMR instruments (which is ∼ 1 µM). Due to their chemical shift similarity, phenylacetylglycine (which is found only in rats and mice) and N-acetylglutamic acid appear to be commonly mistaken for phenylacetylglutamine. We also noticed that, isonicotinic acid (a breakdown product of isoniazid and hydrazine derivatives, which is found only in individuals that have taken isoniazid and other hydrazine derivatives as a drug) appears to be mistaken for trigonelline. Likewise cresol (water-insoluble) appears to be frequently mistaken for cresol-sulfate (water-soluble), while the compounds yellow 7.1, meta-hydroxyphenylpropionic acid and 3-(p-hydroxyphenyl)-propionic acid appear to be commonly mistaken for 3-(3-hydroxyphenyl)-3-hydroxypropanoic acid (HPHPA).
In addition to correcting these compound identification errors, we also observed some significant gender-related effects on creatinine levels in our urine samples. Since males generally have a greater mass of skeletal muscle than females, they tend to have higher urinary levels of creatinine than women. This was clearly evident in our samples as the average male creatinine level was 20 mM while the average female creatinine level was 11 mM. In addition, increased dietary intake of creatine or a protein-rich diet can increase daily creatinine excretion [82].
Quantification and Identification of Urine Metabolites – GC-MS
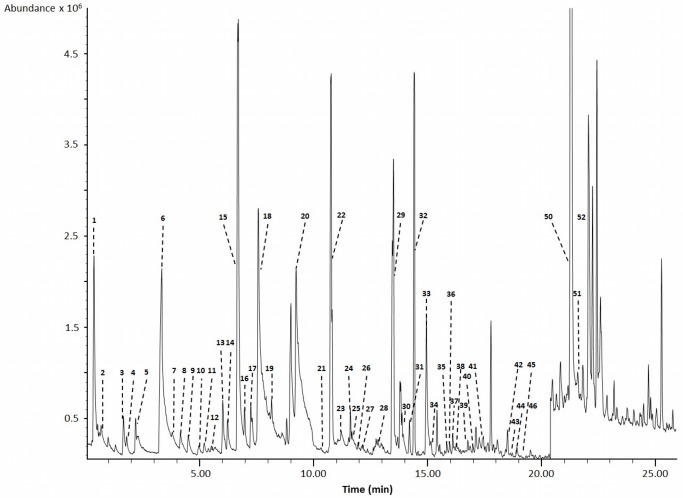
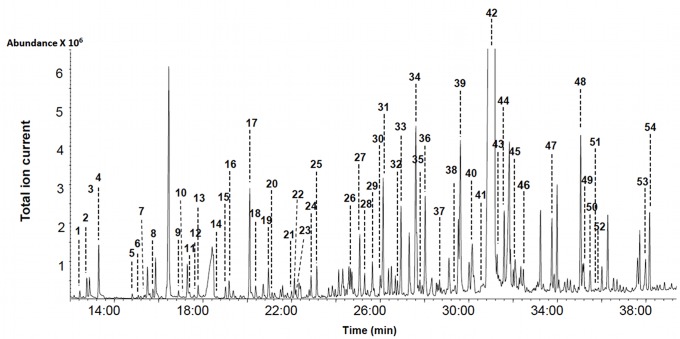
As seen in Table 1, GC-MS methods have long been used to comprehensively characterize the chemical content of human urine. For our studies a total of 4 different GC-MS analyses were performed. The first method employed polar solvent extraction and derivatization to achieve broad metabolite coverage of polar metabolites, the second was more selective and targeted organic acids, the third targeted volatiles, while the fourth targeted bile acids. Representative high-resolution GC-MS total ion chromatograms are shown in Figures 2–4 for each of these analyses (except for the bile acids). Combined, the 4 GC-MS methods allowed us to identify 179 and quantify a total of 85 compounds. Table 4 shows the identified polar, organic acid extracts and bile acids (127 in total), Table 5 shows the identified volatile metabolites (52 in total) while Table 6 shows the 85 fully quantified compounds from all 4 techniques. These numbers actually represent the highest number of urine metabolites both identified and quantified by GC-MS to date. As seen in Table 1, previous GC-MS studies have reported up to 258 unique compounds being identified (but none quantified) [83] and approximately 95 compounds quantified in human urine [84]. Relative to NMR (see previous section) and other methods used to analyze human urine (Table 1), it appears that a multi-pronged GC-MS analysis is an excellent approach to characterize this biofluid. However, unlike NMR where nearly all detectable peaks are identifiable, metabolite coverage by GC-MS tends to be relatively incomplete. As seen in Figure 2, only 60% of the peaks could be identified using as reference the 2008 NIST library and other home-made GC-MS metabolite libraries. Likewise, in Figure 3, we see that only 65% of the organic acid peaks could be identified while in Figure 4, just 60% of the volatile compound peaks could be identified.
Figure 2. Typical total ion chromatogram of water-soluble metabolites extracted from human urine.
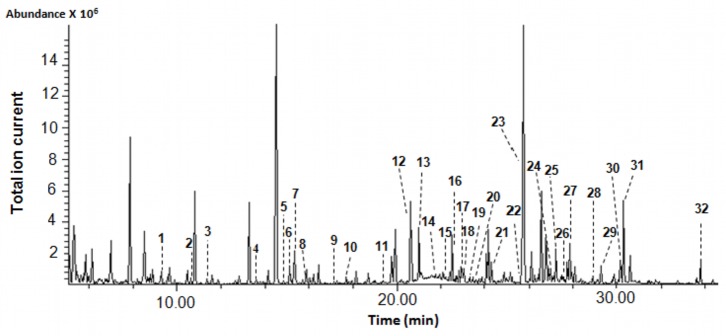
Numbers indicate the following metabolites (quantified compounds): 1: L-valine; 2: oxalic acid; 3∶3-hydroxyisobutyric acid; 4: L-serine; 5: glycine; 6: succinic acid; 7: glyceric acid; 8∶4-deoxythreonic acid; 9∶2,4-dihydroxybutanoic acid; 10∶3,4-dihydroxybutanoic acid; 11: adipic acid; 12: creatinine; 13: threonic acid; 14: L-phenylalanine; 15: p-hydroxyphenylacetic acid; 16: L-ornithine; 17: L-asparagine; 18: L-arabinose; 19: D-xylitol; 20: D-xylulose; 21: Cis/Trans-aconitic acid; 22: hippuric acid; 23: isocitric acid; 24: D-galactose; 25: D-glucose; 26: L- tyrosine; 27: sorbitol; 28: gluconic acid; 29: scyllitol; 30: myoinositol; 31: uric acid; 32: pseudouridine.
Figure 4. Typical gas chromatogram of volatile organic compounds (VOC) from a pooled human urine sample.
Numbers indicate the following metabolites: 1∶3-methyl sulfolane; 2∶3-hexanone; 3∶2-pentanone; 4∶1-hydroxy-2-pentanone; 5: allyl methylsulphide; 6: dimethyl disulfide; 7∶4-heptanone; 8∶1-methylcyclohexanol; 9; 2-hexanone; 10∶3,4-dimethylthiophene; 11: diallyl sulphide; 12∶5-methyl-2-hexanone; 13∶1,3-dithio cyclohexane; 14: dimethyl trisulfide; 15: phenol; 16: o-cymene; 17: p-cymene; 18: m-cymene; 19∶1,4-cineol; 20: p-cresol; 21: linalool oxide; 22: iso-menthol; 23: Alpha-p-dimethylstyrene; 24: L-menthol; 25: undecane; 26: ledene oxide (II); 27: salicylic acid methyl ester; 28: Beta-carvone; 29: piperitone; 30: o-thymol; 31: Beta-cyclocitrol; 32∶4,7-dimethyl-benzofuran; 33: cuminal: 34∶2,6,10,10 Tetramethyl-oxa-spiro-4,5-dec-6-ene; 35∶4-(2,6,6-trimethyl-1-cyclohexen-1-yl)-2-butanone; 36∶1,2,3,4-tetrahydro-1,1,6-trimethyl naphthalene; 37: Alpha-cedrene; 38∶1,2,3,4-tetrahydro-1,5,7-trimethylnapthalene; 39∶1,2-diydro-1,1,6-trimethyl-napthalene; 40: Beta-guaiene; 41: Beta-damascenone; 42∶2,5-cyclohexadiene-1,4-dione-2,6-di-tert-butyl; 43: himachalene; 44∶4-(2,6,6-trimethylcyclohexa-1,3-dienyl)-but-3-en-2-one: 45∶1-(2,6,6-trimethyl-1-cyclohexen-1-yl)-1-penten-3-one; 46∶2,4-Bis (1,1-dimethylethyl)-phenol; 47∶1-(2,3,6-trimethyl phenyl)-3-buten-2-one; 48: L-calamenene; 49: Beta-vatirenene; 50∶1,6,7-trimethylnaphthalene; 51: azulol; 52∶3,3,5,6-tetramethyl-1-indanone.
Table 4. List of metabolites identified in human urine extracts by GC-MS (except volatile compounds).
| Amino acids | HMDB ID | Organic acids | HMDB ID |
| Beta-Alaninea | HMDB00056 | Azelaic acidb | HMDB00784 |
| Glycinea | HMDB00123 | Benzoic acidb | HMDB01870 |
| L-Alaninea | HMDB00161 | Butyric acida | HMDB00039 |
| L-Asparaginea | HMDB00168 | Cis-Aconitic acida,b | HMDB00072 |
| L-Aspartic acida | HMDB00191 | Citraconic acidb | HMDB00634 |
| L-Glutaminea | HMDB00641 | Citramalic acidb | HMDB00426 |
| L-Histidinea | HMDB00177 | Citric acida | HMDB00094 |
| L-Isoleucinea | HMDB00172 | Erythronic acida | HMDB00613 |
| L-Leucinea | HMDB00687 | Ethylmalonic acidb | HMDB00622 |
| L-Lysinea | HMDB00182 | Gluconic acida | HMDB00625 |
| L-Ornithinea | HMDB00214 | Glutaric acidb | HMDB00661 |
| L-Phenylalaninea | HMDB00159 | Glyceric acida | HMDB00139 |
| L-Serinea | HMDB00187 | Glycolic acida,b | HMDB00115 |
| L-Threoninea | HMDB00167 | Glyoxalic acida | HMDB00119 |
| L-Tryptophana | HMDB00929 | Hippuric acida,b | HMDB00714 |
| L-Tyrosinea | HMDB00158 | Homovanillic acidb | HMDB00118 |
| L-Valinea | HMDB00883 | HPHPAb | HMDB02643 |
| Bile acids | HMDB ID | Hydroxypropionic acidb | HMDB00700 |
| Cholenic acidc | HMDB00308 | Indoleacetic acidb | HMDB00197 |
| Deoxycholic acidc | HMDB00626 | Indolelactic acid b | HMDB00671 |
| Lithocholic acidc | HMDB00761 | Isobutyrylglycineb | HMDB00730 |
| Ursodeoxycholic acidc | HMDB00946 | Isocitric acida | HMDB00193 |
| Misc | HMDB ID | Isovalerylglycineb | HMDB00678 |
| 3-Methylhistidinea | HMDB00479 | Kynurenic acidb | HMDB00715 |
| 5-Hydroxyindoleb | HMDB59805 | Levulinic acidb | HMDB00720 |
| Creatininea | HMDB00562 | L-Lactic acidb | HMDB00190 |
| Dimethylglycinea | HMDB00092 | Malic acida | HMDB00744 |
| D-Xylitola | HMDB02917 | Malonic acida,b | HMDB00691 |
| Glycerol 3-phosphatea | HMDB00126 | m-Chlorobenzoic acidb | HMDB01544 |
| Glycerola | HMDB00131 | m-Coumaric acidb | HMDB01713 |
| Mannitola | HMDB00765 | Methylmalonic acidb | HMDB00202 |
| Myoinositola | HMDB00211 | Methylsuccinic acidb | HMDB01844 |
| Pseudouridinea | HMDB00767 | o-Hydroxyhippuric acidb | HMDB00840 |
| Scyllitola | HMDB06088 | o-Hydroxyphenylacetic acidb | HMDB00669 |
| Sorbitola | HMDB00247 | Oleic acidb | HMDB00207 |
| Ureaa | HMDB00294 | Oxalic acida | HMDB02329 |
| Organic acids | HMDB ID | Oxoglutaric acidb | HMDB00208 |
| 2,4-Dihydroxybutanoic acida | HMDB00360 | Palmitic acidb | HMDB00220 |
| 2-Ethylhydracrylic acidb | HMDB00396 | Pantothenic acidb | HMDB00210 |
| 2-Furoylglycineb | HMDB00439 | Phosphoric acidb | HMDB02142 |
| 2-Hydroxy-2-methylbutyric acidb | HMDB01987 | p-Hydroxyphenylacetic acida | HMDB00020 |
| 2-Hydroxyglutaric acida | HMDB00694 | Pimelic acidb | HMDB00857 |
| 2-Hydroxyvaleric acida | HMDB01863 | Pyroglutamic acida,b | HMDB00267 |
| 2-Ketobutyric acidb | HMDB00005 | Pyruvic acidb | HMDB00243 |
| 2-Methyl-3-hydroxybutyric acidb | HMDB00354 | Quinolinic acidb | HMDB00232 |
| 3,4-Dihydroxybenzeneacetic acidb | HMDB01336 | Stearic acidb | HMDB00827 |
| 3,4-Dihydroxybutanoic acida,b | HMDB00337 | Suberic acidb | HMDB00893 |
| 3,4-Dihydroxyhydrocinnamic acidb | HMDB00423 | Succinic acida,b | HMDB00254 |
| 3-Aminoisobutanoic acida,b | HMDB03911 | Sumiki’s acidb | HMDB02432 |
| 3-Hydroxyhippuric acidb | HMDB06116 | Tartaric acida | HMDB00956 |
| 3-Hydroxyisobutyric acida | HMDB00023 | Threonic acida | HMDB00943 |
| 3-Hydroxyisovaleric acida,b | HMDB00754 | Trans-Aconitic acida | HMDB00958 |
| 3-Hydroxymethylglutaric acidb | HMDB00355 | Trans-Ferulic acidb | HMDB00954 |
| 3-Hydroxyphenylacetic acidb | HMDB00440 | Uric acida | HMDB00289 |
| 3-Hydroxysebacic acidb | HMDB00350 | Vanillyllactic acidb | HMDB00913 |
| 3-Methyladipic acidb | HMDB00555 | Vanillylmandelic acidb | HMDB00291 |
| 3-Methylglutaconic acidb | HMDB00522 | Sugars | HMDB ID |
| 4-Deoxythreonic acida,b | HMDB02453 | D-Fructosea | HMDB00660 |
| 4-Hydroxybenzoic acidb | HMDB00500 | D-Galactosea | HMDB00143 |
| 4-Hydroxyphenyllactic acidb | HMDB00755 | D-Glucosea | HMDB00122 |
| 5-Hydroxyhexanoic acidb | HMDB00525 | D-Xylosea | HMDB00098 |
| 5-Hydroxyindoleacetic acidb | HMDB00763 | D-Xylulosea | HMDB01644 |
| Adipic acida,b | HMDB00448 | L-Arabinosea | HMDB00646 |
| Alpha-Hydroxyisobutyric acidb | HMDB00729 | L-Fucosea | HMDB00174 |
| Aminomalonic acida | HMDB01147 | Sucrosea | HMDB00258 |
| Ascorbic acida | HMDB00044 |
Polar metabolites.
Targeted organic acids.
Targeted bile acids. HPHPA: 3-(3-hydroxyphenyl)-3-hydroxypropanoic acid.
Table 5. Compound names, HMDB identification numbers, unique masses, mean mass spectral match quality, retention times, and NIST retention indices for volatile compounds in urine analyzed by HS-SPME-GC/MS.
| Peak # | Compound | HMDB ID | Uniquemass | MSmatch | RT(min) | RI(Calc) | RI(NIST) |
| 1 | 3-Methyl sulfolane | HMDB59667 | 68 | 867 | 0.83 | 725 | 654 |
| 2 | 3-Hexanone | HMDB00753 | 43 | 781 | 0.99 | 732 | 788 |
| 3 | 2-Pentanone | HMDB34235 | 43 | 894 | 1.61 | 758 | 654 |
| 4 | 1-Hydroxy-2-pentanone | HMDB59678 | 43 | 780 | 1.74 | 763 | 897 |
| 5 | Allyl methylsulphide | HMDB31653 | 88 | 793 | 1.83 | 767 | 701 |
| 6 | Dimethyl disulfide | HMDB05879 | 94 | 940 | 2.26 | 785 | 763 |
| 7 | 4-Heptanone | HMDB04814 | 71 | 919 | 3.34 | 831 | 883 |
| 8 | 1-Methylcyclohexanol | HMDB59693 | 71 | 821 | 3.81 | 850 | 931 |
| 9 | 2-Hexanone | HMDB05842 | 43 | 821 | 3.82 | 851 | 889 |
| 10 | 3,4-Dimethylthiophene | HMDB29304 | 111 | 906 | 4.15 | 865 | 888 |
| 11 | Diallyl sulphide | HMDB36491 | 45 | 827 | 4.86 | 895 | 848 |
| 12 | 5-Methyl-2-hexanone | HMDB31549 | 43 | 816 | 5.29 | 913 | 836 |
| 13 | 1,3-Dithio cyclohexane | HMDB31473 | 120 | 841 | 5.85 | 936 | 1027 |
| 14 | Dimethyl trisulfide | HMDB13780 | 126 | 873 | 6.01 | 943 | 950 |
| 15 | Phenol | HMDB00228 | 94 | 859 | 6.34 | 957 | 998 |
| 16 | o-Cymene | HMDB37050 | 119 | 928 | 7.19 | 993 | 1014 |
| 17 | p-Cymene | HMDB05805 | 119 | 928 | 7.33 | 998 | 1018 |
| 18 | m-Cymene | HMDB13806 | 119 | 928 | 7.43 | 1009 | 1022 |
| 19 | 1,4-Cineol | HMDB31455 | 111 | 912 | 8.71 | 1057 | 1016 |
| 20 | p-Cresol | HMDB01858 | 107 | 930 | 8.99 | 1068 | 1082 |
| 21 | Linalool oxide | HMDB31440 | 59 | 783 | 10.49 | 1131 | 1074 |
| 22 | Iso-menthol | HMDB35764 | 57 | 923 | 10.99 | 1152 | 1141 |
| 23 | Alpha-p-dimethylstyrene | HMDB29641 | 132 | 915 | 11 | 1153 | 1088 |
| 24 | L-Menthol | HMDB03352 | 71 | 924 | 11.18 | 1160 | 1140 |
| 25 | Undecane | HMDB31445 | 57 | 910 | 11.35 | 1168 | 1100 |
| 26 | Ledene Oxide (II) | HMDB60295 | 43 | 743 | 11.54 | 1176 | 1201 |
| 27 | Salicylic acid methyl ester | HMDB34172 | 120 | 857 | 11.65 | 1180 | 1193 |
| 28 | Beta-Carvone | HMDB35824 | 82 | 859 | 12.81 | 1229 | 1247 |
| 29 | Piperitone | HMDB34975 | 150 | 873 | 12.96 | 1235 | 1184 |
| 30 | o-Thymol | HMDB35770 | 135 | 904 | 13.9 | 1275 | 1305 |
| 31 | Beta-Cyclocitrol | HMDB41011 | 41 | 884 | 14.1 | 1283 | 1219 |
| 32 | 4,7-Dimethyl-benzofuran | HMDB59897 | 146 | 861 | 14.4 | 1296 | 1244 |
| 33 | Cuminal | HMDB02214 | 133 | 861 | 14.82 | 1313 | 1235 |
| 34 | 2,6,10,10 Tetramethyl-oxa-spiro-4,5-dec-6-ene | HMDB36823 | 138 | 911 | 15.91 | 1359 | 1370 |
| 35 | 4-(2,6,6-Trimethyl-1-cyclohexen-1-yl)-2-butanone | HMDB32913 | 121 | 795 | 15.97 | 1362 | 1433 |
| 36 | 1,2,3,4-Tetrahydro-1,1,6-trimethyl naphthalene | HMDB59826 | 159 | 855 | 16.02 | 1364 | 1349 |
| 37 | Alpha-Cedrene | HMDB59695 | 119 | 821 | 16.1 | 1367 | 1415 |
| 38 | 1,2,3,4,Tetrahydro-1,5,7-trimethylnapthalene | HMDB59696 | 159 | 795 | 16.59 | 1388 | 1310 |
| 39 | 1,2-Diydro-1,1,6-trimethyl-napthalene | HMDB40284 | 157 | 947 | 16.82 | 1398 | 1354 |
| 40 | Beta-Guaiene | HMDB38157 | 161 | 879 | 17 | 1405 | 1500 |
| 41 | Beta-Damascenone | HMDB13804 | 69 | 889 | 17.35 | 1420 | 1365 |
| 42 | 2,5-Cyclohexadiene-1,4-dione-2,6-di-tert-butyl | HMDB13817 | 177 | 843 | 18.51 | 1469 | 1462 |
| 43 | Himachalene | HMDB38168 | 119 | 823 | 18.62 | 1473 | 1489 |
| 44 | 4-(2,6,6-Trimethylcyclohexa-1,3-dienyl)-but-3-en-2-one | HMDB37139 | 136 | 883 | 18.73 | 1478 | 1497 |
| 45 | 1-(2,6,6-Trimethyl-1-cyclohexen-1-yl)-1-penten-3-one | HMDB38130 | 121 | 788 | 19.05 | 1491 | 1507 |
| 46 | 2,4-Bis (1,1-Dimethylethyl)-phenol | HMDB13186 | 191 | 863 | 19.5 | 1510 | 1519 |
| 47 | 1-(2,3,6-Trimethyl phenyl)-3-Buten-2-one | HMDB59697 | 132 | 918 | 19.8 | 1523 | 1528 |
| 48 | L-Calamenene | HMDB59910 | 159 | 920 | 20.25 | 1533 | 1534 |
| 49 | Beta-Vatirenene | HMDB59676 | 202 | 806 | 20.45 | 1550 | 1545 |
| 50 | 1,6,7-Trimethylnaphthalene | HMDB59701 | 170 | 899 | 20.82 | 1564 | 1559 |
| 51 | Azulol | HMDB59686 | 183 | 823 | 21.18 | 1581 | 1572 |
| 52 | 3,3,5,6-Tetramethyl-1-indanone | HMDB59683 | 173 | 686 | 21.28 | 1585 | 1579 |
Table 6. Concentrations of metabolites in human urine as measured by GC-MS.
| Compounds | Observed Concentration(µM/mM creatinine) | Literature Values(µM/mM creatinine) |
| 2,4-Dihydroxybutanoic acid | 1.0 (0.3–1.8) | 2.0±2.0 |
| 2-Ethylhydracrylic acid | 2.1 (1.3–2.9) | NA** |
| 2-Furoylglycine | 3.6 (0.7–5.8) | 9.95 (2.0–18.66) |
| 2-Hydroxy-2-methylbutyric acid | 0.8 (0.4–1.5) | 2.8±1.5 |
| 2-Methyl-3-hydroxybutyric acid | 4.0 (1.3–6.2) | 5.5 (0.0–11.0) |
| 3,4-Dihydroxybenzeneacetic acid | 0.9 (0.6–1.3) | 1.16±0.50 |
| 3,4-Dihydroxybutanoic acid | 42.2 (21.9–56.1) | 34.9 (12.5–57.2) |
| 3-Aminoisobutanoic acid | 25.8 (1.4–155.6) | 2.91–116.43 |
| 3-Hydroxyhippuric acid | 6.1 (4.1–7.8) | 2.5 (0.2–5.0) |
| 3-Hydroxyisobutyric acid | 29.0 (11.8–59.8) | 17.5 (2.0–33.0) |
| 3-Hydroxyisovaleric acid | 7.4 (4.1–17.2) | 8.5±3.2 |
| 3-Hydroxymethylglutaric acid | 3.2 (1.1–5.2) | 3 (0–10) |
| 3-Hydroxyphenylacetic acid | 5.6 (1.4–15.0) | 6.1±4.1 |
| 3-Hydroxysebacic acid | 2.9 (1.7–3.8) | NA** |
| 3-Methyladipic acid | 2.1 (0.9–5.6) | 2.9 (0.7–10.5) |
| 3-Methylglutaconic acid | 6.2 (2.8–8.3) | 4.5 (0.0–9.0) |
| 4-Deoxythreonic acid | 18.2 (5.6–23.9) | 25±10 |
| 4-Hydroxybenzoic acid | 2.4 (1.3–3.6) | 1.8±1.8 |
| 4-Hydroxyphenyllactic acid | 2.8 (2.6–3.0) | 1.1 (0.2–2.6) |
| 5-Hydroxyhexanoic acid | 2.7 (0.8–5.7) | 3.5 (0.0–7.0) |
| 5-Hydroxyindoleacetic acid | 2.9 (0.4–5.8) | 2.4±0.40 |
| Adipic acid | 3.0 (1.0–12.9) | 5.1 (0.8–35.0) |
| Alpha-Hydroxyisobutyric acid | 2.9 (1.3–4.6) | 3.8 |
| Azelaic acid | 1.9 (1.2–3.2) | 4.8 (1.3–15.0) |
| Benzoic acid | 3.6 (1.4–7.5) | 4.2 (1.9–6.5) |
| Cholenic acid | 0.015 (0.004–0.030) | NA** |
| Cis/Trans-Aconitic acid | 15.3 (5.2–21.8) | NA** |
| Citraconic acid | 1.6 (0.9–2.1) | 2.2 |
| Creatinine* | 12,246±8,369 | 10,090±310 |
| Deoxycholic acid | 0.055 (0.050–0.060) | NA** |
| D-Galactose | 3.3 (1.3–8.5) | 4.4 (0.0–31.0) |
| D-Glucose | 35.6 (10.3–56.7) | 31.12 |
| D-Xylitol | 2.3 (0.9–8.2) | 3.50±2. 37 |
| D-Xylulose | 19.7 (6.4–32.6) | NA** |
| Ethylmalonic acid | 2.4 (0.9–4.0) | 2.0 (0.5–3.9) |
| Gluconic acid | 13.3 (8.2–26.4) | NA** |
| Glutaric acid | 1.4 (0.5–2.3 ) | 1.3 (0.6–2.6) |
| Glyceric acid | 14.4 (7.8–28.8) | 4.5 (0.0–9.0) |
| Glycine | 75.7 (21–205) | 151 (233–248) |
| Glycolic acid | 39.4 (2.9–78.1) | 35 (18–55) |
| Hippuric acid | 217 (28–610) | 257 (20–770) |
| Homovanillic acid | 4.3 (0.9–8.9) | 4.9±3.4 |
| HPHPA | 4.2 (0.9–15.3) | 0.55–54.93 |
| Hydroxypropionic acid | 8.1 (3.1–11.8) | 6.5 (3.0–10.0) |
| Indoleacetic acid | 2.6 (0.6–5.4) | 2.0±1.0 |
| Indolelactic acid | 0.9 | 0.460 (0.098–0.980) |
| Isocitric acid | 56.8 (19.4–119.1) | 58 (36–84) |
| Kynurenic acid | 2.6 (0.9–4.1) | 1.34±0.30 |
| L-Arabinose | 8.8 (0.8–19.4) | 8.75 (6.12–11.84) |
| L-Asparagine | 9.5 (3.0–26.0) | 10.0 (4.6–16.3) |
| Levulinic acid | 1.7 (0.6–2.5) | 1.28 (0.33–2.75) |
| Lithocholic acid | 0.16 (0.10–0.20) | NA** |
| L-Lactic acid | 7.1 (0.9–16.4) | 12.3±6.2 |
| L-Ornithine | 5.3 (2.0–8.8) | 5.0 (2.0–8.0) |
| L-Phenylalanine | 7.8 (5.0–11.3) | 4.50±1.15 |
| L-Serine | 19.0 (10.4–35.8) | 26.0±10.0 |
| L-Tyrosine | 15.1 (6.1–23.2) | 8.8±4.5 |
| L-Valine | 5.5 (2.7–9.8) | 3.0±1.0 |
| Malonic acid | 2.3 (1.2–3.1) | 1 (0–2) |
| m-Chlorobenzoic acid | 0.7 | NA** |
| Methylmalonic acid | 1.3 (0.5–2.1) | 1.6 (0.0–3.6) |
| Methylsuccinic acid | 4.7 (1.1–6.2) | 2.3 (0.8–10.8) |
| Myoinositol | 12.6 (5.1–15.3) | 18.8±2.9 |
| o-Hydroxyphenylacetic acid | 2.9 (1.4–3.7) | 7.71±3.65 |
| Oxalic acid | 8.2 (3.9–14.0) | 1.11–33.34 |
| Oxoglutaric acid | 3.4 (1.4–12.5) | 2.98 (0.42–18.30) |
| Palmitic acid | 6.7 (2.6–24.3) | 11 (6–23) |
| Phosphoric acid | 784 (425–1170) | 1000–4900 |
| p-Hydroxyphenylacetic acid | 6.0 (2.4–9.7) | 9.68 |
| Pimelic acid | 1.9 (0.5–3.5) | NA** |
| Pseudouridine | 29.8 (21.8–47.3) | 26.02±4.62 |
| Pyroglutamic acid | 18.6 (4.5–24.9) | 28.8 (3.4–54.2) |
| Pyruvic acid | 2.1 (1.0–3.7) | 2.13 (0.54–8.67) |
| Quinolinic acid | 3.9 (0.9–15.1) | 2.5±1.1 |
| Scyllitol | 3.6 (1.9–6.9) | NA |
| Sorbitol | 3.9 (1.9–5.1) | 3.50±2.24 |
| Stearic acid | 3.9 (2.3–7.7) | 2.9 (1.6–6.6) |
| Suberic acid | 1.2 (0.5–1.9) | 0.5 (0.0–2.9) |
| Succinic acid | 6.2 (2.5–13.5) | 5.6±3.8 |
| Sumiki’s acid | 1.6 | NA** |
| Threonic acid | 14.8 (8.6–36.1) | 10.0±7.0 |
| Trans-Ferulic acid | 1 | 36.3±11.2 |
| Uric acid | 186 (93–329) | 188 (79–296) |
| Ursodeoxycholic acid | 0.022 (0.013–0.030) | NA** |
| Vanillylmandelic acid | 1.6 (0.6–2.8) | 1.2±0.5 |
Concentration of metabolite (Mean ± SD) is expressed by µM. HPHPA: 3-(3-hydroxyphenyl)-3-hydroxypropanoic acid; NA: not available; NA**: not available for healthy adults.
Figure 3. Typical GC-MS total ion chromatogram of organic acids extracted from human urine.
Numbers indicate the following metabolites (quantified compounds): 1: pyruvic acid; 2: L-lactic acid; 3: alpha-hydroxyisobutyric acid; 4: glycolic acid; 5: levulinic acid; 6∶3-hydroxyisovaleric acid; 7∶2-hydroxy-2-methylbutyric acid; 8: hydroxypropionic acid; 9∶2-methyl-3-hydroxybutyric acid; 10: malonic acid; 11∶3-hydroxyisovaleric acid; 12: methylmalonic acid; 13∶2-ethylhydracrylic acid; 14: benzoic acid; 15: phosphoric acid; 16: ethylmalonic acid; 17: succinic acid; 18: methylsuccinic acid; 19∶4-deoxythreonic acid; 20∶5-hydroxyhexanoic acid; 21: citraconic acid; 22: glutaric acid; 23: m-chlorobenzoic acid; 24; 3,4-dihydroxybutanoic acid; 25∶3-methylglutaconic acid; 26: adipic acid; 27: pyroglutamic acid; 28∶3-methyladipic acid; 29: sumiki’s acid; 30: o-hydroxyphenylacetic acid; 31: oxoglutaric acid; 32: pimelic acid; 33∶3-hydroxymethylglutaric acid; 34∶3-hydroxyphenylacetic acid; 35∶4-hydroxybenzoic acid; 36∶2-furoylglycine; 37: suberic acid; 38: quinolinic acid; 39: Cis/Trans-aconitic acid; 40: homovanillic acid; 41: azelaic acid; 42: hippuric acid; 43∶3,4-dihydroxybenzeneacetic acid; 44∶3-(3-hydroxyphenyl)-3-hydroxypropanoic acid (HPHPA); 45: vanillylmandelic acid; 46∶4-hydroxyphenyllactic acid; 47: indoleacetic acid; 48: palmitic acid; 49: kynurenic acid; 50∶3-hydroxyhippuric acid; 51∶3-hydroxysebacic acid; 52: Trans-ferulic acid; 53∶5-hydroxyindoleacetic acid; 54: stearic acid.
Incomplete compound identification is a common problem with global or untargeted GC-MS metabolomics. This may be due to any number of factors including spectral overlap due to incomplete separation, poor signal to noise for low intensity peaks, the lack of reference GC-MS spectral data for certain metabolites (especially unusual dietary sources), or the presence of spectral artefacts such as derivatization by-products or degraded metabolites in the GC-MS spectrum. For our GC-MS studies we used the NIST library supplemented with a home-made GC-MS reference library of known urine compounds assembled from the Human Metabolome Library [16]. No doubt the use of other commercially available reference GC-MS libraries such as the Fiehn GC-MS library from Agilent or the GOLM metabolome database library [85] might have allowed us to further increase our coverage. Likewise the use of a faster scan rate and/or a more sensitive GC-TOF instrument (instead of a slower scanning quadrupole GC-MS) certainly would have increased overall coverage.
Nearly all of the non-volatile metabolites (87) identified by our GC-MS analyses were also identified by NMR. Some of the exceptions were oxalic acid, phosphate and uric acid, each of which was detected by GC-MS but not by NMR. These compounds do not have NMR-detectable protons at physiological pH, making them essentially “NMR invisible”. Other compounds seen by GC-MS but not by NMR included metabolites that were generally below the detection limit of NMR (∼2 µM/mM creatinine) such as indolelactic acid and 2,4-dihydroxybutanoic acid. For our non-targeted GC-MS analysis, the lower limit of detection was 1 µM/mM creatinine (for 2,4-dihydroxybutanoic acid), while for our targeted organic acid GC-MS analysis the lower limit of detection was 0.7 µM/mM creatinine (for m-chlorobenzoic acid). Overall, our data suggests that the sensitivity of a standard single quadrupole GC-MS instrument is perhaps 1.5–2X better than a 500 MHz NMR instrument for water-soluble metabolites. It is also important to note that the level of water-soluble, non-volatile metabolite coverage obtained by GC-MS is not as great as seen with NMR (127 cmpds vs. 209 cmpds). The limited coverage of GC-MS is partly due to the fact that not all compounds can be readily extracted, easily derivatized or routinely separated on a GC column. Furthermore, when analyzing urine by GC-MS there is a need to pretreat the sample with urease (to reduce urea levels) that can diminish the abundance of some metabolites [86]. While GC-MS may not be the best method for analyzing water-soluble metabolites, it certainly excels at the detection of volatile metabolites. Indeed, only one of the volatile metabolites identified by GC-MS is identified by NMR (phenol). This certainly underlines a key strength of GC-MS relative to other metabolomics platforms. When comparing NMR to GC-MS we found that NMR is capable of detecting 121 compounds that the 4 combined GC-MS methods cannot detect while the combined GC-MS methods can detect 91 compounds that NMR cannot routinely detect. Overall, these data suggest that GC-MS and NMR appear to be complementary methods for the identification and quantification of small molecules in urine.
Among the 58 metabolites quantified by both GC-MS and NMR we found very good overall agreement, with the majority of measured concentration values falling within 20 ± 11% of each other. The concentration patterns and rankings of the most abundant to the least abundant compounds were also largely identical for the two platforms. A total of 12 metabolites exhibited somewhat larger concentration discrepancies between GC-MS and NMR (i.e; L-arabinose, L-serine (lower in GC-MS vs. NMR), 4-hydroxybenzoic acid and tyrosine (higher in GC-MS vs NMR). Some of these concentration differences may be due to the extraction or derivatization process needed to conduct GC-MS analyses. This can lead to unspecified compound losses, unusual derivatives or unrecognized fragmentation patterns. Therefore we would have expected at least a few GC-MS concentration values to be slightly lower than those seen by NMR. Likewise, it is important to remember that there are inherent errors (5–10%) in measuring peak areas (i.e. compound concentrations) both in GC-MS and NMR due to peak overlap, uneven baselines and spectral noise.
Nearly all of the compounds we detected or quantified in human urine by GC-MS have been previously described or mentioned in the GC-MS literature. One compound (scyllitol), however, appears not to have been previously detected by GC-MS. The identification of this compound by our GC-MS method was aided by its prior identification by NMR (see previous section). Additionally, a careful literature analysis also indicated the scyllitol is a normal constituent of human urine and has previously been detected in human urine via other methods.
As we noted with our NMR studies earlier, there are a few previously reported GC-MS detectable metabolites in human urine that appear to be artefacts. These artefactual metabolites may arise from extractions with different solvents, pre-treatment with urease, and chemical derivatization. For example, Shoemaker et al [84], reported the presence of bisethane in human urine. We also detected bisethane, but it appears to be artefact of chemical derivatization and is not a urine metabolite.
Quantification and Identification of Urine Metabolites – Combined Direct Flow Injection and LC-MS/MS Assay
Direct flow injection (DFI) MS/MS or DFI-MS/MS is another commonly used global metabolic profiling method [87]. When isotopic standards are used along with multiple reactions monitoring (MRM), it is also possible to perform targeted metabolomics with very accurate concentration measurements. For our urine studies, we employed a combined DFI/LC-MS/MS approach, based on the commercially available AbsoluteIDQ p180 Kit (BIOCRATES Life Sciences AG, Innsbruck). When applied to urine, we were able to identify and quantify a total of 127 metabolites or metabolite species, including 34 acylcarnitines, 21 amino acids, 15 biogenic amines, creatinine, hexose, 35 phospatidylcholines, 15 sphingomyelins and 5 lysophosphatidylcholines. The amino acids and biogenic amines are analyzed by an LC-MS/MS method, whereas all other metabolites are analyzed by DFI-MS/MS as indicated in Table 7. DFI-MS/MS identifies lipid species (as opposed to specific lipids) using their total acyl/alkyl chain content (i.e. PC (38∶4)) rather than their unique structure. Therefore each lipid species identified by the BIOCRATES kit typically corresponds to 5–10 possible unique lipid structures. Consequently, the total number of phosphatidylcholines, sphingolipids and lysophosphatidylcholines structures identified by this method was 458, 19 and 6, respectively. Therefore, combining these probable lipid structures (483 in total, based on the known fatty acid and lipid composition in human serum) with the other 72 confirmed non-lipid metabolites, the DFI-MS/MS method yields 555 confirmed and probable metabolites or metabolite structures. All of these compounds, along with their corresponding estimated concentrations have all been entered into the UMDB.
Table 7. Concentrations of acylcarnitines, amino acids, biogenic amines, lysophosphatidylcholines, phosphatidylcholines and sphingomyelins in human urine by combined DFI/LC-MS/MS (BIOCRATES kit).
| Acylcarnitines (DFI-MS/MS) | Mean (µM/mM creatinine) | Biogenic amines (LC-MS/MS) | Mean (µM/Mm creatinine) |
| Acetyl-L-carnitine | 2.0 (0.4–5.9) | Kynurenine | 0.41 (0.08–1.31) |
| Butenyl-L-carnitine | 0.014 (0.008 –0.021) | L-Dopa | 0.02 (0.01–0.04) |
| Butyryl-L-carnitine | 0.76 (0.12–1.81) | Methioninesulfoxide | 0.31 (0.14–0.66) |
| Decadienyl-L-carnitine | 0.10 (0.04–0.30) | Phenylethylamine | 0.011 (0.003–0.067) |
| Decanoyl-L-carnitine | 0.04 (0.02–0.06) | Putrescine | 0.20 (0.04–0.40) |
| Decenoyl-L-carnitine | 0.14 (0.06–0.32) | Sarcosine | 1.6 (0.2–9.4) |
| Dodecanoyl-L-carnitine | 0.02 (0.01–0.04) | Serotonin | 0.08 (0.04–0.12) |
| Dodecenoyl-L-carnitine | 0.028 (0.011–0.042) | Symmetric dimethylarginine | 2.8 (2.0–9.4) |
| Glutaconyl-L-carnitine | 0.04 (0.01–0.06) | Lysophosphatidylcholines (DFI-MS/MS) | Mean (µM/Mm creatinine) |
| Glutaryl-L-carnitine | 0.04 (0.03–0.10) | lysoPC a C16∶0 | 0.014 |
| Hexadecadienyl-L-carnitine | 0.0010 (0.0005–0.0040) | lysoPC a C17∶0 | 0.0032 (0.0008–0.0087) |
| Hexadecanoyl-L-carnitine | 0.0022 (0.0020–0.0030) | lysoPC a C18∶0 | 0.0040 (0.0013–0.0100) |
| Hexanoyl-L-carnitine | 0.05 (0.03–0.07) | lysoPC a C20∶4 | 0.0020 (0.0005–0.0038) |
| Hexenoyl-L-carnitine | 0.019 (0.009–0.050) | lysoPC a C6∶0 | 0.0045 (0.0022–0.0074) |
| Hydroxyhexadecanoyl-L-carnitine | 0.0015 (0.0006–0.0050) | Phosphatidylcholines (DFI-LC/MS) | Mean (µM/Mm creatinine) |
| Hydroxypropionyl-L-carnitine | 0.008 (0.004–0.012) | PC aa C28∶1 | 0.006 (0.003–0.020) |
| Hydroxytetradecadienyl-L-carnitine | 0.003 (0.001–0.008) | PC aa C30∶2 | 0.0005 (0.0001–0.0022) |
| Hydroxytetradecenoyl-L-carnitine | 0.0020 (0.0008–0.0040) | PC aa C32∶0 | 0.0035 (0.0013–0.0071) |
| L-Carnitine | 4.5 (0.6–15.2) | PC aa C32∶3 | 0.0009 (0.0003–0.0025) |
| Malonyl-L-carnitine | 0.02 (0.01–0.04) | PC aa C34∶1 | 0.0077 (0.0023–0.0500) |
| Methylglutaryl-L-carnitine | 0.030 (0.016–0.052) | PC aa C34∶2 | 0.0047 (0.0016–0.0240) |
| Methylmalonyl-L-carnitine | 0.09 (0.05–0.16) | PC aa C34∶3 | 0.002 |
| Nonayl-L-carnitine | 0.20 (0.07–0.46) | PC aa C34∶4 | 0.0023 (0.0002–0.0180) |
| Octadecadienyl-L-carnitine | 0.0007 (0.0003–0.0020) | PC aa C36∶1 | 0.0034 (0.0006–0.0250) |
| Octanoyl-L-carnitine | 0.05 (0.03–0.09) | PC aa C36∶2 | 0.0160 (0.0021–0.0450) |
| Octenoyl-L-carnitine | 0.50 (0.08–1.20) | PC aa C36∶3 | 0.0024 (0.0005–0.0240) |
| Pimelyl-L-carnitine | 0.03 (0.01–0.11) | PC aa C36∶4 | 0.0018 (0.0005–0.0083) |
| Propenoyl-L-carnitine | 0.003 (0.001–0.006) | PC aa C36∶6 | 0.0037 |
| Propionyl-L-carnitine | 0.07 (0.01–0.20) | PC aa C38∶3 | 0.0056 |
| Tetradecadienyl-L-carnitine | 0.0023 (0.0010–0.0050) | PC aa C38∶4 | 0.0022 (0.0005–0.0065) |
| Tetradecanoyl-L-carnitine | 0.018 (0.001–0.088) | PC aa C38∶5 | 0.0007 (0.0002–0.0020) |
| Tetradecenoyl-L-carnitine | 0.003 (0.001–0.006) | PC aa C38∶6 | 0.0010 (0.0005–0.0036) |
| Tiglyl-L-carnitine | 0.10 (0.05–0.20) | PC aa C40∶3 | 0.00035 (0.0–0.00140) |
| Valeryl-L-carnitine | 0.220 (0.037–0.440) | PC aa C42∶2 | 0.0009 (0.0003–0.0024) |
| Creatinine | Mean | PC aa C42∶4 | 0.0008 (0.0003–0.0025) |
| Creatinine* | 12,475±7,955 | PC ae C34∶1 | 0.0030 (0.0003–0.0110) |
| Hexose | Mean (µM/Mm creatinine) | PC ae C34∶2 | 0.0012 (0.0002–0.0075) |
| Hexose | 110 (37–501) | PC ae C36∶1 | 0.0046 (0.0012–0.0080) |
| Amino acids (LC-MS/MS) | Mean (µM/Mm creatinine) | PC ae C36∶2 | 0.0016 (0.0002–0.0050) |
| Glycine | 101.0 (37.0–250.6) | PC ae C36∶3 | 0.0009 (0.0001–0.0040) |
| L-Alanine | 21.7 (7.2–47.4) | PC ae C36∶4 | 0.0022 |
| L-Arginine | 7.4 (3.2–14.6) | PC ae C38∶2 | 0.0015 (0.0007–0.0023) |
| L-Asparagine | 8.8 (4.6–17.7) | PC ae C38∶3 | 0.0020 (0.0012–0.0028) |
| L-Aspartic acid | 10.9 (1.9–26.8) | PC ae C38∶4 | 0.0006 (0.0–0.0024) |
| L-Citrulline | 0.8 (0.2–1.7) | PC ae C38∶6 | 0.0065 (0.0027–0.0170) |
| L-Glutamic acid | 6.4 (0.6–17.5) | PC ae C40∶2 | 0.0012 (0.0006–0.0020) |
| L-Glutamine | 39.9 (18.4–72.5) | PC ae C40∶5 | 0.0006 (0.0001–0.0025) |
| L-Histidine | 45.6 (21.7–71.5) | PC ae C42∶3 | 0.0007 (0.0001–0.0020) |
| L-Isoleucine | 1.3 (0.5–2.7) | PC ae C44∶3 | 0.0010 (0.0002–0.0031) |
| L-Leucine | 3.0 (1.6–6.0) | Sphingomyelins (DFI-MS/MS) | Mean (µM/Mm creatinine) |
| L-Lysine | 17.9 (3.6–56.1) | SM (OH) C14∶1 | 0.0028 (0.0008–0.0049) |
| L-Methionine | 0.8 (0.4–1.6) | SM (OH) C16∶1 | 0.0006 (0.0001–0.0023) |
| L-Ornithine | 4.4 (1.2–22.1) | SM (OH) C22∶1 | 0.0023 (0.0009–0.0058) |
| L-Phenylalanine | 5.9 (2.8–10.8) | SM (OH) C22∶2 | 0.0007 (0.0001–0.0028) |
| L-Proline | 1.1 (0.1–2.5) | SM (OH) C24∶1 | 0.0010 (0.00034–0.0025) |
| L-Serine | 24.5 (12.3–50.2) | SM C16∶0 | 0.0100 (0.0016–0.0700) |
| L-Threonine | 14.6 (6.6–29.3) | SM C16∶1 | 0.0009 (0.0004–0.0023) |
| L-Tryptophan | 5.6 (9.3–2.1) | SM C18∶0 | 0.0024 (0.0011–0.0070) |
| L-Tyrosine | 8.7 (3.0–20.1) | SM C18∶1 | 0.0004 (0.0001–0.0013) |
| L-Valine | 4.3 (2.4–10.4) | SM C20∶2 | 0.0003 (0.0–0.0016) |
| Biogenic amines (LC-MS/MS) | Mean (µM/Mm creatinine) | SM C22∶3 | 0.0011 (0.0008–0.0014) |
| Acetyl-ornithine | 1.3 (0.5–2.8) | SM C24∶0 | 0.0085 (0.0047–0.0200) |
| Alpha-aminoadipic acid | 7.2 (2.5–16.4) | SM C24∶1 | 0.0030 (0.0007–0.0120) |
| Asymmetric dimethylarginine | 2.7 (1.4–4.2) | SM C26∶0 | 0.0014 (0.0002–0.0043) |
| Carnosine | 1.2 (0.2–4.1) | SM C26∶1 | 0.0005 (0.0001–0.0012) |
| Dopamine | 0.4 (0.2–0.7) | ||
| Histamine | 0.03 (0.01–0.10) | ||
| Hydroxykynurenine | 0.15 (0.06–0.34) |
Concentration of metabolite (Mean±SD) is expressed by µM. aa: diacyl; ae: acyl-alkyl.
Our results show very good agreement with the previous studies conducted by BIOCRATES on human urine samples (Biocrates Application Note 1005-1). We found that the lower limit of quantification by DFI MS/MS based on the AbsoluteIDQ kit was 0.1 nM/mM creatinine for certain phosphatidylcholine species (i.e. PC aa C40∶3) and 0.1 nM/mM creatinine for certain sphingomyelin species (i.e. SM 26∶1). Comparison of our lipid results with literature data was difficult as relatively few papers report urine lipid concentration data. Indeed, the presence of lipids in urine is a little unexpected, but not entirely unreasonable. It is likely that urea, a well known chaotrope, facilitates the dissolution of small amounts of fatty acids and other lipid species in urine.
Many of the compounds we measured with this kit assay appear to be normal constituents of human urine but have not been previously reported (quantified and/or detected) in the scientific literature (with the exception of the BIOCRATES Application Note). In total, 53 compounds are being reported here for the first time as being normal constituents of human urine, while 68 compounds are being robustly quantified in human urine for the first time. The vast majority of these compounds are lipids.
Based on our results, the combined DFI/LC-MS/MS method detected 98 compounds or compound species that GC-MS and NMR methods could not detect, while GC-MS detected 161 compounds and NMR detected 181 compounds that DFI/LC-MS/MS could not detect. The 3 methods were able to detect a common set of 17 compounds including creatinine, L-glutamine, L-tryptophan, L-tyrosine and L-valine. Interestingly, the concentrations measured by DFI/LC-MS/MS, NMR and GC-MS (across the shared set of 17 compounds) showed generally good agreement (within 19 ± 7% of each other). The relatively small overlap, in terms of compound coverage, between the 3 platforms is a bit of a surprise and certainly serves to emphasize the tremendous chemical diversity that must exist in urine. Overall, by combining these 3 platforms, we were able to identify 295 and quantify 231 unique or non-overlapping metabolites or metabolite species. These data suggest that DFI/LC-MS/MS, GC-MS and NMR are highly complementary techniques for the identification and quantification of metabolites in human urine.
Quantification and Identification of Urinary Trace Metals – ICP-MS
To determine the trace elemental composition of urine, we used inductively coupled plasma mass spectrometry (ICP-MS). ICP-MS is widely considered to be one of the best techniques for the characterization of the trace element composition of biological samples [88], [89]. Indeed, none of the other methods (1H-NMR, GC-MS and DFI/LC-MS/MS) are suited for measuring trace element composition or concentrations. Our multi-elemental analysis of urine using ICP-MS provided quantitative results for a total of 40 metals or trace minerals (Table 8). Based on their frequency of occurrence and overall abundance, all 40 trace elements appear to be normal constituents of human urine. Of these, 2 have previously not been quantified for healthy adults.
Table 8. Concentrations of ions (µmol/mmol creatinine) as determined by ICP-MS.
| Compound Name | HMDB | Concentration (µM/mM creatinine) | Litterature values(µM/mM creatinine) |
| Aluminum (Al) | HMDB01247 | 0.31 (0.05–1.21) | 0.667±0.037 |
| Arsenic (As) | HMDB02290 | 0.10 (0.01–0.52) | 0.16±0.16 |
| Barium (Ba) | HMDB04142 | 0.0070 (0.0001–0.0655) | 0.0129–0.0320 |
| Cadmium (Cd) | HMDB03638 | 0.0004 (0.0002–0.0010) | 0.00060 (0.00035–0.00100) |
| Calcium (Ca) | HMDB00464 | 200.0 (16.9–520.0) | 299.0±99.0 |
| Cerium (Ce) | HMDB13672 | 0.00048 (0.00008–0.00180) | 0.0 – 0.0007 |
| Cesium (Cs) | HMDB13669 | 0.0039 (0.0005–0.0110) | 0.0061±0.0011 |
| Chromium (Cr) | HMDB00599 | 0.0084 (0.0001–0.0460) | 0.0100±0.0065 |
| Cobalt (Co) | HMDB00608 | 0.0014 (0.0001–0.0140) | 0.000810±0.00053 |
| Copper (Cu) | HMDB00657 | 0.0163 (0.0006–0.1099) | 0.0190 (0.0092–0.038) |
| Gallium (Ga) | HMDB01478 | 0.0056 (0.0012–0.0150) | 0.0003–0.0006 |
| Hafnium (Hf) | HMDB29201 | 0.00031 (0.00006–0.00110) | 0.00027±0.00012 |
| Iron (Fe) | HMDB00692 | 0.0890 (0.0025–0.4579) | 0.11 (0.0–0.14) |
| Lanthanum (La) | HMDB13711 | 0.00020 (0.00003–0.00079) | 0.00052±0.00039 |
| Lead (Pb) | HMDB04628 | 0.0026 (0.0007–0.0110) | 0.011 (0.0–0.020) |
| Lithium (Li) | HMDB05949 | 0.5 (0.2–0.9) | 0.504 (0.058–3.414) |
| Magnesium (Mg) | HMDB00547 | 262 (42–1,189) | 289.0±92.0 |
| Manganese (Mn) | HMDB01333 | 0.013 (0.002–0.051) | 0.011 (0.0–0.023) |
| Molybdenum (Mo) | HMDB01302 | 0.049 (0.006–0.295) | 0.06±0.03 |
| Neodymium (Nd) | HMDB13715 | 0.00017 (0.00004–0.00070) | 0.0026±0.0013 |
| Nickel (Ni) | HMDB02457 | 0.0080 (0.0015–0.0450) | 0.00420 (0.00302–0.00572) |
| Palladium (Pd) | HMDB13670 | 0.0110 (0.0011–0.1410) | < 0.00001–0.2538 |
| Platinum (Pt) | HMDB13671 | 0.000155 (0.000031–0.000560) | 0.0000656 |
| Posphorus (P) | HMDB01315 | 1,799 (221–4,936) | 1,666.67–4,333.33 |
| Potassium (K) | HMDB00586 | 3,593 (553–8,079) | 4,605 (2,631–6,578) |
| Rhenium (Re) | HMDB13719 | 0.000096 (0.000013–0.000340) | NA |
| Rubidium (Rb) | HMDB01327 | 1.354 (0.187–3.673) | 1.84±0.46 |
| Ruthenium (Ru) | HMDB13668 | 0.0002 (0.0001–0.0009) | NA |
| Selenium (Se) | HMDB01349 | 0.149 (0.028–0.345) | 0.11346 (0.07724–0.15010) |
| Sodium (Na) | HMDB00588 | 12,477 (1,863–37,249) | 14,737 (5,263–36,842) |
| Strontium (Sr) | HMDB03642 | 0.163 (0.030–0.415) | 0.1333 |
| Tantalum (Ta) | HMDB04143 | 0.00020 (0.00004–0.00074) | 0.00008±0.00006 |
| Thallium (Tl) | HMDB13724 | 0.0003 (0.0001–0.0013) | < 0.00659 |
| Thorium (Th) | HMDB29215 | 0.00020 (0.00002–0.00100) | < 0.00584 |
| Tin (Sn) | HMDB01960 | 0.00110 (0.00016–0.00690) | < 0.00552 |
| Titanium (Ti) | HMDB01966 | 0.0013 (0.0005–0.0030) | 0.0142 |
| Tungsten (W) | HMDB01989 | 0.0060 (0.0011–0.0280) | 0.010±0.022 |
| Vanadium (V) | HMDB02503 | 0.033 (0.004–0.083) | 0.00162±0.00144 |
| Zinc (Zn) | HMDB01303 | 0.460 (0.058–0.948) | 0.52±0.25 |
| Zirconium (Zr) | HMDB01475 | 0.021 (0.004–0.083) | 0.0012–0.0074 |
As far as we are aware, this is the first multi-elemental study of urine that has been performed by ICP-MS. As seen in Table 8, there is generally good agreement between the values measured by ICP-MS and those previously reported in literature, with differences generally being less than 22 ± 10%. Larger differences are seen for gallium (Ga), lead (Pb), Neodymium (Nd), titanium (Ti) and vanadium (V), but these may be due to the effects of age, diet, local environment (minerals in local water) or diurnal variation. Alternately they may reflect real differences in the sensitivity or accuracy of the instruments being used. As a general rule, ICP-MS is considered as a gold standard for the identification and quantification of trace metals [18], so we would tend to place higher confidence in the values derived via ICP-MS over those measured by other technologies. By our measurements, the most abundant metals/salts are sodium (Na) (12.5 ± 10.6 mM/mM creatinine) and potassium (K) (3.6 ± 2.5 mM/mM creatinine) – as expected, while the least abundant was rhenium (Re) with a lower limit of quantification by ICP-MS of 96 pM/mM creatinine.
Quantification and Identification of Urine Metabolites – Targeted HPLC Assays
The inventory of metabolites we detected and quantified by 1H NMR, GC-MS, DFI/LC-MS/MS and ICP-MS covers a significant portion of all chemical classes. However, these methods sometimes lack the necessary sensitivity, the appropriate instrumental configuration or detection capabilities and therefore fail to detect/quantify a variety of important compound classes. This includes a number of molecules that are normal constituents of urine such as thiols and isoflavones. To identify and quantify these 2 classes of metabolites we decided to employ High Performance Liquid Chromatography (HPLC). HPLC assays are the method of choice for detecting isoflavones and thiols as they are sensitive, precise and can be easily coupled with sensitive detection methodologies such as fluorescence and ultraviolet detection. In our studies, fluorescence and ultraviolet detection were used for the identification and quantification of urinary thiols and isoflavones, respectively.
Biological thiols, or mercaptans, are very active metabolic products of sulfur and play a central role in redox metabolism, cellular homeostasis and a variety of physiological and pathological processes. In urine, the most important thiols are L-cysteine and L-cysteinylglycine [90], [91], [92], [93]. Isoflavones or phytoestrogens form or constitute another important class of urinary metabolites [90]. Humans are exposed to these biologically active phytochemicals mainly through food intake via vegetables, fruit and wheat/bread products [94]. For the detection of thiols we developed assays to measure L-cysteine, L-cysteineglycine, L-glutathione and L-homocysyeine, while for isoflavones we developed assays to measure biochanin A, coumesterol, daidzein, equol, formonentin and genistein. Using these HPLC assays, we measured a total of 4 thiols and 6 isoflavones in urine (Table 9).
Table 9. Concentrations of isoflavones and thiols as determined by HPLC.
| Compound Name | HMDB ID | Mean(µM/mM creatinine) | Oc (%) | Literature values (µM/mM creatinine) |
| Biochanin Aa | HMDB02338 | 0.0088 (0.0027–0.0200) | 77 | NA |
| Coumesterola | HMDB02326 | 0.0081 (0.0033–0.0120) | 45 | NA |
| Daidzeina | HMDB03312 | 0.026 (0.002–0.064) | 88 | 0.036 ± 0.008 |
| Equola | HMDB02209 | 0.06 (0.01–0.10) | 67 | 0.13 ± 0.12 |
| Formonenetina | HMDB05808 | 0.0007 (0.0002–0.0011) | 33 | NA |
| Genisteina | HMDB03217 | 0.0280 (0.0077–0.0640) | 88 | 0.032 ± 0.007 |
| L-Cysteineb | HMDB00574 | 81.0 (36.7–147.6) | 100 | 33.4 ± 15.5 |
| L-Cysteinylglycineb | HMDB00078 | 0.6 (0.28–1.54) | 100 | 0.48 ± 0.15 |
| L-Glutathioneb | HMDB00125 | 0.030 (0.005–0.085) | 100 | 0.065 |
| L-Homocysteineb | HMDB00742 | 1.1 (0.6–1.7) | 100 | 1.95 ± 1.47 |
Isoflavone metabolites measured by HPLC/UV.
Thiol metabolites measured by HPLC/FD.
Oc: occurrence;
NA: not available.
As seen in Table 9, there is generally good agreement between the values measured by these targeted HPLC assays and those previously reported in literature, with differences generally being less than ∼30%. Only one of these metabolites (L-cysteine) was measured independently on one of our other platforms (NMR) and the NMR concentration was found to be 25% lower than the HPLC assay. A possible explanation for this discrepancy is that cysteine measured by HPLC-FD yields total L-cysteine including the free form and L-cystine reduced to L-cysteine during the reaction [95]. On the other hand, NMR can distinguish between L-cysteine and L-cystine.
The Composition of Human Urine – Comparison with Other Biofluids
By combining a systematic computer-aided literature survey with an extensive, quantitative multiplatform metabolomic analysis we have been able to comprehensively characterize the human urine metabolome. Our data suggests that there are at least 3079 detectable metabolites in human urine, of which 1350 have been quantified. At least 72 of these compounds are of microbial origin, 1453 are endogenous while 2282 are considered exogenous (note some compounds can be both exogenous and endogenous), coming from diet, drugs, cosmetics or environmental exposure. Using a chemical classification system developed for the HMDB [15] we found that human urinary metabolites fall into 230 different chemical classes (25 “super classes”, Table 2). Given that there are only 356 chemical classes in the entire human metabolome [16], this certainly demonstrates the enormous chemical diversity found in urine. As might be expected, most urinary metabolites are very hydrophilic, although there are clearly trace amounts of lipids and fatty acids that contribute a significant number of chemicals to the urinary metabolome (836 fatty acids and lipids). This is in rather striking contrast to the composition of serum [20] which is particularly rich in lipids (i.e. >17000 lipids and fatty acids). Relative to other biofluids such as CSF [18] or saliva [19], urine contains significantly more compounds (5–10X) and exhibits significantly more chemical diversity (2–3X). On the other hand, we know that every compound that is found in human urine is also found in human blood. In other words, the human urine metabolome is a subset of the human serum metabolome, both in terms of composition and chemical diversity [20]. However, more than 484 compounds we identified in urine (either experimentally or via literature review) were not previously reported to be in blood. The fact that so many compounds seem to be unique to urine likely has to do with the fact that the kidneys do an extraordinary job of concentrating certain metabolites from the blood. Consequently compounds that are far below the limit of detection in blood (using today’s instrumentation) are well above the detection limit in urine. In fact, concentration differences between the two biofluids sometimes exceed 1000X for certain compounds, such as histamine, androsterone, normetanephrine, testosterone 13, 14-dihydro-15-keto-PGE2, m-tyramine and aldosterone. So, while the number of water-soluble compounds in blood and urine may be almost identical, the concentrations of these compounds are often profoundly different. This difference, combined with the ability of the kidney to handle abnormally high or abnormally low concentrations of metabolites, makes urine a particularly useful biofluid for medical diagnostics. In fact, according to our data in the UMDB, urinary metabolites have been used to characterize nearly 220 diseases. Furthermore, the ability of the kidneys to filter toxins or xenobiotics makes urine a particularly useful biofluid for diet and drug monitoring and for assessing chemical or pollutant exposure [96].
Method Comparison
One of the central motivations behind this work was to ascertain the strengths and weaknesses of several common metabolomic platforms for characterizing human urine. We employed 6 different analytical platforms: NMR; GC-MS; DFI/LC-MS/MS; HPLC/UV; HPLC/FD and ICP-MS. Using our literature-derived knowledge about the composition of human urine, along with custom-derived spectral libraries and targeted assays we were able to “push the limits” in terms of number of compounds that could be identified and/or quantified via each platform. In total, we identified 445 and quantified 378 distinct metabolites using these 6 different systems. According to Table 1, this is the largest number of urine metabolites ever identified and/or quantified in a single study. NMR spectroscopy was able to identify and quantify 209 compounds; GC-MS was able to identify 179 and quantify 85 compounds; DFI/LC-MS/MS identified and quantified 127 compounds; ICP-MS identified and quantified 40 compounds; while customized HPLC assays (with UV or FD detection) identified and quantified 10 compounds. The number of urinary metabolites we identified/quantified for NMR, GC-MS, DFI/LC-MS/MS and ICP-MS all represent “records” for these platforms.
In terms of platform overlap and compound complementarity, NMR and GC-MS were able to identify a common set of 88 metabolites; NMR and DFI/LC-MS/MS were able to identify and quantify a common set of 28 metabolites, while NMR, GC-MS and DFI/LC-MS/MS were able to identify a common set of 17 metabolites (15 amino acids, creatinine and hexose/glucose). All of these results are summarized in a Venn diagram (Figure 5). As might be expected, metabolite coverage differs from one analytical technique to another. These are difference mostly due to the intrinsic nature of the devices or platforms used. In particular, significant differences exist between these platforms in terms of their sensitivity or separation and/or extraction efficiency. Likewise the use of targeted vs. non-targeted methods along with issues related to compound stability, solubility and volatility led to some significantly different platform-dependent results.
Figure 5. Venn diagram showing the overlap of urine metabolites detected by NMR, GC-MS, DFI/LC-MS/MS, ICP-MS, HPLC coupled to UV detection and HPLC coupled to fluorescence detection compared to the detectable urine metabolome.
Given that the known, quantifiable urine metabolome consists of ∼2651 known metabolites and metabolite species (corresponding to 3079 distinct structures), we can calculate that NMR is able to measure ∼8% (209/2651) of the human urine metabolome; GC-MS is able to measure ∼7% (179/2651); DFI/LC-MS/MS is able to measure ∼5% (127/2651); ICP-MS/MS is able to measure ∼1.5% (40/2651); HPLC/UV is able to measure ∼0.2% (6/2651); while HPLC/FD is able to measure ∼0.15% (4/2651) of the urine metabolome. When combined, the 6 analytical techniques are able to cover >16% of the known urinary metabolome (>445/2651). If we re-evaluate this fraction in terms of total metabolite structures (corresponding to known and highly probable metabolites), the urine metabolome consists of 3079 compounds. From this total we can calculate that NMR is able to measure ∼7% (209/3079) of the human urine metabolome; GC-MS is able to measure ∼6% (179/3079); DFI/LC-MS/MS is able to measure ∼18% (555/3079); ICP-MS/MS is able to measure ∼1.3% (40/3079); HPLC/UV is able to measure ∼0.2% (6/3079); while HPLC/FD is able to measure ∼0.13% (4/3079) of the urine metabolome. When combined, the 6 analytical techniques are able to cover >28% of the known and probable or putative urinary metabolome (>873/3079). In terms of chemical class coverage, NMR detects compounds from 15 of the 25 major chemical superclasses in urine, GC-MS detects compounds from 14 of the 25, DFI/LC-MS/MS detects 6 of the 25, ICP-MS detects 1 of the 25 while the targeted methods for thiols and isoflavones detect just 1 of each.
From these data we can conclude that NMR is currently the best method for identifying and quantifying urinary compounds. Not only does it permit measurement of the largest number of metabolites (209) but it also yields the greatest chemical diversity. Furthermore, NMR is non-destructive so that the same sample can be subsequently re-used for GC-MS, LC-MS or ICP-MS analyses. The minimal sample preparation and relatively rapid data collection for NMR also make it much more appealing for urine metabolomics, although the spectral analysis can be quite slow (∼1–2 hours per sample). While GC-MS is a close second in terms of overall coverage (179 metabolites, 14 chemical superclasses), these numbers represents the result of 4 different analyses performed on 2 different GC-MS instruments. Many labs would not have these multiple configurations available or the resources to routinely run these types of analyses. Likewise each sample required many hours of preparation, sample collection and data analysis. In this regard, multi-platform GC-MS is definitely not a high-throughput metabolomics technique. Relative to NMR and GC-MS, DFI/LC-MS/MS also performs well, with 127 compounds being quantified. However, DFI/LC-MS/MS provides very limited chemical diversity (only 6 chemical superclasses). On the other hand, DFI/LC-MS/MS requires very little sample volume (10 µL) and it is a very low-cost, largely automated, high-throughput route for measuring metabolites. The other techniques (HPLC, ICP-MS) we employed in this study, while useful, do not come close to matching the coverage or diversity of NMR, GC-MS or DFI/LC-MS/MS.
While we certainly went to considerable lengths to use current or cutting edge technologies to characterize the urine metabolome, it is also important to note that there is always potential for future improvement. Using higher field (900–950 MHz) NMR instruments, employing newer model GC-MS instruments or more sensitive GC-TOF instruments, using more than 3 derivatization or extraction steps for our GC-MS analyses, employing the latest version of the NIST database or a larger collection of GC-MS databases, implementing more sophisticated or targeted detection and separation techniques, using various commercial immunodetection kits or employing the latest LC-MS/MS techniques coupled to FT-MS or orbitraps – all of these could have added to the quantity and diversity of metabolites detected or quantified. However, like many laboratories, our resources are somewhat limited. Furthermore, in this study we wanted to address the question of how well a cross-section of commonly accessible metabolomic methods or platforms could perform in identifying and quantifying metabolites in urine.
While being able to quantitatively compare metabolite coverage and chemical diversity amongst the major metabolomics platforms (NMR, GC-MS and DFI/LC-MS/MS) is important, it is also useful to compare their consistency or reproducibility in terms of metabolite quantification. In particular we decided to assess the 3 major platforms in terms of their ability to identify and quantify a common group of compounds, namely the amino acids. Overall we found that the measured concentrations are in relatively good agreement (Table 10). However, a few exceptions are evident. For example, the NMR and DFI/LC-MS/MS concentrations of glycine and serine are higher than the GC-MS values (note that glycine exhibits the highest concentration among urinary amino acids). For serine, after the silylation reaction using MSTFA, we obtained serine-2TMS (13.9 min) and serine-3TMS derivatives (16.6 min). The chromatographic peak corresponding to serine-2TMS is weak and overlaps slightly with the urea peak. This overlap and the corresponding difficulty in peak integration may explain the quantitation differences compared to other analytical assays. Neither L-glutamine nor L-glutamic acid could be accurately quantified or identified by GC-MS. In our case, the glutamine peak co-elutes with glycerol-3-phosphate. Other investigators have noted that glutamine can also be hydrolyzed and converted to glutamic acid [97], or to pyroglutamic acid during derivatization [95]. As a result, only pyroglutamic acid could be identified in our GC-MS assay. The identification of glutamic acid and pyroglutamic acid can be complicated [97], which explains our failure to identify glutamic acid by GC-MS. L-arginine could not be detected by GC-MS, because it is converted to ornithine during derivatization [98], which probably explains the slightly higher concentration of ornithine measured by GC-MS, compared to the one determined by DFI/LC-MS/MS. Finally, L-cysteine and L-cystine can only be identified and quantified by NMR and targeted HPLC/FD because the identification and quantification of theses metabolites is not possible with the Biocrates kit. With GC-MS it has often been noted that L-cystine can be converted to L-cysteine during derivatization, while L-cysteine might be oxidized to L-cystine during prolonged storage of the standard solution [95], confounding the identification of L-cystine and L-cysteine.
Table 10. Comparison of urine concentrations of 23 quantified amino acids by different assays: DFI/LC-MS/MS (BIOCRATES kit), NMR and GC-MS.
| Amino acids* | DFI/LC-MS/MS | NMR | GC-MS |
| Glycine | 101.0±53.5 | 106±66 | 75.7±51.7 |
| L-Alanine | 21.7±10.6 | 21.8±10.3 | NA |
| L-Arginine | 7.4±3.0 | 8.6±3.6 | NA |
| L-Asparagine | 8.8±3.5 | 10.1±4.0 | 9.5±5.5 |
| L-Aspartic acid | 10.9±7.7 | 10.9±5.8 | NA |
| L-Citrulline | 0.8±0.4 | NA | NA |
| L-Cysteine | NA | 65.8±24.0 | NA |
| L-Cystine | NA | 12.5±5.4 | NA |
| L-Glutamic acid | 6.4±4.9 | 8.5±3.6 | NA |
| L-Glutamine | 39.9±14.3 | 37.2±14.8 | NA |
| L-Histidine | 45.6±15.6 | 43.9±19.5 | NA |
| L-Isoleucine | 1.3±0.4 | 1.3±0.5 | NA |
| L-Leucine | 3.0±1.0 | 2.8±0.9 | NA |
| L-Lysine | 17.9±12.2 | 17.2±11.5 | NA |
| L-Methionine | 0.8±0.3 | 1.4±0.6 | NA |
| L-Ornithine | 4.4±5.0 | NA | 5.3±1.9 |
| L-Phenylalanine | 5.9±2.0 | 6.4±2.3 | 7.8±2.0 |
| L-Proline | 1.1±0.6 | NA | NA |
| L-Serine | 24.5±9.2 | 25.3±9.7 | 19.0±7.0 |
| L-Threonine | 14.6±6.1 | 13.3±5.3 | NA |
| L-Tryptophan | 5.6±2.1 | 6.3±2.2 | NA |
| L-Tyrosine | 8.7±4.2 | 9.5±4.5 | 15.1±5.4 |
| L-Valine | 4.3±1.9 | 3.4±1.4 | 5.5±2.0 |
Concentration (Mean ± SD) is expressed by µM/mM creatinine. NA: not available.
Conclusion
Using a combination of multiple experimental assays supplemented with an extensive computer–assisted literature survey we were able to identify a total of 2651 metabolites or metabolite species (corresponding to 3079 distinct structures) that can be or have been identified and/or quantified in human urine using today’s technology. This information, which includes both normal and abnormal (disease or exposure-associated) metabolites has been placed into a publicly accessible web-enabled database called the Urine Metabolome Database (UMDB). To assess the validity of the literature data and to further investigate the capabilities of existing metabolomics technologies we conducted a comprehensive, quantitative analysis of human urine from 22 healthy volunteers. A total of 6 different platforms: NMR, GC-MS, DFI/LC-MS/MS, ICP-MS, HPLC/UV and HPLC/FD were used in this analysis. From this experimental work we were able to identify a total of 445 and quantify 378 metabolites or metabolite species. This corresponds to 873 unique structures (identified) and 806 unique structures (quantified). A total of 53 compounds or compound species are being reported here for the first time as being normal constituents of human urine, while 77 compounds or compound species are being robustly quantified in human urine for the first time. All of the metabolites that we experimentally identified and/or quantified have been added to the UMDB. Based on the information in the UMDB, our experimentally acquired data corresponds to 16% of the human urine metabolome (or 28% if we include probable or putative metabolites).
Considering the level of coverage, the diversity of chemical species and the ease with which analyses can be performed, we have determined that NMR spectroscopy appears to be the method of choice for global or untargeted metabolomic analysis of urine. On the other hand, the kit-based combined DFI/LC-MS/MS methods appear to be optimal for a targeted metabolomic approach. Using a multi-pronged GC-MS approach for urine metabolomics appears to be very promising in terms of coverage, but is not ideal for high-throughput analyses. All methods used in this study appear to be quite complementary with relatively little compound overlap. This strongly suggests that if sufficient time and resources are available, multiple methods should be used in urine metabolomic studies.
If additional resources had been available, we would have liked to assess other technologies (GCxGC-MS, FT-MS, isotope labeled-LC-MS) and to compare the level of metabolite coverage and chemical diversity attainable with these methods. However, this study is not intended to be the “final” word on urine or urine metabolome. Rather, it should be viewed as a starting point for future studies and future improvements in this field. Indeed, our primary objective for undertaking these studies and compiling this data was to help advance the fields of quantitative metabolomics, especially with regard to clinically important biofluids such as urine. Experimentally, our data should serve as a useful benchmark from which to compare other technologies and to assess coming methodological improvements in human urine characterization. From a clinical standpoint, we think the information contained in the human urine metabolome database (UMDB) should provide metabolomic researchers as well as clinicians and clinical chemists with a convenient, centralized resource from which to learn more about human urine and its unique chemical constituents.
Methods
Ethics Statement
All samples were collected in accordance with the ethical guidelines mandated by the University of Alberta as approved by the University’s Health Research Ethics Board. All individuals were over 18 years of age. All were approached using approved ethical guidelines and those who agreed to participate in this study were required to sign consent forms. All participants provided written consent.
Collection of Urine Samples
Human urine samples (first pass, morning) were collected from 22 healthy adult volunteers (14 male, 8 female) in 120 mL sterile urine specimen cups. The average age of the volunteers was 30 (range 19–65) for females, and 32 (range 21–67) for males. Upon receipt (typically within 1 hour of collection), all samples were immediately treated with sodium azide to a final concentration of 2.5 mM. After centrifugation at 4000 rpm for 10 min to remove particulate matter, the urine samples were stored in 2 mL aliquots in falcon tube at −20°C until further use. Prior to each analysis, the samples were thawed at room temperature for 30 minutes and filtered for a second time via centrifugation.
NMR Compound Identification and Quantification
All 1H-NMR spectra were collected on a 500 MHz Inova (Varian Inc., Palo Alto, CA) spectrometer using the first transient of the tnnoesy-presaturation pulse sequence. The resulting 1H-NMR spectra were processed and analyzed using the Chenomx NMR Suite Professional software package version 7.0 (Chenomx Inc., Edmonton, AB), as previously described [17]. Additional NMR spectra for 39 compounds were added to the Chenomx Spectral Reference Library using the company’s recommended spectral acquisition and formatting protocols. Further details on the NMR sample preparation, NMR data acquisition and the customized spectral library are provided in Method S1.
GC-MS Compound Identification and Quantification
Twenty-two urine samples were extracted separately to obtain separate pools of polar, organic acid, bile acid (3 of the 22 urine samples were chosen for analysis and aliquots from these provided an additional “pooled normal” sample) and volatile metabolites using different protocols. The polar metabolites were extracted with cold HPLC grade methanol and double-distilled water after pretreatment with urease, followed by derivatization with MSTFA (N-Methyl-N-trifluoroacetamide) with 1% TMCS (trimethylchlorosilane). For organic acids, the ketoacids were converted first to methoxime derivatives, followed by derivatization with BSTFA (N,O-Bis(trimethylsilyl)trifluoroacetamide) after two successive extractions by ethyl acetate and diethyl ether. The bile acids were eluted with methanol through a SPE column (Bond Elute C18), followed by two different derivatization steps. First the bile acid extracts were esterified using 2% sulfuric acid in methanol, then after phase separation, the esterified bile acids were converted to the corresponding methyl ester-trimethylsilyl ether derivatives using MSTFA with 1% TMCS. Each set of extracted/derivatized metabolites (polar metabolites, organic acids, bile acids) was separated and analyzed using an Agilent 5890 Series II GC-MS operating in electron impact (EI) ionization mode.
The volatile compound extraction and analysis by GC-MS was far different from the other protocols. The pooled urine samples were acidified using HCl and transferred to a SPME (solid phase microextraction) vial (75 µm Carboxen/PDMS from Supelco), and then introducing the SPME fiber assembly into the GC-MS injector port according to procedures described elsewhere [99], [100]. The fibers were conditioned prior to use according to the manufacturer’s instructions by inserting them into the GC injector port. Further details on the extraction, derivatization, separation and GC-MS data analysis for the 4 separate groups of urine metabolites are provided in Method S2.
Direct Flow Injection/LC-MS/MS Compound Identification and Quantification
To assess the performance of direct flow injection DFI-MS/MS methods in urine metabolomics and to determine the concentration ranges of a number of metabolites not measurable by other methods, we used the commercially available Absolute-IDQ p180 Kit (BIOCRATES Life Sciences AG - Austria). The kit, in combination with an ABI 4000 Q-Trap (Applied Biosystems/MDS Sciex) mass spectrometer, can be used for the targeted identification and quantification of 187 different metabolites or metabolite species including amino acids, biogenic amines, creatinine, acylcarnitines, glycerophospholipids, sphingolipids and hexoses. This method involves derivatization and extraction of analytes from the biofluid of interest, along with selective mass spectrometric detection and quantification via multiple reactions monitoring (MRM). Isotope-labeled internal standards are integrated into the kit plate filter to facilitate metabolite quantification. Metabolite concentrations were expressed as ratios relative to creatinine to correct for dilution, assuming a constant rate creatinine excretion for each urine sample (see Method S3 for additional information).
Trace Element Analysis Using ICP-MS
Before trace elemental analysis by ICP-MS was performed, 22 urine samples were processed as described previously [101]. The concentrations of trace elements were determined on a Perkin-Elmer Sciex Elan 6000 quadrupole ICP-MS operating in a dual detector mode. Blank subtraction was applied after internal standard correction (see Method S4 for additional information). The accuracy of the ICP-MS analytical protocol was periodically evaluated via the analysis of certified reference standard materials (whole rock powders) BE-N and DR-N available from the SARM laboratory at the CRPG (Centre de Recherches Pétrographiques et Géologiques).
Characterization of Isoflavones from Urine
We processed 22 urine samples as described previously [102], [103]. The isoflavones were isolated and concentrated by solid-phase extraction (Bond Elut C18 column). The elutes were hydrolyzed enzymatically as the urinary isoflavones occur predominantly as glucuronate and sulfate conjugates. The analysis were performed on an Agilent 1100 HPLC system using NovaPak C18 reversed-phase column connected to Agilent G1315B diode array detector with signals scanned between 190 and 400 nm (see Method S5 for additional information).
Characterization of Thiols from Urine
To extract urinary thiols, we derivatized all 22 urine samples as described previously [104]. A mixture of reagent was used for the reduction and derivatization (with bromobimane) of thiols. The derivatized thiols were injected immediately into a hypersil-ODS HPLC Column connected to Agilent fluorometer operating at an excitation wavelength of 485 nm and emission wavelength of 510 nm (see Method S6 for additional information).
Supporting Information
NMR Compound Identification and Quantification.
(DOC)
GC-MS Compound Identification and Quantification.
(DOC)
Direct Flow Injection/LC-MS/MS Compound Identification and Quantification.
(DOC)
Trace Element Analysis Using ICP-MS.
(DOC)
Characterization of Isoflavones from Urine.
(DOC)
Characterization of Thiols from Urine.
(DOC)
Acknowledgments
The authors wish to thank Kruti Chaudhary and Hetal Chaudhary for adding concentration values from the literature to the UMDB, Rolando Perez-Pineiro for the synthesis of HPHPA and Beomsoo Han for converting all figures to high resolution images.
Funding Statement
Funding for this research has been provided by Genome Canada, Genome Alberta, The Canadian Institutes of Health Research, Alberta Innovates BioSolutions, Alberta Innovates Health Solutions, The National Research Council, The National Institute of Nanotechnology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Bundy JG, Davey MP, Viant MR (2009) Environmental metabolomics: a critical review and future perspectives. Metabolomics 5: 3–21. [Google Scholar]
- 2. Montoliu I, Genick U, Ledda M, Collino S, Martin FP, et al. (2013) Current status on genome-metabolome-wide associations: an opportunity in nutrition research. Genes Nutr 8: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nicholson JK, Holmes E, Kinross JM, Darzi AW, Takats Z, et al. (2012) Metabolic phenotyping in clinical and surgical environments. Nature 491: 384–392. [DOI] [PubMed] [Google Scholar]
- 4. Viant MR (2008) Recent developments in environmental metabolomics. Mol Biosyst 4: 980–986. [DOI] [PubMed] [Google Scholar]
- 5. Brindle JT, Antti H, Holmes E, Tranter G, Nicholson JK, et al. (2003) Rapid and noninvasive diagnosis of the presence and severity of coronary heart disease using 1H-NMR-based metabonomics. Nat Med 9: 477–477. [DOI] [PubMed] [Google Scholar]
- 6. Dunne VG, Bhattachayya S, Besser M, Rae C, Griffin JL (2005) Metabolites from cerebrospinal fluid in aneurysmal subarachnoid haemorrhage correlate with vasospasm and clinical outcome: a pattern-recognition H-1 NMR study. NMR Biomed 18: 24–33. [DOI] [PubMed] [Google Scholar]
- 7. Kaddurah-Daouk R, Beecher C, kristal B, Maston W, Bogdanov M (2004) Bioanalytical advances for metabolomics and metabolic profiling. Pharmagenomics 4: 46–52. [Google Scholar]
- 8. Mendrick DL, Schnackenberg L (2009) Genomic and metabolomic advances in the identification of disease and adverse event biomarkers. Biomark Med 3: 605–615. [DOI] [PubMed] [Google Scholar]
- 9. Ng DJY, Pasikanti KK, Chan ECY (2011) Trend analysis of metabonomics and systematic review of metabonomics-derived cancer marker metabolites. Metabolomics 7: 155–178. [Google Scholar]
- 10. Wishart DS (2008) Applications of metabolomics in drug discovery and development. Drugs R D 9: 307–322. [DOI] [PubMed] [Google Scholar]
- 11. Lim C, Kim J, Choi JN, Ponnusamy K, Jeon Y, et al. (2010) Identification, fermentation, and bioactivity against xanthomonas oryzae of antimicrobial metabolites isolated from phomopsis longicolla S1B4. J Microbiol Biotechn 20: 494–500. [PubMed] [Google Scholar]
- 12. Metz TO, Zhang QB, Page JS, Shen YF, Callister SJ, et al. (2007) Future of liquid chromatography-mass spectrometry in metabolic profiling and metabolomic studies for biomarker discovery. Biomark Med 1: 159–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rousu T, Pelkonen O, Tolonen A (2009) Rapid detection and characterization of reactive drug metabolites in vitro using several isotope-labeled trapping agents and ultra-performance liquid chromatography/time-of-flight mass spectrometry. Rapid Commun Mass Spectrom 23: 843–855. [DOI] [PubMed] [Google Scholar]
- 14. Zhang W, Saif MW, Dutschman GE, Li X, Lam W, et al. (2010) Identification of chemicals and their metabolites from PHY906, a Chinese medicine formulation, in the plasma of a patient treated with irinotecan and PHY906 using liquid chromatography/tandem mass spectrometry (LC/MS/MS). J Chromatogr A 1217: 5785–5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, et al. (2013) HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res 41: D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wishart DS, Knox C, Guo AC, Eisner R, Young N, et al. (2009) HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res 37: D603–D610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wishart DS, Lewis MJ, Morrissey JA, Flegel MD, Jeroncic K, et al. (2008) The human cerebrospinal fluid metabolome. J Chromatogr B 871: 164–173. [DOI] [PubMed] [Google Scholar]
- 18. Mandal R, Guo AC, Chaudhary KK, Liu P, Yallou FS, et al. (2012) Multi-platform characterization of the human cerebrospinal fluid metabolome: a comprehensive and quantitative update. Genome Med 4: 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Takeda I, Stretch C, Barnaby P, Bhatnager K, Rankin K, et al. (2009) Understanding the human salivary metabolome. NMR Biomed 22: 577–584. [DOI] [PubMed] [Google Scholar]
- 20. Psychogios N, Hau DD, Peng J, Guo AC, Mandal R, et al. (2011) The Human Serum Metabolome. Plos One 6 (2): e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Echeverry G, Hortin GL, Rai AJ (2010) Introduction to urinalysis: historical perspectives and clinical application. Methods Mol Biol 641: 1–12. [DOI] [PubMed] [Google Scholar]
- 22. Kouba E, Wallen EM, Pruthi RS (2007) Uroscopy by hippocrates and theophilus: prognosis versus diagnosis. J Urology 177: 50–52. [DOI] [PubMed] [Google Scholar]
- 23. Pardalidis N, Kosmaoglou E, Diamantis A, Sofikitis N (2008) Uroscopy in byzantium. J Urology 179: 1271–1276. [DOI] [PubMed] [Google Scholar]
- 24.Rigby D, Gray K (2005) Understanding urine testing. 60–62 p. [PubMed] [Google Scholar]
- 25. Nicholson JK, Lindon JC (2008) Systems biology - metabonomics. Nature 455: 1054–1056. [DOI] [PubMed] [Google Scholar]
- 26. Garrod AE (1996) The incidence of alkaptonuria: a study in chemical individuality. Mol Med 2: 274–282. [PMC free article] [PubMed] [Google Scholar]
- 27. Issaq HJ, Nativ O, Waybright T, Luke B, Veenstra TD, et al. (2008) Detection of bladder cancer in human urine by metabolomic profiling using high performance liquid chromatography/mass spectrometry. J Urology 179: 2422–2426. [DOI] [PubMed] [Google Scholar]
- 28. Kim K, Aronov P, Zakharkin SO, Anderson D, Perroud B, et al. (2009) Urine metabolomics analysis for kidney cancer detection and biomarker discovery. Mol Cell Proteomics 8: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kind T, Tolstikov V, Fiehn O, Weiss RH (2007) A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal Biochem 363: 185–195. [DOI] [PubMed] [Google Scholar]
- 30. Pasikanti KK, Esuvaranathan K, Ho PC, Mahendran R, Kamaraj R, et al. (2010) Noninvasive urinary metabonomic diagnosis of human bladder cancer. J Proteome Res 9: 2988–2995. [DOI] [PubMed] [Google Scholar]
- 31. Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, et al. (2010) Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc 5: 1005–1018. [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y, Tobias HJ, Auchus RJ, Brenna JT (2011) Comprehensive 2-dimensional gas chromatography fast quadrupole mass spectrometry (GC x GC-qMS) for urinary steroid profiling: Mass spectral characteristics with chemical ionization. Drug Test Anal 3: 857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim H, Ahn E, Moon MH (2008) Profiling of human urinary phospholipids by nanoflow liquid chromatography/tandem mass spectrometry. Analyst 133: 1656–1663. [DOI] [PubMed] [Google Scholar]
- 34. Law WS, Huang PY, Ong ES, Ong CN, Li SFY, et al. (2008) Metabonomics investigation of human urine after ingestion of green tea with gas chromatography/mass spectrometry, liquid chromatography/mass spectrometry and H-1 NMR spectroscopy. Rapid Commun Mass Spectrom 22: 2436–2446. [DOI] [PubMed] [Google Scholar]
- 35. Luo JL, Lamb JH, Lim CK (1997) Analysis of urinary and faecal porphyrin excretion patterns in human porphyrias by fast atom bombardment mass spectrometry. J Pharmaceut Biomed 15: 1289–1294. [DOI] [PubMed] [Google Scholar]
- 36. Nielen MWF, Bovee TFH, van Engelen MC, Rutgers P, Hamers ARM, et al. (2006) Urine testing for designer steroids by liquid chromatography with androgen bioassay detection and electrospray quadrupole time-of-flight mass spectrometry identification. Anal Chem 78: 424–431. [DOI] [PubMed] [Google Scholar]
- 37. Wadhier MC, Almstetter MF, Nurnberger N, Gruber MA, Dettmer K, et al. (2011) Improved enantiomer resolution and quantification of free D-amino acids in serum and urine by comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry. J Chromatogr A 1218: 4537–4544. [DOI] [PubMed] [Google Scholar]
- 38. Bollard ME, Stanley EG, Lindon JC, Nicholson JK, Holmes E (2005) NMR-based metabonomic approaches for evaluating physiological influences on biofluid composition. NMR Biomed 18: 143–162. [DOI] [PubMed] [Google Scholar]
- 39. Lindon JC, Keun HC, Ebbels TMD, Pearce JMT, Holmes E, et al. (2005) The consortium for metabonomic toxicology (comet): aims, activities and achievements. Pharmacogenomics 6: 691–699. [DOI] [PubMed] [Google Scholar]
- 40. Yang WJ, Wang YW, Zhou QF, Tang HR (2008) Analysis of human urine metabolites using SPE and NMR spectroscopy. Sci China Ser B 51: 218–225. [Google Scholar]
- 41. Minkler PE, Hoppel CL (1993) Quantification of carnitine and specific acylcarnitines by high-performance liquid-chromatography - application to normal human urine and urine from patients with methylmalonic aciduria, isovaleric acidemia or medium-chain acyl-coa dehydrogenase-deficiency. J Chromatogr-Biomed 613: 203–221. [DOI] [PubMed] [Google Scholar]
- 43. Giron AJ, Deventer K, Roels K, Van Eenoo P (2012) Development and validation of an open screening method for diuretics, stimulants and selected compounds in human urine by UHPLC-HRMS for doping control. Anal Chim Acta 721: 137–146. [DOI] [PubMed] [Google Scholar]
- 44. Yanamandra R, Vadla CS, Puppala U, Patro B, Murthy YL, et al. (2012) A new rapid and sensitive stability-Indicating UPLC assay method for tolterodine tartrate: application in pharmaceuticals, human plasma and urine samples. Sci Pharm 80: 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kanehisa M (2002) The KEGG database. Novart Fdn Symp 247: 91–103. [PubMed] [Google Scholar]
- 46. Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, et al. (2006) Database resources of the national center for biotechnology information. Nucleic Acids Res 34: D173–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Caspi R, Foerster H, Fulcher CA, Hopkinson R, Ingraham J, et al. (2006) MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res 34: D511–D516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Degtyarenko K, De Matos P, Ennis M, Hastings J, Zbinden M, et al. (2008) ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res 36: D344–D350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Maccarone MC, Tripicano M, Digesu V, Tegolo D (1993) PDB - a Pictorial Database Oriented to data-analysis. Software Pract Exper 23: 55–73. [Google Scholar]
- 50. Bairoch A, Bougueleret L, Altairac S, Amendolia V, Auchincloss A, et al. (2008) The Universal Protein Resource (UniProt). Nucleic Acids Res 36: D190–D195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ouellette BFF (1998) The GenBank sequence database. Bioinformatics 39: 16–45. [DOI] [PubMed] [Google Scholar]
- 52. Safran M, Dalah I, Alexander J, Rosen N, Stein TI, et al. (2010) GeneCards Version 3: the human gene integrator. Database-Oxford 10: 1093/baq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kapushesky M, Adamusiak T, Burdett T, Culhane A, Farne A, et al. (2012) Gene Expression Atlas update-a value-added database of microarray and sequencing-based functional genomics experiments. Nucleic Acids Res 40: D1077–D1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, et al. (2008) The HGNC database in 2008: a resource for the human genome. Nucleic Acids Res 36: D445–D448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Frolkis A, Knox C, Lim E, Jewison T, Law V, et al. (2010) SMPDB: The Small Molecule Pathway Database. Nucleic Acids Res 38: D480–D487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guneral F, Bachmann C (1994) Age-related reference values for urinary organic-acids in a healthy turkish pediatric population. Clin Chem 40: 862–868. [PubMed] [Google Scholar]
- 57. Psihogios NG, Gazi IF, Elisaf MS, Seferiadis KI, Bairaktari ET (2008) Gender-related and age-related urinalysis of healthy subjects by NMR-based metabonomics. NMR Biomed 21: 195–207. [DOI] [PubMed] [Google Scholar]
- 58. Slupsky CM, Rankin KN, Wagner J, Fu H, Chang D, et al. (2007) Investigations of the effects of gender, diurnal variation, and age in human urinary metabolomic profiles. Anal Chem 79: 6995–7004. [DOI] [PubMed] [Google Scholar]
- 59. Zuppi C, Messana I, Forni F, Rossi C, Pennacchietti L, et al. (1997) H-1 NMR spectra of normal urines: Reference ranges of the major metabolites. Clin Chim Acta 265: 85–97. [DOI] [PubMed] [Google Scholar]
- 60. Hao J, Astle W, De Iorio M, Ebbels TMD (2012) BATMAN-an R package for the automated quantification of metabolites from nuclear magnetic resonance spectra using a Bayesian model. Bioinformatics 28: 2088–2090. [DOI] [PubMed] [Google Scholar]
- 61. Kern SM, Bennett RN, Mellon FA, Kroon PA, Garcia-Conesa MT (2003) Absorption of hydroxycinnamates in humans after high-bran cereal consumption. J Agr Food Chem 51: 6050–6055. [DOI] [PubMed] [Google Scholar]
- 62. Rios LY, Gonthier MP, Remesy C, Mila L, Lapierre C, et al. (2003) Chocolate intake increases urinary excretion of polyphenol-derived phenolic acids in healthy human subjects. Am J Clin Nutr 77: 912–918. [DOI] [PubMed] [Google Scholar]
- 63. Lloyd AJ, Beckmann M, Fave G, Mathers JC, Draper J (2011) Proline betaine and its biotransformation products in fasting urine samples are potential biomarkers of habitual citrus fruit consumption. Br J Nutr 106: 812–824. [DOI] [PubMed] [Google Scholar]
- 64. Atkinson W, Downer P, Lever M, Chambers ST, George PM (2007) Effects of orange juice and proline betaine on glycine betaine and homocysteine in healthy male subjects. Eur J Nutr 46: 446–452. [DOI] [PubMed] [Google Scholar]
- 65. Hucker HB, Miller JK, Hochberg A, Brobyn RD, Riordan FH, et al. (1967) Studies on the absorption, excretion and metabolism of dimethylsulfoxide (DMSO) in man. J Pharm Exp Ther 155: 309–317. [PubMed] [Google Scholar]
- 66. Winning H, Roldan-Marin E, Dragsted LO, Viereck N, Poulsen M, et al. (2009) An exploratory NMR nutri-metabonomic investigation reveals dimethyl sulfone as a dietary biomarker for onion intake. Analyst 134: 2344–2351. [DOI] [PubMed] [Google Scholar]
- 67. Kumari K, Augusti KT (2007) Lipid lowering effect of S-methyl cysteine sulfoxide from Allium cepa Linn in high cholesterol diet fed rats. J Ethnopharmacol 109: 367–371. [DOI] [PubMed] [Google Scholar]
- 68. Myint T, Fraser GE, Lindsted KD, Knutsen SF, Hubbard RW, et al. (2000) Urinary 1-methylhistidine is a marker of meat consumption in Black and in White California Seventh-day Adventists. Am J Epidemiol 152: 752–755. [DOI] [PubMed] [Google Scholar]
- 69. Sjolin J, Hjort G, Friman G, Hambraeus L (1987) Urinary excretion of 1-methylhistidine: a qualitative indicator of exogenous 3-methylhistidine and intake of meats from various sources. Metabolism 36: 1175–1184. [DOI] [PubMed] [Google Scholar]
- 70. Sjolin J, Stjernstrom H, Henneberg S, Hambraeus L, Friman G (1989) Evaluation of urinary 3-methylhistidine excretion in infection by measurements of 1-methylhistidine and the creatinine ratios. Am J Clin Nutr 49: 62–70. [DOI] [PubMed] [Google Scholar]
- 71. Rubin D (1979) Free glucuronic acid in urine as estimate of tumor load. Clin Chem 25: 1867–1868. [PubMed] [Google Scholar]
- 72.Dutton GJ ( 1966) Glucuronic Acid, Free and Combined: Chemistry, Biochemistry, Pharmacology, and Medicine. Madison: Academic Press. 629.
- 73. Smith LM, Maher AD, Cloarec O, Rantalainen M, Tang H, et al. (2007) Statistical correlation and projection methods for improved information recovery from diffusion-edited NMR spectra of biological samples. Anal Chem 79: 5682–5689. [DOI] [PubMed] [Google Scholar]
- 74. Rodriguez-Concepcion M, Boronat A (2002) Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol 130: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Dittrich P, Angyal SJ (1988) 2-C-methyl-D-erythritol in leaves of Liriodendron tulipifera. Phytochemistry 27: 935. [Google Scholar]
- 76. Kang SM, Park JC, Shin MJ, Lee H, Oh J, et al. (2011) H nuclear magnetic resonance based metabolic urinary profiling of patients with ischemic heart failure. Clin Biochem 44: 293–299. [DOI] [PubMed] [Google Scholar]
- 77. Engelke UF, Liebrand-van Sambeek ML, De Jong JG, Leroy JG, Morava E, et al. (2004) N-acetylated metabolites in urine: proton nuclear magnetic resonance spectroscopic study on patients with inborn errors of metabolism. Clin Chem 50: 58–66. [DOI] [PubMed] [Google Scholar]
- 78. Rezzi S, Vera FA, Martin FPJ, Wang S, Lawler D, et al. (2008) Automated SPE-RP-HPLC fractionation of biofluids combined to off-line NMR spectroscopy for biomarker identification in metabonomics. J Chromatogr B 871: 271–278. [DOI] [PubMed] [Google Scholar]
- 79. Saude EJ, Slupsky CM, Sykes BD (2006) Optimization of NMR analysis of biological fluids for quantitative accuracy. Metabolomics 2: 113–123. [Google Scholar]
- 80. Salek RM, Maguire ML, Bentley E, Rubtsov DV, Hough T, et al. (2007) A metabolomic comparison of urinary changes in type 2 diabetes in mouse, rat, and human. Physiol Genomics 29: 99–108. [DOI] [PubMed] [Google Scholar]
- 81. Pechlivanis A, Kostidis S, Saraslanidis P, Petridou A, Tsalis G, et al. (2010) H-1 NMR-Based Metabonomic Investigation of the Effect of Two Different Exercise Sessions on the Metabolic Fingerprint of Human Urine. J Proteome Res 9: 6405–6416. [DOI] [PubMed] [Google Scholar]
- 82.Taylor EH (1989) Clinical Chemistry. New York: John Wiley and Sons. 293 p. [Google Scholar]
- 83. Roux A, Xu Y, Heilier JF, Olivier MF, Ezan E, et al. (2012) Annotation of the human adult urinary metabolome and metabolite identification using ultra high performance liquid chromatography coupled to a linear quadrupole ion trap-Orbitrap mass spectrometer. Anal Chem 84: 6429–6437. [DOI] [PubMed] [Google Scholar]
- 84. Shoemaker JD, Elliott WH (1991) Automated screening of urine samples for carbohydrates, organic and amino-acids after treatment with urease. J Chromatogr-Biomed 562: 125–138. [DOI] [PubMed] [Google Scholar]
- 85. Lawson AM, Chalmers RA, Watts RWE (1976) Urinary organic-acids in man. I. normal patterns. Clin Chem 22: 1283–1287. [PubMed] [Google Scholar]
- 86. Pasikanti KK, Ho PC, Chan ECY (2008) Development and validation of a gas chromatography/mass spectrometry metabonomic platform for the global profiling of urinary metabolites. Rapid Commun Mass Sp 22: 2984–2992. [DOI] [PubMed] [Google Scholar]
- 87.Röhring C, Bogumil R, Dammeier S. Analysis of human urine using the absolute IDQ™ Kit Application Note 1003–1 Innsbruck, Austria: Biocrates Life Sciences Available At: http://wwwbiocratescom/images/stories/pdf/biocrates_applnote_1005-1pdf Accessed Accessed 2013 May 24.
- 88. Heitland P, Koster HD (2006) Biomonitoring of 37 trace elements in blood samples from inhabitants of northern Germany by ICP-MS. J Trace Elem Med Bio 20: 253–262. [DOI] [PubMed] [Google Scholar]
- 89. Benkhedda K, Infante HG, Ivanova E, Adams FC (2000) Trace metal analysis of natural waters and biological samples by axial inductively coupled plasma time of flight mass spectrometry (ICP-TOFMS) with flow injection on-line adsorption preconcentration using a knotted reactor. J Anal Atom Spectrom 15: 1349–1356. [Google Scholar]
- 90.Harborne JB (1988) The Flavonoids. London: Chapman & Hall. 531 p. [Google Scholar]
- 91. Kuimierek K, Bald E (2008) Measurement of reduced and total mercaptamine in urine using liquid chromatography with ultraviolet detection. Biomed Chromatogr 22: 441–445. [DOI] [PubMed] [Google Scholar]
- 92. Kusmierek K, Chwatko G, Bald E (2008) Redox status of main urinary sulfur fmino acids evaluation by liquid chromatography. Chromatographia 68: S91–S95. [Google Scholar]
- 93. Kusmierek K, Glowacki R, Bald E (2006) Analysis of urine for cysteine, cysteinylglycine, and homocysteine by high-performance liquid chromatography. Anal Bioanal Chem 385: 855–860. [DOI] [PubMed] [Google Scholar]
- 94. Moure A, Cruz JM, Franco D, Dominguez JM, Sineiro J, et al. (2001) Natural antioxidants from residual sources. Food Chem 72: 145–171. [Google Scholar]
- 95. Woo KL, Lee DS (1995) Capillary gas-chromatographic determination of proteins and biological amino-acids as n(o)-tert-butyldimethylsilyl derivatives. J Chromatogr B Biomed Appl 665: 15–25. [DOI] [PubMed] [Google Scholar]
- 96. Holmes E, Loo RL, Cloarec O, Coen M, Tang HR, et al. (2007) Detection of urinary drug metabolite (Xenometabolome) signatures in molecular epidemiology studies via statistical total correlation (NMR) spectroscopy. Anal Chem 79: 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang QF, Wysocki VH, Scaraffia PY, Wells MA (2005) Fragmentation pathway for glutamine identification: Loss of 73 Da from dimethylformamidine glutamine isobutyl ester. J Am Soc Mass Spectr 16: 1192–1203. [DOI] [PubMed] [Google Scholar]
- 98. Corso G, Esposito M, Gallo M, Dellorusso A, Antonio M (1993) Transformation of arginine into ornithine during the preparation of Its tert-butyldimethylsilyl derivative for analysis by gas chromatography/mass spectrometry. Biol Mass Spectrom 22: 698–702. [Google Scholar]
- 99. Arthur CL, Pawliszyn J (1990) Solid-phase microextraction with thermal-desorption using fused-silica optical fibers. Anal Chem 62: 2145–2148. [Google Scholar]
- 100. Jia CR, Luo YZ, Pawliszyn J (1998) Solid phase microextraction combined with HPLC for determination of metal ions using crown ether as selective extracting reagent. J Microcolumn Sep 10: 167–173. [Google Scholar]
- 101. Cava-Montesinos P, Nilles K, Cervera ML, de la Guardia M (2005) Non-chromatographic speciation of toxic arsenic in fish. Talanta 66: 895–901. [DOI] [PubMed] [Google Scholar]
- 102. Franke AA, Custer LJ (1994) High-performance liquid chromatographic assay of isoflavonoids and coumestrol from human urine. J Chromatogr B Biomed Appl 662: 47–60. [DOI] [PubMed] [Google Scholar]
- 103. Franke AA, Custer LJ, Wang W, Shi CY (1998) HPLC analysis of isoflavonoids and other phenolic agents from foods and from human fluids. Proc Soc Exp Biol Med 217: 263–273. [DOI] [PubMed] [Google Scholar]
- 104. Fiskerstrand T, Refsum H, Kvalheim G, Ueland PM (1993) Homocysteine and other thiols in plasma and urine - automated-determination and sample stability. Clin Chem 39: 263–271. [PubMed] [Google Scholar]
- 105.David FP (1971) Composition and concentrative properties of human urine. Washington: National Aeronautics and Space Administration. 107 p. [Google Scholar]
- 106. Gates SC, Sweeley CC, Krivit W, Dewitt D, Blaisdell BE (1978) Automated metabolic profiling of organic-acids in human Urine.2. analysis of urine samples from healthy adults, sick children, and children with neuroblastoma. Clin Chem 24: 1680–1689. [PubMed] [Google Scholar]
- 107. Hoffmann G, Aramaki S, Blumhoffmann E, Nyhan WL, Sweetman L (1989) Quantitative-analysis for organic-acids in biological samples - batch isolation followed by gas chromatographic-mass Spectrometric analysis. Clin Chem 35: 587–595. [PubMed] [Google Scholar]
- 108. Liebich HM, Forst C (1990) Basic profiles of organic-acids in urine. J Chromatogr-Biomed 525: 1–14. [DOI] [PubMed] [Google Scholar]
- 109. Matsumoto I, Kuhara T (1996) A new chemical diagnostic method for inborn errors of metabolism by mass spectrometry - Rapid, practical, and simultaneous urinary metabolites analysis. Mass Spectrom Rev 15: 43–57. [DOI] [PubMed] [Google Scholar]
- 110. Goulle JP, Mahieu L, Castermant J, Neveu N, Bonneau L, et al. (2005) Metal and metalloid multi-elementary ICP-MS validation in whole blood, plasma, urine and hair - Reference values. Forensic Sci Int 153: 39–44. [DOI] [PubMed] [Google Scholar]
- 111. Guo K, Li L (2009) Differential C-12/C-13-isotope dansylation labeling and fast liquid chromatography/mass spectrometry for absolute and relative quantification of the metabolome. Anal Chem 81: 3919–3932. [DOI] [PubMed] [Google Scholar]
- 112. Shaykhutdinov RA, MacInnis GD, Dowlatabadi R, Weljie AM, Vogel HJ (2009) Quantitative analysis of metabolite concentrations in human urine samples using C-13{H-1} NMR spectroscopy. Metabolomics 5: 307–317. [Google Scholar]
- 113. Kvitvang HFN, Andreassen T, Adam T, Villas-Boas SG, Bruheim P (2011) Highly Sensitive GC/MS/MS Method for Quantitation of Amino and Nonamino Organic Acids. Anal Chem 83: 2705–2711. [DOI] [PubMed] [Google Scholar]
- 114. Silva CL, Passos M, Camara JS (2011) Investigation of urinary volatile organic metabolites as potential cancer biomarkers by solid-phase microextraction in combination with gas chromatography-mass spectrometry. Brit J Cancer 105: 1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Guo K, Peng J, Zhou RK, Li L (2011) Ion-pairing reversed-phase liquid chromatography fractionation in combination with isotope labeling reversed-phase liquid chromatography-mass spectrometry for comprehensive metabolome profiling. J Chromatogr A 1218: 3689–3694. [DOI] [PubMed] [Google Scholar]
- 116. Saude EJ, Skappak CD, Regush S, Cook K, Ben-Zvi A, et al. (2011) Metabolomic profiling of asthma: Diagnostic utility of urine nuclear magnetic resonance spectroscopy. J Allergy Clin Immun 127: 757–U408. [DOI] [PubMed] [Google Scholar]
- 117. Zhang SC, Liu LY, Steffen D, Ye T, Raftery D (2012) Metabolic profiling of gender: Headspace-SPME/GC-MS and H-1 NMR analysis of urine. Metabolomics 8: 323–334. [Google Scholar]
- 118. van der Kloet FM, Tempels FWA, Ismail N, van der Heijden R, Kasper PT, et al. (2012) Discovery of early-stage biomarkers for diabetic kidney disease using ms-based metabolomics (FinnDiane study). Metabolomics 8: 109–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Cheng Y, Xie GX, Chen TL, Qiu YP, Zou X, et al. (2012) Distinct urinary metabolic profile of human colorectal cancer. J Proteome Res 11: 1354–1363. [DOI] [PubMed] [Google Scholar]
- 120. Rocha SM, Caldeira M, Carrola J, Santos M, Cruz N, et al. (2012) Exploring the human urine metabolomic potentialities by comprehensive two-dimensional gas chromatography coupled to time of flight mass spectrometry. J Chromatogr A 1252: 155–163. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NMR Compound Identification and Quantification.
(DOC)
GC-MS Compound Identification and Quantification.
(DOC)
Direct Flow Injection/LC-MS/MS Compound Identification and Quantification.
(DOC)
Trace Element Analysis Using ICP-MS.
(DOC)
Characterization of Isoflavones from Urine.
(DOC)
Characterization of Thiols from Urine.
(DOC)